Abstract
Background
Ginsenoside Rh2 is well known for many pharmacological activities, such as anticancer, antidiabetes, antiinflammatory, and antiobesity properties. Glycosyltransferases (GTs) are ubiquitous enzymes present in nature and are widely used for the synthesis of oligosaccharides, polysaccharides, glycoconjugates, and novel derivatives. We aimed to synthesize new ginsenosides from Rh2 using the recombinant GT enzyme and investigate its cytotoxicity with diverse cell lines.
Methods
We have used a GT gene with 1,224-bp gene sequence cloned from Lactobacillus rhamnosus (LRGT) and then expressed in Escherichia coli BL21 (DE3). The recombinant GT protein was purified and demonstrated to transform Rh2 into two novel ginsenosides, and they were characterized by nuclear magnetic resonance (NMR) techniques and evaluated by 3-(4, 5-dimethylthiazol-2-yl)-2-5-diphenyltetrazolium bromide assay.
Results
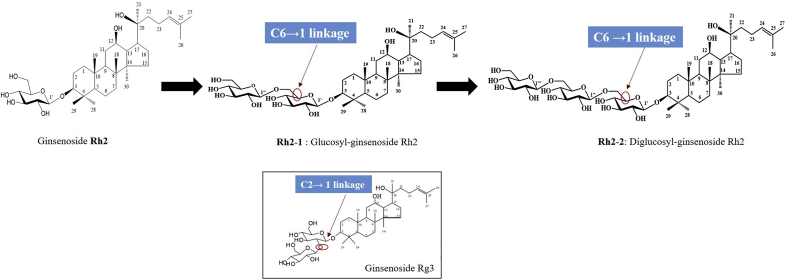
Two novel ginsenosides with an additional glucopyranosyl (6→1) and two additional glucopyranosyl (6→1) linked with the C-3 position of the substrate Rh2 were synthesized, respectively. Cell viability assay in the lung cancer (A549) cell line showed that glucosyl ginsenoside Rh2 inhibited cell viability more potently than ginsenoside Rg3 and Rh2 at a concentration of 10 μM. Furthermore, glucosyl ginsenoside Rh2 did not exhibit any cytotoxic effect in murine macrophage cells (RAW264.7), mouse embryo fibroblasts cells (3T3-L1), and skin cells (B16BL6) at a concentration of 10 μM compared with ginsenoside Rh2 and Rg3.
Conclusion
This is the first report on the synthesis of two novel ginsenosides, namely, glucosyl ginsenoside Rh2 and diglucosyl ginsenoside Rh2 from Rh2 by using recombinant GT isolated from L. rhamnosus. Moreover, diglucosyl ginsenoside Rh2 might be a new candidate for treatment of inflammation, obesity, and skin whiting, and especially for anticancer.
Keywords: Cell viability, Ginsenoside Rh2, Glycosyltransferase, Novel ginsenosides
1. Introduction
Ginseng (Panax ginseng Meyer) has been used for more than 2,000 years and considered one of the most important traditional oriental medicines in Asian countries [1]. P. ginseng has been used as a healing drug and health tonic in Korea, China, Japan, and other East Asian countries [2]. As the major effective ingredients in ginseng [3,4], the minor ginsenosides are well known for their various pharmacological activities such as boosting immunity [[5], [6], [7], [8]]; anticancer [[9], [10], [11], [12]], antiinflammatory [13,14], antitumor [15], antioxidative activities; and antiaging effects. So far, a total of 131 different ginsenosides have been characterized by a sensitive and practical high-performance liquid chromatography (HPLC)–mass spectrometry (MS)/MSn method [16]. Ginsenosides are classified into dammarane- and oleanane-type saponins, which are derived from dammarenediol and β-amyrin, respectively. Among the reported ginsenosides, ginsenoside Rh2 with a dammarane skeleton is one of the characteristic components in red ginseng with various potential bioactivities [17], which have been proven to possess therapeutic effects on inflammation [17], various cancers [[18], [19], [20], [21], [22], [23], [24]], and enteritis. Since 2006, ginsenoside Rh2 has been approved as a food supplement in China [25]. Nowadays, many physical and chemical methods have been used for the transformation of ginsenosides; however, there are limitations in specificity and problems such as environmental pollution. Thus, biotransformation is considered as the most promising method because of the high selectivity and productivity [26].
Glycosyltransferases (GTs; EC 2.4.x.y) are enzymes that can transfer the glucosyl donor to various specific acceptors, including hormones and secondary metabolites [27]. GTs are important for the biosynthesis of glycosylation of unnatural and natural products [28]. GTs have been classified into 97 families by amino acid sequence similarities [29] (available at http://afmb.cnrs-mrs.fr/CAZY). Furthermore, GT family 2 is known as the principal and evolutionarily the most ancient of the inverting enzyme families, which can use diverse sugar moieties, including uridine diphosphate (UDP) donors [30]. UDP GTs (UGTs) are a superfamily of enzymes that can catalyze functional groups including hydroxyl, carboxyl, amine, thiol, and carbon groups using UDP-activated sugar molecules as donors [31]. Glycosylation of natural compounds in plants is modified by GTs, leading to increase in the hydrophilicity, stability, solubility, and subcellular localization and thus the chemical properties and bioactivity of the natural products [32]. Moreover, the related GTs are the key enzymes in the last important step of ginsenoside biosynthesis [33]. Compared with UGTs from plants, a group of microbial UGTs shows high catalytic proficiency and contributes to high expression in the engineered bacteria and yeast. A recent report showed that a UGT from Bacillus licheniformis was conducted for the glycosylation mediating of phloretin and obtained five glucosides [34]. At present, ginsenoside aglycones and compound K have been obtained by synthetic biology [33]. Furthermore, complexity and diversity of ginsenosides are produced by modification of ginsenoside aglycones by GTs, which have more wide ranges of further pharmacological activity [35]. As the last step of ginsenoside biosynthesis, glycosylation of ginsenoside aglycones has been studied intensively in recent years [36]. Probiotic strains are generally preferred to be of human origin and possess satisfying antibiotic resistance along with providing beneficial metabolic activities toward the host [37]. A type of probiotic strains, namely, Lactobacillus rhamnosus, which is commonly found in the human body, has shown its capability to survive through the gastrointestinal tract [38]. Nonetheless, L. rhamnosus has also been proven with regard to its promising effect of maintaining good balance in a human's intestinal microbial ecosystem [39]. Apart from common understanding of the benefit toward the human body, L. rhamnosus also has promising commercial value especially in the production of functional food [40]. Therefore, we investigated and characterized a gene (LRGT) encoding GTs derived from L. rhamnosus ATCC 8530 in this study. Subsequently, two novel ginsenosides, glucosyl ginsenoside Rh2 and diglucosyl ginsenoside Rh2, were biotransformed, with ginsenoside Rh2 as an acceptor and UDP glucose (UDPG) as an active donor using LRGT. Finally, we investigated the cytotoxicity of the main product of glucosyl ginsenoside Rh2 in normal and cancer cells [41].
