Abstract
Objective: The objective of this study was to investigate the effect on the proliferation, invasion, and apoptosis of bladder cancer cells through miR-502-5p of the Circ_0000735 circular RNA. Methods: Circ_0000735 and miR-502-5p expression of bladder cancer patients in malignant and paracancerous tissues was identified using qRT-PCR. Nucleoplasm isolation assay and RNase R enzymatic assay were used to classify Circ_0000735 subcellular origin and stability. Dual luciferase reporter assay and RIP assay were used to confirm Circ_0000735 and miR-502-5p targeting relationships. Cell proliferation, apoptosis, and invasion capacity were identified using CCK8, flow cytometry, and transwell assays. To confirm the effect of Circ_0000735 on tumorigenesis in nude mice, in vivo experiments were conducted. Results: Circ_0000735 expression was increased in bladder cancer tissues and cells compared with paraneoplastic tissues and normal cells, and miR-502-5p expression was reduced (both P<0.05). In the cytoplasm, Circ_0000735 was largely clustered and could not be digested by the RNase R enzyme, and ceRNA may play a role in bladder cancer cells. Circ_0000735 silencing prevented cell proliferation and invasion and facilitated apoptosis (all P<0.05). The incorporation of miR-502-5p inhibitor rescued the effect on bladder cancer cells of Circ_0000735 silencing. In vitro experiments showed that inhibition of Circ_0000735 expression was beneficial in suppressing tumorigenic ability in nude mice. Conclusion: Circ_0000735 can adsorb miR-502-5p to promote bladder cancer cell proliferation and invasion and inhibit apoptosis. Circ_0000735 may be an effective molecular target for bladder cancer therapy.
Keywords: circ_0000735, miR-502-5p, bladder cancer, cell proliferation, invasion, apoptosis
Introduction
Bladder cancer is a common urinary system malignant tumor [1]. Bladder cancer has the highest morality among urinary system tumors according to Chinese cancer statistics [2]. Despite improving earlier diagnosis and systemic treatment of bladder cancer, many patients get a recurrence or metastasis [3]. There are two sub-types of bladder cancer based on the tumor’s penetration depth: non-muscle-invasive tumors and tumors with muscle invasion [4]. Invasive tumors comprise about 20-30 percent of all primary bladder tumors and have a worse prediction due to a higher likelihood of metastasis [5]. Invasive tumors are more common. Therefore its mechanism of action is important to understand and new therapeutic targets can be identified.
Circular RNAs (circRNAs), unlike traditional linear RNAs, are a class of endogenous circular non-coding RNAs [6]. circRNAs originate from exonic or intronic regions of genes and are stably present in the cell. circRNAs are abundant in mammalian cells, unlike linear RNAs [7]. Once thought to be a byproduct of gene mis-transcription, circRNAs are now considered important regulators of a variety of intracellular processes, such as the development and progression of cancer cells [8].
Different RNAs (mRNAs, pseudogenes, lncRNAs) may function as competitive endogenous RNAs (ceRNAs) by competitively binding microRNAs (miRNAs) in the cytoplasm [9]. As a novel non-coding RNA, circRNAs were recognized as a form of ceRNA acting as miRNA sponges, resulting in a loss of miRNA function [10]. CircRNA ciRS-7, for example, was the most commonly known and well-studied circRNA, which can facilitate the metastases of various tumors by sponging miR-7 [11-13]. CircFBXW7 is regulated in tumor tissues and prevents the malignant progression of cancer cells by regulating miR-197-3p and encoding a 185-AA protein [14]. CircKIF4A is identified as a tumor-promoting factor for bladder cancer by competing with endogenous RNAs [15]. circ_0000735 is transcribed from the gene P2RX1 and is predominantly located at chr17:3802927-3808661 (hg19). Circ_0000735 is reported to be upregulated in prostate cancer, and promotes the proliferation and colony formation and inhibits apoptosis of cancer cells by sponging miR-7 [16]. Besides, Circ_0000735 expression is increased in non-small cell lung cancer. It also sponges miR-1179/1182, which promotes the malignant progression of cancer cells [17]. We speculate that Circ_0000735 may also promote the development of bladder cancer.
