Abstract
Osteosarcoma is the most common primary bone malignancy and is a neoplasm thought to be derived from the bone‐forming mesenchymal stem cells. Aberrant activation of oncogenes and inactivation of tumour suppressor genes by somatic mutations and epigenetic mechanisms play a pivotal pathogenic role in osteosarcoma. Aside from alterations in these protein‐coding genes, it has now been realized that dysregulation of non‐coding RNAs (ncRNAs), including microRNAs (miRNAs), long non‐coding RNAs (lncRNAs) and the recently discovered circular RNAs (circRNAs), is crucial to the initiation and progression of osteosarcoma. CircRNAs are single‐stranded RNAs that form covalently closed loops and function as an important regulatory element of the genome through multiple machineries. Recently, an increasing number of studies suggested that circRNAs also played critical roles in osteosarcoma. This review summarizes recent development and progression in circRNA transcriptome analysis and their functions in the modulation of osteosarcoma progression.
Keywords: CircRNAs, CircTCF25, ncRNAs, osteosarcoma
The representative diagram of circRNA mediated ceRNA network and oncogene signaling pathways. The current study in osteosarcoma highlights the regulatory relationship of the circRNA‐miRNA‐mRNA network for different signaling pathways. Multiple identified circRNAs function as miRNA sponge and subsequently upregulate relevant target genes expression level.
1. INTRODUCTION
Osteosarcoma, a neoplasm thought to be derived from the bone‐forming mesenchymal stem cells, is the most common primary bone malignancy, predominantly involving metaphyseal regions of the long bones (eg, proximal end of tibia or humerus and distal end of femur) which are the most rapidly growing parts of the skeleton in children and adolescents. 1 , 2 , 3 , 4 , 5 The incidence of osteosarcoma follows a bimodal age distribution with ages between 10 and 30 primarily affected. 6 , 7 , 8 , 9 , 10 The second peak appears in the elderly in which osteosarcoma very often comes as a later cancer secondary to radiation exposure or is associated with Paget disease (a disorder of bone remodelling resulting in structural weakening). 11 , 12 , 13 , 14 , 15 , 16 With the advent of neoadjuvant and adjuvant chemotherapy with cisplatin, doxorubicin and high‐dose methotrexate and the advances in surgery, the 5‐year survival rate of patients with osteosarcoma has improved from ~20% before the 1980s to currently ~ 70%. 17 , 18 , 19 , 20 , 21 , 22 Nevertheless, half of the patients still do not survive for longer than 10 year. 23 , 24 , 25 Thus, it is important to identify new therapeutic targets for tackling this devastating and potentially fatal disease (Figure 1).
Figure 1.
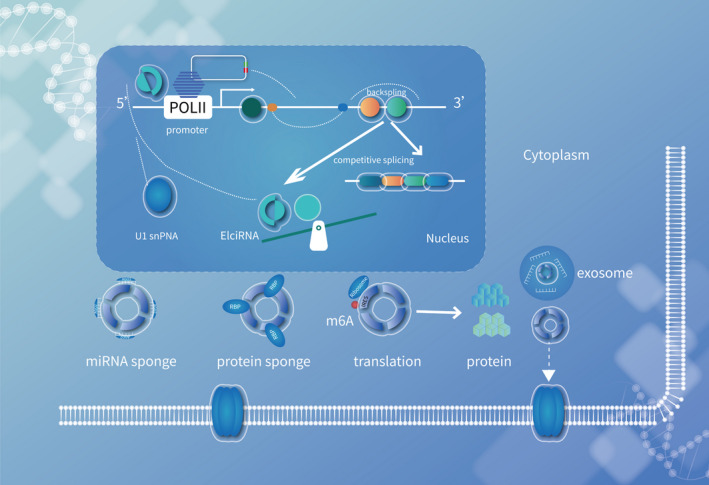
Potential function mechanisms of circRNAs. CircRNA can play as miRNA sponges and subsequently modulate its target gene expression. CircRNA can bind to several proteins to mediate their actions. Abundant m6A modification and IRES can promote a lot of circRNAs to translation
Aberrant activation of oncogenes (eg, COPS3, PIK3CA, CARD11, MYC) and inactivation of tumour suppressor genes (eg, TP53, RB1, ATRX, DLG2, BRCA1/2) by somatic mutations and epigenetic mechanisms play a pivotal pathogenic role in osteosarcoma. 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 Aside from alterations in these protein‐coding genes, it has now been realized that dysregulation of non‐coding RNAs (ncRNAs), including microRNAs (miRNAs), long non‐coding RNAs (lncRNAs) and the recently discovered circular RNAs (circRNAs), is crucial to the initiation and progression of osteosarcoma. 2 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 CircRNAs are single‐stranded RNAs that form covalently closed loops and function as an important regulatory element of the genome through multiple machineries, including transcription regulation, modulation of alternative splicing, sponging of miRNAs and direct interactions with RNA‐binding proteins. 47 , 48 , 49 , 50 , 51 , 52 A subset of circRNAs also retains protein‐coding capacity and was found to be translated in vivo. 53 , 54 , 55
A growing body of evidence now indicates that circRNA expression is dysregulated in most, if not all, types of cancer, including glioma, gastric cancer, bladder cancer, hepatocellular carcinoma, lung cancer, gallbladder carcinoma and renal cell carcinoma, in which circRNA deregulation perturbs cancer‐related phenotypes, such as cell survival, proliferation, differentiation, metabolism and invasion/metastasis. 49 , 51 , 52 , 54 , 56 , 57 , 58 , 59 , 60 , 61 , 62 Recently, an increasing number of studies suggested that circRNAs also played critical roles in osteosarcoma. 63 , 64 , 65 For instance, circLRP6 was shown to promote osteosarcoma progression through inhibiting APC and KLF2 expression. 66 Zhao et al 67 also reported that circSAMD4A increased osteosarcoma growth by sponging miR‐1244 and thereby derepressing MDM2 expression. Previously, Zhang et al 68 elegantly summarized the roles of circRNAs in osteosarcoma. As circRNA research is a rapidly evolving field, over a dozen of original articles related to circRNAs in osteosarcoma have been published since then. It is therefore timely to update the recent progress.
In this review, we first summarize circRNA profiling studies on osteosarcoma so as to provide a comprehensive curation of available datasets which may facilitate subsequent studies, followed by in‐depth discussion of functions and mechanisms of action of newly discovered osteosarcoma‐related circRNAs not covered by the review of Zhang et al. 68 The potential diagnostic, prognostic and therapeutic utilities of circRNAs in the clinical management of osteosarcoma are also addressed.
2. circRNA EXPRESSION PROFILING AND INTEGRATIVE ANALYSIS IN OSTEOSARCOMA
Profiling circRNA expression using microarray or whole‐transcriptome sequencing followed by validation with reverse transcription‐quantitative PCR (RT‐qPCR) is the most frequent method to identify and confirm deregulated expression of circRNAs in specific disease states. 69 , 70 , 71 , 72 In this connection, attempts have been made to profile circRNAs specific to osteosarcoma or related to its resistance to chemotherapy.
Liu et al 65 extracted total RNA from the human osteosarcoma cell lines U2OS, MG63, HOS and 143B and the human normal osteoblast hFOB1.19, and then subjected the RNA samples to digestion with RNase R to remove linear RNAs. The enriched circRNAs were then amplified and transcribed into fluorescent complementary RNA for hybridization onto a human circRNA array. A total of 252 circRNAs were found to be differentially expressed between the human osteosarcoma cell lines and hFOB1.19 (71 upregulated and 181 downregulated in osteosarcoma cell lines). Nevertheless, among the 12 selected differentially expressed circRNAs, only the upregulation of circRNA_103801 and the downregulation of circRNA_104980 were successfully validated in both osteosarcoma cell lines and human osteosarcoma tissues by RT‐qPCR, suggesting a low validity of using cell line models to identify osteosarcoma‐specific circRNAs. For the validated circRNA_103801, its expression was found to be strongly correlated with TANC1 (tetratricopeptide repeat, ankyrin repeat and coiled‐coil containing 1), an oncogene previously reported in rhabdomyosarcoma. In another study, Xi et al 73 used the Illumina HiSeq platform to profile circRNA expression in three pairs of osteosarcoma and para‐cancerous tissues. A total of 259 circRNAs were found to be differentially expressed (32 upregulated and 127 downregulated in osteosarcoma). Among the five selected differentially expressed circRNAs identified by RNA sequencing, significant upregulation of circ_2137 and circ_20403 and downregulation of circ_32279 and circ_24831 were successfully verified in 10 independent pairs of osteosarcoma and para‐cancerous tissues, suggesting an overall high validity of the sequencing findings. Pathway enrichment analysis of genes hosting the differentially expressed circRNAs further indicated the potential involvement of phosphatidylinositol signalling, an oncogenic pathway frequently engaged in tumorigenesis. A recent study used PCR array to profile circRNAs in 30 frozen samples of osteosarcoma and their corresponding adjacent tissues. 74 The authors identified 10 differentially expressed circRNAs (six upregulated and four downregulated in osteosarcoma) with circ_001621 as the top upregulated circRNA.
