SUMMARY
Histone H3.3 glycine 34 to arginine/valine (G34R/V) mutations drive deadly gliomas and show exquisite regional and temporal specificity, suggesting a developmental context permissive to their effects. Here, we show that 50% of G34R/V-tumours (n=95) bear activating PDGFRA mutations that display strong selection pressure at recurrence. While considered gliomas, G34R/V-tumours actually arise in GSX2/DLX-expressing interneuron progenitors, where G34R/V-mutations impair neuronal differentiation. The lineage-of-origin may facilitate PDGFRA co-option through a chromatin loop connecting PDGFRA to GSX2 regulatory elements, promoting PDGFRA overexpression and mutation. At the single-cell level, G34R/V-tumours harbour dual neuronal/astroglial identity and lack oligodendroglial programs, actively repressed by GSX2/DLX-mediated cell-fate specification. G34R/V may become dispensable for tumour maintenance, while mutant-PDGFRA is potently oncogenic. Collectively, our results open novel research avenues in deadly tumours. G34R/V-gliomas are neuronal malignancies, where interneuron progenitors are stalled in differentiation by G34R/V-mutations, and malignant gliogenesis is promoted by co-option of a potentially targetable pathway, PDGFRA signalling.
Keywords: H3.3 G34R/V, oncohistones, gliomas, PDGFRA, GSX2, interneuron progenitors
INTRODUCTION
High-grade gliomas (HGGs) are deadly primary brain tumours and a leading cause of mortality in children and young adults. These tumours frequently harbour somatic mutations in genes encoding histone 3 (H3) variants or epigenetic modifiers with remarkable neuroanatomical and age specificity (Fontebasso et al., 2014b; Khuong-Quang et al., 2012; Schwartzentruber et al., 2012; Sturm et al., 2012; Wu et al., 2012). HGGs in adolescents and young adults (12-35 years old) primarily occur in cerebral hemispheric lobes and a large number are considered epigenetic disorders (Fontebasso et al., 2014a; Fontebasso et al., 2013; Parsons et al., 2008; Verhaak et al., 2010). More than 30% of these HGGs bear heterozygous mutations in the non-canonical H3.3 variant leading to glycine 34 to arginine or valine (G34R/V) amino acid substitutions (Fontebasso et al., 2014b; Schwartzentruber et al., 2012; Wu et al., 2012; Wu et al., 2014). G34R/V tumours are understudied and likely underestimated, since their unique histopathological heterogeneity, with dual neuronal-glial compartments present at variable degrees, leads to misdiagnosis. Indeed, close to 30% of central nervous system primitive neuroectodermal tumours (CNS-PNETs), a now-obsolete entity of mixed high-grade neuronal tumours (Sturm et al., 2016), have been shown to be G34R/V-mutant HGGs (Gessi et al., 2013; Korshunov et al., 2016). At the molecular level, on the other hand, G34R/V HGGs show unifying features: they almost invariably carry mutations in ATRX and TP53 (Korshunov et al., 2016; Liu et al., 2012; Schwartzentruber et al., 2012), lack immunoreactivity for the oligodendroglial marker OLIG2 (Sturm et al., 2012), and cluster distinctly from other glioma entities based on DNA methylation (Sturm et al., 2012). In contrast to K27M or K36M mutations that affect both canonical H3.1/2 and H3.3 variants, G34R/V occur exclusively on the non-canonical H3.3 and have been suggested to act in cis, with the bulky amino acid replacement preventing post-translational modification of the neighbouring H3.3K36 residue (Lewis et al., 2013) and/or its recognition by specific readers (Wen et al., 2014).
Limited information exists on oncogenic or developmental pathways in these tumours, hampering their modelling and development of therapeutic strategies. Compiling comprehensive cohorts to study their molecular landscape has been challenging as this age group spans paediatric and adult care, in addition to presenting a high rate of misdiagnosis. To address this knowledge gap, we assembled the largest cohort of G34R/V HGG tumours to date (n=95) including for primary/relapse pairs, comprehensively profiled them at the genomic, epigenomic, and transcriptomic (bulk and single-cell) levels and developed in vitro and in vivo models. We delineate the unique molecular characteristics of G34R/V tumours, define their developmental origins, characterize inter- and intra-tumour heterogeneity, and uncover specific vulnerabilities that may be amenable to targeted therapies in this deadly cancer type.
RESULTS
High-frequency of PDGFRA mutations in G34R/V HGGs
To define the genetic landscape of G34R/V HGGs, we assembled a multi-institutional cohort of G34R (72 at diagnosis, 8 recurrences) and G34V tumours (9 at diagnosis, 6 recurrences). We compared G34R/V mutation profiles to midline K27M HGGs and to the other hemispheric HGG subgroups occurring in children or adults younger than 50 years old, namely IDH1- and/or SETD2-mutant HGGs or tumours wild-type for these mutations (Figure 1A, Table S1). In addition to the known genetic alterations in TP53 and ATRX (95% and 84% respectively), we observed a specific, previously unappreciated high frequency of mutations in PDGFRA in G34R/V HGGs (46/95), which also showed a high level of PDGFRA expression (Figure S1A). The high frequency at diagnosis (44%) was further enriched at recurrence (81%, p = 0.02, Fisher exact test). Altogether, co-occurrence of G34R/V & PDGFRA mutations was 7- to 8-fold higher than in any other subgroup, where the highest observed PDGFRAMUT frequency was 7% (p = 2.8E-55, chi-square test). No other recurrent mutations (frequency > 10%) were identified beyond PDGFRA (Figure S1B). In contrast to K27M HGGs (Khuong-Quang et al., 2012; Sturm et al., 2012; Wu et al., 2014), amplification of PDGFRA was relatively rare in G34R/V tumours (6/47, 13%) and similar in proportion to other hemispheric HGG subgroups, including IDH1-mutant and IDH1/H3-wild-type (p = 0.64, chi-square test).
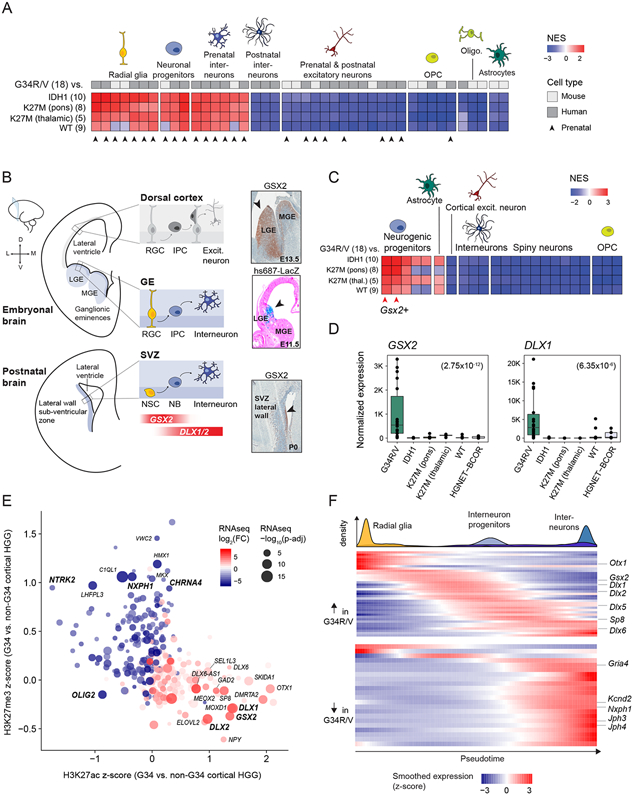
Figure 1. PDGFRA is frequently mutated in G34R/V tumours.
A. Oncoprint showing frequent occurrence of PDGFRA mutations in G34R/V (n = 95), relative to K27M (n = 53), IDH1/SETD2 (n = 80), and H3/IDH1-WT (n = 28) HGG subgroups, details in Table S1. Lines linking consecutive G34R/V samples indicate primary/recurrent tumours from the same patient. B. Spectrum and frequency of PDGFRA somatic mutations identified in G34R/V HGGs. Bold face indicates mutations observed in 2 or more patients. C. Immunohistochemistry staining of total PDGFRA and phosphorylated ERK1/2 in G34R/V HGGs, separated by PDGFRA mutation status. A primary PDGFRAWT and recurrence PDGFRAMUT tumour pair (P-1190) are indicated. See also Figure S1 and Table S1.
PDGFRA is a class III receptor tyrosine kinase (RTK) with five extracellular immunoglobulin-like domains. Within G34R/V HGGs, 85% of PDGFRA mutations occurred in the 2nd-4th extracellular immunoglobulin-like domains (Figure 1B). In contrast, we rarely observed the D842V/Y mutations affecting an autoinhibition site in the kinase domain (n = 2), prevalent and well-characterized in gastrointestinal stromal tumours (Heinrich et al., 2003). Most of these extracellular mutations have been mainly reported in brain tumours, particularly paediatric gliomas (Paugh et al., 2013) and are presumed to be activating (Ip et al., 2018; Paugh et al., 2013) (Table S1). We performed in silico modelling of two extracellular mutations (Figure S1C), the cysteine 235 to tyrosine/phenylalanine (C235Y/F), the most prevalent in our cohort (n = 15), and lysine 385 to methionine (K385M), which was recently reported as the sole recurrent mutation in rare myxoid glioneuronal tumours (Solomon et al., 2018). Both mutations were predicted to lead to receptor dimerization and constitutive activation, analogous to the well characterized Y288C mutation (Ip et al., 2018). Accordingly, we observed marked ERK phosphorylation indicating downstream MAPK/ERK-pathway activation in G34R/V-PDGFRAMUT tumours, while PDGFRAWT tumours showed limited ERK phosphorylation in cells not expressing this RTK (Figure 1C). Moreover, in one available primary/relapsed pair where the tumour acquired PDGFRAK385M at relapse, ERK phosphorylation was only present at relapse (Figure 1C). Collectively, these results suggest that PDGFRA mutations in G34R/V-tumours promote constitutive activation of this RTK and its downstream signalling pathways.
Transcriptional and epigenetic programs in G34R/V gliomas indicate an interneuron progenitor origin
The high co-occurrence of PDGFRA mutations with G34R/V suggests a G34R/V-context dependency, likely linked to development as was recently demonstrated for several related entities (Jessa et al., 2019; Vladoiu et al., 2019). To define the cellular lineage at the origin of G34R/V tumours, we first assessed cell type-specific signatures derived from single-cell RNA-seq (scRNA-seq) atlases of forebrain development (Jessa et al., 2019; Nowakowski et al., 2017; Velmeshev et al., 2019) using gene set enrichment analysis (GSEA). Compared to other HGG entities, transcriptomes of G34R/V gliomas showed strong enrichment of the cortical interneuron lineage, including radial glia, neuronal progenitor and interneuron gene programs, and were depleted of excitatory neuron and oligodendroglial signatures (Figure 2A, Figure S2A-B, Table S2). Consistent with the presence of a neuronal compartment, astrocyte gene programs were decreased relative to other gliomas, but still highly enriched when compared to non-glioma entities, such as high-grade neuroepithelial tumour with BCOR alteration (HGNET-BCOR) (Figure S2C). Importantly, G34R/V tumours showed enrichment of prenatal, but not postnatal, interneuron signatures.
Figure 2. Transcriptional and epigenetic programs in G34R/V gliomas indicate an interneuron progenitor origin.
A. Heatmap of enrichment scores of forebrain cell type signatures in G34R/V compared to other HGG subgroups, by gene set enrichment analysis (GSEA). Normalized Enrichment Scores (NES) are shown for all signatures significantly enriched (adjusted p-value < 0.01) in G34R/V vs. IDH1. Number of tumour samples is indicated in parentheses. OPC: oligodendrocyte progenitor cells. B. Left: Schematic of the embryonic and postnatal forebrain, coronal section. LGE: lateral ganglionic eminence; MGE: medial ganglionic eminence, RGC: radial glia cell; IPC: intermediate neuronal progenitor cell, NSC: neural stem cell, NB: neuroblast. Right: Immunohistochemistry staining of Gsx2 in sagittal sections of E13.5 mouse ganglionic eminences (GE) and P0 (SVZ), and enhancer reporter activity of hs687-LacZ in coronal sections of E11.5 mouse embryos profiled by VISTA enhancer browser. C. Heatmap of enrichment scores of striatal sub-ventricular zone (SVZ) cell type signatures in G34R/V compared to other HGG subgroups, by GSEA. NES scores are shown for all signatures significantly enriched (adjusted p-value < 0.01) in G34R/V vs. IDH1. Number of samples is indicated in parentheses. D. Expression levels of interneuron markers GSX2 and DLX1 in tumour subgroups. Adjusted p-values (Negative Binomial Wald test) for the comparison of G34R/V to other entities are indicated in parentheses. E. Promoter-associated H3K27ac and H3K27me3 for genes significantly differentially expressed between G34R/V and IDH1 HGG by bulk RNA-seq. Genes relevant to glioma (OLIG2), or with high G34R/V-enrichment of either mark (z-score > 0.9) and RNA-seq absolute log2 fold change > 3 are labelled. F. Gene expression levels of G34R/V-specific genes along the interneuron differentiation trajectory (Jessa et al., 2019). Top panel: density of each cell type along pseudotime. Bottom panel: expression of genes identified as specific to G34R/V gliomas by epigenome and transcriptome analyses. Up in G34R/V: genes in the leading edge of the most enriched human interneuron signature, or genes upregulated by RNAseq and enriched for H3K27ac in G34R/V. Down in G34R/V: genes in the leading edge of the most depleted human interneuron signature, or genes downregulated by RNAseq and enriched for H3K27me3 in G34R/V (see Methods). Expression is z-scored across pseudotime. See also Figure S2, and Tables S2 and S3.
Cortical interneurons are generated during embryonic development in transient progenitor domains of the ventral telencephalon termed ganglionic eminences (GEs) (Hansen et al., 2013; Ma et al., 2013) (Figure 2B). A network of transcription factors (TF) including GSX2 and DLX1/2 patterns these domains to specify interneuron fate. GSX2 is expressed throughout the GE, but most highly in the lateral and caudal GE, where it acts upstream of DLX1/2. All three factors (GSX2, DLX1/2) are required for interneuron specification and repression of oligodendroglial fate (Chapman et al., 2013; Petryniak et al., 2007). Postnatally, a Gsx2+ neural stem cell niche, derived from its GE counterpart and regulated by a similar TF hierarchy, persists in the murine brain, within the sub-ventricular zone (SVZ) (Chapman et al., 2018; Lopez-Juarez et al., 2013). We thus extended our analysis to include two recently reported scRNA-seq SVZ datasets (Anderson et al., 2020; Mizrak et al., 2019), confirming that G34R/V HGGs significantly upregulated signatures of Gsx2+ SVZ progenitors (Figure 2C, Figure S2D). Consistent with GSEA results, G34R/V HGGs significantly upregulated GSX2, the human radial glia marker MOXD1 (Pollen et al., 2015), interneuron TFs including DLX1/2 (Figure 2D, Figure S2E), and interneuron markers GAD2, SCGN, NPY, and CALB2 (Ma et al., 2013; Raju et al., 2018). In contrast, they lacked expression of TFs specifying excitatory neurons such as EOMES and NEUROD2 (Figure S2E). This transcriptional profile was unique to G34R/V tumours; expression of forebrain interneuron TFs and gene signatures was absent not only in midline K27M tumours, but also in other hemispheric HGGs. Thus, we conclude that, at the transcriptional level, G34R/V HGGs resemble GSX2/DLX+ progenitors of the interneuron lineage.