We explored the role and function of Circ_0000735 in bladder cancer in this research. In bladder cancer tissues and cells, the expression of Circ_0000735 was increased. The silencing of Circ_0000735 substantially prevented the development of bladder cancer, so we consider that Circ_0000735 may be an oncogene in bladder cancer. RIP and dual luciferase reporter assays have verified that Circ_0000735 will bind to miR-502-5p, which is the target gene for Circ_0000735. In conclusion, Circ_0000735 may sponge miR-502-5p to facilitate the progression of bladder cancer.
Materials and methods
OC organization collection
Primary bladder cancer tissues and paraneoplastic tissues were collected from Affiliated Hospital of Xiangnan University. Immediately after the resection, the tissue was frozen. This study was approved by the Ethics Committee of the Affiliated Hospital of Xiangnan University and was in accordance with the Helsinki Declaration. Informed consent was obtained from all patients. Animal experiments have been performed in line with the recommendations of the Animal Treatment and Experiment Committee of the Affiliated Hospital of Xiangnan University.
Cell culture and transfection
Both cell lines used in this research were purchased from ATCC, USA, namely SV-HUC, 5637, RT-112 and BIU-87. Cells were grown in a medium comprising 10% FBS in RPMI 1640 (31870082, Thermo Fisher Scientific, Inc., USA) and deposited in an incubator at 37°C and 5% CO2.
The cultured cells were randomly divided into Blank (control group), si-NC (negative control sequence transfection), si-circ (si-circ transfection), si-circ+inhibitor-NC (si-circ and inhibitor-NC transfection), si-NC+miR-502-5p (si-NC and miR-502-5p inhibitor transfection), and si-circ+miR-502-5p (si-circ and miR-502-5p inhibitor transfection). Shanghai Genepharma Pharmaceutical Technology Co. engineered and synthesized the sequences. Using the LipofectamineTM 3000 transfection reagent, the sequences were transfected into BIU-87 cells and according to the kit instructions.
Nucleoplasm isolation assay
500 μL of CER reagent was applied to every 1×107 cells or every 100 μL (cell volume after centrifugation) of cell precipitate using the Nucleus-Cytoplasmin-Cytoplasmic Membrane Preparation Kit (P1201, Beijing Apply Gene Technology Co., China), according to the manufacturer’s instructions. Then the mixture was resuspended by shaking and bathed for 2 minutes with ice. In an ice bath, the cell suspensions were manually homogenized 20-30 times. To acquire the nucleus and cytosol-cytoplasmic supernatant at the bottom of the tube, frozen tissue was homogenized and lysed by centrifugation. The supernatant and the precipitate were separately transferred.
RNase R enzymatic tests
Total RNA extracted from tissues and cells (2 μg) was incubated at 37°C for 20 minutes, and 3 U/μg RNase R was then added. Using the RNeasy MinElute Purification Kit (74204, Hangzhou Watson Biotechnology Co., Ltd., China), the resulting RNA was then purified, and the final qRT-PCR analysis was carried.
Analysis of dual luciferase reports
BIU-87 bladder cancer cells were inoculated into 96-well plates with 3×103 cells per well. The plasmids Circ_0000735-wt and Circ_0000735-mut were constructed, transfected for 48 h with miRNA mimics or inhibitors, and a dual luciferase reporter kit (RG027, Shanghai Beyotime Biotechnology Co., Ltd., China) was used to detect the relative activity of luciferase.
Quantitative real-time polymerase chain reaction
Using the Trizol kit (15596026, Thermo Fisher Scientific, Inc., USA), total RNA was extracted from cells and fresh tissues as directed by the manufacturer, and cDNA was synthesized using a reverse transcription kit (Thermo Scientific Technologies, USA), accompanied by Real-time Quantitative Fluorescence PCR. GAPDH and U6 were used as internal references. The primer sequences are shown in Table 1.
