Abstract
The therapeutic effect of retinal gene therapy using CRISPR/Cas9-mediated genome editing and knockout applications is dependent on efficient and safe delivery of gene-modifying tool kits. Recently, transient administration of single guide RNAs (sgRNAs) and SpCas9 proteins delivered as ribonucleoproteins (RNPs) has provided potent gene knockout in vitro. To improve efficacy of CRISPR-based gene therapy, we delivered RNPs containing SpCas9 protein complexed to chemically modified sgRNAs (msgRNAs). In K562 cells, msgRNAs significantly increased the insertion/deletion (indel) frequency (25%) compared with unmodified counterparts leading to robust knockout of the VEGFA gene encoding vascular endothelial growth factor A (96% indels). Likewise, in HEK293 cells, lipoplexes containing varying amounts of RNP and EGFP mRNA showed efficient VEGFA knockout (43% indels) and strong EGFP expression, indicative of efficacious functional knockout using small amounts of RNP. In mice, subretinal injections of equivalent lipoplexes yielded 6% indels in Vegfa of isolated EGFP-positive RPE cells. However, signs of toxicity following delivery of lipoplexes containing high amounts of RNP were observed. Although the mechanism resulting in the varying efficacy remains to be elucidated, our data suggest that a single subretinal injection of RNPs carrying msgRNAs and SpCas9 induces targeted retinal indel formation, thus providing a clinically relevant strategy relying on nonviral delivery of short-lived nuclease activity.
Keywords: vascular endothelial growth factor A, gene knockout, ribonucleoproteins, chemically modified single guide RNAs, Cas9, nonviral gene therapy, short-lived nuclease activity, retina pigment epithelium, age-related macular degeneration
Graphical Abstract

The therapeutic effect of retinal gene therapy using CRISPR-Cas9-mediated knockout applications is dependent on efficient and safe delivery of gene-modifying tools. For the first time, Holmgaard et al. demonstrate targeted knockout of the Vegfa gene in the retina using RNPs containing SpCas9 protein complexed with chemically modified sgRNAs, suggesting that delivery of gene-disrupting agents relying on short-lived nuclease activity is a clinically relevant strategy.
Introduction
Gene therapies based on CRISPR/Cas9-mediated genome editing and gene knockout are at advanced stages of (pre)-clinical development to treat a broad range of monogenic diseases, such as sickle cell disease1 and Duchenne muscular dystrophy.2 The successful application of genome editing for treatment is strongly dependent on delivery and safety of the technology.3 Following adaptation of the CRISPR system from the adaptive immune system in prokaryotes, CRISPR/Cas9 genome-editing technologies have been optimized to induce gene knockout with unprecedented efficacy and specificity.4, 5, 6, 7 Streptococcus pyogenes (Sp) Cas9 demonstrates high efficacy and target flexibility because of a protospacer adjacent motif (PAM), which is common in genomes.8 To increase targeting options, alterations in SpCas9 have been introduced to relax PAM specificity, and novel CRISPR effectors, such as CjCas9, have been discovered.9, 10, 11, 12
Genome surgery of retinal targets has been shown to result in the precise introduction of double-stranded breaks, even with allele-specific targeting,13,14 as illustrated by the current clinical trial utilizing CRISPR/Cas9 to treat Leber congenital amaurosis type 10 (LCA10) caused by mutations in the CEP290 gene (ClinicalTrials.gov: NCT03872479).15 Vectors based on adeno-associated virus (AAV) packaged in serotypes 1, 2, 5, and 9 have been used to deliver CRISPR components targeting genes (primarily Vegfa and Rho) in retinal pigment epithelium (RPE)16, 17, 18 and photoreceptor (PR) cells14,19,20 using either SpCas9 or smaller nucleases in all-in-one AAVs.16,18 Several of these studies relied on fluorescence-activated cell sorting (FACS)-based isolation of edited cells, and in a recent paper, we demonstrated that lentiviral vector (LV) delivery of SpCas9 and single guide RNA (sgRNA) expression cassettes induced insertions/deletions (indels) in transduced EGFP-positive RPE cells.21 Knockdown in the retina has also been achieved by electroporation of subretinally injected plasmids encoding SpCas9 and sgRNAs targeting RHO in transgenic mice.22 However, as genome editing approaches clinical application,23 safety issues have been raised regarding increased off-target activity with prolonged SpCas9 expression.24
Consequently, this has spawned a massive interest in exploiting nonviral CRISPR-based therapeutics that can induce knockout. Direct delivery of the Cas9 RNP is being carefully studied as a therapeutic strategy for generating knockout and has great potential for clinical translation, mainly because of proven procedures for producing recombinant proteins and well-documented clinical applications of therapeutics based on proteins. Hence various methodologies have been developed for delivering Cas9 ribonucleoproteins (RNPs) in vitro and in vivo.25,26 Lipofectamine has been successful in delivering Cas9 RNP to the ear27 and into the eye.17 In the latter study, Kim et al.17 demonstrated retinal targeting of Vegfa and Rosa26 using RNPs delivered by Lipofectamine 2000-based lipoplexes. Extensive amounts of RNPs (8 μg Cas9 and 4 μg sgRNA) were delivered by a single subretinal injection. Up to 25% indel formation was obtained in RPE cells isolated from the injected area, while reduced choroidal neovascularization (CNV) in a mouse model was observed as a result of Vegfa knockout. Analysis of predicted off-target sites revealed minimal off-target activity, thus emphasizing the potential use of RNPs for clinical application.
To increase on-target efficacy, 2′O-methyl-3′phosphorothiate modifications on the terminal three nucleotides of sgRNAs have demonstrated increased sgRNA stability and flexibility in vitro.28 As a further improvement of previous studies relying on electroporation of plasmid-based CRISPR/Cas922 or transfection of RNPs carrying unmodified sgRNAs,17 transient delivery of chemically modified sgRNAs (msgRNAs) and Cas9 combined in RNPs could thus provide a clinically attractive option. In this study, our findings demonstrate, to our knowledge for the first time, in vivo gene knockout in the murine retina facilitated by msgRNAs and SpCas9 targeted to the Vegfa gene in the RPE cells. Moreover, the present study includes two additional key evaluations that have not been previously investigated: assessment of the efficiency of chemically msgRNAs compared with unmodified sgRNAs targeting Vegfa and evaluation of the efficacy of different transfection reagents in delivering Vegfa-targeting RNPs both in vitro and in vivo. Due to the hit-and-run nature of RNPs, the present study further supports the notion that nonviral gene knockout using delivery of target gene-disrupting RNPs containing Cas9 protein and sgRNAs can become clinically relevant for applications relying on safe gene knockout in the retina.
Results
Verification of sgRNA Efficacy in Electroporated K562 Cells
To investigate RNP-based targeting of Vegfa in both murine and human cells, we surveyed Vegfa genes from both species for conserved Cas9 target sites. Three sgRNAs (sgRNA1–3) spanning a homologous 32-bp region in exon 3 were designed using the Broad Institute’s online CRISPRko tool (Figure 1A). Improved sgRNAs with chemical modifications of the terminal three nucleotides (msgRNAs) were compared with unmodified sgRNAs. To validate electroporation parameters, we delivered 2 μg EGFP mRNA to 2 × 105 K562 cells (a human erythroleukemic cell line). One day after electroporation, fluorescence microscopy showed robust EGFP expression, suggesting efficient electroporation-based delivery of mRNA (Figure 1B). Next, the capacity of msgRNA versus unmodified sgRNA to support Cas9-directed indel formation was evaluated in K562 cells. A total of 2 × 105 K562 cells were electroporated with 3 μg Cas9 protein complexed with 1.5 μg of either a msgRNA (msgRNA1–3) or an unmodified sgRNA (sgRNA1–3). Indel quantification by tracking of indels by decomposition (TIDE) analysis performed 2 days after electroporation demonstrated high frequencies of indel formation using msgRNA1–3 (Figures 1C and 1D). With 96% ± 1% indel formation, msgRNA1 significantly outperformed both msgRNA2 (78% ± 10%) and msgRNA3 (84% ± 2%). In parallel, sgRNA1 displayed a higher indel frequency compared with both sgRNA2 and sgRNA3. Approximately 20% indel formation was observed in the K562 cells electroporated with sgRNA2 and sgRNA3, whereas up to 76% ± 8% indel formation was obtained using sgRNA1. The efficacies of both modified and unmodified sgRNA1–3 were in accordance with the Broad Institute’s CRISPRko prediction scores. sgRNA1 was ranked the highest among 24 predicted sgRNAs for human VEGFA exon 3. Altogether, msgRNAs significantly increased indel frequencies in electroporated K562 cells compared with the unmodified counterparts (Figure 1C). For this reason, only msgRNAs were used in the following experiments.
Figure 1.
sgRNA Design and Effect of sgRNA Modification
(A) Part of the human and murine vascular endothelial growth factor A (Vegfa) exon 3 sequence. Homologous sequence used for design of single guide RNAs (sgRNAs) targeting both human and murine Vegfa shown in bold. Mismatches are indicated in red font. Three sgRNAs targeting the Vegfa gene were designed and obtained with and without chemical modifications of the first and last three nucleotides of the sgRNA. sgRNAs were named according to CRISPRko on-target prediction scores, with sgRNA1 ranked highest. sgRNA sequences were indicated with underlined protospacer adjacent motif (PAM) sequences. (B) Fluorescence analysis of EGFP expression 1 day post-electroporation of K562 cells. Electroporation of approximately 2 × 105 K562 cells with 2 μg EGFP mRNA was used to evaluate electroporation efficacy. As judged from the EGFP and phase contrast images, almost all of the cells expressed EGFP. Representative images were shown (n = 3). Scale bar, 200 μm. (C) Tracking of indels by decomposition (TIDE) analysis of indels following electroporation of modified and unmodified synthetic sgRNAs. A total of 3 μg Cas9 protein and 1.5 μg sgRNA were used for electroporation of approximately 2 × 105 K562 cells. Two days post-electroporation, genomic DNA (gDNA) was isolated and used as the template for PCR amplification. PCR products were isolated following gel electrophoresis and sequenced. Sequencing results were analyzed for indels by TIDE. sgRNA-Irr (irrelevant) was used as negative control (CTRL) for TIDE analysis. Indel formation frequencies are shown as mean ± SD (n = 3). ∗Statistically significant. (D) Representative sequencing chromatogram for each modified sgRNA (msgRNA) and sgRNA. Cut site 3 bp upstream to the PAM was indicated by the red dotted line. PC, phase contrast.
