Abstract
Background
Lung cancer has a high incidence worldwide, and most lung cancer-associated deaths are attributable to cancer metastasis. Although several medicinal properties of Panax ginseng Meyer have been reported, the effect of ginsenosides Rk1 and Rg5 on epithelial-mesenchymal transition (EMT) stimulated by transforming growth factor beta 1 (TGF- β1) and self-renewal in A549 cells is relatively unknown.
Methods
We treated TGF-β1 or alternatively Rk1 and Rg5 in A549 cells. We used western blot analysis, real-time polymerase chain reaction (qPCR), wound healing assay, Matrigel invasion assay, and anoikis assays to determine the effect of Rk1 and Rg5 on TGF-mediated EMT in lung cancer cell. In addition, we performed tumorsphere formation assays and real-time PCR to evaluate the stem-like properties.
Results
EMT is induced by TGF-β1 in A549 cells causing the development of cancer stem-like features. Expression of E-cadherin, an epithelial marker, decreased and an increase in vimentin expression was noted. Cell mobility, invasiveness, and anoikis resistance were enhanced with TGF-β1 treatment. In addition, the expression of stem cell markers, CD44, and CD133, was also increased. Treatment with Rk1 and Rg5 suppressed EMT by TGF-β1 and the development of stemness in a dose-dependent manner. Additionally, Rk1 and Rg5 markedly suppressed TGF-β1-induced metalloproteinase-2/9 (MMP2/9) activity, and activation of Smad2/3 and nuclear factor kappa B/extra-cellular signal regulated kinases (NF-kB/ERK) pathways in lung cancer cells.
Conclusions
Rk1 and Rg5 regulate the EMT inducing TGF-β1 by suppressing the Smad and NF-κB/ERK pathways (non-Smad pathway).
Keywords: Epithelial-mesenchymal transition, Ginsenosides, Lung cancer, Panax ginseng Meyer, Transforming growth factor β1
1. Introduction
Lung cancer is one of the leading causes for mortality. Non-small cell lung carcinoma (NSCLC) is the most common lung malignancy and contributes to approximately 80% of all lung cancers [[1], [2], [3]]. One of the major challenges associated with lung cancer management is cancer metastasis. Even with advancements in cancer treatment and management, there are no effective treatments for lung cancer metastasis [4]. Therefore, understanding the metastasis mechanisms is necessary for successful treatment of NSCLC.
Cancer metastasis is a multistep process. One of the prerequisites for the initiation of metastasis is the migratory and invasive behavior of cancer [5]. Epithelial-mesenchymal transition (EMT) is crucial in cancer metastasis because it is the earliest processes. During EMT, cancer cells undergo morphological changes where the epithelial phenotype is converted to a mesenchymal phenotype by decreasing adhesion between cell-to-cell junctions, which leads to acquiring migratory and invasive capabilities [6,7]. During EMT, the expression of E-cadherin decreases, promoting cell motility, invasiveness, and imparting anoikis resistance owing to the dysregulated activity of several transcriptional repressors such as Zeb2 (SIP1), Twist, Snail, and Slug (Snail2). The transcription factors regulating EMT have been identified in numerous malignant cancers [[8], [9], [10]]. Transforming growth factor-beta (TGF-β), which suppresses cell growth, differentiation, and death, is known to be particularly relevant to EMT [11,12]. It has a vital role in cancer metastasis and promotes cancer invasiveness and metastasis during EMT [13].
According to recent studies, EMT is associated with the formation of stem-like cells from differentiated cancer cells. Cancer stem-like cells (CSCs) play a significant role in metastasis, chemotherapy resistance, and recurrence. CSCs have pluripotent and self-renewal properties and can seed metastatic tumors at distant sites [14,15]. Understanding the properties of CSCs is necessary to develop effective treatment strategies for metastatic cancers.
Panax ginseng Meyer, the root of P. ginseng has been used as functional food and herbal medicine for thousands of years worldwide, especially in Korea and China, owing to its medicinal properties [16,17]. The biologically active components of ginseng are called ginsenosides. For example, it has been recently evaluated for the antiarthritic effects of Rg3, Rk1, and Rg5, using a collagen-induced mouse arthritis model [18]. There are several methods for enhancing the bioactive properties of ginseng. In an earlier study, we had described the conversion of ginsenosides by microwave-thermal processing to improve the bioactive properties of ginseng [19]. Microwave-thermal processing has several advantages over conventional methods, such as autoclaving. Microwave-processed ginseng extract, developed by microwave-assisted processing, contains higher quantities of ginsenosides Rg3, Rk1, and Rg5 [20,21]. Several studies have reported the anticancer properties of Rg3; however, the effects of Rk1 and Rg5 on cancer progression are relatively unknown. In particular, to the best of our knowledge, there are no studies on the role of Rk1 and Rg5 in TGF-β1-mediated EMT in human NSCLC. This study investigated the anticancer effects of Rk1 and Rg5 on TGF-β1-promoted EMT in A549 cells.
2. Materials and methods
2.1. Preparation of processed ginseng – Korea Institute of Science and Technology-maximized ginsenoside using microwave irradiation
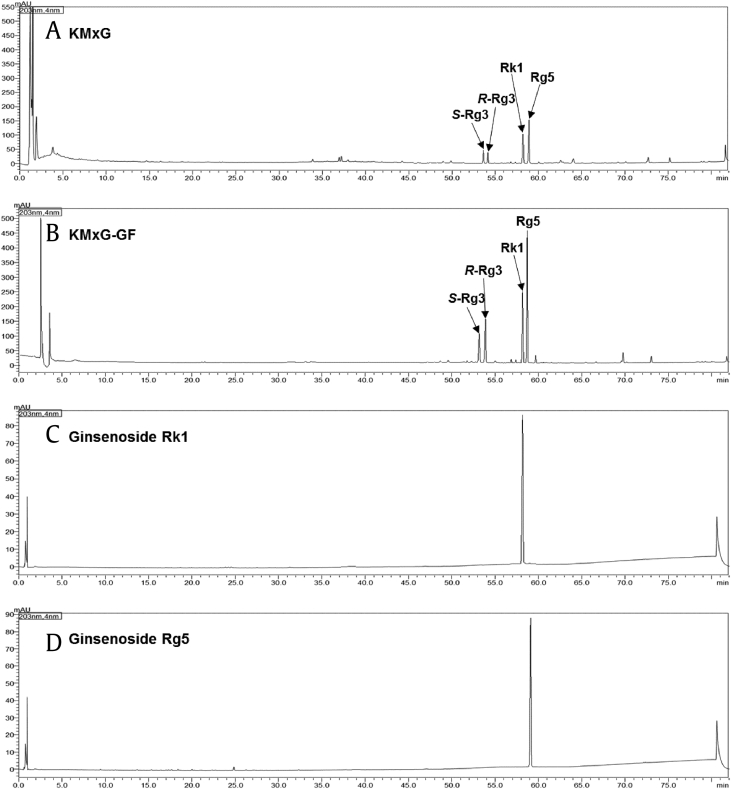
P. ginseng (1 kg) was extracted three times under reflux with 50% ethanol (EtOH) (7 L) at 80 °C for 2 h and then filtered. The filtrate was evaporated under vacuum to obtain the white ginseng extract (220 g). The white ginseng extract (WG) was processed using microwave irradiation, as reported previously [20]. In short, WG (15 g) was added to 30 mL 50% EtOH in an 80-mL vessel of a microwave reactor (Discover, CEM Co., USA). The WG was processed with microwave reactor in the sealed vessel at a wattage of 100 W (a frequency of 2450 MHz) and temperature of 150 °C for 1 h. The microwave-processed dry ginseng extract, i.e., KIST-maximized ginsenoside (KMxG) was freeze-dried to yield the microwave-processed extracts (Fig. 1A). Microwave process was performed at a pressure of 15 atm. The pressure of reaction was automatically controlled by the microwave equipment.
Fig. 1.
UHPLC chromatogram of KMxG, KMxG-GF, ginsenoside Rk1, and Rg5.
2.2. Purification of ginsenoside fractions – KIST-maximized ginsenoside-ginsenoside fractionation
KMxG extract (10 g) was treated with flash column chromatography on a Diaion HP-20 (Mitsubishi Chemical Co., Ltd., Japan) and eluted stepwise with 1 L of distilled water and methanol (MeOH). The water and MeOH fractions were evaporated under a vacuum to obtain the water fraction (KMxG−H2O, 7.6 g) and the MeOH fraction (KMxG−GF, 2.1 g) (Fig. 1B).
2.3. Purification of Rk1 and Rg5
KMxG−GF (1.0 g) was separated by preparative HPLC using an acetonitrile/water mobile phase (40/60 to 80/20; v/v, 60 min) and Phenomenex Luna C18 (2) column (250 × 21.2 mm, 5 μm) at a flow rate of 8 mL/min to yield four peaks. Purification of peak 1, 2, 3, and 4 was performed by semipreparative HPLC using a MeOH/water mobile phase (50/50 to 100/0; v/v, 60 min) and Phenomenex Gemini C6 Phenyl column (250 × 10 mm, 5 μm) at a flow rate of 4 mL/min to obtain ginsenosides S-Rg3 (86.4 mg), R-Rg3 (82.6 mg), Rk1 (176.1 mg, Fig. 1C), and Rg5 (361.7 mg, Fig. 1D). The chemical structures of ginsenosides were determined by comparing with previously reported reference compounds [21].
2.4. Analysis of ginsenosides
Ginsenosides were identified and quantified using a previously reported method [21]. An analytical reversed phase Shimadzu Nexera X2 UHPLC system comprising of a solvent degassing unit (DGU−20A), binary pump (LC−30AD), autosampler (SIL−30AC), system controller unit (CBM−20A), photodiode array detector (SPD−M20A), and column oven unit (CTO−20AC) was used for qualitative and/or quantitative analysis. Electrospray ionization (ESI)-mass spectrometry (MS) was performed using a Shimadzu LCMS-2020 system for qualitative analysis. A Phenomenex Luna Omega polar C18 column (150 × 2.1 mm, 1.6 μm) was used. The mobile phase consisted of binary gradients of solvent A (water) and solvent B (acetonitrile) The flow rate was set to 0.3 mL/min. The gradient flow program was as follows – initial: 15% B, 28.5 min: 30% B, 30.5 min: 32% B, 36.5 min: 38% B, 47.5 min: 43% B, 54 min: 55% B, 62 min: 55% B, 70 min: 70% B, 76 min: 90% B, 79 min: 95% B, and 82 min: 15% B. The detection wavelength was 203 nm. The analysis was performed three times. The ginsenoside standard samples were purchased from Ambo Institute (Daejeon, Korea). Standard solutions containing 10−250 ppm of each ginsenoside were injected into the column, with all calibration curves showing good linearity (R2 ≥ 0.9992, Supplementary Data Figure S11). The contents of ginsenosides in 1g of KMxG-GF were 102.2 mg/g (S-Rg3), 108.1 mg/g (R-Rg3), 258.2 mg/g (Rk1), and 410.7 mg/g (Rg5), respectively.
