Abstract
Background
Abnormality in chromatin regulation is a major determinant in the progression of multiple neoplasms. Astrocytoma is a malignant histologic morphology of glioma that is commonly accompanied by chromatin dysregulation. However, the systemic interpretation of the expression characteristics of chromatin-regulating genes in astrocytoma is unclear.
Methods
In this study, we investigated the expression profile of chromatin regulation genes in 194 astrocytoma patients sourced from The Cancer Genome Atlas (TCGA) database. The relevance of gene expression and postoperative survival outcomes was assessed.
Results
Based on the expression patterns of chromatin regulation genes, two primary clusters and three subclusters with significantly different survival outcomes were identified. The patients in cluster_1 (or subcluster_1) had a poorer prognosis than the other groups, and this particular cohort were older, with a more advanced grade of tumor and isocitrate dehydrogenase-wildtype distribution. Detection of the differentially expressed genes revealed that the group with poor prognosis was characterized by downregulation of H2AFY2, WAC, HDAC5, ZMYND11, TET1, SATB1, and MYST4, and overexpression of EYA4. Moreover, all eight genes were significantly correlated with overall survival (OS) in astrocytoma. Age-associated genes were investigated and the expression levels of EYA4, TET1, SATB1, WAC, ZMYND11, and H2AFY2 were found to be closely correlated with advanced age. Regression analysis suggested that the expression levels of H2AFY2, HILS1, EYA1, EYA4, and KDM5B were independently associated with IDH mutation status. The differential expressions of 34 common genes were significantly associated with age, grade, and IDH mutant.
Conclusions
The study revealed that the expression pattern of chromatin regulation genes was significantly associated with postoperative prognosis in astrocytoma. Moreover, the differential expression of particular genes was strongly associated with clinical characteristics such as age, grade, and IDH subtype. These results suggest that the genes involved in chromatin regulation play important roles in the biological process of astrocytoma progression, and these molecules could potentially serve as therapeutic targets in astrocytoma.
Keywords: Chromatin regulation, prognosis, IDH mutation, astrocytoma
Introduction
Diffuse gliomas are characterized by a high incidence rate, poor prognosis, high rates of mortality, and treatment insensitivity (1). Astrocytoma is the main type of low-grade glioma and has relatively high malignancy (2,3). Despite recent advances in multimodality therapy incorporating surgery, chemotherapy, and radiotherapy as the commonly accepted treatments for astrocytoma, little improvement has been seen in patient outcomes (4,5). According to the recent World Health Organization (WHO) classification, grade II–III astrocytomas are divided into isocitrate dehydrogenase-wildtype (IDH-wt) and IDH-mutant (IDH-mut) groups, with the former being significantly more aggressive and associated with poorer outcomes (6). Exploration of the molecular characteristics and classification in specific populations is crucial to improving therapy and prognosis for patients with astrocytoma (7). Genetic analyses of glioma tissues have revealed that the development of glioma is closely related to various epigenetic phenomena, including chromatin remodeling (8,9). Due to the reversibility of epigenetic modifications, the various proteins and genes regulating these changes have become potential new targets in the treatment of glioma (2,10).
Chromatin remodeling involves changing the configuration of chromatin structure and is crucial in regulating gene expression, apoptosis, and DNA replication and repair (11). In the process of chromatin remodeling, chromatin regulator proteins act to control alterations in the structure of chromatin. Dysfunctions in chromatin-regulating proteins have been associated with the development and progression of brain tumors (12,13). Multiple studies have shown that specific molecular variations, such as ATRX mutations, have clinical implications for the molecular diagnosis of gliomas and can provide diagnostic and prognostic information (14). However, the expression patterns of genes that regulate chromatin remodeling have not been well studied. To comprehensively understand the biological heterogeneity of astrocytoma, the gene expressions coding chromatin-regulating proteins in astrocytoma are in urgent need of systematic study. Given that the deregulation of chromatin structure can be reversed by DNA methylation and histone deacetylation inhibitors, it is possible that identified subgroups with typical gene expression could be treated with targeted therapy to promote the expression of tumor suppressor genes or to suppress the expression of tumor driver genes.
In this study, we focused on the expression distributions of chromatin regulation genes in surgically excised astrocytoma tissue and assessed their relevance to prognosis and clinical outcome. To examine variations associated with clinical features, we identified genes that were differentially expressed or commonly varied. Our findings suggest that chromatin regulation genes have the potential to be promising therapeutic targets. We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7229).
Methods
Samples and database
The transcriptome data and corresponding clinical data of low-grade glioma patients were obtained from The Cancer Genome Atlas database (TCGA, cbioportal.org) (15). The data were filtered based on whether the messenger RNA (mRNA) z-score data and clinical features, including overall survival (OS), progression free survival (PFS), disease free survival (DFS), and disease-specific survival (DSS), were complete. Patients who had a postoperative pathological diagnosis and detailed records, including age, sex, grade of tumor, and IDH subtype were enrolled in the current study. All the samples were diagnosed as astrocytoma according to the histological records and RNA sequencing. Finally, 194 astrocytoma samples were enrolled in the study. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). All utilized public omics datasets had been generated in previous studies and obtained with prior ethical approval.
Bioinformatics
Chromatin remodeling genes included in the current analysis were derived from the Uniport-keyword database (KW-0156, uniport.org/keywords) as published (16). A total of 295 genes coding for reviewed proteins associated with chromatin regulation were arrayed, and the expression data (RNA-Seq V2 RSEM) were obtained from the transcriptome of astrocytoma originating from TCGA data. A cluster analysis of the expressions of the 295 genes in the 194 astrocytoma tissues was performed to distinguish samples based on their gene expression profiles. Subjects with similar gene expression patterns were identified from the total sample. The transcription levels were expressed as mRNA z-scores and clustered using the hierarchical clustering algorithm using a Stanford program (17). A cluster heat map and pattern were generated with the Java Treeview program according to the tumor stage (18).
Prognostic relevance analyses
The prognostic role of the chromatin remodeling genes was investigated by comparing the survival outcomes of different groups. Data on overall survival (OS), progression-free survival (PFS), disease-free survival (DFS), and disease-specific survival (DSS) were accessed using GraphPad Prism (GraphPad Software, Inc., California, US; Version 8). Comparisons of survival between different clusters were conducted to determine the relevance of gene expression profiles to prognosis. Additionally, an analysis of the difference in OS between cohorts with low and high expression levels of individual genes was conducted using GraphPad Prism 8.