Transfection of Vegfa-Targeting RNPs in HEK293 Cells
We next investigated the efficacy of lipoplex-based delivery of RNPs to human embryonic kidney (HEK293) cells using Lipofectamine 3000 (LF3000). Previously, the delivery of RNPs has been demonstrated using LF2000.17 LF3000 is an enhanced version of LF2000, exhibiting increased delivery efficacy and reduced toxicity.29 As recommended by the supplier of LF3000, Cas9 and msgRNA amounts relative to cell numbers were decreased compared with the experiments involving electroporation of K562 cells, and a total volume of 50 μL was used for the formation of lipoplexes. Hence 1 × 105 HEK293 cells were transfected with LF3000 lipoplexes containing 1.5 μg Cas9 RNP (the number indicates the amount of Cas9 protein used together with the sgRNA in a 2:1 mass ration (1:2.5 molar ratio) in the RNPs; see Materials and Methods) guided by msgRNA1, msgRNA2, or msgRNA3. Two days post-transfection, cells were harvested, and VEGFA PCR amplicons generated from extracted genomic DNA (gDNA) were subjected to TIDE analysis. In accordance with the results obtained in the electroporated K562 cells, msgRNA1 demonstrated significantly higher efficacy in transfected HEK293 cells with 43% ± 9% indel frequency compared with msgRNA2 (13% ± 7%) and msgRNA3 (15% ± 2%) (Figure 2A). Consequently, msgRNA1 was used in the subsequent experiments.
Figure 2.
Comparison of msgRNAs and Validation of EGFP mRNA Transfection
(A) Comparison of indel frequencies following Cas9 ribonucleoprotein (RNP) transfection into human embryonic kidney (HEK)293 cells using msgRNAs. Approximately 1 × 105 HEK293 cells were seeded and transfected the next day with RNP complexes consisting of 1,500 ng Cas9 protein and 750 ng msgRNA1–3, using Lipofectamine (LF)3000 in a total volume of 50 μL. Two days post-transfection, indel formation efficacy of msgRNA1–3 was evaluated. Indel formation frequencies are shown as mean ± SD (n = 3). Statistical tests in (A) are Student’s t test: ∗p < 0.05. (B) Gating used for flow cytometry analysis of EGFP+ cells. Approximately 2 × 104 HEK293 cells were seeded and transfected the next day with increasing amounts of EGFP mRNA from 25 to 500 ng in a total volume of 50 μL using LF3000. The four upper plots represent non-transfected cells (negative) cells. The two lower plots represent cells treated with 25 or 500 ng EGFP mRNA. (C and D) Gating of all the samples analyzed in (C) and (D) is shown in Figure S1. Foreward scatter (FSC), side scatter (SSC), and applied wavelength and detector are indicated on the plots. (C and D) Quantification of EGFP signals in HEK293 cells transfected with 25–500 ng EGFP mRNA in a total volume of 50 μL using flow cytometry as described in (B). EGFP mRNA was delivered by either LF2000 (blue) or LF3000 (red). Mean values ± SD of EGFP+ cells are shown in (C). EGFP intensity median values ± SD are shown in (D). Statistical tests in (C) and (D) are Student’s t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (LF2000 versus LF3000). (E) Analysis of EGFP expression 1 day post-transfection by fluorescence microscopy. Approximately 1 × 105 HEK293 cells were seeded and transfected the next day with 25 or 500 ng EGFP mRNA in a total volume of 50 μL using LF3000. Representative images are shown (n = 3). Scale bar: 20 μm. PI, propidium iodide.
Tracking Delivery of RNPs by Adding EGFP mRNA to Lipoplexes
The robust in vitro indel formation with LF3000 lipoplex-based RNP delivery encouraged us to investigate whether this strategy could also be used in vivo. To evaluate transfection efficacy, we used lipoplexes containing EGFP mRNA. Importantly, addition of EGFP mRNA also allowed in vivo identification of transfected cells, which could be exploited for FACS.21 To determine the required amount of EGFP mRNA, we prepared lipoplexes containing 25, 50, 100, 250, and 500 ng EGFP mRNA with either LF2000 or LF3000 prior to HEK293 transfection. Analysis by flow cytometry 1 day post-transfection revealed elevated EGFP expression as a result of increased EGFP mRNA amounts formulated in the lipoplexes (Figures 2B–D). Lipoplexes carrying 25 ng EGFP mRNA resulted in approximately 60% EGFP+ cells, and the number of cells expressing EGFP gradually increased with increasing amounts of EGFP mRNA to almost 100% when applying 500 ng EGFP mRNA (Figure 2C). Similarly, the EGFP intensity gradually increased with increasing amounts of EGFP mRNA (Figure 2D; Figure S1), consistent with the fluorescence microscopy analysis (Figure 2E). Delivery by LF2000 and LF3000 both resulted in increasing EGFP expression with increasing amounts of EGFP mRNA. In the case of LF3000, a small, but statistically significant increase relative to LF2000 in the number of EGFP+ cells was observed for 100, 250, and 500 ng EGFP mRNA (Figure 2C). LF2000 conversely produced a small, statistically significantly increase in the EGFP intensity for all amounts applied compared with LF3000 (Figure 2D). Because high EGFP protein levels can result in cell toxicity,30 and the number of EGFP+ cells almost reached a maximum at 100 ng EGFP mRNA (approximately 93% EGFP+ cells with LF3000), this dose was chosen for the following experiments.
In Vitro Indel Formation: Downscaling the Transfection Volume for Subretinal Delivery
Subretinal injection in mice restricts the injection volume to 2 μL. Because standard protocols for transfection experiments use a minimum of 50 μL for complex formation, downscaling the volume from 50 to 2 μL was necessary to allow subretinal injection of lipoplexes containing EGFP mRNA and RNP. The volume used for lipoplex formation is essential for subsequent transfection efficacy.31 Therefore, the Cas9 RNP amount was titrated to optimize efficacy. Without altering the amount of EGFP mRNA (fixed to 100 ng) or concentration of LF3000 (20% v/v) in a total volume of 2 μL, 2 × 104 HEK293 cells were transfected with lipoplexes containing either 250, 500, 1,000, 2,000, 3,000, or 4,000 ng Cas9 RNP. To compare LF3000 with LF2000, we prepared similar LF2000-based lipoplexes in parallel. Fluorescence microscopy revealed decreased EGFP expression as a result of increased amounts of Cas9 RNPs (Figure 3A). This phenomenon was observed for lipoplex formulations based on both LF2000 and LF3000 (Figure 3A). The most prominent expression of EGFP was observed using the lowest amount of Cas9 RNP (250 ng). Expression of EGFP was severely affected by 3,000 ng Cas9 RNP, and only traces of EGFP expression were observed following transfection with 4,000 ng Cas9 RNP. Cells were harvested 2 days post-transfection, and in most cases, TIDE analyses revealed increased indel frequencies as a result of increased amounts of Cas9 RNPs (Figure 3B). For LF3000-based lipoplexes, the titration of Cas9 RNP resulted in an increase in indel formation from 12.5% ± 4.3% for 250 ng Cas9 RNP to 36.9% ± 6.8% with 3,000 ng Cas9 RNP. Following transfection with 4,000 ng Cas9 RNP, less indel formation was observed (21.8% ± 7.4%). In comparison, lipoplexes based on LF2000 demonstrated a similar pattern. No statistically significant difference in indel frequency using the two transfection reagents was observed, except for the 3,000-ng samples. Here, significantly lower indel formation (6.7% ± 5.4%) was obtained using LF2000 compared with LF3000. This discrepancy most likely reflects a technical issue related to the samples prepared with 3,000 ng Cas9 RNP, because the 4,000-ng samples revealed higher efficacy following delivery by both LF2000 and LF3000. To quantify the EGFP expression, we performed flow cytometry (Figure 4A; Figure S2). Clearly detectable EGFP expression was observed even at 250 ng Cas9 RNP, whereas the EGFP expression level was severely affected by 4,000 ng Cas9 RNP (Figures 4B–4D). The decrease in EGFP expression was independent of the sgRNA used in the RNP complexes, and complexes with msgRNA1 and msgRNA-Irr (irrelevant control sgRNA without target sites in the human genome) resulted in comparable expression levels (Figure 4B). In one case (LF2000, msgRNA1, 2,000 ng Cas RNP), we detected an unexpected increase in EGFP expression. Meanwhile, a decline of the EGFP expression intensity was observed when the amount of RNP was increased (Figure 4C), consistent with the fluorescence microscopy analysis (Figure 3A). On the contrary, the indel frequency increased with increasing amount of RNP. Taken together, LF3000-based lipoplexes demonstrated reliable indel frequencies with peak efficacy using 3,000 ng Cas9 RNP, and similar efficacy was obtained using LF2000-based lipoplexes, although less consistently (Figure 3B). No signs of sgRNA-dependent toxicity were observed in the transfected cells. In both cases, the EGFP expression was negatively affected by increasing amounts of Cas9 RNPs (Figure 4D), in a manner that did not depend on the sgRNA target site (Figures 4B and 4C).
Figure 3.
Evaluation of EGFP Expression and Indel Formation in 2 μL Transfection Volume
Approximately 2 × 104 HEK293 cells were seeded and transfected the next day with 100 ng EGFP mRNA and increasing amounts of RNP. Cas9 and sgRNAs were complexed in a 2:1 mass ratio, with Cas9 RNP amounts of 250, 500, 1,000, 2,000, 3,000, and 4,000 ng. LF2000 and LF3000 were used to deliver EGFP mRNA and RNPs containing msgRNA1 or msgRNA-Irr. (A) Assessment of the EGFP expression 1 day post-transfection by fluorescence microscopy revealed decreasing EGFP expression as a result of increasing amounts of RNPs containing msgRNA1. Representative images are shown (n = 3). Scale bar, 200 μm. (B) Two days post-transfection, gDNA was isolated from the cells shown in (A), and PCR amplification provided samples for sequencing. TIDE analysis was performed to analyze indel frequency to compare indel formation following LF2000 (blue columns) or LF3000 (red columns) transfection. Indel formation frequencies are shown as mean ± SD (n = 3). Statistical tests in (B) are Student’s t test: ∗p < 0.05. n.s., not significant.
Figure 4.
Quantification of EGFP Expression in 2 μL Transfection Volume
Quantification of the EGFP expression in HEK293 cells transfected with 100 ng EGFP mRNA and increasing amounts of RNP was performed by flow cytometry 1 day after transfection. Cas9 and sgRNAs were complexed in a 2:1 mass ratio, with Cas9 RNP amounts of 250, 500, 1,000, 2,000, 3,000, and 4,000 ng. LF2000 and LF3000 were used to deliver EGFP mRNA and RNPs containing msgRNA1 or msgRNA-Irr to HEK293 cells. Approximately 2 × 104 HEK293 cells were seeded 1 day before transfection. (A) Settings used to gate the EGFP+ cells. The four upper plots represent non-transfected (negative) cells. The four lower plots represent cells treated with 250 or 4,000 ng Cas9 containing chemically modified sgRNA (msgRNA 1 or msgRNA-Irr). (B and C) Gating of all the samples analyzed in (B) and (C) are shown in Figure S2. (B) Quantification of EGFP+ cells by flow cytometry using the gating shown in (A). The number of EGFP+ cells treated with RNPs containing msgRNA1 or msgRNA-Irr following LF2000-based delivery are shown in dark blue and light blue, respectively (mean values ± SD). The number of EGFP+ cells treated with RNPs containing msgRNA1 or msgRNA-Irr following LF3000-based delivery are shown in dark red and light red, respectively. (C) EGFP intensity median values ± SD from cells analyzed in (B). (D) Representation of the relationship between the obtained EGFP expression and indel formation in HEK293 cells as a result of increased amounts of LF3000-based delivery of Cas9 RNPs containing msgRNA1. As observed by flow cytometry (shown in C), the EGFP expression intensity was decreased as a result of increased RNP load. In contrast, the indel frequency increased with increasing amount of RNP. The indel formation frequency is represented by a red line. The EGFP intensity median is presented by a green line, and the number of EGFP+ cells is displayed by a green dotted line. AU, arbitrary units.
