Abstract
The epithelial–mesenchymal transition (EMT) plays a pivotal role in the differentiation of vertebrates and is critically important in tumorigenesis. Using this evolutionarily conserved mechanism, cancer cells become drug-resistant and acquire the ability to escape the cytotoxic effect of anti-cancer drugs. In addition, these cells gain invasive features and increased mobility thereby promoting metastases. In this respect, the process of EMT is critical for dissemination of solid tumors including breast cancer. It has been shown that miRNAs are instrumental for the regulation of EMT, where they play both positive and negative roles often as a part of a feed-back loop. Recent studies have highlighted a novel association of p53 and EMT where the mutation status of p53 is critically important for the outcome of this process. Interestingly, p53 has been shown to mediate its effects via the miRNA-dependent mechanism that targets master-regulators of EMT, such as Zeb1/2, Snail, Slug, and Twist1. This regulation often involves interactions of miRNAs with lncRNAs. In this review, we present a detailed overview of miRNA/lncRNA-dependent mechanisms that control interplay between p53 and master-regulators of EMT and their importance for breast cancer.
Subject terms: Breast cancer, Long non-coding RNAs, miRNAs
Facts
Epithelial-to-mesenchymal transition (EMT) is a highly orchestrated transcriptional program that takes place during the development and is mediated by several EMT-specific transcription factors (TFs): Zeb1/Zeb2, Snail, Slug, and Twist1.
EMT is frequently re-activated during tumorigenesis, including breast cancer, and is responsible for drug resistance and metastasis.
p53 hinders EMT by augmenting the expression of miRNA-200 that in turn, attenuates Zeb1/Zeb2 levels. The opposite is also true, thus forming a regulatory feedback loop.
Open questions
How does mutant p53 affect EMT in breast cancer?
What are the relationships between p53, long non-coding RNAs, and miRNAs?
Do p53-dependent miRNAs that target EMT:-TFs identified in various tumors also operate in breast cancer?
Introduction
Breast cancer
Breast cancer is the world’s leading cause of cancer-related death in women, impacting >2.0 million women each year worldwide. It is characterized by a high degree of heterogeneity on the clinical, morphological, and molecular levels. Despite its heterogeneity, breast cancer can be broadly categorized into four subtypes: luminal A (ER+, and/or PR+, HER2−), luminal B (ER+, PR+, HER2+), HER2 enriched (ER–, PR–, HER2+), and triple-negative breast cancers (TNBC) comprising basal-like (ER–, PR–, HER2–) and claudin-low breast cancers. The TNBC group is considered to be the most aggressive and highly metastatic type of breast cancer. Despite high incidences, the 5-year survival rate of women with breast cancer is comparatively favorable estimated at ~80–90%, but it rapidly decreases to around 24% in cases where distant metastases are detected. It is also predicted that 30% of early-stage breast cancer patients will progress to the severe forms of the disease, developing remote organ metastases1.
The aggressive, basal forms of breast cancer that are highly metastatic have undifferentiated, stem cell-like features correlating to a worse prognosis compared to other subtypes. This process of dedifferentiation and acquisition of an invasive phenotype is mediated by a molecular mechanism called epithelial-to-mesenchymal transition (EMT). The EMT process has been strongly correlated with tumor migration, invasion, and metastases. Since metastases are responsible for almost 90% of deaths in breast cancer, it is not surprising that a lot of effort has been made to study the molecular mechanisms governing the control of EMT and invasion.
Non-coding RNAs in cancer
Non-coding RNAs (ncRNAs) are RNA products transcribed from the non-protein-coding parts of the genome. Although originally believed to be “junk DNA,” the ever-growing list of publications has defined the central role of expressed ncRNAs in development, differentiation, stress, and pathogenesis. There exists a vast variety of ncRNAs, including miRNAs, lncRNAs, piRNAs, and circRNAs, which efficiently modulate gene expression and importantly, regulate expression levels of each other. In cancers, these ncRNAs are reported to function either as oncogenes or tumor-suppressors depending on the cell-type and cellular context.
miRNAs are 18–24-nt long ncRNAs that primarily work by binding to short complementary sequences in 3′UTRs of their target transcripts. This complementary binding usually results in the degradation or sequestration, and translation repression of the respective base-paired mRNAs. However, some evidence suggests that miRNAs can also be positive regulators of translation. By forming complexes with special proteins, such miRNAs on the contrary, have the potential to stabilize or upregulate the translation of their target transcripts in response to specific conditions and cues. Moreover, because of a short seed sequence, a single miRNA has the capacity to target mRNAs of multiple genes either from the same pathway or across diverse pathways, leading to global changes in the expression patterns, which are typically observed in malignancies. To date, there is a grate list of miRNAs, both intracellular and circulating, that were repeatedly demonstrated to be the biomarkers of BC progression, metastasis formation, drug resistance, and outcome prediction, e.g. miR-21, miR-155, or miR-1452.
LncRNAs are longer transcripts (>200 nt), and have diverse mechanisms of action. While the majority of lncRNAs are retained in the nucleus to perform tasks like epigenetic modifications, cis or trans regulation of gene expression, and splicing control, a number of lncRNAs are also translocated to the cytoplasm. These cytoplasmic lncRNAs act as competing endogenous RNAs (ceRNAs) and mediate the sponging of miRNAs via miRNA response elements (MRE), thereby eliminating the inhibition on miRNA target genes. They can also regulate translation by complexing with other proteins. The ability of lncRNAs to form secondary and tertiary structures allows them to exert various roles as scaffolds or decoys for their respective target proteins. In addition, lncRNAs can target other RNAs and metabolites, consequently interfering with gene expression. Like miRNAs, there has been a multitude of studies linking the dysregulation of lncRNAs to cancer. Thus, it was repeatedly shown that abnormal expression of numerous lncRNAs among which, for example, MALAT1, HOTAIR, and DANCR, contributes to BC aggressiveness, metastasis formation, and hence BC patients’ prognosis outcome3. Therefore, owing to their clinical implications and networks of cross-talk, both miRNAs and lncRNAs are explored as therapeutic and diagnostic options in cancer and subjected to extensive research.
Epithelial-to-mesenchymal transition (EMT)
The epithelial-to-mesenchymal transition (EMT) is an evolutionary conserved reversible biological process that typically takes place during the development of organisms and results in a switch from epithelial phenotype to the mesenchymal phenotype. Morphologically, EMT is manifested in loss of cell polarity, adhesive, and tight contacts leading to the detachment of cells from the basement membrane. This is paralleled by massive cytoskeletal rearrangements that consequently change the morphology of epithelial cells to fibroblast-like, spindle-shaped cells and render them motile with an increased potential to invade the surrounding tissue and migrate to distant sites motile with an increased potential to invade the surrounding tissue and migrate to distant sites4. While EMT is utmost essential during embryo-development (classified as EMT type-1) and wound healing (EMT type-2), it plays an equally important role in cancer progression and dissemination (EMT type-3), consequently determining the prognosis. Mesenchymal cells produced through EMT have been found to be similar to tumor-initiating CSCs with a high expression of CD44, exhibiting the drug-, apoptosis-, and anoikis-resistant phenotypes5,6. Further, these cells are able to avoid oncogene-induced senescence and are immunosuppressive in nature7.
It is also worth noting that EMT has a large degree of plasticity, i.e. it can be reversed. This process is called mesenchymal-to-epithelial transition (MET). These evidences support the notion that EMT not only helps cancer cells to invade and metastasize but also confers an enhanced endurance and survival, enabling them to establish new tumors at distant sites.
EMT is activated and stabilized in response to a number of paracrine signals driven by stromal cells, such as fibroblasts, myofibroblasts, and mesenchymal stem cells. These signals and growth factors trigger cascades inside the target cells that culminate in the activation of gene expression pathways that initiate EMT. Cancer cells exploit this program to their benefit by modulating a handful of core transcription factors involved in the direct regulation of EMT, conveniently known as the EMT-TFs. These core EMT-TFs, which include Zeb1, Zeb2, Snail, Slug, and Twist1, are pleiotropic in nature and act in various combinations in different cell types to kickstart EMT. In recent times, non-coding RNAs (miRNAs and lncRNAs) have been discovered to act as critical downstream mediators by modulating a large number of target genes and pathways together to either promote or suppress EMT8.
It has also been discovered that breast cancer cells of the basal phenotype are more primed transcriptionally to respond quickly to extracellular EMT-inducing signals like TGF-β, thereby becoming more mesenchymal and dedifferentiated as compared to luminal cells which resist this transition9. This indicates that the phenotype of the cell is also an important determinant of its response to EMT-inducers and may explain why basal-like breast cancers are so aggressively metastatic with a bleak prognosis.
The master-regulators of EMT in breast cancer
As mentioned above, the EMT program is largely regulated at the level of gene expression by several transcription factors, including Zeb1 (Zfhx1a, Bzp, Zfhep, dEF1, TCF8, Nil-2, and AREB6)10, Zeb2 (Zfh1, CIP1)11, Snail (SNAI1), and Slug (SNAI2)12, as well as Twist113. Twist1 belongs to the family of Helix-Loop-Helix (HLH) factors that bind specific DNA sequences called E-box (5′-CANNTG-3′) that are located in the regulatory regions of their target genes. In response to specific signaling cues, they repress transcription of the epithelial genes and promote the expression of mesenchymal genes.
The expression of Zeb1 is undetected in luminal breast cell lines but is observed in basal ones with the highest expression in CD44high cancer stem cells (CSC)9. Similarly, while Snail is detected in all subtypes, its expression is also the highest in basal-like cells14. A study shows that di-acetylated Twist1 is responsible for activating WNT5A and directly contributes to the invasion and tumorigenicity of basal-like breast cells15. Likewise, Slug is also known to suppress the expression of ER-alpha by binding to the E-box motif and is responsible for the migration of triple-negative MDA-MB-231 cells16. Cells overexpressing Slug also display the basal-like phenotype17. These studies reinforce the concept that the EMT pathway is majorly responsible for driving the malignancy of basal-like and TNBC cells, leading to dismal outcomes.
Association of p53 and EMT
The p53 tumor suppressor, known as the “genome guardian”, is known to play a key role in preventing tumor development18. The p53-coding gene TP53 is mutated in the vast majority of human tumors (in more than 50% of cases). p53 plays the role of a transcription factor that acts in response to various stress signals, causing cell cycle arrest, cell aging, and apoptosis, as well as controlling the metabolism and antioxidant status of cells19. Functioning in the development, p53 limits the plasticity of epithelial cells during EMT. For example, p53 can interfere with the delamination of the neural crest, which usually occurs as a result of triggering EMT20.