Table 1.
qRT-PCR sequences
| Names | Sequence (5’-3’) | |
|---|---|---|
| Circ_0000735 | Forward | GTGGAGTGGTTGGCATCACC |
| Reverse | GAGAAACACCCACCTGAAGTTGA | |
| miR-502-5p | Forward | ATCCTTGCTATCTGGGTGCTA |
| Reverse | CGAATTCTAGAGCTCGAGGCAGG | |
| GAPDH | Forward | ACCACAGTCCATGCCATCAC |
| Reverse | TCCACCACCCTGTTGCTGTA | |
| U6 | Forward | GCUUCGGCAGCACAUAUACUAAAAU |
| Reverse | CGCUUCACGAAUUUGCGUGUCAU |
RIP
According to the guidance of the maker, the Magna RIP suite (17-700, Shanghai Hu Zhen Industrial Co., China) and Ago2 antibody (hz-50683-R036) were applied. A total of 107 transfected cells were washed twice in a buffer of equivalent volume of ice-cold PBS and RIP lyses, and 2 μg of primary antibody were incubated for 2 hours at 4°C. Each sample was subsequently applied with 50 μg of prepared magnetic beads and overnight incubated at 4°C. The beads were briefly rinsed 5 times with RIP buffer and were resuspended in Trizol. Real-time qPCR was used to detect binding products.
CCK-8
A total of 5×103 cells were inoculated in 96-well plates. In each of these wells, 10 μL of CCK-8 was added. The absorbance at 450 nm was measured to detect the proliferation of cells after incubation at 37°C over 1 h.
Transwell
A total of 24 wells were used to test cell invasion in BD Transwell chambers. Cells were inoculated in 500 μL serum-free medium and then it was applied as a chemoattractant to the lower chamber with a 10% FBS. On the lower surface of the inserts we have set and stained the invading cells with 1% crystalline violet for the first day after incubation to 37°C. The cells were eventually counted and pictured.
Flow cytometry analysis
300 g of suspendedcells and cells attached to the tube were collected after digestion with trypsin without EDTA and centrifugation at 4°C for 5 min, respectively; cells were washed twice with 300 g pre-cooled PBS, and were centrifuged at 4°C for 5 min. 1×105 to 5×105 were collected. PBS was removed and 100 μL of 1× Binding Buffer was applied to the resuspended cells; 5 μL of Annexin V-FITC and 10 μL of PI Staining Solution were applied and gently combined; the sample was set at room temperature for 10-15 min; 400 μL of 1× Binding Buffer was added, combined and deposited on ice, and samples were identified by flow cytometry within 1 hour. With a passband filter at 515 nm, FITC fluorescence was observed by excitation at 488 nm and PI was observed with a passband filter at 560 nm. The lower left quadrant is for living cells, the upper right quadrant is for necrotic cells and the lower right quadrant is for apoptotic cells in the bivariate flow cytometry two-dimensional scatter plot.
Mouse xenograft model
The dorsal side of BALB/c nude mice (4 weeks old, female) was subcutaneously injected with 5×107 BIU-87 bladder cancer cells. Mice were randomly clustered (5 mice/group) and intratumorally injected with cholesterol-modified si-NC or si-circ_0000735 at 20 mg/kg every 4 days after a tumor became palpable. The mice were euthanized, weighed, and tumors were registered after 4 weeks.
Statistical analysis
In this research, every statistical analysis was done with SPSS 22.0 (SPSS, Inc., Chicago, IL, USA). Quantitative data were expressed as mean ± standard deviation (x̅ ± sd). The t-test was used to compare the groups. P<0.05 was considered significant.
Results
Circ_0000735 expression was enhanced in bladder cancer tissues and cells
The expression of Circ_0000735 was observed by qRT-PCR in bladder cancer tissues, and Circ_0000735 was highly expressed in cancer tissues (P<0.0001) compared to adjacent healthy tissues (n=30) (Figure 1A). In addition, Circ_0000735 was significantly upregulated relative to SV-HUC cell lines in 5637, RT-112, and BIU-87 cell lines (all P<0.05) (Figure 1B). For subsequent experiments, the BIU-87 cell line will be chosen. The findings showed that in bladder cancer tissues and cells, Circ_0000735 was highly expressed.
Figure 1.

Relative expression of Circ_0000735. A: Circ_0000735 expression in tissues; B: Circ_0000735 expression in cells. Compared with SV-HUC, *P<0.05.