We next assessed epigenomic profiles of G34R/V tumours, which were also consistent with an interneuron origin. GSX2 and DLX1/2 displayed an active chromatin conformation, enriched for the activating H3K27ac mark and lacking the repressive H3K27me3 mark (Figure 2E, Table S3). On the other hand, gene sets enriched for H3K27me3 were related to neuronal synapse, axon and ion-channel associated functions (e.g. NTRK2, NXPH1, CHRNA4) (Figure S2F). H3K27me3 and H3K27ac marks were absent on the promoter of the oligodendrocyte lineage factor OLIG2 in G34R/V gliomas, in keeping with previous data showing it to be silenced through DNA methylation in this entity (Sturm et al., 2012) (Figure 2E). Furthermore, G34R/V HGGs lacked activation of a core module of transcription factors necessary for oligodendrocyte specification (Figure S2G).
Finally, to confirm the match to immature neurons, we extracted the genes driving the transcriptional enrichment of signatures in G34R/V tumours, as well as genes displaying differential H3K27ac/me3 deposition, and profiled their expression along a normal interneuron differentiation trajectory (Figure 2F). G34R/V-upregulated genes were primarily expressed in interneuron progenitors. Conversely, genes down-regulated and H3K27me3-enriched in G34R/V peaked in expression late in normal interneuron maturation. Altogether, the transcriptomic and epigenomic profiles of G34R/V HGGs indicate that they arise within ventrally-derived progenitors committed to the interneuron lineage, which appear restricted from progressing to a mature neuronal state.
G34R/V mutations promote H3K27me3 retention at genic promoters and silencing of mature neuronal genes
H3K27me3 patterns in G34R/V gliomas may either reflect their developmental origin or derive from G34R/V-mediated epigenetic dysregulation. H3.3G34R/V substitutions were previously shown to impair the catalytic deposition of H3K36me3 by SETD2 (Lewis et al., 2013; Zhang et al., 2017), a mark coupled to active transcription and known to directly antagonize H3K27 methylation (Schmitges et al., 2011). Using liquid-chromatography/mass spectrometry (LC/MS) of histones extracted from patient-derived cell lines, we observed H3K36me3 loss coupled to reciprocal gain of H3K27me3 specifically on H3.3G34R/V-mutant histone peptides (Figure S3A), confirming the cis-effect of these mutations. At the genome-wide level, chromatin immunoprecipitation (ChIP-Seq) showed H3K27me3 deposition to be significantly enriched at a large number of genic promoters (n = 3,901) in G34R/V HGGs compared to non-G34R/V cortical HGGs (Figure S3B). We observed specific H3K27me3 enrichment at promoters of several genes encoding brain-specific proteins implicated in neuronal function in G34R/V samples. This gain in H3K27me3 contrasted with the relatively balanced changes observed in H3K27ac deposition where only ~100 promoters showed differential enrichment in G34R/V (Figure S3C).
To determine whether H3K27me3 enrichment is a direct consequence of the G34R/V oncohistone, we targeted the mutation for removal in the HSJD-GBM002 (H3F3AG34R) cell line using CRISPR-Cas9 (Figure S3D). There were limited differences in histone marks by ChIP-seq or the transcriptome by RNA-seq between G34R and edited clones (Table S2-3, Figure S3E-G), suggesting that the global epigenomic landscape is not actively maintained by the G34R/V mutant H3.3 in transformed cells. Expression of the DLX family of interneuron transcription factors was maintained in both edited and unedited lines (Figure 3A, top row), suggesting that specification of the interneuron transcriptional program in G34R/V gliomas reflects their lineage of origin. Notably, when assessing genomic bins showing the greatest H3K36me3 loss in G34R, these concurrently displayed increased PRC2-SUZ12/H3K27me3 enrichment and decrease of transcriptional activity, in keeping with the presumed effect of G34R/V on these marks (Figure 3B, S3E-G). When edited clones were subjected to serum differentiation, we observed up-regulation of DLX1, but notably also of DLX5, which is normally induced later in interneuron differentiation (Figure 3A, bottom row). These edited clones showed enrichment of interneuron gene programs, and depletion of radial glia cell gene signatures, consistent with further progression into neuronal differentiation upon removal of the mutation (Figure 3C).
Figure 3. G34R/V HGGs aberrantly express PDGFRA, unlike normal interneuron progenitors.
A. Expression of selected interneuron transcription factors in the patient-derived cell line HSJD-GBM002 in stem cell media (top, n = 6) or differentiation media (bottom, n = 2). Green: unedited or parental clones containing G34R. Blue: edited clones with the G34R mutation removed by CRISPR. Horizontal bar indicates the median. B. Parallel coordinate plot depicting epigenomic and transcriptomic changes in HSJD-GBM002 G34R and edited clones. Genomic bins (5kb) were stratified into 6 quantiles (Q1 to Q6) based on H3K36me3 difference upon CRISPR editing, and changes for the other histone marks and transcription were computed for each quantile. Solid line = median, shaded area = 25% and 75% percentile. C. Single-sample gene set enrichment (ssGSEA) score for radial glia cell (RGC) and interneuron gene signatures in transcriptomes of HSJD-GBM002 clones in differentiation media. D. Pdgfra, Gsx2, and Dlx1/2 expression in mouse scRNA-seq developmental forebrain atlas (left) and postnatal striatal sub-ventricular zone (SVZ) atlas (right). Mean expression and proportion of cells expressing the gene are indicated. OPC: oligodendrocyte progenitor cells. E. 5kb genomic bins ranked by H3K27ac z-score, showing the top-ranking loci with H3K27ac enrichment in G34R/V vs. non-G34 HGGs. See also Figures S3 and S4, and Tables S2 and S3.
Finally, to confirm the G34R-mediated epigenomic and transcriptomic effects in vivo, we utilized a previously described in-utero electroporation (IUE) murine model (Pathania et al., 2017). E13.5 neocortices were electroporated with exogenous empty vector (EV), H3.3WT or H3.3G34R in combination with PdgfraWT, Atrx shRNA, and sgRNA for Trp53, along with episomal vectors for Cas9 and pBase. We profiled ex vivo expanded neural precursor cells derived from sorted GFP+TdTomato+ cells after 72h and in >1-year-old mice (Figure S4A). Minimal changes were observed 72h after electroporation, while marked accumulation of H3K27me3 at genic promoters was observed in adult G34R-expressing cells. Importantly, and consistent with H3K27me3 patterns in human G34R/V tumours and CRISPR lines, the IUE cells also exhibited G34R-specific H3K27me3 retention at genic promoters such as Jph4 and Foxp2, a transcription factor expressed in mature forebrain neurons (Figure S4B-C). By leveraging model systems to decouple the effect of G34 mutations from existing lineage-of-origin programs, we conclude that G34R/V oncohistones may directly impede terminal neuronal differentiation through aberrant H3K27me3 retention at specific neuronal maturation loci.
Active chromatin conformation facilitates PDGFRA co-option in G34R/V tumours
We next investigated the mechanism underlying the association of G34R/V interneuron progenitor programs with PDGFRA. The PDGFRA gene is located immediately adjacent to the interneuron transcription factor GSX2. In G34R/V tumours and cell lines, PDGFRA expression was positively correlated with GSX2 (R2 = 0.48, p-value = 0.0002) (Figure S5A), while in the other subtypes, correlation was lower (R2 = 0.2) and GSX2 was very lowly expressed (Figure S1A). This transcriptional coupling observed in G34R/V tumours is likely aberrant, since PDGFRA and GSX2 are normally expressed in distinct cell lineages (Figure 3D, S5B), suggesting that PDGFRA hijacks surrounding cis regulatory regions to promote its ectopic expression in G34R/V HGGs.
To determine whether epigenetic dysregulation underlies this transcriptional coupling, we first examined H3K27ac profiles in G34R/V tumours. Indeed, the top genomic bins showing differential H3K27ac deposition in G34R/V included the PDGFRA promoter, as well as a distal enhancer element, hs687, located near GSX2/PDGFRA (Visel et al., 2013) (Figure 3E). This VISTA-validated enhancer has been shown to drive reporter expression in the lateral GE of the embryonal telencephalon, coincident with Gsx2 expression, and it is thus likely involved in its regulation (Figure 2B). We next examined PDGFRA adjacent cis regulatory elements and observed specific activation in the form of discrete H3K27ac peaks throughout a 350 kb region on chr4 encompassing PDGFRA, GSX2 and hs687, that is unique to G34R/V HGGs (Figure 4A, bottom tracks, Figure S5C).
Figure 4. Active chromatin conformation facilitates PDGFRA-GSX2 co-option in G34R/V tumours.
A. Top, Hi-C heatmaps depicting sub-TAD structure at the PDGFRA-GSX2-hs687 locus in glioma cell lines. Small black triangle illustrates a TAD between PDGFRA and the hs687 enhancer enriched for contacts in G34R/V lines. Large dashed triangle demarcates the TAD formed by contact to a known distal insulator. Heatmap represents the log2 ratios of observed interactions relative to expected interactions at a 5kb resolution. Middle, virtual 4C plots representing the average intensity of PDGFRA-anchored contacts in G34R/V and non-G34 glioma cell lines. Black bar denotes virtual 4C anchor region. Red bar and * denotes regions with significantly increased contact in G34R/V relative to non-G34 lines (p-value < 0.05). Bottom, composite CTCF and H3K27ac ChIP-seq of primary cortical glioma G34R/V, IDH1/SETD2, and WT subgroups. Teal bars and * denotes significantly H3K27ac-enriched regions in G34R/V (z-score > 0.5, p-value < 0.05). B. Top, Hi-C heatmaps from murine embryonic stem cells (ESCs), E13.5 cortex and ganglionic eminences (GE) depicting sub-TAD structure at the Pdgfra-Gsx2-hs687 locus, as in (A). Middle, virtual 4C plots representing the average intensity of Pdgfra-anchored contacts in ESCs, E13.5 cortex and ganglionic eminences. Black bar denotes virtual 4C anchor region. Red bar and * denotes regions with significantly increased contact in GE relative to cortex (p-value < 0.05). Grey bar and * denotes region with significantly increased contact in ESC, relative to cortex (p-value < 0.05). Bottom, PRC2 ChIA-PET in ESCs illustrating PRC2-bound chromatin contacts. Underneath, H3K27me3, H3K27ac and H3K4me3 ChIP-seq for ESCs, murine adult cortex and E13.5 GE. C. Schematic illustrating chromosome conformation and chromatin landscape at the Pdgfra-hs687 locus. The active hs687 enhancer (yellow) is in close contact with poised Pdgfra selectively in the ganglionic eminences. See also Figure S5, and Table S4.
To determine whether these H3K27ac peaks correspond to distal regulatory elements used to promote ectopic PDGFRA expression, we performed Hi-C chromosome conformation capture and CTCF ChIP-Seq on G34R/V, K27M and WT primary cell lines (Figure 4A, Figure S5D, Table S4). We confirm the presence of a topologically associated domain (TAD) encompassing the PDGFRA promoter (dashed triangle), known to be present in non-cancerous somatic cells (Flavahan et al., 2016). In addition to this non-specific TAD, we uncovered a novel interaction loop in G34R/V cell lines (Figure 4A, solid triangle), linking the PDGFRA promoter with hs687, the putative GSX2 enhancer element. In fact, overall throughout the region spanning PDGFRA, GSX2 and hs687, the TAD in G34R/V glioma cell lines harboured significantly more interactions in G34R/V compared to H3-WT and K27M lines (p = 0.04, one-tailed t-test), supporting a conformation with increased potential for ectopic activation of PDGFRA in G34R/V HGGs through active enhancer/promoter contacts.
PDGFRA expression in PDGFRAMUT G34R/V HGGs is higher than in PDGFRAWT tumours (Figure S1A), suggesting that additional cis regulatory elements may augment expression of this RTK. We observed that PDGFRAMUT tumours further increased H3K27ac activation of regulatory elements within the neighbouring region, including at the GSX2 promoter and hs687, as well as new distal regulatory elements (e.g. chr4: ~ 53,100,000) (Figure S5E). Consistent with the H3K27ac profile, Hi-C interaction matrices showed more PDGFRA-anchored distal interactions in the PDGFRAMUT cell line (Figure S5F, black boxes). Altogether, this suggests that further activation of the existing chromatin conformation, particularly at key hs687 and GSX2 regulatory elements, is used to promote higher PDGFRA expression in samples carrying a mutation of this RTK.
The chromatin conformation of G34R/V tumours, bringing the PDGFRA promoter in close proximity to active GSX2 regulatory elements, may be transiently present during normal differentiation of the interneuron lineage. We thus investigated the dynamics of this region during murine development, comparing the epigenomic landscape and chromosome conformation of embryonic stem cells (ESC) and E13.5 forebrain tissue (Davis et al., 2018; Lindtner et al., 2019). In ESCs, a strong promoter-promoter interaction is present between Pdgfra and Gsx2, where both genic promoters are bivalently marked with active H3K4me3 and repressive H3K27me3 marks (Figure 4B, bottom tracks). Chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) confirms that this loop between Pdgfra and Gsx2 promoters is anchored by PRC2-mediated contacts, indicating that this interaction is present while both genes are repressed (Ngan et al., 2020). Moreover, no interactions were observed in ESCs between Pdgfra and hs687. At E13.5, in turn, a loop between hs687 and Gsx2 was detected in GE (p=0.03), confirming the function of this enhancer element as a driver of Gsx2 expression in the lineage (Figure S5G). More importantly, we observed that a second strong loop between Pdgfra and hs687 is formed uniquely in these GE. In addition, we identify a TAD anchored at the Pdgfra promoter and hs687, analogous to the one found in G34R/V tumours, which is significantly enriched in GEs compared to the dorsal cortex (p = 0.04) and ESCs (p = 0.02). This increase in contact frequency is concurrent with increased H3K27ac at the hs687 enhancer and Gsx2, confirming their active conformation in this developmental window. Pdgfra remains transcriptionally repressed in the GE, retaining bivalent H3K27me3 and H3K4me3 marks, consistent with aberrant oncogenic expression of this RTK in G34R/V gliomas.