2. Materials and methods
2.1. Materials
Ginsenoside Rh2 was provided by Ginseng Resource Bank, Kyung Hee University, South Korea. UDPG was purchased from Sigma-Aldrich (St. Louis, MO, USA). The analytical grade solvents (water and acetonitrile) were purchased from SK Chemicals (Ulsan, Korea). B16BL6 melanoma cells, murine macrophage cells (RAW264.7), and A549 human lung adenocarcinoma cells were obtained Korean Cell Line Bank (Seoul, Korea). 3T3-L1 fibroblast cells were purchased from American Type Culture Collection (ATCC, USA).
2.2. Isolation and cloning analysis of the LRGT gene from L. rhamnosus
The genomic DNA was extracted from L. rhamnosus American Type Culture Collection 8530 using the genomic DNA Reagent kit (GeneAll, Seoul, Korea). The target gene encoding the UGTs LRGT was amplified from the chromosomal DNA by polymerase chain reaction using Pfu DNA polymerase (GeneAll) using the genomic DNA as the template. The fragment was amplified at EcoRV and BamH1 sites using the designed primers: LRGT-F (5′-CGATATCATGGCAGGTCATGAAGCAG-3′) and LRGT-R (5′-GGGATCCAGACTCCTTTTTAACATTCTCATCT-3′). The amplified DNA fragment was digested with the aforementioned primer and then ligated into a pMAL-c5X vector to generate a maltose-binding protein (MBP)-LRGT gene fusion using the EzFusion Kit (Enzynomics, Korea). Then, it was sequenced and confirmed at the Genotech facility (Daejeon, Korea).
Moreover, the E. coli BL21 (DE3) cells harboring the recombinant pMAL-LRGT was cultured in Luria-Bertani (containing ampicillin) broth at 37°C until the absorbance reached 0.4–0.6 at 600 nm, and then, the protein was expressed by 0.3–0.5 mM isopropyl-β-D-thiogalactopyranoside induction at 16°C for 8–9 h. The cells were harvested by centrifugation (5,000 × g, 20 min, 4°C), washed, and resuspended in 20 mM sodium phosphate buffer (pH 7.0) containing 1 mM EDTA and NaCl. The cells were disrupted by 5-min periods of sonications at 5-s interval with 10 repetitions on an ultrasonic processor at 80% output with cooling on ice. The cell debris was separated by centrifugation (12,000 × g, 20 min, 4°C). The tagged fusion protein was purified in a preequilibrated amylose column (New England BioLabs, France, UK) and eluted using 10 mM elution buffer. The recombinant LRGT was confirmed by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
2.3. Application of metabolite biosynthesis
Glycosylation assays containing 0.5 mM Rh2, 2.5 mM UDPG, and 0.1 mg/mL of purified overexpressed LRGT enzyme (pH 7.0) were carried out at 30°C for 12 h (200 rpm). In addition, the assays were carried out for the following groups: with Control 1 containing ginsenoside Rh2 and LRGT (C1), Control 2 containing LRGT and UDPG (C2), and Control 3 containing ginsenoside Rh2 and UDPG (C3).
2.4. Kinetic properties of LRGT
The kinetic parameters of the purified LRGT enzyme were detected in the reactants of UDPG concentrations (from 0.5 mM to 5 mM) [41]. The duplicate reactions were repeated under the same conditions. Vmax (μmol/L*min) and the Michaelis–Menten constant, Km (mol/L) were calculated using the Michaelis–Menten plot of specific activities. The values of Vmax and Km were determined by means of the Lineweaver–Burk plot.
2.5. Analysis of LRGT activities based on optimal temperature, pH, and metal ions in synthesis of ginsenoside Rh2
The reaction mixtures (1 mL) containing 0.5 mM ginsenoside Rh2 and 2.5 mM UDPG as the substrate were incubated for the initial synthesis experiment. The optimal temperature was tested at various temperatures ranging from 20 to 60°C. The optimal pH of purified LRGT was studied in 20 mM buffer at various pH ranging from 3 to 10. Activity of enzyme with metal ions was evaluated in the presence of 10 mM CoCl2, MgCl2, FeCl3, NaCl, CuSO4, NH4Cl, KCl, CaCl2, and ZnSO4 and then compared with a control without metal ions under the optimum ratio of temperature and pH. The samples were terminated by adding equal volume of water-saturated n-butanol; then, the mixtures were centrifuged at 13000 g for 5 min, and the supernatant was evaporated and dissolved with methanol for thin layer chromatography (TLC), HPLC, and high-resolution MS analysis.
2.6. TLC analysis
The transfer products were detected by TLC analysis. One microliter of reactants was spotted on the silica gel 60F254 plates by the solvent system of CHCl3/CH3OH/H2O (65/35/10) in a developing tank. The plate was kept to air-dry and developed by soaking in methanol solvent with 10 % (v/v) sulfuric acid. After that, the plate was heated at 110°C for 10 min to obtain the reaction spots [41].
2.7. Metabolite 1 and 2 analyses by HPLC, nuclear magnetic resonance, and high-resolution MS
The reactants were terminated by adding water-saturated butanol and examined by HPLC (Agilent 1260 system) consistent with the gradient protocol in our laboratory [30]. The separation was performed on a C18 column (5 μm, 250 × 4.6 mm) using acetonitrile and water as the mobile phases, and the time and ratios of the procedure were reported in our previous study. The flow rate was set at 1.6 mL/min, and the metabolites were monitored at 203 nm of UV wavelength [41]. To identify the molecular weight, the products were detected by MS analysis using a Finnigan LCQ-Advantage mass spectrometer (Thermo Fisher Scientific, CA, USA) (ion mode: FAB-; elements: C 43/0, H 72/0, O 14/0; mass tolerance: 1000 ppm, 10 mmu if m/z < 10, 20 mmu if m/z > 20; unsaturation: −0.5-50.0). The structure of Compound 1 and Compound 2 was identified via proton and carbon nuclear magnetic resonance (NMR). The metabolites were purified by preparative HPLC and recorded on the basis of 1H-NMR and 13C-NMR spectra at 100 MHz using a Bruker AV 600 NMR spectrometer (Bruker, Germany) with pyridine-d5 as the solvent.
2.8. In vitro cytotoxic test of Metabolite 1
With the aforementioned analysis, Metabolite 1 was the main product in our study. Therefore, we selected it for future work. Murine macrophage cells (RAW264.7), human lung adenocarcinoma cells (A549), fibroblast cells (3T3-L1), and melanoma cells (B16BL6) were cultured in a 5% CO2 atmosphere at 37°C. Cytotoxicity of ginsenoside Rh2, ginsenoside Rg3, and the new metabolite, Metabolite 1, was evaluated by 3-(4, 5-dimethylthiazol-2-yl)-2-5-diphenyltetrazolium bromide (MTT) assay following our previous procedure. The cell lines were seeded at a density of 1–1.2 × 105 cells/mL in a 96-well plate (NEST®, Nest Scientific USA Inc.,USA) in triplicate. After culturing for 24-h, the cells were treated with Rh2, Rg3, and Metabolite 1 (1, 10, 50, and 100 μM). After treatment was finished, 10 μL of MTT solution (5 mg/mL) was mixed to each well, and the plates were incubated for another 4 h. The media containing MTT reagent was then removed from each well, and 100 μL of dimethyl sulfoxide was added. After completion of this step, the absorbance of the reaction solution was measured at 570 nm/630 nm using a multimodel plate reader (BioTek Instruments, Vermont, USA).