2.5. Reagents and antibodies
Ginsenoside, KMxG crude, KMxG-GF, Rk1, and Rg5; prepared 50 mg/mL stock solution in dimethyl sulfoxide (DMSO) and placed at −20°C. Recombinant human TGF-β1 was purchased from ProSpec (EST Brunswick, NJ). Anti-E-cadherin antibody was obtained from BD Biosciences (San Jose, CA). Anti-Cleaved PARP, p-Smad2, Smad2, p-Smad3, Smad3, Nuclear factor kappa B (NF-κB), p-ERK, and extra-cellular signal regulated kinases (ERK), Snail, Slug were obtained from Cell Signaling Technology (Beverly, MA). Anti-vimentin, Tubulin, and Actin, Santa Cruz Biotechnology (Dallas, TX), were used. All secondary antibodies were obtained from Pierce (Rockford, IL).
2.6. Cell culture
Human lung cancer A549 cells were purchased from the American Type Culture Collection (Manassas, VA), which was grown in Roswell Park Memorial Institute (RPMI 1640) medium with 10% fetal bovine serum (FBS), 1% streptomycin/penicillin (Logan, UT) in 37°C incubator with 5% CO2. TGF (5 ng/mL) was treated with RPMI-1640 complete media when cell confluence reached 30–40% and ginsenoside, KMxG crude, KMxG-GF, Rk1, or Rg5 at different concentrations were also treated with TGF for 48 h.
2.7. Analysis of cell proliferation rate
To investigate the cell proliferation, Water Soluble Tetrazolium Salts (WST)-based Cell Proliferation Assay (EZ-cytox, Dogen Bio, Seoul, Korea) was used. A549 cells (5 × 103 cells/well) were seeded into 96-well plates triplicated for each group and incubated at 37°C overnight. The next day, the cells were treated with various concentrations of ginsenoside, KMxG crude, KMxG-GF, Rk1, or Rg5 for 24 or 48 h. After incubation, cells were reached with 10 μL EZ-cytox solution and incubated for 2 h. Cell proliferation was detected at 450 nm absorbance using microplate reader.
2.8. Western blot analysis
Cells were lysed using radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany) for 10 min under ice conditions and centrifuged at 10,000 rpm for 20 min. To quantify protein content, the Bradford assay (Bio7 Rad, Hercules, CA) was employed using the supernatant, which is a protein layer. The quantified proteins were 8 mixed with 2 × sample buffer (Bio-Rad, Hercules, CA) and denatured to a linear structure by linear heat treatment at 95°C for 5 min and used as a loading sample for sodium dodecyl sulfate - polyacrylamide gel electrophoresis (SDS-PAGE). A constant amount of protein (10–30 μg) was loaded at 120 V, and the protein was separated by molecular weight and 8–12% acrylamide gel was used. Then, transfer step was performed using a polyvinylidene difluoride (PVDF) membrane at 90 V for 2 h and reacted with the primary antibody at 4°C for overnight. After incubation, the transferred membrane was washed with TBST buffer for 5 min three times and reacted with the secondary antibody for 1–2 h at RT. Then, washing step was performed with Tris Buffered Saline with Tween (TBST) buffer, exposed to enhanced chemiluminescence (ECL) solution, and visualized with a chemiluminescence reader (Vilber Lourmat, Collégien, France). The expression was analyzed by using FuisonCapt software program (Vilber Lourmat, Collégien, France).
2.9. RNA isolation and quantitative real-time polymerase chain reaction
Total RNA isolation from the lung cancer cells were performed using RNA isoPlus reagent (Takara Bio, Shiga, Japan). The cells were suspended in the reagent, mixed with the reagent and chloroform at a ratio of 5:1, vortexed, and centrifuged at 13,000 rpm for 10 min. After centrifugation, the highest layer was taken and mixed in the equal ratio as isopropanol. The mixture was reacted at room temperature (RT) for 10 min and then centrifuged at 13,000 rpm for 5 min. The precipitated protein was centrifuged at 10,000 rpm for 5 min with diethyl pyrocarbonate (DEPC)-ethanol, washed, air-dried, and dissolved with autoclaved DEPC- water. Subsequently, cDNA synthesis was performed to cDNA Reverse Transcription kit (Applied biosystem, Carlsbad, CA). When quantitative real-time PCR (qRT-PCR) was performed, 2 × Master Mix (ELPIS Bio, South Korea) was used. The information on the primer sequence of the target genes is given as follows. E-cadherin (CDH1): forward: 5′-AAAGGCCCATTTCCTAAAAACC-3′ and reverse: 5′-TGCGTTCTCTATCCAGAGGCT-3′; vimentin (VIM): forward: 5′-TGTCCAAATCGATGTGGATGTTTC-3′ and reverse: 5′-TTGTACCATTCTTCT-GCCTCCTG-3′; Snail (SNAI1): forward: 5′-CCCCAATCGGAAGCCTAACT-3′ and reverse: 5′-GCTGGAAGGTAAACTCTGGATTAG-3′; Slug (SNAI2): forward: 5′-GAGCATACAGCCCCATCAC-T-3′ and reverse: 5′-GGGTCTGAAAGCTT-GGACTG-3′; MMP-2: forward: 5′-ACATCAAGGGCATTCAGGAG-3′ and reverse: 5′-GCCTCGTATACCGCATCAAT-3′; MMP-9: forward: 5′-CATCGTCATCCAG-TTTGGTG-3′ and reverse: 5′-TCGAAGATGAAGGGGAAGTG-3′; CD44: forward: 5′-TCACAGGTGGAAGAAGAGAC-3′ and reverse: 5′-CATTGCCACTGTTGATC-ACT-3′; Oct-4: forward: 5′-CTTGAATCCCGAATGGAAAGGG-3′ and reverse: 5′-GTGTATATCCCAGGGTGATCCTC-3′; CD133: forward: 5′-CAACTACATGGT-TTACATGTTC-3′ and reverse: 5′-GCCAGTGGACTCCACGAC-3′; Nanog: forward: 5′-AGCTGGGTGGAAGAGAACACAG-3′ and reverse: 5′-CTATGGCG-AACACTTGCAGATTAC-3′; SOX-2: forward: 5′-GCACATGAACGGCTGGAGC-AAC-3′ and reverse: 5′-TGCTGCGAGTAGGACATGCTGT-3′; GAPDH: forward: 5′-CGCGGGGCTCTCCAGAACATCATCC-3′ and reverse: 5′-CTCCGACGCCT-GCTTCACCACCTTCTT-3′. The mRNA expression was normalized by comparing with the expression of GAPDH). All reactions were done in triplicate. Relative mRNA expression levels and standard deviations were obtained through comparing those of the control by setting the value to 1.
2.10. Wound healing assay and invasion assay
Cells were seeded at an appropriate concentration to reach 90% cell confluency until the next day, and then scratch was made. Cells were cultured for 48 h with transforming growth factor (TGF) (5 ng/mL) plus Rk1 or Rg5 or not. All experiments were carried out in triplicate.
Cell invasion kit containing transwell (Cell Biolabs, San Diego, CA) was employed to examine the invasive ability of A549 cells, and the procedure was carried out according to the protocol of manufacturer. TGF with or without Rk1 or Rg5 were pretreated for 48 h, and cells were recovered using trypsin/EDTA. After cell counting, the equal number of cells were seeded in Matrigel-coated transwells and then incubated with serum-free medium plus TGF with or without Rk1 or Rg5 for 48 h. Among all the cells cultured under serum-free conditions, those with invasive potential will be able to migrate to the bottom well contacting the medium supplemented with FBS that acts as an attractant. After incubation, the remaining cells that did not pass through the membranes contained in the transwell were removed slowly. Only the cells that invaded on the lower surface of the membrane were taken and stained with a cell staining solution, one of the kit components, for 10 min. The membrane was washed with tap water, and specimens were observed under a microscope. The dyed membrane was treated with 10% acetic acid solution to dissolve the dye. Optical density of the dye was measured at 560 nm to analyze the cell invasive ability. The relative cell invasiveness of each experimental group was compared with the that of control group converted to 1. All experiments were performed three times.
2.11. gelatin zymography
Zymography was performed to measure the extracellular (MMPs) activity of A549 cells. To illustrate, cells were treated with TGF in presence or absence of Rk1 or Rg5 in serum-free medium for 48 h. After incubation, the culture medium containing MMP-2 and MMP-9 was obtained and concentrated through Centrifugal Filter (Millipore, Temecula, CA) to increase the concentration of MMP-2/9. The concentrated supernatant was incubated with 2 × sample buffer not containing β-mercaptoethanol for 5 min at RT and then separated according to molecular weight through electrophoresis at 125 V for 90 min in SDS-PAGE gel containing gelatin. Then, the gel washed three times and renatured for 30 min and developed at 37°C for overnight. After the enzymatic reaction was completed, Coomassie blue staining was performed to identify the degraded substrate in the gel, and the band region was captured on the light box.
2.12. Confocal microscopy
Cells were seeded in confocal dish at an appropriate concentration to reach 90% confluency until the next day in confocal dish. It was fixed and permeabilized with 2% paraformaldehyde with 0.1% Triton X-100 for 10 min and washed three times by 0.02 M glycine-PBS-T 300 μL/well. Cells blocked with 1–3% BSA in PBS-T and incubated with primary antibodies overnight at 4°C. After washing, the secondary antibody is attached for 2 h at RT. The antibodies, anti-E-cadherin, anti-vimentin, goat anti-rabbit IgG-FITC, were obtained from Santa Cruz Biotechnology (Dallas, TX). DAPI (Sigma–Aldrich, St. Louis, MO) was used to stain the nuclei.
2.13. Anoikis assay and sulforhodamine B assay
Anoikis assay was employed using poly-hydroxyethyl methacrylate (poly-HEMA) coated plate. Poly-HEMA stock solution was prepared by dissolving poly-HEMA in 100% ethanol. Poly-HEMA coated plates was prepared by adding 0.5 mL of poly-HEMA working solution (8 mL poly-HEMA stock solution, 30.4 mL 100% ethanol, and 1.6 mL distilled water) into each well of 6-well plate and air-dried for overnight, and this procedure was repeated twice. Then, the poly-HEMA coated plate was washed with phosphate buffered saline (PBS) and stored at room temperature until subsequent experiments. A549 cells were pretreated with TGF in presence or absence of Rk1 or Rg5 for 48 h, then these cells were detached and resuspended as single cell suspension with complete medium. To induce anoikis, single cells were cultured in suspended conditions using poly-HEMA plates with TGF with or without Rk1 or Rg5 for the indicated time points at the density of 3 × 104 cells/well. Cell suspensions were collected, and media supernatants were removed by centrifuge at 10,000 rpm for 1 min. Cells were resuspended with fresh media, transferred to 24-well adhesive plates, cultured overnight, and followed by cell survival of suspended cells was determined by Crystal violet staining. Briefly, cells were washed twice with PBS, fixed with formalin, and stained with crystal violet solution. The stained cells were washed, air-dried, photographed, and dissolved with 10% acetic acid, and optical density was measured at 570 nm. Protein content of suspended cells was examined by sulforhodamine B (SRB) colorimetric assay to measure cell proliferation. Briefly, after collection of cell suspension, medium supernatant was removed by centrifuge at 10,000 rpm for 1 min. Then, cells were fixed with ice-cold 10% trichloroacetic acid at 4°C for 30 min, washed five times with distilled water, and air-dried for 5 min. The dried cells were stained with 50 μL of 0.4% SRB solution in 1% acetic acid for 30 min. The stained cells were washed four times with 1% acetic acid, air-dried, and dissolved with 10 mM Tris solution (pH 10.5). After centrifugation, supernatants were collected, and optical density at 570 nm was read using microplate reader.
