Abstract
Background
As a new member of the vasculoprotective gasotransmitter family, hydrogen sulfide (H2S) functions similar to nitric oxide (NO) and carbon monoxide (CO). Endothelial cell (EC) death and autophagy enable cells to cope with the progression of cardiovascular diseases. However, the impacts and underlying mechanisms of H2S in the autophagic process in ECs are not completely understood. Here, we investigated the effects of H2S on autophagy in human vascular ECs.
Methods
Human umbilical vein endothelial cells (HUVECs) were exposed to different concentrations (0, 50, 100, 200, 500 and 1,000 µmol/L) GYY4137 (H2S donor) for indicated times (0, 0.5, 1, 2, 4 and 8 h), with or without pre-treatment with the autophagy inhibitor 3-methyladenine (3-MA) or bafilomycin A1. HUVECs were transfected with sirtuin 1 (Sirt1) overexpression plasmids (PIRES-Sirt1), Sirt1-siRNAs or forkhead box O1 (FoxO1)-siRNA using Lipofectamine 2000. Cell autophagy was evaluated via Western blotting and fluorescence microscopy. Co-immunoprecipitation assay was used to measure acetylation level of FoxO1. The distribution of FoxO1 in the cytoplasm and nucleus was observed using Western blotting and immunofluorescence. Western blotting, flow cytometric analysis, and cell count kit-8 assay were conducted to evaluate the effect of H2S on the oxidized low-density lipoprotein (Ox-LDL) induced apoptosis of HUVECs.
Results
Using both gain- and loss-of-function experiments, we showed that Sirt1-dependent activation of FoxO1, including its nuclear translocation and deacetylation, was critical for mediating H2S-induced autophagy in ECs. Furthermore, H2S-induced autophagy protected ECs from Ox-LDL-induced apoptosis by activating Sirt1.
Conclusions
These results suggest that Sirt1-mediated autophagy in ECs is a novel mechanism by which H2S exerts vascular-protective actions.
Keywords: Atherosclerosis, autophagy, deacetylation, GYY4137, sirtuin 1 (Sirt1)
Introduction
Vascular endothelial cells (ECs) are essential for maintaining the functional and structural homeostasis of blood vessels. Endothelial dysfunction, including increased oxidative stress, inflammation and impaired autophagy, represents an early step in the onset of atherosclerosis (1,2). Autophagy is a dynamic process involving the formation of biphasic membrane autophagosomes that isolate cytoplasmic components and fuse with lysosomes, eventually degrading the cargo in autolysosomes; this process has been named as autophagy flux (3). Multiple studies have shown that impaired autophagy is closely related to EC dysfunction, apoptosis, inflammation, and atherosclerosis progression (4-7).
H2S is a gas signal molecule involved in many systems of the body, including the cardiovascular system. It is mainly synthesized by cystathionine-γ-lyase in the cardiovascular system (8), and it can protect ECs by inhibiting excessive autophagy in patients with diabetes (9) and antagonize low shear stress-induced autophagy disorder of HUVECs (10). In contrast, exogenous H2S promotes mitophagy to protect rat aortic ECs against apoptosis under conditions of high glucose and palmitate levels (11,12). Different stimuli and experimental environments that induce autophagy in ECs have different cellular outcomes (9,13,14). H2S inhibits or promotes autophagy through different mechanisms in different cell and animal models, such as by alleviating kidney injury in rats by inhibiting autophagy via regulation of NO production (12), alleviating obstructive nephropathy in mice via inhibition of reactive oxidative stress (ROS)-adenosine monophosphate activated protein kinase (AMPK)-mediated autophagy (15). H2S protects arterial ECs by suppressing excessive autophagy which induced by oxidative stress through the nuclear factor erythroid-2 related factor 2 (Nrf2)-ROS-AMPK signalling pathway (9). Therefore, investigation of the effects and regulatory mechanisms of H2S on EC autophagy in the physiological state is essential for developing strategies to protect normal endothelial functions.
Sirtuin 1 (Sirt1), a member of the sirtuin protein family (Sirt1-7), is a highly conserved nicotinamide adenine dinucleotide-dependent histone deacetylase (16). Sirt1 exerts anti-atherosclerotic effects by promoting endothelial survival and endothelial function (17) via the following routs: regulation of endothelial NO synthase, angiotensin II type 1 receptor, p53, and FoxO1 activity; repression of vascular smooth muscle cell migration and proliferation; induction of cellular autophagy (18). Sirt1 prevents atherosclerosis by adjusting autophagy to match the cellular needs with the current metabolic state (19).
Although both H2S and Sirt1 have been shown to play important roles in endothelial homeostasis, their interactions in EC autophagy are unknown. In this study, we investigated the effect of H2S on the autophagy of ECs and role of the Sirt1-FoxO1 signalling pathway in this process.
This study was conducted in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-3647).
Methods
Cell culture and treatments
HUVECs were obtained from Wuhan Union Hospital Regenerative Medicine Center, Hubei, China, cultured in DMEM (Hyclone, Logan, UT, USA) containing 10% foetal bovine serum, 100 IU/mL streptomycin, and 100 mg/mL penicillin and incubated at 37 °C in humidified air containing 5% CO2. Cells were treated with 0.25% trypsin for 2–3 min after reaching confluence, centrifuged at 1,000 rpm for 5 min, and then diluted and seeded into culture dishes. The HUVECs were incubated with 0, 12.5, 25, 50, 100, and 200 µmol/L GYY4137 (a water-soluble slow-release H2S donor, Sigma, St. Louis, MO, USA) for 4 h or incubated with 50 µmol/L GYY4137 for 0, 0.5, 1, 2, 4, and 8 h when the cells were 60–70% confluent. To elucidate the role of H2S in autophagy, we pre-treated the HUVECs with the autophagy inhibitor 3-MA (5 mmol/L) or bafilomycin A1 (Baf; 100 nmol/L) for 30 min, followed by treatment with 50 µmol/L GYY4137 for 4 h.
