Main Text
The development of immunotherapeutic strategies against cancer relies on the identification and validation of optimal target antigens, which should be specific to the tumor and be able to elicit a swift and potent anti-tumor immune response. Strategies based on tumor-associated antigens (TAAs) or tumor-specific antigens (TSAs) have a series of limitations that hamper the efficacy of the anti-tumor response or its implementation on a large scale.1
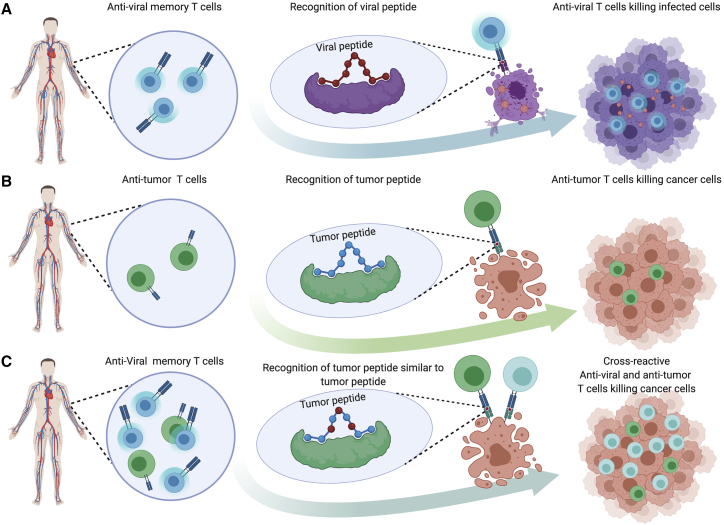
Surprisingly, a new class of effective tumor antigen (TuA) may be coopted from microorganisms that enter the body during our lifetime, referred to as microorganism antigens (MoAs). Indeed, scattered data suggest that MoAs may converge with TuAs to elicit cross-reacting CD8+ T cell responses that possibly drive the fate of cancer development, progression, and response to therapy. In fact, if a presented TuA shares sufficient molecular similarity with a MoA, we might suppose that cross-reactive T cells would be engaged to fight the cancer (Figure 1).
Figure 1.
Schematic Representing Antiviral and Antitumor Crossreactive T Cells Engaged against Cancer
(A) Antiviral memory T cell recognizing a viral peptide presented on MHC class I and killing infected tissue. (B) Antitumor T cell recognizing a tumor-specific peptide presented on MHC class I and killing tumor cells. (C) Crossreactive antiviral memory T cells and antitumor T cell recognizing the same tumor-specific peptide and killing both tumor cells.
In some cases, cancer patients expressing TuAs exhibiting a high degree of similarity to those from pathogens have been reported to experience a better clinical outcome. For example, long-term survival of pancreatic cancer patients has been associated with greater similarity of neoantigens to MoAs, as opposed to their net amount.2 Similar observations have been made in non-small cell lung cancer (NSCLC)3 and melanoma4 patients.
We have previously shown in a cohort of hepatocellular cancer (HCC) patients that predicted mutated neoantigens with high affinity to histocompatibility leukocyte antigen (HLA) may show >50% sequence similarity to MoAs and the central T cell receptor (TCR)-facing residues can even be identical. For such “paired” epitopes, bioinformatics modeling shows a very similar conformation when bound to the HLA molecule and an identical spatial conformation of the residues protruding toward the TCR. Paired TuAs and MoAs were shown to elicit crossreacting T cells in immunized mice and peripheral blood mononuclear cells (PBMCs) from a long-term surviving HCC patient crossreacted against both the tumor-specific mutated neoantigens identified in the tumor lesion and the paired pathogen epitopes.5
Another study in a preclinical model has recently reported the presence of antiviral T cells within the tumor that influence the anti-tumor immune response and immunotherapy outcome.6 Furthermore, in a controlled murine model experiment, we have shown that mice pre-vaccinated with viral epitopes with a high degree of similarity to tumor epitopes respond more robustly to treatment compared to naive mice.7
Very recently, antigens derived from the extracellular commensal intestinal microbiota, which are known to be presented to the immune system via the major histocompatibility complex (MHC) class II pathway, have also been shown to be crosspresented through the intracellular MHC class I pathway. Specific CD8+ T cells crossreact with neoantigens and are able to control tumor growth in an animal model.8
Taken together, these scattered data reported by different research groups show that MoAs may share high sequence similarity with TuAs and suggest that memory immunity primed against such MoAs may crossreact with TuAs and, therefore, represent a “natural” anti-cancer vaccination. Indeed, cancer cells arising in any tumor type, even without an etiopathogenic correlation to pathogens, may, in a stochastic manner, express TuAs similar or identical to microorganisms. If such a condition occurs, cancer cells might exhibit a greater chance of being promptly targeted and cleared by the immune system, provided that a memory CD8+ T cell specific for the pathogen’s epitope had already been established in the host.