Taken together, the epigenomic profiles of embryonic stem cells suggest that the promoters of Gsx2 and Pdgfra are physically anchored by the PRC2 complex and are poised for activation (Figure 4C). In the neocortex, where progenitors give rise to excitatory neurons, the Gsx2-Pdgfra contact is weakened and the Gsx2 promoter no longer possesses the potential to become activated, similar to what is observed in non-G34 HGGs. In contrast, in the GE where interneurons are born, Gsx2 contacts its distal enhancer hs687 for activation and remains bound to H3K27me3-repressed Pdgfra, bringing Pdgfra in close proximity to this active chromatin conformation. Collectively, the transcriptomic and epigenomic evidence suggests that the chromatin architecture of G34R/V HGGs resembles that of a GSX2+ progenitor, with PDGFRA expression reflecting an oncogenic event. Altogether, these results provide a mechanistic link for the interneuron progenitor-context dependency of PDGFRA mutations in G34R/V HGGs, where GSX2-associated cis-regulatory elements are recruited to induce aberrant PDGFRA expression in G34R/V tumours, and possibly to a greater extent in PDGFRA mutants.
G34R/V HGGs are neuronal tumours devoid of oligodendroglial cells where PDGFRA mutations expand aberrant astrocytic compartments
To then evaluate the effect of PDGFRA mutations on cellular heterogeneity of G34R/V HGGs, we profiled 6 PDGFRAWT and 10 PDGFRAMUT tumours by scRNA-seq (Figure 5A-B, Figure S6A, Table S5). Projection of malignant cells to a reference developmental brain atlas (Jessa et al., 2019) revealed that G34R/V tumours were predominantly comprised of neuron- and astrocyte-like cells, consistent with the reported heterogeneous histopathology. We observed a striking absence of oligodendroglial-like cells in all G34R/V tumours, in contrast to other paediatric and adult HGGs entities where they were readily detectable (Filbin et al., 2018; Neftel et al., 2019; Venteicher et al., 2017), and limited numbers of immune cells, similar to K27M gliomas (Filbin et al., 2018) (Figure 5C, Figure S6B). At the level of individual cells, G34R/V astrocytic and neuronal cells were highly dysplastic, with most cells displaying abnormal co-expression of both interneuron and astrocytic gene signatures and indefinite segregation between compartments (Figure 5D, Figure S6C). Across patients, G34R/V HGGs exhibited significant inter-tumour heterogeneity, with varying proportions of neuronal and astrocytic-like cells (Figure 5B, Figure S6A). Importantly, G34R/V tumours mutant for PDGFRA displayed an expansion of the astrocytic compartment when compared to wild type tumours (p=0.028, wilcoxon rank sum test), suggesting that acquisition of a PDGFRA mutation promotes an astrocytic state at the expense of the neuronal component.
Figure 5. G34R/V tumours are devoid of oligodendrocytes, and PDGFRA mutant tumours exhibit expanded astrocytic compartments.
A. UMAP embedding of scRNA-seq G34R/V malignant cells from patient samples colored by patient of origin (left) or consensus cell type projection (middle, right). The two primary/recurrence tumour pairs are highlighted. B. Pie charts depicting the proportion of each cell type in individual tumours. C. Radar plots depicting the proportion of cells projected to a certain cell type within each tumour entity. Each line represents one sample, color coded as in (A) for G34R/V tumours. Outer circle: 100%, middle circle: 50%, inner circle: 0%. D. Mean expression of foetal interneuron and astrocyte gene signatures in individual cells from mouse scRNA-seq developmental forebrain atlas (left), and in cells from G34R/V tumours (right). For tumours, only cells called malignant and projected as neurons or astrocytes were included. See also Figure S6, and Table S5.
We next examined the role of PDGFRA mutations in G34R/V tumour evolution from three available matched primary and recurrent tumour pairs (Figure 6A, Table S6). Phylogenetic trees inferred from allele frequencies revealed a remarkably high mutation burden (n = 4,218) consistent with a temozolomide treatment signature in P-1978 recurrence (G34R; PDGFRAWT). In contrast, relatively few new mutations appeared in recurrences of patients P-1190 and P-3200 (43 and 124 mutations respectively), with PDGFRA being the only gene commonly mutated in recurrences of both patients. Moreover, there was little evidence of clonal heterogeneity (Figure 6A), suggesting strong sweeping selection for the new PDGFRA mutations acquired. At the transcriptomic level, scRNAseq data from both patients showed an expansion of the astrocytic compartment, with concomitant decrease of the neuron-like compartment (Figure 6B). Moreover, MAPK/ERK pathway was significantly more active in tumours mutant for PDGFRA (Figure 1C). Together, these findings suggest that the acquisition of PDGFRA mutations in G34R/V activate downstream MAPK signalling and lead to clonal selection, astrocytic expansion and potentially oncogenic addiction upon recurrence.
Figure 6. Mutant Pdgfra is a strong oncogenic driver while G34R/V may be dispensable for tumour maintenance.
A. Left, fish plots depicting tumour clonal structure of G34R/V primary and matched recurrence samples from patients P-1978, P-1190, and P-3200. Right, phylogeny of G34R/V primary and matched recurrence samples from patients P-1978, P-1190, and P-3200. Scale bar: 10 mutations. Dashed lines: potentially germline mutations. B. Doughnut plots representing the proportion of cells projected to each cell type in two primary/recurrence pairs, highlighting the increased proportion of astrocyte-like cells in the PDGFRAMUT recurrence (outer circle) compared to the PDGFRAWT primary tumour (inner circle). C. Immunohistochemical GFP staining of coronal forebrain sections from in-utero electroporated mice. All mice received shAtrx, sgTp53 in addition to PdgfraWT or PdgraD842V, and H3.3 WT/G34R. D. Kaplan-Meier curve depicting survival of in-utero electroporated mice. All mice received shAtrx, sgTp53 in addition to PdgfraWT or PdgraD842V, and empty vector/H3.3 WT/G34R. E. Immunofluorescence validation of CRISPR-mediated removal of G34V in KNS-42 clones. F. Kaplan-Meier survival curves of KNS-42 clones. Teal depicts the parental line carrying G34V, dashed blue depicts edited clones. G. Immunofluorescence validation of CRISPR-mediated repair/removal of G34R in HSJD-GBM002 clones. H. Kaplan-Meier survival curves of HSJD-GBM002 clones. Teal depicts the parental and unedited clones carrying G34R, dashed blue depicts edited clones. See also Figure S7, and Table S6.
G34R/V mutations may be dispensable for tumour maintenance, while mutant-PDGFRA is a potent oncogenic driver
To delineate individual effects of PDGFRA and G34R/V mutations on gliomagenesis, we compared latency to tumour formation in the murine IUE and CRISPR cell line models (Pathania et al., 2017). Consistent with known oncogenic properties of PDGFRAD842V, immunohistochemical GFP staining showed extensive tumour growth in PdgfraD842V-electroporated mice in as little as one month (Figure 6C). Addition of the G34R mutation did not impact latency in this model as both H3f3aWT and H3f3aG34R IUE mice showed similar tumour growth and survival (Figure 6D). In contrast, GFP staining at two months upon electroporation of PdgfraWT was largely negative. This finding is further reinforced in the time to tumour formation; at 12 months post-IUE, H3f3aG34R;PdgfraWT formed tumours with ⅔ penetrance compared to EV;PdgfraWT with similar latency and ⅓ penetrance. In contrast, PdgfraD842V was strongly oncogenic and fully penetrant regardless of G34R presence, decreasing tumour latency for the entire cohort to 50 days.
Consistent with IUE results, CRISPR-editing of H3.3G34V and G34R mutations in the respective patient-derived cell lines KNS-42 and HSJD-GBM002 showed limited effect on tumorigenicity in orthotopic xenograft models (Figure 6E-H). Edited lines exhibited comparable latency to tumour formation as G34-mutant lines (median survival G34V/Edited: 148/166 days; G34R/Edited: 365/323 days), and developed tumours with similar histopathology (Figure S7A-B). These findings suggest that G34R/V mutations may become dispensable for oncogenic maintenance, and instead probably co-opt PDGFRA mutations to promote gliomagenesis.
DISCUSSION
We comprehensively profiled the molecular landscape of H3.3G34R/V HGGs in the largest multi-institutional cohort to date, which features several rare cases of matched primary and recurrence tumours. We show an enrichment of activating PDGFRA mutations (44%), concurrent with elevated expression, in G34R/V primary tumours relative to all other HGG subtypes. This frequency is higher in the recurrence setting (81%), as tumours initially wildtype acquired PDGFRA mutations with strong selective pressure for the mutant clone at recurrence. PDGFRA is an important RTK in glial development and a recurrent driver in HGGs (Mackay et al., 2017; Sturm et al., 2012; Verhaak et al., 2010), which show distinct mechanisms for its co-option in the different subgroups. Indeed, PDGFRA amplifications are commonly observed in K27M-mutant midline gliomas (40%) (Khuong-Quang et al., 2012; Mackay et al., 2017; Sturm et al., 2012) and IDH-WT pro-neural HGGs (Mackay et al., 2017; Sturm et al., 2012). Conversely, IDH1-mutant HGGs use a distal enhancer through DNA methylation of a proximal CTCF insulator to overexpress PDGFRA (Flavahan et al., 2016), while activating mutations are more frequently observed in non-brainstem gliomas occurring in older children (14%) (Koschmann et al., 2016; Paugh et al., 2013).
The high frequency of PDGFRA mutations unique to G34R/V gliomas is likely due to their distinct differentiation program. Our data suggest G34R/V tumours originate in GSX2+ interneuron progenitors (Figure 7), where the oncohistone impedes terminal neuronal differentiation through altered H3K36me3 deposition and aberrant H3K27me3 retention at specific genes, a pattern suggested by prior biochemical analyses performed on G34R/V-mutant nucleosomes (Lewis et al., 2013). This progenitor state likely maintains the lineage-specific GSX2 enhancer in an active chromatin conformation state, rendering the locus permissive to transcriptional exploitation and aberrant PDGFRA expression (Figure 7). The spatial contact between GSX2 and PDGFRA seems to be an early event, present in embryonic stem cells where expression of both genes is repressed by PRC2/H3K27me3. As cells undergo neuronal differentiation it is uniquely preserved in Gsx2+ interneuron progenitors in E13.5 GEs, while it is lost in other neuronal populations as our data indicate. In ganglionic eminences, Pdgfra remains in a poised state in close physical proximity with Gsx2-associated enhancer machinery (including hs687), presenting an opportunity for oncogenic co-option, as the epigenetic landscape favours activation of this lineage factor. We speculate that PDGFRA-mutant tumours may form additional contacts with distal regulatory elements to maintain high PDGFRA expression, fuelling their oncogenic addiction and dependence on PDGFRA, given the selective advantage of this mutant RTK. Whether the G34R/V mutation occurs during this embryonic window of development or in postnatal GSX2+ interneuron progenitors in the subventricular zone, remains to be determined.
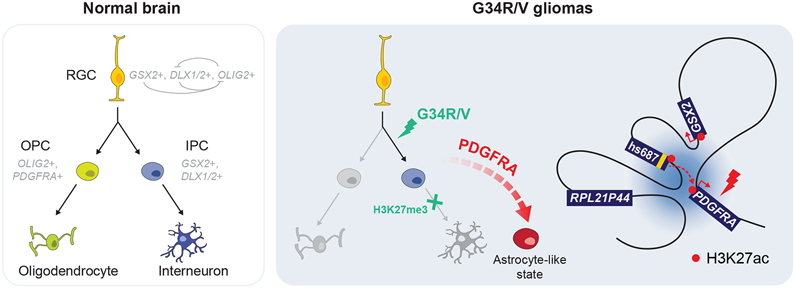
Figure 7. Model of aberrant development in G34R/V gliomas, compared to normal development in the ventral forebrain.
Left: During normal development, radial glial cells (RGC) in the ventral forebrain give rise to oligodendrocyte precursor cells (OPC) which differentiate into oligodendrocytes, and intermediate neuronal progenitor cells (IPC) which differentiate into cortical interneurons. Right: G34R/V gliomas retain molecular features of a committed interneuron progenitor (GSX2, DLX1/2+) and oncohistone-mediated H3K27me3 gain may impede terminal neuronal differentiation. G34R/V tumours exhibit dual neuronal and astrocytic components. Elevated expression and oncogenic PDGFRA mutation may promote the abnormal astrocyte-like state. G34R/V HGGs display a topologically associated domain (TAD) which brings the GSX2 associated cis-regulatory elements (hs687) into proximity with the PDGFRA promoter to induce overexpression.
The ectopic PDGFRA expression and subsequent acquisition of activating mutations likely promote astroglial features in the G34R/V-stalled interneuron progenitors. Notably, a Nestin-inducible K27M/Tp53cKO mouse model aiming to model K27M midline gliomas led to neuronal high-grade tumours, which became HGGs only upon co-expression of PDGFRAV544ins (Larson et al., 2019), in keeping with a major role for PDGFRA mutations in driving gliogenesis. Specific induction of downstream MAPK signalling is probably necessary for the expansion of astrocytic lineage programs in G34R/V tumours, as increasing ERK activation has been shown to promote glial specification at the expense of neuronal lineages (Li et al., 2014). Finally, persistent high levels of GSX2 and DLX1/2 are likely responsible for the absence of oligodendroglial lineage programs in G34R/V HGGs, as down-regulation of these transcription factors is required for the transition from neurogenesis to oligodendrocyte formation in the ventral forebrain (Chapman et al., 2018; Chapman et al., 2013; Petryniak et al., 2007).
Our data suggest that PDGFRA mutants are robust glioma drivers in G34R/V HGGs and that, in contrast to K27M (Harutyunyan et al., 2019; Krug et al., 2019), G34R/V mutations may be dispensable for tumour maintenance. We show that ~80% of PDGFRA mutations occurred in the extracellular domain, with the glioma-specific C235Y/F mutations alone accounting for 30% of all PDGFRA mutations in G34R/V. These neomorphic mutations constitutively activate this RTK, as also suggested by downstream activation of the MAPK pathway observed in PDGFRAMUT G34R/V HGGs. These mutants may have additional biochemical properties that warrant specific investigation, as alluded to by one study indicating that they may be resistant to standard PDGFRA inhibitors (Ip et al., 2018). Initially targeting the downstream MAPK pathway may be of benefit in this context, while further investigations are needed to identify therapeutic vulnerabilities in tumours with extracellular-domain PDGFRA mutations.
In sum, we highlight a novel mechanism where PDGFRA expression is ectopically promoted by the interneuron transcription factor GSX2, potentially leading to the high rate of activating mutations of this RTK specifically in G34R/V HGGs. Cells which acquire a PDGFRA mutation undergo sweeping clonal selection, and potently activate MAPK signalling to promote gliomagenesis. Altogether, these mechanisms have important therapeutic implications as G34R/V HGGs are classified as gliomas based on morphology, while they seem neuronal in origin based on genetic and molecular markers. These tumours are invariably lethal and PDGFRA mutations and downstream MAPK activation are potentially actionable targets, providing hope for novel therapeutic opportunities in this deadly cancer.