The published studies of the past decade highlighted the novel role of p53 in regulation of metastasis. Wild-type p53 (WTp53) was shown to prevent EMT and the associated stem cell-like phenotype across multiple cancers. As a transcriptional factor, p53 can repress EMT by helping the cells maintain the epithelial gene signature. Furthermore, p53 was shown to induce the attenuation of EMT-TFs levels21 via the augmentation of the expression of EMT-suppressing miRNAs. In this respect, it is important to note that p53 induces the expression of miRNAs that target the Zeb, Snail, and Twist families of transcription factors4 (Figs. 1, 2). In turn, EMT regulators are shown to attenuate the p53 functions. For example, one of the key EMT inducers Snail) is known to bind to and repress wild-type p53 directly. This Snail-mediated inhibition of p53 was found to be essential for tumor-initiation and growth in breast cancer models22.
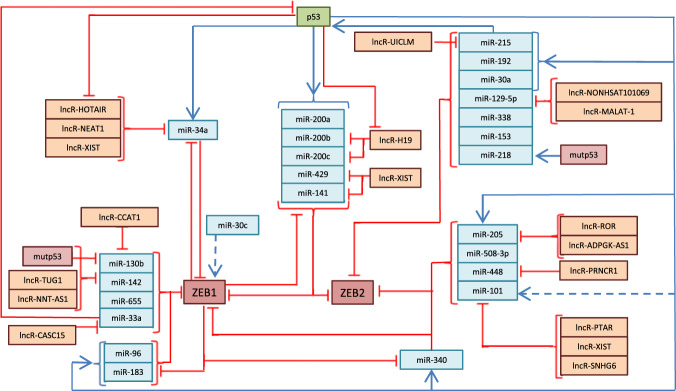
Fig. 1. Cross-regulation of p53 and Zeb1/2 EMT-TFs in breast cancer (BC) via miRNAs and lncRNAs.
р53 mediates expression of the indicated miRs (denoted with blue color) that in turn target Zeb1/2 EMT-TFs (denoted as burgundy-colored boxes). Long non-coding RNAs (lncRNAs) denoted with pink-color act as competing endogenous RNAs (ceRNAs) to counteract the effect of miRs. Interactions between all of these players form simple links and feedback loops (indicated with arrows and bars). Positive influence is denoted by blue arrows and negative is denoted by red bars.
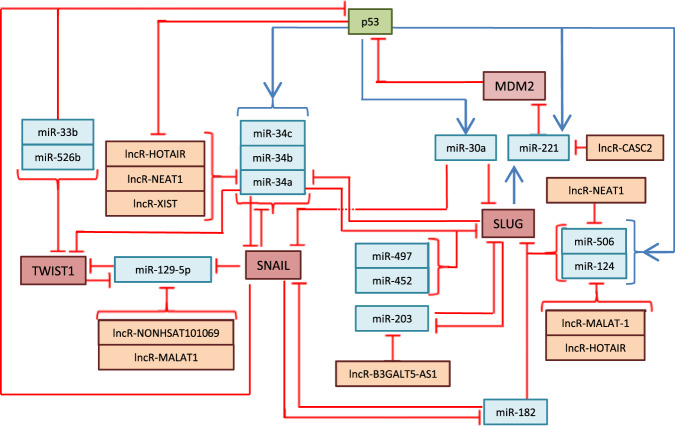
Fig. 2. Interactions between p53 and MDM2 in respect to EMT-TFs Twist, Snail, and Slug, which are mediated by miRNAs and lncRNAs in breast cancer.
The color-coding is the same as in Fig. 1.
The importance of p53 in averting cancer metastasis is evident in cases where p53 is lost or worse, mutated. Loss of WTp53 in breast epithelial cells triggers EMT with a parallel increase in the number of cancer stem cells and is associated with a higher tumor grade. This has been attributed to the decrease in expression of tumor-suppressing microRNAs such as miR-200c, which are directly transactivated by p5323.
On the other hand, the presence of gain-of-function (GOF) mutant p53 is enough to unleash EMT and makes the cells more migratory in nature. Mutant p53 upregulates the expression of EMT-TFs like Twist1 and Zeb124. This likely occurs because many forms of mutant p53 retain their transcriptional activity albeit with different sequence-specificity25. Furthermore, GOF mutants of p53 can mediate its oncogenic effects via the regulation of onco-miRs. This also explains why only the mutant p53 is able to activate multiple oncogenic miRNAs like miR-155 in breast cancer.
Control over the epithelial cell phenotype between p53 and EMT-TFs is a dynamic two-way process. To counteract the repressive effect of p53 on EMT, the EMT-TFs themselves negatively regulate p53 and its regulated miRNAs. For example, Twist1 is known to attenuate the effects of p53 and inhibit cellular apoptosis. Likewise, Snail directly binds and represses p5322. As a result of such opposing effects, multiple feedback loops have evolved that include intricate networks of p53, EMT-TFs, and non-coding RNAs. In this review, we attempt to portrait this complex circuitry.
p53-regulated miRNAs in control of EMT TFs
p53 has been shown to induce the expression of a number of miRNAs that suppress EMT via targeting various EMT-regulating transcription factors, such as Zeb1, Zeb24, and Snail26. A list of various miRNAs is shown in Tables 1–3.
Table 2.
miRNAs targeting SNAIL and SLUG.
| miRNA | Targets | Cancer subtype | Expression changes | Effect | References | p53 regulation |
|---|---|---|---|---|---|---|
|
miR-30a mir-30c |
Snail | N/S | ↓ | Reduced expression is associated with an unfavorable outcome, including late tumor stage, lymph node metastasis, and worse progression | 131,132 | Induced by p53 |
| Zeb1, Snail, Twist* (*indirectly activates) |
FBT TN |
↑ ↑ |
Elevated expression in cancer compared to normal tissue is a characteristic of familiar BC Promotes invasive phenotype |
133,134 | ||
|
miR-34a,c mir-34a |
Snail | N/S | ↑ | Associated with positive nodal status, high tumor grade, ER-negativity, and Her2-positivity | 135 | Induced by p53 |
| TN | ↓ | Associated with higher tumor grade and reduced patients survival | 136 | |||
| miR-203 | Snail | TN | ↓ | Low level is a characteristic of triple-negative BC subtype | 109 | Induced by p53 in colon and lung cancer |
| LA | ↓ | Low level is a characteristic of luminal A BC subtype | 137 | |||
| N/S | ↑ | High level is a marker of presence circulating tumor cells and metastasis | 106 | |||
| miR-124 | Slug, CDK4/6 | N/S | ↓ | Downregulated in human breast cancer specimens and the reduced expression is associated with advanced clinical stage and positive lymph node metastasis in breast cancer patients | 138 | Induced by p53 |
| miR-452 | Slug | LA | ↓ | Downregulated in adriamycin-resistant MCF7 cells compared with the parental cell line | 139 | |
| TN, LA | ↓ | Low expression was associated with elevated migration, invasion, and metastasis formation | 140 | |||
| miR-497 | Slug | N/S | ↓ | Low expression was associated with higher differentiation grade, positive Her-2 expression, higher incidence of lymph node metastasis, and advanced clinical stage | 141 | |
| miR-506 | Slug | N/S | ↓ | Downregulated in malignant breast tissues and its expression is inversely correlated with tumor stage | 142 | Induced by p53 |
| miR-182 | Slug, Snail | TN | ↑ | Upregulated in TN breast cancer tissues, promotes the proliferation, and invasion | 143 | |
| TN, HER2+ | ↑ | High expression of circulating mir-182 was the marker of ER- and PR-negative BC | 144 |
BC breast cancer, LA luminal A subtype, LB luminal B subtype, TN triple-negative (basal-like) subtype, HER2+ HER2-positive subtype, MBC metaplastic breast cancer, MTC metastatic breast carcinomas, N/S subtype is not specified, FBT familiar breast tumors, ER estrogen receptor, PR progesterone receptor.
Table 1.
miRNAs targeting Zeb1 and Zeb2.