Qiu et al 75 performed an integrative analysis of publicly available gene expression datasets on osteosarcoma downloaded from the Gene Expression Omnibus (GEO) database. They identified 15 downregulated circRNAs, 136 upregulated miRNAs and 52 downregulated mRNAs in osteosarcoma, among which 14 circRNAs, 24 miRNAs and 52 mRNAs formed a circRNA‐miRNA‐mRNA network. Pathway analysis revealed these mRNAs were enriched in ‘phosphoinositide 3‐kinase‐Akt signalling pathway’, ‘proteoglycans in cancer’, ‘cell cycle’ and ‘FoxO signalling pathway’. Importantly, low expression of four mRNAs (ARID5B, ELL2, PHC2 and STAT4) in the circRNA‐regulated network was associated with significantly shortened overall survival of osteosarcoma patients.
Aside from osteosarcoma‐specific circRNAs, efforts have been made to identify circRNAs pertinent to chemosensitivity in osteosarcoma. Zhu et al compared the circRNA expression profiles of three chemoresistant osteosarcoma cell lines (MG63/DXR, U2OS/DXR, KHOS/DXR) that were insensitive to cisplatin, doxorubicin and methotrexate with their cognate chemosensitive osteosarcoma cell lines (MG63, U2OS, KHOS) using next‐generation RNA sequencing. 76 A total of 80 circRNAs were found to be differentially expressed (57 upregulated and 23 downregulated in chemoresistant cell lines). Validation with RT‐qPCR indicated that nine out of 10 selected circRNAs showed consistent direction of deregulation as shown by RNA sequencing. Pathway analyses of the differentially expressed circRNAs and their parental genes revealed biological processes pertinent to chemoresistance, including glycolysis/gluconeogenesis, ABC transporters, and vascular endothelial growth factor signalling (Table 1).
Table 1.
circRNAs expression profiles in osteosarcoma
| Num | Method | Sample | Upregulated | Downregulated | Reference |
|---|---|---|---|---|---|
| 1 |
Microarray RT‐PCR |
Osteosarcoma cell lines and hFOB1.19 |
71 LncRNAs circRNA_103801 |
181 LncRNAs circRNA_104980 |
65 |
| 2 |
Microarray RT‐PCR |
Osteosarcoma tissues and para‐cancerous tissues |
32 LncRNAs circ_2137 circ_20403 |
127 LncRNAs circ_32279 circ_24831 |
73 |
| 3 |
Microarray RT‐PCR |
Osteosarcoma tissues and para‐cancerous tissues | 6 lncRNAs circ_001621 |
4 lncRNAs |
74 |
| 4 | Bioinformatics | Gene expression datasets |
15 lncRNAs |
75 | |
| 5 |
Microarray RT‐PCR |
Chemoresistant and chemosensitive osteosarcoma cell line |
57 lncRNAs |
23 lncRNAs X |
76 |
3. FUNCTIONS AND MECHANISMS OF ACTION OF NEWLY DISCOVERED circRNAs IN OSTEOSARCOMA
3.1. Oncogenic circRNAs
3.1.1. CircTCF25
circTCF25 was first identified as an oncogenic circRNA in bladder carcinoma. 77 Wang et al reported that the expression of circTCF25 was significantly higher in osteosarcoma tissues as compared with morphologically normal bone tissues from the same patients. Functionally, enforced expression of circTCF25 promoted osteosarcoma cell proliferation, migration and invasion, accompanied by corresponding alterations of phenotype‐related proteins (upregulation of cyclin D1 and CDK6 for increased cell proliferation; upregulation of MMP2, MMP9 and Vimentin and downregulation of TIMP‐1 for enhanced cell migration and invasion). Mechanistically, overexpression of circTCF25 reduced the levels of miR‐206 in osteosarcoma cells, where transfection of miR‐206 mimic abrogated circTCF25‐induced pro‐tumorigenic phenotypes and the associated protein expression. In this regard, the MEK/ERK and AKT/mTOR pathways were found to be the downstream pathways inhibited by miR‐206. 78
3.1.2. CircPVT1
CircPVT1 is a circRNA derived from the genomic region that also encodes the oncogenic lncRNA PVT1. 79 Liu et al 80 found that circPVT1 increased the invasiveness and metastatic capacity of osteosarcoma cells in vitro via promoting epithelial‐mesenchymal transition through sponging miR‐205‐5p and thereby derepressing c‐FLIP. Yan et al 81 also showed that circPVT1 played as one oncogene in development of osteosarcoma. These studies suggested that circPVT1 is an oncogenic circRNA in osteosarcoma, where it contributes to metastasis through upregulating c‐FLIP.
3.1.3. CircMMP9
CircMMP9 has been shown to promote glioblastoma. 82 Pan et al 83 found that the expression of circMMP9 was higher in osteosarcoma tissues, where high expression was associated with larger tumours and more advanced TNM staging. The overall survival of osteosarcoma patients with high tumoral expression of circMMP9 was also shorter than those with low circMMP9 expression. Functionally, knockdown of circMMP9 reduced the viability, colony‐forming ability, migration and invasion of osteosarcoma cells, suggesting an oncogenic role for circMMP9. The pro‐tumorigenic actions of circMMP9 were found to be mediated through sponging of miR‐1265 and the subsequent depression of CHI3L1 (chitinase‐3‐like protein 1). 83
3.1.4. Circ_001621
Circ_001621 was identified as the top upregulated circRNA in osteosarcoma by PCR array, and its high expression was associated with more advanced TNM staging and shortened overall survival. 74 In this regard, miR‐578 was found to be the target of circ_001621 in osteosarcoma where their expression exhibited strong negative correlation. Functional experiments and mechanistic studies revealed that circ_001621 promoted the proliferation and migration of osteosarcoma cells in vitro via attenuating miR‐558‐mediated repressing of VEGF and the downstream expression of CDK4 and MMP9. Importantly, circ_001621 enhanced the metastatic capacity of osteosarcoma to the lung and liver in vivo accompanied by the upregulating of VEGF, CDK4 and MMP9. 74 Collectively, circ_001621 is an oncogenic circRNA promoting the metastasis of osteosarcoma by regulating the miR‐558‐VEGF pathway.
3.1.5. Circ‐0000285
Circ‐0000285 is another circRNA generated from HIPK3. Zhang et al found that overexpression of circ‐0000285 increased the colony‐forming ability, proliferation and migration of osteosarcoma cells in vitro and accelerated the growth of osteosarcoma xenografts in vivo whereas knockdown of circ‐0000285 produced the opposite effects. Mechanistic investigations suggested that circ‐0000285 bound to miR‐599 to derepress TGF‐β2 (transforming growth factor‐beta 2). 84 In this connection, TGF‐β signalling is known to contribute to osteosarcoma progression by promoting angiogenesis, bone remodelling, cell migration and immune evasion. 85
3.1.6. Circ_0001658
Circ_0001658 was found to be upregulated in osteosarcoma tissues as compared with normal bone tissues. Functional experiments suggested that circ_0001658 promoted the proliferation, migration, and invasion of osteosarcoma cells and impeded apoptosis. These pro‐tumorigenic effects were mediated by sponging miR‐382‐5p and the subsequent depression of YB‐1. Taken together, circ_0001658 is an oncogenic circRNA via modulating the miR‐382‐5p/YB‐1 axis. 86
3.1.7. Circ_0102049
The expression of circ_0102049 was found to be higher in osteosarcoma tissue samples as compared with their paired non‐cancerous tissues. 87 In patients with osteosarcoma, high tumoral expression of circ_0102049 was associated with larger tumour size, lung metastasis and shortened overall survival. Gain‐of‐function and loss‐of‐function experiments indicated that circ_0102049 acted as an oncogene by promoting osteosarcoma cell proliferation, migration and invasion and inhibiting apoptosis. Mechanistically, circ_0102049 was found to upregulate MDM2 (an E3 ubiquitin‐protein ligase responsible degradation of p53) through sponging miR‐1304‐5p. To this end, knockdown of MDM2 attenuated the stimulatory effects of circ_0102049 overexpression on osteosarcoma cell viability, migration and invasion, 87 suggesting the miR‐1304‐5p/MDM2 axis is an effector pathway for the oncogenic action of circ_0102049.