2.9. Sequence analysis
The search of GT was carried out using the BLAST program at the National Center for Biotechnology Information server. The BlasTn program was used with the expectation value (E-value) lower than 10−4. By comparing the other GT, we identified and selected a sequence that was isolated from L. rhamnosus on the basis of the open reading frames encoding the specific protein via the National Center for Biotechnology Information BlastX program [42]. The sequence analysis of nucleotides and amino acids were carried out using the DNASIS program (Hitachi, Japan). The multiple alignments of GT isolated from L. rhamnosus were analyzed using ClustalX (http://www.ebi.ac.uk/tools/clustalw2). We constructed the phylogenetic tree using the neighbor-joining method. And for the reliability, each node was established using MEGA6 software by bootstrap methods [43,44]. The protein properties were estimated using ProtParam (https://web.expasy.org/protparam, SIB: Swiss Institute of Bioinformatics) [39], and the hydropathy value was estimated by the method described by Kyte and Doolittle [45]. Conserved motifs were identified using MEME (http://meme-suite.org, Multiple EM for Motif Elicitation) [43]. Another databases, such as SOPMA (http://npsa-pbil.ibcp.fr), HMMTOP (http://www.enzim.hu), MotifScan (http://myhits.isbsib.ch), and PSORT (http://psort.hgc.jp/form.html), were also used to analyze the full-length GT gene. A three-dimensional (3D) model was established using GT as a template on an SWISS-MODEL workspace in automated mode [6,26]. The generated 3D structure was visualized using the UCSF Chimera package (Resource for biocomputing, visualization, and informatics, University of California).
2.10. Statistical analysis
All our experiments were repeated in triplicate, independently. Statistical significance of all data was analyzed using the Student t test using Excel 2013 software. The p-value (less than 0.05) was considered significant statistically.
3. Results and discussion
3.1. Cloning and expression of recombinant LRGT
The designed primers and amplification were used to amplify the candidate gene. And then, the tagged gene from the L. rhamnosus genome was inserted into a pMAL-C5X vector and heterologously expressed in E. coli BL21 (DE3). The length of the sequence was 1224 bp, encoding a protein of 407 amino acid residues, the gene of which was homogeneous to GT family 2 (GenBank accession number: KY021433). To obtain the maximize yields of the fusion protein, diverse concentrations of protein induction were examined. As a result, 0.5 mM isopropyl β-D-thiogalactopyranoside produced the maximum amount of enzyme at 20°C for 9 h. The active protein was eluted using an elution buffer containing maltose [7]. The recombinant MBP-LRGT was purified by affinity chromatography using amylose resin and confirmed by SDS-PAGE analysis (Coomassie blue staining) (Fig. 1). What's more, the molecular mass of 81.2 kDa was calculated.
Fig. 1.
SDS-PAGE analysis of recombinant LRGT. M, molecular mass maker; 1, crude extract of induced recombinant BL 21 (DE3) cells carrying pMAL-LRGT; 2, pMAL-LRGT after purification with the MBP-bound agarose resin; MBP, maltose-binding protein; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
3.2. Biosynthesis of metabolites
Glycosylation contributes to the last step of biosynthesis of ginsenosides for functional diversity along with the capability to identify the diversity of ginsenosides. Generally, UGTs are capable of regulating the properties of ginsenosides (i.e., bioactivity and solubility) by transferring glycosyl residues from activated sugars to aglycones of ginesonides. Nonetheless, such an approach has been actively adopted for the screening of UGT genes in the ginsenoside biosynthesis by various researchers [[46], [47], [48]]. Here, in this report, we aimed to synthesize novel ginsenosides from 2.5 mM UDPG (donor) and 0.5 mM ginsenoside Rh2 (acceptor) using recombinant GT LRGT (0.1 mg/mL). After the reactants were extracted, two different obvious spots of Metabolites 1 and 2 were shown in the TLC plate except for Rh2 (Fig. 2). Moreover, no new spots were produced in the control reaction. The two signals of Metabolites 1 and 2 were observed by HPLC spectrum analysis (Fig. 3A); Metabolite 1 was the major product in the reaction mixtures, and we selected it for further analysis.
Fig. 2.
Thin layer chromatography (TLC) analysis of glycosylated products transformed by LRGT. C1, Control 1—enzyme and Rh2; C2, Control 2—enzyme and UDP glucose; C3, Rh2 and UDP glucose; R, result of synthesis; S, ginsenoside standard; UDP,
Fig. 3.
The kinetics of biosynthesis toward Metabolite 1 by LRGT were determined using the Michaelis–Menten model, the results of which are presented as follows: (A) HPLC spectra of the glycosylated metabolites transformed by LRGT. (B) The Lineweaver–Burk plot of LRGT. HPLC, high-performance liquid chromatography.
3.3. Kinetic parameters of LRGT
The kinetics parameters of LRGT with a diverse concentration of substrates were determined at 30°C and pH 7. The kinetic parameters of the products were analyzed by HPLC. The values were calculated using the Michaelis–Menten model. The results are shown in Fig. 3B. The values of Vmax and Km were estimated to be 0.10 μmol/L*min and 2.9 μmol/L as per the Lineweaver–Burk plot, respectively.
3.4. Optimal condition of temperature, pH, and metal ions of LRGT enzyme for synthesizing Metabolite 1
The optimal temperature was 30°C, which showed stronger activity than other temperature conditions (Fig. 4A). The optimal pH of LRGT activity was pH 7, which was more effective of catalytic activity than others (Fig. 4B). The effect of metal ions on LRGT enzyme activity was measured (Fig. 4C). The results showed enzyme activity was significantly stimulated by CoCl2, MgCl2, FeCl3, NaCl, CuSO4, NH4Cl, KCl, CaCl2, and ZnSO4. In addition, the optimum condition of metal ion regarding LRGT activity was intensively promoted by CuSO4.
Fig. 4.
Effect of glycosylated metabolites under diverse conditions including temperature, pH, and metal ions. (A) Effect of temperature on recombinant LRGT activity in synthesis of Metabolite 1. The most significant activity has been detected at the optimal temperature of 30°C. (B) Effect of pH on recombinant LRGT activity in synthesis of Metabolite 1. The optimal pH of LRGT activity was identified at pH 7, which was more effective while comparing catalytic activity with others. (C) Effect of metal ions on recombinant LRGT activity in synthesis of Metabolite 1. The optimal condition of metal ion regarding LRGT activity was intensively promoted by CuSO4.
3.5. Biosynthesis pathway of ginsenoside Rh2 by LRGT
Ginsenoside Rh2 (0.5 mM), 2.5 mM UDPG, and 10 mM CuSO4 with LRGT enzyme (pH 7, 0.1 mg/mL) were incubated at 30°C for 24 h in a shaking incubator. Then, the mass metabolites were separated and purified using open silica column chromatography. At the same time, the solvent system of CHCl3/CH3OH/H2O (65/35/10, lower phase) was used. After that, the first separated products were purified for further study by Waters Prep HPLC (Milford, U.S.A.) using a Nova-pak C18 column (3.9 × 300 mm) [15]. After harvesting, the results revealed that the yield of Metabolite 1 was 45.2%, whereas the yield of Metabolite 2 was 9.3%.