| miRNA | Targets | Cancer subtype | Expression changes | Effect | References | p53 regulation |
|---|---|---|---|---|---|---|
| miR-142-3p | Zeb1 | N/S | ↓ | Downregulated in BC. Reduced expression is associated with tumor size and lymph node metastasis | 98 | Downregulated by mutant p53 |
| Attenuates breast CSC characteristics | 99 | |||||
| miR-96-183 (miR183-96-182 cluster) | Zeb1 | N/S | ↑ | High expression is a prognostic biomarker for BC, and correlated with local relapse, distant metastasis, and poor clinical outcomes | 100 | Induced by p53 |
| LA | ↓ | Downregulated in LA BC-induced overexpression of miR-183 inhibited migration of breast cancer cells | 101 | |||
| ↑ | miR-96 was upregulated in BC, enhanced proliferation, migration, and tumor growth | 102 | ||||
| miR-141 (miR-200 family) |
Zeb1 Zeb2 |
N/S | ↑ | The level was significantly higher in the blood of patients with stage I–III, lymph node metastasis, and Her2 negative tumors. Low expression of miR-141 was associated with unfavorable overall survival | 103 | Induced by p53 |
| TN | | Overexpression promotes migratory and invasive ability of TN BC cells | 104 | |||
| TN | | Ectopic expression enhanced brain metastatic colonization | 105 | |||
| N/S | ↑ | High level is a marker of circulating tumor cells presence and metastasis | 106 | |||
| miR-200a-200b-200c (miR-200 family) |
Zeb1 Zeb2 |
TN | | High levels block angiogenesis in tumor | 107 | Induced by p53 |
| MBC | ↑ | High levels are associated with metaplastic breast cancer and upregulated Zeb1, Zeb2, Snail, and Slug | 108 | |||
| N/S | ↑ | High levels are markers of circulating tumor cells presence and metastasis | 106 | |||
| TN | ↓ | Low level is a characteristic of triple-negative BC subtype | 109 | |||
| LA, LB | ↓ | Low expression indicated reduced survival in PR-positive BC cases | 110 | |||
| HER2+, TN | ↑ | High expression in tumors and increased content in blood were associated with shortened relapse-free survival of patients with in PR-negative BC | 103 | |||
| miR-429 (miR-200 family) |
Zeb1 Zeb2 |
TN | ↓ | Low level is a marker of BC compared to normal breast tissue. TN breast cancer is characterized by the lowest miR-429 compared to other BC types | 111 | Induced by p53 |
| TN | - | Upregulated in TN BC (MDA-MB-231 and MDA-MB-468) treated with δ-tocotrienol and induces apoptosis | 112 | |||
| TN | | High expression suppresses metastasis formation and migration of MDA-MB-231cells | 113 | |||
| miR-33a | Zeb1 | TN, MTC | ↓ | Downregulated in TN breast cancer | 114 | Suppresses p53 in hematopoetic stem cells |
| N/S | – | Expression is negatively associated with lymph node metastasis and clinical stage, expression lower in metastatic cell lines | 65 | |||
| miR-655 | Zeb1 | TN | ↓ | Reduced expression in triple-negative BC compared to normal tissue and other cancer types. Lower expression is associated with higher lymph-node status | 115 | |
| miR-153 | Zeb2 | LA, TN | | Ectopic expression inhibited tumor growth and impair the migration and invasion of breast cancer cells | 116 | |
| TN | | Expression was induced by Mifepristone and resulted in suppressed tumor growth of the TN BC cell lines and patient-derived xenografts | 117 | |||
| miR-192 | Zeb2 | N/S | ↓ | Decreased in BC tissues | 118 | Induced by p53 |
| N/S | ↓ | Decreased in BC tissues and known to inhibit proliferation | 119 | |||
| miR-215 | Zeb2 | N/S | ↓ | Downregulated in BC and targets Sox9 lower levels contributed to unfavorable prognosis | 120 | Induced by p53, activates p53 |
| N/S | ↓ | Low expression correlated with higher tumor grade, Her2 positive status, lymph node metastasis, and low expression led to lower 5-year DSS (Disease-specific survival) | 121 | |||
| miR-218 | Zeb2 | N/S | ↓ | Lower expression of mir-218 is associated with a higher tumor grade, lymph node metastasis and poor prognosis | 122 | |
| miR-338p | Zeb2 | N/S | ↓ | Downregulated in BC, expression is inversely correlated with lymph node metastasis and TNM stage | 123 | |
| miR-101 | Zeb1, Zeb2 | MTC | ↓ | Downregulated in metastatic BC cells, expression varies amongst different subtypes | 124 | Induced by p53 |
| miR-205 | Zeb1, Zeb2 | N/S | ↓ | Downregulated in BC | 125 | Induced by p53 |
| N/S | ↓ | Lowest levels in basal-like and invasive TN breast cancers, inhibits CSC-like phenotype by downregulating ITGA5 | 126 | |||
| miR-340 | Zeb1, Zeb2 | N/S | | BC patients with high miR-340 expression unlikely to achieve pathologic complete response | 127 | Stabilizes p53 |
| TN | | Inhibits BC progression and metastasis | 128 | |||
| miR-448 | Zeb1, Zeb2 | N/S | ↓ | Downregulation is observed in BC patients with chemotherapy-induced EMT | 129 | |
| miR-508-3p | Zeb1, Zeb2 | TN | ↓ | Expression was decreased in TN BC. Lower miR-508-3p expression significantly associated with lymph node metastasis and distant metastasis | 130 |
BC breast cancer, LA luminal A subtype, LB luminal B subtype, TN triple-negative (basal-like) subtype, HER2+ HER2-positive subtype, MBC metaplastic breast cancer, MTC metastatic breast carcinomas, N/S subtype is not specified, FBT familiar breast tumors, ER estrogen receptor, PR progesterone receptor, CSC cancer stem cells.
Table 3.
miRNAs targeting Twist1.
| miRNA | Targets | Cancer subtype | Expression changes | Effect | References | p53 regulation |
|---|---|---|---|---|---|---|
| miR-129-5p | Twist1 | HER2+ | | High level is associated with sensitivity of BC cells to trastuzumab | 145 | Has potential p53 response elements in promoter |
| N/S | ↓ | Low level is associated with breast cancer diagnosis; Suppresses the proliferation of BC cells | 146 | |||
| TN | | Overexpression sensitized MDA-MB-231 cells to irradiation | 147 | |||
| LA | – | Knockdown of miR-129-5p reduced radiosensitivity of MCF-7 cells | ||||
| miR-526b | Twist1 | N/S | | Associated with reduced patients survival and higher tumor grade | 148 | |
| miR-33b | Twist1 | N/S | ↓ | Downregulated in BC, its expression negatively correlates with lymph node metastasis and tumor stage | 66 | May repress p53 in hematopoetic stem cells |
BC breast cancer, LA luminal A subtype, TN triple-negative (basal-like) subtype, HER2+ HER2-positive subtype, N/S subtype is not specified.
miR 200 family
Kim et al. were the first to show that p53 brings about the suppression of EMT by inhibiting the expression of Zeb1 and Zeb2 through induction of the miR-200 family of miRNAs in hepatocellular carcinoma4. Later, Chang and colleagues found that the p53 family directly controls the expression of the miR-200 family in breast cancer23.
Several studies have now established that miR-200 family acts a critical mediator of the p53-regulated suppression of EMT by targeting several EMT-TFs including Zeb1 to maintain the epithelial phenotype. All five members of the miR-200 family (miR-200a, miR-200b, miR-200c, miR-141, and miR-429), as well as miR-205, were shown to suppress EMT via targeting Zeb127. The increase in ectopic expression of the genes of this family (miR-200a, miR-200b) leads to morphological changes in cells, which change from a spindle-shaped mesenchymal back to a round epithelial form with concomitant association of cells into groups. In turn, ablation of miR-200 promoted chemoresistance of breast cancer cells during EMT28. Loss of the miR-200 family in regions of metaplastic breast cancer specimens was shown to be paralleled with loss of E-cadherin. Thus, downregulation of the members of this miR family is likely to be an important step in the tumor progression.
Regulation by EMT-TFs: Zeb1 and Snail
Zeb1, along with Snail, plays a critical role in the regulation of p53-regulated miR-200 family members. Zeb1 and Snail have been reported to bind the promoters of miR-200b-200a-429, miR-200c-149, and curb their expression29. Additionally, Snail is also involved in promoting the methylation of miR-200 locus, which is essential for sustaining the mesenchymal phenotype in cancer cells.
Regulation of miR-200 family by lncRNAs
Several p53-suppressed long non-coding RNAs are also involved in the regulation of the miR-200 family, including lncRNA H19 and lncRNA XIST30,31. Both lncRNA H19 and lncRNA XIST act as so-called competing endogenous RNAs (ceRNAs), which physically associate with specific miRs through miRNA response elements (MREs) on their target transcripts32. Thus, lncRNA H19 and lncRNA XIST inhibit miR-200b and miR-200c33, miR-14130 and miR-42931, respectively.
Overall, the expression of the miR-200 family seems to be under tight regulation by TFs, epigenetic regulators and lncRNAs, and displays a strong effect on the EMT process by targeting the EMT TFs.
miR-34
It has recently been shown that miR-34a, another target of p5334 can directly suppress Zeb1. p53 augments the expression of miR-34a both in cultured cells, as well as in irradiated mice35. Notably, Zeb1 can reciprocally inhibit the transcription of miR-34 by binding to the E-boxes located in the promoter regions of miR-34a-b-c36. In addition to targeting Zeb1, miR-34a also downregulates Snail, Slug, Twist1, and Notch in metastatic breast cancers37, as well as other stemness-associated factors like CD133, CD44, BMI1, and c-Myc36 via direct binding 3′UTRs of the respective genes and thereby attenuating the process of EMT and metastasis. Given that miR-34 targets several critical EMT-TFs, it can be considered as a “universal weapon” against EMT.
Regulation of miR-34 by EMT-TFs: Zeb1, Snail, and Slug
Similar to the miR-200 regulatory mechanism described above, Zeb1 and Snail regulate miR-34a. Siemens and colleagues have established the presence of a negative feedback loop, wherein miR-34a and Snail repress each other by direct binding. This interplay is mediated via the p53-dependent activation of miR-34a during the regulation of EMT36. Another member of the Snail family, SLUG has also been reported by De Carolis et al. to directly bind to the promoter of miR-34a and repress its transcription in breast cancer cells exposed to hypoxia, and upregulate the expression of Carbonic Anhydrase isoenzyme 9 (CAI9). This enabled the cells to acquire a stemness like phenotype with an increased ability to form mammospheres38.
Regulation of miR-34 by lncRNAs
It has been noted that several lncRNAs, including lncRNA XIST39 and the p53-inducible LncRNA NEAT140 act as ceRNA for miR-34a, which leads to the activation of EMT. The physiological meaning of LncRNA NEAT1 activation by p53, especially in breast cancer, needs to be investigated further.
miR-192/215 family
Kim et al. demonstrated that the p53-regulated miR-192 was able to prevent EMT in hepatocellular carcinoma by suppressing Zeb2 at the post-transcriptional level41. Similarly, in the same study, another p53-dependent miR-215 was shown to significantly reduce levels of mRNA and protein of Zeb2 in cells of the colorectal cancer lines SW620 and DLD-1. Importantly, the observation of EMT-suppressing activity of miR-215 was confirmed in breast cancer cells42. However, in this case, the authors focused on Sox-9 as the main target of miR-215. The question of whether miR-215-dependent attenuation of Zeb2 contributed to this process remains to be answered.