Statistical analysis
Survival curves of different groups were plotted and compared using the log-rank (Mantel-Cox) test in GraphPad Prism 8 (GraphPad Software, Inc., California, US; Version 8). Differences in clinical characteristics and the variables associated with each cluster were assessed with Fisher’s exact test and Spearman’s correlation. Differences in the expression levels of genes between clusters were detected with analysis of variance (ANOVA). Regression analyses were performed to determine correlations between variables. All tests were performed with SPSS 24.0 (IBM, Inc., New York, US). A P value <0.05 was considered statistically significant.
Results
The expression profile of chromatin remodeling genes was significantly associated with prognosis
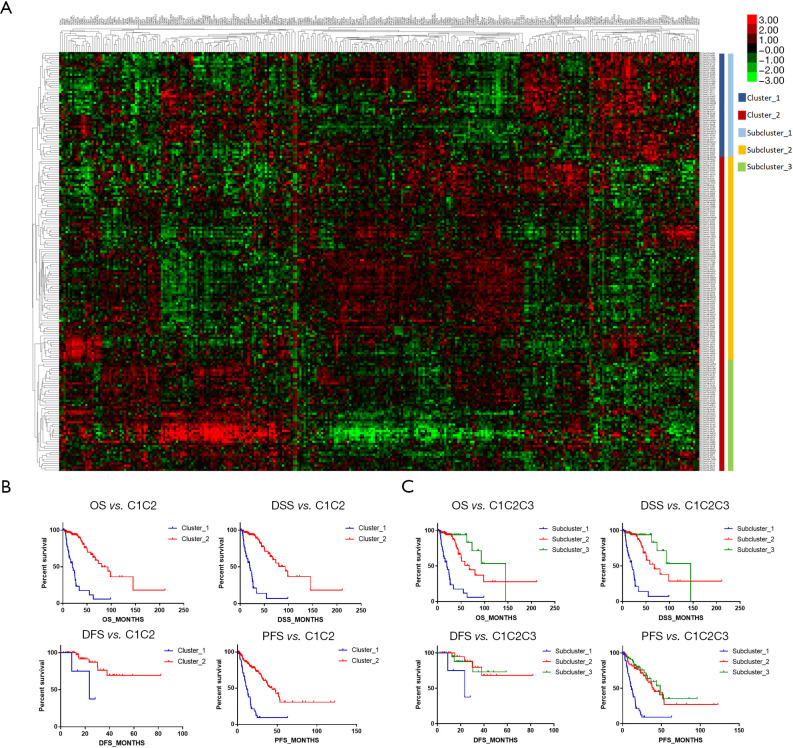
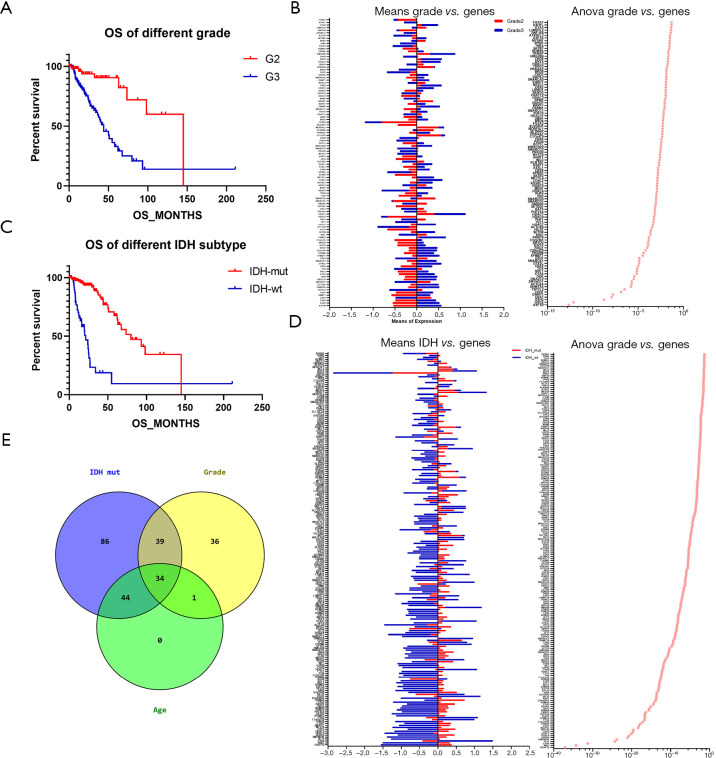
All patient samples were sorted into clusters based on the transcriptional array of chromatin remodeling genes. The mRNA expression levels of 295 protein coding genes involved in controlling the alteration of chromatin structure were assessed (Box S1). A total of 194 patients with complete clinical records were recruited in the cluster analysis. The initial cluster analysis defined 50 patients as primary cluster_1 and 144 patients as primary cluster_2. Cluster_2 was subsequently subdivided into two clusters. The three clusters are shown in Figure 1A. The difference in prognosis between groups was also assessed. In the dichotomous grouping, cluster_1 showed significantly poorer prognosis than cluster_2, specifically in OS (19.89 vs. 79.99 months, P<0.0001), DSS (19.89 vs. 93.20 months, P<0.0001), DFS (23.44 vs. undefined months, P=0.0179), and PFS (11.51 vs. 43.53 months, P<0.0001) (Table 1 and Figure 1B). In the trichotomous grouping, subcluster_1 showed the worst outcomes in OS (P<0.0001), DSS (P<0.0001), and PFS (P<0.0001). Subcluster_3 showed a significant survival advantage of 145.05 months in both OS and DSS (Table 2). No significant difference was detected in DFS between the subclusters (Table 2 and Figure 1C).
Figure 1.
The identified subgroups in relation to the different distributions of chromatin regulation genes in astrocytoma. (A) The different distributions of genes in the dichotomous and trichotomous groups; (B) the overall survival (OS), progression free survival (PFS), disease free survival (DFS) and disease-specific survival (DSS) of cluster 1 and cluster 2 in the dichotomous groups; (C) the OS, DSS, DFS, and PFS of each subcluster in the trichotomous group.
Table 1. Comparison of survival in the dichotomous grouping.
| Median survival of cluster_1 | Median survival of cluster_2 | Hazard ratio | 95% CI of ratio | P value | |
|---|---|---|---|---|---|
| Entry name | 19.89 | 79.99 | 6.405 | 3.128 to 13.12 | <0.0001*** |
| TET1 | 19.89 | 93.20 | 6.731 | 3.158 to 14.35 | <0.0001*** |
| PRDM6 | 23.44 | Undefined | 5.307 | 0.273 to 103.2 | 0.0179* |
| RAG1 | 11.51 | 43.53 | 4.119 | 2.293 to 7.397 | <0.0001*** |
*, P<0.05; ***, P<0.001.