Fluorescence microscopy
HUVECs were transduced with the Ad-mRFP-GFP-LC3 virus (synthesized by Obio Technology, Shanghai, China; multiplicity of infection =50) for 48 h, and subsequently, the medium was exchanged with 50 µmol/L GYY4137-containing medium for 4 h. In some experiments, HUVECs were infected with Ad-mRFP-GFP-LC3 virus and then transfected with scrambled short interfering RNA (siRNA), Sirt1-siRNA, FoxO1-siRNA, or PIRES-Sirt1 plasmids for 48 or 72 h, and then GYY4137 for another 4 h, to determine the role of Sirt1 and FoxO1 in H2S-induced autophagy. HUVECs were fixed in 4% paraformaldehyde, subjected to 4',6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA, USA) staining to detect the nucleus, and observed via fluorescence microscopy. At 60× objective magnification, green fluorescent protein (GFP) and red fluorescent protein (RFP) were randomly counted in at least 4 samples per group for at least 50 cells per sample. The number of GFP or RFP fluorescent spots in each cell was obtained by dividing the total number of spots by the number of nuclei.
For immunofluorescence staining, HUVECs cultured on coverslips were fixed with 4% paraformaldehyde and incubated with anti-FoxO1 antibodies (1:500 dilution; ab70382, Abcam, Cambridge, UK, AB_1268912). DAPI was used to stain the nuclei. A Cy3-conjugated secondary antibody (Invitrogen, Carlsbad, CA, USA) was used to detect indirect fluorescence. Fields were selected randomly from various sections to ensure the objectivity of sampling. The fluorescence intensity was quantified using ImageJ software (RRID:SCR_003070, NIH, Bethesda, MD, USA).
Western blotting
HUVECs were lysed in modified RIPA buffer (Beyotime Institute of Biotechnology, Shanghai, China) containing 1 mmol/L phenylmethylsulphonyl fluoride. The cytosol and nuclear fractions were isolated according to the kit instructions (NanJing KeyGen Biotech, Nanjing, China). The protein content was determined via bicinchoninic acid (BCA) assay. Proteins were separated by sodium dodecyl sulphide (SDS)-polyacrylamide gel electrophoresis (PAGE) and then electro-transferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). The membranes were incubated with primary antibodies against GAPDH (10494-1-AP, Proteintech, 1:10,000, AB_2263076), Histone-H3 (17168-1-AP, Proteintech, 1:2,000, AB_2716755), Tubulin (10068-1-AP, Proteintech, 1:2,000, AB_2303998), LC3 (12741, Cell Signalling Technology, Danvers, MA, USA, 1:1,000, AB_2617131), p62 (18420-1-AP, Proteintech, 1:1,000, AB_10694431), ATG5 (12994, Cell Signalling Technology, 1:1,000, AB_2630393), Beclin-1 (11306-1-AP, Proteintech, 1:1,000, AB_2259061), Sirt1 (8469, Cell Signalling Technology, 1:1,000, AB_10999470), cleaved-caspase 3 (AF7022, Affinity Biosciences, Solon, OH, USA, 1:1,000, AB_2835326), and cleaved-PARP (ab32064, Abcam, 1:1,000, AB_777102) overnight at 4 °C. The membranes were incubated with goat anti-mouse or goat anti-rabbit secondary antibodies (1:10,000, Abbkine, Redlands, CA, USA) for 1–2 h at 20–30 °C. Protein bands were checked using a horseradish peroxidase (HRP) substrate, Luminol Reagent (Merck Millipore, Billerica, MA, USA). The relative intensities of the protein bands were analysed using ImageJ software. Detailed information regarding the antibodies used is shown in Table 1.
Table 1. Primary antibodies used.
| Antigen | Description of immunogen | Source, host species, catalog no. | Concentration used | RRIDs |
|---|---|---|---|---|
| Anti-GAPDH | GAPDH fusion protein Ag0766 | Proteintech, rabbit polyclonal antibody, 10494-1-AP | 1:10,000 (WB) | AB_2263076 |
| Anti-LC3 | Residues surrounding Leu44 of human LC3B protein (conserved in LC3A) | Cell Signalling Technology, rabbit monoclonal antibody, #12741 | 1:1,000 (WB) | AB_2617131 |
| Anti-p62 | P62/SQSTM1 fusion protein Ag13131 | Proteintech, rabbit polyclonal antibody, 18420-1-AP | 1:1,000 (WB) | AB_10694431 |
| Anti-ATG5 | Residues surrounding Leu265 of human Atg5 protein | Cell Signaling Technology, rabbit monoclonal antibody, #12994 | 1:1,000 (WB) | AB_2630393 |
| Anti-beclin-1 | Beclin 1 fusion protein Ag1843 | Proteintech, rabbit polyclonal antibody, 11306-1-AP | 1:1,000 (WB) | AB_2259061 |
| Anti-Sirt1 | A recombinant protein representing the central region of human SirT1 protein | Cell Signaling Technology, mouse monoclonal antibody, #8469 | 1:1,000 (WB) | AB_10999470 |
| Anti-cleaved-caspase 3 | Synthesized peptide derived from human caspase 3 | Affinity Biosciences, rabbit polyclonal antibody, AF7022 | 1:1,000 (WB) | AB_2835326 |
| Anti-cleaved-PARP | Synthetic peptide within human cleaved PARP1 aa 150–250 | Abcam, rabbit monoclonal antibody, ab32064 | 1:1,000 (WB) | AB_777102 |
| FoxO1 | Synthetic peptide mapping to a region between residues 600 and the C-terminus (residue 652) of human FOXO1A | Abcam, rabbit polyclonal antibody, ab70382 | 1:500 (IF), 1:1,000 (WB) | AB_1268912 |
| Anti-acetylated-lysine | A synthetic acetylated lysine-containing peptide | Cell Signalling Technology, rabbit antibody, #9441 | 1:1,000 (WB) | AB_331805 |
| Anti-histone-H3 | Histone-H3 fusion protein Ag10644 | Proteintech, rabbit polyclonal antibody, 17168-1-AP | 1:3,000 (WB) | AB_2716755 |
| Anti-tubulin | Beta-tubulin fusion protein Ag0117 | Proteintech, rabbit polyclonal antibody, 10068-1-AP | 1:3,000 (WB) | AB_2303998 |
Co-immunoprecipitation assay
Cells were lysed on ice in co-immunoprecipitation cell lysis buffer (Beyotime, P0031). After removing non-specifically bound antibodies using Pierce Protein A/G Agarose Beads (Thermo Fisher Scientific, Rockford, IL, USA), the protein samples were centrifuged at 14,000 rpm for 4 s at 4 °C to collect the supernatant, the protein concentration in which was quantified by BCA assay. Next, 500 µg protein in 250 µL supernatant was incubated with 10 µg of anti-FoxO1 antibody overnight at 4 °C, followed by incubation with Protein A/G Agarose Beads at a ratio of 6:1 (v/v) for 2 h at 20–30 °C to allow the antibody and beads to form complexes. Proteins were eluted by boiling the sample for 5 min in 2× SDS-loading buffer followed by Western blotting using anti-FoxO1 and anti-acetylated-lysine antibodies (#9441, 1:1,000, Cell Signalling Technology, AB_331805). Immunoreactive bands were visualized via chemiluminescence.