2.14. Tumorsphere formation assay
A549 cells were incubated by serum-free medium (SFM) to isolate lung cancer stem cells. Poly-HEMA coated 6-well plates were used to culture cell in SFM. SFM condition is Nutrient Mixture F-12 (Dulbecco's Modified Eagle Medium (DMEM)/F-12), % B27 Supplement, 20 ng/mL human basic fibroblast growth factor (bFGF), and 20 ng/mL epidermal growth factor (EGF). The number and size of the tumorspheres were estimated on day 14 (sphere size was counted on >50 μm in diameter)
2.15. Reporter assay
To investigate promoter activity, luciferase reporter assays are used by measuring luminescence generated by the activity of luciferase, showed under the control of the target promoter. The Smad promoter activity was determined by transfecting with the pSBE4-Luc reporter plasmid in A549 cells. A β-galactosidase luciferase reporter plasmid was used the control. Luciferase activity was progressed by the manufacturer's instructions (Promega, Madison, WI). Approximately 100 μL luciferase assay reagent was added to each well, and the luminescence was measured for 10 s with a delay of 2 s. β-Gal luciferase was used by normalizing activities and three independent experiments were performed.
2.16. Statistical analyses
All experiments were repeated at least three times, and the results were expressed as mean and standard deviation. Statistical significance was analyzed using t-test. p value was considered to be significant within a range of less than 0.05.
3. Results
3.1. KMxG crude, but not crude ginseng, inhibits TGF-β1 signaling pathway
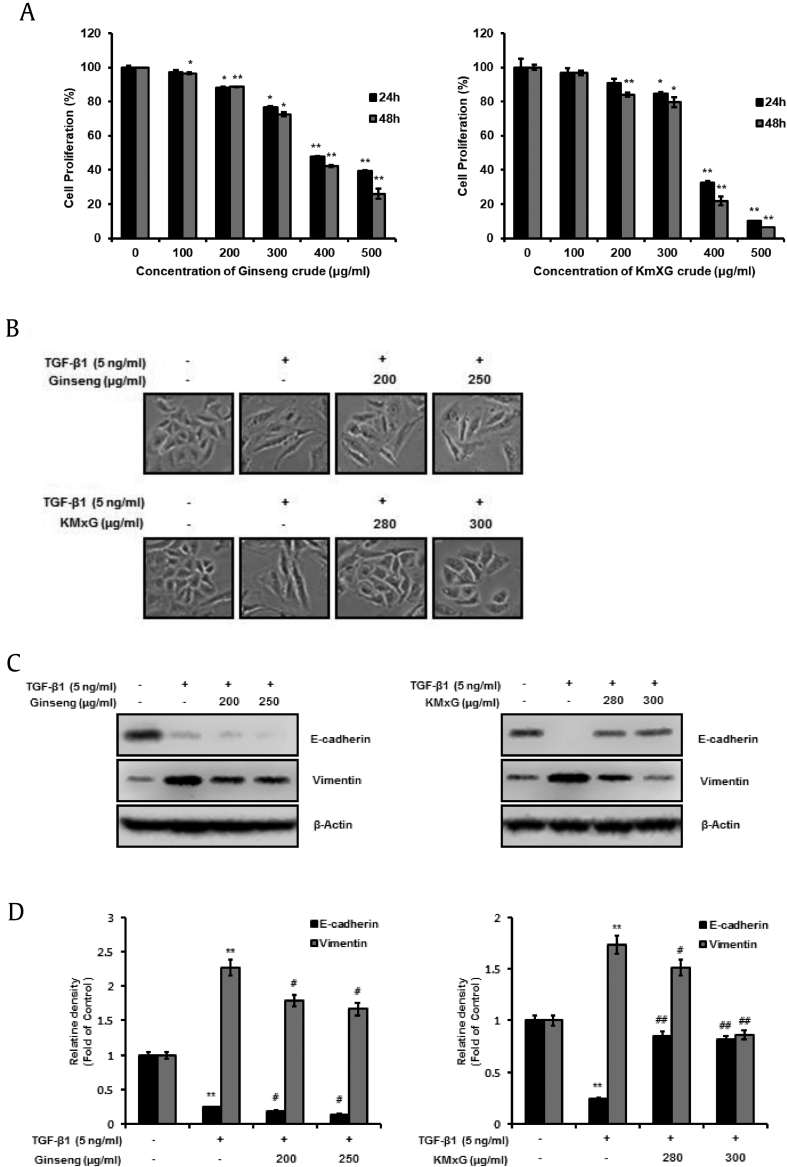
We studied the effects of ginseng and KMxG to investigate whether crude ginseng and KMxG could suppress EMT employing TGF-β1 in A549 cells in vitro. First, to investigate whether crude ginseng and KMxG inhibit A549 cell proliferation, we evaluated change of cell proliferation at a variant concentration (0, 100, 200, 300, 400, and 500 μg/mL) for 24–48 h and examined by WST-8 assay. Crude ginseng at concentrations less than 250 μg/mL and crude KMxG at concentrations less than 300 μg/mL did not influence A549 cell proliferation. At these concentrations, the cell proliferation rate was over 80% (Fig. 2A). For all subsequent studies, these sublethal doses (≤250 μg/mL crude ginseng crude and ≤ 300 μg/mL crude KMxG) were used. When we treated A549 cells with TGF-β1, morphological changes, similar to the mesenchymal phenotype, were observed. However, treatment of KMxG inhibited morphological changes. On the contrary, treatment with 200 and 250 μg/mL crude ginseng did not show suppression of TGF-β1-induced mesenchymal phenotype (Fig. 2B), suggesting that crude KMxG suppresses TGF-β1-mediated EMT, while crude ginseng does not.
Fig. 2.
KMxG crude is a TGF-β1 inhibitor and inhibits TGF-β1 signaling pathway. A. Cell proliferation effect of Ginseng and KMxG in A549 cells. The cells were treated with various concentrations of Ginseng and KMxG for 24 h or 48 h on Roswell Park Memorial Institute (RPMI-1640) media. Cell proliferation was measured using CCK-8 assay. CCK-8, Cell Counting Kit-8; DMSO, dimethyl sulfoxide. B. Control A549 cells treated with DMSO appears original epithelial morphology (pebble-like shape and tight cell-cell adhesion). TGF-β1 treatment induces mesenchymal morphology. However, A549 cells treated with KMxG at concentrations of 280 μg/mL and 300 μg/mL plus 5 ng/mL TGF-β1 for 48 h show the epithelial phenotype. C. Changes in protein of the epithelial marker, E-cadherin and the mesenchymal marker vimentin were analyzed by Western blot analysis and densitometric quantification. β-Actin was used as a loading control. ∗∗p < 0.01, ∗p < 0.05 versus control and ##p < 0.01 #p < 0.05 versus TGF- β1 treated control. The data are expressed as mean ± SD for triplicates.
TGF-β1-mediated EMT is characterized by molecular alteration of EMT markers such as E-cadherin and vimentin, followed by morphological changes that enable cell migration [22]. To determine the suppressive effect of crude ginseng and crude KMxG on TGF-β1-mediated EMT, we determined the level of E-cadherin and vimentin, and the protein levels of A549 cells were measured by western blotting. The results revealed that TGF stimulation considerably decreased the protein level of E-cadherin, and whereas it notably increased the that of vimentin compared with control cells and these alterations were inhibited through KMxG treatment (Fig. 2C and D, left). In contrast, treatment with crude ginseng did not affect the expression of E-cadherin and vimentin inducing TGF- β1-mediated EMT (Fig. 2C and D, right). These data revealed that crude KMxG, but not crude ginseng crude, strongly inhibits EMT in A549 cells.
3.2. KMxG-GF shows higher suppression of TGF-β1-mediated EMT compared to KMxG
Recently, we found a novel ginseng extract using microwave-assisted processing. Our studies showed that this novel extract (KMxG) has higher ginsenoside Rg3, Rg5, and Rk1 concentrations [20]. The KMxG-GF fraction is solely composed of Rg3, Rk1, and Rg5. To investigate the effect of KMxG-GF on EMT employing TGF-β1, we evaluated the suppressive effects of KMxG and KMxG-GF. We measured the antiproliferative effect of KMxG and KMxG-GF at various concentrations (0, 100, 120, 140, 160, 180 μg/mL). As shown in Fig. S1A, KMxG <180 μg/mL and KMxG-GF of <140 μg/mL did not affect A549 cell proliferation. Cell proliferation rate at these concentrations was >80% (Fig. S1A). Therefore, these concentrations (≤140 μg/mL KMxG or ≤140 μg/mL KMxG-GF) were used for subsequent experiments to compare the suppressive effects of KMxG and KMxG-GF on TGF-β1-induced EMT. KMxG (100 and 140 μg/mL) and KMxG-GF (100 and 140 μg/mL) inhibited TGF-β1-induced mesenchymal phenotype. However, when KMxG-GF was treated, we observed a more pronounced inhibition of EMT (Fig. S1B). Next, we confirmed that KMxG-GF restored E-cadherin protein level and attenuated the increment of vimentin expression more effectively than KMxG under similar treatment conditions (Fig. S1C and S1D). Taken together, these results suggested that KMxG-GF, which is solely composed of Rg3, Rk1, and Rg5, is a stronger suppressor compared to KMxG on EMT mediated by TGF-β1.
In an earlier study, we had shown that ginsenoside 20-Rg3 inhibits TGF-β1-stimulated EMT in lung cancer cells [23]. However, to the best of our knowledge, there are no studies on the effect of ginsenosides Rk1 and Rg5 on EMT. Therefore, following experiments were conducted using Rk1 and Rg5.