Table 2. Comparison of survival in the trichotomous grouping.
| Median survival of cluster_1 | Median survival of cluster_2 | Median survival of cluster_3 | P value | |
|---|---|---|---|---|
| OS | 19.89 | 67.46 | 145.05 | <0.0001*** |
| DSS | 19.89 | 67.46 | 145.05 | <0.0001*** |
| DFS | 23.44 | Undefined | Undefined | 0.0597 |
| PFS | 11.51 | 38.93 | 48.49 | <0.0001*** |
***, P<0.001.
The expression of chromatin remodeling genes differed significantly in different clusters
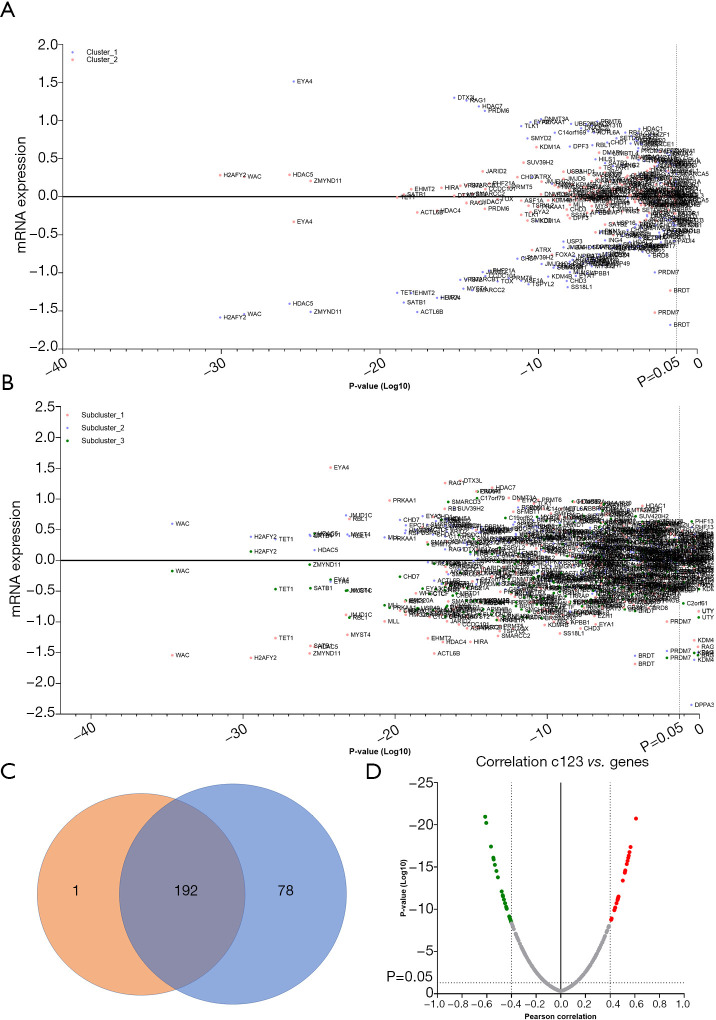
To study the gene variations associated with prognosis, the expression levels of chromatin remodeling genes were statistically analyzed between each cluster. A comparison of gene expressions between the two primary clusters revealed that the expression of 193 genes varied significantly (P<0.05, Figure 2A). Additional analysis of the differences in gene expression between the three subclusters showed 270 discrepant genes (P<0.05, Figure 2B). Among these, 192 genes were verified as overlaps in the two different gene sets (Figure 2C). Compared to the other clusters, the cohort with a poor prognosis (cluster_1 or subcluster_1) was significantly characterized by low expression levels of H2AFY2, WAC, HDAC5, ZMYND11, TET1, SATB1, and MYST4, and high expression levels of EYA4 (Figure 2A,B).
Figure 2.
Differentially expressed chromatin regulation genes in astrocytoma. (A) Differentially expressed genes between the dichotomous groups; (B) differentially expressed genes between the trichotomous groups; (C) the overlap of the dichotomous and trichotomous groups; (D) the gene expressions correlated with subcluster changes.
In the comparison of survival across individual subclusters, both OS and DSS extended gradually from subcluster_1 to subcluster_3. To uncover the genes correlated with prognosis in astrocytoma, changes in the expression of chromatin regulation genes across the three subclusters were assessed. The expressions of 24 genes, represented by HDAC5, were found to be positively correlated with subcluster in order of improved prognosis (r>0.4, P<0.001). Conversely, the expressions of 26 genes, represented by PRKAA1 and RBL1, were negatively correlated with subcluster order, from the worst to the best prognosis (r<−0.4, P<0.001) (Figure 2D and Table 3).
Table 3. The survival correlation of individual chromatin-regulating genes.
| Low expression | High expression | P value | Hazard ratio | 95% CI of ratio | |