Metabolite 1 was a white and amorphous powder. The negative HRFABMS and 13C-NMR (DEPT) spectroscopic data showed that the molecular formula was C42H72O13 as deduced by a molecular ion peak m/z 783.4879 ([M-H]-, scaled for C42H71O13, 783.4895) (Fig. 5). There were absorption bands at 3401 cm−1 and 1649 cm−1 due to the OH group and double bond as per the IR spectrum, respectively. The 1H- and 13C-NMR spectra of Metabolite 1 confirmed an exception of the presence of an additional hexose sugar attached to Rh2. The observation of 13C-NMR signals of the glucosyl moiety characterized a hemiacetal (δC 105.0), four oxygenated methines (δC 78.2, 78.1, 75.3, 71.8), and an oxygenated methylene (δC 62.9), which indicated the glucosyl moiety to be a β-glucopyranose. What's more, the coupling constant of the anomer proton signal (δH 4.40, d, J = 7.8 Hz) confirmed the configuration was anomer hydroxy. A notably significant downfield with glycosylation shift at δ 69.9 (∼6.9 ppm, C6) indicated that a glucosyl moiety was attached to the C6–OH of Rh2 owing to the glycosidation shift. Similarly, the HMBC spectrum exhibited that the position of two sugars was confirmed to be C-6 and C-6’. The correlations of two anomer proton signals H-1’ (δH 4.30, d, J = 7.8 Hz) and H-1″ demonstrated the cross peaks with the oxygenated methine carbon signal (δC 90.7, C-3) and the oxygenated methylene carbon signal (δC 69.6, C-6′), respectively. Taken together, Fig. 6 depicted that the metabolite was identified to be (20S)-3β, 12β, 20-trihydroxydammar-24-ene-3-O-β-D-glucopyranosyl (1→6)-β-D-glucopyranoside [10,11]. Metabolite 1 has never been reported so far; therefore, it is a novel ginsenoside which is now named glucosyl ginsenoside Rh2 (Table 1).
Fig. 5.
MS spectrum of Metabolites 1 and 2 after transformation by recombinant LRGT. (A) Metabolite 1 molecular ion peak was m/z 783.4 ([M-H]-, scaled for C42H71O13, 783.4). (B) Metabolite 2 molecular ion peak was m/z 969.54014 ([M+Na]-, scaled for C48H82O18Na, 969.53934). MS, mass spectrometry.
Fig. 6.
Biosynthesic pathway of ginsenoside Rh2 to glucosyl ginsenoside Rh2 and diglucosyl ginsenoside Rh2 using LRGT.
Table 1.
1H- (600 MHz) and 13C-NMR (150 MHz) spectrum of glucosyl ginsenoside Rh2 and diglucosyl ginsenoside Rh2
| No of C | δC |
δH |
||
|---|---|---|---|---|
| 1 | 2 | 1 | 2 | |
| 1 | 40.4 | 40.3 | 1.71, 1.07, overlapped | 1.72, 1.09, overlapped |
| 2 | 27.6 | 27.4 | 1.86, 1.57, m | 1.87, 1.58, m |
| 3 | 90.7 | 90.4 | 3.20, overlapped | 3.21, overlapped |
| 4 | 41.1 | 40.4 | - | - |
| 5 | 57.7 | 57.4 | 0.79, br d, J = 11.7 Hz | 0.81, br d, J = 11.4 Hz |
| 6 | 19.4 | 19.3 | 1.57, 1.48, overlapped | 1.57, 1.47, overlapped |
| 7 | 36.1 | 35.9 | 1.54, 1.32, overlapped | 1.55, 1.32, overlapped |
| 8 | 38.1 | 38.0 | - | - |
| 9 | 51.5 | 51.2 | 1.49, overlapped | 1.49, overlapped |
| 10 | 40.5 | 40.9 | - | - |
| 11 | 32.2 | 32.1 | 1.26, 1.09, m | 1.26, 1.09, m |
| 12 | 71.6 | 71.4 | 3.33, overlapped | 3.33, overlapped |
| 13 | 49.7 | 49.6 | 1.73, dd, J = 10.8, 2.0 Hz | 1.74, overlapped |
| 14 | 52.8 | 52.6 | - | - |
| 15 | 32.2 | 32.0 | 1.89, 1.38, overlapped | 1.89, 1.38, overlapped |
| 16 | 27.4 | 27.3 | 1.96, 1.94, m | 1.98, 1.96, m |
| 17 | 55.3 | 55.1 | 2.11, overlapped | 2.06, overlapped |
| 18 | 16.3 | 16.4 | 1.01, s | 1.02, s |
| 19 | 17.0 | 17.1 | 0.92, s | 0.93, s |
| 20 | 74.5 | 74.2 | - | - |
| 21 | 26.6 | 26.8 | 1.14, s | 1.16, s |
| 22 | 36.5 | 36.3 | 1.52, overlapped, 1.34, ddd, J = 6.5, 6.5, 5.2 Hz, |
1.59, overlapped 1.40, ddd, J = 6.4, 6.4, 5.3 Hz |
| 23 | 23.4 | 23.2 | 2.16, 2.02, overlapped | 2.16, 2.03, overlapped |
| 24 | 126.3 | 126.4 | 5.13, m | 5.16, m |
| 25 | 132.1 | 131.9 | - | - |
| 26 | 26.1 | 26.3 | 1.68, s | 1.71, s |
| 27 | 17.9 | 18.1 | 1.61, s | 1.64, s |
| 28 | 28.5 | 28.6 | 1.04, s | 1.06, s |
| 29 | 17.0 | 17.1 | 0.85, s | 0.86, s |
| 30 | 17.3 | 17.4 | 0.91, s | 0.93, s |
| C-1′ | 106.9 | 106.7 | 4.30, d, J = 7.8 Hz | 4.33, d, J = 7.9 Hz |
| C-2′ | 75.8 | 75.5 | 3.19, overlapped | 3.20, overlapped |
| C-3′ | 78.3 | 78.0 | 3.28, overlapped, | 3.25, overlapped |
| C-4′ | 72.3 | 72.0 | 3.57, dd, J = 11.2, 5.0 Hz | 3.55, overlapped |
| C-5′ | 77.1 | 76.8 | 3.43, m | 3.44, overlapped |
| C-6′ | 69.9 | 69.9 | 4.11, br. d, J = 11.7 Hz, 3.79, dd, J = 11.7, 5.5 Hz |
4.16, br d, J = 11.3 Hz, 3.76, dd, J = 11.3, 5.3 Hz |
| C-1″ | 105.0 | 104.9 | 4.40, d, J = 7.8 Hz | 4.36, d, J = 7.8 Hz |
| C-2″ | 75.3 | 75.0 | 3.21, overlapped | 3.22, overlapped |
| C-3″ | 78.2 | 78.0 | 3.33, overlapped | 3.33, overlapped |
| C-4″ | 71.8 | 72.5 | 3.31, overlapped | 3.33, overlapped |
| C-5″ | 78.1 | 77.0 | 3.36, overlapped | 3.45, overlapped |
| C-6″ | 62.9 | 70.2 | 3.87, br. d, J = 11.7 Hz, 3.67, dd, J = 11.7, 5.4 Hz |
4.08, br d, J = 11.6 Hz, 3.80, dd, J = 11.6, 5.9 Hz |
| C-1‴ | - | 104.9 | 4.40, d, J = 6.7 Hz | |
| C-2‴ | - | 74.2 | 3.22, overlapped | |
| C-3‴ | - | 77.9 | 3.33, overlapped | |
| C-4‴ | - | 71.5 | 3.27, overlapped | |
| C-5‴ | - | 77.9 | 3.37, overlapped | |
| C-6‴ | - | 62.7 | 3.87, br d, J = 11.8 Hz, 3.67, dd, J = 11.8, 5.3 Hz |
|
NMR, nuclear magnetic resonance.