Regulation of miR-215 by lncRNAs
Interestingly, LncRNA-UICLM was shown to compete with miR-215 thereby regulating the expression of Zeb2. lncRNA-CDC6, which is overexpressed in breast cancer tissues, also negatively regulates miR-215 by sponging it. This inhibition of miR-215 promoted the proliferation and migration ability of breast cancer cells43.
miR-30 family
The miR-30 family, which includes five members (miR-30a-e), is involved in the pathogenesis of various types of tumors. It has also been demonstrated that p53 binding to the miR-30a promoter induces the transcription of both miRNA chains—5p and 3p—which are able to interact with Zeb244. Interestingly, the same family members can be either cancer-promoting or tumor-suppressing. For example, miR-30d45 and miR-30a have been identified as anti-metastatic factors in different tumors. In an experiment conducted by Mahsa and colleagues using MCF-7 breast cancer cells, an inverse correlation was found between miR-30c and Zeb1. This suggests that miR-30c likely activates Zeb1, although this has not been confirmed by direct experiments46. Furthermore, reduced miR-30 expression has been reported to be critical for the maintenance of self-renewal and inhibition of apoptosis in breast tumor-initiating cells.
miR-124
Mutation in or deletion of the TP53 gene affects the expression of several miRNAs, among which miR-124 was the most strongly downregulated. This effect correlated with an upregulation of the anti-apoptotic gene, iASPP, suggesting that the latter was the target for miR-124. Consistent with this notion, p53 was shown to bind the promoter of the miR-124 gene to facilitate its expression, which consequently inhibited iASPP expression. Overexpression of miR-124 led to suppression of the CDK4 protein expression and attenuated cell viability, proliferation, and cell cycle progression in MCF-7 and MDA-MB-435S breast cancer cells in vitro47.
miR-124 targets SLUG
The expression of miR-124 is reduced in human breast cancer tissues and its levels have been inversely correlated with the tumor grade. miR-124 attenuated the migration of metastatic breast cancer cell line MDA-MB-231 and reversed the morphology from spindle-shaped to epithelial cobblestone-like, with a parallel increase in the expression of E-cadherin. It was found that miR-124 mediated direct targeting of Slug’s 3′UTR was responsible for the reversal of EMT characteristics48.
Regulation of miR-124 by lncRNAs
LncRNA MALAT-1 serves as a ceRNA for miR-124 in breast cancer47 and in the development of non-small cell lung cancer. MALAT1 eliminates the suppressive effect of miR-124 on CDK4 and increases cell proliferation. In addition, lncRNA HOTAIR can also act as ceRNA for miR-124. The authors suggest that HOTAIR activation may enhance the EMT process through inhibition of miR-12449.
miR-203
An important tumor-suppressive miRNA, miR-203, is known to inhibit the invasiveness and migration of breast cancer cells. Two independent studies have shown that miR-203 also targets Slug directly through its 3′UTR50. Ding et al. described the presence of a double-negative feedback loop, wherein the promoter of miR-203 itself is targeted and repressed by Slug. In a similar manner, Slug also directly suppresses the expression of members of the miR-200 family51. Recently, miR-124 and miR-203 were shown to inhibit the expression of Zeb2 at the post-transcriptional level in human kidney carcinoma cells52. It will be interesting to see whether miR-203 can also attenuate the expression of Zeb2 and potentially Zeb1 in metastatic breast cancer cell models.
Studies on keratinocytes have demonstrated the expression of miR-203 to be dependent on p53. In these cells, the knockdown or HPV-mediated degradation of p53 significantly decreased the level of mir-203, while activation of p53 by doxorubicin resulted in the opposite effect53. This observation of mir-203’s dependence on p53 was also confirmed in lung cancer cells, wherein the reintroduction of p53 in p53-null cells increased miR-203, while mutant p53 failed to do so. Consequently, the overexpressed miR-203 augmented the sensitivity of both colon and lung cancer cells to gemcitabine-induced apoptosis, possibly by positively regulating the expression of Puma.54. Although the association between p53 and miR-203 has not been explored in breast cancer yet, it is very tempting to speculate that such regulation also takes place in BC cells.
Regulation of miR-203 by lncRNA
LncRNA B3GALT5-AS1, which promoted colon cancer invasion, inhibited miR-203 directly by interacting with the miR-203 promoter55. The overexpression of this lncRNA thereby increased the cellular levels of Zeb2 and Slug and promoted EMT.
miR-129-5p
Tan et al. have reported that the promoter region of miR-129-5p could be potentially targeted by p53, whose malfunction is frequently detected in human cancers56. High levels of miR-129-5p increased the expression of E-cadherin by interacting with the 3′UTR region of Zeb257. miR-129-5p levels were significantly decreased in breast cancer cell lines. In a study by Yu et al., Twist1 was shown to be a direct target of miR-129-5p and was repressed on its overexpression. The promoter of miR-129-5p reportedly contains three E-box motifs, and the ChIP analysis confirmed the enrichment of both SNAIL and TWIST1 to these sites, correlating with a decrease in the promoter activity. This suggests the presence of reciprocal negative regulation between Twist1 and miR-129-5p. This downregulation of miR-129-5p via the Twist1-Snail feedback loop stimulates EMT and is associated with poor prognosis in breast cancer58.
Regulation of miR-129 by lncRNAs
The lncRNA NONHSAT101069 was found to function as ceRNA by directly binding to miR-129-5p in breast cancer cells59. lncRNA MALAT1 also targets miR-129-5p by directly binding and sponging it, resulting in greater cell invasion and migration. MALAT1 and miR-129-5p were found to have an inverse correlation of expression in TNBC tissues60. There have been numerous other reports of MALAT1 regulating multiple modulators and enhancing the EMT and stemness phenotype, which underlines its importance in breast cancer progression61. Interestingly, it was found that mutant gain-of-function p53, along with other factors ID4 and SRSF1, associates with MALAT1, and represses the production of an anti-angiogenic splicing isoform of VEGFA, hence promoting angiogenesis in breast cancer62.
The miR-33 family
miR-33a-5p targets Zeb1
Surprisingly, miR-33a-5p, which was shown to target p53 in stem cells63, also inhibits the expression of Zeb1 by interacting with its 3′UTR region64. The overexpression of miR-33a in metastatic breast cancer cells remarkably decreases cell proliferation and invasion in vitro and significantly inhibits tumor growth and lung metastasis in vivo. Conversely, its knockdown in non-metastatic breast cancer cells considerably enhances cell proliferation and invasion in vitro and promotes tumor growth and lung metastasis in vivo, which strongly supports the idea of miR-33 as a tumor suppressor65. In hematopoietic stem cells, it is known that miR-33 negatively regulates p53 by binding to two conserved motifs in the 3′UTR of its mRNA. This downregulation of p53 and p53-activated apoptosis is responsible for maintaining the stemness phenotype, and reveals the molecular mechanisms of how miR-33 plays opposing roles depending on the cell type63.
miR-33b targets Twist1
Twist1 is overexpressed in aggressive breast cancers and is also known to promote breast cancer metastasis to the bone. Lin et al. reported that the levels of miR-33b in human breast cancer tissues are significantly reduced and correlate inversely with node metastasis and tumor stage66. MiR-33b was shown to directly bind to the 3′UTR of Twist1 and suppress it. Furthermore, ectopic overexpression of miR-33b in metastatic breast cancer cell lines decreased the number of cancer stem cells (CSCs) and their invasive properties. Their ability to metastasize was also found to be reduced in mice models in vivo, suggesting that this miR plays a vital role in maintaining the stemness and invasion properties of breast cancer cells66.
Regulation of miR33a-5p by lncRNA
Using pull-down analysis with biotin-labeled miR-33a-5p in gastric cancer cells, it was found that miR-33a-5p directly interacted with lncRNA CASC15, establishing its role as a ceRNA for miR-33a-5p64. RT-PCR and Western blot analyzes proved that the inhibition of lncRNA CASC15, as well as the induced expression of miR-33a-5p, reduced levels of Zeb1 expression in AGS and SGC7901 gastric cancer cells64.
Other miRNAs
miR-101
miR-101 is one of the potential targets of p53 in induced pluripotent stem cells67. Several studies implicate miR-101 as a regulator of breast cancer, which targets several important oncogenes. By binding to the 3′UTR of the Zeb1 and Zeb2 mRNA sites, it reduces the levels of Zeb1 and Zeb2, which leads to the attenuation of EMT68.
The lncRNA PTAR (a pro-transition associated RNA, which is upregulated in mesenchymal subtype cells) has been discovered to inhibit miR-101 activity, acting as ceRNA, and eventually promoting EMT48. In addition, lncRNA SNHG669 and lncRNA XIST70 also inactivate miR-101 by the same mechanism.
Regulation of miR-101 by lncRNAs
The lncRNA PTAR (a pro-transition associated RNA) has been discovered to inhibit miR-101 activity, acting as ceRNA and eventually promoting EMT71. In addition, another lncRNA SNHG669 and lncRNA XIST70 also inactivate miR-101 by the same mechanism.
miR-205
In breast cancer, p53-inducible miR-205 was shown to possess tumor-suppressive functions72. A team of scientists led by Lee73 showed that miR-205 inhibits both Zeb1 and Zeb2 mRNAs in the breast cancer cell lines MCF7, MDA231, and SK-BR-3. However, under hypoxic conditions miR-205 in lung cancer promoted EMT by targeting apoptosis-stimulating protein of p53-2 (ASPP2)74. This fact indicates that depending on the environment, miR-205 can play either a positive or negative role in the regulation of EMT.
miR-205-5p is under the regulatory control of lncRNAs ROR75 and ADPGK-AS176 that inhibit miR-205-5p by acting as ceRNA, promoting cell proliferation, migration, and invasion through stabilizing mRNA of Zeb1. LncRNA-ROR is upregulated in breast tumors, and forced expression of lncRNA-ROR in breast epithelial cells leads to visible changes in morphology, increases mesenchymal markers, activates EMT, and promotes invasion, further generating stem-cell-like cells (CD44hi/CD24lo) with advanced mammosphere forming ability75.
miR-221
miR-221 is well-known as a basal subtype-specific miRNA and is overexpressed in TNBC cells. It targets the tumor suppressor p27KIP77. Noteworthy, in metastatic MDA-MB-231 cells, the miR-221 gene expression was upregulated by Slug, as it was recruited to the E-boxes located in the miR-221 promoter. Conversely, repression of Slug led to decreased levels of miR-221, as well as decreased cell motility78.
miR-221 was reported to activate the p53/mdm2 axis by inhibiting Mdm2. In turn, p53 activation was shown to enhance miR-221 expression79.
It is important to note that this effect was evident only upon activation of p53 by the cytotoxic drug, doxorubicin. Future studies should elucidate whether the p53-activating effect of miR-221 is still valid in metastatic EMT cells.
In hepatocellular carcinoma, miR-221 (as well as miR-24) is negatively regulated by the sponging action of CASC2, which is a well-established tumor-suppressive lncRNA80. It was found that by inhibiting miR-221, CASC2 could mediate the sensitivity of cancer cells to apoptosis induced by anti-cancer agent TRAIL and upregulated Caspase-3, which is a direct target of miR-221.
miR-506
MiR-506 was shown to target the Slug gene directly and decrease EMT characteristics in breast cancer cell lines81. The upstream region of miR-506 has a putative p53 response element. p53 was shown to directly target and upregulate its expression in lung cancer cells82, but no such study has been carried out yet for breast cancer.