To assess whether complexation and transfection were affected by decreasing the volume from 50 to 2 μL, we transfected 2 × 104 HEK293 cells with LF2000 or LF3000 lipoplexes carrying EGFP mRNA (100 ng) and Cas9 RNP (250–2,000 ng) in a 50 μL volume. The cells were transfected in parallel to the cells analyzed in Figure 3. Similar to the experiment using 2 μL volume (compare Figures 3A and 5A), fluorescence microscopy carried out 1 day post-transfection revealed a significant decrease in EGFP expression as a result of increasing the amount of RNP (Figures 5A and 5B). Quantification of the EGFP signal revealed a decrease from 28.1% ± 8.0% to 3.3% ± 0.1% when comparing cells treated with 250 and 2,000 ng Cas9 RNP in LF2000 lipoplexes, respectively (Figure 5B). The decline was particularly evident for cells transfected with 1,000 and 2,000 ng Cas9 RNP (250 versus 1,000 ng: p < 0.01; 250 versus 2,000 ng: p < 0.0001). The decrease in EGFP expression was independent of complexation volume, because a similar tendency was observed using 2 or 50 μL. A similar EGFP expression pattern was observed using LF3000 lipoplexes. The EGFP signals decreased from 22.6% ± 1.2% to 0.6% ± 0.2% when comparing cells treated with 250 and 2,000 ng Cas9 RNP, respectively (Figure 5B). As in the case of LF2000, the decline was particularly evident for cells transfected with 1,000 and 2,000 ng Cas9 RNP (250 ng Cas9 RNP versus 1000 ng Cas9 RNP, p < 0.05; 250 ng Cas9 RNP versus 2000 ng Cas9 RNP, p < 0.001) (Figure 5B). TIDE analysis revealed a tendency for a higher genome-editing efficacy using 500, 1,000, or 2,000 ng Cas9 RNP following transfection with LF2000 lipoplexes compared with LF3000 (Figure 5C). The indel efficacy reached a plateau at 500 ng Cas9 RNP using LF2000 lipoplexes (up to 62.4%), whereas peak indel formation frequency was obtained at 1,000 ng Cas9 RNP using LF3000. Decreasing the transfection volume from 50 to 2 μL clearly affected indel frequency (compare Figures 3B and 5C). This is illustrated by LF2000 lipoplexes carrying 2,000 ng Cas9 RNP, which displayed indel frequencies of 62.4% ± 4.1% and 29.0% ± 1.5% upon formation of lipoplexes in 50 or 2 μL volume, respectively (Figure S3A). Overall, a volume reduction from 50 to 2 μL decreased indel frequency by 68% ± 10.7% and 54.7% ± 10.5% for LF2000 and LF3000 samples, respectively (Figure S3B). In conclusion, comparable indel formation was obtained by LF2000- and LF3000-based RNP lipoplexes. Because LF3000 has been described to show lower toxicity in vivo compared with LF2000, the former was chosen for further application of RNP delivery to the retina.29
Figure 5.
In Vitro Validation of Lipoplex Formation in 50 μL Transfection Volume
Approximately 2 × 104 HEK293 cells were seeded and transfected the next day with LF2000 or LF3000. The cells were transfected in parallel to the cells analyzed in Figure 3. Increasing amounts of RNP, containing Cas9 protein and msgRNA1 or Cas9 protein and msgRNA-Irr, were co-packaged with 100 ng EGFP mRNA into lipoplexes. Cas9 and msgRNAs were complexed in mass ratio of 2:1, with 250–2,000 ng Cas9 RNP. (A) Fluorescence analysis of EGFP expression 1 day post-transfection demonstrates decreasing EGFP expression upon titration of Cas9 RNP (containing msgRNA1) from 250 to 2,000 ng. Representative images are shown (n = 3). Scale bar, 200 μm. (B) Quantification of EGFP signals. Determination of the area intensity was used to quantify the EGFP signals presented in (A) (n = 3). Mean values ± SD are indicated by horizontal lines. LF2000 (blue) and LF3000 (red). Statistical tests in (B) are one-way ANOVAs (multiple comparison): ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 (250 versus 1,000 ng and 250 versus 2,000 ng). (C) Two days post-transfection, gDNA was isolated and used as the template for PCR. Resulting sequences from samples transfected with LF2000 (blue columns) and LF3000 (red columns) were analyzed using TIDE. Samples transfected with LF2000 induced indels at a significantly higher frequency than LF3000 for cells transfected with 500, 1,000, and 2,000 ng Cas9 RNP. Indel formation frequencies are shown as mean ± SD (n = 3). Statistical tests in (C) are Student’s t test: ∗p < 0.05.
Validation of Functional Knockout of VEGFA Production
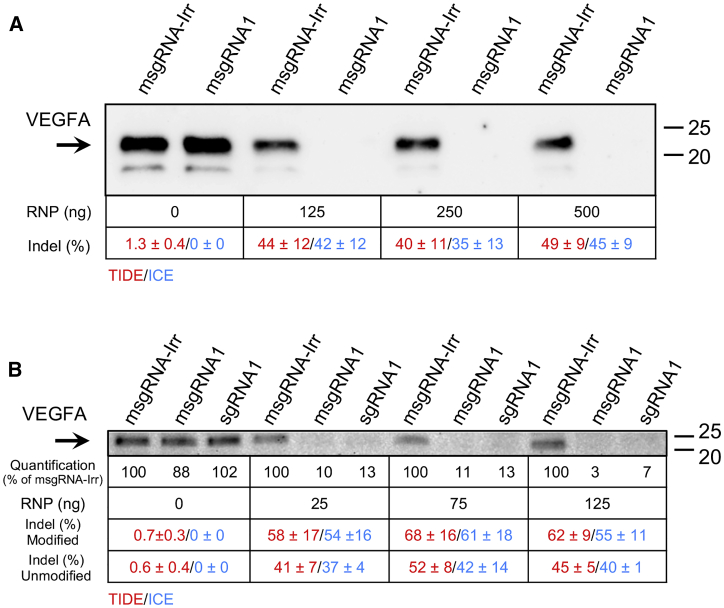
To validate the efficacy of msgRNA1 for functional knockout of vascular endothelial growth factor A (VEGFA) production, we used Flip-In HEK293 cells stably expressing human VEGFA (HEK293-VEGFA) from a cDNA sequence inserted in the Flip recombination target (FRT) locus. This cell line was chosen because the endogenous level of VEGFA is low in HEK293, K562, and RPE cell lines. Following LF3000-based delivery of 0, 125, 250, and 500 ng Cas9 RNP containing either msgRNA1 or msgRNA-Irr, the HEK293-VEGFA cells were maintained for 1 or 3 days. Cells intended for TIDE and Inference of CRISPR edits (ICE) analysis were harvested 24 h post-transfection, and for western blotting, the cultivation medium was replaced 24 h post-transfection and harvested at the end of the experiment (72 h post-transfection). Western blotting revealed almost complete elimination of VEGFA even after delivery of 125 ng Cas9 RNP containing msgRNA1 in the HEK293-VEGFA cells, consistent with detection of indel formation in the VEGFA gene using primers selectively amplifying VEGFA located in the FRT locus (Figure 6A). Comparable indel frequencies were obtained by TIDE and ICE analysis. In contrast, only a minor reduction in the VEGFA level was observed in the cells receiving 125 ng Cas9 RNPs containing msgRNA-Irr, compared with the 0-ng Cas9 RNP sample. A similar elimination pattern was observed following delivery of 250 and 500 ng Cas9 RNP. To investigate whether the modification of sgRNA1 and the dose-dependent SpCas9 indel rate correlated with the efficiency in suppressing the expression of VEGFA, we delivered 0, 25, 75, and 125 ng Cas9 RNP containing sgRNA1, msgRNA1, or msgRNA-Irr to HEK293-VEGFA (Figure 6B). Delivery of 25 ng Cas9 RNP carrying either msgRNA1 or sgRNA1 almost abolished VEGFA production as in the case of higher RNP amounts. As previously observed, higher indel frequencies were obtained when RNPs were complexed with msgRNA1 compared with RNPs containing unmodified sgRNA1. This may also explain why higher residual signs of VEGFA seemed to be present following delivery of 25 ng unmodified sgRNA1 compared with msgRNA1 (Figure 6B). A similar trend was also observed following delivery of 75 and 125 ng Cas9 RNP (Figure 6B).
Figure 6.
Functional Knockout of VEGFA Protein following Increased Delivery of RNP Levels
Validation of VEGFA knockout effects on the protein level was performed by western blotting of media samples collected from HEK293-VEGFA cells following LF3000-based delivery of increasing amounts of RNP. Approximately 2 × 104 HEK293-VEGFA cells were seeded and transfected the next day. The cells were maintained for 2 (TIDE and inference of CRISPR edits [ICE]) or 4 days (western blotting). Twenty-four hours after transfection, cells intended for TIDE and ICE analysis were harvested, and the cultivation medium was replaced on cells intended for western blot analysis. These cells were harvested 72 h after transfection. Each lane contains medium pooled from three independent replicates (n = 3). (A) Analysis of VEGFA suppression in HEK293 cells receiving 0, 125, 250, and 500 ng Cas9 RNP containing either msgRNA1 or msgRNA-Irr. (B) Analysis of VEGFA suppression in HEK293 cells receiving 0, 25, 75, and 125 ng Cas9 RNP containing either msgRNA1, sgRNA1, or msgRNA-Irr. Quantification of VEGFA is indicated below the blot shown in (B) (% of msgRNA-Irr). The corresponding indel formation frequencies obtained by TIDE (red) and ICE (blue) analysis are shown as mean ± SD below the blots (n = 3). Molecular sizes in kilodaltons are indicated on the right. The arrow indicates VEGFA.