3.3. Rk1 and Rg5 inhibit TGF-β1-mediated EMT
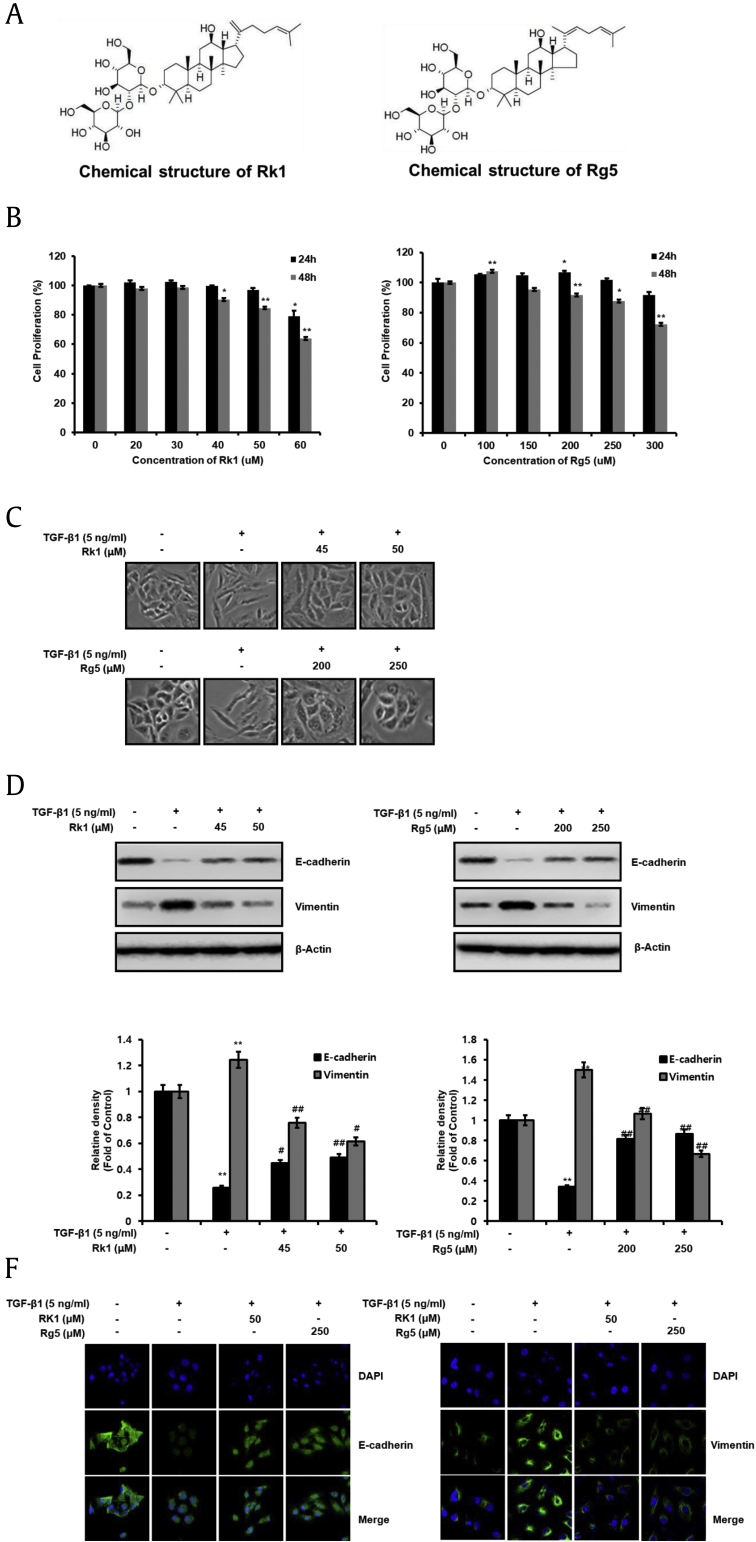
Next, we examined whether ginsenosides Rk1 and Rg5 (Fig. 3A) could inhibit EMT employing TGF. As shown in Fig. 3B, Rk1 <50 μM and Rg5 <250 μM did not affect A549 cell proliferation. The cell proliferation rate at these concentrations were >80% (Fig. 3B). Therefore, these concentrations (Rk1: 45 μM and 50 μM or Rg5: 200 μM and 250 μM) were used for subsequent experiments. TGF-β1-treated cells pretreated with Rk1 or Rg5 showed decreased transition to a mesenchymal phenotype (Fig. 3C). Further, treatment with Rk1 and Rg5 prominently rescued the TGF-β1-triggered decrease in E-cadherin expression (Fig. 3D and E). Additionally, TGF-β1-triggered increase in vimentin by was attenuated by treatment with Rk1 and Rg5 (Fig. 3D and E). Immunofluorescence was also performed and showed that E-cadherin protein levels in cell membrane and cytoplasm was decreased by TGF treatment whereas Rk1 and Rg5 suppressed the decrease in E-cadherin expression (Fig. 3F, left). However, vimentin, founded in the cytoplasm of mesenchymal, was increased by TGF treatment compared with TFG-untreated cells whereas Rk1 and Rg5 decreased the increase in vimentin expression (Fig. 3F, right). In addition, E-cadherin mRNA levels were restored, and vimentin mRNA level was reduced by Rk1 and Rg5 (Fig. S2A). These results strongly suggest that Rk1 and Rg5 promoted E-cadherin expression and suppressed vimentin expression, and thus, inhibited TGF-β1-induced EMT.
Fig. 3.
Rk1 and Rg5 inhibit TGF-β1-induced EMT. A. Chemical structure of Rk1 and Rg5. A. Chemical structure of Rk1 and Rg5. B. The antiproliferative effects of Rk1 and Rg5 on cell proliferation were determined by CCK-8 assay. A549 cells were treated with Rk1 or Rg5 at concentrations of 0 (DMSO was used as a control), 20 μM, 30 μM, 40 μM, 50 μM, and 60 μM and 0 μM, 100 μM, 150 μM, 200 μM, 250 μM, and 300 μM for 24 h or 48 h. C. Control A549 cells treated with DMSO appears to be of original epithelial morphology. TGF-β1 treatment induces mesenchymal morphology. However, A549 cells treated with Rk1 at concentrations of 45 μM and 50 μM of Rk1 or Rg5 at concentrations of 200 μMand 250 μM plus 5 ng/mL of TGF-β1 for 48 h show the epithelial phenotype. D and E. Changes in protein of the epithelial marker, E-cadherin and the mesenchymal marker vimentin were analyzed by Western blot analysis and densitometric quantification. β-Actin was used as a loading control. F. Rk1 or Rg5 increased E-cadherin expression and decreased vimentin expression in TGF-β1-stimulated A549 cells. The expression of E-cadherin and vimentin were detected by using immunofluorescence analysis with primary anti-E-cadherin and anti-vimentin. A549 cells were cultured in the TGF-β1 with Rk1 or Rg5 or without Rk1 or Rg5 for 48 h. Images show nucleus stained by (DAPI). ∗∗p < 0.01, ∗p < 0.05 versus control and ##p < 0.01 #p < 0.05 versus TGF-β1 treated control. The data are expressed as mean ± SD for triplicates.
3.4. Rk1 and Rg5 suppress TGF-β1-induced snail expression
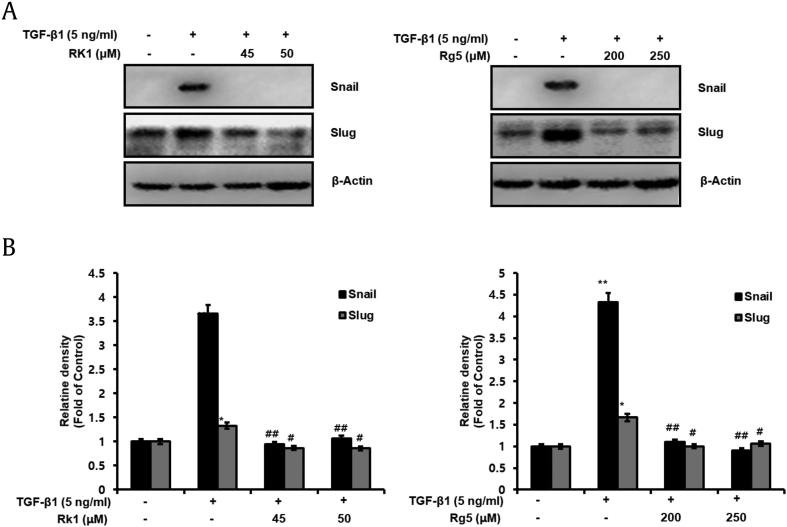
Snail (encoded by SNAI1) is a transcription factor containing zinc finger ion that has been established as an E-cadherin transcriptional repressor (encoded by CDH1) and an EMT stimulator. It certainly represses E-cadherin expression, which is the hallmark of EMT and is considered to suppress tumor progression [24]. We used western blot analyses and qRT-PCR to examine whether Rk1 and Rg5 suppressed the TGF-β1-mediated increased expression of Snail and Slug. As shown in Fig. 4A and B and Fig. S2B, increased protein and mRNA level of Snail and Slug induced by TGF were observed compared to that in untreated cells. However, Rk1 and Rg5 significantly decreased the level of Snail and Slug in TGF-β1-treated A549 cells. Therefore, these data showed that Rk1 and Rg5 suppress Snail and Slug expression, which retained E-cadherin expression.
Fig. 4.
Rk1 and Rg5 suppress TGF-β1-induced Snail and Slug, EMT-induced transcription factor. A549 cells were treated with the indicated concentration of Rk1 or Rg5 and stimulated with TGF-β1 (5 ng/mL) for 48 h. A and B. Change in protein expression of Snail and Slug was analyzed by Western blot analysis and densitometric quantification. β-Actin was used as a loading control. ∗∗p < 0.01, ∗p < 0.05 versus control and ##p < 0.01 #p < 0.05 versus TGF-β1 treated control. The data are expressed as mean ± SD for triplicates.
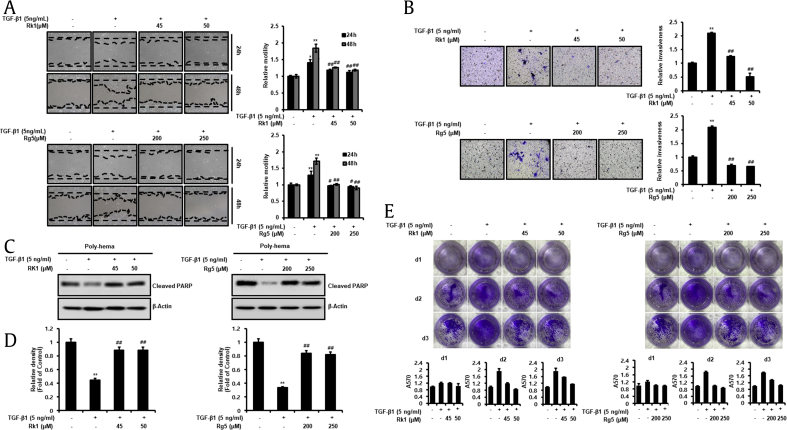
3.5. Rk1 and Rg5 suppress migration, invasion, and anoikis resistance by inhibiting TGF-β1-induced EMT
Numerous findings indicated that EMT is required for metastatic cascade during malignant progression including cell motility, invasiveness, and anoikis resistance [9]. Therefore, it is important to explore the effect of Rk1 and Rg5 on TGF-βh-mediated cell motility. Wound healing assay was employed to verify whether Rk1 and Rg5 inhibit motility of A549 cells, and we observed that Rk1 and Rg5 prevented TGF-β1-induced migration by approximately two-fold (Fig. 5A). Next, invasion assay was employed using Boyden chamber method. We then treated the inner chamber with TGF-β1 and Rk1 or Rg5 for 48 h. The pretreated cells were transferred to inner chamber and incubated with TGF plus Rk1 or Rg5 or kept untreated for 48 h. The infiltrated cells, located at the bottom surface of the membrane by infiltrating ECM-like membrane, were stained, photographed, and analyzed by measuring OD at 570 nm (Fig. 5B). The results revealed that Rk1 and Rg5 prevented invasion ability of TGF-β1-treated A549 cells.