siRNA and Sirt1 overexpression plasmid transfection
HUVECs were transfected with a Sirt1 overexpression plasmid (PIRES-Sirt 1), Sirt1-siRNA, or FoxO1-siRNA using Lipofectamine 2000 (#11668-019, Invitrogen) after reaching 60–70% confluence for 48 or 72 h according to the manufacturer’s instructions. PIRES-Sirt1 and siRNAs were synthesized by Guangzhou Ribobio (Guangzhou, China). The sequences of Sirt1-siRNA and FoxO1-siRNA were as follows: 5'-GATGAAGTTGACCTCCTCA-3' and 5'-CCAGCTATAAATGCACATTTA-3', respectively. Gene silencing and overexpression were assessed using Western blotting.
Flow cytometric analysis
Cell apoptosis was analysed using an Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) staining kit according to the manufacturer’s instructions (MultiSciences, Hangzhou, China). Briefly, HUVECs were digested and centrifuged at 300 ×g for 5 min, followed by incubation with Annexin V-FITC and PI for 5 min in the dark. The percentages of apoptotic cells were quantified using flow cytometry with a FACSCalibur (BD Biosciences, Franklin Lakes, NJ, USA). FlowJo software (TreeStar, Ashland, OR, USA) was used for data analysis.
Cytotoxicity assays
Cell viability was determined using CCK-8 assay (CK04, Dojindo, Kumamoto, Japan) assay. In a 96-well cell culture plate, the cells were pre-treated with 3-MA (5 mmol/L, 30 min), resveratrol (10 µmol/L, 4 h), Ex-527 (10 µmol/L, 4 h), and/or H2S (50 µmol/L, 4 h) followed by treatment with Ox-LDL. To measure cell viability, 10 µL CCK-8 solution was added to 100 µL medium per well and incubated for another 4 h. The optical density of the wells was measured, and the percentage of living cells was determined as the ratio of optical density of the experimental wells to that of the control wells.
Statistical analysis
Data are presented as the means ± SEM. Differences between groups were analysed using one-way ANOVA or MNOVA. Individual group statistical comparisons were analysed by unpaired Student’s t-test. A value of P<0.05 was considered as statistically significant.
Results
H2S increases autophagic flux in cultured HUVECs
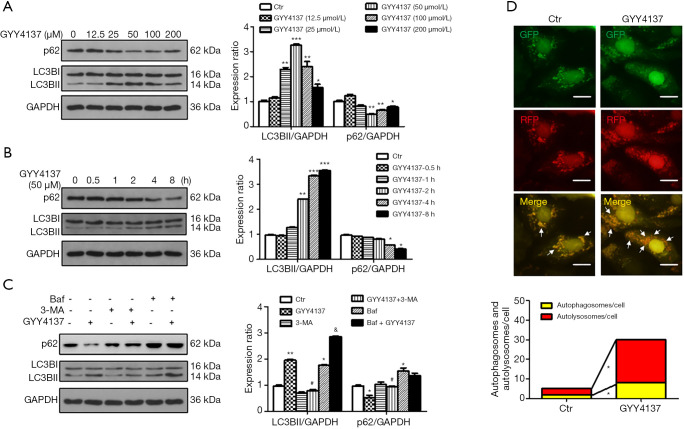
Western blot analysis (Figure 1A) showed that LC3BII was markedly increased in a dose-dependent manner following GYY4137 (12.5, 25, 50, 100, and 200 µmol/L) treatment for 4 h, with a peak at 50 µmol/L followed by a gradual decrease at 100 and 200 µmol/L (Figure 1A) but the values remained higher than those of the control. Thus, we selected 50 µmol/L GYY4137 for subsequent experiments.
Figure 1.
H2S increases autophagic flux in cultured HUVECs. HUVECs were exposed to indicated concentrations of GYY4137 for 4 h (A) and 50 µmol/L GYY4137 for indicated time (B). After pre-treatment with 5 mmol/L 3-MA or 100 nmol/L Baf for 30 min, HUVECs were exposed to 50 µmol/L GYY4137 for 4 h, and the expression of LC3 and p62 was measured (C). Western blots indicated the expression of LC3 and p62. Expression in control (Ctr) group cells was assigned a value of 1 (n≥3). *, P<0.05; **, P<0.01; ***, P<0.001 versus Ctr. #, P<0.05 versus GYY4137. &, P<0.05 versus Baf. Data were expressed as the mean ± SEM. (D) HUVECs were transduced with Ad-mRFP-GFP-LC3 and then treated with 50 µmol/L GYY4137 for 4 h. Representative images of fluorescent LC3 puncta. Arrows indicate autophagosomes (yellow puncta: red puncta overlaid with green puncta) and autolysosomes (represented by red puncta) in merged images. Scale bar =50 µm. The results represent the means from at least 4 independent experiments. *, P<0.05. HUVECs, human umbilical vein endothelial cells; 3-MA, 3-methyladenine; Baf, bafilomycin A1.