STAR*METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Nada Jabado (nada.jabado@mcgill.ca).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
ChIP-seq, RNA-seq, Hi-C sequencing data for cell lines and murine samples (Table S7) have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE146731. WES, bulk RNA-seq, ChIP-seq, and scRNA-seq sequencing data for human tumours (Table S7) have been deposited in the European Genome-phenome Archive (EGA) under accession number EGAS00001004301. Bulk RNA-seq sequencing data for non-G34R/V tumour entities have been previously deposited in EGA under dataset accession number EGAD00001005131.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Patient samples and clinical information
Protocols for this study involving collection of patient samples and information were approved by the Research Ethics and Review Board of McGill University and affiliated Hospitals Research Institutes. Informed consent was obtained from all research participants. Clinical information (age, sex) and mutation status of primary HGG tumour samples are presented in Table S1.
Primary cell lines
We thank Drs. Angel M. Carcaboso and Dinesh Rakheja for generously sharing primary HGG-derived cell lines. The HSJD-GBM002 cell line was cultured in NeuroCult NS-A proliferation media (StemCell Technologies) supplemented with bFGF (10 ng/mL), rhEGF (20 ng/mL), and heparin (0.0002%) (StemCell Technologies) and on plates coated with poly-L-ornithine (0.01%) (Sigma) and laminin (0.01 mg/mL) (Sigma). KNS-42, PS10-801 and CMC1118G8 cell lines were cultured in DMEM containing 4.5 g/L glucose, L-glutamine, phenol red, and 10% FBS (Wisent). All lines were tested monthly for mycoplasma contamination (MycoAlert Mycoplasma Detection kit by Lonza), and STR fingerprinting was regularly performed. Clinical information (age, sex) and mutation status of primary HGG cell lines are presented in Table S7.
Animal models
All mice were housed, bred, and subjected to listed procedures according to the McGill University Health Center Animal Care Committee and in compliance with Canadian Council on Animal Care guidelines, and Animal Welfare and Ethical Review Body (AWERB) and UK Home Office guidelines (Project license 70/8240, 70/7428 and 80/2325). Live colonies were maintained and genotyped as per Jackson Laboratories guidelines and protocols. Mice were housed together and monitored daily for neurological symptoms of brain tumours (weight loss, epilepsy, altered gait, lethargy) and euthanized immediately at clinical endpoint when recommended by veterinary and biological services staff members.
Female NOD SCID (RRID:IMSR_JAX:005557) mice (4-6 weeks, 18-25g) were used for xenograft experiments. Tissue from ganglionic eminences and cortex was isolated from embryos at gestation day E13 (plug day considered as day 0) using B6C3F1/J (RRID:IMSR_CRL:031) or FoxG1-cre (B6.129T(SJL)-Foxg1tm1.1(cre)Ddmo/J) (RRID:IMSR_JAX:029690) pregnant mice (6-8 weeks, 20-25g). In utero electroporations were performed on pregnant C57BL/6J (RRID:IMSR_JAX:000664) female mice at E13.5.
METHOD DETAILS
Immunohistochemistry for patient tumours
Immunohistochemistry (IHC) was performed at Histology Platform (RI-MUHC) and the Segal Cancer Centre Research Pathology Facility (Jewish General Hospital). G34R/V high-grade glioma samples were cut at 4-6 μm, placed on SuperFrost/Plus slides (Fisher) and dried overnight at 37°C, before IHC processing. After de-paraffinization and epitope retrieval, sections were incubated with primary antibodies: phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204, 4376 Cell Signaling, RRID:AB_331772) in 1:100; and PDGFRA (AF-307-NA, R&D Systems, 1:100, RRID:AB_354459). Slides were then loaded onto the Discovery XT Autostainer or Ventana Discovery Ultra Instrument (Ventana Medical Systems). Slides were counterstained with hematoxylin, blued with Bluing Reagent, washed, dehydrated through graded alcohols, cleared in xylene, and mounted with mounting medium (Eukitt, Fluka Analytical) or Leica CV 5030 coverslipper. Sections were analyzed by conventional light microscopy or scanned using the Aperio AT Turbo Scanner (Leica Biosystems).
CRISPR/Cas9 gene editing
CRISPR-Cas9 editing of KNS-42 cell line was performed as described in Ran et al (Ran et al., 2013). The pSpCas9(BB)-2A-Puro (PX459 V2.0) was a gift from Feng Zhang (Addgene plasmid #62988). A sgRNA targeting the H3F3A G34V mutation was cloned into the plasmid, and the construct was transfected using lipofectamine 2000 (Thermo Fisher Scientific) as per manufacturer’s protocol. Transfected cells were selected with puromycin for 3 days. CRISPR-Cas9 editing of HSJD-GBM002 cell line was performed by designing a synthetic ALT-R crRNA (IDT) guide targeting the H3F3A G34R mutation. Nucleofection was performed using program A-023 on the Amaxa Nucleofector 2b (Lonza) device, following manufacturer’s instructions to introduce crRNA, tracrRNA and recombinant Alt-R S.p. Cas9 Nuclease V3 (IDT). Single cell clones were isolated by limiting dilution, expanded and screened for editing events at the target locus by Sanger sequencing and confirmed through targeted deep sequencing using Illumina MiSeq.
Guide RNA sequences:
H3F3A G34V gRNA: TTCTTCACCACTCCAGTAG
H3F3A G34R crRNA: TTCTTCACCCTTCCAGTAG
Immunofluorescence
HSJD-GBM002 and KNS-42 CRISPR clones were cultured in 8-well chambers. Cells were fixed with 4% paraformaldehyde and 15% sucrose in PBS solution for 20 minutes at 4°C. Permeabilization was performed with 0.1% Triton X-100 for 3 mins on ice, followed by incubation in blocking buffer of 5% goat serum for 1 h at room temperature. Cells were incubated overnight at 4°C with primary antibodies specific for Histone H3.3 G34R or G34V (RevMab 31-1120 RRID:AB_2716433 and 31-1193 RRID:AB_2716435), diluted 1:200 in 1% BSA/PBS solution. Incubation with secondary anti-rabbit Alexa Fluor 488 or 594 antibodies (ThermoFisher Scientific) was performed at 1:1000 dilution in PBS under light protection for 90 minutes. ProLong Diamond Antifade Mountant with blue DNA DAPI stain (ThermoFisher Scientific) was applied. Fluorescent signal was captured the following day.
Histone post-translational modification by nLC/MS
The complete workflow for histone extraction, LC/MS, and data analysis was described in detail(Karch et al., 2016; Sidoli et al., 2016). Briefly, cell pellets (~1 × 106 cells) were lysed on ice in nuclear isolation buffer supplemented with 0.3% NP-40 alternative. Isolated nuclei were incubated with 0.4 N H2SO4 for 3 h at 4 °C with agitation. 100% trichloroacetic acid (w/v) was added to the acid extract to a final concentration of 20% and samples were incubated on ice overnight to precipitate histones. The resulting histone pellets were rinsed with ice cold acetone + 0.1% HCl and then with ice cold acetone before resuspension in water and protein estimation by Bradford assay. Approximately 20 μg of histone extract was then resuspended in 100 mM ammonium bicarbonate and derivatized with propionic anhydride. 1 μg of trypsin was added and samples were incubated overnight at 37 °C. After tryptic digestion, a cocktail of isotopically-labeled synthetic histone peptides was spiked in at a final concentration of 250 fmol/μg and propionic anhydride derivatization was performed for second time. The resulting histone peptides were desalted using C18 Stage Tips, dried using a centrifugal evaporator, and reconstituted using 0.1% formic acid in preparation for nanoLC-MS analysis.
nanoLC was performed using a Thermo ScientificTM Easy nLCTM 1000 equipped with a 75 μm × 20 cm in-house packed column using Reprosil-Pur C18-AQ (3 μm; Dr. Maisch GmbH, Germany). Buffer A was 0.1% formic acid and Buffer B was 0.1% formic acid in 80% acetonitrile. Peptides were resolved using a two-step linear gradient from 5 to 33% B over 45 min, then from 33 to 90% B over 10 min at a flow rate of 300 nL/min. The HPLC was coupled online to an Orbitrap Elite mass spectrometer operating in the positive mode using a Nanospray FlexTM Ion Source (Thermo Scientific) at 2.3 kV. Two full MS scans (m/z 300–1100) were acquired in the orbitrap mass analyzer with a resolution of 120,000 (at 200 m/z) every 8 DIA MS/MS events using isolation windows of 50 m/z each (e.g., 300–350, 350–400, …,650–700). MS/MS spectra were acquired in the ion trap operating in normal mode. Fragmentation was performed using collision-induced dissociation (CID) in the ion trap mass analyzer with a normalized collision energy of 35. AGC target and maximum injection time were 10e6 and 50 ms for the full MS scan, and 10e4 and 150 ms for the MS/MS can, respectively. Raw files were analyzed using EpiProfile.
Mouse orthotopic xenograft
Female NOD SCID mice (4-6 weeks) were used for xenograft experiments. Mice were injected with the following cell lines at a density of 7x105 cells in the parietal cortex, using the following coordinates from bregma: anteroposterior −1.94 mm, mediolateral 2.5 mm, dorsoventral 1.4 mm. For KNS-42: (1) Parental cells (H3F3A+/G34V, n=3), Clone 1-9 (H3F3A+/−, n=3), Clone 2-2 (H3F3A+/−, n=3). For HSJD-GBM002: (1) Parental cells (H3F3A+/G34R, n=2), Clone F06 (H3F3A+/G34R, n=4), Clone A10 (H3F3A+/G34R, n=4), Clone C08 (H3F3A+/−, n=4), Clone A09 (H3F3A+/+, n=4). Stereotaxic injections were performed using the Robot Stereotaxic machine from Neurostar. At clinical endpoint, brains were removed and fixed in formalin for histological analysis. Kaplan-Meier survival curves were generated using Graphpad Prism software.
Ganglionic eminences and cortices isolation
Tissue from ganglionic eminences (GEs) and cortex was isolated from embryos at gestation day 13 (plug day considered as day 0) using B6C3F1/J or FoxG1-cre (B6.129T(SJL)-Foxg1tm1.1(cre)Ddmo/J) pregnant mice. Mice were anesthetized, and euthanized by CO2 exposure followed by cervical dislocation. Uterine horns containing the embryos were removed and placed in cold 1X HBSS buffer (containing 1% Penicillin/streptomycin). Embryos were removed and dissected under the microscope. The brain was removed from the skull, forebrain was separated, and placed in a new Petri dish containing cold 1X HBSS buffer. Cortices of each hemisphere were cut using micro-scissors to expose the GEs. GEs were then isolated and placed in 12-well plates until cell dissociation. Matching cortices were recovered and similarly processed. Cells were dissociated by pipetting the tissue into a single-cell suspension that was processed as per requirements for downstream experiments. In addition, whole brains at E13 and P0 were fixed in formalin for histological analyses.
IHC for PDOX and mouse brain tissue
Immunohistochemistry (IHC) was performed at Histology Platform (RI-MUHC) and the Segal Cancer Centre Research Pathology Facility (Jewish General Hospital). PDOX and mouse brain tissue samples were cut at 4-6 μm, placed on SuperFrost/Plus slides (Fisher) and dried overnight at 37°C, before IHC processing. After de-paraffinization and epitope retrieval, sections were incubated with primary antibodies: Gsx2 (ABN162, Millipore Sigma RRID:AB_11203296) in 1:100; H3.3 G34R (RM240 , RevMAb Biosciences, RRID:AB_2716433) in 1:50; and H3.3 G34V (RM307, RevMAb Biosciences RRID:AB_2716435) in 1:40. Slides were then loaded onto the Discovery XT Autostainer or Ventana Discovery Ultra Instrument (Ventana Medical Systems). Slides were counterstained with hematoxylin, blued with Bluing Reagent, washed, dehydrated through graded alcohols, cleared in xylene, and mounted with mounting medium (Eukitt, Fluka Analytical) or Leica CV 5030 coverslipper. Sections were analyzed by conventional light microscopy or scanned using the Aperio AT Turbo Scanner (Leica Biosystems).
Mouse in utero electroporation (IUE)
Vector Construction
The piggyBac donor and helper vector system (CAG-PBase, PBCAG-GFP) was used to transduce NPCs in utero as described previously(Chen and LoTurco, 2012). Two ATRX shRNA vectors, cDNA encoding murine Pdgfra and cDNA encoding C-terminal HA-tagged Drosophila His3.3A (highly conserved ortholog of mammalian H3f3a) were cloned into the PBCAG-GFP vector to generate shAtrx-PdgfraWT-H3f3aWT constructs, previously described in Pathania et al(Pathania et al., 2017). CRISPR/Cas9 pX330 vectors containing Trp53-targeted gRNA were similarly used. G34R and PdgfraD842V mutants were generated using Quikchange Lightning site-directed mutagenesis (Agilent).
shRNA and sgRNA sequences:
shATRX vector1: 5’-TTCATTTACATTCTCATCCGTG-3’
shATRX vector2: 5’-TCATTTACATTCTCATCCG-3’
Trp53 sgRNA: 5’-ACAGCCATCACCTCACTGCA-3’
In Utero Electroporation
In utero electroporations were performed using sterile technique on isoflurane/oxygen-anesthetized pregnant C57BL/6J female mice at E13.5 (cortex) as described(Nitarska et al., 2016). Analgesic support was provided pre-emptively (subcutaneous delivery of Vetergesic and Carpofen at 0.1 mg/kg and 5 mg/kg, respectively). Uterine horns were exposed through a 1 cm incision and individual embryos were digitally manipulated into the correct orientation for intraventricular injection. Pulled borosilicate capillaries were loaded with endotoxin-free DNA and Fast Green dye (0.05%, Sigma) for visualization, and a microinjector (Eppendorf) was used to inject either the lateral or fourth ventricles with the DNA-dye mixture. 3-5 plasmids were injected simultaneously, each at a final concentration of 2 μg/μl and 1-2 μl of total solution was injected per embryo. DNA was electroporated into cortical neural progenitors using 5 mm tweezertrodes (BTX), or into lower rhombic lip progenitors using 3 mm tweezertrodes, applying 5 square pulses at 35 V, 50 ms each with 950 ms intervals. The embryos were returned into the abdominal cavity, the muscle and skin were sutured and the animal was monitored until fully recovered from the procedure.