|---|---|---|---|---|---|
| H2AFY2 | 24.39 | 79.99 | <0.0001**** | 4.16 | 2.198 to 7.871 |
| WAC | 26.93 | 79.99 | <0.0001**** | 3.21 | 1.837 to 5.611 |
| HDAC5 | 29.13 | 73.48 | <0.0001**** | 3.06 | 1.746 to 5.361 |
| ZMYND11 | 31.59 | 93.2 | <0.0001**** | 2.86 | 1.653 to 4.933 |
| MYST4 | 29.13 | 79.99 | <0.0001**** | 2.81 | 1.640 to 4.820 |
| TE1 | 37.38 | 67.46 | 0.0024** | 2.20 | 1.312 to 3.696 |
| SATB1 | 33.96 | 67.46 | 0.0106* | 1.92 | 1.123 to 3.285 |
| EYA4 | 93.20 | 24.39 | <0.0001**** | 0.21 | 0.112 to 0.383 |
****, P<0.0001; **, P<0.01; *, P<0.05.
The prognostic role of individual chromatin-regulating genes in astrocytoma
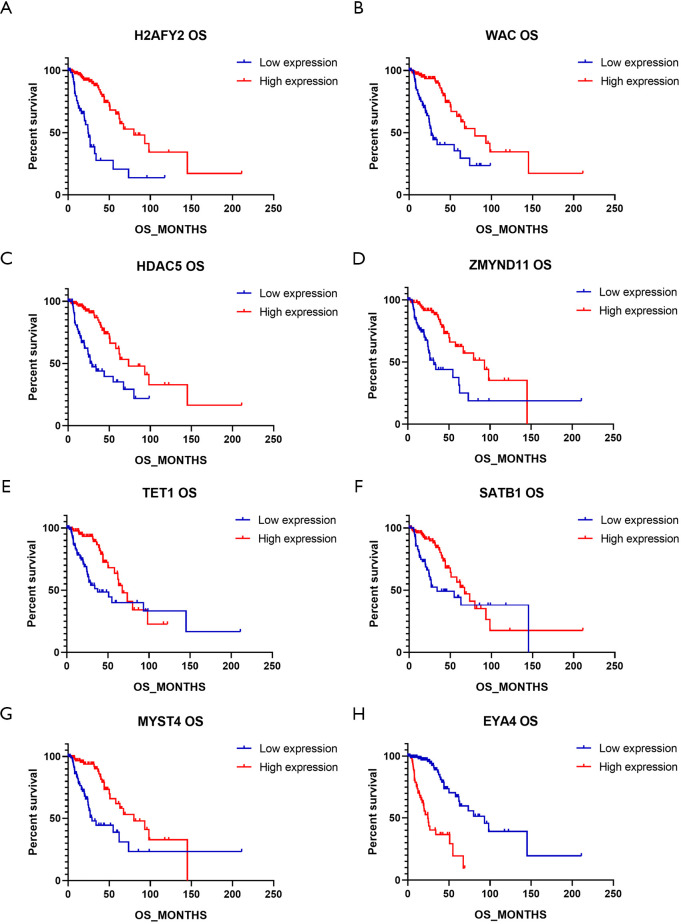
Comparison between the different clusters and subclusters revealed that the expression profile of chromatin regulated genes was closely related to prognosis. Additionally, analysis of the differentially expressed genes showed that eight genes were significantly and commonly changed in both the dichotomous and trichotomous groups. To investigate the effect of individual genes on the prognosis of astrocytoma, the patients were divided into two groups, the high- and low-expression groups, according to the expression levels of these eight genes. Comparisons of OS between the paired groups revealed that low expression levels of H2AFY2, WAC, HDAC5, ZMYND11, TET1, SATB1, and MYST4 were significantly associated with shorter OS (P<0.05, Figure 3). Among these genes, the high expression of H2AFY2 was positively correlated with OS (24.39 vs. 79.99 months, P<0.0001; HR =4.16, 95% CI: 2.198–7.871) (Table 3). In contrast, high expression of EYA4 was significantly correlated with poorer prognosis (P<0.0001, Figure 4). Patients with low expression of EYA4 had a median OS of 93.20 months, compared to 24.39 months for those with high expression (HR =0.21, 95% CI: 0.112–0.383, Table 3).
Figure 3.
Comparison of overall survival (OS) in patients with different gene expression. (A,B,C,D,E,F,G,H) The OS of patients with high gene expression levels versus those with low gene expression levels.
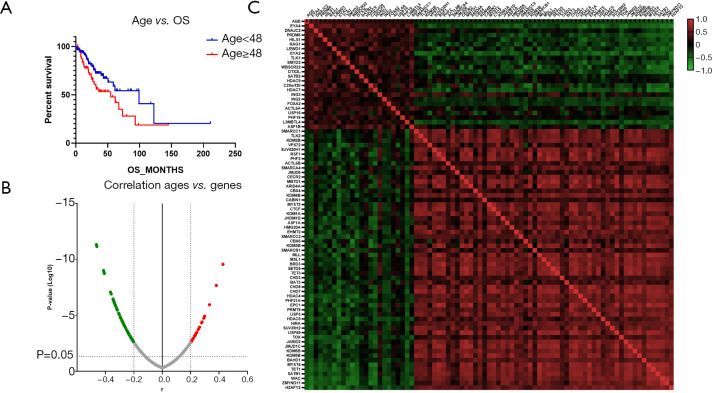
Figure 4.
Differential expressions of chromatin regulation genes correlated with increasing age in astrocytoma. (A) Discrepancies in overall survival (OS) between different ages; (B) the gene expressions correlated with age change; (C) the heat map of Pearson’s correlation analysis between age change and gene expression.
The chromatin regulation gene variations associated with clinical characteristics in astrocytoma
To assess the correlation between chromatin remodeling genes and the clinical characteristics of astrocytoma, comparisons of age, sex, grade, and IDH-mut status were performed between each cluster and subcluster. In the dichotomous group, there were no differences in sex (P>0.05); however, significant differences were found in age, grade, and IDH subtype (P<0.001). Cluster_1 was significantly older (52.3±12.53 vs. 38.17±10.49 years, P<0.001), with more advanced-grade tumors (92.0% vs. 59.0% of G3, P<0.001) and more IDH-wt subtype distribution (88.0% vs. 9.0%, P<0.001) than cluster_2 (Table 4). Similarly, subcluster_1 was significantly older (52.3±12.53 vs. 37.21±9.61 and 39.40±11.49 years, P<0.001), with more advanced-grade tumors (92.0% vs. 65.4% and 50.8% of G3, P<0.001) and higher levels of the IDH-wt genotype (88.0% vs. 6.2% and 12.7% of WT, P<0.001) than subcluster_2 and subcluster_3. No difference in sex distribution was detected in the trichotomous group (P>0.05) (Table 5).
Table 4. Comparison of clinical characteristics in the dichotomous grouping.
| Cluster_1 (n=50) | Cluster_2 (n=144) | P value | |
|---|---|---|---|
| Age (years) | 52.3±12.53 | 38.17±10.49 | 4.02E-13**** |
| Sex | 0.96 | ||
| Female | 22 (44.0%) | 64 (44.4%) | |
| Male | 28 (56.0%) | 80 (55.6%) | |
| Grade | 1.79E-05**** | ||
| G2 | 4 (8.0%) | 59 (41.0%) | |
| G3 | 46 (92.0%) | 85 (59.0%) | |
| IDH subtype | 3.63E-24**** | ||
| Mutant | 6 (12%) | 129 (89.6%) | |
| WT | 44 (88.0%) | 13 (9.0%) | |
| NA | 0 (0) | 2 (1.4%) |
****, P<0.0001. IDH, isocitrate dehydrogenase.
Table 5. Comparison of clinical characteristics in the trichotomous grouping.