We also found that LC3BII was markedly increased in a time-dependent manner following 50 µmol/L GYY4137 treatment for 0.5, 1, 2, 4, and 8 h; the changes at 4 h were greater than those observed at shorter time points (0.5, 1, and 2 h), and no significant difference was detected between 4 and 8 h (Figure 1B). Changes in LC3BII were accompanied by a loss of p62, which functions as an autophagy substrate (20). Thus, cells treated with 50 µmol/L GYY4137 for 4 h were considered as optimal for subsequent experiments. Our data revealed a low level of autophagy in HUVECs in the basal state, and that H2S promotes the autophagic response of HUVECs in time- and concentration-dependent manners.
To explore whether H2S-induced autophagy was due to changes in the stages of autophagy, HUVECs were pre-treated with 3-MA and Baf. As shown in Figure 1C, 3-MA inhibited the increase in LC3BII and decrease in p62 that had been induced by GYY4137 treatment. When autolysosome formation was inhibited by Baf, LC3BII was increased significantly, whereas the decrease in p62 was not significant, indicating that H2S promotes autophagosome formation and autolysosome degradation. Therefore, H2S increases autophagic flux in cultured HUVECs.
To separately evaluate the effect of H2S on autophagosomes and autolysosomes, we transfected HUVECs with Ad-mRFP-GFP-LC3, and analysed the cells via fluorescence microscopy (21,22). In the acidic environment of lysosomes, GFP loses its fluorescence, whereas monomeric red fluorescent protein (mRFP) retains its fluorescence. Thus, green LC3 puncta mainly indicate autophagosomes, whereas red LC3 puncta indicate both autophagosomes and autolysosomes in individual images. Red puncta were overlaid with green puncta and appeared as yellow in the merged images, indicating autophagosomes, with free red puncta in the merged images indicating autolysosomes. As shown in Figure 1D, when autophagy was induced by GYY4137 (50 µmol/L), red and yellow puncta markedly accumulated in HUVECs. These results demonstrate that H2S activated autophagic flux in HUVECs.
Sirt1 is required for H2S-induced autophagy in HUVECs
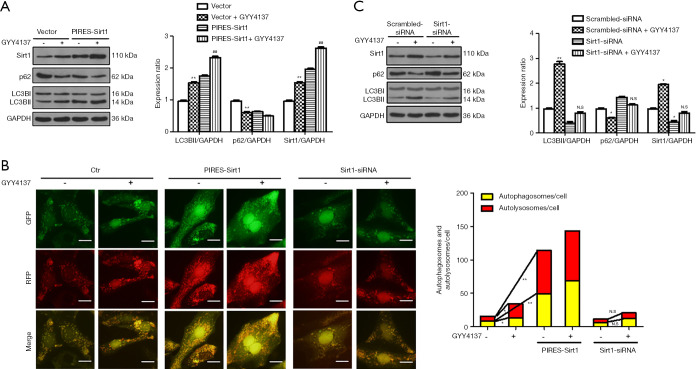
As shown in Figure 2A, Sirt1 was significantly upregulated by H2S in HUVECs. Sirt1 overexpression significantly upregulated LC3BII expression and downregulated p62 expression (Figure 2A). To further evaluate the effect of Sirt1 on autophagic flux, HUVECs were transduced with Ad-mRFP-GFP-LC3. Compared to the control group, Sirt1 overexpression significantly increased both red and yellow puncta (Figure 2B), suggesting that Sirt1 stimulates autophagic flux. Our results suggest that Sirt1 mimics H2S exposure to stimulate basal rates of autophagy in HUVECs.
Figure 2.
Sirt1 is required for H2S-induced autophagy in HUVECs. (A) HUVECs were transduced with vector or PIRES-Sirt1 and then treated with 50 µmol/L GYY4137 for 4 h. Immunoblot analyses showed Sirt1, LC3, p62, and GAPDH expression. Expression in vector group cells was assigned a value of 1 (n≥3). **, P<0.01 versus vector. ##, P<0.01 versus vector + GYY4137. (B) Representative images of fluorescent LC3 puncta after Ad-mRFP-GFP-LC3 transduction in HUVECs treated with PIRES-Sirt1 or Sirt1-siRNA and GYY4137. The mean number of autolysosomes represented by red puncta and autophagosomes represented by yellow puncta in merged images per cell. Scale bar =50 µm. Results represent the means from at least 4 independent experiments. *, P<0.05; **, P<0.01. N.S, not significant. (C) HUVECs were transduced with scrambled-siRNA or Sirt1-siRNA and treated with 50 µmol/L GYY4137 for 4 h. Immunoblot analyses showed Sirt1, LC3, p62, and GAPDH expression. Expression in the scrambled-siRNA group cells was assigned a value of 1 (n≥3). *, P<0.05, **, P<0.01 versus scrambled-siRNA. N.S, not significant versus Sirt1-siRNA. Data were expressed as the mean ± SEM. HUVECs, human umbilical vein endothelial cells.
To evaluate whether Sirt1 is necessary for H2S-induced autophagy, HUVECs were transfected with Sirt1-siRNA. Knockdown of Sirt1 weakened H2S-induced stimulation of autophagic flux, as evidenced by the non-significant increase in LC3BII and decrease in p62 (Figure 2C). Furthermore, Sirt1-siRNA inhibited H2S-induced increases in autophagosome and autolysosome formation (Figure 2B), indicating that Sirt1 is required for H2S activated autophagic flux in HUVECs.