Fig. 5.
Effect of Rk1 and Rg5 on A549 cell migration, invasion and anoikis resistance during inhibition of TGF-β1-induced EMT. A. Rk1 and Rg5 antagonized the TGF-β1-induced migration of A549 cells. When cell confluence had reached about 90%, the cells were scratched with a 200 μL-pipette tip and washed with culture media to remove any free-floating cells and debris. Then culture media was added, and the culture plates were incubated with TGF-β1or with TGF-β1 and Rk1 or Rg5. The cells were photographed under a microscope 48 h after scratching. Relative motility was quantified by measuring the cell surface area with Image J program. B. Rk1 and Rg5 inhibited TGF-β1-induced invasion. Effect of Rk1 and Rg5 on A549 cell invasion in a 200 × light scope after staining by Matrigel invasion assay as described in Section 2.10. In vitro invasiveness of A549 cells were measured by counting cells that migrated through the extracellular matrix layer of invasion chambers. C and D. Rk1 and Rg5 suppressed TGF-β1-induced anoikis resistance. A549 cells were pretreated with the indicated concentration of Rk1 or Rg5 for 2 h and then stimulated with TGF-β1 (5 ng/mL) for 48 h. The cells were then cultured on poly(hydroxyethyl methacrylate) [poly-HEMA]-coated plates for 48 h at TGF-β1 (5 ng/mL) with or without Rk1 or Rg5 condition. The cells were harvested and then lysed. Cleaved pPARP)-1, apoptosis marker, was detected by Western blotting. Densitometric analysis of band intensity of cleaved PARP-1was performed. E. Rk1and Rg5 lead to increased cell survival of suspended cells. A549 cells (1 × 106) were grown in suspension for 0 to 3 d in treated with TGF-β1 and/or Rk1 or Rg5, and subsequently transferred into six-well adhesive plates followed by Coomassie staining. Columns mean of three separate experiments; bars. ∗∗p < 0.01, ∗p < 0.05 vs control and ##p < 0.01 #p < 0.05 versus TGF-β1 treated control. The data are expressed as mean ± SD for triplicates.
Anoikis is a type of apoptosis that occurs upon anchorage-independent condition. Anoikis resistance of tumor cells is a representative property of EMT and is required for tumor cells to successfully metastasize through blood circulation. Moreover, TGF activation by its ligands suppresses anoikis in various cancers [25]. Hence, to determine whether Rk1 or Rg5 inhibits anoikis resistance by TGF, we examined the apoptotic property of A549 cells in the suspended condition using poly-HEMA plates. A549 cells underwent anoikis after 48 h, whereas TGF stimulation prevented the anoikis of A549 cells. In contrast, Rk1 or Rg5 increased anoikis of A549 cells even after treatment with TGF. The results revealed that TGF treatment reduced the cleavage of (PARP), which occurs by anoikis in A549 cells, whereas the decreased expression was significantly inhibited by Rk1 or Rg5 treatment (Fig. 5C and D). We also measured inhibitory effects of Rk1 or Rg5 against TGF-induced anoikis resistance in A549 cells employing SRB assay (Fig. S3A). This method relies on the stoichiometric binding of SRB dye to proteins under mild acidic conditions and its subsequent extraction under basic conditions. The amount of dye extracted is a proxy for cell mass and thus the number of cells in a sample. Next, we performed reattachment experiments. To measure survival rate during detachment status, cells were prevented from attachment for 1–3 d and subsequently transferred to adhesive plates. Strikingly, Coomassie staining showed that Rk1 and Rg5 led to a significantly increased cell survival compared to that of TGF-treated cells after 48 h in suspension (Fig. 5E). The results revealed that A549 cells died in anchorage-independent condition for 48 h, whereas TGF treatment induced anoikis resistance, which is significantly prevented by Rk1 or Rg5 treatment.
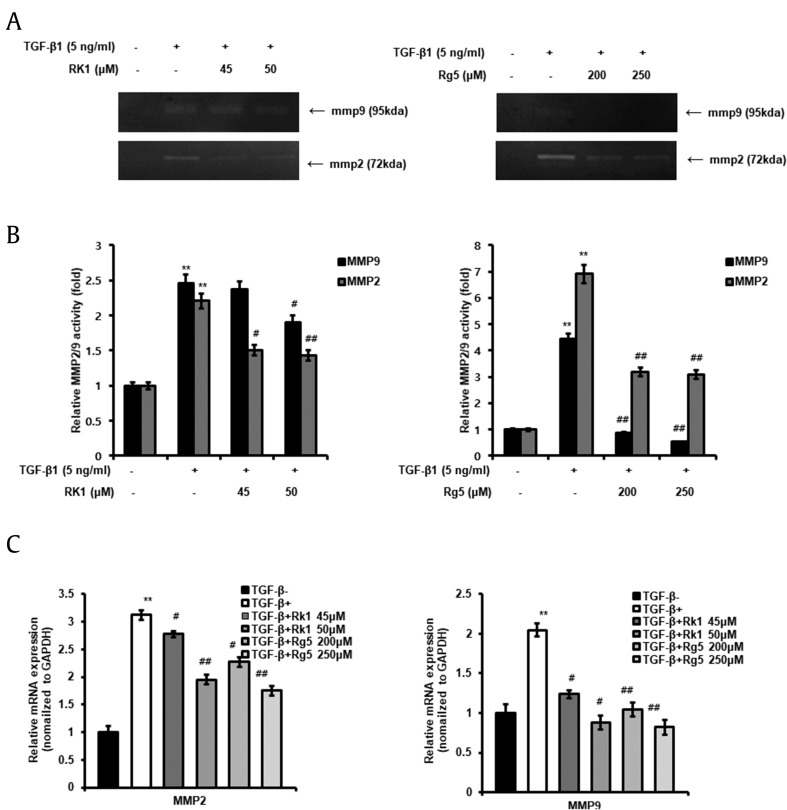
3.6. MMP-2 and MMP-9 expression in TGF-β1-treated cells was decreased by Rk1 and Rg5
MMP-2 and MMP-9 play an important role in tumor cell migration and invasion. TGF-β1 increases the synthesis of MMP-2 and MMP-9 and enhances their activity in tumor cells [26]. We used zymography and qRT-PCR to determine whether Rk1 and Rg5 inhibit the TGF-β1-induced activation of MMP-2/9 in A549 cells. As shown in Fig. 6A and B, we observed that TGF-β1-activated cells showed higher MMP-2/9 expression compared to that in control cells. Rk1 and Rg5 significantly suppressed the TGF-β1-induced increase in MMP-2/9 activity (Fig. 6A and B). Additionally, Rk1 and Rg5 reduced the TGF-β1-triggered increase in transcriptional expression of MMP-2/9 (Fig. 6C). These results indicated that Rk1 and Rg5 inhibit TGF-β1-induced MMP-2 and MMP-9 expression and activity in A549 cells.
Fig. 6.
Effect of Rk1 and Rg5 on mRNA expression and activity of (MMP–2) activated by TGF-β1. A. Chemical structure of Rk1 and Rg5A and B. A549 cells were serum starved in medium containing 0.2% fetal bovine serum (FBS) overnight. The cells were treated with Rk1 or Rg5 and stimulated with TGF-β1 (5 ng/mL) for 48 h. The conditioned media were collected and MMP-2 and MMP-9 activity in the media was analyzed by gelatin zymography. The relative MMP-2 and MMP-9 activity was expressed as fold changes with the density of untreated cells designated as 1 and shown as mean ± SD of three independent experiments. C. The expression of MMP-2 and MMP-9 mRNA was analyzed by quantitative real-time polymerase chain reaction (qRT-PCR). Experiments were repeated at least three times (n = 3). Fold change was calculated by 2−ΔΔCt relative quantitative analysis. ∗∗p < 0.01, ∗p < 0.05 vs control and ##p < 0.01 #p < 0.05 vs TGF-β1 treated control. The data are expressed as mean ± SD for triplicates.
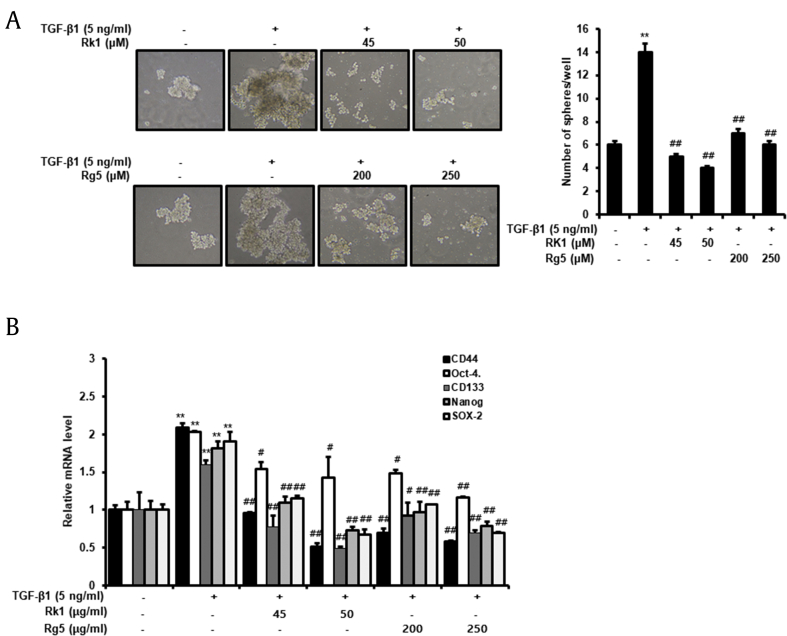
3.7. Rk1 and Rg5 inhibit TGF-β1-induced development of stem-like properties
CSCs perform an important role in the cancer development. They have self-renewal properties and can form three-dimensional structures and spheres. They can also differentiate into different phenotypes. In addition, the characteristics and metastasis of CSC are known to be controlled by EMT as well as hypoxic environments [27]. Mammosphere formation assay in SFM is routinely used to evaluate the stemness of cancer cells. We incubated A549 cells in SFM for 14 days and monitored the development of tumorspheres. As shown in Fig. 7A, cells treated with TGF-β1 formed larger spheres compared to untreated cells. Rk1 and Rg5 suppressed TGF-β1-induced formation of tumorspheres; the size and number of tumorspheres decreased. Next, we evaluated the lung CSC markers mRNA levels, CD44 and CD133, and transcriptional regulators in pluripotent stem cells, Nanog, Oct-4, and SOX-2. The mRNA levels of CD44, CD133, Nanog, Oct4, and SOX-2 were upregulated in TGF-β1-treated cell, and treatment with Rk1 and Rg5 downregulated their expression (Fig. 7B). These results revealed that Rk1 and Rg5 inhibit TGF-β1-induced development of stem-like properties.