Knockdown of circ_0000735 inhibited the proliferation and invasion of bladder cancer cells and induced apoptosis
In order to observe the possible role of circ_0000735 in cells, circ_0000735 was first extracted in BIU-87 cells (Figure 2A) and si-circ#2 with the highest knockdown efficacy was chosen for the follow-up assay; CCK-8 revealed that the knockdown of circ_0000735 substantially reduced the spread of bladder cancer (P<0.05) (Figure 2B); Flow cytometry analysis showed that once circ_0000735 was downregulated in BIU-87 cells, the number of apoptotic cells increased rapidly (P<0.05) (Figure 2C). As shown in Figure 2D, relative to the si-NC group, the number of invading cells was significantly reduced in cells transfected with si-circ_0000735 with BIU-87 (P<0.05).
Figure 2.

Effects of knocking out circ_0000735 on cell proliferation, invasion, and apoptosis. A: circ_0000735 knockdown results; B: Knockdown of circ_0000735 inhibited cell proliferation; C: Flow cytometry detection of apoptosis; D: Cell invasion results by Transwell method (400×). Compared with si-NC group, #P<0.05.
circ_0000735 is predominantly cytoplasmic and cannot be degraded by RNase R
Circ_0000735 was mainly localized in the cytoplasm as detected by nucleoplasm isolation experiments (Figure 3A). RNase R usually degrades the phosphodiester bond at the end of the RNA by exonuclease and can digest most linear RNA molecules, but cannot easily digest circular RNA. The results showed that circ_0000735 could not be digested by RNase R enzyme (P<0.05), indicating that circ_0000735 was resistant to RNase R enzyme, and circ_0000735 had a cyclic structure (Figure 3B).
Figure 3.
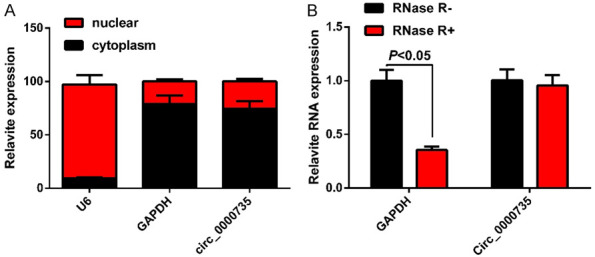
Nature of circ_0000735. A: Nucleus isolation assay detects circ_0000735 localization to the cytoplasm. B: qRT-PCR detects circ_0000735 resistance to RNase R.
miR-502-5p is down-regulated in bladder cancer tissues and cells and has a target-binding relationship with circ_0000735
In cancer tissues, miR-502-5p was under-expressed (P<0.05) (Figure 4A) relative to adjacent healthy tissues (n=30). In addition, miR-502-5p was significantly down-regulated relative to SV-HUC cell lines (all P<0.05) in 5637, RT-112, and BIU-87 cell lines (Figure 4B). Correlation analysis showed that circ_0000735 levels were negatively correlated with miR-502-5p levels (P<0.0001, r=-0.6634) (Figure 4C) in bladder cancer tissues. Several binding sites between circ_0000735 and miR-502-5p (Figure 4D) were analyzed by the bioinformatics tool circinteractome (https:/circinteractome.nia.nih.gov). We conducted a dual luciferase reporter assay to validate the relationship between them. The results showed that the luciferase activity of 293 T cells transfected with circ_0000735-wt (P<0.05) was significantly reduced by miR-502-5p transfection but did not alter the luciferase activity of 293 T cells transfected with circ_0000735-mut (Figure 4E). The circ_0000735 knockdown group was more successful than the control group in terms of the luciferase assay (P<0.05). The Ago2 antibody was effectively precipitated by miR-502-5p (Figure 4F). Such data indicate that miR-502-5p is actually a circ_0000735 target, and circ_0000735 affects its level of expression in bladder cancer cells.
Figure 4.

Relative expression of miR-502-5p and its targeting relationship with circ_0000735. A: miR-502-5p expression in tissues; B: miR-502-5p expression in cells; C: miR-502-5p and circ_0000735 are negatively correlated. D: The website predicts the presence of complementary sequences between miR-502-5p and circ_0000735; E: Dual luciferase reports the results of the experiment. F: RIP experimental results. Compared with SV-HUC, *P<0.05.