On the other hand, Metabolite 2 was also a white and amorphous powder. The negative HRFABMS and 13C-NMR (DEPT) spectroscopic data showed that the molecular formula was C48H82O18 as deduced by a molecular ion peak m/z 969.54014 ([M+Na]-, scaled for C48H82O18Na, 969.53934) (Fig. 5). There were absorption bands at 3401 cm−1 and 1649 cm−1 owing to the OH group and double bond as per the IR spectrum, respectively. The NMR data were similar to those of glucosyl ginsenoside Rh2 (Metabolite 1), with the exception of the presence of an additional hexose sugar. The observation of 13C-NMR signals of the glucosyl moiety characterized a hemiacetal (δC 104.9), four oxygenated methines (δC 77.9. 77.9. 74.2, 71.5), and an oxygenated methylene (δC 62.7), which indicated the glucosyl moiety to be a β-glucopyranose. Moreover, the coupling constant of the anomer proton signal (δH 4.40, d, J = 6.7 Hz) confirmed the configuration was anomer hydroxy. The glucosyl moiety was attached to the hydroxy of C-6″, an oxygenated methylene, from the chemical shift of C-6” (δC 70.2), which was downfield shifted by 7.3 ppm comparing to that of glucosyl ginsenoside Rh2 (Metabolite 1) owing to the glycosidation shift. In addition, the HMBC spectrum confirmed the position of three glucosyl moieties to be C-3, C-6′, and C-6”. The anomer proton signals H-1’ (δH 4.33, d, J = 7.9 Hz), H-1” (δH 4.36, d, J = 7.8 Hz), and H-1‴ (δH 4.40, d, J = 6.7 Hz) showed the cross peaks with the oxygenated methine carbon signal (δC 90.4, C-3), the oxygenated methylene carbon signal (δC 69.9, C-6‘), and the oxygenated methylene carbon signal (δC 70.2, C-6″), respectively. Therefore, Metabolite 2 was identified to be (20S)-3β,12β,20-trihydroxydammar-24-ene-3-O-β-D-glucopyranosyl (1→6)-β-D-Glucopyranosyl (1→6)-β-D-glucopyranoside (Fig. 6) [10,11]. Metabolite 2 has never been reported so far; therefore, it is a novel ginsenoside which is now named diglucosyl ginsenoside Rh2 (Table 1).
3.6. In vitro evaluation assay of glucosyl ginsenoside Rh2 on murine macrophage cell lines, preadipocyte cell lines, melanoma cell lines, and lung cancer cell lines
Ginsenosides derived from P. ginseng are well known for their biological activities, such as antiinflammation, antioxidant, and anticancer properties [3]. The ginsenoside Rh2 was reported for prominent antitumor effects against pancreatic cancer, leukemia, skin squamous cell carcinoma, prostatic cancer, and glioblastoma [8,12,49,50]. Furthermore, ginsenoside Rg3, the main metabolite, can be isolated from 9-times steam-processed red ginseng. Previous studies indicated that it was feasible to reduce the viability of cancer cells, induce apoptosis, and decrease angiogenesis through different mechanisms of many cancer cell lines, including A549 lung cancer cells [37]. Thus, glucosyl ginsenoside Rh2, which is derived from Rh2 and possesses the same formula and molecular weight but has a different glycosylated group at the C-3 position compared with Rg3, was used to treat RAW264.7 macrophage cells, B16BL6 melanoma cells, 3T3-L1, and A549 lung cancer cells to determinate its cytotoxicity using MTT assay.
In our study, it was found that glucosyl ginsenoside Rh2 exhibited less toxicity than Rh2 and Rg3 in RAW264.7, 3T3-L1, and B16BL6 cell lines up to a concentration of 50 μM after 24, 72, and 120 hours of treatment, respectively (Fig. 7A, C and D). On the other hand, glucosyl ginsenoside Rh2 was more toxic than ginsenoside Rh2 and Rg3 from a concentration of 10 μM in A549 lung cancer cells after 24 h of treatment (Fig. 7B). In previous studies, it was reported that Rh2 and Rg3 exhibited an activity for the treatment of inflammation, obesity, skin whitening, and cancer [4,38]. Because glucosyl ginsenoside Rh2 possesses less toxicity in inflammation-, obesity-, and skin whitening–related cell lines but higher toxicity in lung cancer cell lines, we hypothesize that glucosyl ginsenoside Rh2 might be a candidate for the treatment of these diseases, although further experiments are needed to confirm these properties.
Fig. 7.
Cytotoxicity assay of ginsenoside Rh2, Rg3, and glucosyl ginsenoside Rh2 on murine macrophage (RAW264.7), lung cancer (A549), preadipocyte (3T3-L1), and melanoma (B16BL6) cell lines. (A) Cell viability (%) of RAW264.7, exposed to ginsenoside Rh2, Rg3, and glucosyl ginsenoside Rh2. (B) Cell viability (%) of A549, exposed to ginsenoside Rh2, Rg3, and glucosyl ginsenoside Rh2. (C) Cell viability (%) of 3T3-L1, exposed to ginsenoside Rh2, Rg3, and glucosyl ginsenoside Rh2. (D) Cell viability (%) of B16BL6, exposed to ginsenoside Rh2, Rg3, and glucosyl ginsenoside Rh2. Such cell viability was determined by 3-(4,5-dimethyl-thiazol-2yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Data are expressed as a percentage of sample-treated control and presented as mean ± SD of three separate experiments. *P < 0.05, **P < 0.01, and ***P <0.001 vs. control. SD, standard deviation.