Analysis of Predicted Off-Target Sites
To analyze whether the chemical modification of the sgRNA affects off-target activity, we searched for off-target sites for sgRNA1 using in silico prediction tools as shown in Figure 7A.28,32,33 For the top three predicted sites, we found near-background off-target activity using ICE analysis despite high on-target activity demonstrated by both msgRNA1 and sgRNA1 in K562 cells (Figure 7B). Consequently, the top three predicted off-target sites did not demonstrate off-target activity exceeding the 1% detection limit of ICE. In the case of TIDE analysis, we observed slightly increased indels scores, although no −1/+1 indels were identified, indicating that background noise may have caused the increased indel score (Figure 7C). In support of this, TIDE analysis revealed a 1.8% indels score in untransfected K562 cells in comparison with 0% obtained by ICE analysis (Figures 7B and 7C).
Figure 7.
Assessment of Off-Target Effects Mediated by sgRNA1
(A) Predicted off-target loci using the Benchling in silico bioinformatics tool.32,33 Benchling score presents the target compatibility of sgRNA1 (ON), with 100.0 representing a perfect match, and declining values decrease targeting probability. The top three computationally predicted off-target sites (OFF1–3) are highlighted in bold. Mismatches indicated by red font and PAM sequences are underlined. (B and C) Specificity of targeted cleavage in K562 cells mediated by RNPs containing 3 μg Cas9 protein and 1.5 μg of either msgRNA1 or sgRNA1 using ICE (B) and TIDE (C) analysis. Indel frequencies were measured following Sanger sequencing of PCR amplicons of the targeted genomic locus (VEGFA) and the top three bioinformatically predicted off-target sites. Bars represent average values + SD (n = 3). OFF1, off-target 1.
In Vivo Indel Formation in Isolated RPE Cells following Subretinal Injection of LF3000-Based RNP Lipoplexes
Our in vitro data demonstrated decreased EGFP expression with increasing amounts of RNP. However, because of uncertainty of the EGFP expression level delivered to the retinas of treated animals, different amounts of RNP were evaluated in vivo. C57BL/6J mice were divided into 10 groups. LF3000 lipoplexes were prepared with 100 ng EGFP mRNA and increasing amounts of Cas9 RNP with msgRNA1: 0 ng (n = 10), 150 ng (n = 10), 300 ng (n = 10), 500 ng (n = 5), 1,000 ng (n = 5), 1,650 ng (n = 5), 2,000 ng (n = 5), 3,300 ng (n = 5), 4,000 ng (n = 5), and 8,000 ng (n = 5). Once formed, 2 μL lipoplexes were delivered to the RPE cells by unilateral subretinal injection. The experiment was conducted as outlined in Figure 8A. Immediately after subretinal injection, optical coherence tomography (OCT) confirmed targeting of the subretinal space (Figures 8B and 8C). In parallel, the genome-editing capacity of the injected RNP lipoplexes was confirmed in vitro by transfection of HEK293 cells. In all cases, EGFP+ cells were observed (data not shown), and increasing indel formation with increasing amounts of RNP was observed (Figure S4). Mice were euthanized 4 days post-injection, and RPE cells were isolated after retinal dissection of the eye cup. All isolated RPE cells were pooled within each group and subjected to FACS analysis, after which the cells were sorted using the GFP detector (Figures 8D and S5). The FACS analysis did not provide a uniform population of distinct EGFP+ cells. Instead, a small increase (from 1.1% to 2.5%–7.0%) in EGFP+ RPE cells with low to medium signals was observed in retinas injected with EGFP mRNA and EGFP mRNA + Cas9 RNP, compared with non-injected eyes (CTRL) (Figure 8D). Subsequent TIDE analysis of EGFP+ RPE cells (populations shown in Figure 8D) revealed 2.4% ± 0.7% and 5.5% ± 0.5% indel formation in EGFP+ RPE cells isolated from retinas injected with lipoplexes carrying 1,650 and 3,300 ng Cas9 RNP, respectively (Figures 8E and 8F). To further substantiate these results, 4.2% indels displayed a +1 insertion, which is similar to the indel profile observed in in vitro experiments with msgRNA1 (data not shown) and our recent findings using LV-based delivery of sgRNA1.21 By further analysis of the FACS data obtained from mice injected with EGFP mRNA and 8 μg Cas9 RNP, EGFP+ RPE cells also displayed signals in cyanine5 (Cy5), phycoerythrin (PE), and peridinin chlorophyll protein complex (PerCP) detectors, indicating autofluorescence (Figure 8G). This was observed only in mice receiving Cas9 RNP and was not evident in EGFP+ RPE cells from animals injected with only EGFP mRNA (100 ng). In addition, an increased number of RPE cells showed EGFP expression and higher EGFP intensity, even though 100 ng EGFP mRNA was used for all preparations of RNP lipoplexes (compare columns 2 and 3 in Figure 8G).
Figure 8.
In Vivo Indel Assessment and EGFP Expression Analysis following Subretinal Injection of RNP Lipoplexes
Subretinal injection of C57BL/6J mice with LF3000 lipoplexes containing 100 ng EGFP mRNA combined with 0, 150, 300, 500, 1,000, 1,650, 2,000, 3,300, 4,000, and 8,000 ng Cas9 RNP formulated in 2 μL. (A) Overall timeline of in vivo evaluation of indel formation. Four days post-injection, mice were euthanized, retinas dissected, and isolated EGFP+ RPE cells were sorted using fluorescence activated cell sorting (FACS). (B) Optical coherence tomography (OCT) imaging analysis of non-injected eye. (C) OCT imaging analysis confirmed delivery of RNP lipoplexes to the subretinal space. (D) FACS analysis of selected samples used for discriminating EGFP+ RPE cells. All isolated RPE cells were pooled within each group and subjected to FACS analysis, during which the cells were: (1) identified due to FSC and SSC, (2) excluded due to FSC doublets, (3) excluded due to SSC doublets, and (4) isolated according to EGFP levels. Gating of the EGFP+ RPE cells is presented in Figure S5. Samples were isolated from mice injected with 0, 3.3, or 8 μg Cas9 RNP, all combined with 100 ng EGFP mRNA. Non-injected contralateral eyes were used as CTRL. (E) Genomic DNA isolated from EGFP+ RPE cells from mice injected with increasing amounts of Cas9 RNP (0–8,000 ng) was used as a template for PCR amplification prior to sequencing. Sequencing results were analyzed by TIDE. (F) Graphical illustration of aberrant sequences beginning at the expected cut site obtained from mice injected with 3.3 μg Cas9 RNP (green columns). No aberrant sequences were observed in mice injected with non-targeting irrelevant sgRNAs (CTRL sample, black columns). The expected cut site is indicated by the blue line. (G) FACS analysis of RPE cells isolated from animals injected with 100 ng EGFP mRNA or 100 ng EGFP mRNA combined with 8 μg Cas9 RNP. Non-injected contralateral eyes were included as CTRL. Analysis of fluorescence in detectors beside the EGFP detector revealed increased non-specific median fluorescence in the phycoerythrin (PE), PE-Cy5 (cyanine5), peridinin chlorophyll protein complex (PerCP), and allophycocyanin (APC) channels with the addition of 8 μg Cas9 RNP. (H–U) For evaluation of the EGFP+ RPE cells following subretinal delivery of Cas9 RNP lipoplexes, injected C57BL/6J mice were divided into four groups (n = 3): 100 ng EGFP mRNA and either 0 ng Cas9 RNP (K and N), 300 ng Cas9 RNP containing msgRNA1 (L), 1,650 ng Cas9 RNP containing msgRNA1 (M), 3,300 ng Cas9 RNP containing msgRNA1 (O–Q and U), or 3,300 ng Cas9 RNP containing msgRNA-Irr (R–T) were subretinally delivered by LF3000 lipoplexes. Non-injected contralateral eyes were used as CTRL (H–J). Five days post-injection, wholemounts were prepared and analyzed by fluorescence microscopy for EGFP expression in the GFP filter (I, K–N, P, S, and U) and unspecific autofluorescence in the Texas Red-X (TRX) filter (H, O, and R). (J), (Q), and (T) are mergers of (H) and (I), (O) and (P), and (R) and (S), respectively. (N) and (U) represent higher magnification of (K) and (P) as indicated by the pink squares, respectively. Efficacy of the injected lipoplexes was evaluated for indel formation in HEK293 cells (see Figure S4). All images are presented in Figures S6 and S7. Scale bars, 50 μm (H–M and O–T); 20 μm (N and U). ∗Injection bleb. SR, subretinal.
To further evaluate the EGFP+ cells and relate the degree of autofluorescence in retinas injected with RNP lipoplexes to the amount of RNP, we initiated an additional mouse study to analyze wholemounts of sclera/choroid/RPE. Of several samples analyzed for indels, the two samples showing indel formation (1,650 and 3,300 ng), as well as samples treated with 300 ng Cas9, were chosen for evaluation by whole mounts. This allowed us to analyze samples containing indels as a result of either a high or low amount of RNPs. C57BL/6J mice were divided into four groups of three animals. LF3000 lipoplexes carrying 100 ng EGFP mRNA and 0, 300, 1,650, or 3,300 ng Cas9 RNP were formed and administered by unilateral subretinal injection. Non-injected contralateral eyes served as controls (Figure S6). Four days post-injection, wholemounts were prepared. Retinas from the group receiving lipoplexes carrying EGFP mRNA alone displayed a high number of EGFP+ RPE cells, all showing the hexagonal shape, which is characteristic of healthy RPE cells (Figures 8K and 8N), whereas increasing amounts of Cas9 RNP significantly reduced the EGFP expression (Figures 8K–8M). Hence EGFP+ RPE cells were detected in the wholemounts from mice receiving 300 ng Cas9 RNP lipoplexes, but in a small number and with lower intensity compared with wholemounts from mice injected with lipoplexes containing EGFP mRNA alone (compare Figures 8K and 8L). Following delivery of 1,650 ng Cas9 RNP lipoplexes, only a few RPE cells were EGFP+ (Figure 8M), and a further increase of the Cas9 RNP amount to 3,300 ng significantly altered the distribution of EGFP and the morphology of the EGFP+ cells (Figures 8P and 8U). Instead, in confined areas of wholemounts from mice injected with 3,300 ng Cas9 RNP lipoplexes, we observed small irregular EGFP+ areas without the hexagonal shape of RPE cells. A similar pattern was observed in wholemounts from mice receiving increasing amounts of RNPs containing msgRNA-Irr (Figures 8S and S7V). In addition to the EGFP signal, fluorescence microscopy also displayed signals in the Texas Red-X (TRX) and DAPI filters, indicating autofluorescence following delivery of high levels of RNP containing either msgRNA1 (Figures 8O, 8Q, and S6) or msgRNA-Irr (Figures 8R, 8T, and S7). Autofluorescence was not observed in wholemounts from animals that did not receive RNPs (Figures 8H, 8J, S6A, S6B, and S6D). To allow FACS-based isolation of EGFP+ cells, we evaluated 100, 250, and 500 ng EGFP mRNA in vivo in order to increase the EGFP signal. However, retinal wholemounts demonstrated the most robust EGFP expression following transfection with 100 ng EGFP mRNA (Figure S8). By increasing the amount of EGFP mRNA in the lipoplexes, a decrease in both the number and intensity of EGFP+ cells was observed following in vivo delivery.