The effect of si-circ_0000735 on bladder cancer cells can be partially rescued by miR-502-5p inhibitor
The CCK-8 and Transwell tests showed that circ_0000735 knockdown could significantly inhibit cell proliferative activity and invasion number relative to the si-NC group (P<0.05), while miR-502-5p inhibitor could partly inhibit the effect of si-circ_0000735 on the spread and invasion of bladder cancer cells (Figure 5A and 5C). Flow cytometry analysis found that in the si-circ_0000735 and si-circ_0000735+NC inhibitor groups, apoptosis was considerably induced compared to the si-NC classes (both P<0.05). The reversed miR-502-5p inhibitor was partly caused by the si-circ_0000735 induced apoptosis rate (Figure 5B). In conclusion, the effects of si-circ_0000735 on bladder cancer cell proliferation, invasion and apoptosis were partially rescued by miR-502-5p inhibitor.
Figure 5.
miR-502-5p inhibitor can partially rescue the effects of si-circ_0000735 on cells. A: CCK8 method to detect up-regulation of cell proliferation; B: Flow cytometry to detect apoptosis; C: Transwell method to detect cell invasion (400×). Compared with si-NC group, #P<0.05; compared with si-circ_0000735+inhibitor-NC group, ^P<0.05.
Circ_0000735 knockdown inhibits tumorigenesis in nude mice
A mouse xenograft model was created to further evaluate the role of circ_0000735 in vivo. The results showed that silencing circ_0000735 could reduce tumor volume and weight relative to the si-NC group as determined by the BIU-87 bladder cancer cell line (both P<0.05) (Figure 6). The results showed that circ_0000735 knockdown could inhibit the tumorigenic ability innude mice.
Figure 6.
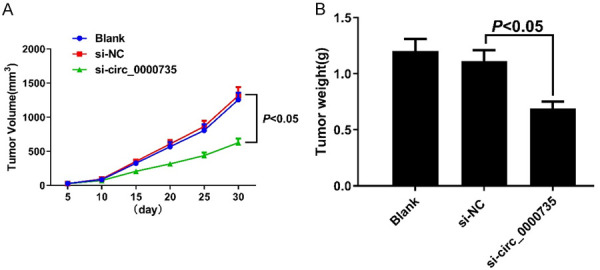
Knocking out circ_0000735 inhibits tumorigenic ability in nude mice. A: Tumor volume of nude mice at different time points; B: Tumor weight of nude mice in each group.
Discussion
For the first time, this study showed the role of Circ_0000735 in tissues and cells of bladder cancer. Circ_0000735 knockdown functionally inhibited bladder cancer cell proliferation, invasion, and tumor formation, and promoted apoptosis. Circ_0000735 mechanistically has a target-binding relationship with miR-502-3p. Our study showed that miR-502-5p sponge adsorption by Circ_0000735 facilitated bladder cancer cell proliferation and invasion and inhibited their apoptosis.
As the most prevalent genitourinary tract disease in China [18,19], bladder cancer is vulnerable to metastasis and recurrence, carrying high medical costs and putting a heavy burden on patients and society.
Cyclic RNAs are common, diverse, stable, and conserved and have functions such as nuclear protein translation transcriptional regulation [20-22]. Recent studies have shown that circRNA molecules are rich in miRNA-binding sites and serve as molecular sponges in cells, thereby relieving miRNAs’ inhibitory effect on their target genes and playing an important role in regulating disease through miRNA interactions associated with diseases [23,24]. CircRNAs are involved in a range of tumorigenesis as regulatory factors, influencing biologic processes such as proliferation, apoptosis, and cell differentiation [25-27]. Circ_0000735 has been shown to be highly expressed in prostate cancer and non-small cell lung cancer and has a favorable impact on cancer cells, but in bladder cancer, Circ_0000735 has not been studied. In this experiment, Circ_0000735 was found to be highly expressed in tissues and cells of bladder cancer, and Circ_0000735 knockdown could effectively inhibit the proliferation and invasion potential of bladder cancer cells and promote apoptosis, implying that Circ_0000735 gives a decent opportunity of targeting bladder cancer.