3.7. The protein properties estimated by ProtParam
The protein properties were estimated using ProtParam (http://web.expasy.org/protparam/) as described by Gasteiger et al [39]. From the BLASTn analysis, the sequence was found to have originated from L. rhamnosus (Supplementary Fig. S1). Protein sequence was identified based on their ORF encoding protein via BLASTx analysis. The sequence was found to belong to GT family 2. Multiple alignments of LRGT was analyzed using ClustalX [42]. Conserved motifs on LRGT were predicted using MEME [43] (Supplementary Fig. S2). As stated before, GTs are expected to be divided into two major types owing to the configuration of the anomeric functional group of the glycosyl moiety and the glycoconjugate metabolites: a kind of retaining GTs transfer sugar residue with the retention of anomeric configuration and another kind of inverting GTs transfer sugar residue with the inversion of anomeric configuration. One inverting GT with a conserved domain is responsible for the recognition of the UDP or thymidine diphosphate portion of a donor sugar–nucleotide molecule, which is Nucleotide Recognition Domain 1 beta (NRD1 beta), belonged to 140 GTs. The significant portion of NRD1 beta is intensely similar to the retaining GTs. On the basis of stereochemistry and similarity of glucosyl moiety transfer, the NRD1 alpha was designated to 77 GTs. Besides, there is a homologous region of the two families that probably has a catalytic function. In addition, a third conserved domain named NRD2 is present in 98 GTs and membrane-bound GTs. These three identified NRDs are presented in archaebacterial, eubacterial, viral, and eukaryotic GTs. The hydropathy value was determined following the method used by Kyte and Doolittle [45] using BioEdit software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The secondary structure of LRGT was predicted using SOPMA (self-optimized prediction method, https://npsa-prabi.ibcp.fr/cgi-bin) [6]. The SWISS-MODEL workspace in automated mode was used to predict a 3D model using the LRGT amino acid sequence as a template [26]; then, by using the UCSF Chimera package, the 3D structure was visualized (Fig. 8). The number of amino acids was 349, the molecular weight was 39631.23, and the theoretical pI was 6.26. The amino acid composition of LRGT was analyzed as follows: Ala (A), 36–10.3%; Arg (R), 21–6.0%; Asn (N), 15–4.3%; Asp (D), 26–7.4%; Cys (C), 1–0.3%; Gln (Q), 17–4.9%; Glu (E), 17–4.9%; Gly (G), 18–5.2%; His (H), 8–2.3%; Ile (I), 16–4.6%; Leu (L), 40–11.5%; Lys (K), 19–5.4%; Met (M), 10–2.9%; Phe (F), 15–4.3%; Pro (P), 12–3.4%; Ser (S), 22–6.3%; Thr (T), 11–3.2%; Trp (W), 5–1.4%; Tyr (Y), 17–4.9%; and Val (V), 23–6.6%. The number of negatively charged residues (Asp, Glu) was 43, whereas the number of positively charged residues (Arg, Lys) was 40. The atomic compositions are carbon (C-1783), hydrogen (H-2774), nitrogen (N-484), oxygen (O-518), and sulfur (S-11). It was established that the formula of LRGT is C1783H2774N484O518S11. The total number of atoms was 5570, the instability index was calculated to be 33.40, and this classified the protein to be stable. The aliphatic index was 92.01, and the grand average of hydropathicity was −0.247.
Fig. 8.
The predicted three-dimensional model using LRGT amino acid sequence as a template then by using the UCSF Chimera package.
4. Conclusion
In our study, the GT LRGT was identified from L. rhamnosus. The MBP-LRGT was purified, and the molecular weight was characterized as 81.2 kDa. The 3D structure of the protein was estimated using ProtParam. As shown by the results, the LRGT sequence was found to belong to GT family 2. Moreover, the optimal conditions for synthesis using LRGT enzyme were under 30°C and pH 7.0. Meanwhile, the optimal metal ion with regard to LRGT activity was intensively affected by CuSO4. Under optimal conditions, the mass products were produced from Rh2 using UDPG and LRGT. As shown by the results, ginsenoside Rh2 was transformed into two novel ginsenosides: glucosyl ginsenoside Rh2, which had the same formula and molecular weight as those of Rg3 but one glucose attached at the C-3 position of Rh2 with C6→1 linkage and is the isomer of Rg3 (C2→1 linkage); diglucosyl ginsenoside Rh2, which has two glucosyl moieties attached at the C-3 position of Rh2 with C6→1 linkage. Our study suggested that the diversity of ginsenosides could be generated by specific and efficient recombinant GT enzymatic biotransformation. As a result, the LRGT enzyme is critical for successful synthesis of mass production of potential promising active novel ginsenosides in vitro.
Ginsenosides are unique, famous, and the main active ingredient with various biological activities and considerable economic value. Recently, the uncontrolled and excessive collection led to the number of wild ginseng resources have become reduced. Ginsenosides are exceptionally difficult to convert, and their structure is complicated. Through years of efforts of scientists, the biosynthetic pathway almost has been completed. However, the glycosylation of ginsenoside aglycone is still in its initial phase. As a multigene family, the large number of types, specific sequence, and selectivity of GT have brought challenges of research of ginsenosides aglycone glycosylation. So far, the number of GT which has been completed to be identified is extremely limited. But what can be expected is that, it will be a new generation of ginsenosides production pathway using synthetic biotechnology to produce engineering bacterial along with the deepening of the GT enzyme combined with biological fermentation method [14]. In our study, we synthesized two novel ginsenosides by recombinant GT enzymatic biotransformation and focused on the main product of diglucosyl ginsenoside Rh2 for in vitro applications.
Several studies already found that ginsenosides Rh2 and Rg3 which inhibit tumor cells also show toxicity to other cells. In the present study, the cytotoxicity of the main product, diglucosyl ginsenoside Rh2, was determined in vitro in murine macrophage cell lines, preadipocyte cells, melanoma cell lines, and lung cancer cells and was conjointly compared with that of Rh2 and Rg3, which are well-known saponins with anticancer activity. We found that diglucosyl ginsenoside Rh2 showed less toxicity than Rh2 and Rg3 in normal cells but more toxicity than Rh2 and Rg3 from a concentration of 10 μM in A549 lung cancer cells. Thus, diglucosyl ginsenoside Rh2 might be a new candidate for treatment of inflammation, obesity, and skin whitening, and especially for anticancer.
Conflicts of interest
The authors have no conflicts of interest to report.
Acknowledgments
This work was supported by the grant from Kyung Hee University in 2020 (KHU-20202298); Basic Science Research Program through the National Research Foundation of Korea (2019R1A2C1010428); and the grant by the Natural Science Foundation of Shandong Province, (ZR2018BH035), in China.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgr.2019.11.004.
Contributor Information
Yeon-Ju Kim, Email: yeonjukim@khu.ac.kr.
Deok-Chun Yang, Email: dcyang@khu.ac.kr.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. S1.
A neighbor-joining–based phylogenetic tree was constructed using MEGA6 software with the bootstrap method. GT, glycosyltransferase.
Supplementary Fig. S2.
Multiple alignments of LRGT and predicted conserved motifs on LRGT by using MEME. GT, glycosyltransferase.