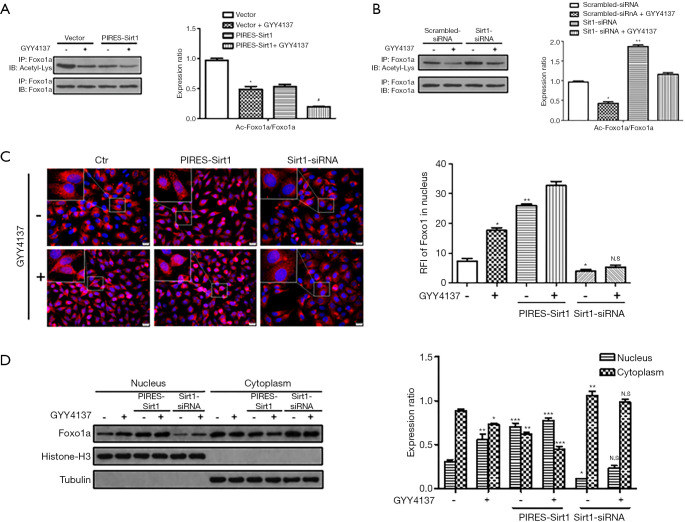
H2S increases Sirt1 production and deacetylase activity and promotes FoxO1 translocation into the nucleus
FoxO family of transcription factors are important downstream effectors of Sirt1 (23). We hypothesized that Sirt1 deacetylates FoxO1 during H2S treatment and promotes translocation of FoxO1 from the cytoplasm to the nucleus, thereby inducing autophagy in HUVECs. As shown in Figure 3, nuclear translocation of FoxO1was significantly increased following H2S treatment (Figure 3C,D), which induced deacetylation of FoxO1 according to co-immunoprecipitation analysis (Figure 3A). These effects were augmented when HUVECs overexpressed Sirt1, indicating a close interaction between Sirt1 and FoxO1 under H2S stimulation. These effects were absent when Sirt1 was downregulated by Sirt1-siRNA, as revealed by increased acetylation of FoxO1 (Figure 3B), and nuclear translocation was significantly attenuated (Figure 3C,D). Taken together, these results indicate that H2S upregulates and activates Sirt1, which in turn induces nuclear translocation and deacetylation of FoxO1.
Figure 3.
H2S increases Sirt1 deacetylase activity and promotes FoxO1 translocation into the nucleus. (A) HUVECs were transduced with vector or PIRES-Sirt1 and then treated with 50 µmol/L GYY4137 for 4 h. Co-immunoprecipitation was performed to detect acetylated FoxO1 (Ac-FoxO1). Expression in vector group cells was assigned a value of 1 (n≥3). *, P<0.05 versus vector; #, P<0.05 versus PIRES-Sirt1. Data were expressed as the mean ± SEM. (B) HUVECs were transduced with scrambled-siRNA or Sirt1-siRNA and then treated with 50 µmol/L GYY4137 for 4 h. Co-immunoprecipitation was performed to detect acetylated FoxO1 (Ac-FoxO1). Expression in scrambled-siRNA group cells was assigned a value of 1 (n≥3). *, P<0.05; **, P<0.01 versus scrambled-siRNA. Data were expressed as the mean ± SEM. (C) Immunofluorescence images of FoxO1 in HUVECs treated with GYY4137, Sirt1 overexpression plasmids and siRNA. Nuclei were counter-stained with DAPI (blue). Insets show higher magnification. Scale bar =20 µm. Statistical analysis revealed the relative fluorescence intensity (RFI) of FoxO1 in the nucleus, which was quantified using ImageJ software. Values represent the means ± SEM of 3 independent experiments. *, P<0.05; **, P<0.01 versus Ctr. N.S, not significant versus Sirt1-siRNA. (D) Western blot analysis of FoxO1 in the nucleus and cytoplasm of HUVECs treated under the same conditions as in (C) (n≥3). *, P<0.05; **, P<0.01; ***, P<0.01 versus Ctr. N.S, not significant versus Sirt1-siRNA. Data were expressed as the mean ± SEM. HUVECs, human umbilical vein endothelial cells.
Deacetylated FoxO1 promotes autophagy-related gene expression
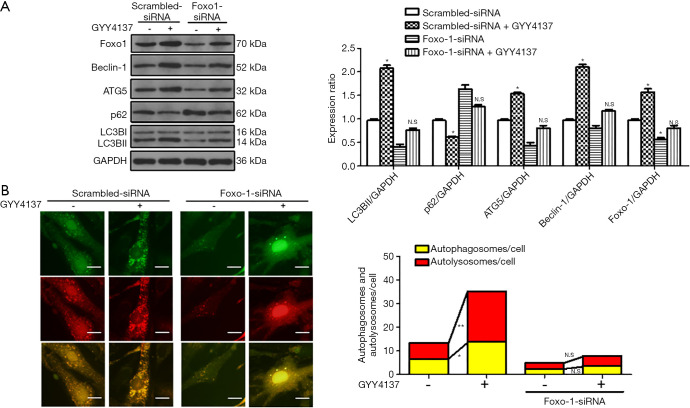
As shown in Figure 4A, we found that knockdown of FoxO1 inhibited H2S-induced stimulation of autophagic flux, as demonstrated by the observation that after H2S stimulation, the decrease in p62 and increases in ATG5, Beclin-1, and LC3BII were not significantly different from those in the FoxO1-siRNA group. However, compared to the FoxO1-siRNA group, these indicators showed increasing (ATG5, Beclin-1, and LC3BII) and decreasing (p62) tendencies in the FoxO1-siRNA + GYY4137 group, indicating that H2S also affects autophagy through other pathways (24). Furthermore, FoxO1-siRNA inhibited H2S-induced increases in autophagosome (yellow dots) and autolysosome (red dots) formation (Figure 4B), indicating that FoxO1 is involved in H2S-induced autophagy to some extent.
Figure 4.
FoxO1 participates in H2S-induced autophagy. HUVECs were transduced with scrambled-siRNA or FoxO1-siRNA and treated with 50 µmol/L GYY4137 for 4 h. (A) Immunoblot analyses showing FoxO1, Beclin-1, ATG5, LC3BII, p62, and GAPDH expression. Expression in scrambled-siRNA group cells was assigned a value of 1 (n≥3). *, P<0.05 versus scrambled-siRNA, N.S, not significant versus FoxO1-siRNA. Data were expressed as the mean ± SEM. (B) Representative images of fluorescent LC3 puncta in different groups. Scale bar =50 µm. Statistical analysis showed the mean number of autolysosomes (red puncta) and autophagosomes (yellow puncta) in the merged images per cell. The results represent the means from at least 4 independent experiments. *, P<0.05; **, P<0.01. N.S, not significant.