Finally, we further explored the performance of msgRNAs in vivo, by investigating whether the transfection reagent Viromer RED could be used as an alternative to LF3000. Viromer RED-based delivery resulted in approximately 60%–80% indel formation in HEK293 cells using preformed RNPs containing msgRNA1–3 (Figure S9A). Additionally, a similar EGFP expression (Figure S9B), as well as comparable indel frequencies (Figure S9C), was observed using either Cas9 mRNA or Cas9 protein (50 μL transfection volumes). Furthermore, Viromer RED provided robust EGFP expression, as well as significant indel formation, even at low RNP amounts using 2 μL transfection volumes in vitro (Figures S9D and S9E). However, subretinal injection of Viromer RED-based complexes in C57BL/6J mice did not generate detectable indels in FACS-isolated RPE cells (data not shown). Instead, the FACS analysis revealed autofluorescence following injection of transfection reagent only without RNPs and EGFP mRNA (Figure S9F). Autofluorescence was not observed in RPE cells from non-injected mice or mice injected with LF3000 transfection reagent only (Figure S9F).
Discussion
In a previous study, we demonstrated CRISPR/Cas9-mediated indel formation in the Vegfa gene in murine RPE cells following subretinal injection of all-in-one LV vectors encoding SpCas9, EGFP, and sgRNAs, including sgRNA1, designed to target exon 3.21 Several other studies have used AAV for Cas9-based retinal genome editing,15,16,18,34,35 whereas nonviral RNP delivery has been scarcely studied.17 In the present study, we show: (1) increased indel formation in vitro using chemically msgRNAs delivered by electroporation; (2) robust indel formation and EGFP expression in vitro induced by LF3000 lipoplexes containing RNP and EGFP mRNA; (3) efficacious functional knockout of VEGFA protein using chemically msgRNA1 at low Cas9 RNP levels compatible with murine subretinal injection; (4) undetectable off-target activity using both msgRNA1 and sgRNA1; (5) up to 6% indel formation in FACS-isolated EGFP+ RPE cells following subretinal delivery of lipoplexes containing RNP with msgRNA1 and EGFP mRNA; and (6) reduced EGFP expression in vitro and increased autofluorescence in RPE cells in vivo as sign of toxicity following delivery of lipoplexes containing high amounts of RNP. These data collectively demonstrate the feasibility of using synthetic msgRNAs, albeit with varying efficacy, as guides for in vivo RNP-mediated gene knockout.
In our validation and comparison of sgRNAs, we delivered RNPs by electroporation, which is an efficient delivery method for in vitro target validation.36 With 96% indel frequency based on TIDE analysis, msgRNA demonstrated remarkable efficacy. Similarly, high indel frequencies of >90% have been reported in previous studies using electroporation to deliver Cas9 RNP with chemically msgRNAs.37 Notably, indel frequencies may even approach 100%, because in two out of three samples using msgRNA1, the TIDE software did not identify any wild-type VEGFA alleles. Overall, our study demonstrates that msgRNAs significantly outperformed unmodified sgRNAs as part of pre-complexed, electroporated RNPs.
Because our ultimate aim was to achieve in vivo RNP delivery, it was essential to optimize lipoplex formation in vitro. To validate lipoplex formation and demonstrate efficient delivery, we transfected HEK293 cells with lipoplexes containing Cas9 RNP and EGFP mRNA. Our data demonstrated highly efficacious lipoplex delivery of EGFP mRNA and RNPs guided by msgRNA. Interestingly, 500 ng Cas9 RNP generated indels in 58% and 42% of analyzed alleles using LF2000 and LF3000, exceeding the 30%–50% indel formation demonstrated by Yu et al.26 targeting HEK293 cells with an equal amount of RNP consisting of Cas9 complexed with unmodified sgRNAs. Another study utilized Lipofectamine RNAiMAX or LF3000 to target HEK293 cells with maximal efficacy of 51% indel formation using RNPs guided by sgRNAs.38 Thus, utilizing msgRNAs at standard transfection conditions increased efficacy. To evaluate conditions relevant for in vivo subretinal administration, lipoplexes were formed in 2 μL volumes, resulting in peak indel formation of 29% (2,000 ng Cas9 RNP) and 37% (3,000 ng Cas9 RNP) using LF2000 and LF3000, respectively. Besides significant indel formation, EGFP expression was evident in the transfected cells, although diminished EGFP intensity, and to some extent reduced number of EGFP+ cells, was observed as a result of increasing the amount of Cas9 RNP in the lipoplexes, especially when applying high levels of RNP. Notably, this trend was sgRNA independent. To our knowledge, the tradeoff between EGFP expression from mRNA and the amount of Cas9 RNPs formulated in lipoplexes has not been studied previously.
To further explore the performance of msgRNA1 in human cells, we turned to a Flip-In HEK293 cell line encoding VEGFA from a genomic locus. We observed almost complete elimination of VEGFA production after delivery of 125 ng Cas9 RNP containing msgRNA1, compared with cells receiving RNPs with msgRNA-Irr. This demonstrates that delivery of RNPs results in efficacious functional knockout and is consistent with the robust indel formation of the VEGFA gene obtained in cells treated with msgRNA1. A similar effect on the VEGFA level was observed in cells receiving increased amounts of RNPs. A minor reduction in the VEGFA level was observed in cells receiving msgRNA-Irr, compared with the cells not receiving RNPs, which possibly reflects toxicity introduced by delivery of RNPs by LF3000. Notably, as little as 25 ng Cas9 RNP containing msgRNA1 almost abolished VEGFA production. A similar effect was observed following delivery of 25 ng Cas9 RNP containing unmodified sgRNA1. However, our data suggest that modification of sgRNA1 tends to have higher functional knockout of VEGFA compared with unmodified sgRNA. It should also be noted that higher indel frequencies were observed in the HEK293-VEGFA cells compared with transfected HEK293. This observation most likely reflects that a 5-fold lower number of HEK293-VEGFA cells was used. Another explanation could be higher accessibility of the VEGFA locus in the HEK293-VEGFA cells compared with the endogenous VEGFA in HEK293 cells. Because delivery of even small amounts of RNP results in efficacious functional knockout, we did not observe a direct correlation between the indel rate formation and the efficiency in suppressing the expression of VEGFA.
Robust in vitro indel formation and pronounced functional efficacy following lipoplex-based RNP delivery stimulated us to explore whether this strategy could also be applied for in vivo targeting of Vegfa. As demonstrated in several previous studies, EGFP expression allows FACS-based isolation of treated cells.19,21,22,35 By subretinal injection of lipoplexes containing EGFP mRNA and 150–8,000 ng Cas9 RNP formulated in 2 μL, we were able to achieve indel formation of 6% in the EGFP+ RPE cells isolated by FACS. The indel formation frequency peaked at 3,300 ng Cas9 RNP, which is similar to the in vitro peak efficacy at 3,000 ng. This is the first demonstration of retinal indel formation using RNPs guided by msgRNAs. In vivo delivery of preformed RNPs is a relatively new concept, and to our knowledge, only one previous study has tested the indel efficacy of retinal RNP delivery.17 In the study by Kim et al.,17 LF2000 was used to deliver RNPs containing 8 μg Cas9 protein with in-vitro-transcribed sgRNA targeting Vegfa.17 In addition, both functional and genomic knockout was demonstrated with 25% indel formation in RPE cells, which might be comparable with the present findings considering the 3-fold difference in the applied RNP amounts. Another important difference between the study by Kim et al.17 and the present study is the addition of EGFP mRNA and FACS-based isolation of EGFP+ RPE cells. In the paper by Kim et al.,17 genomic knockout was observed in RPE cells isolated from the injected area.
Notably, indel formation was observed in EGFP+ RPE cells isolated from retinas injected with lipoplexes carrying 1,650 and 3,300 ng Cas9 RNP. However, indels could not be detected in a relatively high number of injected animals. This could reflect: (1) that no changes were introduced in Vegfa, (2) that the indel formation frequency was under the detection level, or (3) that autofluorescence occurred as a result of apoptosis in RPE cells caused by the injection or the presence of immune cells in the retina (or some combination of these factors).
Lipoplexes containing EGFP mRNA and 3,300 ng Cas9 RNP induced indels in EGFP+ RPE cells. However, retinal wholemounts demonstrated a different RPE cell phenotype compared with animals receiving lower RNP amounts. The characteristic hexagonal structure of RPE cells observed in retinas injected with lipoplexes containing EGFP mRNA was not observed when Cas9 RNP containing msgRNA1 or msgRNA-Irr was included. Instead, the cells were highly fluorescent in EGFP, TRX, and DAPI filters and contained numerous small circular structures. The change in phenotype could therefore reflect either altered cell function based on gene knockout or toxicity caused by the injection of lipoplexes.
Knockout or truncation of Vegfa may result in cellular stress and altered health. Hence if indels caused the phenotypical change, on-target indel formation is considered to be the most likely event. However, this notion is not supported by our previous studies using LV-based delivery of sgRNAs, including sgRNA1, targeting Vegfa.21 Although off-target cleavage also can be detrimental to cellular health, it is unlikely to cause a relatively homogeneous pattern in multiple cells. This is supported by the fact that for the top three predicted sites, we observed near-background off-target activity for msgRNA1, despite detecting high levels of on-target activity of msgRNA1.
We also observed that the msgRNA1 and sgRNA1 did not demonstrate detectable off-target activity at three predicted off-target sites based on Sanger sequencing. The observed discrepancy between the off-target indel frequencies obtained by TIDE and ICE analysis is most likely due to lack of sensitivity, and this notion is substantiated by the 0% indel scores obtained from all analyzed off-target samples using ICE. In addition, a small increase of unspecific indel scores was also obtained by TIDE analysis compared with ICE analysis in the HEK293 cells. NGS-based off-target detection analysis of indels below 1% may resolve this issue. The presented data are supported by recent findings showing that chemically msgRNAs targeting, e.g., IL2RG and HBB sites tend to have lower off-target activity, although with variable impact, suggesting that the sequence of the sgRNA and likely also other elements, including delivery system, cell type, and the applied methodology, seem to play a role.28 In addition, transient Cas9 expression by nanoparticle-based mRNA delivery was shown to substantially reduce off-targeting in cells compared with long-term LV-based Cas9 expression emphasizing the potential of strategies relying on nonviral delivery of short-lived nuclease activity.39
Because autofluorescence was observed in wholemounts from animals receiving RNPs containing msgRNA-Irr, whereas wholemounts from untreated animals display only autofluorescence to a very low extent, our findings collectively suggest that induction of the observed phenotypic changes in vivo is independent of the msgRNA delivered to the RPE cells.