3.1.8. CircEPSTI1
Tan et al 88 reported that circEPSTI1 was significantly upregulated in osteosarcoma where knockdown of this circRNA suppressed cell proliferation and migration, suggesting an oncogenic role. Mechanistically, circEPSTI1 upregulated the expression of MCL1 (myeloid cell leukaemia 1) via reducing the availability of miR‐892b. 88 These data suggested an important role for circEPSTI1‐miR‐892b‐MCL1 axis in the progression of osteosarcoma.
3.1.9. Circ‐0060428
Cao and Liu reported that Circ‐0060428 was expressed at significantly higher levels in osteosarcoma cell lines (U2OS, 143B, SAOS‐2, HOS) as compared with the human normal osteoblast hFOB1.19. 89 Functionally, knockdown of circ‐0060428 induced apoptosis of U2OS and HOS osteosarcoma cells, suggesting this circRNA functioned as an oncogenic circRNA. Mechanistic studies found that circ‐0060428 bound to miR‐375 and thereby depressing RBPJ (a transcription factor positively regulating Notch signalling). In this connection, miR‐375 inhibitor nullified the pro‐apoptotic effect of circ‐0060428 knockdown. 89 These findings suggest that upregulation of circ‐0060428 could confer apoptosis resistance to osteosarcoma cells by sponging miR‐375. Nevertheless, whether RBPJ is functionally involved in the oncogenic action of circ‐0060428 remains unclear. Moreover, the upregulation of circ‐0060428 needs to be confirmed in human osteosarcoma tissues.
3.1.10. Circ_ANKIB1
Du et al 90 illustrated that circ_ANKIB1 directly sponged miR‐19b in the osteosarcoma cell and Circ_ANKIB1 enhanced expression of miR‐19b and suppressed SOCS3 expression, which is its downstream gene. Knockdown of miR‐19b or circ_ANKIB1 suppressed cell invasion and growth and induced cell apoptosis via modulating STAT3 pathway. These data indicated that circ_ANKIB1 acted as one oncogene and potential treatment target for osteosarcoma.
3.1.11. CircMYO10
Chen et al 91 illustrated that miR‐370‐3p was decreased and circMYO10 was overexpressed in osteosarcoma samples and cells. Inhibition expression of circMYO10 suppressed EMT development and growth through regulating miR‐370‐3p/RUVBL1/β‐catenin/LEF1 axis. These results revealed that circMYO10 played as one oncogene role in progression of osteosarcoma.
3.1.12. CircLRP6
Previous study noted that circLRP6 level was overexpressed in osteosarcoma samples and was correlated with poor overall survival and shorter disease‐free survival. 92 Knockdown of circLRP6 inhibited cell migration, invasion and growth and induced cell cycle arrested and apoptosis. The interaction between EZH2 and LSD1 with circLRP6 regulates their binding to promoter regions of APC and KLF2. Inhibition expression of circLRP6 decreased binding abilities of EZH2, LSD1 to APC, KLF2. Furthermore, the oncogene effect of circLRP6 on osteosarcoma can be reversed with APC. These data indicated that circLRP6 served as one oncogene through binding to EZH2 and LSD1 to suppress APC and KLF2 expression.
3.1.13. CircSAMD4A
Zhao et al 67 noted that circSAMD4 was upregulated in osteosarcoma specimens compared with adjacent non‐cancerous specimens. Ectopic expression of circSAMD4A induced osteosarcoma cell growth in vitro and in vivo and increased cells stemness characteristics. Moreover, they illustrated that circSAMD4A sponged miR‐1244 and identified MDM2 was one direct target gene of miR‐1244. Their data illustrated that miR‐1244/MDM2/ circSAMD4A modulator loop may be one treatment target for osteosarcoma.
3.1.14. Circ‐0001785
Previous reference noted that circ‐0001785 was overexpressed in cell lines of osteosarcoma. 93 Downregulation expression of circ‐0001785 inhibited cell growth and enhanced osteosarcoma cell apoptosis. In addition, they showed that circ‐0001785 can sponge miR‐1200 expression and suppress HOXB2 expression, which is one target gene of miR‐1200. They showed that circ‐0001785 modulated Bcl‐2 pathway and Akt/PI3K signalling in the osteosarcoma. These results suggested that circ‐0001785 modulates osteosarcoma pathogenesis via regulating miR‐1200 to enhance HOXB2 expression.
3.1.15. Circ_ORC2
Li et al 94 illustrated that circ_ORC2 was distributed in cell cytoplasm and overexpressed in several cell lines of osteosarcoma. Circ_ORC2 can sponge miR‐19a expression, and knockdown of circ_ORC2 suppressed miR‐19a and enhanced its target gene PTEN expression. Knockdown of circ_ORC2 inhibited cell invasion and growth and enhanced cell apoptosis. Thus, they indicated that circ_ORC2 binds with the miR‐19a and increased its expression, then enhancing Akt pathway and suppressing its downstream gene PTEN expression to induce cell invasion and growth. These data provided a new oncogene circ_ORC2 for osteosarcoma.
3.1.16. CircRNA_100876
Jin et al 95 showed that circRNA_100876 level was overexpressed in osteosarcoma tissues and was associated with differentiation degree and size of tumour. CircRNA_100876 knockdown suppressed tumour growth in vivo and in vitro. Inhibition expression of circRNA_100876 suppressed cell growth and induced cell apoptosis and decreased cell cycle. Moreover, the expression of circRNA_100876 was negatively correlated with miR‐136 expression. Knockdown of miR‐136 can reverse the inhibition of cell growth induced with silencing circRNA_100876. These data indicate that circRNA_100876 may act as one promising treatment target and biomarker for osteosarcoma.
3.1.17. CircTADA2A
Wu et al 96 revealed that circTADA2A was overexpressed in osteosarcoma cells and samples and inhibition of circTADA2A suppressed tumour metastasis and tumorigenesis in vivo and inhibited cell invasion, growth and migration in vitro. Moreover, they noted that circTADA2A sponged miR‐203a‐3p expression and enhanced CREB3 expression, which was found as one driver gene of osteosarcoma. CREB3 overexpression or knockdown of miR‐203a‐3p can reverse circTADA2A silencing‐leading impairment of tumour behaviour. These results suggested that circTADA2A acted as one oncogene through regulating miR‐203a‐3p/CREB3 axis in osteosarcoma.
3.1.18. Circ_0000885
Zhu et al 97 indicated that circ_0000885 expression was upregulated in serum samples and tissue from osteosarcoma patients and higher expression of circ_0000885 was correlated with Enneking stage. Cases with high tumour and serum levels of the circ_0000885 had the lower rates of overall survival and disease‐free survival. These results suggested that circ_0000885 may act as one diagnostic biomarker for the osteosarcoma.
3.2. Tumour‐suppressing circRNAs
3.2.1. Circ0021347
The expression of circ0021347 was lower in human osteosarcoma tissues and cell lines than their normal counterparts. In patients with osteosarcoma, the low expression of circ0021347 was associated with more advanced tumour‐node‐metastasis (TNM) staging and shortened overall survival, suggesting circ0021347 functions as a tumour suppressor. 98 Nevertheless, the mechanism of action of circ0021347 is presently unclear.