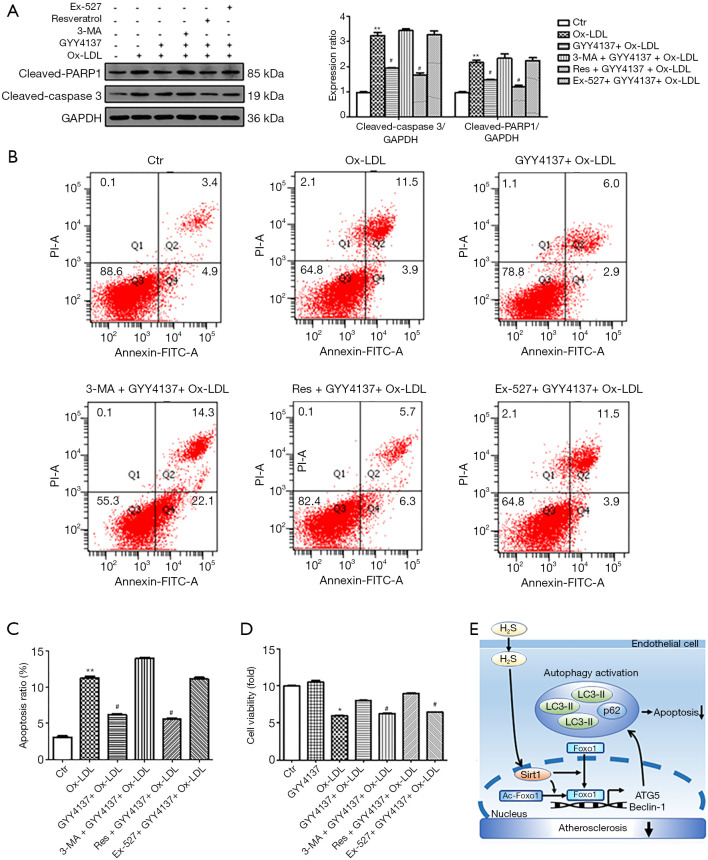
H2S-induced autophagy was cytoprotective in HUVECs
Ox-LDL plays an important role in atherosclerosis by inducing ROS generation and direct cytotoxicity (25). To understand the functional importance of autophagy induced by H2S in cell homeostasis, we pre-treated HUVECs with 3-MA, as well as the Sirt1 activator resveratrol and inhibitor Ex-527, and induced cellular apoptosis using Ox-LDL. We found that Ox-LDL induced HUVEC apoptosis, as evidenced by the elevated expression levels of cleaved-caspase 3 and cleaved-PARP (Figure 5A), increased apoptosis ratio (Figure 5B,C) and decreased cell viability (Figure 5D); whereas, H2S significantly reduced the toxicity of Ox-LDL towards HUVECs. Resveratrol further enhanced the effects of H2S. However, when autophagy and Sirt1 were inhibited by 3-MA and Ex-527 respectively, the protective effect of H2S was abolished, indicating that autophagy and Sirt1 are involved in the cytoprotective role of H2S. Thus, we predicted that H2S suppressed Ox-LDL induced cell apoptosis by activating autophagy via upregulation of Sirt1.
Figure 5.
H2S-induced autophagy via Sirt1 protects against Ox-LDL-induced apoptosis in HUVECs. HUVECs were pre-treated with or without 3-MA, resveratrol, Ex-527, and GYY4137 for the indicated times followed by treatment with Ox-LDL. (A) Immunoblot analyses showing cleaved-caspase-3 and cleaved-PARP. Expression in control (Ctr) group cells was assigned a value of 1 (n≥3). **, P<0.01 versus Ctr; #, P<0.05 versus Ox-LDL. (B,C) Flow cytometric analysis to detect the apoptosis of HUVECs (n≥3). **, P<0.01 versus Ctr; #, P<0.05 versus Ox-LDL. (D) Cell viability was measured using the CCK-8 assay. Cell viability in control (Ctr) group was assigned a value of 1 (n=6). *, P<0.01 versus Ctr; #, P<0.05 versus Ox-LDL. Data were expressed as the mean ± SEM. (E) Schematic representation of the effects and mechanisms of H2S on autophagy and apoptosis in HUVECs. HUVECs, human umbilical vein endothelial cells; 3-MA, 3-methyladenine; Ox-LDL, oxidized low-density lipoprotein.
Therefore, our data indicate that H2S increases endothelial autophagic flux through the Sirt1-FoxO1 signalling pathway and protects HUVECs from Ox-LDL-induced apoptosis, which may be another important mechanism by which H2S mitigates atherosclerosis (Figure 5E).
Discussion
The results of our study provide insight into the mechanisms of H2S-induced autophagy in ECs and reveal that exogenous H2S effectively protects against Ox-LDL-induced EC apoptosis. Our results indicate that (I) exogenous H2S activates the autophagy of vascular ECs by increasing autophagic flux; (II) exogenous H2S increases Sirt1 production and deacetylase activity, which is attributed to the promotion of FoxO1 translocation into the nucleus and enhancement of autophagy-related gene (ATG5, Beclin-1, and LC3) expression; and (III) exogenous H2S ameliorates Ox-LDL-induced apoptosis by activating Sirt1-mediated EC autophagy.
We found that H2S activates HUVEC autophagy in time- and concentration-dependent manners. H2S not only stimulates autophagosome formation but also enhances autolysosome formation and stimulates autophagic flux. It was reported that alterations in EC autophagy are highly correlated with atherosclerosis acceleration (26), and thus, H2S intervention may significantly retard the occurrence and development of atherosclerosis.