As an alternative to on- and off-targeting effects as an explanation for the observed in vivo phenotypic alterations, lipoplex delivery could affect the cells by two mechanisms. First, because the phenotype presents with high Cas9 RNP levels, toxicity resulting in autofluorescence, and hence low indel rates, could be caused by the increased level of Cas9 RNP delivered into the cells. In vitro, increasing amounts of RNP clearly reduce EGFP expression in HEK293 cells. In comparison, increased signals were revealed by the GFP detector in RPE cells from mice injected with increasing amounts of RNP. Additionally, RPE cells of wholemounts from eyes treated with EGFP mRNA, but without RNP, are green, resulting in approximately 2.5% EGFP+ cells. Hence one could argue that at least 1.5% (EGFP minus CTRL; Figure 8D) of the obtained EGFP+ RPE cells represent a “true EGFP signal,” whereas a part of the detected signals above this limit could potentially represent autofluorescence. Second, increased Cas9 RNP could alter the preparation of lipoplexes. The decline of EGFP+ cells may possibly be supported by competition between EGFP mRNA and Cas9 RNP during lipoplex formation, resulting in less EGFP mRNA packaged into lipoplexes. Additionally, at a certain amount of Cas9 RNP, lipoplexes cannot contain all of the available EGFP mRNA and Cas9 RNP. Instead, excess EGFP mRNA and Cas9 RNP would be co-injected with the lipoplexes, possibly resulting in an immune response caused by injection of naked mRNA and Cas9 protein. Likewise, a surplus of msgRNA from RNP complexation could participate in the initiation of an immune response. Because the retina is a sensitive tissue, the transfection reagent itself might also harm cells in the vicinity of the injection site, leading to autofluorescence. However, because no visible changes in the RPE phenotype were observed in retinas receiving only EGFP mRNA and LF3000, the transfection reagent or the injection itself do not seem to introduce inflammation or other forms of damage in the current setup.
In this context, it should be mentioned that signs of transfection-reagent-induced toxicity were observed using the Viromer RED transfection reagent as an alternative to LF3000. Even though Viromer RED-based delivery resulted in robust indel formation in HEK293 cells using preformed RNPs containing msgRNA1–3, thereby confirming our previous findings, subretinal injection of Viromer RED-based complexes in C57BL/6J mice did not generate detectable indels in isolated RPE cells. Instead, FACS analysis using the EGFP detector revealed autofluorescence following injection of Viromer RED without RNPs or EGFP mRNA, suggesting that the transfection reagent might induce toxicity.
Taken together, including the short time frame between RNP delivery and isolation of RPE cells, as well as the results obtained from the wholemounts, we believe that the apparent low in vivo indel formation frequency in the EGFP+ RPE cells most likely is explained by toxicity induced by the highly concentrated RNP solution delivered to the subretinal space. Despite robust indel formation and pronounced functional knockout effects in vitro, our findings further address the delivery challenge of efficacious clinical genome knockout approaches, which continues to be a severe challenge for clinical translation of genome-editing therapies.3
For the first time, we demonstrate RNP-mediated indel formation in the Vegfa gene in vivo using chemically msgRNAs. Additionally, complexation of such msgRNAs with SpCas9 in RNPs induces higher indel frequencies in vitro by lipoplex delivery compared with unmodified sgRNAs. Even though further studies are needed to optimize vehicles that can deliver Cas9 RNP and transiently induce genome surgery in the retina, the present findings suggest that RNP-based delivery of targeted gene-disrupting agents in vivo is a potential strategy for future treatment of acquired or autosomal dominant inherited retinal diseases, such as age-related macular degeneration and retinitis pigmentosa.
Materials and Methods
Design of Synthetic sgRNAs
sgRNAs were designed to target the Vegfa gene utilizing the SpCas9. Because we intended to validate efficacy in human cells, and ultimately target murine RPE cells, a 32-bp homologous region was identified to target both human and murine Vegfa. Three Vegfa targeting sgRNAs were designed: sgRNA1, 5′-CUCCUGGAAGAUGUCCACCA-3′; sgRNA2, 5′-ACUCCUGGAAGAUGUCCACC-3′; and sgRNA3, 5′-GACCCUGGUGGACAUCUUCC-3′. The designed sgRNAs were evaluated by the online design tool CRISPRko for on-target efficacy (sgRNA1 > sgRNA2 > sgRNA3). Of note, sgRNA1 in the present study is identical to sgRNA3 used in our previous study.21 A non-targeting sgRNA, designated sgRNA-Irr, was used as control: sgRNA-Irr, 5′-ACGGAGGCUAAGCGUCGCAA-3′. Synthetic sgRNAs, with an 80-mer SpCas9 scaffold, were ordered from Synthego (Menlo Park, CA, USA) with or without 2′O-methyl-3′phosphorothiate modifications of the first and last three nucleotides.
Cell Culture and RNP Complexation
K562 (CLL-243; ATCC) cells were maintained in RPMI 1640 medium (Lonza, Basel, Schweiz). HEK293 (CRL-1573; ATCC) cells were maintained in DMEM (Lonza). Both cell types were supplemented with 10% fetal calf serum (Sigma-Aldrich, Broendby, Denmark), 0.29 mg/mL glutamine (Sigma-Aldrich), 0.06 mg/mL penicillin (FarmaPlus, Oslo, Norway), and 0.1 mg/mL streptomycin (Sigma-Aldrich). Cells were cultured in tissue culture flasks and 6-, 12-, and 24-well plates (Sarstedt, Nürnbrecht, Germany) at 37°C with 5% (v/v) CO2. RNP complexes were formed by mixing SpCas9 protein (Integrated DNA Technologies, Leuven, Belgium) and sgRNAs (Synthego) in a 2:1 mass ratio (1:2.5 molar ratio) in a PCR tube that was placed in a thermocycler for 15 min at 25°C.
Stable VEGFA-Expressing Cell Line
The human VEGFA sequence was cloned from pVax1-hVEGF165 (a gift from Dr. Loree Heller, plasmid #74466; Addgene),40 into the FRT site of the pcDNA5/FRT expression vector containing a cytomegalovirus (CMV) promoter (Thermo Fisher Scientific, Hvidovre, Denmark) using BamHI and NotI (Thermo Fisher Scientific). Flip-In HEK293 cells (Thermo Fisher Scientific) were cotransfected with the pcDNA5/FRT-VEGFA construct and the pOG44 vector encoding the Flp recombinase. This recombinase catalyzes the homologous recombination between the FRT sites of the Flp expression vector and the genome of the Flp expression cell. Cotransfection and subsequent selection of cells through hygromycin and zeocin resistance genes were performed following the supplier’s recommendations, and cells were seeded at a density of 0.5 cell/well in a 96-well plate in order to isolate a cell line of single-cell origin. Stable expression of VEGFA was evaluated by immunostaining and western blotting (data not shown) as previously shown.21,41 Stable VEGFA-expressing HEK293 cells are designated HEK293-VEGFA. HEK293-VEGFA cells were maintained in DMEM (Lonza) supplemented with 10% fetal calf serum (Sigma-Aldrich, Broendby, Denmark), 0.29 mg/mL glutamine (Sigma-Aldrich), 0.06 mg/mL penicillin (FarmaPlus, Oslo, Norway), and 0.1 mg/mL streptomycin (Sigma-Aldrich).
Electroporation and Transfection
For efficient in vitro target validation of modified and unmodified sgRNAs, RNPs were delivered to K562 cells by electroporation for transfection reagent independent analysis.36 Approximately 2 × 105 K562 cells were electroporated on the 4D-Nucleofector system (Lonza) with OptiMEM (Thermo Fisher Scientific) as the electroporation buffer. A total of 3 μg SpCas9 protein (Integrated DNA Technologies) and 1.5 μg sgRNA or msgRNA were complexed and mixed with K562 cells in Nucleocuvette strips (Lonza). Electroporation with 2 μg EGFP mRNA (TriLink Biotechnologies, San Diego, CA, USA) was used as control. Following the addition of preheated RPMI 1640 growth medium, cells were grown in 24-well plates for 2 days. Prior to transfection, complexed RNPs were mixed with EGFP mRNA and either OptiMEM (Lonza) or Hank’s balanced salt solution (HBSS; Sigma-Aldrich). OptiMEM was used for initial HEK293 transfection of sgRNA1–3, whereas HBSS was used for titration of Cas9 RNPs. Approximately 2 × 104 or 1 × 105 HEK293 cells were seeded and transfected the following day using LF2000 (Thermo Fisher Scientific) or LF3000 (Thermo Fisher Scientific) according to the manufacturer’s protocol. In brief, the RNP-EGFP solution was transferred to Lipofectamine combined with either HBSS or OptiMEM and incubated at room temperature (RT) for 5 min. In some experiments (shown in Figures S9A–S9E), RNP delivery was performed using the Viromer RED transfection reagent (LabLife Nordic, Danderyd, Sweden) according to the manufacturer’s guidelines. In summary, 10% (v/v) Viromer RED was used for delivery of 250 ng EGFP mRNA combined with either 1.5 μg Cas9 RNP or 500 ng Cas9 mRNA and 750 ng msgRNA. A total of 50 μL transfection volume was used for in vitro validation, while 2 μL was used for both in vitro validation and in vivo subretinal injections. In the 2 μL transfection mixes, 20% (v/v) Lipofectamine or 10% (v/v) Viromer RED was used. RNPs contained either sgRNA1–3, msgRNA1–3, or msgRNA-Irr. Transfection efficacy was evaluated based on EGFP expression using either a Leica DM IRBE fluorescence microscope (Leica Microsystems, Wetzlar, Germany) and images captured with a Leica DFC 320 camera, or a Olympus IX83 microscope (Olympus, Ballerup, Denmark), with a ×10 air objective, bandpass filter for Alexa Fluor 488, and a Hamamatsu camera (C8484-05G).
Quantification of EGFP Signals
Quantitative image acquisition of EGFP signals was conducted either by flow cytometry (Figures 2 and 4) or area intensity analysis (Figures 5 and S10). For flow cytometry, transfected HEK293 cells were washed in PBS with 0.3 mM EDTA, trypsinized, and centrifuged at 350 × g for 3 min at 4°C. Trypsin was removed and cells resuspended in 80 μL PBS with 1% BSA and 0.5 μL propidium iodide (1 mg/mL) and transferred to a 96-well plate for analysis. Data were acquired on a NovoCyte flow cytometer (ACEA Biosciences, San Diego, FL, USA) by collecting 2.5 × 104 cells. NovoExpress software (v.1.4.1; ACEA Biosciences) was used for acquisition, and FlowJo software (v.10.6.1; Ashland, OR, USA) was used for data analysis of EGFP signals. The detailed gating strategy is presented in Figures 2 and 4. Cells were initially separated using forward scatter (FSC) and side scatter (SSC), and FSC doublets were excluded prior to identification of EGFP. The number of dead cells was identified by propidium iodide staining.