| Cluster 1 (n=50) | Cluster 2 (n=81) | Cluster 3 (n=63) | P value | |
|---|---|---|---|---|
| Age | 52.3±12.53 | 37.21±9.61 | 39.40±11.49 | 2E-12**** |
| Sex | 0.94 | |||
| Female | 22 (44.0%) | 35 (43.2%) | 29 (46.0%) | |
| Male | 28 (56.0%) | 46 (56.8%) | 34 (54.0%) | |
| Grade | 1.79E-05**** | |||
| G2 | 4 (8.0%) | 28 (34.6%) | 31 (49.2%) | |
| G3 | 46 (92.0%) | 53 (65.4%) | 32 (50.8%) | |
| IDH subtype | 1.45E-22**** | |||
| Mutant | 5 (12%) | 71 (91.4%) | 55 (87.3%) | |
| WT | 44 (88.0%) | 5 (6.1%) | 8 (12.7%) | |
| NA | 0 (0) | 2 (2.5%) | 0 (0) |
****, P<0.0001. IDH, isocitrate dehydrogenase.
The differences in chromatin regulation gene expression are associated with age, grade, and IDH subtype distribution in astrocytoma
The differences in gene expression were assessed in order to detect which genes contributed to the differences in clinical characteristics. OS was compared between patients of different ages, and younger patients (aged <48 years) were found to have significantly prolonged OS compared to older patients (aged ≥48 years) (99.06 vs. 54.77 months, P=0.035, Figure 3A). Correlation analysis revealed that the variations of 79 molecules were significantly correlated with age difference in astrocytoma (r>0.2 or <−0.2, P<0.05) (Figure 3B). Among them, the expressions of 24 genes were positively correlated with increased age, and 57 gene expressions were negatively correlated with increased age (Figure 3C). A strong positive correlation was found between EYA4 and age, while TET1, SATB1, WAC, ZMYND11, and H2AFY2 showed a strong negative correlation (r>0.4 or <−0.4, P<0.0001).
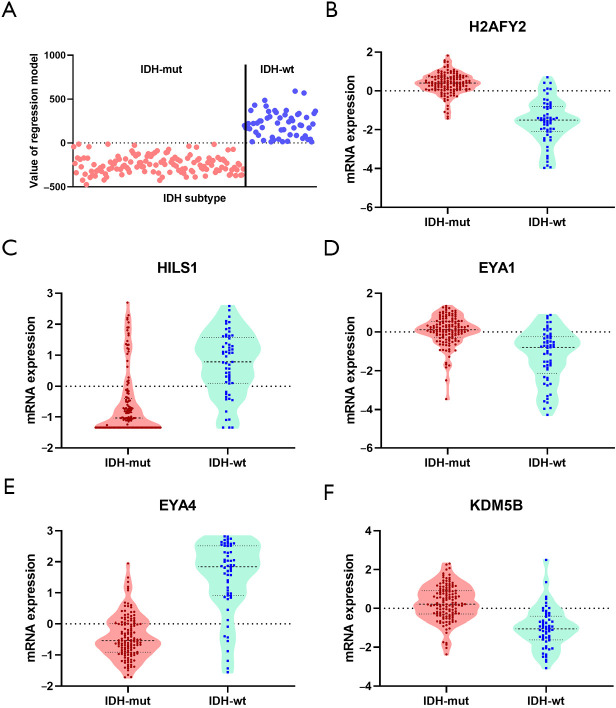
Additionally, prognostic comparisons of different grades of tumor revealed that patients with G2 grade disease showed significantly favorable OS compared to G3 patients (145.10 vs. 43.89 months, P<0.0001) (Figure 5A). An assessment of grade-correlated genes showed that 110 genes involved in chromatin change varied significantly between G2 and G3 grades (Figure 5B). The prognostic differences between the IDH subtypes (WT and mutant) were investigated; patients with IDH-mut type showed significantly prolonged OS (79.99 months), while those with IDH-wt type showed much worse OS (21.30 months, P<0.0001) (Figure 5C). A comparison of gene expressions between the two types indicated that 203 genes were significantly associated with IDH-mut (Figure 5D). To detect the overlap of gene sets with regard to age, grade, and IDH subtype, 34 common elements associated with differences in clinical characteristics were identified in astrocytoma (Figure 5E) and these genes are listed in Table 6. A regression analysis was conducted to verify the relevance of chromatin regulation genes and IDH-mut, with the results indicating that 5 genes (H2AFY2, HILS1, EYA1, EYA4, and KDM5B) were independently related to IDH-mut status. Moreover, the attribution of IDH subtypes was assessed, and a 5-gene model was found to be capable of distinguishing between the IDH-wt and IDH-mut subtypes (Figure 6A). Among these genes, the expression levels of H2AFY2, EYA1, and KDM5B were significantly higher in the IDH-mut type, whereas, the expression levels of HILS1 and EYA4 were significantly higher in the IDH-wt type (Figure 6B,C,D,E,F).
Figure 5.
Differences in the expression levels of genes correlated with grade and IDH mutation in astrocytoma. (A) The survival differences between G2 and G3 grades; (B) the differentially expressed genes relevant to differences in grade (C) the survival differences between the IDH-mut and IDH-wt subtypes; (D) the differentially expressed genes relevant to IDH-mut status; (E) the overlap of differentially expressed genes associated with age, tumor grade, and IDH-mut. IDH, isocitrate dehydrogenase.
Table 6. Genes involved in chromatin regulation which are significantly associated with clinical features in astrocytoma.
| Entry name | Protein names | Gene names |
|---|---|---|
| TET1 | Methylcytosine dioxygenase TET1 (EC 1.14.11.n2) (CXXC-type zinc finger protein 6) (Leukemia-associated protein with a CXXC domain) (Ten-eleven translocation 1 gene protein) | TET1, CXXC6, KIAA1676, LCX |
| PRDM6 | Putative histone-lysine N-methyltransferase PRDM6 (EC 2.1.1.361) (PR domain zinc finger protein 6) (PR domain-containing protein 6) | PRDM6, PFM3 |
| RAG1 | V(D)J recombination-activating protein 1 (RAG-1) (RING finger protein 74) [Includes: Endonuclease RAG1 (EC 3.1.-.-); E3 ubiquitin-protein ligase RAG1 (EC 2.3.2.27) (RING-type E3 ubiquitin transferase RAG1)] | RAG1, RNF74 |
| PF21A | PHD finger protein 21A (BHC80a) (BRAF35-HDAC complex protein BHC80) | PHF21A, BHC80, KIAA1696, BM-006 |
| WAC | WW domain-containing adapter protein with coiled-coil | WAC, KIAA1844 |
| SATB2 | DNA-binding protein SATB2 (Special AT-rich sequence-binding protein 2) | SATB2, KIAA1034 |
| SATB1 | DNA-binding protein SATB1 (Special AT-rich sequence-binding protein 1) | SATB1 |
| HDAC9 | Histone deacetylase 9 (HD9) (EC 3.5.1.98) (Histone deacetylase 7B) (HD7) (HD7b) (Histone deacetylase-related protein) (MEF2-interacting transcription repressor MITR) | HDAC9, HDAC7, HDAC7B, HDRP, KIAA0744, MITR |
| PHF19 | PHD finger protein 19 (Polycomb-like protein 3) (hPCL3) | PHF19, PCL3 |
| HDAC7 | Histone deacetylase 7 (HD7) (EC 3.5.1.98) (Histone deacetylase 7A) (HD7a) | HDAC7, HDAC7A |