References
- 1.Kim Y.J., Jeon J.N., Jang M.G., Oh J.Y., Kwon W.S., Jung S.K., Yang D.C. Ginsenoside profiles and related gene expression during foliation in Panax ginseng Meyer. J Ginseng Res. 2014;38:66–72. doi: 10.1016/j.jgr.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Radad K., Gille G., Liu L., Rausch W.D. Use of ginseng in medicine with emphasis on neurodegenerative disorders. J Pharmacol Sci. 2006;100:175–186. doi: 10.1254/jphs.crj05010x. [DOI] [PubMed] [Google Scholar]
- 3.Leung K.W., Wong A.S.-T. Pharmacology of ginsenosides: a literature review. Chin Med. 2010;5:20. doi: 10.1186/1749-8546-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park E.J., Yi J., Chung K.H., Ryu D.Y., Choi J., Park K. Oxidative stress and apoptosis induced by titanium dioxide nanoparticles in cultured BEAS-2B cells. Toxicol Lett. 2008;180:222–229. doi: 10.1016/j.toxlet.2008.06.869. [DOI] [PubMed] [Google Scholar]
- 5.Choi K.T. Botanical characteristics, pharmacological effects and medicinal components of Korean Panax ginseng C A Meyer. Acta Pharmacol Sin. 2008;29:1109–1118. doi: 10.1111/j.1745-7254.2008.00869.x. [DOI] [PubMed] [Google Scholar]
- 6.Geourjon C., Deleage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 7.Quan L.H., Min J.W., Yang D.U., Kim Y.J., Yang D.C. Enzymatic biotransformation of ginsenoside Rb1 to 20 (S)-Rg3 by recombinant β-glucosidase from Microbacterium esteraromaticum. Appl Microbiol Biotechnol. 2012;94:377–384. doi: 10.1007/s00253-011-3861-7. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Q., Hong B., Wu S., Niu T. Inhibition of prostatic cancer growth by ginsenoside Rh2. Tumor Biol. 2015;36:2377–2381. doi: 10.1007/s13277-014-2845-5. [DOI] [PubMed] [Google Scholar]
- 9.Chae S., Kang K.A., Chang W.Y., Kim M.J., Lee S.J., Lee Y.S., Kim H.S., Kim D.H., Hyun J.W. Effect of compound K, a metabolite of ginseng saponin, combined with γ-ray radiation in human lung cancer cells in vitro and in vivo. J Agric Food Chem. 2009;57:5777–5782. doi: 10.1021/jf900331g. [DOI] [PubMed] [Google Scholar]
- 10.Fu B.D., Bi W.Y., He C.L., Zhu W., Shen H.Q., Yi P.F., Wang L., Wang D.C., Wei X.B. Sulfated derivatives of 20 (S)-ginsenoside Rh2 and their inhibitory effects on LPS-induced inflammatory cytokines and mediators. Fitoterapia. 2013;84:303–307. doi: 10.1016/j.fitote.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 11.Li L., Chen X., Zhou J., Zhong D. In vitro studies on the oxidative metabolism of 20 (s)-ginsenoside Rh2 in human, monkey, dog, rat, and mouse liver microsomes, and human liver s9. Drug Metab and Dispos. 2012;40:2041–2053. doi: 10.1124/dmd.112.046995. [DOI] [PubMed] [Google Scholar]
- 12.Chung K.S., Cho S.H., Shin J.S., Kim D.H., Choi J.H., Choi S.Y., Rhee Y.K., Hong H.D., Lee K.T. Ginsenoside Rh2 induces cell cycle arrest and differentiation in human leukemia cells by upregulating TGF-β expression. Carcinogenesis. 2013;34:331–340. doi: 10.1093/carcin/bgs341. [DOI] [PubMed] [Google Scholar]
- 13.Ahn S., Siddiqi M.H., Noh H.-Y., Kim Y.-J., Kim Y.-J., Jin C.-G., Yang D.-C. Anti-inflammatory activity of ginsenosides in LPS-stimulated RAW 264.7 cells. Sci Bull. 2015;60:773–784. [Google Scholar]
- 14.Mathiyalagan R., Subramaniyam S., Kim Y.J., Kim Y.C., Yang D.C. Ginsenoside compound K-bearing glycol chitosan conjugates: synthesis, physicochemical characterization, and in vitro biological studies. Carbohydr Polym. 2014;112:359–366. doi: 10.1016/j.carbpol.2014.05.098. [DOI] [PubMed] [Google Scholar]
- 15.Mathiyalagan R., Kim Y.H., Kim Y.J., Kim M.K., Kim M.J., Yang D.C. Enzymatic formation of novel ginsenoside Rg1-α-glucosides by rat intestinal homogenates. Appl Biochem Biotechnol. 2015;177:1701–1715. doi: 10.1007/s12010-015-1847-0. [DOI] [PubMed] [Google Scholar]
- 16.Wang H.P., Zhang Y.B., Yang X.W., Zhao D.Q., Wang Y.P. Rapid characterization of ginsenosides in the roots and rhizomes of Panax ginseng by UPLC-DAD-QTOF-MS/MS and simultaneous determination of 19 ginsenosides by HPLC-ESI-MS. J Ginseng Res. 2016;40:382–394. doi: 10.1016/j.jgr.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye H., Wu Q., Zhu Y., Guo C., Zheng X. Ginsenoside Rh2 alleviates dextran sulfate sodium-induced colitis via augmenting TGFβ signaling. Mol Biol Rep. 2014;41:5485–5490. doi: 10.1007/s11033-014-3422-0. [DOI] [PubMed] [Google Scholar]
- 18.Choi S., Kim T.W., Singh S.V. Ginsenoside Rh2-mediated G1 phase cell cycle arrest in human breast cancer cells is caused by p15 Ink4B and p27 Kip1-dependent inhibition of cyclin-dependent kinases. Pharm Res. 2009;26:2280–2288. doi: 10.1007/s11095-009-9944-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kikuchi Y., Sasa H., Kita T., Hirata J., Tode T., Nagata I. Inhibition of human ovarian cancer cell proliferation in vitro by ginsenoside Rh2 and adjuvant effects to cisplatin in vivo. Anti-cancer Drugs. 1991;2:63–68. doi: 10.1097/00001813-199102000-00009. [DOI] [PubMed] [Google Scholar]
- 20.Li B., Zhao J., Wang C.Z., Searle J., He T.C., Yuan C.S., Du W. Ginsenoside Rh2 induces apoptosis and paraptosis-like cell death in colorectal cancer cells through activation of p53. Cancer Lett. 2011;301:185–192. doi: 10.1016/j.canlet.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J., Shimizu K., Yu H., Zhang C., Jin F., Kondo R. Stereospecificity of hydroxyl group at C-20 in antiproliferative action of ginsenoside Rh2 on prostate cancer cells. Fitoterapia. 2010;81:902–905. doi: 10.1016/j.fitote.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 22.Nakata H., Kikuchi Y., Tode T., Hirata J., Kita T., Ishii K., Kudoh K., Nagata I., Shinomiya N. Inhibitory effects of ginsenoside Rh2 on tumor growth in nude mice bearing human ovarian cancer cells. Jpn J Cancer Res. 1998;89:733–740. doi: 10.1111/j.1349-7006.1998.tb03278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang X.P., Tang G.D., Fang C.Y., Liang Z.H., Zhang L.Y. Effects of ginsenoside Rh2 on growth and migration of pancreatic cancer cells. World J Gastroenterol. 2013;19:1582–1592. doi: 10.3748/wjg.v19.i10.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tode T., Kikuchi Y., Kita T., Hirata J., Imaizumi E., Nagata I. Inhibitory effects by oral administration of ginsenoside Rh2 on the growth of human ovarian cancer cells in nude mice. J Cancer Res Clin Oncol. 1993;120:24–26. doi: 10.1007/BF01200720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi J., Cao B., Zha W.B., Wu X.L., Liu L.S., Xiao W.J., Gu R.R., Sun R.B., Yu X.Y., Zheng T. Pharmacokinetic interactions between 20 (S)-ginsenoside Rh2 and the HIV protease inhibitor ritonavir in vitro and in vivo. Acta Pharmacol Sin. 2013;34:1349–1358. doi: 10.1038/aps.2013.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnold K., Bordoli L., Kopp J., Schwede T. The SWISS-MODEL Workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 27.Hu Y., Walker S. Remarkable structural similarities between diverse glycosyltransferases. Chem Biol. 2002;9:1287–1296. doi: 10.1016/s1074-5521(02)00295-8. [DOI] [PubMed] [Google Scholar]