3.2.2. Circ_0000190
Circ_0000190 has been shown to inhibit the progression of multiple myeloma through sponging miR‐767‐5p. Li et al reported that circ_0000190 was expressed at significantly lower levels in human osteosarcoma tissues and cell lines as compared with para‐cancerous normal tissues and hFOB1.19 normal osteoblasts, respectively. 99 Enforced expression of circ_0000190 suppressed osteosarcoma cell proliferation, migration and invasion. Mechanistically, circ_0000190 bound to miR‐767‐5p and thereby derepressing the downstream target TET1. The levels of circ_0000190 in extracellular nanovesicles isolated from plasma of osteosarcoma patients were also significantly lower than those from healthy subjects, suggesting the potential diagnostic use of circulating circ_0000190. In this regard, low abundance of circ_0000190 in extracellular nanovesicles could distinguish cases from controls with an AUROC of 0.8894. 99
3.2.3. Circ‐LARP4
Hu et al 100 showed that circ‐LARP4 level was decreased in osteosarcoma tissues compared to non‐tumour samples and circ‐LARP4 was associated with Enneking stage. Patients with high circ‐LARP4 level indicated enhanced tumour cell necrosis rates to chemotherapy after resection. The higher expression of circ‐LARP4 was associated with prolonged overall survival and disease‐free survival. Ectopic expression of circ‐LARP4 enhanced MG63 cells chemosensitivity to doxorubicin and cisplatin but not the methotrexate. Moreover, overexpression of miR‐424 decreased the chemosensitivity effect in circ‐LARP4 overexpression handled MG63 cells. Their data suggested that high expression of circ‐LARP4 was associated with prolonged survival profiles and decreased Enneking stage and overexpression of circ‐LARP4 enhanced chemosensitivity to doxorubicin and cisplatin through sponging miR‐424 in development of osteosarcoma.
3.3. Circ‐ITCH, a circRNA with an ambiguous role
Circ‐ITCH was found to act as a tumour‐suppressing circRNA in oesophageal squamous cell carcinoma, 101 colorectal cancer 102 and lung cancer. 103 In contrast with its inhibitory role in these cancer types, Li et al 104 reported that cir‐ITCH was expressed at significantly higher levels in a panel of osteosarcoma cell lines as compared with hFOB1.19 normal osteoblasts. Importantly, enforced expression of cir‐ITCH promoted osteosarcoma cell proliferation, migration and invasion whereas knockdown of cir‐ITCH produced the opposite effects. The oncogenic action of cir‐ITCH was mediated by the sponging of miR‐7 and the subsequent enhancement of epidermal growth factor receptor (EGFR) expression and activation the downstream MEK‐ERK cascade. To this end, erlotinib (an EGFR tyrosine kinase inhibitor) nullified the pro‐migratory and pro‐invasive effects of cir‐ITCH. 104 Ren et al 105 also found that circ‐ITCH played asa tumour suppressor in progression of osteosarcoma by modulating miR‐22 (Tables 2 and 3; Figures 2 and 3).
Table 2.
Dysregulated circRNAs in osteosarcoma
| Name | Dysregulation | Sponge target | Function | Related gene | Role | Reference |
|---|---|---|---|---|---|---|
| circTCF25 | Upregulated | miR‐206 | Proliferation, migration and invasion |
MEK/ERK AKT/mTOR |
Oncogenic | 78 |
| CircPVT1 | Upregulated |
miR‐205‐5p miR‐526b |
Invasion EMT |
c‐FLIP FOXC2 | Oncogenic | 80, 81 |
| CircMMP9 | Upregulated | miR‐1265 | Proliferation, colony, migration, invasion | CHI3L1 | Oncogenic | 83 |
| Circ_001621 | Upregulated | miR‐578 | Proliferation migration |
VEGF CDK4 MMP9 |
Oncogenic | 74 |
| Circ‐0000285 | Upregulated | miR‐599 | Colony proliferation migration | TGF‐β | Oncogenic | 84 |
| Circ_0001658 | Upregulated | miR‐382‐5p | Proliferation migration invasion | YB‐1 | Oncogenic | 86 |
| circ_0102049 | Upregulated | miR‐1304‐5p | Proliferation migration invasion apoptosis | MDM2 | Oncogenic | 87 |
| circEPSTI1 | Upregulated | miR‐892b | Proliferation migration | MCL1 | Oncogenic | 88 |
| Circ‐0060428 | Upregulated | miR‐375 | Apoptosis | RBPJ | Oncogenic | 89 |
| circ_ANKIB1 | Upregulated | miR‐19b | Invasion growth apoptosis | SOCS3 STAT3 | Oncogenic | 90 |
| circMYO10 | Upregulated | miR‐370‐3p |
EMT growth |
RUVBL1 β‐catenin LEF1 |
Oncogenic | 91 |
| circLRP6 | Upregulated |
Migration invasion growth cell cycle apoptosis |
EZH2 LSD1 APC KLF2 |
Oncogenic | 92 | |
| circSAMD4 | Upregulated | miR‐1244 |
Growth stemness |
MDM2 | Oncogenic | 67 |
| circ‐0001785 | Upregulated | miR‐1200 | Growth apoptosis |
Akt/PI3K Bcl‐2 HOXB2 |
Oncogenic | |
| circ_ORC2 | Upregulated | miR‐19a | Invasion growth apoptosis |
PTEN Akt |
Oncogenic | 94 |
| circRNA_100876 | Upregulated | miR‐136 | Growth apoptosis cycle | Oncogenic | 95 | |
| circTADA2A | Upregulated | miR‐203a‐3p | Metastasis invasion growth migration | CREB3 | Oncogenic | 96 |
| circ_0000885 | Upregulated | Oncogenic | 97 | |||
| circ0021347 | Downregulated | Tumour suppressor | 98 | |||
| Circ_0000190 | Downregulated | miR‐767‐5p | Proliferation migration invasion | TET1 | Tumour suppressor | 99 |
| circ‐LARP4 | Downregulated | miR‐424 | Chemosensitivity | Tumour suppressor | 101 | |
| Circ‐ITCH | Downregulated |
miR‐7 miR‐2 |
Proliferation migration invasion |
MEK ERK |
Tumour suppressor | 104, 105 |
Table 3.
CircRNAs as diagnostic and prognostic biomarkers in Osteosarcoma
| Name | Deregulation | Function | Clinicopathological association | Reference |
|---|---|---|---|---|
| circMMP9 | Upregulated | Independent prognostic indicator | Larger tumours advanced TNM staging overall survival | 83 |
| Circ_001621 | Upregulated | Independent prognostic indicator | TNM staging overall survival | 74 |
| circ_0102049 | Upregulated | Independent prognostic indicator | Tumour size lung metastasis overall survival | 87 |
| circLRP6 | Upregulated | Independent prognostic indicator | Overall survival disease‐free survival | 92 |
| circRNA_100876 | Upregulated | Independent prognostic indicator | Differentiation degree tumour size | 95 |
| circ_0000885 | Upregulated | Independent prognostic Indicator and Diagnosis biomarker | Enneking stage overall survival disease‐free survival | 97 |
| circ0021347 | Downregulated | Independent prognostic indicator and diagnosis biomarker | Advanced TNM staging overall survival | 98 |
| circ‐LARP4 | Downregulated | Independent prognostic indicator and diagnosis biomarker | Enneking stage overall survival and disease‐free survival | 101 |
Figure 2.
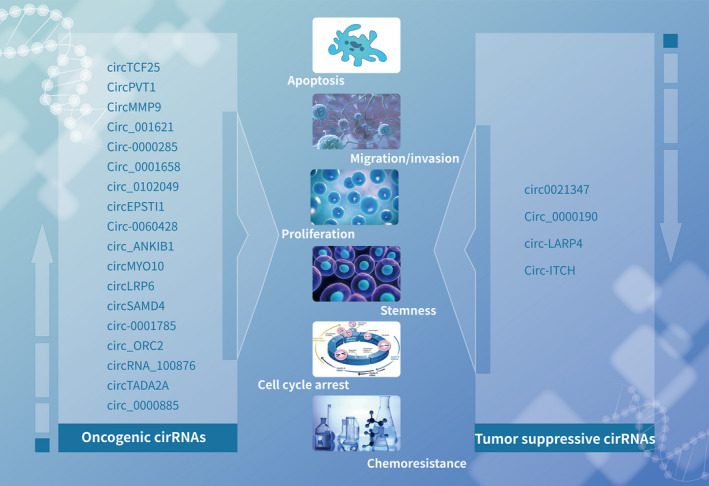
Oncogenic and tumour suppressor circRNAs in osteosarcoma and their influence on biological processes, including proliferation, apoptosis, chemoresistance, migration/invasion, cell cycle arrest and stemness. The left panel shows oncogenic circRNAs with upregulated expression, while the right panel indicates tumour‐suppressive circRNAs with downregulated expression in osteosarcoma
Figure 3.