Ex vivo NPC Isolation and Culture
Animals were euthanized by CO2 exposure and/or cervical dislocation and cortices were rapidly dissected in ice-cold dissociation medium containing: 20 mM glucose, 81.8 mM Na2SO4, 30 mM K2SO4, 5.8 mM MgCl2, 250 μM CaCl2, 1 mM HEPES, 160 μM NaOH, 0.8 mM kynurenic acid, 50 μM D-APV, 100 U/mL penicillin, 100 μg/mL streptomycin, 5 μg/ml plasmocin and 100 μg/ml primocin. GFP+ regions were microdissected under an epifluorescence stereomicroscope in the same medium and enzymatically digested into a single-cell suspension using the Papain Dissociation System (Worthington Biochemicals) followed by mechanical trituration through a series of fire-polished Pasteur pipettes. The dissociated cell solution was then separated on an OptiPrep density gradient to remove debris and GFP+TdTomato+ cells were sorted using a FACSAria III or LSRFortessa X-20 (BD Biosciences) into Hibernate-E media (Life Technologies) containing 2% B-27, 2 mM Glutamax, 0.8 mM kynurenic acid, 50 μM D-APV, 100 U/mL penicillin, 100 μg/mL streptomycin, 5 μg/ml plasmocin and 100 μg/ml primocin. Sorted cells were plated into NeuroCult NSC proliferation media (STEMCELL Technologies) containing 10 ng/ml of epidermal growth factor (EGF, Peprotech) and basic fibroblast growth factor (bFGF, Peprotech) on laminin-coated culture vessels (Sigma).
Model Figures
Graphical abstract was created with the aid of BioRender software.
QUANTIFICATION AND STATISTICAL ANALYSIS
Description of statistical details for each experiment can be found in figure legends.
Structural molecular modeling
The PDGFRA mutants Y288C and C235Y, in the Ig-like domain 3, were modelled using the structure of PDGFRB (PDB ID: 3MJG), with a 29% sequence identity to PDGFRA in this region, where the residues of interest are conserved(Shim et al., 2010). The PDGFRA mutant K385M, in the Ig-like domain 4, was modelled using the structure of Vascular Endothelial Growth Factor Receptor (VGFR1) (PDB ID: 5T89), with a 29% sequence identity to PDGFRA in this region(Markovic-Mueller et al., 2017). The corresponding three residues were mutated and subsequently modelled in PyMOL.
Whole exome- and RNA-sequencing
WES sample and library preparation
DNA was extracted from tumours using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen) following manufacturer instructions. The Nextera Rapid Capture Exome kit (Illumina) or SureSelect Reagent Exome kit (Agilent) were used to prepare libraries according to manufacturer’s instructions. Paired-end sequencing (100 bp) was performed on Illumina HiSeq 2500/4000 platform.
RNA-seq sample and library preparation
Total RNA was extracted from cell pellets using the Aurum Total RNA Mini Kit (Bio-Rad) and tumours using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen) according to instructions from the manufacturers. Library preparation was performed with ribosomal RNA (rRNA) depletion according to instructions from the manufacturer (Epicentre) to achieve greater coverage of mRNA and other long non-coding transcripts. Paired-end sequencing (100 bp) was performed on Illumina HiSeq 2500/4000 or NovaSeq 6000 platforms.
Data processing
Raw reads were trimmed using Trimmomatic v0.32(Bolger et al., 2014). Adaptors and other Illumina-specific sequences were removed using palindrome mode. Next, a four-nucleotide sliding window was used to remove the bases once the average quality within the window fell below 30. Finally, reads shorter than 30 base pairs were dropped. For WES, cleaned reads were aligned to the human reference genome build hg19 using BWA-MEM(Li and Durbin, 2009) v0.7.17 with default settings. Read duplicates were removed using Picard tools v2.10.7. For RNA-seq, cleaned reads were aligned to the human reference genome build hg19 using STAR(Dobin et al., 2013) v2.3.0e with default settings. Multiple control metrics (Table S2a) were obtained using FASTQC (v0.11.2), samtools(Li et al., 2009) (v0.1.20), BEDtools(Quinlan and Hall, 2010) (v2.17.0) and custom scripts. For visualization, normalized Bigwig tracks were generated using BEDtools and UCSC tools. Integrative Genomic Viewer(Thorvaldsdóttir et al., 2013) was used for data visualization.
Variant calling
GATK(DePristo et al., 2011) v3.2-2 split'N'Trim was used to split reads in splice junction and rescale the mapping quality from 255 to 60. Then, GATK v3.2-2 IndelRealigner was used to realign the indels. Single-nucleotide variants were called using samtools(Li et al., 2009) (v0.1.20) mpileup and Annovar(Wang et al., 2010) v2016Feb01 was used for annotation (Table S1). More specifically, variants were annotated using hg19 build with RefSeq gene, dbSNP (snp138), 1000 Genome Project (1000g2015aug_all), SIFT, PolyPhen, LRT, MutationTaster, GERP++ and PhastCons. Variants with a total read count of less than 10, an alternative allele read count of less than three, a SNV ratio of less than 0.2, an indel read ratio of less than 0.15, a variant quality of less than 20, a mapping quality less than 15, a strand bias p-value lower than 0.001 and a minor allele frequency higher than 0.001 were filtered out. Variants in repetitive regions (RepeatMasker, InterruptedRpts, segmentalDups, simpleRepeats and retroposons) or annotated as benign or likely benign from ClinVar(Landrum et al., 2018) were also filtered out. In addition, only variants of the following types were kept for downstream analyses: nonsynonymous SNV, splicing, splicing-extended, stop-gain and stop-loss. For gene amplification inference from WES, copy number variants (CNVs) were inferred from calculating RPKM coverage over annotated ENSEMBL genes. Samples were denoted as being positive for genic amplification if RPKM coverage was > 2-fold of the median RPKM.
Reconstruction of tumour phylogeny
For reconstruction of tumour phylogeny (Table S6), somatic variant calling was performed as previously described in Suzuki et al(Suzuki et al., 2019). Sequencing reads were aligned to human reference genome ‘hs37d5’ by 1000 Genomes Project Phase II, using Burrows Wheeler aligner (BWA) – MEM(Li and Durbin, 2009) (v0.7.9) with ‘-T 0’ option. Duplicates were marked using biobambam v.0.0.148. Samples without matched controls were run with a pseudo-control generated from 25 randomly selected WES samples from 1000 Genomes Project. Fastq files were mapped with the same settings and subsampled using the samtools view function to adjust sequencing coverage around 200x.
Variants were called using eight variant callers: MuTect2(Cibulskis et al., 2013), EBCall(Shiraishi et al., 2013), Varscan2(Koboldt et al., 2012), Strelka(Saunders et al., 2012), SomaticSniper(Larson et al., 2012), Virmid(Kim et al., 2013), Platypus(Rimmer et al., 2014) and Seurat(Christoforides et al., 2013). Each caller except for Strelka was run as previously described in Suzuki et al(Suzuki et al., 2019). Because the previously described method was for whole genome sequencing, Strelka was run with ‘isSkipDepthFilters = 1’. Variants were identified if called by at least two callers, with ≥ 2 variant reads, ≥ 7 total reads and ≥ 0.05 variant allele frequency in the tumour and ≤ 1 variant read in the control, as calculated using the realignment function of Genomon-MutationFilter v.0.2.1.
For samples without matched controls, variants with a frequency ≥ 0.01 in 1000 Genomes, dbSNP138, Exome Aggregation Consortium database, NHLBI-ESP project, Kaviar Genomic Variant Database, Haplotype Reference Consortium database, Greater Middle East Variome, or Brazilian genomic variants database were discarded. Missense, synonymous mutations or non-frameshift indels registered in any SNP databases listed above a frequency of 0.01 and registered with fewer than 10 samples in COSMIC v87 were also discarded. Variants with SIFT score ≥ 0.05, PolyPhen-2 HDIV ≤ 0.908, PolyPhen-2 HVAR ≤ 0.956, “polymorphism” or “polymorphism_automatic” by MutationTaster, and variants “predicted non-functional” by MutationAssessor were discarded. Phylogenetic trees were constructed based on shared mutations detected in exons, splice sites, or UTRs. Fish-plots were generated using R package fish-plot (v0.5).
Gene expression quantification from RNA-seq data
Gene expression levels were estimated by quantifying reads uniquely mapped to exonic regions defined by ensGene annotation set from Ensembl (GRCh37, n = 60,234 genes) using featureCounts(Liao et al., 2014) (v1.4.4). Normalization (mean-of-ratios), variance-stabilized transformation of the data and differential gene expression analysis were performed using DESeq2(Love et al., 2014) (v1.14.1). Unless otherwise stated, all reported p-values have been adjusted for multiple testing using the Benjamini-Hochberg procedure. For tumours, comparisons were performed between G34R/V samples and each other tumour entity, with batch included as a covariate.
Assessment of developmental programs in tumours
To evaluate developmental gene programs in tumour samples, a reference panel of 167 forebrain cell-type specific signatures was assembled using data from five published scRNAseq atlases(Anderson et al., 2020; Jessa et al., 2019; Mizrak et al., 2019; Nowakowski et al., 2017; Velmeshev et al., 2019), spanning embryonal mouse forebrain, P9 striatum, adult mouse sub-ventricular zone, fetal huma n telencephalon, and pediatric and adult human cortex. For the mouse forebrain dataset, 100-gene signatures were used as reported in the original study. For the mouse striatum and the two human datasets, we derived 100-gene signatures from the published cluster markers by filtering out genes encoding ribosomal proteins (defined as having a gene symbol matching “Rps”, “Rpl”, “Mrps”, “Mrpl”), and selecting the top 100 genes ranked by adjusted p-value. Finally, for the mouse SVZ dataset, we called markers by performing differential gene expression analysis for the clusters reported by the authors in the original study, and subsequently derived the 100-gene signatures. Proliferating clusters were identified as those either annotated by the authors of the original studies, or with high expression of the proliferation marker Top2a (mean cluster expression > 0.4). Signatures from these clusters were excluded from analysis to prevent spurious enrichment due to cell cycle genes. Gene set enrichment analysis (GSEA) was performed with these signatures as input, applied to expressed genes in tumour transcriptomes (bulk RNA-seq) ranked using the Negative Binomial Wald test statistic from differential expression analysis, using the fgsea package(Sergushichev, 2016) (v1.8.0). With fgsea, leading edge genes were obtained and normalized enrichment scores (NES) were computed by normalizing enrichment to the average enrichment of 10,000 random gene samples. P-values were adjusted using the Benjamini-Hochberg procedure; signatures with adjusted p-value < 0.01 were considered significantly enriched or depleted.
In the case of cell line datasets, the sample size was not adequate to perform GSEA, and thus enrichment of human radial glia and interneuron gene signatures was evaluated using single-sample GSEA (ssGSEA)(Barbie et al., 2009) using the GSVA(Hanzelmann et al., 2013) package (v1.27.0) as described previously(Jessa et al., 2019).
Gene expression profiling in normal development
Gene expression patterns of individual genes during human and mouse brain development were assessed across three previously reported scRNAseq reference developmental datasets(Anderson et al., 2020; Jessa et al., 2019; Nowakowski et al., 2017). Dendrograms of gene expression in neural cell types displayed in Figure 3 and Figure S5 were constructed as follows: normalized gene expression, cell type annotations (cluster assignments) and cluster gene signatures (cluster markers) were obtained from original publications(Jessa et al., 2019; Nowakowski et al., 2017). For each cluster and each gene, the mean expression across cells from the cluster was computed, as well as the proportion of cells in which the gene was detected (UMI counts > 0). Genes appearing in any of the clusters’ signatures were selected for unsupervised hierarchical clustering. Spearman’s rank correlation was used as distance metric on the cluster mean gene expression profiles, and complete linkage as agglomeration method. For visualization purposes, mean gene expression was scaled across clusters to [0,1].
Gene expression along interneuron differentiation
To profile gene expression during normal interneuron differentiation, displayed in Figure 2F, we used a previously reported reconstruction of the interneuron differentiation trajectory(Jessa et al., 2019), in which ventral radial glial cells, interneuron progenitors, and interneurons from the E12.5, E15.5, and P0 mouse forebrain were ordered in pseudotime using Monocle(Qiu et al., 2017a; Qiu et al., 2017b). For visualization, Monocle (v2.0.1) was used to fit, for each gene, a smooth spline to the nonlinear expression dynamics as a function of pseudotime. Smoothed expression was then z-scored across pseudotime and extreme upper and lower values were set to 3 and −3 respectively. The genes displayed in the heatmap are the union of genes identified as specific to G34R/V gliomas by RNA-seq and ChIP-seq. Genes enriched in G34R/V were defined as those 1) in the leading edge of the human interneuron gene signature most significantly enriched in G34R/V (ranked by NES, GSEA analysis), or 2) significantly upregulated in G34R/V (adj. p-value < 0.01 and log2 fold-change > 2, RNA-seq differential expression analysis) and enriched for promoter H3K27ac in G34R/V (z-score > 0.6, ChIP-seq analysis). Conversely, genes were defined as depleted in G34R/V were genes 1) in the leading edge of the human interneuron gene signature most significantly depleted in G34R/V (ranked by NES, GSEA analysis), or 2) significantly downregulated in G34R/V (adj. p-value < 0.01 and log2 fold-change < −2, RNA-seq differential expression analysis) and enriched for promoter H3K27me3 in G34R/V (z-score > 0.6).
Chromatin immunoprecipitation sequencing
Sample and library preparation
ChIP-seq was performed as previously described in Harutyunyan et al(Harutyunyan et al., 2019). Briefly, cells (cell lines or dissociated tumour cells) were fixed with 1% formaldehyde (Sigma). Fixed cell preparations were washed, pelleted and stored at −80 °C. Sonication of lysed nuclei (lysed in a buffer containing 1% SDS) was performed on a BioRuptor UCD-300 for 60 cycles, 10s on 20s off. Samples were checked for sonication efficiency using the criteria of 150–500 bp by gel electrophoresis. After sonication, the chromatin was diluted to reduce SDS level to 0.1% and before ChIP reaction 2% of sonicated Drosophila S2 cell chromatin was spiked-in the samples.
ChIP reaction for histone modifications was performed on a Diagenode SX-8G IP-Star Compact using Diagenode automated Ideal ChIP-seq Kit. Twenty-five microliter Protein A beads (Invitrogen) were washed and then incubated with antibodies (anti-H3K27me3 (1:40, CST 9733 RRID:AB_2616029), (anti-H3K27ac (1:80, Diagenode C15410196 RRID:AB_2637079)), (anti-H3K36me3 (1:100, Active Motif 61021 RRID:AB_2614986)), (anti-H3.3 (1:66, Millipore 09–838)), and 2 million cells of sonicated cell lysate combined with protease inhibitors for 10 h, followed by 20 min wash cycle with provided wash buffers. ChIP reactions for SUZ12 and CTCF were performed RRID:AB_10845793 as follows: anti-SUZ12 (1:150, CST 3737 RRID:AB_2196850) or anti-CTCF (1 μg/reaction, Diagenode C15410210 RRID:AB_2753160) antibodies were conjugated by incubating with 40 μl protein A beads at 4 °C for 6 h, then chromatin from ~4 million cells was added in RIPA buffer, incubated at 4 °C o/n, washed using buffers from Ideal ChIP-seq Kit (one wash with each buffer, corresponding to RIPA, RIPA + 500 mM NaCl, LiCl, TE), eluted from beads by incubating with Elution buffer for 30 min at room temperature.