Area intensity analysis was performed by using the Olympus cellSens Dimension software (v.2.19). Background subtraction was performed first, because the segmentation process requires locally homogeneous intensities. Segmentation was performed using a manual threshold model identifying two phases, “all cells” (low-high intensity fluorescence based on autofluorescence and EGFP+ cells) and “EGFP+ cells” (high-intensity only), respectively. The manual thresholds were applied similarly for all quantifications. The ratio of segmented phases (in %) represents the EGFP+ cells. In Figure S10, a representative phase contrast image is shown together with the EGFP images and the resulting segmented images of EGFP+ cells, as well as all cells to visually verify the estimation of the “all cells” phase.
Indel Analysis by TIDE and ICE
DNeasy Blood and Tissue Kit (QIAGEN, Hilden, Germany) was applied for gDNA isolation from electroporated K562 or transfected HEK293 cells following the manufacturer’s protocol. QIAamp DNA Micro Kit (QIAGEN) was used to isolate gDNA from FACS isolated EGFP+ RPE cells. gDNA was quantified using a NanoDrop One spectrophotometer (Thermo Fisher Scientific) prior to PCR amplification using the Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific). Primers used for amplification of the human VEGFA exon 3 were 5′-CATACTCAGACTGTCCTCTG-3′ and 5′-GAATGAAGCATCTTCTCCAC-3′. Primers for amplification of the murine Vegfa exon 3 were 5′-AGAGCTTCGGCAGGGAAGTACA-3′ and 5′-TATTTGATGAGTGGCTGTTGGCCT-3′. In the case of the HEK293-VEGFA cells 5′-CTCCACCATGCCAAGTGGTC-3′ and 5′-CCTCGGCTTGTCACATCTGC-3′ primers were used to selectively amplify human VEGFA located in the FRT site. PCR products were purified by gel electrophoresis using MinElute Gel Extraction Kit (QIAGEN) and subsequently sequenced (Eurofins Genomics, Ebersberg, Germany). Sequence results were applied for TIDE analysis (http://shinyapps.datacurators.nl/tide/) and ICE (https://www.synthego.com/) for quantification of indels as previously described.21 In brief, the indel frequencies were assessed by comparing the samples receiving editing sgRNAs (unmodified or modified) with the corresponding control samples treated with non-targeting irrelevant sgRNAs.
Functional Knockout: Assessment of the VEGFA Protein Level by Western Blotting following RNP Delivery
Western blot analysis was performed on media samples from HEK293-VEGFA cells transfected with 0, 25, 75, 125, 250, or 500 ng Cas9 RNP containing either msgRNA1 or msgRNA-Irr and 100 ng EGFP mRNA. Three days post-transfection, media from three replica samples were harvested and pooled. Media were filtered (0.22 μm), and 40 μL was used for western blotting as previously described with the following modifications.21 Cell medium samples were electrophoresed on a Mini-PROTEAN TGX 4%–15% Stain-Free precast gel (Bio-Rad, Copenhagen, Denmark). Membranes were incubated with rabbit anti-VEGF antibody (ab46154; Abcam, Cambridge, UK) overnight or for 2 days at 4°C in a concentration of 1:2,000 in Tris-buffered saline, 0.1% Tween 20 (TBS-T). Membranes were then washed three times in TBS-T and subsequently incubated with horseradish peroxidase-conjugated goat anti-rabbit (Dako; Agilent Technologies, Santa Clara, CA, USA) for 1 h at RT. Bound antibodies were visualized with Clarity Western ECL Blotting substrate (Bio-Rad) on a ChemiDoc digital imaging system (Bio-Rad). Quantification of VEGFA was performed using the Image Lab 6.0.1 software (Bio-Rad).
Off-Target Activity
Predicted off-target sites were identified using the online Benchling’s CRISPR prediction tool (https://www.benchling.com/), and the top three off-targets were analyzed. Following electroporation of approximately 2 × 105 K562 cells with 3 μg SpCas9 protein complexed with 1.5 μg msgRNA1 or sgRNA1, indel analysis was performed essentially as described above, including gDNA isolation, PCR amplification, sequencing, TIDE, and ICE analysis. Primers used for amplification of the top three off-target sites were: off-target site 1, 5′-CCAGAGAGCTGATTATCCCTTTT-3′ and 5′-ACGATCACTGGACACTTGAAACC-3′; off-target site 2, 5′-TGGTGAAGAAAACGCTAAGGCTA-3′ and 5′-TGTTCTAGCCATATTTCTTCCTG-3′; and off-target site 3, 5′-CTTTATTCATATTAACCTATTAT-3′ and 5′-CTCTGCAACTGAAAAGTCTTAT-3′.
Animals
All animal experiments were approved by the Danish Animal Inspectorate (Case #2016-15-0201-00947). Male C57BL/6J mice, 8–9 weeks of age, were purchased at Janvier Labs (Le Genest-Saint-Isle, France). Mice were kept at a 12-h/12-h light/dark cycle at the Animal Facilities, Department of Biomedicine, Aarhus University. Medetomidine hydrochloride 0.5–1 mg/kg (Cepetor; ScanVet Animal Health, Fredensborg, Denmark) and ketamine 60–100 mg/kg (Ketador, Richer Pharma, Wels, Austria) were combined for anesthesia during subretinal injections and fundoscopy. A drop of 1% tropicamide solution (Mydriacy, Alcon Nordic, Copenhagen, Denmark) was used for pupil dilation. A carbomer eye gel (2 mg/g, Viscotears; Alcon Nordic) was used to lubricate the eyes during subretinal injections and fundoscopy. Atipamezole hydrochloride 0.5–1 mg/kg (Antisedan, Orion Pharma, Espoo, Finland) was used to bring mice out of sedation. Mice were kept warm until mobile, before being transferred back into their cages. To ensure adequate analgesia, mice received subcutaneous injections of the nonsteroidal antiinflammatory drug carprofen 5 mg/kg (Norodyl; ScanVeg Animal Health) 24 h prior to and immediately after subretinal injection. As well, mice were treated with carprofen 5 mg/150 mL via their drinking water from 1 day before the subretinal injections until 3 days after.
Subretinal Injections
C57BL/6J male mice, 8–9 weeks of age, were used for subretinal injections. For wholemounts, mice were divided into groups of three mice. Mice were injected with LF3000 lipoplexes containing either increasing amounts of EGFP mRNA or increasing amounts of RNP (containing either msgRNA1 or msgRNA-Irr) combined with EGFP mRNA in 2 μL. For FACS analysis, mice were divided into groups of 5–10 mice injected with 0 ng (n = 10), 150 ng (n = 10), 300 ng (n = 10), 500 ng (n = 5), 1,000 ng (n = 5), 1,650 ng (n = 5), 2,000 ng (n = 5), 3,300 ng (n = 5), 4,000 ng (n = 5), and 8,000 ng (n = 5) Cas9 RNP, all combined with 100 ng EGFP mRNA. Non-injected contralateral eyes were used as control. In some experiments (shown in Figure S9F) mice were injected with Viromer RED lipoplexes containing either 349 ng Cas9 RNP with 58 ng EGFP mRNA (n = 5), 1,500 ng Cas9 RNP with 250 ng EGFP mRNA (n = 5), 1,650 ng Cas9 RNP and 100 ng EGFP mRNA (n = 10), or 3,300 ng Cas9 RNP and 100 ng EGFP mRNA (n = 10). RNPs contained either msgRNA1 or msgRNA-Irr. Non-injected contralateral eyes and eyes injected with only the transfection reagent were used as controls. Injections were performed using an OPMI 1 FR PRO Surgical microscope (Zeiss, Jena, Germany) as previously described.42,43
Optical Coherence Tomography
To validate the quality and location of the subretinal injections, we analyzed retinas 5 min after injection using a commercial imaging device for rodents, the OCT2 integrated with the Micron IV retinal imaging microscope system and the accompanying Reveal software (all from Phoenix Research Labs, Pleasanton, CA, USA). In brief, injected mice were positioned on the mouse stage, and following fundus imaging, the OCT2 device was used to obtain cross-sectional b-scans of treated animals. Images of retinal detachments were made by a horizontal OCT scan through the center of the bleb guided by real-time fundus view, and the best image quality was obtained with at least 20 averaged automatic real-time images. Motion artifacts were minimized by imaging in contact with the cornea and real-time eye tracking in the device software.
Retinal Dissection, Wholemounts, and FACS
Mice were euthanized by cervical dislocation. For histological analysis of wholemounts, eyes were prepared as previously described.44 In brief, eyes were enucleated, cleaned, and fixed in fresh 4% paraformaldehyde for 2 h at RT. The cornea, lens, and neuroretina were removed. Eight incisions from the periphery to the optic nerve enabled flat mounting of the tissue with the RPE cells facing upward on a glass slide. For FACS analysis, the enucleated eyes were cleansed and placed in 1% trypsin (Sigma-Aldrich) for 1 h at 37°C. Cells were rinsed off the eyecup using a syringe and a 27-gauge needle with HBSS. The resulting single-cell solution was sorted on a FACSAria III high-speed cell sorter (BD Biosciences, San Jose, CA, USA) using the GFP detector. Cells were initially separated using FSC and SSC as indicated in Figure S5. FSC and SSC doublets were excluded prior to sorting of cells according to detected GFP levels. No cell-specific marker proteins were utilized.
Statistical Analysis
Data analysis was performed by using GraphPad Prism software v.8 or Microsoft Excel software v.16.38. Data are presented as mean ± SD, and statistical differences between two groups were evaluated using Student’s t test and one-way ANOVAs for multiple comparison. A p value <0.05 was considered statistically significant.
Author Contributions
Writing – Original Draft: A.B.H., A.L.A., and T.J.C.; Writing – Reviewing & Editing: A.B.H., A.L.A., E.G.J., S.A., R.O.B., J.G.M., and T.J.C.; Supervision: T.J.C., J.G.M., R.O.B., and A.L.A.; Investigation: A.B.H., E.G.J., S.A., and A.L.A.; Methodology: A.B.H., T.J.C., A.L.A., R.O.B., and J.G.M.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The authors would like to thank Tina Hindkjaer and Kamilla Zahll Hornbek for their excellent technical support. The authors also thank Christian Hasselbalch Garm, Bioimaging Core Facility, Aarhus University, Denmark, for helping with the quantification of EGFP signals. Cell sorting was performed at the FACS Core Facility, Aarhus University, Denmark. The Graphical Abstract was created with BioRender.com. This work was supported by the Faculty of Health Sciences (PhD scholarship to A.B.H.), the Danish Eye Research Foundation (T.J.C.), Aase and Ejnar Danielsen’s Foundation (T.J.C.), Bagenkop-Nielsens Myopifond, and Svend Helge Schrøder og hustru Ketty Lydia Larsen Schrøders fond (T.J.C.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2020.09.032.