Fig. 7.
Rk1 and Rg5 impaired TGF-β1- induced sphere formation and stemness properties. A. A549 Cells were cultured in serum-free medium (SFM) in the presence or absence of TGF-β1 (5 ng/mL) with or without Rk1 or Rg5. Representative images were acquired during the culturing under a light microscope. A549 cells were cultured in SFM for 14 d. B. The mRNA levels of lung CSCs markers (CD44, CD133, (Oct-4), (Nanog), and (SOX-2)) were analyzed by quantitative real-time PCR. ∗∗p < 0.01, ∗p < 0.05 vs control and ##p < 0.01 #p < 0.05 vs TGF-β1 treated control. The data are expressed as mean ± SD for triplicates.
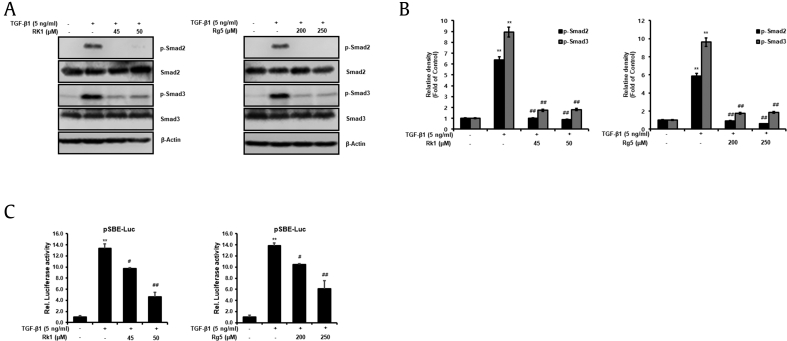
3.8. Rk1 and Rg5 inhibit TGF-β1-activation of Smad2 and Smad3
Smad signaling pathway is mediated by TGF-β1-stimulated EMT, and Smad2/3 are activated. TGF-β1/2 receptor, binding TGF-β type 1 and 2 kinases, can phosphorylate Smad2/3, and then interact with Smad4. Smad complex is formed [9]. We used western blot analyses to investigate whether Rk1 and Rg5 suppress TGF-β1-induced Smad2/3 activation. As shown in Fig. 8A and B, phosphorylation of Smad2/3 inducing TFG-β1 is suppressed by Rk1 and Rg5, while their total protein levels remained unchanged. To uncover the detailed mechanism underlying Rk1 and Rg5 mediated attenuation of TGF-β1-induced EMT, we used a reporter assay with a Smad binding element (SBE) coupled luciferase promoter (pSBE-Luc). We transiently transfected A549 cells with SBE-Luc reporter plasmid and treated with TGF-β1 and/or with Rk1 or Rg5. Thus, these results demonstrated whether Rk1 and Rg5 suppressed TGF-β1-induced transcriptional activation of Smad2 and Smad3 by a SBE-Luc reporter assay, respectively. As shown in Fig. 8C, our results confirmed the inhibitory effect of Rk1 and Rg5 on TGF-β1-stimulated Smad transcriptional activity (Fig. 8C). These data demonstrate that Rk1 and Rg5 inhibit activation of Smad2 and Smad3, which is phosphorylated by TGF-β1 in A549 cells.
Fig. 8.
Rk1 and Rg5 inhibit the Smad2/3 activation of the TGF-β1 signaling pathway in TGF-β1-stimulated A549 cells. A and B. Change in protein expression of p-Smad2/3 was analyzed by Western blot analysis and densitometric quantification. Smad2/3 was used as a loading control. C. Rk1 and Rg5 inhibit the transcription activity of Smad by TGF-β1. A549 cells were transiently transfected with the pSBE-Luc reporter gene, and treated with TGF-β1 or with TGF-β1 and Rk1, or TGF-β1 and Rg5. ∗∗p < 0.01, ∗p < 0.05 versus control and ##p < 0.01 #p < 0.05 versus TGF-β1 treated control. The data are expressed as mean ± SD for triplicates.
3.9. Rk1 and Rg5 suppress TGF-β1-stimulated activation of ERK and NF-κB
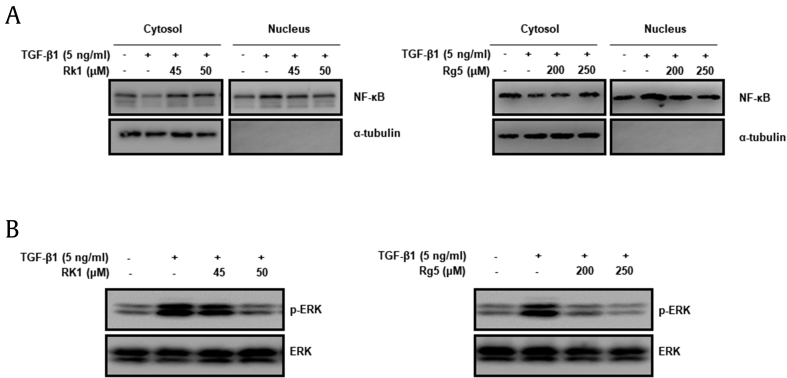
NF-κB and transcriptional factors enhanced the activity of MMP-2/9, which improve cancer cell migration. It has been reported that IκB (group of inhibitory proteins) bind to NF-κB in the cytoplasm and prevent NF-κB from translocating to the nucleus, inactivating NF-κB and decreasing downstream targets [28]. Our results showed that Rk1 and Rg5 suppressed NF-κB activation inducing TGF-β1 (Fig. 9A). The TGF-β signaling pathways, which are mediated by Smad-activated NF-κB, JNKs), ERK, and p38 MAPK pathway controlling EMT as important regulators [29]. It has been reported that ERK signaling regulates MMPs and induces cancer migration and invasion [30]. Thus, we investigated whether Rk1 and Rg5 antagonizes against the activation of ERK by TGF. The results suggested that ERK is phosphorylated in TGF-β1-treated condition (Fig. 9B), but Rk1 and Rg5 decreased TGF-induced phosphorylation of ERK. These results consistent with inhibitory effects of Rk1 and Rg5 translocation of NF-κB and ERK triggered by TGF-β1 in A549 cells. Taken together, Rk1 and Rg5 could decrease TGF-β1-induced cancer invasiveness by suppressing NF-κB translocation and ERK activation, thereby inhibiting MMP-2/9 expression.
Fig. 9.
Effects of Rk1 and Rg5 on translocation of NF-κB and activation of ERK by TGF-β1. A. A549 cells were treated with Rk1 or Rg5 and treated with TGF-β1 for 48 h. Protein levels of NF-κB translocation from cytosol to the nucleus was assessed using Western blotting using α-tubulin as cytosolic markers. B. A549 cells were incubated with TGF-β1 (5 ng/mL) or with TGF-β1 (5 ng/mL) and Rk1 or Rg5 for 48 h. The cells were harvested and then lysed. Phosphorylation of ERK was analyzed by Western blotting using ERK as loading control. ∗∗p < 0.01, ∗p < 0.05 versus control and ##p < 0.01 #p < 0.05 versus TGF-β1 treated control. The data are expressed as mean ± SD for triplicates.
3.10. Rk1 and Rg5 inhibit TGF-β1-induced activation of p38 and JNK
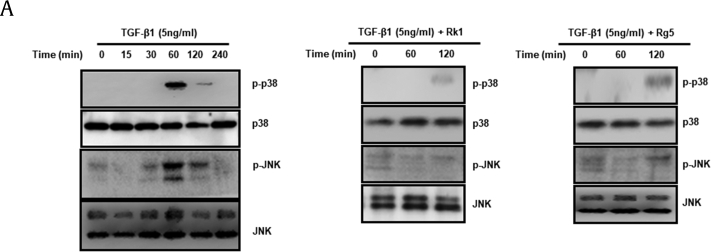
As mentioned above, TGF-β1 phosphorylates and activates JNK, p38 MAPK), and ERK (29). It has been reported that TGF-β1-induced EMT in A549 cells is partially mediated by p38 MAPK and JNK activation [31]. Moreover, the p38 MAPK pathway enhances cell invasion through the TGF-β1-mediated activation of MMP-2 [32]. Therefore, we examined whether Rk1 and Rg5 suppress TGF-β1-induced phosphorylation of JNK and p38 MAPK. TGF-β1 treatment induced substantial phosphorylation of p38 MAPK and JNK in A549 cells. The highest phosphorylation was observed with a 60-min stimulation (Fig. 10A). As shown in Fig. 10A, Rk1 (50 μM) and Rg5 (250 μM) treatment inhibited TGF-β1-induced phosphorylation of p38 MAPK and JNK (Fig. 10A). However, we found that Rk1 (50 μM) and Rg5 (250 μM) treatment could not suppress TGF-β1-induced AKT activation (data not shown). These results indicated that Rk1 and Rg5 inhibit TGF-β1-induced phosphorylation of JNK and p38 MAPK in A549 cells.
Fig. 10.
Effects of Rk1 and Rg5 on p-p38 MAPK and p-JNK expression activated by TGF-β1. A. A549 cells were incubated with TGF-β1 (5 ng/mL) or with TGF-β1 (5 ng/mL) and Rk1 (50 μM), TGF-β1 (5 ng/mL) or Rg5 (250 μM) for various durations. The cells were harvested and then lysed. The phosphorylation of p38 MAPK and JNK were analyzed by western blotting. p38 MAPK and JNK were used loading control.
4. Discussion
Lung cancer is a malignant tumor of the lungs. Metastasis is the leading cause of failure of cancer therapy [[1], [2], [3]]. During metastasis, malignant tumor cells from solid tumors are separated by cell dedifferentiation of the epithelial cells that gain migratory and invasive ability as well as decrease cell-cell junctions. The phenotypic transition of epithelial tumor cells was termed EMT and was confirmed in various cancers [33].