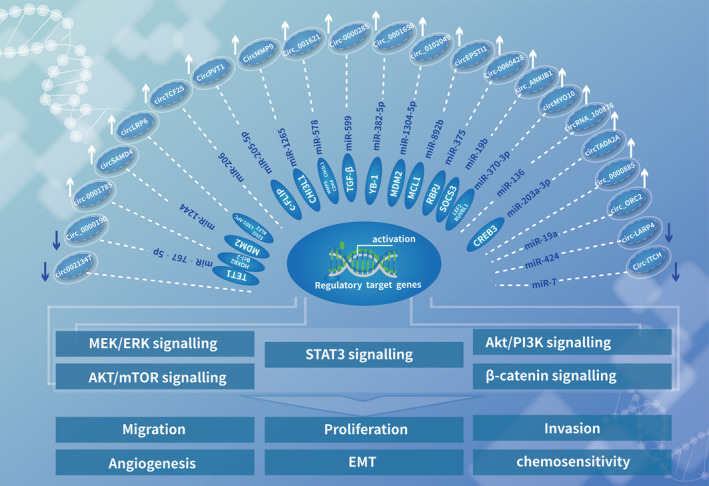
The representative diagram of circRNA mediated ceRNA network and oncogene signalling pathways. The current study in osteosarcoma highlights the regulatory relationship of the circRNA‐miRNA‐mRNA network for different signalling pathways. Multiple identified circRNAs function as miRNA sponge and subsequently upregulate relevant target genes expression level
4. CONCLUSIONS AND FUTURE PERSPECTIVES
Osteosarcoma is one multi‐factor and multi‐step comprehensive disease, pathogenesis mechanism is still unclear, and more efforts are in urgent need. CircRNAs function as an important regulatory element of the genome via several machineries, including transcription regulation, modulation of alternative splicing, sponging of miRNAs and direct interactions with RNA‐binding proteins. CircRNAs may play as tumour suppressors or activators in tumour and have been applied in modulating cancer metastasis and chemoresistance. As summarized in our review, circRNAs are acting critical roles in many biological processes in the procedures in the progression and development of osteosarcoma including cell apoptosis, invasion, growth, differentiation, migration, drug resistance and cachexia. As one crucial biomarker for prognosis and diagnosis of osteosarcoma, circRNA is one potential target for therapy of osteosarcoma. This use of circRNAs as therapeutics has vast tremendously and may supply a new method to treat osteosarcoma.
Here are some future suggestions for circRNAs study. Firstly, more exact mechanisms about how circRNAs participated in progression of osteosarcoma need to further research, potentially via using online databases. Secondly, further study on circRNAs in osteosarcoma needs to use non‐invasive specimens such as saliva, urine and blood. Thirdly, since the purpose of circRNA study is to use circRNAs for clinical application for osteosarcoma, generous clinical markers experiments and cell function mechanism studies are required in future.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
ZL, XYL, DRX, XC and SGL collected the related paper. ZL, XYL and DRX drafted and wrote the manuscript. LZ, MTVC and WKKW revised the manuscript. SGL participated in the design of the review and helped to draft and revise the manuscript. All authors read and approved the final manuscript.
Li Z, Li X, Xu D, et al. An update on the roles of circular RNAs in osteosarcoma. Cell Prolif. 2021;54:e12936 10.1111/cpr.12936
Li, Li and Xu are the co‐first authors.
DATA AVAILABILITY STATEMENT
Research data are not shared.
REFERENCES
- 1. Li Z, Yu X, Shen JX. Long non‐coding RNAs: emerging players in osteosarcoma. Tumor Biol. 2016;37(3):2811‐2816. 10.1007/s13277-015-4749-4 [DOI] [PubMed] [Google Scholar]
- 2. Li Z, Shen JX, Chan MTV, Wu WKK. MicroRNA‐379 suppresses osteosarcoma progression by targeting PDK1. J Cell Mol Med. 2017;21(2):315‐323. 10.1111/jcmm.12966 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3. Zhao J, Gao Z, Zhang C, Wu H, Gu R, Jiang R. Long non‐coding RNA ASBEL promotes osteosarcoma cell proliferation, migration and invasion by regulating microRNA‐21. J Cell Biochem. 2018;119(8):6461‐6469. 10.1002/jcb.26671 [DOI] [PubMed] [Google Scholar]
- 4. Yang C, Wu K, Wang S, Wei G. Long non‐coding RNA XIST promotes osteosarcoma progression by targeting YAP via miR‐195‐5p. J Cell Biochem. 2018;119(7):5646‐5656. 10.1002/jcb.26743 [DOI] [PubMed] [Google Scholar]
- 5. Xu RD, Feng F, Yu XS, Liu ZD, Lao LF. LncRNA SNHG4 promotes tumour growth by sponging miR‐224‐3p and predicts poor survival and recurrence in human osteosarcoma. Cell Prolif. 2018;51(6): 10.1111/cpr.12515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ba ZW, Gu LL, Hao SN, Wang XF, Cheng ZP, Nie GC. Downregulation of lncRNA CASC2 facilitates osteosarcoma growth and invasion through miR‐181a. Cell Prolif. 2018;51(1): 10.1111/cpr.12409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Duan Z, Choy E, Harmon D, et al. MicroRNA‐199a‐3p is downregulated in human osteosarcoma and regulates cell proliferation and migration. Mol Cancer Ther. 2011;10(8):1337‐1345. 10.1158/1535-7163.MCT-11-0096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao G, Cai C, Yang T, et al. MicroRNA‐221 induces cell survival and cisplatin resistance through PI3K/Akt pathway in human osteosarcoma. PLoS One. 2013;8(1):e53906 10.1371/journal.pone.0053906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sun XH, Geng XL, Zhang J, Zhang C. miRNA‐646 suppresses osteosarcoma cell metastasis by downregulating fibroblast growth factor 2 (FGF2). Tumor Biol. 2015;36(3):2127‐2134. 10.1007/s13277-014-2822-z. [DOI] [PubMed] [Google Scholar]
- 10. Wang J, Xu G, Shen F, Kang Y. miR‐132 targeting cyclin E1 suppresses cell proliferation in osteosarcoma cells. Tumor Biol. 2014;35(5):4859‐4865. 10.1007/s13277-014-1637-2. [DOI] [PubMed] [Google Scholar]
- 11. Joo MW, Shin SH, Kang YK, et al. Osteosarcoma in Asian populations over the age of 40 years: a multicenter study. Ann Surg Oncol. 2015;22(11):3557‐3564. 10.1245/s10434-015-4414-6 [DOI] [PubMed] [Google Scholar]
- 12. Parry MC, Laitinen M, Albergo J, et al. Osteosarcoma of the pelvis. Bone Joint J. 2016;98B(4):555‐563. 10.1302/0301-620X.98B3.36583 [DOI] [PubMed] [Google Scholar]
- 13. Liu CL, Tang YW, Li M, et al. Clinical characteristics and prognoses of six patients with multicentric giant cell tumor of the bone. Oncotarget. 2016;7(50):83795‐83805. 10.18632/oncotarget.13057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cox TR, Erler JT, Rumney RMH. Established models and new paradigms for hypoxia‐driven cancer‐associated bone disease. Calcif Tissue Int. 2018;102(2):163‐173. 10.1007/s00223-017-0352-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin E, Senders JT, ter Wengel PV, Smith TR, Broekman MLD. Treatment and survival of osteosarcoma and Ewing sarcoma of the skull: a SEER database analysis. Acta Neurochir. 2019;161(2):317‐325. 10.1007/s00701-018-3754-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Green D, Mohorianu I, McNamara I, Dalmay T, Fraser WD. miR‐16 is highly expressed in Paget's associated osteosarcoma. Endocr Relat Cancer. 2017;24(5):L27‐L31. 10.1530/Erc-16-0487 [DOI] [PubMed] [Google Scholar]
- 17. Li Z, Shen JX, Chan MTV, Wu WKK. The long non‐coding RNA SPRY4‐IT1: an emerging player in tumorigenesis and osteosarcoma. Cell Prolif. 2018;51(4): 10.1111/cpr.12446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Song B, Wang Y, Titmus MA, et al. Molecular mechanism of chemoresistance by miR‐215 in osteosarcoma and colon cancer cells. Mol Cancer. 2010;9(1):96 10.1186/1476-4598-9-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shen P, Cheng YF. Long noncoding RNA lncARSR confers resistance to Adriamycin and promotes osteosarcoma progression. Cell Death Dis. 2020;11(5):Article number: 362. 10.1038/S41419-020-2573-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiang WX, Xia J, Xie SD, et al. Long non‐coding RNAs as a determinant of cancer drug resistance: towards the overcoming of chemoresistance via modulation of lncRNAs. Drug Resist Update. 2020;50:100683 10.1016/J.Drup.2020.100683 [DOI] [PubMed] [Google Scholar]