Atherosclerosis is promoted by Sirt1 deficiency in ECs, vascular smooth muscle cells, and monocyte/macrophages which actively mediate oxidative stress, inflammation, foam cell formation, and impaired autophagy in the vascular wall (19). In our study, Sirt1 expression was significantly upregulated in HUVECs during H2S exposure. However, whether increased Sirt1 mediates the H2S-induced autophagy of HUVECs was unclear. After silencing Sirt1 in HUVECs using siRNA, we found that the effect of H2S on autophagy was eliminated, indicating that Sirt1 is necessary for H2S-induced autophagy.
Recent studies have revealed the importance of fine-tuned epigenetic modulation mediated by Sirt1 within the molecular circuitry underlying endothelial dysfunction in cardiovascular diseases, including atherosclerosis (16). The FoxO family of transcription factors is the main target molecule of Sirt1 (27), and the function of FoxO proteins depends on their complicated post-translational modifications (28). Studies have shown that deacetylation of FoxO1 is important for its interaction with Sirt1 and is required for glucose-deprivation induced autophagy (29). Expression of the FoxO1 (3A/LXXAA) mutant abolishes the interaction between Sirt1 and FoxO1 as well as Sirt1-induced activation of FoxO1-mediated transcription (30). Interestingly, we found that nuclear localization and deacetylation of FoxO1 are enhanced in the presence of H2S and Sirt1 overexpression, whereas these effects were absent after Sirt1 was silenced, suggesting that Sirt1-mediated deacetylation and nuclear localization of FoxO1 contribute to the effects of H2S. Studies have shown that multiple elements contribute to nuclear/cytoplasmic shuttling of FoxO1, and that phosphorylation and 14-3-3 binding regulate the cellular distribution and function of FoxO1 through multiple mechanisms (31,32). Silencing of FoxO1 in HUVECs downregulated the expression of the autophagy-related proteins ATG5, Beclin-1, and LC3 and upregulated the expression of p62. Moreover, we found that GYY4137-induced HUVEC autophagosomes and autolysosomes were significantly reduced. These results demonstrate that FoxO1 mediates the autophagy of ECs induced by H2S by enhancing the expression of autophagy-related proteins.
Autophagy can be triggered by different stress stimuli, and cellular outcomes following autophagy induction in ECs vary depending on the nature of the stimulus and specific experimental settings (13,14). Here, we found that autophagy activated by H2S reduced Ox-LDL-induced HUVEC apoptosis and that H2S induced a beneficial autophagic process to protect ECs. Impaired autophagy in ECs is closely related to the development of atherosclerosis. Therefore, strategies that promote endogenous H2S synthesis and/or provide exogenous H2S in a physiologically relevant manner may slow the development of atherosclerosis by activating EC autophagy.
In conclusion, Sirt1-mediated translocation and deacetylation of FoxO1 play essential roles in H2S-induced EC autophagy. Thus, autophagy activated by H2S exerts vascular-protective actions by activating the Sirt1-FoxO1 signalling pathway, which may be a new mechanism of the antiatherogenic action of H2S. Indeed, previous studies showed that the Sirt1-FoxO axis plays an important role in modulating autophagy in cardiomyocytes and skeletal muscle cells (29,33), although the results are controversial. The mechanisms by which FoxO modulates autophagy are complex (29,34), whereas transcriptional regulation of autophagic genes may not fully explain the effects of FoxO. Therefore, additional studies are needed to determine the mechanisms by which the Sirt1-FoxO1 pathway regulates autophagy induced by H2S in ECs.
Supplementary
The article’s supplementary files as
Acknowledgments
We thank Editage (http://www.editage.cn/) for providing linguistic assistance during the preparation of this manuscript.
Funding: This work was supported by the National Natural Science Foundation of China (grant numbers 81770652, 81901561) and Natural Science Foundation of Hubei Province (grant numbers 2017ACA096, 2017CFB748).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-3647
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-3647
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-3647
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3647). The authors have no conflicts of interest to declare.
References
- 1.Donato AJ, Morgan RG, Walker AE, et al. Cellular and molecular biology of aging endothelial cells. J Mol Cell Cardiol 2015;89:122-35. 10.1016/j.yjmcc.2015.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gimbrone MA, Jr, Garcia-Cardena G. Endothelial Cell Dysfunction and the Pathobiology of Atherosclerosis. Circ Res 2016;118:620-36. 10.1161/CIRCRESAHA.115.306301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshii SR, Mizushima N. Monitoring and Measuring Autophagy. Int J Mol Sci 2017;18:1865. 10.3390/ijms18091865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang F, Yang TL. MicroRNA-126 alleviates endothelial cells injury in atherosclerosis by restoring autophagic flux via inhibiting of PI3K/Akt/mTOR pathway. Biochem Biophys Res Commun 2018;495:1482-9. 10.1016/j.bbrc.2017.12.001 [DOI] [PubMed] [Google Scholar]
- 5.Fetterman JL, Holbrook M, Flint N, et al. Restoration of autophagy in endothelial cells from patients with diabetes mellitus improves nitric oxide signaling. Atherosclerosis 2016;247:207-17. 10.1016/j.atherosclerosis.2016.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiss M, Kost B, Renner-Muller I, et al. Efavirenz Causes Oxidative Stress, Endoplasmic Reticulum Stress, and Autophagy in Endothelial Cells. Cardiovasc Toxicol 2016;16:90-9. 10.1007/s12012-015-9314-2 [DOI] [PubMed] [Google Scholar]