Reverse cross linking took place on a heat block at 65 °C for 4 h. ChIP samples were then treated with 2 μl RNase Cocktail at 65 °C for 30 min followed by 2 μl Proteinase K at 65 °C for 30 min. Samples were then purified with QIAGEN MiniElute PCR purification kit as per manufacturers’ protocol. In parallel, input samples (chromatin from about 50,000 cells) were reverse crosslinked and DNA was isolated following the same protocol.
Library preparation was carried out using Kapa HTP Illumina library preparation reagents, following manufacturer’s instructions. Briefly, 25 μl of ChIP sample was incubated with 45 μl end repair mix at 20 °C for 30 min followed by Ampure XP bead purification. A tailing: bead bound sample was incubated with 50 μl buffer enzyme mix at 30 °C for 30 min, followed by PEG/NaCl purification. Adaptor ligation: bead-bound sample was incubated with 45 μl buffer enzyme mix and 5 μl of TruSeq DNA adapters (Illumina), for 20 °C 15 min, followed by PEG/NaCl purification (twice). Library enrichment: 12 cycles of PCR amplification. Size selection was performed after PCR using a 0.6 × /0.8x ratio of Ampure XP beads (double size selection) set to collect 250–450 bp fragments. ChIP libraries were sequenced on Illumina HiSeq 4000 or NovaSeq 6000 platforms at 50 bp single reads or paired-reads.
Data processing
Murine embryonic stem cell and adult cortex ChIP-seq datasets were obtained from the ENCODE consortium portal(Davis et al., 2018). E13.5 ganglionic eminence ChIP-seq were obtained from Lindtner et al. 2019 (GSE124936)(Lindtner et al., 2019). ChIP-seq datasets were processed using the ChIP-seq module of GenPipes (v3.1.2)(Bourgey et al., 2019). Briefly, raw reads were trimmed using Trimmomatic(Bolger et al., 2014) v0.32 to remove adaptor and sequencing-primer associated reads, then aligned to hg19 or mm10 using bwa-mem(Li and Durbin, 2009) (v0.7.12) with default parameters. PCR duplicate reads as defined by reads with identical mapping coordinates were then collapsed by Picard (v2.0.1) to produce uniquely aligned reads. Reads were then filtered for mapping quality of > 5. For single-end (SE) 50bp datasets, reads were extended by 250bp. Wiggle tracks are generated using uniquely aligned reads using Homer (v4.9.1)(Heinz et al., 2010). RPKM was calculated using VisRSeq (v0.9.40) (Younesy et al., 2015) or SeqMonk (v1.46) at annotated genes. Promoter is defined as a 5kb centered bin on the transcription start site. Median values were generated for promoter-associated H3K27me3 and H3K27ac in the two subgroups - 1) G34R/V tumours and 2) non-G34R/V IDH1, SETD2, and WT cortical tumours. Z-score was calculated from the median RPKM as . Z-score > ∣0.5∣ were designated as significant changes for ChIP-seq datasets (Table S3). Gene ontology analyses were performed using PANTHER.
Core regulatory circuitry analysis
H3K27ac ChIP-sequence reads were aligned to the human genome (HG19) using Bowtie2(Langmead and Salzberg, 2012) (v2.1.0) under default settings. PCR duplicates were removed using Picard tools generating BAM. Significant peaks were identified using Model-Based Analysis for ChIP-seq (MACS) (v1.4)(Zhang et al., 2008) with a p value cutoff of 1e−9. Peaks were annotated using HOMER(Heinz et al., 2010) (v3.12) with promoter regions classified as any peak within +/− 2.5 kb of a transcriptional start site (TSS), and enhancer region greater than 2.5 kb from a TSS. Peaks were also annotated using ChIP-atlas annotating distal enhancers to genes based upon public CHIA-PET datasets. Super enhancers were identified using the ROSE algorithm with exclusion of peaks within +/− 2.5 kb of a TSS and a stitch distance of 12.5 kb.
Core regulatory circuitry (CRC) analysis was used with default parameters to quantify the interaction network of transcription factor (TF) regulation at super enhancers. Briefly, for all promoters within 100 kb, the most acetylated promoter was assigned as the target of the SE (excluding promoters that overlap SEs, which are automatically assigned the target. If there were no active promoters within 100kb, the SE was assigned to the nearest active promoter. All SE-associated promoters annotated to regulate a TF were considered as the node-list for network construction. For any given TF (TFi), the IN degree was defined as the number of TFs with an enriched binding motif at the proximal SE or promoter of TFi. The OUT degree was defined as the number of TF associated SEs containing an enriched binding site for TFi. Within any given SE, enriched TF binding sites were determined at putative nucleosome free regions (valleys) flanked by high levels of H3K27ac. Valleys were calculated using an algorithm adapted from Ramsey et al.(Ramsey et al., 2010). In these regions, we searched for enriched TF binding sites using the FIMO59 algorithm with TF position weight matrices defined in the TRANSFAC database(Matys et al., 2006). An FDR cutoff of 0.01 was used to identify enriched TF binding sites. Hierarchal clustering of core TFs were performed using the heatmap function of R, and Gene Ontology assessment was performed using PANTHER.
Hi-C chromatin conformation capture
Sample and library preparation
In situ Hi-C libraries were generated from patient-derived glioma cell lines (5 million cells each), and murine embryonic brain tissue (1-3 million cells per sample), as described in Rao et al(Rao et al., 2014) with minor modifications. Briefly, in situ Hi-C was performed in 7 steps: (1) crosslinking cells with formaldehyde, (2) digesting DNA using a 4-cutter restriction enzyme (DpnII) within intact permeabilized nuclei, (3) filling in, biotinylating the resulting 5′-overhangs and ligating the blunt ends, (4) shearing the DNA, (5) pulling down biotinylated ligation junctions with streptavidin beads, (6) library amplification and (7) analyzing fragments using paired end sequencing. As quality control (QC) steps, efficient sonication was checked by agarose DNA gel electrophoresis and for appropriate size selection by Agilent Bioanalyzer profiles for libraries. For final QC, we performed superficial sequencing on the Illumina Hiseq 2500 (~30M reads/sample) to assess quality of the libraries using percent of reads passing filter, percent of chimeric reads, and percent of forward-reverse pairs. Hi-C libraries were sequenced (paired-end, 125 bp) on the Illumina HiSeq2500 platform.
Hi-C data analysis
Hi-C reads were trimmed and assessed for quality control using the Trim Galore package(Krueger, 2012) (TrimGalore v0.6.5, Cutadapt v2.6 and Fastqc v0.11.9) (Table S4). Reads were then mapped to hg19 or mm10, and filtered for common Hi-C artefacts using HiCUP(Wingett et al., 2015) (HiCUP v0.7.2, Bowtie2 v2.3.5, R v3.6.0_3.9). Analysis of Hi-C libraries and downloaded files was performed using Juicer and associated Juicer Tools (Durand et al., 2016) (v1.22.01). Contact maps were generated using Juicer with the following parameter: -s DpnII. Map resolution was determined by using Juicer’s “calculate_map_resolution.sh” script. All observed matrices were computed using HOMER, accounting for both the linear distance between two given loci and sequencing depth. Hi-C contact maps and associated annotations were visualized using Juicebox (Durand et al., 2016). We used the HIFI algorithm(Cameron et al., 2020) to process 5-kb resolution Hi-C data to obtain higher accuracy estimates of interaction frequencies, using the following parameters: bandSize=1000, outputNormalized, boundaryKS=1000. Virtual 4C plots were derived using HIFI 5 kb resolution heatmaps. Observed/expected ratios were extracted using the dump function in Juicer Tools from HIFI heatmaps, using the following anchor regions: PDGFRA/Pdgfra promoter (chr4:55090000-55105000 in hg19, chr5:75140000-751550000 in mm10), or GSX2/Gsx2 promoter (chr4:54960000–54970000 in hg19, chr5:75070000–75080000 in mm10). TAD boundaries were calculated using robusTAD tool (Dali et al., 2018) at a 50kb resolution. For loop quantification, we extracted and calculated the mean interaction values for each 5kb bin at our region of interest with PDGFRA/Pdgfra or GSX2/Gsx2 promoters. Quantification of interactions within a TAD was computed as the sum of normalized (observed/expected) interaction scores(Kloetgen et al., 2020) for each cell line, extracted using the dump function from Juicer Tools for the region of interest. Significant differences in intra-TAD activity or interaction loops anchored on PDGFRA or GSX2 promoters were assessed using a one-tailed t-test. All related quantification and statistical analyses are provided in Table S4. Publicly available Hi-C datasets on murine embryonic stem cells were downloaded from GEO (GSE96107)(Bonev et al., 2017), and processed as described above.
Hi-C data visualization
To ensure visual clarity for Hi-C heatmaps, we either plotted observed/expected interaction values or observed interaction values alone. When plotting broader windows for the Hi-C interaction heatmaps (e.g. Figure S5F), we plotted observed values since observed/expected values tend to exaggerate the intensity of long-range interactions. Observed values also allow better visualization of TAD structures across large-scale windows. Similarly, we represented observed values to represent the Gsx2-hs687 interaction in Figure S5G because they are better suited for small distances, unlike observed/expected values that tend to underestimate short interactions due to their proximity in the linear genome. By contrast, we represented observed/expected values for mid-range interactions across the PDGFRA-hs687 locus, as depicted in Figure 4A-B.
Single-cell and single-nuclei RNAseq
Tissue handling and dissociation
Fresh tumours collected after surgery were enzymatically digested and mechanically dissociated using the papain version of the Brain Tumor Dissociation kit (Miltenyi Biotech) or the Worthington Papain dissociation kit (Table S6). Red blood cells were lysed by ammonium chloride treatment for 5 min on ice. Cell viability was assessed with Trypan Blue. For samples with low viability (<60%), dissociated cells were enriched for live cells using the Dead Cell Removal kit (Miltenyi Biotech). 10,000 dissociated cells per sample were loaded on the 10X Genomics Chromium controller. Nuclei were prepared from frozen tissue (5-50 mg) as previously described(Nagy et al., 2019). Nuclei concentration was assessed using the ReadyProbes Cell Viability fluorescence assay (ThermoFisher Scientific). As nuclei capture was 55-60% less efficient than cell capture, we aimed to capture 22,000 nuclei per sample.
Single-cell and single-nuclei library preparation
The Chromium Single Cell 3′ (10X Genomics, Version 3) protocol was strictly followed to prepare libraries. The 10X libraries were sequenced on the Illumina HiSeq4000 or NovaSeq sequencing platforms (Table S6).
Data processing and quality control
Cell Ranger (10X Genomics) was used with default parameters to demultiplex reads and align sequencing reads to the genome, distinguish cells from background, and obtain gene counts per cell. Alignment was performed using the hg19 reference genome build, coupled with the Ensembl transcriptome (v75). For snRNAseq data, intronic counts were included. For each sample, cells were filtered using the following quality control (QC) metrics (Table S6): mitochondrial content (indicative of cell damage), number of genes, and number of unique molecular identifiers (UMIs), using the Seurat(Butler et al., 2018) package (v2.3). Thresholds for each sample were selected according to the distribution of each metric within the sample, which varies with sequencing coverage and the number of cells captured. Data was processed as described previously(Jessa et al., 2019) using Seurat. Briefly, libraries were scaled to 10,000 UMIs per cell and log-normalized. UMI counts and mitochondrial content were regressed from normalized gene counts and the residuals z-scored gene-wise. Dimensionality reduction was performed using principal component analysis (PCA) applied to the most variant genes, and PCA was used as input for projection to two dimensions using uniform manifold approximation and projection(McInnes et al., 2018) (UMAP) and clustering using a shared nearest neighbor modularity optimization algorithm(Waltman and Van Eck, 2013).
Integration of scRNAseq and snRNAseq data
For visualization of single-cell (scRNAseq) and single-nuclei (snRNAseq) datasets in a shared UMAP space as in Figure 5, cells from all G34R/V tumour datasets were integrated using the method implemented in Seurat(Stuart et al., 2019) (v3). The resulting matrix was subjected to z-scoring and dimensionality reduction as described above. Only cells identified as malignant (see below) were included in this visualization.
Projection of cells to a brain reference atlas
Cell type assignment for each cell was based on projection to the most similar cell type in the normal developing brain, with cell type signatures obtained from the mouse brain scRNAseq atlas(Jessa et al., 2019), using three different cell type projection methods, detailed below: (i) ACTINN(Ma and Pellegrini, 2020), (ii) pattern correlation, and (iii) random sampling(Filbin et al., 2018)). Signatures derived from proliferating clusters were excluded from analysis, resulting in 176 gene signatures. Each tumour cell was assigned a cell identity based on the majority vote of the three projection methods. For cells without a majority assignment, the cell identity was imputed based on the assigned cell type for the majority of the ten nearest neighbors obtained using the shared nearest neighbor(Waltman and Van Eck, 2013) method implemented in Seurat.
Neural network ACTINN projection
A neural network with three hidden layers(Ma and Pellegrini, 2020) designed for assigning cell type in scRNAseq data was trained on single-cell gene expression profiles from the mouse scRNAseq reference atlas. In the reference, the training data was the normalized single-cell gene expression profiles for the 1000 most variable genes, and the training labels were the cell types assigned to each individual cell. The trained neural network was applied to normalized single cell gene expression data for tumour samples.
Pattern correlation projection
We adapted a method developed for cell cycle phase assignment in scRNAseq data(Macosko et al., 2015) to cell type projection. Given our set of 176 signatures, S ={s1, s2, … , s176} where si = {gene1, gene2, gene3, … , gene100}, we compute, for each signature si and for each cell cj, the average normalized expression of all the genes in si, defining a score Tsicj. The scores Tsicj are z-scored across cells to yield normalized scores Nsicj. Finally, for each cell, the scores Nsicj are z-scored across signatures to obtain a final score Fsicj. In the reference atlas, each cluster of cells has a gene signature. Many clusters in the atlas are composed of cell types identified at multiple brain regions or timepoints (e.g. astrocyte clusters were identified in both brain regions sampled). We therefore define “cell type identity” vectors, representing the 37 distinct cell types captured. Each cell type identity vector is a binary vector of the same length as the signature set S, which takes on a value of 1 if the corresponding signature belongs to the given cell type, and 0 otherwise. We then compute, for each cell, the correlation between the final scores Fcisk and each of the cell type identity vectors. Finally, each cell is assigned the identity of the cell type with the greatest correlation.
Random sampling projection
We compute a score Tsicj for each cell as described above. For each signature, this score is compared to the score for a control gene set, computed in an identical manner. To define the control gene set, all detected genes are first binned based on their average expression across the dataset. The control gene set is generated by randomly sampling with replacement 100 genes of similar expression level for each gene in the signature. The final score for each signature is the difference between the score Tsicj and the score of the control set. Finally, each cell is assigned the identity of signature with the highest final score.