| ACL6B | Actin-like protein 6B (53 kDa BRG1-associated factor B) (Actin-related protein Baf53b) (ArpNalpha) (BRG1-associated factor 53B) (BAF53B) | ACTL6B, ACTL6, BAF53B |
| ASF1A | Histone chaperone ASF1A (Anti-silencing function protein 1 homolog A) (hAsf1) (hAsf1a) (CCG1-interacting factor A) (CIA) (hCIA) | ASF1A, CGI-98, HSPC146 |
| HIRA | Protein HIRA (TUP1-like enhancer of split protein 1) | HIRA, DGCR1, HIR, TUPLE1 |
| LRWD1 | Leucine-rich repeat and WD repeat-containing protein 1 (Centromere protein 33) (CENP-33) (Origin recognition complex-associated protein) (ORC-associated protein) (ORCA) | LRWD1, CENP-33, ORCA |
| BAHD1 | Bromo adjacent homology domain-containing 1 protein (BAH domain-containing protein 1) | BAHD1, KIAA0945 |
| DTX3L | E3 ubiquitin-protein ligase DTX3L (EC 2.3.2.27) (B-lymphoma- and BAL-associated protein) (Protein deltex-3-like) (RING-type E3 ubiquitin transferase DTX3L) (Rhysin-2) (Rhysin2) | DTX3L, BBAP |
| FOXA2 | Hepatocyte nuclear factor 3-beta (HNF-3-beta) (HNF-3B) (Forkhead box protein A2) (Transcription factor 3B) (TCF-3B) | FOXA2, HNF3B, TCF3B |
| ING3 | Inhibitor of growth protein 3 (p47ING3) | ING3, HSPC301 |
| HDAC4 | Histone deacetylase 4 (HD4) (EC 3.5.1.98) | HDAC4, KIAA0288 |
| HDAC5 | Histone deacetylase 5 (HD5) (EC 3.5.1.98) (Antigen NY-CO-9) | HDAC5, KIAA0600 |
| ZMY11 | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A-binding protein) (Bone morphogenetic protein receptor-associated molecule 1) (Protein BS69) | ZMYND11, BRAM1, BS69 |
| EYA4 | Eyes absent homolog 4 (EC 3.1.3.48) | EYA4 |
| DNJC2 | DnaJ homolog subfamily C member 2 (M-phase phosphoprotein 11) (Zuotin-related factor 1) [Cleaved into: DnaJ homolog subfamily C member 2, N-terminally processed] | DNAJC2, MPHOSPH11, MPP11, ZRF1 |
| CHD3 | Chromodomain-helicase-DNA-binding protein 3 (CHD-3) (EC 3.6.4.12) (ATP-dependent helicase CHD3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc finger helicase) (hZFH) | CHD3 |
| ASF1B | Histone chaperone ASF1B (Anti-silencing function protein 1 homolog B) (hAsf1) (hAsf1b) (CCG1-interacting factor A-II) (CIA-II) (hCIA-II) | ASF1B |
| CBX6 | Chromobox protein homolog 6 | CBX6 |
| SMRC2 | SWI/SNF complex subunit SMARCC2 (BRG1-associated factor 170) (BAF170) (SWI/SNF complex 170 kDa subunit) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2) | SMARCC2, BAF170 |
| TOX | Thymocyte selection-associated high mobility group box protein TOX (Thymus high mobility group box protein TOX) | TOX, KIAA0808 |
| SMYD2 | N-lysine methyltransferase SMYD2 (EC 2.1.1.-) (HSKM-B) (Histone methyltransferase SMYD2) (EC 2.1.1.354) (Lysine N-methyltransferase 3C) (SET and MYND domain-containing protein 2) | SMYD2, KMT3C |
| HILS1 | Putative spermatid-specific linker histone H1-like protein (H1.9 linker histone pseudogene) | H1-9P, H1-9, HILS1 |
| MRGBP | MRG/MORF4L-binding protein (MRG-binding protein) (Up-regulated in colon cancer 4) (Urcc4) | MRGBP, C20orf20 |
| KAT6B | Histone acetyltransferase KAT6B (EC 2.3.1.48) (Histone acetyltransferase MOZ2) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (MYST-4) (Monocytic leukemia zinc finger protein-related factor) | KAT6B, KIAA0383, MORF, MOZ2, MYST4 |
| BUD23 | Probable 18S rRNA (guanine-N(7))-methyltransferase (EC 2.1.1.-) (Bud site selection protein 23 homolog) (Metastasis-related methyltransferase 1) (Williams-Beuren syndrome chromosomal region 22 protein) (rRNA methyltransferase and ribosome maturation factor) | BUD23, MERM1, WBSCR22, HUSSY-03, PP3381 |
| H2AW | Core histone macro-H2A.2 (Histone macroH2A2) (mH2A2) | MACROH2A2, H2AFY2 |
Figure 6.
The chromatin regulation genes independently implicated in IDH-mut. (A) The regression model distinguishing the IDH-mut and IDH-wt types; (B,C,D,E,F) H2AFY2, EYA1, KDM5B, HILS1, and EYA4 exhibited different expression levels in the IDH-mut and IDH-wt types in astrocytoma. IDH, isocitrate dehydrogenase.
Discussion
Glioma is one of the most common primary brain malignancies and has a high mortality rate (19). Current treatment involves surgical resection followed by chemotherapy and radiotherapy. However, patient outcomes remain disappointing (20). Low-grade astrocytoma is a relatively slow-growing histological type of glioma but has an extremely heterogeneous clinical manifestation (21). Many studies have been conducted on specific populations; however, the identification of specific subgroups and the targeted management of astrocytoma remain inadequate. Classifying patients with astrocytoma on the basis of their genetic features is important for predicting their prognoses after surgery and improving treatment strategies. Chromatin remodeling, the changing configuration of chromatin structure, is crucial in the regulation of gene expression, apoptosis, and DNA replication and repair. Dysfunctions in chromatin-remodeling mechanisms have been associated with disease development and, in particular, alterations in chromatin structure can lead to the deregulated expression of tumor suppressor genes or oncogenes, and cancer initiation (22). The present study was conducted to investigate the expression patterns of chromatin regulation genes in samples from different populations, in an effort to distinguish specific gene expression profiles.