MiR-506 is also known to induce the demethylation of the MEG3 promoter of a lncRNA MEG3 via SP1/SP2 and Dnmt1. MEG3 is a lncRNA that inhibits cell growth and metastasis of breast cancer cells83. The expression of lncRNA MEG3 is reduced in breast cancer tissues and also in cell lines MCF7 and MDA-MB-231. Overexpression of MEG3 also led to a decrease in the Mdm2 RNA and protein levels, which subsequently stabilized p53 on the protein levels resulting in activation of its targets including p21, Maspin, and KAI184. Another lncRNA that plays an important role in regulating miR-506 is lncRNA NEAT1. The gene expression profile data and results of dual-luciferase reporter assay in serous ovarian cancer demonstrated that lncRNA NEAT1 functioned as a competing ceRNA for miR-506 to promote cell proliferation and migration85.
The H19/miR-675 locus
Another long non-coding RNA, LncRNA H19, is highly expressed in metastatic breast cells. The promoter region of LncRNA H19 has been shown to be effectively suppressed by the wild-type p53 protein86. Slug upregulates the expression of the H19 locus. Interestingly, this locus also encodes for another miRNA, miR-675. It was found that H19 itself could also upregulate Slug through a mechanism dependent on miR-675. This positive feedback loop increased the invasive properties of cancer cells both in vitro and in vivo87.
miR-10b
MiR-10b is a well-known metastatic miRNA in breast cancer, which is highly expressed in breast cancer tissues and metastatic cell lines. While it was found to have no significant effect on cellular proliferation, the invasiveness of cells, both in vitro and in vivo, significantly increased on miR-10b ectopic expression, and distant metastasis was also promoted. E-box motifs were discovered in the region upstream of miR-10b, and Twist1 was shown to directly bind to one of these regions. Because of this direct binding and their positive correlation of expression, it was suggested that Twist1 drives the expression of miR-10b to promote cancer spread. Downstream, miR-10b targets HOXD10, which is involved in the repression of cell motility genes88. However, miR-10b promoter has a p53 response element, and Bisio et al. showed using CHIP assay that p53 was actively recruited to the miR-10b promoter in MCF7 cells and induced its expression89. This seems counter-intuitive to the role of miR-10b as a “metastamiR”, and functional studies are lacking to further explore the effects of this association.
Hence, the existence of multiple feedback loops between the EMT-TFs and p53-dependent miRNAs may be responsible for driving the final fate of the cell.
miRNAs regulated by mutant p53 in EMT
Since wild-type and mutant p53 control different networks of genes, it is not surprising that they also affect different spectra of miRNAs involved in EMT. A list of mutant p53 regulated miRNAs is presented below:
miR-130b
The mutant p53 control different networks of genes, it is not surprising that they also affect different spectra of miRNAs involved in EMT. For example, mutant p53 exerts oncogenic functions and promotes EMT by directly binding to the promoter of miR-130b (a negative regulator of Zeb1) and inhibiting its transcription. Attenuation of mutant p53 in endometrial cancer cells increased miR-130b expression, leading to repression of Zeb1 and blocking the execution of the EMT program90. In contrast, Jia et al. have shown that miR-130b by targeting the tumor suppressor protein Pten, enhanced multiple drug resistance, proliferation, and tolerance to apoptosis of breast cancer cells thereby promoting oncogenesis91. The nature of this controversy is currently unknown and requires additional investigation.
Interestingly, lncRNA CCAT1, known to interact with p53, also acts as a ceRNA for miR-130b, augmenting the EMT, cell migration, and invasion92. By inhibiting the action of miR-130b, CCAT1 upregulates the expression of Zeb1 and Stat3 in ovarian cancer. While CCAT1 has been observed to be oncogenic in TNBC as well, its association with miR-130b has not been examined in breast cancer yet.
miR-142-3p
An onco-suppressive miR-142-3p inhibits the proliferation, migration, and invasion of breast cancer cells93. It is often downregulated upon overexpression of gain-of-function mutant form of p53 due to hypermethylation of its promoter94. Using luciferase reporter assay, it was shown that miR-142-3p interacts with the 3′UTR region of Zeb1 mRNA and reduces the levels of Zeb1 transcript and protein. This result suggests that miR-142 likely represses EMT.
Multiple lncRNAs are able to counteract the EMT-suppressing effects of miR-142. In HCC, the RNA product of the taurine upregulated gene 1 (TUG1) reduces the effect of miR-142-3p on Zeb1 by competitively binding to miR-142-3p, acting as ceRNA95. Knockdown of lncRNA-TUG1 was consistent with an increase in miR-142-3p levels and limited the invasion of cells. In breast cancer, it was revealed that another lncRNA NNT-AS1, functions as ceRNA specific for miR-142-3p, thereby restoring Zeb1 and blocking EMT96. lncRNA NNT-AS1 was also significantly overexpressed in breast cancer and correlated with poor prognosis.
miR-218
Expression of another miR-218 is specifically upregulated by mutant p53 (R172H) in mesenchymal cells67. The R172H mutant displays a GOF activity during the reprogramming of somatic cells into induced pluripotent stem cells and also increases their oncogenic potential. While miR-218 was upregulated in mutant p53 cells and downregulated in wtp53, the exact role of miR-218 in facilitating this reprogramming has not been detailed yet. Unexpectedly, miR-218 was shown to regulate EMT by inhibiting the expression of Zeb2. Liu et al. investigated the role of miR-214 and miR-218 in breast cancer97. The authors found that the aberrant expression of miR-214 and miR-218 were negatively associated with Ki 67, and the expression of miR-218 expression was positively associated with progesterone receptor (PR) in breast cancer tissues. Upon overexpression, the cell proliferation and migration in vitro were decreased and cell apoptosis was induced in breast cancer cells. The authors concluded that miR-214 and miR-218 function as tumor suppressors in breast cancer.
miR-218 also caused the inhibition of tumor growth and metastasis in lung cancer. Taken together, these reports may suggest that exerts different functions depending on the cellular context, i.e. in epithelial cells it serves as a tumor suppressor and in the mesenchymal ones upon the induction of pluripotency it may behave as an oncogene.
Conclusions
MicroRNAs play a decisive role in EMT, either as effector molecules of major transcription factors or as modulators of their expression. Recently, it has become apparent that miRNAs are among the critical regulators of EMT, with the miR-200 family making the main contribution to the process. This in no way undermines the relevance of other players, however, it should be noted that miRNAs play an important role in the regulation of TFs genes, whose products are master-regulators of the EMT. In addition, due to the large number of miRNAs operating in this process, it seems that the regulation is carried out by the additive principle: even a slight dysregulation in the expression levels of several members of one miRNA family would lead to significant amplification of the effect on the level of protein expression for Zeb/Snail/Twist, subsequently affecting the course of EMT. The regulatory complexity of EMT is further exacerbated by the fact that miRNAs themselves are often regulated by other lncRNAs. These feedback loops and networks of p53 and the EMT TFs, miRNA, and lncRNAs involved in the regulation of EMT are shown in Figs. 1 and 2.
Over the past few years, there has been an increase in the number of articles on the regulation of miRNAs using lncRNA. Based on this, it is likely that in the near future, the list of known miRNA regulators involved in EMT will expand significantly. The p53 tumor suppressor protein is a transcription factor by itself. Thus, perhaps it was not surprising to find out that p53 regulates vital cellular processes, including EMT, by regulating multiple miRNAs. Given the fact that p53 is mutated in more than 50% of all human cancers, the question arises as to whether the mutant p53 regulates different cohorts of miRNAs. In fact, it can be hypothesized that the “onco-miRs” regulated by various mutants of p53 may facilitate EMT, in contrast to wild-type p53. Thus, in order to unravel the complex network of EMT regulation one would take into account the status of p53 and other major tumor suppressors as they may all affect the final outcome of EMT and its reverse process called MET. This knowledge should provide means to consciously intervene with the regulation of EMT as part of the anti-cancer therapy.
Acknowledgements
N.B., O.F., A.D., and R.K. would like to acknowledge the joint funding from Russian Science Foundation (RSF grant #19-45-02011), Russia and Department of Science and Technology (DST), India. S.P., O.F., A.D., and N.B. acknowledge Mega-grant program of the Government of Russian Federation (#14.W03.31.0029). N.B., O.F., and A.D. also appreciate the support of RFBR (grant #18-29-09144). A.S. thanks Council of Scientific and Industrial Research (CSIR), Govt. of India for Junior Research Fellowship.
Author contributions
S.P. and A.S. collected the data and wrote the manuscript. N.B. and R.K. designed and edited the manuscript. O.F. designed illustrations. A.D. collected the data for Tables 1–3.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by E. Candi
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ritu Kulshreshtha, Email: drritukulshreshtha@gmail.com.