- 7.Li W, Sultana N, Siraj N, et al. Autophagy dysfunction and regulatory cystatin C in macrophage death of atherosclerosis. J Cell Mol Med 2016;20:1664-72. 10.1111/jcmm.12859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo W, Gui DD, Yan BJ, et al. Hydrogen Sulfide Switch Phenomenon Regulating Autophagy in Cardiovascular Diseases. Cardiovasc Drugs Ther 2020;34:113-21. 10.1007/s10557-019-06927-4 [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Wu J, Sun A, et al. Hydrogen sulfide decreases high glucose/palmitate-induced autophagy in endothelial cells by the Nrf2-ROS-AMPK signaling pathway. Cell Biosci 2016;6:33. 10.1186/s13578-016-0099-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, Bi X, Chen T, et al. Shear stress regulates endothelial cell autophagy via redox regulation and Sirt1 expression. Cell Death Dis 2015;6:e1827. 10.1038/cddis.2015.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan B, Jin S, Sun J, et al. New method for quantification of gasotransmitter hydrogen sulfide in biological matrices by LC-MS/MS. Sci Rep 2017;7:46278. 10.1038/srep46278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu N, Wu J, Zhang L, et al. Hydrogen Sulphide modulating mitochondrial morphology to promote mitophagy in endothelial cells under high-glucose and high-palmitate. J Cell Mol Med 2017;21:3190-203. 10.1111/jcmm.13223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiang X, Huang J, Song S, et al. 17β-estradiol inhibits H2O2-induced senescence in HUVEC cells through upregulating SIRT3 expression and promoting autophagy. Biogerontology 2020;21:549-57. 10.1007/s10522-020-09868-w [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Song X, Li Z, et al. MicroRNA-103 Protects Coronary Artery Endothelial Cells against H2O2-Induced Oxidative Stress via BNIP3-Mediated End-Stage Autophagy and Antipyroptosis Pathways. Oxid Med Cell Longev 2020;2020:8351342. 10.1155/2020/8351342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Q, Yu S, Zhang K, et al. Exogenous H2S Inhibits Autophagy in Unilateral Ureteral Obstruction Mouse Renal Tubule Cells by Regulating the ROS-AMPK Signaling Pathway. Cell Physiol Biochem 2018;49:2200-13. 10.1159/000493824 [DOI] [PubMed] [Google Scholar]
- 16.Chen C, Zhou M, Ge Y, et al. SIRT1 and aging related signaling pathways. Mech Ageing Dev 2020;187:111215. 10.1016/j.mad.2020.111215 [DOI] [PubMed] [Google Scholar]
- 17.Wang W, Li X, Ren L, et al. MiR-132 relieves vascular endothelial inflammation and improves endothelial function in atherosclerosis rats by regulating SIRT1. Minerva Endocrinol 2020;45:158-61. 10.23736/S0391-1977.19.03074-8 [DOI] [PubMed] [Google Scholar]
- 18.D'Onofrio N, Servillo L, Balestrieri ML. SIRT1 and SIRT6 Signaling Pathways in Cardiovascular Disease Protection. Antioxid Redox Signal 2018;28:711-32. 10.1089/ars.2017.7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitada M, Ogura Y, Koya D. The protective role of Sirt1 in vascular tissue: its relationship to vascular aging and atherosclerosis. Aging (Albany NY) 2016;8:2290-307. 10.18632/aging.101068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han J, Pan XY, Xu Y, et al. Curcumin induces autophagy to protect vascular endothelial cell survival from oxidative stress damage. Autophagy 2012;8:812-25. 10.4161/auto.19471 [DOI] [PubMed] [Google Scholar]
- 21.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007;3:452-60. 10.4161/auto.4451 [DOI] [PubMed] [Google Scholar]
- 22.Hariharan N, Zhai P, Sadoshima J. Oxidative stress stimulates autophagic flux during ischemia/reperfusion. Antioxid Redox Signal 2011;14:2179-90. 10.1089/ars.2010.3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujita Y, Yamashita T. Sirtuins in Neuroendocrine Regulation and Neurological Diseases. Front Neurosci 2018;12:778. 10.3389/fnins.2018.00778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu D, Wang H, Teng T, et al. Hydrogen sulfide and autophagy: A double edged sword. Pharmacol Res 2018;131:120-7. 10.1016/j.phrs.2018.03.002 [DOI] [PubMed] [Google Scholar]
- 25.Kao ES, Tseng TH, Lee HJ, et al. Anthocyanin extracted from Hibiscus attenuate oxidized LDL-mediated foam cell formation involving regulation of CD36 gene. Chem Biol Interact 2009;179:212-8. 10.1016/j.cbi.2009.01.009 [DOI] [PubMed] [Google Scholar]
- 26.Xie L, Gu Y, Wen M, et al. Hydrogen Sulfide Induces Keap1 S-sulfhydration and Suppresses Diabetes-Accelerated Atherosclerosis via Nrf2 Activation. Diabetes 2016;65:3171-84. 10.2337/db16-0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma X, Su P, Yin C, et al. The Roles of FoxO Transcription Factors in Regulation of Bone Cells Function. Int J Mol Sci 2020;21:692. 10.3390/ijms21030692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ioannilli L, Ciccarone F, Ciriolo MR. Adipose Tissue and FoxO1: Bridging Physiology and Mechanisms. Cells 2020;9:849. 10.3390/cells9040849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hariharan N, Maejima Y, Nakae J, et al. Deacetylation of FoxO by Sirt1 Plays an Essential Role in Mediating Starvation-Induced Autophagy in Cardiac Myocytes. Circ Res 2010;107:1470-82. 10.1161/CIRCRESAHA.110.227371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakae J, Cao Y, Daitoku H, et al. The LXXLL motif of murine forkhead transcription factor FoxO1 mediates Sirt1-dependent transcriptional activity. J Clin Invest 2006;116:2473-83. 10.1172/JCI25518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Der Heide LP, Hoekman MF, Smidt MP. The ins and outs of FoxO shuttling: mechanisms of FoxO translocation and transcriptional regulation. Biochem J 2004;380:297-309. 10.1042/bj20040167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao X, Gan L, Pan H, et al. Multiple elements regulate nuclear/cytoplasmic shuttling of FOXO1: characterization of phosphorylation- and 14-3-3-dependent and -independent mechanisms. Biochem J 2004;378:839-49. 10.1042/bj20031450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee D, Goldberg AL. SIRT1 protein, by blocking the activities of transcription factors FoxO1 and FoxO3, inhibits muscle atrophy and promotes muscle growth. J Biol Chem 2013;288:30515-26. 10.1074/jbc.M113.489716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y, Yang J, Liao W, et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol 2010;12:665-75. 10.1038/ncb2069 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as