Two primary clusters and three subclusters of astrocytoma patients were identified. Significant differences in prognosis between each group were revealed by comparisons of OS, DSS, DFS, and PFS. Cluster_1 (or subcluster_1) had the worst survival (19.89 months of OS) of any cluster or subcluster. Further analysis of the different gene expressions between the clusters and subclusters revealed that 191 overlapped genes were significantly varied in astrocytoma (Figure 2C). Moreover, the cohort with poor prognosis demonstrated low expression levels of H2AFY2, WAC, HDAC5, ZMYND11, TET1, SATB1, and MYST4, and high expression levels of EYA4. These results suggest that changes in genes involved in chromatin regulation are potentially relevant to the clinical manifestation and outcomes of astrocytoma. The prognostic roles of eight differentially expressed genes were also investigated. High expression levels of H2AFY2, WAC, HDAC5, ZMYND11, TET1, SATB1, and MYST4 were significantly related to favorable OS in astrocytoma. Strikingly, the hazard ratio of H2AFY2 expression was as high as 4.16. In contrast to other genes, the high expression of EYA4 was significantly correlated with short-term survival, and the hazard ratio was as low as 0.21. These results reveal the significant potential prognostic role that genes involved in chromatin regulation may play in astrocytoma.
There was a gradual improvement in survival from subcluster_1 to subcluster_3. Therefore, we examined the differences in gene expression correlated with these subcluster changes. Importantly, 24 genes were found to be positively correlated with the subcluster order and 26 genes were negatively correlated with the subcluster order (Table S1). The genes associated with these subcluster orders included the eight differently expressed genes mentioned previously, which confirms the potential prognostic roles of these genes in astrocytoma.
When comparing clinical characteristics between the defined clusters in astrocytoma, we discovered differing distributions in age, grade and IDH genotype according to cluster. Compared to patients with a favorable prognosis, the poor prognostic cohort (Cluster_1 or Subcluster_1) was significantly older (mean =52.3 years), had more advanced-grade tumors (92% G3), and exhibited widely distributed non-mutant IDH genotype (88% IDH-wt). Identification of the genes associated with age revealed that EYA4 was strongly and positively correlated with older age, while TET1, SATB1, WAC, ZMYND11, and H2AFY2 showed a strong negative correlation with older age. All of these genes that were correlated with age were also found in the differentially expressed gene inter-cluster noted above. It is common knowledge that advanced tumor grades are associated with worse prognosis in multiple carcinomas. In the current study, we confirmed that G2 astrocytoma shows a considerably favorable prognosis compared to G3 (145.10 vs. 43.89 months). Moreover, the assessment of differentially expressed genes between patients with different tumor grades revealed that 110 genes involved in chromatin changes were significantly associated with grade progression.
IDH mutations are initiating events that define major clinical and prognostic classes of gliomas (23). Previous studies have identified an IDH mutant-enriched subtype of cholangiocarcinoma with low expression levels of chromatin remodeling genes (24). However, the molecular features of the IDH-mut population in astrocytoma are largely unknown. In this study we identified that the poor-survival cohort was significantly associated with IDH subtype. Patients with the IDH-mut subtype were also discovered to exhibit high expression levels of H2AFY2, EYA1, and KDM5B, and low expression levels of HILS1 and EYA4, compared to those with IDH-wt. These results suggest that chromatin modification is linked to basic biological events that are also regulated by IDH mutations in astrocytoma.
In this study, we found that H2AFY2 (core histone macro-H2A.2 coding gene) and EYA4 (Eyes Absent Homologue 4 coding gene) play a significant role in astrocytoma. H2AFY2, which is involved in transcriptional repression, was found to be associated with prognosis and is significantly correlated with increased age and IDH-mut status. However, this gene has not been studied in glioma before, so there is no prior evidence to support the interactions of H2AFY2 and IDH mutation. In previous research, H2AFY2 has been reported to be differentially expressed and co-expressed with noncoding RNAs LOC286002 in breast cancer (25). In another study on nervous system disease, H2AFY2 was specifically downregulated by the HDAC inhibitors (26). The expression changes indicate the relevance of H2AFY2 and HDACs. Our results showed that, in astrocytoma, the expression levels of H2AFY2 were positively correlated with HDAC5 (r=0.629, P<0.0001). Our data also revealed that the expression of HDAC5 was positively implicated with OS in astrocytoma (29.13 vs. 73.48 months, P<0.0001, HR =3.06, 95% CI: 1.746–5.361), and this result is consistent with the findings of previous research (27). In contrast, a previous study reported that HDAC5 expression was significantly upregulated in high-risk medulloblastoma in comparison with low-risk medulloblastoma and is associated with poor survival (28). We attribute this discrepancy to the effect of different histological types. The significant prognostic and negatively correlated gene, EYA4, has been reported to play an important role in tumorigenesis and the progression of various cancers (29,30). In pancreatic ductal adenocarcinoma and hepatocellular carcinoma, EYA4 has been shown to work as a tumor suppressor gene, with its overexpression inhibiting tumor proliferation and invasion (29,30). However, in this study, elevated EYA4 was found to be associated with a shortened OS in astrocytoma. A recently published study supported our result, indicating a reverse predictive role of EYA4, which promoted tumor progression as a result of the downregulation of p27Kip1 in glioma (31). These results indicate that the expression changes of the specific genes involved in chromatin modification were notable in the development of astrocytoma.
In conclusion, this study described the systemic expression pattern of chromatin regulation genes in low-grade astrocytoma and identified a cohort with poor prognosis. Chromatin regulation genes are significantly associated with prognosis and correlated with age, grade, and IDH subtype in astrocytoma. This study has highlighted the prognostic role of chromatin regulation genes and the relevance of gene expression and biological characteristics in astrocytoma. However, further research is necessary to validate the current findings. Our results suggest that chromatin regulation genes have a promising role to play in the prediction of postoperative survival, and that this association with prognosis could make them important therapeutic targets in astrocytoma.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). All utilized public omics datasets had been generated in previous studies and obtained with prior ethical approval.
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7229
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7229). The authors have no conflicts of interest to declare.