Re-analysis of published data
Normalized expression levels for scRNAseq (Smartseq2) datasets of other HGG entities from published studies were downloaded from GEO: IDH1 mutant HGG(Venteicher et al., 2017) (GSE89567), K27M HGG(Venteicher et al., 2017) (GSE102130) and WT HGG(Neftel et al., 2019) (GSM3828672). In the latter case, pediatric and adult samples were separated. For each dataset, normalized expression levels were subjected to z-scoring, dimensionality reduction, clustering and cell type projection as described above. Clusters of cells projected to immune cell types were flagged as normal.
Identification of normal and malignant cells
Malignant cells within a sample were identified as follows. First, cells from all samples, separately from each technology (scRNAseq and snRNAseq) were pooled and subjected to scaling, dimensionality reduction, cell projection and clustering as described above. Clusters composed of cells projecting to a single cell type and originating from multiple patients were labeled as normal. Specifically, we identified normal clusters as those for which a single patient accounted for at most 75% of the cells/nuclei in the cluster, and for which at least 70% of all the cells/nuclei in the cluster projected to the same cell type. Clusters of cells originating from multiple patients but projecting to different cell types were flagged as low quality and removed from the analysis. Next, copy number alterations (CNV) were inferred on a per sample basis as previously described(Filbin et al., 2018). For this, the cells labeled as normal as described above were used as reference. In some samples, a clear CNV distinction between normal and malignant cells was not present. In these cases, we identified groups of cells which, (1) clustered together based on transcriptional profiles and (2) were assigned the same cell type based on projections to the reference atlas and (3) no obvious CNV alteration was observed. The cells fulfilling all three criteria were then labeled as normal. Finally, independent CNV calls were obtained with the inferCNV package (v0.8.2)(Tickle et al., 2020) on a per sample basis, using cells labeled as normal pooled from all samples as a combined normal reference, in order to confirm aberrant CNV profiles in malignant cells.
Supplementary Material
Figure S1. Recurrent somatic mutations in G34R/V HGGs, Related to Figure 1
A. Expression of PDGFRA in HGGs. PDGFRA is significantly upregulated in G34R/V tumours relative to others (adjusted p-value = 0.018, Negative Binomial Wald test). B. Oncoprint depicting recurrently mutated genes (observed in > 3 patients) in G34R/V HGG, grouped by associated gene functions as determined by Gene Ontology. Columns adjacent to gene names represent the number of patients in which mutations were called, in the entire cohort or stratified by PDGFRA mutation status (red = PDGFRAMUT, green = PDGFRAWT). Underlined genes indicate a bias towards PDGFRAWT G34 samples. C. Complex structure diagrams depicting extracellular PDGFRA mutations C235Y/F, Y288C, and K385M. Crystal structures of PDGFRB (PDB: 3MJG) were used to model C235 and Y288 mutations, and VGFR1 (PDB: 5T89) for K385. The C235Y/F mutations destabilize the Ig-like domain, leaving C291 available to form another inter- or intra-molecular disulfide bond, potentially leading to receptor dimerization and activation, analogous to Y288C. Finally, for K385M, in the wildtype protein, the positive charges of two K385 residues at the dimerization interface create an unfavourable repulsive effect. The K385M mutation may promote a small hydrophobic patch adjacent to several salt-bridges, enhancing receptor dimer formation and activation.
Figure S2. G34R/V uniquely exhibits a transcriptome and epigenome program specific to the forebrain interneuron lineage, Related to Figure 2 and Tables S2 and S3
A. Gene set enrichment analysis (GSEA) plots for the most enriched or depleted signature of each cell type. Enrichments are computed for G34R/V compared to IDH1 HGGs. NES: normalized enrichment score. B. Heatmap of expression levels of genes driving the enrichment of interneuron gene signature, or the depletion of the oligodendrocyte signature, in G34R/V tumours compared to other HGG subgroups. Expression is z-scored across samples. C. Heatmap of GSEA NES scores of forebrain astrocyte signatures in G34R/V tumours compared to the HGNET-BCOR tumours (non-gliomas). D. Heatmap of GSEA NES scores of 8-10 week old mouse ventricular/sub-ventricular zone (V-SVZ) cell type signatures in G34R/V compared to IDH1, K27M and WT HGG subgroups, as in (A). Scores are shown for all signatures significantly enriched (adjusted p-value < 0.01) in G34R/V vs. IDH1. aNSC: actively dividing neural stem cells, TAC: transit amplifying cells, NB: neuroblasts, OPC: oligodendrocyte progenitor cells, COP: committed oligodendrocyte precursors. E. Expression levels of the human radial glia marker MOXD1 and markers for cortical inhibitory and excitatory neurons in G34R/V, other HGG subgroups, and HGNET-BCOR. For genes upregulated in G34R/V, adjusted p-values (Negative Binomial Wald test) for comparison of G34R/V to other entities are indicated in parentheses. F. Gene ontology analysis of differentially expressed genes that showed significant H3K27ac or H3K27me3 enrichment in G34R/V tumours (z-score > 0.5). G. Core transcription factors (inferred by H3K27ac enrichment) activated in G34R/V compared to other HGGs, non-glioma brain tumours, and normal brain. Indicated TFs from cluster 2 lack activation signals specifically in G34R/V HGGs. Bolded genes indicate transcription factors annotated as necessary for oligodendrocyte differentiation (FDR = 0.0001) based on Gene ontology enrichment analysis.
Figure S3. G34R/V promotes aberrant H3K27me3 retention at terminal neuronal genes, Related to Figure 3 and Tables S2 and S3
A. Quantification of H3K27 and H3K36 methylation states by histone mass spectrometry on the mutant H3.3G34R or H3.3G34V histone normalized by H3.3 WT abundance in PS10-801 (left, n = 3), HSJD-GBM002 (middle, n = 1) or KNS-42 (right, n = 6) cell lines respectively. * denotes p-value < 0.05 (paired t-test). B. Volcano plot depicting differential H3K27me3 enrichment between G34R/V and non-G34R/V cortical HGGs at all annotated promoters. Blue data points indicate G34R/V-specific H3K27me3+ genes (z-score > 0.5, p-value < 0.05) involved in spontaneous synaptic transmission (FDR = 0.0273), calcium ion-regulated exocytosis of neurotransmitter (FDR = 0.000269), and regulation of GABAergic synaptic transmission (FDR = 0.00332). C. Volcano plot depicting differential H3K27ac enrichment between G34R/V and non-G34R/V cortical HGGs at all annotated promoters. Red data points indicate G34R/V-specific H3K27ac+ genes (z-score > 0.5, p-value < 0.05) involved in neuron fate commitment (FDR = 0.00243). D. Targeted deep sequencing validation of CRISPR-mediated editing of H3F3A G34R in HSJD-GBM002 clones. E. Volcano plot depicting transcriptomic change in CRISPR HSJD-GBM002 clones. Genes significantly up-regulated (log2FC > 1, p-adj < 0.05) in G34R unedited clones are highlighted in teal, whereas genes significantly up-regulated upon G34R removal are highlighted in navy (log2FC > 1, p-adj < 0.05). F. Volcano plot depicting H3K36me3 change at all annotated genic promoters in CRISPR HSJD-GBM002 clones (grey points), highlighting differentially expressed genes from RNA-seq (from panel E, teal = up-regulated in G34R, navy = up-regulated in edited). G. Volcano plot depicting H3K27me3 change at all annotated genic promoters in CRISPR HSJD-GBM002 clones (grey points), highlighting differentially expressed genes from RNA-seq (from panel E, teal = up-regulated in G34R, navy = up-regulated in edited).
Figure S4. Introduction of G34R in murine embryonic forebrain induces H3K27me3 epigenomic reprogramming, Related to Figure 3
A. Volcano plot depicting differential promoter H3K27me3 enrichment between G34R and non-G34R sorted cells 72-hours post-electroporation (left) or >1-year post-electroporation (right). Genic promoters with significantly increased H3K27me3 enrichment in G34R cells are indicated in red. B-C. Genome browser snapshot of ChIP-seq profiles in IUE-derived neural precursor cells showing H3K27me3 profiles associated with G34R-introduction. Some genic promoters, such as Foxp2 (B) and Jph4 (C) show striking H3K27me3 enrichment uniquely in G34R-electroporated cells. EV: empty vector. WT: wildtype H3.3. APP: Atrx shRNA, sgRNA for Trp53, PDGFRAWT. AP: Atrx shRNA, sgRNA for Trp53.
Figure S5. Active chromatin conformation promotes GSX2-PDGFRA co-expression in G34R/V HGGs, Related to Figure 4 and Table S4
A. Normalized expression of PDGFRA and GSX2 in bulk RNA-seq of G34R/V tumours (circles) and cell lines (triangles). Pearson correlation=0.69, R2=0.48, p-value=0.0002. B. PDGFRA, GSX2, and DLX1/2 expression in cell populations from scRNAseq atlas of the developing (5-37 PCW) human telencephalon. For each cell population (tree leaves), mean expression and proportion of cells expressing the gene are indicated. Dendrograms were constructed based on pairwise Spearman correlations of gene expression. MGE: medial ganglionic eminence, OPC: oligodendrocyte progenitor cells. *: GSX2 is not detected in this dataset due to low coverage. C. Top, H3K27ac profiles at the PDGFRA-GSX2-hs687 locus in individual G34R/V (n = 7), IDH1 (n = 5), SETD2 (n = 5), WT (n = 3), and K27M (n = 4) tumours. PDGFRAMUT samples are distinguished by darker teal. Note that PDGFRA is amplified in one K27M sample (first yellow track). Bottom, H3K27me3 profiles at the PDGFRA-GSX2 locus in individual G34R/V tumours (n=5), IDH1 (n=5), SETD2 (n=5), WT (n=3), and K27M (n=2) tumours. D. Hi-C heatmaps from a G34R, H3/IDH WT and K27M glioma cell lines depicting sub-TAD structure at the PDGFRA-GSX2-hs687 locus. Small black triangle illustrates a TAD between PDGFRA and hs687 enriched for contacts in G34R/V lines. Large dashed triangle demarcates the TAD formed by contact to a known distal insulator. Heatmap represents the log2 ratios of observed interactions relative to expected interactions across the PDGFRA-hs687 locus at a 5kb resolution (red = positive, blue = negative). E. Manhattan plot depicting H3K27ac enhancer activation difference between PDGFRAMUT and PDGFRAWT G34R/V HGGs (z-score > 0 indicate H3K27ac enriched in PDGFRAMUT) at far distal regions of PDGFRA-GSX2 locus. Black points depict comparison across different patients (n=5 PDGFRAMUT and n=2 PDGFRAWT), and teal points depict recurrence (PDGFRAMUT) vs. primary (PDGFRAWT) comparison in the same patient P-1190. F. Hi-C heatmaps from two G34R/V cell lines, HSJD-GBM002 (PDGFRAMUT) and KNS-42 (PDGFRAWT), depicting TAD structure at the PDGFRA-GSX2-hs687 locus. Black rectangles illustrate long-distance interactions between PDGFRA promoter and potential enhancers, enriched for contacts in HSJD-GBM002 vs. KNS-42. Heatmap represents the normalized observed interactions across the locus at a 5kb resolution (red = high interaction, white = low interaction). G. Hi-C heatmaps from murine E13.5 cortex and ganglionic eminences (GE) depicting sub-TAD structure at the Pdgfra-Gsx2-hs687 locus. The dashed square represents the interaction between GSX2 promoter and hs687. Heatmap represents the normalized observed interactions across the locus at a 10kb resolution (red = high interaction, white = low interaction).
Figure S6. G34 HGGs are devoid of the oligodendroglial lineage, and co-option of PDGFRAMUT promotes neuronal-to-astrocytic transition, Related to Figure 5 and Table S5
A. Bubble plot representing the composition of cell types in each tumour. Circle size represents the proportion of each cell type per tumour, and circle colour represents the total number of malignant cells within the tumour. B. UMAP embedding of IDH1, K27M, H3/IDH1-WT HGG (pediatric and adult) scRNA-seq datasets, colour coded by patient of origin (top) and cell type projection (bottom). Oligodendrocytes (in green) are clearly detected in all non G34R/V HGG entities. C. Mean expression of foetal interneuron and astrocyte gene signatures in individual cells from the normal human telencephalon. Cells of the interneuron lineage, as well as radial glial cells and astrocytes were included.
Figure S7. G34R/V mutations are dispensable for tumour maintenance, Related to Figure 6
A. Hematoxylin & eosin (H&E) and immunohistochemical analysis of representative KNS-42 parental and edited clone-derived PDOX tumours using a G34V-specific antibody. B. H&E and immunohistochemical analysis of representative HSJD-GBM002 parental and edited clone-derived PDOX tumours using a G34R-specific antibody.