Niсkolai A. Barlev, Email: nick.a.barlev@gmail.com
References
- 1.Redig AJ, McAllister SS. Breast cancer as a systemic disease: a view of metastasis. J. Intern. Med. 2013;274:113–126. doi: 10.1111/joim.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nassar FJ, Nasr R, Talhouk R. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol. Ther. 2017;172:34–49. doi: 10.1016/j.pharmthera.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Huang Q-Y, et al. Long non-coding RNA: dual effects on breast cancer metastasis and clinical applications. Cancers. 2019;11:1802. doi: 10.3390/cancers11111802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim T, et al. p53 regulates epithelial–mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J. Exp. Med. 2011;208:875–883. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frisch SM, Schaller M, Cieply B. Mechanisms that link the oncogenic epithelial–mesenchymal transition to suppression of anoikis. J. Cell Sci. 2013;126:21–29. doi: 10.1242/jcs.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurrey NK, et al. Snail and slug mediate radioresistance and chemoresistance by antagonizing p53‐mediated apoptosis and acquiring a stem‐like phenotype in ovarian cancer cells. Stem Cells. 2009;27:2059–2068. doi: 10.1002/stem.154. [DOI] [PubMed] [Google Scholar]
- 7.Tiwari V. Burn wound: how it differs from other wounds? Indian J. Plast. Surg. 2012;45:364–373. doi: 10.4103/0970-0358.101319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gugnoni M, Ciarrocchi A. Long noncoding RNA and epithelial mesenchymal transition in cancer. Int. J. Mol. Sci. 2019;20:1924. doi: 10.3390/ijms20081924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaffer CL, et al. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell. 2013;154:61–74. doi: 10.1016/j.cell.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eger A, et al. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24:2375–2385. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- 11.Comijn J, et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell. 2001;7:1267–1278. doi: 10.1016/S1097-2765(01)00260-X. [DOI] [PubMed] [Google Scholar]
- 12.Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62:1613–1618. [PubMed] [Google Scholar]
- 13.Yang J, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Muenst, S. et al. Nuclear expression of snail is an independent negative prognostic factor in human breast cancer. Dis. Markers35, 337–344 (2013). [DOI] [PMC free article] [PubMed]
- 15.Shi J, et al. Disrupting the interaction of BRD4 with diacetylated twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell. 2014;25:210–225. doi: 10.1016/j.ccr.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, et al. Slug contributes to cancer progression by direct regulation of ERα signaling pathway. Int. J. Oncol. 2015;46:1461–1472. doi: 10.3892/ijo.2015.2878. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Storci G, et al. The basal‐like breast carcinoma phenotype is regulated by SLUG gene expression. J. Pathol. 2008;214:25–37. doi: 10.1002/path.2254. [DOI] [PubMed] [Google Scholar]
- 18.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 19.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rinon A, et al. p53 coordinates cranial neural crest cell growth and epithelial-mesenchymal transition/delamination processes. Development. 2011;138:1827–1838. doi: 10.1242/dev.053645. [DOI] [PubMed] [Google Scholar]
- 21.Yang-Hartwich Y, et al. p53–Pirh2 complex promotes Twist1 degradation and inhibits EMT. Mol. Cancer Res. 2019;17:153–164. doi: 10.1158/1541-7786.MCR-18-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ni T, et al. Snail1-dependent p53 repression regulates expansion and activity of tumour-initiating cells in breast cancer. Nat. Cell Biol. 2016;18:1221–1232. doi: 10.1038/ncb3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang C-J, et al. p53 regulates epithelial–mesenchymal transition and stem cell properties through modulating miRNAs. Nat. Cell Biol. 2011;13:317–323. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong P, et al. Mutant p53 gain-of-function induces epithelial–mesenchymal transition through modulation of the miR-130b–ZEB1 axis. Oncogene. 2013;32:3286–3295. doi: 10.1038/onc.2012.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fedorova O, et al. Attenuation of p53 mutant as an approach for treatment Her2-positive cancer. Cell Death Discov. 2020;6:1–8. doi: 10.1038/s41420-020-00337-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat. Cell Biol. 2014;16:488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 27.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 28.Yan X, et al. Mesenchymal stem cells promote hepatocarcinogenesis via lncRNA–MUF interaction with ANXA2 and miR-34a. Cancer Res. 2017;77:6704–6716. doi: 10.1158/0008-5472.CAN-17-1915. [DOI] [PubMed] [Google Scholar]
- 29.Burk U, et al. A reciprocal repression between ZEB1 and members of the miR‐200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, et al. Long non-coding RNA XIST promotes TGF-β-induced epithelial-mesenchymal transition by regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer. Cancer Lett. 2018;418:185–195. doi: 10.1016/j.canlet.2018.01.036. [DOI] [PubMed] [Google Scholar]
- 31.Shen J, et al. LncRNA XIST promotes pancreatic cancer migration, invasion and EMT by sponging miR-429 to modulate ZEB1 expression. Int. J. Biochem. Cell Biol. 2019;113:17–26. doi: 10.1016/j.biocel.2019.05.021. [DOI] [PubMed] [Google Scholar]
- 32.Su X, et al. microRNAs and ceRNAs: RNA networks in pathogenesis of cancer. Chin. J. Cancer Res. 2013;25:235. doi: 10.3978/j.issn.1000-9604.2013.03.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou, W. et al. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci. Signal.10, eaak9557 (2017). [DOI] [PubMed]
- 34.Zhang L, Liao Y, Tang L. MicroRNA-34 family: a potential tumor suppressor and therapeutic candidate in cancer. J. Exp. Clin. Cancer Res. 2019;38:53. doi: 10.1186/s13046-019-1059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raver-Shapira N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 36.Siemens H, et al. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- 37.Imani S, et al. MicroRNA-34a targets epithelial to mesenchymal transition-inducing transcription factors (EMT-TFs) and inhibits breast cancer cell migration and invasion. Oncotarget. 2017;8:21362. doi: 10.18632/oncotarget.15214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Carolis S, et al. Carbonic anhydrase 9 mRNA/microRNA34a interplay in hypoxic human mammospheres. J. Cell. Physiol. 2016;231:1534–1541. doi: 10.1002/jcp.25245. [DOI] [PubMed] [Google Scholar]
- 39.Song P, Ye LF, Zhang C, Peng T, Zhou XH. Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene. 2016;592:8–14. doi: 10.1016/j.gene.2016.07.055. [DOI] [PubMed] [Google Scholar]
- 40.Ji Y, Wang M, Li X, Cui F. The long noncoding RNA NEAT1 targets miR-34a-5p and drives nasopharyngeal carcinoma progression via Wnt/β-catenin signaling. Yonsei Med. J. 2019;60:336–345. doi: 10.3349/ymj.2019.60.4.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim T, et al. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J. Exp. Med. 2011;208:875–883. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao JB, Zhu MN, Zhu XL. miRNA-215-5p suppresses the aggressiveness of breast cancer cells by targeting Sox9. FEBS Open Bio. 2019;9:1957–1967. doi: 10.1002/2211-5463.12733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong X, et al. LncRNA–CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA‐215. J. Cell. Physiol. 2019;234:9105–9117. doi: 10.1002/jcp.27587. [DOI] [PubMed] [Google Scholar]
- 44.di Gennaro A, et al. A p53/miR-30a/ZEB2 axis controls triple negative breast cancer aggressiveness. Cell Death Differ. 2018;25:2165–2180. doi: 10.1038/s41418-018-0103-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, et al. Linc HOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 2015;6:e1802–e1802. doi: 10.1038/cddis.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rahimi M, et al. Down-regulation of miR-200c and up-regulation of miR-30c target both stemness and metastasis genes in breast cancer. Cell J. 2020;21:467–478. doi: 10.22074/cellj.2020.6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng T, et al. MiR-124 inhibits cell proliferation in breast cancer through downregulation of CDK4. Tumour Biol. 2015;36:5987–5997. doi: 10.1007/s13277-015-3275-8. [DOI] [PubMed] [Google Scholar]
- 48.Liang YJ, et al. MiR-124 targets Slug to regulate epithelial-mesenchymal transition and metastasis of breast cancer. Carcinogenesis. 2013;34:713–722. doi: 10.1093/carcin/bgs383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou H, et al. LncRNA HOTAIR promotes renal interstitial fibrosis by regulating Notch1 pathway via the modulation of miR-124. Nephrology. 2019;24:472–480. doi: 10.1111/nep.13394. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Z, et al. Epigenetic silencing of miR-203 upregulates SNAI2 and contributes to the invasiveness of malignant breast cancer cells. Genes Cancer. 2011;2:782–791. doi: 10.1177/1947601911429743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ding X, Park SI, McCauley LK, Wang CY. Signaling between transforming growth factor β (TGF-β) and transcription factor SNAI2 represses expression of microRNA miR-203 to promote epithelial-mesenchymal transition and tumor metastasis. J. Biol. Chem. 2013;288:10241–10253. doi: 10.1074/jbc.M112.443655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen J, Zhong Y, Li L. miR-124 and miR-203 synergistically inactivate EMT pathway via coregulation of ZEB2 in clear cell renal cell carcinoma (ccRCC) J. Transl. Med. 2020;18:69. doi: 10.1186/s12967-020-02242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McKenna DJ, McDade SS, Patel D, McCance DJ. MicroRNA 203 expression in keratinocytes is dependent on regulation of p53 levels by E6. J. Virol. 2010;84:10644–10652. doi: 10.1128/JVI.00703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Funamizu N, Lacy CR, Kamada M, Yanaga K, Manome Y. MicroRNA-203 induces apoptosis by upregulating Puma expression in colon and lung cancer cells. Int. J. Oncol. 2015;47:1981–1988. doi: 10.3892/ijo.2015.3178. [DOI] [PubMed] [Google Scholar]