(English Language Editors: D. Fitzgerald and J. Reynolds)
References
- 1.Tan AC, Ashley DM, López GY, et al. Management of glioblastoma: State of the art and future directions. CA Cancer J Clin 2020;70:299-312. 10.3322/caac.21613 [DOI] [PubMed] [Google Scholar]
- 2.Reardon DA, Rich JN, Friedman HS, et al. Recent advances in the treatment of malignant astrocytoma. J Clin Oncol 2006;24:1253-65. 10.1200/JCO.2005.04.5302 [DOI] [PubMed] [Google Scholar]
- 3.Reed LK, Huang JH. Variability of relative cerebral blood volume measurements of recurrent glioma. Ann Transl Med 2019;7:S260. 10.21037/atm.2019.12.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yahanda AT, Patel B, Shah AS, et al. Impact of Intraoperative Magnetic Resonance Imaging and Other Factors on Surgical Outcomes for Newly Diagnosed Grade II Astrocytomas and Oligodendrogliomas: A Multicenter Study. Neurosurgery 2020. [Epub ahead of print]. 10.1093/neuros/nyaa320 [DOI] [PubMed] [Google Scholar]
- 5.Franceschi E, Tosoni A, Bartolini S, et al. Histopathological grading affects survival in patients with IDH-mutant grade II and grade III diffuse gliomas. Eur J Cancer 2020;137:10-7. 10.1016/j.ejca.2020.06.018 [DOI] [PubMed] [Google Scholar]
- 6.Richardson TE, Snuderl M, Serrano J, et al. Rapid progression to glioblastoma in a subset of IDH-mutated astrocytomas: a genome-wide analysis. J Neurooncol 2017;133:183-92. 10.1007/s11060-017-2431-y [DOI] [PubMed] [Google Scholar]
- 7.Choi C, Raisanen JM, Ganji SK, et al. Prospective Longitudinal Analysis of 2-Hydroxyglutarate Magnetic Resonance Spectroscopy Identifies Broad Clinical Utility for the Management of Patients With IDH-Mutant Glioma 2016;34:4030-9. [DOI] [PMC free article] [PubMed]
- 8.Kondo Y, Katsushima K, Ohka F, et al. Epigenetic dysregulation in glioma. Cancer Sci 2014;105:363-9. 10.1111/cas.12379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zang L, Kondengaden SM, Che F, et al. Potential Epigenetic-Based Therapeutic Targets for Glioma. Front Mol Neurosci 2018;11:408. 10.3389/fnmol.2018.00408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez FJ, Brosnan-Cashman JA, Allen SJ, et al. Alternative lengthening of telomeres, ATRX loss and H3-K27M mutations in histologically defined pilocytic astrocytoma with anaplasia. Brain Pathol 2019;29:126-40. 10.1111/bpa.12646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bönisch C, Nieratschker SM, Orfanos NK, et al. Chromatin proteomics and epigenetic regulatory circuits. Expert Rev Proteomics 2008;5:105-19. 10.1586/14789450.5.1.105 [DOI] [PubMed] [Google Scholar]
- 12.Spyropoulou A, Piperi C, Adamopoulos C, et al. Deregulated chromatin remodeling in the pathobiology of brain tumors. Neuromolecular Med 2013;15:1-24. 10.1007/s12017-012-8205-y [DOI] [PubMed] [Google Scholar]
- 13.Martinez R, Esteller M. The DNA methylome of glioblastoma multiforme. Neurobiol Dis 2010;39:40-6. 10.1016/j.nbd.2009.12.030 [DOI] [PubMed] [Google Scholar]
- 14.Xie Y, Tan Y, Yang C, et al. Omics-based integrated analysis identified ATRX as a biomarker associated with glioma diagnosis and prognosis. Cancer Biol Med 2019;16:784-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ata SK, Fang Y, Wu M, et al. Disease Gene Classification with Metagraph Representations. Methods Mol Biol 2018;1807:211-24. 10.1007/978-1-4939-8561-6_16 [DOI] [PubMed] [Google Scholar]
- 17.de Hoon MJ, Imoto S, Nolan J, et al. Open source clustering software. Bioinformatics 2004;20:1453-4. 10.1093/bioinformatics/bth078 [DOI] [PubMed] [Google Scholar]
- 18.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics 2004;20:3246-8. 10.1093/bioinformatics/bth349 [DOI] [PubMed] [Google Scholar]
- 19.Chien LN, Gittleman H, Ostrom QT, et al. Comparative Brain and Central Nervous System Tumor Incidence and Survival between the United States and Taiwan Based on Population-Based Registry. Front Public Health 2016;4:151. 10.3389/fpubh.2016.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castro-Gamero AM, Pezuk JA, Brassesco MS, et al. G2/M inhibitors as pharmacotherapeutic opportunities for glioblastoma: the old, the new, and the future. Cancer Biol Med 2018;15:354-74. 10.20892/j.issn.2095-3941.2018.0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shirahata M, Ono T, Stichel D, et al. Novel, improved grading system(s) for IDH-mutant astrocytic gliomas. Acta Neuropathol 2018;136:153-66. 10.1007/s00401-018-1849-4 [DOI] [PubMed] [Google Scholar]
- 22.Endo A, Ly T, Pippa R, et al. The Chromatin Assembly Factor Complex 1 (CAF1) and 5-Azacytidine (5-AzaC) Affect Cell Motility in Src-transformed Human Epithelial Cells. J Biol Chem 2017;292:172-84. 10.1074/jbc.M116.751024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flavahan WA, Drier Y, Liau BB, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 2016;529:110-4. 10.1038/nature16490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farshidfar F, Zheng S, Gingras MC, et al. Integrative Genomic Analysis of Cholangiocarcinoma Identifies Distinct IDH-Mutant Molecular Profiles. Cell Rep 2017;18:2780-94. 10.1016/j.celrep.2017.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong Y, Zhang T, Li X, et al. Comprehensive analysis of coexpressed long noncoding RNAs and genes in breast cancer. J Obstet Gynaecol Res 2019;45:428-37. 10.1111/jog.13840 [DOI] [PubMed] [Google Scholar]
- 26.Soragni E, Chou CJ, Rusche JR, et al. Mechanism of Action of 2-Aminobenzamide HDAC Inhibitors in Reversing Gene Silencing in Friedreich's Ataxia. Front Neurol 2015;6:44. 10.3389/fneur.2015.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dali-Youcef N, Froelich S, Moussallieh FM, et al. Gene expression mapping of histone deacetylases and co-factors, and correlation with survival time and 1H-HRMAS metabolomic profile in human gliomas. Sci Rep 2015;5:9087. 10.1038/srep09087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milde T, Oehme I, Korshunov A, et al. HDAC5 and HDAC9 in medulloblastoma: novel markers for risk stratification and role in tumor cell growth. Clin Cancer Res 2010;16:3240-52. 10.1158/1078-0432.CCR-10-0395 [DOI] [PubMed] [Google Scholar]
- 29.Zhu XX, Li JH, Cai JP, et al. EYA4 inhibits hepatocellular carcinoma by repressing MYCBP by dephosphorylating β-catenin at Ser552. Cancer Sci 2019;110:3110-21. 10.1111/cas.14159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mo SJ, Liu X, Hao XY, et al. EYA4 functions as tumor suppressor gene and prognostic marker in pancreatic ductal adenocarcinoma through β-catenin/ID2 pathway. Cancer Lett 2016;380:403-12. 10.1016/j.canlet.2016.06.021 [DOI] [PubMed] [Google Scholar]
- 31.Li Z, Qiu R, Qiu X, et al. EYA4 Promotes Cell Proliferation Through Downregulation of p27Kip1 in Glioma. Cell Physiol Biochem 2018;49:1856-69. 10.1159/000493631 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as