We found through the bioinformatics website that Circ_0000735 can act as a sponge for a variety of miRNAs, and for this study, we selected miR-502-5p among these molecules. In the diagnosis and treatment of gastric cancer, osteosarcoma, and tumors of glial origin, miR-502-5p was preliminarily studied [28-30]. MiR-502-5p has been used to prevent the proliferation, migration, invasion, tumor growth and metastasis of gastric cancer cells in the diagnosis and treatment of gastric cancer, osteosarcoma, and glial tumors by targeting PD-L1. In osteosarcoma tissues, miR-502-5p is limited and can be used as a new potential diagnostic marker. In glial-derived tumors, miR-502-5p expression can be used to predict patient prognosis. Previous studies have also confirmed that the development of bladder cancer requires miR-502-5p [31]. In bladder cancer tissues and cells, miR-502-5p was lowly expressed. Overexpression of miR-502-5p inhibited in vitro cancer cell proliferation and invasion and inhibited tumor growth in vivo. Therefore, on this basis, we showed that miR-502-5p could be sponged by Circ_0000735, and miR-502-5p down-regulation facilitated the proliferation and invasion of bladder cancer cells and inhibited apoptosis. The proliferation and invasion potential of bladder cancer cells was weakened after knocking down Circ_0000735, and apoptosis was enhanced, but the effect was reversed after inhibiting miR-502-5p on this basis, suggesting that miR-502-5p could rescue the function of Circ_0000735 in bladder cancer.
In short, this research investigated the biological role of Circ_0000735 in bladder cancer and revealed that by sponging miR-502-5p, Circ_0000735 facilitated the development of bladder cancer. However, the small number of samples obtained in this analysis could affect the reliability of the results and, by further increasing the sample size, the reliability of the data needs to be verified. Secondly, animal experiments only briefly demonstrated the tumorigenic ability in nude mice. Future research should further improve the use of Circ_0000735 in the treatment of bladder cancer. In short, Circ_0000735 might become a new target for bladder cancer treatment.
Disclosure of conflict of interest
None.
References
- 1.Jinsheng W, Peimei Z, Xiaocui W, Xianzhong L. MiRNA-153 regulates cell viability and the cell cycle in bladder cancer. Int J Clin Exp Med. 2019;12:5933–5937. [Google Scholar]
- 2.Fletcher SA, Cole AP, Lu C, Marchese M, Krimphove MJ, Friedlander DF, Mossanen M, Kilbridge KL, Kibel AS, Trinh QD. The impact of underinsurance on bladder cancer diagnosis, survival, and care delivery for individuals under the age of 65 years. Cancer. 2020;126:496–505. doi: 10.1002/cncr.32562. [DOI] [PubMed] [Google Scholar]
- 3.Steinberg RL, Thomas LJ, Brooks N, Mott SL, Vitale A, Crump T, Rao MY, Daniels MJ, Wang J, Nagaraju S, DeWolf WC, Lamm DL, Kates M, Hyndman ME, Kamat AM, Bivalacqua TJ, Nepple KG, O’Donnell MA. Multi-institution evaluation of sequential gemcitabine and docetaxel as rescue therapy for nonmuscle invasive bladder cancer. J Urol. 2020;203:902–909. doi: 10.1097/JU.0000000000000688. [DOI] [PubMed] [Google Scholar]
- 4.Jung A, Nielsen ME, Crandell JL, Palmer MH, Smith SK, Bryant AL, Mayer DK. Health-related quality of life among non-muscle-invasive bladder cancer survivors: a population-based study. BJU Int. 2020;125:38–48. doi: 10.1111/bju.14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akand M, Muilwijk T, Cornelissen J, Van Bruwaene S, Vander Eeckt K, Baekelandt F, Mattelaer P, Van Reusel R, Van Cleynenbreugel B, Joniau S, Van Der Aa F. Development of a prospective data registry system for non-muscle-invasive bladder cancer patients incorporated in the electronic patient file system. Front Oncol. 2019;9:1402. doi: 10.3389/fonc.2019.01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ling L, Tan Z, Zhang C, Gui S, Cui Y, Hu Y, Chen L. CircRNAs in exosomes from high glucose-treated glomerular endothelial cells activate mesangial cells. Am J Transl Res. 2019;11:4667–4682. [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, Zhang X, Liu M, Xu F, Zhang Q, Zhang Y, Weng X, Liu S, Du Y, Zhou X. Direct detection of circRNA in real samples using reverse transcription-rolling circle amplification. Anal Chim Acta. 2020;1101:169–175. doi: 10.1016/j.aca.2019.12.027. [DOI] [PubMed] [Google Scholar]