- 55.Wang L, et al. Long noncoding RNA B3GALT5-AS1 suppresses colon cancer liver metastasis via repressing microRNA-203. Aging. 2018;10:3662–3682. doi: 10.18632/aging.101628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan G, et al. A novel role for microRNA-129-5p in inhibiting ovarian cancer cell proliferation and survival via direct suppression of transcriptional co-activators YAP and TAZ. Oncotarget. 2015;6:8676–8686. doi: 10.18632/oncotarget.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao L, et al. MicroRNA-129-5p modulates epithelial-to-mesenchymal transition by targeting SIP1 and SOX4 during peritoneal dialysis. Lab. Invest. 2015;95:817–832. doi: 10.1038/labinvest.2015.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu Y, et al. Down-regulation of miR-129-5p via the Twist1-Snail feedback loop stimulates the epithelial-mesenchymal transition and is associated with poor prognosis in breast cancer. Oncotarget. 2015;6:34423–34436. doi: 10.18632/oncotarget.5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao N, et al. Long non-coding RNA NONHSAT101069 promotes epirubicin resistance, migration, and invasion of breast cancer cells through NONHSAT101069/miR-129-5p/Twist1 axis. Oncogene. 2019;38:7216–7233. doi: 10.1038/s41388-019-0904-5. [DOI] [PubMed] [Google Scholar]
- 60.Zuo Y, Li Y, Zhou Z, Ma M, Fu K. Long non-coding RNA MALAT1 promotes proliferation and invasion via targeting miR-129-5p in triple-negative breast cancer. Biomed. Pharmacother. 2017;95:922–928. doi: 10.1016/j.biopha.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 61.Wu Y, et al. The role of lncRNAs in the distant metastasis of breast cancer. Front. Oncol. 2019;9:407. doi: 10.3389/fonc.2019.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pruszko M, et al. The mutant p53-ID4 complex controls VEGFA isoforms by recruiting lncRNA MALAT1. EMBO Rep. 2017;18:1331–1351. doi: 10.15252/embr.201643370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Herrera-Merchan A, et al. miR-33-mediated downregulation of p53 controls hematopoietic stem cell self-renewal. Cell Cycle. 2010;9:3277–3285. doi: 10.4161/cc.9.16.12598. [DOI] [PubMed] [Google Scholar]
- 64.Wu Q, et al. Long non-coding RNA CASC15 regulates gastric cancer cell proliferation, migration and epithelial mesenchymal transition by targeting CDKN1A and ZEB1. Mol. Oncol. 2018;12:799–813. doi: 10.1002/1878-0261.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang C, et al. MiR-33a suppresses breast cancer cell proliferation and metastasis by targeting ADAM9 and ROS1. Protein Cell. 2015;6:881–889. doi: 10.1007/s13238-015-0223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin Y, et al. MicroRNA-33b inhibits breast cancer metastasis by targeting HMGA2, SALL4 and Twist1. Sci. Rep. 2015;5:9995. doi: 10.1038/srep09995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grespi F, et al. Differential regulated microRNA by wild type and mutant p53 in induced pluripotent stem cells. Cell Death Dis. 2016;7:e2567. doi: 10.1038/cddis.2016.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo F, et al. MiR-101 suppresses the epithelial-to-mesenchymal transition by targeting ZEB1 and ZEB2 in ovarian carcinoma. Oncol. Rep. 2014;31:2021–2028. doi: 10.3892/or.2014.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meng Q, Yang BY, Liu B, Yang JX, Sun Y. Long non-coding RNA SNHG6 promotes glioma tumorigenesis by sponging miR-101-3p. Int. J. Biol. Markers. 2018;33:148–155. doi: 10.1177/1724600817747524. [DOI] [PubMed] [Google Scholar]
- 70.Cheng Y, et al. LncRNA XIST promotes the epithelial to mesenchymal transition of retinoblastoma via sponging miR-101. Eur. J. Pharmacol. 2019;843:210–216. doi: 10.1016/j.ejphar.2018.11.028. [DOI] [PubMed] [Google Scholar]
- 71.Liang H, et al. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol. Cancer. 2018;17:119. doi: 10.1186/s12943-018-0870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Piovan C, et al. Oncosuppressive role of p53-induced miR-205 in triple negative breast cancer. Mol. Oncol. 2012;6:458–472. doi: 10.1016/j.molonc.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee JY, et al. Loss of the polycomb protein Mel-18 enhances the epithelial-mesenchymal transition by ZEB1 and ZEB2 expression through the downregulation of miR-205 in breast cancer. Oncogene. 2014;33:1325–1335. doi: 10.1038/onc.2013.53. [DOI] [PubMed] [Google Scholar]
- 74.Wang X, et al. Upregulation of MiR-205 under hypoxia promotes epithelial-mesenchymal transition by targeting ASPP2. Cell Death Dis. 2016;7:e2517. doi: 10.1038/cddis.2016.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hou P, et al. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014;5:e1287. doi: 10.1038/cddis.2014.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Song S, et al. LncRNA ADPGK-AS1 promotes pancreatic cancer progression through activating ZEB1-mediated epithelial-mesenchymal transition. Cancer Biol. Ther. 2018;19:573–583. doi: 10.1080/15384047.2018.1423912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Piva R, Spandidos DA, Gambari R. From microRNA functions to microRNA therapeutics: novel targets and novel drugs in breast cancer research and treatment (review) Int. J. Oncol. 2013;43:985–994. doi: 10.3892/ijo.2013.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lambertini E, et al. Correlation between Slug transcription factor and miR-221 in MDA-MB-231 breast cancer cells. BMC Cancer. 2012;12:445. doi: 10.1186/1471-2407-12-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fornari F, et al. p53/mdm2 feedback loop sustains miR-221 expression and dictates the response to anticancer treatments in hepatocellular carcinoma. Mol. Cancer Res. 2014;12:203–216. doi: 10.1158/1541-7786.MCR-13-0312-T. [DOI] [PubMed] [Google Scholar]
- 80.Jin X, et al. CASC2/miR-24/miR-221 modulates the TRAIL resistance of hepatocellular carcinoma cell through caspase-8/caspase-3. Cell Death Dis. 2018;9:318. doi: 10.1038/s41419-018-0350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arora H, Qureshi R, Park WY. miR-506 regulates epithelial mesenchymal transition in breast cancer cell lines. PLoS One. 2013;8:e64273. doi: 10.1371/journal.pone.0064273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yin M, et al. Selective killing of lung cancer cells by miRNA-506 molecule through inhibiting NF-κB p65 to evoke reactive oxygen species generation and p53 activation. Oncogene. 2015;34:691–703. doi: 10.1038/onc.2013.597. [DOI] [PubMed] [Google Scholar]
- 83.Wang XX, et al. miR-506 attenuates methylation of lncRNA MEG3 to inhibit migration and invasion of breast cancer cell lines via targeting SP1 and SP3. Cancer Cell Int. 2018;18:171. doi: 10.1186/s12935-018-0642-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sun L, Li Y, Yang B. Downregulated long non-coding RNA MEG3 in breast cancer regulates proliferation, migration and invasion by depending on p53’s transcriptional activity. Biochem. Biophys. Res. Commun. 2016;478:323–329. doi: 10.1016/j.bbrc.2016.05.031. [DOI] [PubMed] [Google Scholar]
- 85.Yong W, et al. Long noncoding RNA NEAT1, regulated by LIN28B, promotes cell proliferation and migration through sponging miR-506 in high-grade serous ovarian cancer. Cell Death Dis. 2018;9:861. doi: 10.1038/s41419-018-0908-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dugimont T, et al. The H19 TATA-less promoter is efficiently repressed by wild-type tumor suppressor gene product p53. Oncogene. 1998;16:2395–2401. doi: 10.1038/sj.onc.1201742. [DOI] [PubMed] [Google Scholar]
- 87.Matouk IJ, et al. Oncofetal H19 RNA promotes tumor metastasis. Biochim. Biophys. Acta. 2014;1843:1414–1426. doi: 10.1016/j.bbamcr.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 88.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 89.Bisio A, et al. Identification of new p53 target microRNAs by bioinformatics and functional analysis. BMC Cancer. 2013;13:552. doi: 10.1186/1471-2407-13-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dong P, et al. Mutant p53 gain-of-function induces epithelial-mesenchymal transition through modulation of the miR-130b-ZEB1 axis. Oncogene. 2013;32:3286–3295. doi: 10.1038/onc.2012.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miao Y, et al. MicroRNA-130b targets PTEN to mediate drug resistance and proliferation of breast cancer cells via the PI3K/Akt signaling pathway. Sci. Rep. 2017;7:41942. doi: 10.1038/srep41942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cao Y, Shi H, Ren F, Jia Y, Zhang R. Long non-coding RNA CCAT1 promotes metastasis and poor prognosis in epithelial ovarian cancer. Exp. Cell Res. 2017;359:185–194. doi: 10.1016/j.yexcr.2017.07.030. [DOI] [PubMed] [Google Scholar]
- 93.Schwickert A, et al. microRNA miR-142-3p inhibits breast cancer cell invasiveness by synchronous targeting of WASL, Integrin Alpha V, and additional cytoskeletal elements. PLoS ONE. 2015;10:e0143993. doi: 10.1371/journal.pone.0143993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Godfrey JD, Morton JP, Wilczynska A, Sansom OJ, Bushell MD. MiR-142-3p is downregulated in aggressive p53 mutant mouse models of pancreatic ductal adenocarcinoma by hypermethylation of its locus. Cell Death Dis. 2018;9:644. doi: 10.1038/s41419-018-0628-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.He C, et al. lncRNA TUG1-mediated Mir-142-3p downregulation contributes to metastasis and the epithelial-to-mesenchymal transition of hepatocellular carcinoma by targeting ZEB1. Cell. Physiol. Biochem. 2018;48:1928–1941. doi: 10.1159/000492517. [DOI] [PubMed] [Google Scholar]
- 96.Li Y, et al. Long non-coding RNA NNT-AS1 affects progression of breast cancer through miR-142-3p/ZEB1 axis. Biomed. Pharmacother. 2018;103:939–946. doi: 10.1016/j.biopha.2018.04.087. [DOI] [PubMed] [Google Scholar]
- 97.Liu B, et al. Tumor-suppressing roles of miR-214 and miR-218 in breast cancer. Oncol. Rep. 2016;35:3178–3184. doi: 10.3892/or.2016.4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xu T, et al. MiR‐142‐3p functions as a tumor suppressor by targeting RAC1/PAK1 pathway in breast cancer. J. Cell. Physiol. 2020;235:4928–4940. doi: 10.1002/jcp.29372. [DOI] [PubMed] [Google Scholar]
- 99.Troschel FM, et al. miR-142-3p attenuates breast cancer stem cell characteristics and decreases radioresistance in vitro. Tumor Biol. 2018;40:1010428318791887. doi: 10.1177/1010428318791887. [DOI] [PubMed] [Google Scholar]