As for memory immunity induced by prophylactic vaccines targeting an incoming infection, it is desirable that the host’s immune system be primed against TuAs and that specific memory CD8+ T cell responses are already established. This would allow the immune system to promptly recognize, target, and eliminate cancer cells at very early stages (in situ, Tis or stage 0, T0), preventing the tumor from progressing in the context of an immune-suppressive tumor microenvironment.
Such crossreactivity against similar antigens is possible owing to the biological principle that a single TCR may recognize at least 106 different MHC-bound peptides,9 allowing recognition and protection from virtually all possible pathogens and malignancies.10,11 Such TCR degeneracy gives rise to a repertoire of highly crossreactive clonal CD8+ T cells responding to a given non-self pMHC complex,10,12 reducing the possibility of expansion of mutant escape variants.13,14 Indeed, an epitope binds the HLA molecule with specific residues in fixed positions along the sequence (anchor residues), and only the central residues are exposed for recognition by the TCR.15 Therefore, two unrelated antigens sharing the same TCR-facing central residues, or showing conservative variations at those positions, are very likely recognized by the same TCRs, even if the peripheral residues are different, without affecting the structural conformation of the entire epitope. TCR recognition alone is insufficient to trigger a T cell-mediated immune response because many other intracellular and extracellular signals must occur for effective licensing of cytotoxic T cells. This at least partly explains why many tumors presenting TuAs that are highly similar to MoAs can grow without significant immune editing until a proper immune response is triggered against that specific antigen.
In summary, a growing body of evidence suggests that the broadness of the anti-MoA memory CD8+ T cell repertoire—built upon the individual experience of interaction with intracellular pathogens and intestinal microbiota—may strongly influence the fate of tumor progression and prognosis and might represent an unexplored opportunity to develop next generation anti-cancer therapeutic vaccines. Indeed, a broader number of TCRs elicited by MoAs during one’s lifetime is correlated with a greater probability of eliminating cancer cells at very early stages, thereby preventing tumor progression. Therefore, the identification of TuAs exhibiting a high degree of homology with MoAs using high throughput proteogenomics strategies represents a new frontier in the quest for the optimal public antigens for developing effective preventive as well as therapeutic cancer immunotherapies. Likewise, such MoAs would be of highest relevance for screening pre-cancerous patients to identify progressors versus non-progressors. Furthermore, the presence of such a pool of memory T cells specific to MoAs might improve the efficacy of checkpoint inhibitor therapy, unleashing the expansion of effector memory cells targeting TuAs already present in the host. Coopting the anti-microorganism immune response to control tumor growth may represent a turning point in cancer immunotherapy and offer more effective therapies for cancer patients.
Contributor Information
Luigi Buonaguro, Email: l.buonaguro@istitutotumori.na.it.
Vincenzo Cerullo, Email: vincenzo.cerullo@helsinki.fi.