It is reported that EMT has a significant role in tumor recurrence. Many studies indicate that EMT contributes to the migratory, invasive, and anoikis resistance abilities of cancer cells. During the course of EMT, the cells undergo a transition of phenotype and gain migratory and invasive ability as well as decrease in cell-cell junctions. The EMT-initiated metastasis of primary tumor cells can be described as follows; when EMT occurs in tumor cells, detachment from primary tumor takes place, followed by migration, invasion, anoikis resistance, and colonization in distant organs. During this process, the expression of E-cadherin is decreased while the expression of mesenchymal markers such as vimentin is upregulated [34,35]. The several transcription factors such as Snail and ZEB1 are associated with EMT regulation during malignant tumor progression [[36], [37], [38]].
TGF-β1 is the growth factor that plays a key role in EMT, and this induces stem-like characteristics in lung cancer [39,40]. Importantly, the TGF-β1 signaling pathway exerts disruption of desmosomes and adherens junctions leading to dissociation of cancer cells and improved cell migration, invasive ability, anoikis resistance, and stem cell-like properties. TGF-β1 increases the MMP-2/9 activity, thereby enhancing cell motility and invasiveness [41].
The Smad pathway and MAPK pathways (non-Smad signaling pathway) are also activated by TGF-β1-mediated metastasis. In particular, TGF-β1 activates p38 MAPK through non-Smad pathway [42] ZD 2-1 has been known to suppress NF- κB from moving to the nuclear and EMT by TGF-β1 inducing p42/44 MAPK/AP1 signaling pathway [26]. Therefore, MMP activation, Non-Smad signaling pathway-related proteins, and Smads play an important role in EMT during cancer metastasis.
Ginsenoside Rk1 and Rg5, the products from microwave-assisted processing of ginseng extract, have enhanced medicinal properties compared to other ginsenosides and have potential anticancer effects. Rk1 and Rgf5 have been known to be effective against cancer [43,44], but studies on the effect of EMT and CSCs in A549 cells are minimal. Our results revealed that ginsenosides Rk1 and Rg5 inhibit metastasis by EMT and characteristic of cancer stem cell in A549 cells and activation of Smad, nuclear factor kappa B (NF-κB), extracellular signal-regulated kinases 1/2 (ERK1/2), (p38 MAPK), and c-Jun N-terminal kinase (JNK) signaling pathways. In our study, we first examined the effect of Rk1 and Rg5 on EMT inhibition. We observed reduction of epithelial cell polarity and alteration in the level of related EMT markers such as E-cadherin, vimentin, Snail, and Slug. It is often observed that during TGF-β1-mediated EMT, epithelial cells lose E-cadherin expression and express the mesenchymal marker vimentin. In this study, we confirmed that Rk1 and Rg5 inhibited the EMT-mediated mesenchymal cell phenotype, although Rk1 and Rg5 did not affect cell proliferation. In particular, the Snail protein belongs to the zinc finger containing Snail superfamily, which is known as a transcription repressor by linking strongly to the transcription start site of the E-cadherin gene [7]. According to our results, Rk1 and Rg5 inhibited Snail and Slug expression in EMT mediated by TGF. We also demonstrated that treatment with Rk1 and Rg5 dramatically inhibited cell migration, invasion, anoikis resistance, and stem cell-like properties. We found that Rk1 and Rg5 can suppress the TGF-β1-stimulated self-renewal ability of lung cancer cells. We also showed that Rk1 and Rg5 suppressed the stem cell-like properties, CD44, CD133, Nanog), Oct4), and (SOX2); the characteristic marker of lung cancer stem cell. Another characteristic stem cell property is the formation of tumorspheres and three-dimensional structures [45]. Nanog, Oct4, and SOX-2 are represented at a high level, and ALDH1) is a specific CSC marker in the lung CSCs [7,46,47].
Recent studies on lung cancer cells have shown inhibition ability of TGF-β1-stimulated EMT by suppressing Smad signaling pathway, especially Epigallocatechin gallate (EGCG) [48], geraniin, and sanguiin H6 [49,50]. EMT enhances metastasis by increasing migration and invasion of cancer cells. We demonstrated that Rk1 and Rg5 effectively suppressed cell migratory, invasive, and anoikis resistance ability by inhibiting the Smad2/3 and NF-κB/ERK1/2 signaling pathways. TGF-β1/2 receptor, binding TGF-β type 1 and 2 kinases, can phosphorylate Smad2/3, and then interact with Smad4. When Smad complex is formed, it translocates the nucleus and activates as a gene regulator [51]. Suppressing TGF-β/Smad inhibits metastasis in NSCLC. Our results showed that Rk1 and Rg5 reduce the Smad2/3 signaling pathways in TGF-β1-induced NSCLC cells. NF-κB plays an important role as a transcription factor that modulates the onset of EMT and regulates self-renewal capacity of NSCLC [52,53]. We confirmed that activity of NF-κB which is related with activation of ERK is increased in TGF-β1 treat condition. Blocking the NF-κB and ERK pathway affects TGF-β1-mediated EMT. The results of this study revealed that Rk1 and Rg5 inhibit ERk1/2 pathway inducing activation of NF-κB. The p38 MARK and JNK pathways are also activated during TFG-β1-mediated EMT, but we demonstrated that Rk1 and Rg5 prevent p38 and JNK pathway. In addition, p38 MAPK has known to block the MMP-2 activation and eventually suppress cell invasion in prostate cancer [32]. It has also been reported that TGF-β1/p38/JNK/ERK signal pathway plays a crucial role in cancer metastasis in salivary adenoid cystic carcinoma [53]. Thus, we assume that p38 MAPK, ERK, p38 MAPK, and JNK are activated under TGF-β1-stimulated EMT condition in A549 cells.
In previous reports, ginsenoside has been shown to exhibit anticancer activity in in vivo models. Particularly, the antimetastatic effect of the ginsenoside Rg3 was tested in lung metastasis model using B16-BL6 melanoma cells. The intravenous (i.v.) administration of Rg3 after tumor inoculation dramatically inhibited the lung metastasis of B16-BL6 melanoma cells [54]. Therefore, further studies are required to confirm the inhibitory effect of Rk1 and Rg5 on TGF-β1-induced EMT in vivo model.
In conclusion, our finding indicates that Rk1 and Rg5 suppress TGF-β1-mediated EMT and induce stem-like properties by suppressing the activation of Smad2/3, NF-κB, ERK, p38 MAPK, and JNK pathways in NSCLC. Our results also suggest that ginsenosides Rk1 and Rg5 are promising drugs for the treatment of NSCLC.
Conflicts of interest
The authors have no conflicts of interests to disclose.
Acknowledgments
This work was supported by the Korea Institute of Science and Technology (KIST) institutional program (Project No. 2Z06260) and by the Nano Convergence Industrial Strategic Technology Development Program (No. 20000105) funded by the Ministry of Trade, Industry & Energy (MOTIE, Korea).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgr.2020.02.005.
Contributor Information
Hyeonseok Ko, Email: drug9054@naver.com.
Jungyeob Ham, Email: ham0606@kist.re.kr.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Torre L.A., Siegel R.L., Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R., Ma J., Zou Z., Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.Reck M., Heigener D.F., Mok T., Soria J.-C., Rabe K.F. Management of non-small-cell lung cancer: recent developments. The Lancet. 2013;382(9893):709–719. doi: 10.1016/S0140-6736(13)61502-0. [DOI] [PubMed] [Google Scholar]
- 4.Valastyan S., Weinberg R.A. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thiery J.P. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 6.Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Batlle E., Sancho E., Franci C., Dominguez D., Monfar M., Baulida J., Herreros A.G. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2(2):84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 8.Humbert P.O., Grzeschik N.A., Brumby A.M., Galea R., Elsum I., Richardson H.E. Control of tumourigenesis by the Scribble/Dlg/Lgl polarity module. Oncogene. 2008;27(55):6888–6907. doi: 10.1038/onc.2008.341. [DOI] [PubMed] [Google Scholar]
- 9.Birchmeier W., Behrens J. Cadherin expression in carcinomas: role in the formation of cell junctions and the prevention of invasiveness. Biochim Biophys Acta. 1994;1198(1):11–26. doi: 10.1016/0304-419x(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 10.Fong Y.C., Hsu S.F., Wu C.L., Li T.M., Kao S.T., Tsai F.J., Chen W.C., Liu S.C., Wu C.M., Tang C.H. Transforming growth factor-beta1 increases cell migration and beta1 integrin up-regulation in human lung cancer cells. Lung Cancer. 2009;64(1):13–21. doi: 10.1016/j.lungcan.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 11.Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13(10):616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X., Yun F., Shi L., Li Z.-H., Luo N.-R., Jia Y.-F. Roles of signaling pathways in the epithelial-mesenchymal transition in cancer. Asian Pac J Cancer Prev. 2015;16(15):6201–6206. doi: 10.7314/apjcp.2015.16.15.6201. [DOI] [PubMed] [Google Scholar]
- 13.Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monteiro J., Fodde R. Cancer stemness and metastasis: therapeutic consequences and perspectives. Eur J Cancer. 2010;46(7):1198–1203. doi: 10.1016/j.ejca.2010.02.030. [DOI] [PubMed] [Google Scholar]
- 15.Bent S., Ko R. Commonly used herbal medicines in the United States: a review. Am J Med. 2004;116(7):478–485. doi: 10.1016/j.amjmed.2003.10.036. [DOI] [PubMed] [Google Scholar]
- 16.Nocerino E., Amato M., Izzo A.A. The aphrodisiac and adaptogenic properties of ginseng. Fitoterapia. 2000;71(Suppl 1):S1–S5. doi: 10.1016/s0367-326x(00)00170-2. [DOI] [PubMed] [Google Scholar]
- 17.Huang Y.C., Chen C.T., Chen S.C., Lai P.H., Liang H.C., Chang Y., Yu L.C., Sung H.W. A natural compound (ginsenoside Re) isolated from Panax ginseng as a novel angiogenic agent for tissue regeneration. Pharm Res. 2005;22(4):636–646. doi: 10.1007/s11095-005-2500-3. [DOI] [PubMed] [Google Scholar]