- 21. Inkol JM, Poon AC, Mutsaers AJ. Inhibition of copper chaperones sensitizes human and canine osteosarcoma cells to carboplatin chemotherapy. Vet Comp Oncol. 2020; 10.1111/vco.12579 [DOI] [PubMed] [Google Scholar]
- 22. Fanelli M, Tavanti E, Patrizio MP, et al. Cisplatin resistance in osteosarcoma: in vitro validation of candidate DNA repair‐related therapeutic targets and drugs for tailored treatments. Front Oncol. 2020;10:331 10.3389/Fonc.2020.00331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang B, Zhang Y, Li RZ, Li JZ, Lu XC, Zhang Y. The efficacy and safety comparison of first‐line chemotherapeutic agents (high‐dose methotrexate, doxorubicin, cisplatin, and ifosfamide) for osteosarcoma: a network meta‐analysis. J Orthop Surg Res. 2020;15(1):51 10.1186/s13018-020-1576-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen K, Jiao JB, Xue JW, et al. Ginsenoside CK induces apoptosis and suppresses proliferation and invasion of human osteosarcoma cells through the PI3K/mTOR/p70S6K1 pathway. Oncol Rep. 2020;43(3):886‐896. 10.3892/or.2020.7460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rainusso N, Wang LL, Yustein JT. The adolescent and young adult with cancer: state of the art – bone tumors. Curr Oncol Rep. 2013;15(4):296‐307. 10.1007/s11912-013-0321-9 [DOI] [PubMed] [Google Scholar]
- 26. Yan TQ, Wunder JS, Gokgoz N, et al. COPS3 amplification and clinical outcome in osteosarcoma. Cancer. 2007;109(9):1870‐1876. 10.1002/cncr.22595 [DOI] [PubMed] [Google Scholar]
- 27. Zhang F, Yan TQ, Guo W, et al. Novel oncogene COPS3 interacts with Beclin1 and Raf‐1 to regulate metastasis of osteosarcoma through autophagy. J Exp Clin Canc Res. 2018;37:135 10.1186/S13046-018-0791-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Dartel M, Hulsebos TJM. Amplification and overexpression of genes in 17p11.2 similar to p12 in osteosarcoma. Cancer Genet Cytogenet. 2004;153(1):77‐80. 10.1016/j.cancergencyto.2004.03.007 [DOI] [PubMed] [Google Scholar]
- 29. He ML, Wu Y, Zhao JM, Wang Z, Chen YB. AKT gene polymorphisms in susceptibility to osteosarcoma in a Chinese population. Asian Pac J Cancer Prev. 2013;14(9):5117‐5122. 10.7314/APJCP.2013.14.9.5117 [DOI] [PubMed] [Google Scholar]
- 30. Gobin B, Baud'Huin M, Lamoureux F, et al. BYL719, a new alpha‐specific PI3K inhibitor: Single administration and in combination with conventional chemotherapy for the treatment of osteosarcoma. Int J Cancer. 2015;136(4):784‐796. 10.1002/ijc.29040 [DOI] [PubMed] [Google Scholar]
- 31. Gao JJ, Ma S, Yang F, et al. miR‐193b exhibits mutual interaction with MYC, and suppresses growth and metastasis of osteosarcoma. Oncol Rep. 2020;44(1):139‐155. 10.3892/or.2020.7601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Samsa WE, Mamidi MK, Bashur LA, et al. The crucial p53‐dependent oncogenic role of JAB1 in osteosarcoma in vivo. Oncogene. 2020;39(23):4581‐4591. 10.1038/s41388-020-1320-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cabrera‐Andrade A, Lopez‐Cortes A, Jaramillo‐Koupermann G, et al. Gene prioritization through consensus strategy, enrichment methodologies analysis, and networking for osteosarcoma pathogenesis. Int J Mol Sci. 2020;21(3):1053 10.3390/Ijms21031053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Folk WP, Homlar K, Sakamuro D. PD‐0332991/palbociclib facilitates the physical interaction between RB1 and BIN1 to increase the vulnerability of pediatric osteosarcoma cells to PARP inhibition. Can Res. 2018;78(19). 10.1158/1538-7445.PEDCA17-B34 [DOI] [Google Scholar]
- 35. Smolle MA, Heitzer E, Geigl JB, et al. A novel mutation in ATRX associated with intellectual disability, syndromic features, and osteosarcoma. Pediatr Blood Cancer. 2017;64(10): 10.1002/pbc.26522 [DOI] [PubMed] [Google Scholar]
- 36. Yost KE, Soper SFC, Walker RL, et al. Rapid and reversible suppression of ALT by DAXX in osteosarcoma cells. Sci Rep. 2019;9(1):4544 10.1038/S41598-019-41058-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dhir A, Li R, Li GL, Dean J, Robin NH, Alva E. Simultaneous osteosarcoma and renal cell carcinoma with BRCA1 mutation in a young male adult with prior oligodendroglioma. Pediatr Blood Cancer. 2020;67(3): 10.1002/pbc.28116 [DOI] [PubMed] [Google Scholar]
- 38. Tian ZZ, Guo XJ, Zhao YM, Fang Y. Decreased expression of long non‐coding RNA MEG3 acts as a potential predictor biomarker in progression and poor prognosis of osteosarcoma. Int J Clin Exp Pathol. 2015;8(11):15138‐15142. [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Y, Duan G, Feng S. MicroRNA‐301a modulates doxorubicin resistance in osteosarcoma cells by targeting AMP‐activated protein kinase alpha 1. Biochem Biophys Res Commun. 2015;459(3):367‐373. 10.1016/j.bbrc.2015.02.101. [DOI] [PubMed] [Google Scholar]
- 40. Zhou C, Tan W, Lv H, Gao F, Sun J. Hypoxia‐inducible microRNA‐488 regulates apoptosis by targeting Bim in osteosarcoma. Cell Oncol (Dordr). 2016;39(5):463‐471. 10.1007/s13402-016-0288-2 [DOI] [PubMed] [Google Scholar]
- 41. Silva G, Aboussekhra A. p16(INK4A) inhibits the pro‐metastatic potentials of osteosarcoma cells through targeting the ERK pathway and TGF‐beta1. Mol Carcinog. 2016;55(5):525‐536. 10.1002/mc.22299 [DOI] [PubMed] [Google Scholar]
- 42. Xing B, Ren C. Tumor‐suppressive miR‐99a inhibits cell proliferation via targeting of TNFAIP8 in osteosarcoma cells. Am J Transl Res. 2016;8(2):1082‐1090. [PMC free article] [PubMed] [Google Scholar]
- 43. Wang Y, Shi S, Zhang Q, Dong H, Zhang J. MicroRNA‐206 upregulation relieves circTCF25‐induced osteosarcoma cell proliferation and migration. J Cell Physiol. 2020; 10.1002/jcp.29570 [DOI] [Google Scholar]
- 44. Li S, Liu F, Pei Y, Wang W, Zheng K, Zhang X. Long noncoding RNA enhances the malignant characteristics of osteosarcoma by acting as a competing endogenous RNA on microRNA‐376a thereby upregulating dickkopf‐1. Aging. 2019;11(18):7678‐7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yu D, Xu X, Li S, Zhang K. LINC00514 drives osteosarcoma progression through sponging microRNA‐708 and consequently increases URGCP expression. Aging. 2020;12(8):6793‐6807. 10.18632/aging.103043 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46. Gui D, Cao H. Long non‐coding RNA CDKN2B‐AS1 promotes osteosarcoma by increasing the expression of MAP3K3 via sponging miR‐4458. In Vitro Cell Dev Biol Anim. 2020;56(1):24‐33. 10.1007/s11626-019-00415-7 [DOI] [PubMed] [Google Scholar]