References
- 1.Buonaguro L., Tagliamonte M. Selecting Target Antigens for Cancer Vaccine Development. Vaccines (Basel) 2020;8:615. doi: 10.3390/vaccines8040615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balachandran V.P., Łuksza M., Zhao J.N., Makarov V., Moral J.A., Remark R., Herbst B., Askan G., Bhanot U., Senbabaoglu Y., Australian Pancreatic Cancer Genome Initiative. Garvan Institute of Medical Research. Prince of Wales Hospital. Royal North Shore Hospital. University of Glasgow. St Vincent’s Hospital. QIMR Berghofer Medical Research Institute. University of Melbourne, Centre for Cancer Research. University of Queensland, Institute for Molecular Bioscience. Bankstown Hospital. Liverpool Hospital. Royal Prince Alfred Hospital, Chris O’Brien Lifehouse. Westmead Hospital. Fremantle Hospital. St John of God Healthcare. Royal Adelaide Hospital. Flinders Medical Centre. Envoi Pathology. Princess Alexandria Hospital. Austin Hospital. Johns Hopkins Medical Institutes. ARC-Net Centre for Applied Research on Cancer Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551:512–516. doi: 10.1038/nature24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snyder A., Makarov V., Merghoub T., Yuan J., Zaretsky J.M., Desrichard A., Walsh L.A., Postow M.A., Wong P., Ho T.S. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rizvi N.A., Hellmann M.D., Snyder A., Kvistborg P., Makarov V., Havel J.J., Lee W., Yuan J., Wong P., Ho T.S. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrizzo A., Tagliamonte M., Mauriello A., Costa V., Aprile M., Esposito R., Caporale A., Luciano A., Arra C., Tornesello M.L. Unique true predicted neoantigens (TPNAs) correlates with anti-tumor immune control in HCC patients. J. Transl. Med. 2018;16:286. doi: 10.1186/s12967-018-1662-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosato P.C., Wijeyesinghe S., Stolley J.M., Nelson C.E., Davis R.L., Manlove L.S., Pennell C.A., Blazar B.R., Chen C.C., Geller M.A. Virus-specific memory T cells populate tumors and can be repurposed for tumor immunotherapy. Nat. Commun. 2019;10:567. doi: 10.1038/s41467-019-08534-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiaro J., Kasanen H., Whalley T., Capasso C., Gronholm M., Feola S., Peltonen K., Hamdan F.H., Hernberg M., Makela S. Viral Molecular Mimicry Influences the Antitumor Immune Response in Murine and Human Melanoma. medRxiv. 2020 doi: 10.1101/2020.09.09.20191171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bessell C.A., Isser A., Havel J.J., Lee S., Bell D.R., Hickey J.W., Chaisawangwong W., Glick Bieler J., Srivastava R., Kuo F. Commensal bacteria stimulate antitumor responses via T cell cross-reactivity. JCI Insight. 2020;5:e135597. doi: 10.1172/jci.insight.135597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wooldridge L., Ekeruche-Makinde J., van den Berg H.A., Skowera A., Miles J.J., Tan M.P., Dolton G., Clement M., Llewellyn-Lacey S., Price D.A. A single autoimmune T cell receptor recognizes more than a million different peptides. J. Biol. Chem. 2012;287:1168–1177. doi: 10.1074/jbc.M111.289488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sewell A.K. Why must T cells be cross-reactive? Nat. Rev. Immunol. 2012;12:669–677. doi: 10.1038/nri3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antunes D.A., Rigo M.M., Freitas M.V., Mendes M.F.A., Sinigaglia M., Lizée G., Kavraki L.E., Selin L.K., Cornberg M., Vieira G.F. Interpreting T-Cell Cross-reactivity through Structure: Implications for TCR-Based Cancer Immunotherapy. Front. Immunol. 2017;8:1210. doi: 10.3389/fimmu.2017.01210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mason D. A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol. Today. 1998;19:395–404. doi: 10.1016/s0167-5699(98)01299-7. [DOI] [PubMed] [Google Scholar]
- 13.Chan K.F., Gully B.S., Gras S., Beringer D.X., Kjer-Nielsen L., Cebon J., McCluskey J., Chen W., Rossjohn J. Divergent T-cell receptor recognition modes of a HLA-I restricted extended tumour-associated peptide. Nat. Commun. 2018;9:1026. doi: 10.1038/s41467-018-03321-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song I., Gil A., Mishra R., Ghersi D., Selin L.K., Stern L.J. Broad TCR repertoire and diverse structural solutions for recognition of an immunodominant CD8+ T cell epitope. Nat. Struct. Mol. Biol. 2017;24:395–406. doi: 10.1038/nsmb.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L., Udaka K., Mamitsuka H., Zhu S. Toward more accurate pan-specific MHC-peptide binding prediction: a review of current methods and tools. Brief. Bioinform. 2012;13:350–364. doi: 10.1093/bib/bbr060. [DOI] [PubMed] [Google Scholar]