- 100.Song C, et al. High expression of microRNA-183/182/96 cluster as a prognostic biomarker for breast cancer. Sci. Rep. 2016;6:24502. doi: 10.1038/srep24502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lowery AJ, Miller N, Dwyer RM, Kerin MJ. Dysregulated miR-183 inhibits migration in breast cancer cells. BMC Cancer. 2010;10:502. doi: 10.1186/1471-2407-10-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hong Y, et al. miR-96 promotes cell proliferation, migration and invasion by targeting PTPN9 in breast cancer. Sci. Rep. 2016;6:37421. doi: 10.1038/srep37421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Antolín S, et al. Circulating miR-200c and miR-141 and outcomes in patients with breast cancer. BMC Cancer. 2015;15:297. doi: 10.1186/s12885-015-1238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Choi SK, et al. Overexpression of the miR-141/200c cluster promotes the migratory and invasive ability of triple-negative breast cancer cells through the activation of the FAK and PI3K/AKT signaling pathways by secreting VEGF-A. BMC Cancer. 2016;16:570. doi: 10.1186/s12885-016-2620-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Debeb, B. G. et al. miR-141-mediated regulation of brain metastasis from breast cancer. J. Natl. Cancer Inst.108, djw026 (2016). [DOI] [PMC free article] [PubMed]
- 106.Madhavan D, et al. Circulating miRNAs as surrogate markers for circulating tumor cells and prognostic markers in metastatic breast cancer. Clin. Cancer Res. 2012;18:5972–5982. doi: 10.1158/1078-0432.CCR-12-1407. [DOI] [PubMed] [Google Scholar]
- 107.D’Ippolito E, et al. miR-9 and miR-200 regulate PDGFRβ-mediated endothelial differentiation of tumor cells in triple-negative breast cancer. Cancer Res. 2016;76:5562–5572. doi: 10.1158/0008-5472.CAN-16-0140. [DOI] [PubMed] [Google Scholar]
- 108.Castilla, M. A. et al. MicroRNA-200 family modulation in distinct breast cancer phenotypes. PLoS ONE7, e47709 (2012). [DOI] [PMC free article] [PubMed]
- 109.Yang F, Zhang W, Shen Y, Guan X. Identification of dysregulated microRNAs in triple-negative breast cancer. Int. J. Oncol. 2015;46:927–932. doi: 10.3892/ijo.2015.2821. [DOI] [PubMed] [Google Scholar]
- 110.Tuomarila, M. et al. Overexpression of microRNA-200c predicts poor outcome in patients with PR-negative breast cancer. PLoS ONE9, e109508 (2014). [DOI] [PMC free article] [PubMed]
- 111.Li D, et al. The microRNAs miR-200b-3p and miR-429-5p target the LIMK1/CFL1 pathway to inhibit growth and motility of breast cancer cells. Oncotarget. 2017;8:85276. doi: 10.18632/oncotarget.19205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang C, Ju H, Shen C, Tong Z. miR-429 mediates δ-tocotrienol-induced apoptosis in triple-negative breast cancer cells by targeting XIAP. Int. J. Clin. Exp. Med. 2015;8:15648. [PMC free article] [PubMed] [Google Scholar]
- 113.Ye Z-B, et al. miR-429 inhibits migration and invasion of breast cancer cells in vitro. Int. J. Oncol. 2015;46:531–538. doi: 10.3892/ijo.2014.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Weihua Z, Guorong Z, Xiaolong C, Weizhan L. MiR-33a functions as a tumor suppressor in triple-negative breast cancer by targeting EZH2. Cancer Cell Int. 2020;20:1–12. doi: 10.1186/s12935-020-1160-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lv ZD, et al. miR‐655 suppresses epithelial‐to‐mesenchymal transition by targeting Prrx1 in triple‐negative breast cancer. J. Cell. Mol. Med. 2016;20:864–873. doi: 10.1111/jcmm.12770. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 116.Li W, Zhai L, Zhao C, Lv S. miR-153 inhibits epithelial–mesenchymal transition by targeting metadherin in human breast cancer. Breast Cancer Res. Treat. 2015;150:501–509. doi: 10.1007/s10549-015-3346-y. [DOI] [PubMed] [Google Scholar]
- 117.Liu R, et al. Mifepristone suppresses basal triple-negative breast cancer stem cells by down-regulating KLF5 expression. Theranostics. 2016;6:533. doi: 10.7150/thno.14315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tavakolian S, Goudarzi H, Torfi F, Faghihloo E. Evaluation of microRNA-9 and-192 expression levels as biomarkers in patients suffering from breast cancer. Biomed. Rep. 2020;12:30–34. doi: 10.3892/br.2019.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen P, et al. MicroRNA-192 inhibits cell proliferation and induces apoptosis in human breast cancer by targeting caveolin 1. Oncol. Rep. 2019;42:1667–1676. doi: 10.3892/or.2019.7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gao JB, Zhu MN, Zhu XL. miRNA‐215‐5p suppresses the aggressiveness of breast cancer cells by targeting Sox9. FEBS Open Bio. 2019;9:1957–1967. doi: 10.1002/2211-5463.12733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhou S-w, et al. Aberrant miR-215 expression is associated with clinical outcome in breast cancer patients. Med. Oncol. 2014;31:259. doi: 10.1007/s12032-014-0259-2. [DOI] [PubMed] [Google Scholar]
- 122.Ahmadinejad F, et al. Lower expression of miR-218 in human breast cancer is associated with lymph node metastases, higher grades, and poorer prognosis. Tumor Biol. 2017;39:1010428317698362. doi: 10.1177/1010428317698362. [DOI] [PubMed] [Google Scholar]
- 123.Jin Y, et al. MicroRNA-338-3p functions as tumor suppressor in breast cancer by targeting SOX4. Int. J. Oncol. 2015;47:1594–1602. doi: 10.3892/ijo.2015.3114. [DOI] [PubMed] [Google Scholar]
- 124.Wang R, et al. MiR-101 is involved in human breast carcinogenesis by targeting Stathmin1. PLoS ONE. 2012;7:e46173. doi: 10.1371/journal.pone.0046173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wu H, Zhu S, Mo Y-Y. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009;19:439–448. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xiao Y, et al. Integrin α5 down-regulation by miR-205 suppresses triple negative breast cancer stemness and metastasis by inhibiting the Src/Vav2/Rac1 pathway. Cancer Lett. 2018;433:199–209. doi: 10.1016/j.canlet.2018.06.037. [DOI] [PubMed] [Google Scholar]
- 127.Raychaudhuri M, et al. MicroRNAs miR-7 and miR-340 predict response to neoadjuvant chemotherapy in breast cancer. Breast Cancer Res. Treat. 2017;162:511–521. doi: 10.1007/s10549-017-4132-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shi Z, et al. MiR-340 inhibits triple-negative breast cancer progression by reversing EZH2 mediated miRNAs dysregulated expressions. J. Cancer. 2017;8:3037. doi: 10.7150/jca.19315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li Q, et al. Involvement of NF-κ B/miR-448 regulatory feedback loop in chemotherapy-induced epithelial–mesenchymal transition of breast cancer cells. Cell Death Differ. 2011;18:16–25. doi: 10.1038/cdd.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Guo S, et al. MiR-508-3p inhibits cell invasion and epithelial-mesenchymal transition by targeting ZEB1 in triple-negative breast cancer. Eur. Rev. Med. Pharm. Sci. 2018;22:6379–6385. doi: 10.26355/eurrev_201810_16050. [DOI] [PubMed] [Google Scholar]
- 131.Cheng C-W, et al. MicroRNA-30a inhibits cell migration and invasion by downregulating vimentin expression and is a potential prognostic marker in breast cancer. Breast Cancer Res. Treat. 2012;134:1081–1093. doi: 10.1007/s10549-012-2034-4. [DOI] [PubMed] [Google Scholar]
- 132.Kawaguchi T, et al. Overexpression of suppressive microRNAs, miR-30a and miR-200c are associated with improved survival of breast cancer patients. Sci. Rep. 2017;7:1–12. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tanic M, et al. Deregulated miRNAs in hereditary breast cancer revealed a role for miR-30c in regulating KRAS oncogene. PLoS ONE. 2012;7:e38847. doi: 10.1371/journal.pone.0038847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dobson JR, et al. hsa-mir-30c promotes the invasive phenotype of metastatic breast cancer cells by targeting NOV/CCN3. Cancer Cell Int. 2014;14:73. doi: 10.1186/s12935-014-0073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Peurala H, et al. MiR-34a expression has an effect for lower risk of metastasis and associates with expression patterns predicting clinical outcome in breast cancer. PLoS ONE. 2011;6:e26122. doi: 10.1371/journal.pone.0026122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zeng Z, Chen X, Zhu D, Luo Z, Yang M. Low expression of circulating microRNA-34c is associated with poor prognosis in triple-negative breast cancer. Yonsei Med. J. 2017;58:697–702. doi: 10.3349/ymj.2017.58.4.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sandhu R, Rivenbark AG, Mackler RM, Livasy CA, Coleman WB. Dysregulation of microRNA expression drives aberrant DNA hypermethylation in basal-like breast cancer. Int. J. Oncol. 2014;44:563–572. doi: 10.3892/ijo.2013.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li L, et al. Microrna-124 targets flotillin-1 to regulate proliferation and migration in breast cancer. Mol. Cancer. 2013;12:163. doi: 10.1186/1476-4598-12-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang T-FM, Ji H, Lv M-M, Zhao J-H, Tang J-H. Down-regulation of miRNA-452 is associated with adriamycin-resistance in breast cancer cells. Asian Pac. J. Cancer Prev. 2014;15:5137–5142. doi: 10.7314/APJCP.2014.15.13.5137. [DOI] [PubMed] [Google Scholar]
- 140.Kim M, et al. VEGFA links self-renewal and metastasis by inducing Sox2 to repress miR-452, driving Slug. Oncogene. 2017;36:5199–5211. doi: 10.1038/onc.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang S, Li H, Wang J, Wang D. Retracted article: expression of microRNA-497 and its prognostic significance in human breast cancer. Diagn. Pathol. 2013;8:172. doi: 10.1186/1746-1596-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 142.Sun G, Liu Y, Wang K, Xu Z. miR-506 regulates breast cancer cell metastasis by targeting IQGAP1. Int. J. Oncol. 2015;47:1963–1970. doi: 10.3892/ijo.2015.3161. [DOI] [PubMed] [Google Scholar]
- 143.Liu H, et al. Expression and regulatory function of miRNA-182 in triple-negative breast cancer cells through its targeting of profilin 1. Tumor Biol. 2013;34:1713–1722. doi: 10.1007/s13277-013-0708-0. [DOI] [PubMed] [Google Scholar]
- 144.Wang PY, et al. Higher expression of circulating miR-182 as a novel biomarker for breast cancer. Oncol. Lett. 2013;6:1681–1686. doi: 10.3892/ol.2013.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lu X, et al. MiR-129-5p sensitizes the response of Her-2 positive breast cancer to trastuzumab by reducing Rps6. Cell. Physiol. Biochem. 2017;44:2346–2356. doi: 10.1159/000486122. [DOI] [PubMed] [Google Scholar]
- 146.Meng R, et al. miR-129-5p suppresses breast cancer proliferation by targeting CBX4. Neoplasma. 2018;65:572–578. doi: 10.4149/neo_2018_170814N530. [DOI] [PubMed] [Google Scholar]
- 147.Luo J, Chen J, He L. mir-129-5p attenuates irradiation-induced autophagy and decreases radioresistance of breast cancer cells by targeting HMGB1. Med. Sci. Monit. 2015;21:4122. doi: 10.12659/MSM.896661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Majumder M, Landman E, Liu L, Hess D, Lala PK. COX-2 elevates oncogenic miR-526b in breast cancer by EP4 activation. Mol. Cancer Res. 2015;13:1022–1033. doi: 10.1158/1541-7786.MCR-14-0543. [DOI] [PubMed] [Google Scholar]