- 47. Zhang PF, Gao C, Huang XY, et al. Cancer cell‐derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti‐PD1 therapy in hepatocellular carcinoma. Mol Cancer. 2020;19(1):110 10.1186/s12943-020-01222-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lu Q, Liu TY, Feng HJ, et al. Circular RNA circSLC8A1 acts as a sponge of miR‐130b/miR‐494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer. 2019;18(1):111 10.1186/S12943-019-1040-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang XX, Li Z, Zhang Q, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR‐198 suppression. Mol Cancer. 2019;18(1):71 10.1186/s12943-019-0969-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu K, Liao X, Gong YL, et al. Circular RNA F‐circSR derived from SLC34A2‐ROS1 fusion gene promotes cell migration in non‐small cell lung cancer. Mol Cancer. 2019;18(1):98 10.1186/s12943-019-1028-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ding LX, Zhao YY, Dang SW, et al. Circular RNA circ‐DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation o transcription factor SOX4. Mol Cancer. 2019;18(1):45 10.1186/s12943-019-1006-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang SH, Zhang YJ, Cai Q, et al. Circular RNA FOXP1 promotes tumor progression and Warburg effect in gallbladder cancer by regulating PKLR expression. Mol Cancer. 2019;18(1):145 10.1186/S12943-019-1078-Z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang QG, Wang WW, Zhou QB, et al. Roles of circRNAs in the tumour microenvironment. Mol Cancer. 2020;19(1):14 10.1186/s12943-019-1125-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang F, Hu AP, Li D, et al. Circ‐HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation. Mol Cancer. 2019;18(1):158 10.1186/S12943-019-1094-Z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kong Y, Li MT, Luo YM, et al. circNFIB1 inhibits lymphangiogenesis and lymphatic metastasis via the miR‐486‐5p/PIK3R1/VEGF‐C axis in pancreatic cancer. Mol Cancer. 2020;19(1):82 10.1186/S12943-020-01205-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. He JH, Huang ZY, He ML, et al. Circular RNA MAPK4 (circ‐MAPK4) inhibits cell apoptosis via MAPK signaling pathway by sponging miR‐125a‐3p in gliomas. Mol Cancer. 2020;19(1):17 10.1186/s12943-019-1120-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jie MM, Wu YR, Gao MY, et al. CircMRPS35 suppresses gastric cancer progression via recruiting KAT7 to govern histone modification. Mol Cancer. 2020;19(1):56 10.1186/S12943-020-01160-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu P, Li X, Guo X, et al. Circular RNA DOCK1 promotes bladder carcinoma progression via modulating circDOCK1/hsa‐miR‐132‐3p/Sox5 signalling pathway. Cell Prolif. 2019;52(4):e12614 10.1111/cpr.12614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wei Y, Zhang Y, Meng Q, Cui L, Chang X. Hypoxia‐induced circular RNA has_circRNA_403658 promotes bladder cancer cell growth through activation of LDHA. Am J Transl Res. 2019;11(11):6838‐6849. [PMC free article] [PubMed] [Google Scholar]
- 60. Tian F, Yu C, Wu M, Wu X, Wan L, Zhu X. MicroRNA‐191 promotes hepatocellular carcinoma cell proliferation by has_circ_0000204/miR‐191/KLF6 axis. Cell Prolif. 2019;52(5):e12635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Huang X, He M, Huang S, et al. Circular RNA circERBB2 promotes gallbladder cancer progression by regulating PA2G4‐dependent rDNA transcription. Mol Cancer. 2019;18(1):166 10.1186/S12943-019-1098-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li JF, Huang CC, Zou YF, Ye J, Yu J, Gui YT. CircTLK1 promotes the proliferation and metastasis of renal cell carcinoma by sponging miR‐136‐5p. Mol Cancer. 2020;19(1):103 10.1186/s12943-020-01225-2 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63. Soghli N, Qujeq D, Yousefi T, Soghli N. The regulatory functions of circular RNAs in osteosarcoma. Genomics. 2020;112(4):2845‐2856. 10.1016/j.ygeno.2020.03.024 [DOI] [PubMed] [Google Scholar]
- 64. Wang CC, Jing JH, Cheng L. Emerging roles of non‐coding RNAs in the pathogenesis, diagnosis and prognosis of osteosarcoma. Invest New Drug. 2018;36(6):1116‐1132. 10.1007/s10637-018-0624-7 [DOI] [PubMed] [Google Scholar]
- 65. Liu WH, Zhang JJ, Zou CY, et al. Microarray expression profile and functional analysis of circular RNAs in Osteosarcoma. Cell Physiol Biochem. 2017;43(3):969‐985. 10.1159/000481650 [DOI] [PubMed] [Google Scholar]
- 66. Zheng SN, Qian ZY, Jiang F, et al. CircRNA LRP6 promotes the development of osteosarcoma via negatively regulating KLF2 and APC levels. Am J Transl Res. 2019;11(7):4126‐4138. [PMC free article] [PubMed] [Google Scholar]
- 67. Zhao YB, Zhang J. CircSAMD4A accelerates cell proliferation of osteosarcoma by sponging miR‐1244 and regulating MDM2 mRNA expression. Biochem Biophys Res Comm. 2019;516(1):102‐111. 10.1016/j.bbrc.2019.05.182 [DOI] [PubMed] [Google Scholar]
- 68. Zhang Y, Li JL, Wang YS, Jing JH, Li J. The roles of circular RNAs in osteosarcoma. Med Sci Monitor. 2019;25:6378‐6382. 10.12659/Msm.915559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vo J, Cieslik M, Zhang Y, et al. The landscape of circular RNA in Cancer. Cell. 2019;176(4):869‐81.e13. 10.1016/j.cell.2018.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N‐methyladenosine. Cell Res. 2017;27(5):626‐641. 10.1038/cr.2017.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. You X, Vlatkovic I, Babic A, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18(4):603‐610. 10.1038/nn.3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen Y, Chen C, Mai T, et al. Genome‐wide, integrative analysis of circular RNA dysregulation and the corresponding circular RNA‐microRNA‐mRNA regulatory axes in autism. Genome Res. 2020;30(3):375‐391. 10.1101/gr.255463.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Xi YZ, Fowdur M, Liu Y, Wu H, He ML, Zhao JM. Differential expression and bioinformatics analysis of circRNA in osteosarcoma. Biosci Rep. 2019;39: 10.1042/Bsr20181514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ji X, Shan L, Shen P, He M. Circular RNA circ_001621 promotes osteosarcoma cells proliferation and migration by sponging miR‐578 and regulating VEGF expression. Cell Death Dis. 2020;11(1):18 10.1038/s41419-019-2204-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Qiu Y, Pu C, Li YC, Qi BC. Construction of a circRNA‐miRNA‐mRNA network based on competitive endogenous RNA reveals the function of circRNAs in osteosarcoma. Cancer Cell Int. 2020;20:48 10.1186/s12935-020-1134-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhu KP, Ma XL, Zhang L, et al. Screening circular RNA related to chemotherapeutic resistance in osteosarcoma by RNA sequencing. Epigenomics. 2018;10(10):1327‐1346. 10.2217/epi-2018-0023 [DOI] [PubMed] [Google Scholar]
- 77. Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25‐miR‐103a‐3p/miR‐107‐CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919 10.1038/srep30919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang Y, Shi S, Zhang Q, Dong H, Zhang J. MicroRNA‐206 upregulation relieves circTCF25‐induced osteosarcoma cell proliferation and migration. J Cell Physiol. 2020; 10.1002/jcp.29570 [DOI] [Google Scholar]