Supplemental Information
References
- 1.Dever D.P., Bak R.O., Reinisch A., Camarena J., Washington G., Nicolas C.E., Pavel-Dinu M., Saxena N., Wilkens A.B., Mantri S. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Min Y.-L., Li H., Rodriguez-Caycedo C., Mireault A.A., Huang J., Shelton J.M., McAnally J.R., Amoasii L., Mammen P.P.A., Bassel-Duby R., Olson E.N. CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci. Adv. 2019;5:eaav4324. doi: 10.1126/sciadv.aav4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Haasteren J., Li J., Scheideler O.J., Murthy N., Schaffer D.V. The delivery challenge: fulfilling the promise of therapeutic genome editing. Nat. Biotechnol. 2020;38:845–855. doi: 10.1038/s41587-020-0565-5. [DOI] [PubMed] [Google Scholar]
- 4.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrington L.B., Burstein D., Chen J.S., Paez-Espino D., Ma E., Witte I.P., Cofsky J.C., Kyrpides N.C., Banfield J.F., Doudna J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362:839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishimasu H., Shi X., Ishiguro S., Gao L., Hirano S., Okazaki S., Noda T., Abudayyeh O.O., Gootenberg J.S., Mori H. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 2018;361:1259–1262. doi: 10.1126/science.aas9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sapranauskas R., Gasiunas G., Fremaux C., Barrangou R., Horvath P., Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walton R.T., Christie K.A., Whittaker M.N., Kleinstiver B.P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020;368:290–296. doi: 10.1126/science.aba8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bakondi B., Lv W., Lu B., Jones M.K., Tsai Y., Kim K.J., Levy R., Akhtar A.A., Breunig J.J., Svendsen C.N., Wang S. In vivo CRISPR/Cas9 gene editing corrects retinal dystrophy in the S334ter-3 rat model of autosomal dominant retinitis pigmentosa. Mol. Ther. 2016;24:556–563. doi: 10.1038/mt.2015.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li P., Kleinstiver B.P., Leon M.Y., Prew M.S., Navarro-Gomez D., Greenwald S.H., Pierce E.A., Joung J.K., Liu Q. Allele-Specific CRISPR-Cas9 genome editing of the single-base P23H mutation for rhodopsin-associated dominant retinitis pigmentosa. CRISPR J. 2018;1:55–64. doi: 10.1089/crispr.2017.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruan G.X., Barry E., Yu D., Lukason M., Cheng S.H., Scaria A. CRISPR/Cas9-mediated genome editing as a therapeutic approach for Leber congenital amaurosis 10. Mol. Ther. 2017;25:331–341. doi: 10.1016/j.ymthe.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim E., Koo T., Park S.W., Kim D., Kim K., Cho H.Y., Song D.W., Lee K.J., Jung M.H., Kim S. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017;8:14500. doi: 10.1038/ncomms14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim K., Park S.W., Kim J.H., Lee S.H., Kim D., Koo T., Kim K.E., Kim J.H., Kim J.S. Genome surgery using Cas9 ribonucleoproteins for the treatment of age-related macular degeneration. Genome Res. 2017;27:419–426. doi: 10.1101/gr.219089.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koo T., Park S.W., Jo D.H., Kim D., Kim J.H., Cho H.Y., Kim J., Kim J.H., Kim J.S. CRISPR-LbCpf1 prevents choroidal neovascularization in a mouse model of age-related macular degeneration. Nat. Commun. 2018;9:1855. doi: 10.1038/s41467-018-04175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giannelli S.G., Luoni M., Castoldi V., Massimino L., Cabassi T., Angeloni D., Demontis G.C., Leocani L., Andreazzoli M., Broccoli V. Cas9/sgRNA selective targeting of the P23H Rhodopsin mutant allele for treating retinitis pigmentosa by intravitreal AAV9.PHP.B-based delivery. Hum. Mol. Genet. 2018;27:761–779. doi: 10.1093/hmg/ddx438. [DOI] [PubMed] [Google Scholar]
- 20.Matsuda T., Oinuma I. Optimized CRISPR/Cas9-mediated in vivo genome engineering applicable to monitoring dynamics of endogenous proteins in the mouse neural tissues. Sci. Rep. 2019;9:11309. doi: 10.1038/s41598-019-47721-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmgaard A., Askou A.L., Benckendorff J.N.E., Thomsen E.A., Cai Y., Bek T., Mikkelsen J.G., Corydon T.J. In vivo knockout of the Vegfa gene by lentiviral delivery of CRISPR/Cas9 in mouse retinal pigment epithelium cells. Mol. Ther. Nucleic Acids. 2017;9:89–99. doi: 10.1016/j.omtn.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latella M.C., Di Salvo M.T., Cocchiarella F., Benati D., Grisendi G., Comitato A., Marigo V., Recchia A. In vivo editing of the human mutant rhodopsin gene by electroporation of plasmid-based CRISPR/Cas9 in the mouse retina. Mol. Ther. Nucleic Acids. 2016;5:e389. doi: 10.1038/mtna.2016.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Askou A.L., Jakobsen T.S., Corydon T.J. Retinal gene therapy: an eye-opener of the 21st century. Gene Ther. 2020 doi: 10.1038/s41434-020-0168-2. Published online October 20, 2020. [DOI] [PubMed] [Google Scholar]
- 24.Lattanzi A., Meneghini V., Pavani G., Amor F., Ramadier S., Felix T., Antoniani C., Masson C., Alibeu O., Lee C. Optimization of CRISPR/Cas9 delivery to human hematopoietic stem and progenitor cells for therapeutic genomic rearrangements. Mol. Ther. 2019;27:137–150. doi: 10.1016/j.ymthe.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu X., Liang X., Xie H., Kumar S., Ravinder N., Potter J., de Mollerat du Jeu X., Chesnut J.D. Improved delivery of Cas9 protein/gRNA complexes using lipofectamine CRISPRMAX. Biotechnol. Lett. 2016;38:919–929. doi: 10.1007/s10529-016-2064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao X., Tao Y., Lamas V., Huang M., Yeh W.H., Pan B., Hu Y.J., Hu J.H., Thompson D.B., Shu Y. Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature. 2018;553:217–221. doi: 10.1038/nature25164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendel A., Bak R.O., Clark J.T., Kennedy A.B., Ryan D.E., Roy S., Steinfeld I., Lunstad B.D., Kaiser R.J., Wilkens A.B. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rust A., Hassan H.H., Sedelnikova S., Niranjan D., Hautbergue G., Abbas S.A., Partridge L., Rice D., Binz T., Davletov B. Two complementary approaches for intracellular delivery of exogenous enzymes. Sci. Rep. 2015;5:12444. doi: 10.1038/srep12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ansari A.M., Ahmed A.K., Matsangos A.E., Lay F., Born L.J., Marti G., Harmon J.W., Sun Z. Cellular GFP toxicity and immunogenicity: Potential confounders in in vivo cell tracking experiments. Stem Cell Rev. Rep. 2016;12:553–559. doi: 10.1007/s12015-016-9670-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross P.C., Hui S.W. Lipoplex size is a major determinant of in vitro lipofection efficiency. Gene Ther. 1999;6:651–659. doi: 10.1038/sj.gt.3300863. [DOI] [PubMed] [Google Scholar]
- 32.Cradick T.J., Qiu P., Lee C.M., Fine E.J., Bao G. COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas Off-target Sites. Mol. Ther. Nucleic Acids. 2014;3:e214. doi: 10.1038/mtna.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hung S.S., Chrysostomou V., Li F., Lim J.K., Wang J.H., Powell J.E., Tu L., Daniszewski M., Lo C., Wong R.C. AAV-mediated CRISPR/Cas gene editing of retinal cells in vivo. Invest. Ophthalmol. Vis. Sci. 2016;57:3470–3476. doi: 10.1167/iovs.16-19316. [DOI] [PubMed] [Google Scholar]
- 35.McCullough K.T., Boye S.L., Fajardo D., Calabro K., Peterson J.J., Strang C.E., Chakraborty D., Gloskowski S., Haskett S., Samuelsson S. Somatic gene editing of GUCY2D by AAV-CRISPR/Cas9 alters retinal structure and function in mouse and macaque. Hum. Gene Ther. 2019;30:571–589. doi: 10.1089/hum.2018.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laustsen A., Bak R.O. Electroporation-based CRISPR/Cas9 gene editing using Cas9 protein and chemically modified sgRNAs. Methods Mol. Biol. 2019;1961:127–134. doi: 10.1007/978-1-4939-9170-9_9. [DOI] [PubMed] [Google Scholar]
- 37.Bak R.O., Porteus M.H. CRISPR-mediated integration of large gene cassettes using AAV donor vectors. Cell Rep. 2017;20:750–756. doi: 10.1016/j.celrep.2017.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang X., Potter J., Kumar S., Zou Y., Quintanilla R., Sridharan M., Carte J., Chen W., Roark N., Ranganathan S. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 39.Yin H., Song C.Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marshall W.G., Jr., Boone B.A., Burgos J.D., Gografe S.I., Baldwin M.K., Danielson M.L., Larson M.J., Caretto D.R., Cruz Y., Ferraro B. Electroporation-mediated delivery of a naked DNA plasmid expressing VEGF to the porcine heart enhances protein expression. Gene Ther. 2010;17:419–423. doi: 10.1038/gt.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Askou A.L., Aagaard L., Kostic C., Arsenijevic Y., Hollensen A.K., Bek T., Jensen T.G., Mikkelsen J.G., Corydon T.J. Multigenic lentiviral vectors for combined and tissue-specific expression of miRNA- and protein-based antiangiogenic factors. Mol. Ther. Methods Clin. Dev. 2015;2:14064. doi: 10.1038/mtm.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Askou A.L., Alsing S., Benckendorff J.N.E., Holmgaard A., Mikkelsen J.G., Aagaard L., Bek T., Corydon T.J. Suppression of choroidal neovascularization by AAV-based dual-acting antiangiogenic gene therapy. Mol. Ther. Nucleic Acids. 2019;16:38–50. doi: 10.1016/j.omtn.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bemelmans A.P., Kostic C., Crippa S.V., Hauswirth W.W., Lem J., Munier F.L., Seeliger M.W., Wenzel A., Arsenijevic Y. Lentiviral gene transfer of RPE65 rescues survival and function of cones in a mouse model of Leber congenital amaurosis. PLoS Med. 2006;3:e347. doi: 10.1371/journal.pmed.0030347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Askou A.L., Pournaras J.A., Pihlmann M., Svalgaard J.D., Arsenijevic Y., Kostic C., Bek T., Dagnaes-Hansen F., Mikkelsen J.G., Jensen T.G., Corydon T.J. Reduction of choroidal neovascularization in mice by adeno-associated virus-delivered anti-vascular endothelial growth factor short hairpin RNA. J. Gene Med. 2012;14:632–641. doi: 10.1002/jgm.2678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.