- 18.Kim K.R., Chung T.Y., Shin H., Son S.H., Park K.K., Choi J.H., Chung W.Y. Red ginseng saponin extract attenuates murine collagen-induced arthritis by reducing pro-inflammatory responses and matrix metalloproteinase-3 expression. Biol. Pharm. Bull. 2010;33(4):604–610. doi: 10.1248/bpb.33.604. [DOI] [PubMed] [Google Scholar]
- 19.Choi P., Park J.Y., Kim T., Park S.-H., Kim H-k, Kang K.S., Ham J. Improved anticancer effect of ginseng extract by microwave-assisted processing through the generation of ginsenosides Rg3, Rg5 and Rk1. J Funct Foods. 2015;14:613–622. [Google Scholar]
- 20.Kim Y.J., Yamabe N., Choi P., Lee J.W., Ham J., Kang K.S. Efficient thermal deglycosylation of ginsenoside Rd and its contribution to the improved anticancer activity of ginseng. J Agric Food Chem. 2013;61(38):9185–9191. doi: 10.1021/jf402774d. [DOI] [PubMed] [Google Scholar]
- 21.Ko H., Kim S., Yang K., Kim K. Phosphorylation-dependent stabilization of MZF1 upregulates N-cadherin expression during protein kinase CK2-mediated epithelial-mesenchymal transition. Oncogenesis. 2018;7(3):27. doi: 10.1038/s41389-018-0035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y.J., Choi W.I., Jeon B.N., Choi K.C., Kim K., Kim T.J., Ham J., Jang H.J., Kang K.S., Ko H. Stereospecific effects of ginsenoside 20-Rg3 inhibits TGF-beta1-induced epithelial-mesenchymal transition and suppresses lung cancer migration, invasion and anoikis resistance. Toxicology. 2014;322:23–33. doi: 10.1016/j.tox.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Lim W.C., Kim H., Kim Y.J., Choi K.C., Lee I.H., Lee K.H., Kim M.K., Ko H. Dioscin suppresses TGF-beta1-induced epithelial-mesenchymal transition and suppresses A549 lung cancer migration and invasion. Bioorg Med Chem Lett. 2017;27(15):3342–3348. doi: 10.1016/j.bmcl.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 24.Frisch S.M., Schaller M., Cieply B. Mechanisms that link the oncogenic epithelial-mesenchymal transition to suppression of anoikis. J Cell Sci. 2013;126(Pt 1):21–29. doi: 10.1242/jcs.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim Y.J., Jeon Y., Kim T., Lim W.C., Ham J., Park Y.N., Kim T.J., Ko H. Combined treatment with zingerone and its novel derivative synergistically inhibits TGF-beta1 induced epithelial-mesenchymal transition, migration and invasion of human hepatocellular carcinoma cells. Bioorg Med Chem Lett. 2017;27(4):1081–1088. doi: 10.1016/j.bmcl.2016.12.042. [DOI] [PubMed] [Google Scholar]
- 26.Hayden M.S., Ghosh S. Regulation of NF-kappaB by TNF family cytokines. Semin Immunol. 2014;26(3):253–266. doi: 10.1016/j.smim.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hadler-Olsen E., Winberg J.-O., Uhlin-Hansen L.J.T.B. Matrix metalloproteinases in cancer: their value as diagnostic and prognostic markers and therapeutic targets. 2013;34(4):2041–2051. doi: 10.1007/s13277-013-0842-8. [DOI] [PubMed] [Google Scholar]
- 28.Smith A.L., Robin T.P., Ford H.L. Molecular pathways: targeting the TGF-beta pathway for cancer therapy. Clin Cancer Res. 2012;18(17):4514–4521. doi: 10.1158/1078-0432.CCR-11-3224. [DOI] [PubMed] [Google Scholar]
- 29.Hsieh H.L., Wang H.H., Wu W.B., Chu P.J., Yang C.M. Transforming growth factor-beta1 induces matrix metalloproteinase-9 and cell migration in astrocytes: roles of ROS-dependent ERK- and JNK-NF-kappaB pathways. J Neuroinflammation. 2010;7:88. doi: 10.1186/1742-2094-7-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H.H., Zhou X.L., Shi Y.L., Yang J. Roles of p38 MAPK and JNK in TGF-beta1-induced human alveolar epithelial to mesenchymal transition. Arch Med Res. 2013;44(2):93–98. doi: 10.1016/j.arcmed.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Huang X., Chen S., Xu L., Liu Y., Deb D.K., Platanias L.C., Bergan R.C. Genistein inhibits p38 map kinase activation, matrix metalloproteinase type 2, and cell invasion in human prostate epithelial cells. Cancer Res. 2005;65(8):3470–3478. doi: 10.1158/0008-5472.CAN-04-2807. [DOI] [PubMed] [Google Scholar]
- 32.Ma L., Teruya-Feldstein J., Weinberg R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 33.Stetler-Stevenson W.G., Aznavoorian S., Liotta L.A. Tumor cell interactions with the extracellular matrix during invasion and metastasis. Annu Rev Cell Biol. 1993;9:541–573. doi: 10.1146/annurev.cb.09.110193.002545. [DOI] [PubMed] [Google Scholar]
- 34.Weiss L. Metastatic inefficiency. Adv Cancer Res. 1990;54:159–211. doi: 10.1016/s0065-230x(08)60811-8. [DOI] [PubMed] [Google Scholar]
- 35.Asiedu M.K., Ingle J.N., Behrens M.D., Radisky D.C., Knutson K.L. TGFbeta/TNF(alpha)-mediated epithelial-mesenchymal transition generates breast cancer stem cells with a claudin-low phenotype. Cancer Res. 2011;71(13):4707–4719. doi: 10.1158/0008-5472.CAN-10-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernando R.I., Castillo M.D., Litzinger M., Hamilton D.H., Palena C. IL-8 signaling plays a critical role in the epithelial-mesenchymal transition of human carcinoma cells. Cancer Res. 2011;71(15):5296–5306. doi: 10.1158/0008-5472.CAN-11-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yadav A., Kumar B., Datta J., Teknos T.N., Kumar P. IL-6 promotes head and neck tumor metastasis by inducing epithelial-mesenchymal transition via the JAK-STAT3-SNAIL signaling pathway. Mol Cancer Res. 2011;9(12):1658–1667. doi: 10.1158/1541-7786.MCR-11-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Massague J., Blain S.W., Lo R.S. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103(2):295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 39.FOXM1 confers to epithelial-mesenchymal transition, stemness and chemoresistance in epithelial ovarian carcinoma cells. Oncotarget. 2014;6 doi: 10.18632/oncotarget.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sehgal I., Thompson T.C. Novel regulation of type IV collagenase (matrix metalloproteinase-9 and -2) activities by transforming growth factor-beta1 in human prostate cancer cell lines. Mol Biol Cell. 1999;10(2):407–416. doi: 10.1091/mbc.10.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hedges J.C., Dechert M.A., Yamboliev I.A., Martin J.L., Hickey E., Weber L.A., Gerthoffer W.T. A role for p38(MAPK)/HSP27 pathway in smooth muscle cell migration. J Biol Chem. 1999;274(34):24211–24219. doi: 10.1074/jbc.274.34.24211. [DOI] [PubMed] [Google Scholar]
- 42.Liang L.D., He T., Du T.W., Fan Y.G., Chen D.S., Wang Y. GinsenosideRg5 induces apoptosis and DNA damage in human cervical cancer cells. Mol Med Rep. 2015;11(2):940–946. doi: 10.3892/mmr.2014.2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu Q., Zeng K.W., Ma X.L., Jiang Y., Tu P.F., Wang X.M. Ginsenoside Rk1 suppresses pro-inflammatory responses in lipopolysaccharide-stimulated RAW264.7 cells by inhibiting the Jak2/Stat3 pathway. Chin J Nat Med. 2017;15(10):751–757. doi: 10.1016/S1875-5364(17)30106-1. [DOI] [PubMed] [Google Scholar]
- 44.He X., Ota T., Liu P., Su C., Chien J., Shridhar V. Downregulation of HtrA1 promotes resistance to anoikis and peritoneal dissemination of ovarian cancer cells. Cancer Res. 2010;70(8):3109–3118. doi: 10.1158/0008-5472.CAN-09-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu J., Wang S., Chen Y., Li X., Jiang Y., Yang X., Li Y., Wang X., Meng Y., Zhu M. miR-19 targeting of GSK3beta mediates sulforaphane suppression of lung cancer stem cells. J Nutr Biochem. 2017;44:80–91. doi: 10.1016/j.jnutbio.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 46.Wu Y.C., Tang S.J., Sun G.H., Sun K.H. CXCR7 mediates TGFbeta1-promoted EMT and tumor-initiating features in lung cancer. Oncogene. 2016;35(16):2123–2132. doi: 10.1038/onc.2015.274. [DOI] [PubMed] [Google Scholar]
- 47.Ko H., So Y., Jeon H., Jeong M.H., Choi H.K., Ryu S.H., Lee S.W., Yoon H.G., Choi K.C. TGF-beta1-induced epithelial-mesenchymal transition and acetylation of Smad2 and Smad3 are negatively regulated by EGCG in human A549 lung cancer cells. Cancer Lett. 2013;335(1):205–213. doi: 10.1016/j.canlet.2013.02.018. [DOI] [PubMed] [Google Scholar]
- 48.Ko H., Jeon H., Lee D., Choi H.K., Kang K.S., Choi K.C. Sanguiin H6 suppresses TGF-beta induction of the epithelial-mesenchymal transition and inhibits migration and invasion in A549 lung cancer. Bioorg Med Chem Lett. 2015;25(23):5508–5513. doi: 10.1016/j.bmcl.2015.10.067. [DOI] [PubMed] [Google Scholar]
- 49.Ko H. Geraniin inhibits TGF-beta1-induced epithelial-mesenchymal transition and suppresses A549 lung cancer migration, invasion and anoikis resistance. Bioorg Med Chem Lett. 2015;25(17):3529–3534. doi: 10.1016/j.bmcl.2015.06.093. [DOI] [PubMed] [Google Scholar]
- 50.Pang L., Li Q., Wei C., Zou H., Li S., Cao W., He J., Zhou Y., Ju X., Lan J. TGF-beta1/Smad signaling pathway regulates epithelial-to-mesenchymal transition in esophageal squamous cell carcinoma: in vitro and clinical analyses of cell lines and nomadic Kazakh patients from northwest Xinjiang, China. PLoS One. 2014;9(12) doi: 10.1371/journal.pone.0112300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feng H., Lu J.J., Wang Y., Pei L., Chen X. Osthole inhibited TGF beta-induced epithelial-mesenchymal transition (EMT) by suppressing NF-kappaB mediated Snail activation in lung cancer A549 cells. Cell Adh Migr. 2017;11(5–6):464–475. doi: 10.1080/19336918.2016.1259058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie L., Law B.K., Chytil A.M., Brown K.A., Aakre M.E., Moses H.L. Activation of the Erk pathway is required for TGF-beta1-induced EMT in vitro. Neoplasia. 2004;6(5):603–610. doi: 10.1593/neo.04241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang Y., Ye J., Jiao J., Zhang J., Lu Y., Zhang L., Wan D., Duan L., Wu Y., Zhang B. Down-regulation of miR-125a-5p is associated with salivary adenoid cystic carcinoma progression via targeting p38/JNK/ERK signal pathway. Am J Transl Res. 2017;9(3):1101–1113. [PMC free article] [PubMed] [Google Scholar]
- 54.Mochizuki M., Yoo Y.C., Matsuzawa K., Sato K., Saiki I., Tono-oka S., Samukawa K., Azuma I. Inhibitory effect of tumor metastasis in mice by saponins, ginsenoside-Rb2, 20(R)- and 20(S)-ginsenoside-Rg3, of red ginseng. Biol Pharm Bull. 1995;18:1197–1202. doi: 10.1248/bpb.18.1197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.