- 8.Jeyaraman S, Hanif EAM, Ab Mutalib NS, Jamal R, Abu N. Circular RNAs: potential regulators of treatment resistance in human cancers. Front Genet. 2020;10:1369. doi: 10.3389/fgene.2019.01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang M, Han Y, Zhai Y, Ma X, An X, Zhang S, Li Z. Integrative analysis of circRNAs, miRNAs, and mRNAs profiles to reveal ceRNAs networks in chicken intramuscular and abdominal adipogenesis. BMC Genomics. 2020;21:594. doi: 10.1186/s12864-020-07000-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sang MJ, Wu M, Meng LJ, Zheng Y, Gu LN, Liu F, Sang MX. Identification of epithelial-mesenchymal transition-related circRNA-miRNA-mRNA ceRNA regulatory network in breast cancer. Pathol Res Pract. 2020;216:153088. doi: 10.1016/j.prp.2020.153088. [DOI] [PubMed] [Google Scholar]
- 11.Han JY, Guo S, Wei N, Xue R, Li W, Dong G, Li J, Tian X, Chen C, Qiu S, Wang T, Xiao Q, Liu C, Xu J, Chen KS. ciRS-7 promotes the proliferation and migration of papillary thyroid cancer by negatively regulating the miR-7/epidermal growth factor receptor axis. Biomed Res Int. 2020;2020:9875636. doi: 10.1155/2020/9875636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, Liu FB, Huang M, Xie K, Xie QS, Liu CH, Shen MJ, Huang Q. Circular RNA ciRS-7 promotes the proliferation and metastasis of pancreatic cancer by regulating miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat Dis Int. 2019;18:580–586. doi: 10.1016/j.hbpd.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Huang H, Wei L, Qin T, Yang N, Li Z, Xu Z. Circular RNA ciRS-7 triggers the migration and invasion of esophageal squamous cell carcinoma via miR-7/KLF4 and NF-κB signals. Cancer Biol Ther. 2019;20:73–80. doi: 10.1080/15384047.2018.1507254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye F, Gao G, Zou Y, Zheng S, Zhang L, Ou X, Xie X, Tang H. circFBXW7 inhibits malignant progression by sponging miR-197-3p and encoding a 185-aa protein in triple-negative breast cancer. Mol Ther Nucleic Acids. 2019;18:88–98. doi: 10.1016/j.omtn.2019.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi YR, Wu Z, Xiong K, Liao QJ, Ye X, Yang P, Zu XB. Circular RNA circKIF4A Sponges miR-375/1231 to Promote Bladder Cancer Progression by Upregulating NOTCH2 Expression. Front Pharmacol. 2020;11:605. doi: 10.3389/fphar.2020.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao Y, Liu J, Huan J, Che F. Downregulation of circular RNA hsa_circ_0000735 boosts prostate cancer sensitivity to docetaxel via sponging miR-7. Cancer Cell Int. 2020;20:334. doi: 10.1186/s12935-020-01421-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, Jiang W, Liu T, Lv J, Guan J. Enhanced expression of circ_0000735 forecasts clinical severity in NSCLC and promotes cell progression via sponging miR-1179 and miR-1182. Biochem Biophys Res Commun. 2019;510:467–471. doi: 10.1016/j.bbrc.2019.01.134. [DOI] [PubMed] [Google Scholar]
- 18.Zhu G, Wang K, Qin H, Zhao X, Chen W, Xu L, Cao W, Guo H. Internal cross-linked polymeric nanoparticles with dual sensitivity for combination therapy of muscle-invasive bladder cancer. J Nanobiotechnology. 2020;18:124. doi: 10.1186/s12951-020-00686-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodgson A, van Rhijn BWG, Kim SS, Ding C, Saleeb R, Vesprini D, Liu SK, Yousef GM, van der Kwast TH, Xu B, Downes MR. Reassessment of p53 immunohistochemistry thresholds in invasive high grade bladder cancer shows a better correlation with TP53 and FGFR3 mutations. Pathol Res Pract. 2020;216:153186. doi: 10.1016/j.prp.2020.153186. [DOI] [PubMed] [Google Scholar]