- 79. Zheng F, Xu R. CircPVT1 contributes to chemotherapy resistance of lung adenocarcinoma through miR‐145‐5p/ABCC1 axis. Biomed Pharmacother. 2020;124:109828 10.1016/j.biopha.2020.109828 [DOI] [PubMed] [Google Scholar]
- 80. Liu YP, Wan J, Long F, Tian J, Zhang C. circPVT1 Facilitates invasion and metastasis by regulating miR‐205‐5p/c‐FLIP Axis in osteosarcoma. Cancer Manag Res. 2020;12:1229‐1240. 10.2147/Cmar.S231872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yan M, Gao H, Lv Z, et al. Circular RNA PVT1 promotes metastasis via regulating of miR‐526b/FOXC2 signals in OS cells. J Cell Mol Med. 2020;24(10):5593‐5604. 10.1111/jcmm.15215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang R, Zhang S, Chen X, et al. EIF4A3‐induced circular RNA MMP9 (circMMP9) acts as a sponge of miR‐124 and promotes glioblastoma multiforme cell tumorigenesis. Mol Cancer. 2018;17(1):166 10.1186/s12943-018-0911-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pan GJ, Hu T, Chen XW, Zhang C. Upregulation of circMMP9 promotes osteosarcoma progression via targeting miR‐1265/CHI3L1 Axis. Cancer Manag Res. 2019;11:9225‐9231. 10.2147/Cmar.S226264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhang Z, Pu F, Wang B, Wu Q, Liu J, Shao Z. Hsa_circ_0000285 functions as a competitive endogenous RNA to promote osteosarcoma progression by sponging hsa‐miRNA‐599. Gene Ther. 2020;27(5):186‐195. 10.1038/s41434-019-0112-5 [DOI] [PubMed] [Google Scholar]
- 85. Verrecchia F, Rédini F. Transforming growth factor‐β signaling plays a pivotal role in the interplay between osteosarcoma cells and their microenvironment. Front Oncol. 2018;8:133 10.3389/fonc.2018.00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wang L, Wang P, Su X, Zhao B. Circ_0001658 promotes the proliferation and metastasis of osteosarcoma cells via regulating miR‐382‐5p/YB‐1 axis. Cell Biochem Funct. 2020;38(1):77‐86. 10.1002/cbf.3452 [DOI] [PubMed] [Google Scholar]
- 87. Jin Y, Li L, Zhu T, Liu G. Circular RNA circ_0102049 promotes cell progression as ceRNA to target MDM2 via sponging miR‐1304‐5p in osteosarcoma. Pathol Res Pract. 2019;215(12):152688 10.1016/j.prp.2019.152688 [DOI] [PubMed] [Google Scholar]
- 88. Tan X, Tan D, Li H, Lin Y, Wen Z, Zeng C. circEPSTI1 Acts as a ceRNA to Regulate the Progression of Osteosarcoma. Curr Cancer Drug Targets. 2020;20(4):288‐294. 10.2174/1568009619666191107140948 [DOI] [PubMed] [Google Scholar]
- 89. Cao J, Liu X. Circular RNA 0060428 sponges miR‐375 to promote osteosarcoma cell proliferation by upregulating the expression of RPBJ. Gene. 2020;740:144520 10.1016/j.gene.2020.144520 [DOI] [PubMed] [Google Scholar]
- 90. Du Y, Guo L, Pan H, Liang Y, Li X. Circ_ANKIB1 stabilizes the regulation of miR‐19b on SOCS3/STAT3 pathway to promote osteosarcoma cell growth and invasion. Hum Cell. 2020;33(1):252‐260. 10.1007/s13577-019-00298-6 [DOI] [PubMed] [Google Scholar]
- 91. Chen J, Liu G, Wu Y, et al. CircMYO10 promotes osteosarcoma progression by regulating miR‐370‐3p/RUVBL1 axis to enhance the transcriptional activity of β‐catenin/LEF1 complex via effects on chromatin remodeling. Mol Cancer. 2019;18(1):150 10.1186/s12943-019-1076-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zheng S, Qian Z, Jiang F, et al. CircRNA LRP6 promotes the development of osteosarcoma negatively regulating KLF2 and APC levels. Am J Transl Res. 2019;11(7):4126‐4138. [PMC free article] [PubMed] [Google Scholar]
- 93. Li S, Pei Y, Wang W, Liu F, Zheng K, Zhang X. Circular RNA 0001785 regulates the pathogenesis of osteosarcoma as a ceRNA by sponging miR‐1200 to upregulate HOXB2. Cell Cycle. 2019;18(11):1281‐1291. 10.1080/15384101.2019.1618127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Li X, Sun X, Xu H, Pan H, Liu Y, He L. Circ_ORC2 enhances the regulatory effect of miR‐19a on its target gene PTEN to affect osteosarcoma cell growth. Biochem Biophys Res Comm. 2019;514(4):1172‐1178. 10.1016/j.bbrc.2019.04.188 [DOI] [PubMed] [Google Scholar]
- 95. Jin J, Chen A, Qiu W, et al. Dysregulated circRNA_100876 suppresses proliferation of osteosarcoma cancer cells by targeting microRNA‐136. J Cell Biochem. 2019;120(9):15678‐15687. 10.1002/jcb.28837 [DOI] [PubMed] [Google Scholar]
- 96. Wu Y, Xie Z, Chen J, et al. Circular RNA circTADA2A promotes osteosarcoma progression and metastasis by sponging miR‐203a‐3p and regulating CREB3 expression. Mol Cancer. 2019;18(1):73 10.1186/s12943-019-1007-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zhu K, Niu L, Wang J, et al. Circular RNA hsa_circ_0000885 levels are increased in tissue and serum samples from patients with osteosarcoma. Med Sci Monitor. 2019;25:1499‐1505. 10.12659/msm.914899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wang L, Zhang G, Kang F, Zhang L, Zhang Y. hsa_circ0021347 as a potential target regulated by B7–H3 in modulating the malignant characteristics of osteosarcoma. Biomed Res Int. 2019;2019:9301989 10.1155/2019/9301989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Li S, Pei Y, Wang W, Liu F, Zheng K, Zhang X. Extracellular nanovesicles‐transmitted circular RNA has_circ_0000190 suppresses osteosarcoma progression. J Cell Mol Med. 2020;24(3):2202‐2214. 10.1111/jcmm.14877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Hu Y, Gu J, Shen H, et al. Circular RNA LARP4 correlates with decreased Enneking stage, better histological response, and prolonged survival profiles, and it elevates chemosensitivity to cisplatin and doxorubicin via sponging microRNA‐424 in osteosarcoma. J Clin Lab Anal. 2020;34(2):e23045 10.1002/jcla.23045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β‐catenin pathway. Oncotarget. 2015;6(8):6001‐6013. 10.18632/oncotarget.3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. cir‐ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/β‐catenin pathway. PLoS One. 2015;10(6):e0131225 10.1371/journal.pone.0131225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wan L, Zhang L, Fan K, Cheng Z, Sun Q, Wang J. Circular RNA‐ITCH suppresses lung cancer proliferation via inhibiting the Wnt/β‐catenin pathway. Biomed Res Int. 2016;2016:1579490 10.1155/2016/1579490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Li H, Lan M, Liao X, Tang Z, Yang C. cir‐ITCHCircular RNA promotes osteosarcoma migration and invasion through /miR‐7/EGFR Pathway. Technol Cancer Res Treat. 2020;19:1533033819898728 10.1177/1533033819898728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ren C, Liu J, Zheng B, Yan P, Sun Y, Yue B. The circular RNA circ‐ITCH acts as a tumour suppressor in osteosarcoma via regulating miR‐22. Artif Cells Nanomed Biotechnol. 2019;47(1):3359‐3367. 10.1080/21691401.2019.1649273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Research data are not shared.