- 28.Taniguchi N., Honke K., Fukuda M. Springer Science & Business Media; 2011. Handbook of glycosyltransferases and related genes. [Google Scholar]
- 29.Coutinho P.M., Deleury E., Davies G.J., Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 30.Tarbouriech N., Charnock S.J., Davies G.J. Three-dimensional structures of the Mn and Mg dTDP complexes of the family GT-2 glycosyltransferase SpsA: a comparison with related NDP-sugar glycosyltransferases. J Mol Biol. 2001;314:655–661. doi: 10.1006/jmbi.2001.5159. [DOI] [PubMed] [Google Scholar]
- 31.Mackenzie P.I., Bock K.W., Burchell B., Guillemette C., Ikushiro Si, Iyanagi T., Miners J.O., Owens I.S., Nebert D.W. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics. 2005;15:677–685. doi: 10.1097/01.fpc.0000173483.13689.56. [DOI] [PubMed] [Google Scholar]
- 32.Bowles D., Lim E.-K., Poppenberger B., Vaistij F.E. Glycosyltransferases of lipophilic small molecules. Annu Rev Plant Biol. 2006;57:567–597. doi: 10.1146/annurev.arplant.57.032905.105429. [DOI] [PubMed] [Google Scholar]
- 33.Yan X., Fan Y., Wei W., Wang P., Liu Q., Wei Y., Zhang L., Zhao G., Yue J., Zhou Z. Production of bioactive ginsenoside compound K in metabolically engineered yeast. Cell Res. 2014;24:770–773. doi: 10.1038/cr.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pandey R.P., Li T.F., Kim E.H., Yamaguchi T., Park Y.I., Kim J.S., Sohng J.K. Enzymatic synthesis of novel phloretin glucosides. Appl Environ Microbiol. 2013;79:3516–3521. doi: 10.1128/AEM.00409-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang H.C., Wang Q.H., Gong T., Du G.H., Yang J.L., Zhu P. The basic strategies and research advances in the studies on glycosyltransferases involved in ginsenoside biosynthesis. Acta Pharm Sin. 2015;50:148–153. [PubMed] [Google Scholar]
- 36.Dai L., Liu C., Li J., Dong C., Yang J., Dai Z., Zhang X., Sun Y. One-pot synthesis of ginsenoside Rh2 and bioactive unnatural ginsenoside by coupling promiscuous glycosyltransferase from Bacillus subtilis 168 to sucrose synthase. J Agric Food Chem. 2018;66:2830–2837. doi: 10.1021/acs.jafc.8b00597. [DOI] [PubMed] [Google Scholar]
- 37.Chen M., Ni L., Zhao X., Niu X. The inhibition of 20 (R)-ginsenoside Rg3 on the expressions of angiogenesis factors proteins in human lung adenocarcinoma cell line A549 and HUVEC304 cell. Zhongguo Zhong Yao Za Zhi. 2005;30:357–360. [PubMed] [Google Scholar]
- 38.Lee S.J., Lee W.J., Chang S.E., Lee G.-Y. Antimelanogenic effect of ginsenoside Rg3 through extracellular signal-regulated kinase-mediated inhibition of microphthalmia-associated transcription factor. J Ginseng Res. 2015;39:238–242. doi: 10.1016/j.jgr.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. In: The proteomics protocols handbook, protein identification and analysis tools on the ExPASy server. John M.W., editor. Humana Press; 2005. pp. 571–607. [Google Scholar]
- 40.Kant R., Rintahaka J., Yu X., Sigvart-Mattila P., Paulin L., Mecklin J.-P., Saarela M., Palva A., Ossowski I. A comparative pan-genome perspective of Niche-adaptable cell-surface protein phenotypes in Lactobacillus rhamnosus. PLOS ONE. 2014;9(7) doi: 10.1371/journal.pone.0102762. e102762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang D.D., Jin Y., Wang C., Kim Y.J., Perez Z.E.J., Baek N.I., Mathiyalagan R., Markus J., Yang D.C. Rare ginsenoside Ia synthesized from F1 by cloning and overexpression of the UDP-glycosyltransferase gene from Bacillus subtilis: synthesis, characterization, and in vitro melanogenesis inhibition activity in BL6B16 cells. J Ginseng Res. 2018;42:42–49. doi: 10.1016/j.jgr.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson J.D., Gibson T., Higgins D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002;6:2–3. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- 43.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 46.Jacobsen C.N., Rosenfeldt Nielsen V., Hayford A.E., Møller P.L., Michaelsen K.F., Pærregaard A., Sandstrom B., Tvede M., Jakobsen M. Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl Environ Microbiol. 1999;65:4949–4956. doi: 10.1128/aem.65.11.4949-4956.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bertazzoni Minelli E., Benini A., Marzotto M., Sbarbati A., Ruzzenente O., Ferrario R., Hendrinks H., Dellaglio F. Assessment of novel probiotic Lactobacillus casei strains for the production of functional dairy foods. Int Dairy J. 2004;14:723–736. [Google Scholar]
- 48.Luo H.M., Sun C., Sun Y.Z., Wu Q., Li Y., Song J.Y., Niu Y.Y., Cheng X., Xu H.X., Li C.Y. Analysis of the transcriptome of Panax notoginseng root uncovers putative triterpene saponin–biosynthetic genes and genetic markers. BMC Genom. 2011;12:S5. doi: 10.1186/1471-2164-12-S5-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li S., Gao Y., Ma W., Guo W., Zhou G., Cheng T., Liu Y. EGFR signaling-dependent inhibition of glioblastoma growth by ginsenoside Rh2. Tumor Biol. 2014;35:5593–5598. doi: 10.1007/s13277-014-1739-x. [DOI] [PubMed] [Google Scholar]
- 50.Liu S., Chen M., Li P., Wu Y., Chang C., Qiu Y., Cao L., Liu Z., Jia C. Ginsenoside rh2 inhibits cancer stem-like cells in skin squamous cell carcinoma. Cellular Physiol Biochem. 2015;36:499–508. doi: 10.1159/000430115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.