- 20.Song J, Chen ZH, Zheng CJ, Song KH, Xu GY, Xu S, Zou F, Ma XS, Wang HL, Jiang JY. Exosome-Transported circRNA_0000253 Competitively adsorbs microRNA-141-5p and increases IDD. Mol Ther Nucleic Acids. 2020;21:1087–1099. doi: 10.1016/j.omtn.2020.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma HB, Yao YN, Yu JJ, Chen XX, Li HF. Extensive profiling of circular RNAs and the potential regulatory role of circRNA-000284 in cell proliferation and invasion of cervical cancer via sponging miR-506. Am J Transl Res. 2018;10:592–604. [PMC free article] [PubMed] [Google Scholar]
- 22.Jin H, Li C, Dong P, Huang J, Yu J, Zheng J. Circular RNA cMTO1 promotes PTEN expression through sponging miR-181b-5p in liver fibrosis. Front Cell Dev Biol. 2020;8:714. doi: 10.3389/fcell.2020.00714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Wu T, Wang P, Yang L, Li Q, Wang J, Zhao R, Zhang J, Liu M, Cao J, Tian L, Yu B, Sun Y. Circular RNA 103862 promotes proliferation and invasion of laryngeal squamous cell carcinoma cells through the miR-493-5p/GOLM1 axis. Front Oncol. 2020;10:1064. doi: 10.3389/fonc.2020.01064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dell’Orco M, Oliver RJ, Perrone-Bizzozero N. HuD binds to and regulates circular RNAs derived from neuronal development- and synaptic plasticity-associated genes. Front Genet. 2020;11:790. doi: 10.3389/fgene.2020.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo Q, Wang J, Sun R, He Z, Chen Q, Liu W, Wu M, Bao J, Liu Z, Wang J, Zhang Y. Comprehensive construction of a circular RNA-associated competing endogenous RNA network identified novel circular RNAs in hypertrophic cardiomyopathy by integrated analysis. Front Genet. 2020;11:764. doi: 10.3389/fgene.2020.00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nahand JS, Jamshidi S, Hamblin MR, Mahjoubin-Tehran M, Vosough M, Jamali M, Khatami A, Moghoofei M, Baghi HB, Mirzaei H. Circular RNAs: new epigenetic signatures in viral infections. Front Microbiol. 2020;11:1853. doi: 10.3389/fmicb.2020.01853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang HG, Yan H, Wang C, Li MM, Lv XZ, Wu HD, Fang ZH, Mo DL, Zhang ZY, Liang B, Lai KG, Bao JY, Yang XJ, Zhao HJ, Chen S, Fan YM, Tong XG. circAFF1 aggravates vascular endothelial cell dysfunction mediated by miR-516b/SAV1/YAP1 axis. Front Physiol. 2020;11:899. doi: 10.3389/fphys.2020.00899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.You W, Liu X, Yu Y, Chen C, Xiong Y, Liu Y, Sun Y, Tan C, Zhang H, Wang Y, Li R. miR-502-5p affects gastric cancer progression by targeting PD-L1. Cancer Cell Int. 2020;20:395. doi: 10.1186/s12935-020-01479-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang C, Wang Q, Ma S, Sun Y, Vadamootoo AS, Jin C. A four serum-miRNA panel serves as a potential diagnostic biomarker of osteosarcoma. Int J Clin Oncol. 2019;24:976–982. doi: 10.1007/s10147-019-01433-x. [DOI] [PubMed] [Google Scholar]
- 30.Drusco A, Fadda P, Nigita G, Fassan M, Bottoni A, Gardiman MP, Sacchi D, Calore F, Carosi M, Antenucci A, Casini B, Kelani H, Pescarmona E, Di Leva G, Zanesi N, Berger MS, Croce CM. Circulating micrornas predict survival of patients with tumors of glial origin. EBioMedicine. 2018;30:105–112. doi: 10.1016/j.ebiom.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ying Y, Li J, Xie H, Yan H, Jin K, He L, Ma X, Wu J, Xu X, Fang J, Wang X, Zheng X, Liu B, Xie L. CCND1, NOP14 and DNMT3B are involved in miR-502-5p-mediated inhibition of cell migration and proliferation in bladder cancer. Cell Prolif. 2020;53:e12751. doi: 10.1111/cpr.12751. [DOI] [PMC free article] [PubMed] [Google Scholar]