Table S1. PDGFRA mutations in G34R/V, K27M, IDH1/SETD2, and WT HGG subgroups, Related to Figure 1
Table S2. Analysis of bulk RNA-seq profiles in G34R/V HGGs, Related to Figures 2, 3, S2 and S3
Table S3. Analysis of H3K27me3 and H3K27ac ChIP-seq profiles in G34R/V HGGs, Related to Figures 2, 3, S2 and S3
Table S4. Analysis of Hi-C profiles in G34R/V and other HGG cell lines, and E13.5 murine cortex and GE samples, Related to Figure 4, and S5
Table S6. G34R/V primary/recurrence mutation comparison for P-1190, P-3200, and P-1978, Related to Figure 6
Table S7. Extended information on in-house G34R/V HGG samples, G34R/V cell lines and murine IUE samples characterized by pathology, transcriptomics, and epigenomics, Related to STAR Methods
ACKNOWLEDGEMENTS
We thank the patients and their families for their invaluable contributions to this research, without whom it would be impossible. This work was supported by funding from: A Large-Scale Applied Research Project grant from Genome Quebec, Genome Canada, the Government of Canada, and the Ministère de l'Économie, de la Science et de l’Innovation du Québec, with the support of the Ontario Institute for Cancer Research through funding provided by the Government of Ontario to NJ, MDT, CLK. Fondation Charles Bruneau to NJ, US National Institutes of Health (NIH grant P01-CA196539 to NJ, R01CA148699 and R01CA159859 to MDT); the Canadian Institutes for Health Research (CIHR grant MOP-286756 and FDN-154307 to NJ and PJT-156086 to CLK); the Canadian Cancer Society (CCSRI grant 705182) and the Fonds de Recherche du Québec en Santé (FRQS) salary award to CLK; the Fonds de recherche du Québec - Santé, as well as from Génome Québec and the Cancer Research Society to CLK; NSERC (RGPIN-2016-04911) to CLK; CFI Leaders Opportunity Fund 33902 to CLK, Genome Canada Science Technology Innovation Centre, Compute Canada Resource Allocation Project (WST-164-AB); Data analyses were enabled by compute and storage resources provided by Compute Canada and Calcul Québec. NJ is a member of the Penny Cole Laboratory and holds a Canada Research Chair Tier 1 in Paediatric Oncology from CIHR. This study was partly funded by the ERC Consolidator Award H3.3Cancer to PS. This work was performed within the context of the International CHildhood Astrocytoma INtegrated Genomic and Epigenomic (ICHANGE) consortium, and the Stand Up to Cancer (SU2C) Canada Cancer Stem Cell Dream Team Research Funding (SU2C-AACR-DT-19-15 to MDT, NJ), with funding from Genome Canada and Genome Quebec. MDT is supported by The Paediatric Brain Tumour Foundation, The Terry Fox Research Institute, The Canadian Institutes of Health Research, The Cure Search Foundation, b.r.a.i.n.child, Meagan’s Walk, Genome Canada, Genome BC, Genome Quebec, the Ontario Research Fund, Worldwide Cancer Research, V-Foundation for Cancer Research, Canadian Cancer Society Research Institute Impact grant and the Garron Family Chair in Childhood Cancer Research at the Hospital for Sick Children and the University of Toronto. The Children’s Cancer Centre Tissue Bank at RCH Melbourne runs thanks to the generous support of Cancer in Kids @ RCH, Leukaemia Auxiliary at RCH, the Murdoch Children’s Research Institute, and the RCH Foundation. CCLC is supported by fellowship from RI-MUHC sponsored by Toronto Dominion Bank and Alex’s Lemonade Stand Foundation. SJ is supported by fellowships from CIHR and FRQS. NDJ is a recipient of a fellowship from FRQS and RMGA. TG is a recipient of the Henry R. Shibata Fellowship. PS is supported by DZNE, the Wilhelm Sander Foundation, the Helmholtz Aging and Metabolic Programming (AMPro). JV was supported by the ÚNKP-20-4-II grant of the Ministry for Innovation and Technology, Hungary. SCM is supported by Cancer Prevention Research Institute of Texas (CPRIT) scholar in Cancer Research award (RR170023), Alex’s Lemonade Stand Foundation (ALSF) A award. M. Zapotocky was supported by the PRIMUS/19/MED/06 Grant Agency of Charles University in Prague. A. Klekner was supported by the 2017-1.2.1-NKP-2017-00002 National Brain Research Program NAP 2.0. We also acknowledge support from the We Love You Connie Foundation (PB, NJ), the Pediatric Brain Tumor Foundation (PB) and NIH 4R00CA201592-03 (PB), as well as the German Childhood Cancer Foundation (Molecular Neuropathology 2.0), the German Cancer Consortium, the German Federal Ministry of Education and Research and the Scheu family (INFORM).
Footnotes
DECLARATION OF INTERESTS
PB and RB receive grant funding from the Novartis Institute of Biomedical Research for an unrelated project. JRH has received compensation for consultation from Bayer for unrelated work.
REFERENCES
- Anderson AG, Kulkarni A, Harper M, and Konopka G (2020). Single-Cell Analysis of Foxp1-Driven Mechanisms Essential for Striatal Development. Cell reports 30, 3051–3066.e3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. (2009). Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England) 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Mendelson Cohen N, Szabo Q, Fritsch L, Papadopoulos GL, Lubling Y, Xu X, Lv X, Hugnot J-P, Tanay A, et al. (2017). Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell 171, 557–572.e524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgey M, Dali R, Eveleigh R, Chen KC, Letourneau L, Fillon J, Michaud M, Caron M, Sandoval J, Lefebvre F, et al. (2019). GenPipes: an open-source framework for distributed and scalable genomic analyses. GigaScience 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature biotechnology 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron CJ, Dostie J, and Blanchette M (2020). HIFI: estimating DNA-DNA interaction frequency from Hi-C data at restriction-fragment resolution. Genome biology 21, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman H, Riesenberg A, Ehrman LA, Kohli V, Nardini D, Nakafuku M, Campbell K, and Waclaw RR (2018). Gsx transcription factors control neuronal versus glial specification in ventricular zone progenitors of the mouse lateral ganglionic eminence. Dev Biol 442, 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman H, Waclaw RR, Pei Z, Nakafuku M, and Campbell K (2013). The homeobox gene Gsx2 controls the timing of oligodendroglial fate specification in mouse lateral ganglionic eminence progenitors. Development 140, 2289–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, and LoTurco J (2012). A method for stable transgenesis of radial glia lineage in rat neocortex by piggyBac mediated transposition. Journal of neuroscience methods 207, 172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforides A, Carpten JD, Weiss GJ, Demeure MJ, Von Hoff DD, and Craig DW (2013). Identification of somatic mutations in cancer through Bayesian-based analysis of sequenced genome pairs. BMC genomics 14, 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, and Getz G (2013). Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature biotechnology 31, 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dali R, Bourque G, and Blanchette M (2018). RobusTAD: A Tool for Robust Annotation of Topologically Associating Domain Boundaries. bioRxiv, 293175. [Google Scholar]
- Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, Hilton JA, Jain K, Baymuradov UK, Narayanan AK, et al. (2018). The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic acids research 46, D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, Del Angel G, Rivas MA, and Hanna M (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics 43, 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, and Aiden EL (2016). Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell systems 3, 99–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filbin MG, Tirosh I, Hovestadt V, Shaw ML, Escalante LE, Mathewson ND, Neftel C, Frank N, Pelton K, Hebert CM, et al. (2018). Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science (New York, NY) 360, 331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suva ML, and Bernstein BE (2016). Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontebasso AM, Gayden T, Nikbakht H, Neirinck M, Papillon-Cavanagh S, Majewski J, and Jabado N (2014a). Epigenetic dysregulation: a novel pathway of oncogenesis in pediatric brain tumors. Acta Neuropathol 128, 615–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontebasso AM, Papillon-Cavanagh S, Schwartzentruber J, Nikbakht H, Gerges N, Fiset PO, Bechet D, Faury D, De Jay N, Ramkissoon LA, et al. (2014b). Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nature genetics 46, 462–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontebasso AM, Schwartzentruber J, Khuong-Quang DA, Liu XY, Sturm D, Korshunov A, Jones DT, Witt H, Kool M, Albrecht S, et al. (2013). Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol 125, 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessi M, Gielen GH, Hammes J, Dorner E, Muhlen AZ, Waha A, and Pietsch T (2013). H3.3 G34R mutations in pediatric primitive neuroectodermal tumors of central nervous system (CNS-PNET) and pediatric glioblastomas: possible diagnostic and therapeutic implications? J Neurooncol 112, 67–72. [DOI] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Flandin P, Yoshikawa K, Rubenstein JL, Alvarez-Buylla A, and Kriegstein AR (2013). Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat Neurosci 16, 1576–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzelmann S, Castelo R, and Guinney J (2013). GSVA: gene set variation analysis for microarray and RNA-seq data. BMC bioinformatics 14, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harutyunyan AS, Krug B, Chen H, Papillon-Cavanagh S, Zeinieh M, De Jay N, Deshmukh S, Chen CCL, Belle J, Mikael LG, et al. (2019). H3K27M induces defective chromatin spread of PRC2-mediated repressive H3K27me2/me3 and is essential for glioma tumorigenesis. Nat Commun 10, 1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich MC, Corless CL, Demetri GD, Blanke CD, von Mehren M, Joensuu H, McGreevey LS, Chen CJ, Van den Abbeele AD, Druker BJ, et al. (2003). Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol 21, 4342–4349. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip CKM, Ng PKS, Jeong KJ, Shao SH, Ju Z, Leonard PG, Hua X, Vellano CP, Woessner R, Sahni N, et al. (2018). Neomorphic PDGFRA extracellular domain driver mutations are resistant to PDGFRA targeted therapies. Nat Commun 9, 4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessa S, Blanchet-Cohen A, Krug B, Vladoiu M, Coutelier M, Faury D, Poreau B, De Jay N, Hebert S, Monlong J, et al. (2019). Stalled developmental programs at the root of pediatric brain tumors. Nature genetics 51, 1702–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch KR, Sidoli S, and Garcia BA (2016). Identification and quantification of histone PTMs using high-resolution mass spectrometry. In Methods in enzymology (Elsevier), pp. 3–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuong-Quang DA, Buczkowicz P, Rakopoulos P, Liu XY, Fontebasso AM, Bouffet E, Bartels U, Albrecht S, Schwartzentruber J, Letourneau L, et al. (2012). K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol 124, 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Jeong K, Bhutani K, Lee J, Patel A, Scott E, Nam H, Lee H, Gleeson JG, and Bafna V (2013). Virmid: accurate detection of somatic mutations with sample impurity inference. Genome biology 14, R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloetgen A, Thandapani P, Ntziachristos P, Ghebrechristos Y, Nomikou S, Lazaris C, Chen X, Hu H, Bakogianni S, Wang J, et al. (2020). Three-dimensional chromatin landscapes in T cell acute lymphoblastic leukemia. Nature genetics 52, 388–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, and Wilson RK (2012). VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome research 22, 568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunov A, Capper D, Reuss D, Schrimpf D, Ryzhova M, Hovestadt V, Sturm D, Meyer J, Jones C, Zheludkova O, et al. (2016). Histologically distinct neuroepithelial tumors with histone 3 G34 mutation are molecularly similar and comprise a single nosologic entity. Acta Neuropathol 131, 137–146. [DOI] [PubMed] [Google Scholar]
- Koschmann C, Zamler D, MacKay A, Robinson D, Wu YM, Doherty R, Marini B, Tran D, Garton H, Muraszko K, et al. (2016). Characterizing and targeting PDGFRA alterations in pediatric high-grade glioma. Oncotarget 7, 65696–65706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F (2012). Trim Galore: a wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files, with some extra functionality for MspI-digested RRBS-type (Reduced Representation Bisufite-Seq) libraries. URL http://www bioinformatics babraham ac uk/projects/trim_galore/(Date of access: 28/04/2016). [Google Scholar]
- Krug B, De Jay N, Harutyunyan AS, Deshmukh S, Marchione DM, Guilhamon P, Bertrand KC, Mikael LG, McConechy MK, Chen CCL, et al. (2019). Pervasive H3K27 Acetylation Leads to ERV Expression and a Therapeutic Vulnerability in H3K27M Gliomas. Cancer cell 35, 782–797 e788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, and Jang W (2018). ClinVar: improving access to variant interpretations and supporting evidence. Nucleic acids research 46, D1062–D1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nature methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DE, Harris CC, Chen K, Koboldt DC, Abbott TE, Dooling DJ, Ley TJ, Mardis ER, Wilson RK, and Ding L (2012). SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics (Oxford, England) 28, 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson JD, Kasper LH, Paugh BS, Jin H, Wu G, Kwon CH, Fan Y, Shaw TI, Silveira AB, Qu C, et al. (2019). Histone H3.3 K27M Accelerates Spontaneous Brainstem Glioma and Drives Restricted Changes in Bivalent Gene Expression. Cancer cell 35, 140–155 e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, and Allis CD (2013). Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science (New York, NY) 340, 857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics (Oxford, England) 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R (2009). The sequence alignment/map format and SAMtools. Bioinformatics (Oxford, England) 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Mattar P, Dixit R, Lawn SO, Wilkinson G, Kinch C, Eisenstat D, Kurrasch DM, Chan JA, and Schuurmans C (2014). RAS/ERK signaling controls proneural genetic programs in cortical development and gliomagenesis. J Neurosci 34, 2169–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics (Oxford, England) 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lindtner S, Catta-Preta R, Tian H, Su-Feher L, Price JD, Dickel DE, Greiner V, Silberberg SN, McKinsey GL, McManus MT, et al. (2019). Genomic Resolution of DLX-Orchestrated Transcriptional Circuits Driving Development of Forebrain GABAergic Neurons. Cell reports 28, 2048–2063.e2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XY, Gerges N, Korshunov A, Sabha N, Khuong-Quang DA, Fontebasso AM, Fleming A, Hadjadj D, Schwartzentruber J, Majewski J, et al. (2012). Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol 124, 615–625. [DOI] [PubMed] [Google Scholar]
- Lopez-Juarez A, Howard J, Ullom K, Howard L, Grande A, Pardo A, Waclaw R, Sun YY, Yang D, Kuan CY, et al. (2013). Gsx2 controls region-specific activation of neural stem cells and injury-induced neurogenesis in the adult subventricular zone. Genes Dev 27, 1272–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F, and Pellegrini M (2020). ACTINN: automated identification of cell types in single cell RNA sequencing. Bioinformatics (Oxford, England) 36, 533–538. [DOI] [PubMed] [Google Scholar]
- Ma T, Wang C, Wang L, Zhou X, Tian M, Zhang Q, Zhang Y, Li J, Liu Z, Cai Y, et al. (2013). Subcortical origins of human and monkey neocortical interneurons. Nat Neurosci 16, 1588–1597. [DOI] [PubMed] [Google Scholar]
- Mackay A, Burford A, Carvalho D, Izquierdo E, Fazal-Salom J, Taylor KR, Bjerke L, Clarke M, Vinci M, Nandhabalan M, et al. (2017). Integrated Molecular Meta-Analysis of 1,000 Pediatric High-Grade and Diffuse Intrinsic Pontine Glioma. Cancer cell 32, 520–537 e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, and Martersteck EM (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovic-Mueller S, Stuttfeld E, Asthana M, Weinert T, Bliven S, Goldie KN, Kisko K, Capitani G, and Ballmer-Hofer K (2017). Structure of the Full-length VEGFR-1 Extracellular Domain in Complex with VEGF-A. Structure 25, 341–352. [DOI] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, and Hornischer K (2006). TRANSFAC® and its module TRANSCompel®: transcriptional gene regulation in eukaryotes. Nucleic acids research 34, D108–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L, Healy J, and Melville J (2018). Umap: Uniform manifold approximation and projection for dimension reduction. arXiv. [Google Scholar]
- Mizrak D, Levitin HM, Delgado AC, Crotet V, Yuan J, Chaker Z, Silva-Vargas V, Sims PA, and Doetsch F (2019). Single-Cell Analysis of Regional Differences in Adult V-SVZ Neural Stem Cell Lineages. Cell reports 26, 394–406.e395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy C, Maitra M, Tanti A, Suderman M, Théroux J-F, Mechawar N, Ragoussis J, and Turecki G (2019). Single-nucleus RNA sequencing shows convergent evidence from different cell types for altered synaptic plasticity in major depressive disorder. bioRxiv, 384479. [Google Scholar]
- Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, Richman AR, Silverbush D, Shaw ML, Hebert CM, et al. (2019). An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 178, 835–849.e821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngan CY, Wong CH, Tjong H, Wang W, Goldfeder RL, Choi C, He H, Gong L, Lin J, Urban B, et al. (2020). Chromatin interaction analyses elucidate the roles of PRC2-bound silencers in mouse development. Nature genetics 52, 264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitarska J, Smith JG, Sherlock WT, Hillege MM, Nott A, Barshop WD, Vashisht AA, Wohlschlegel JA, Mitter R, and Riccio A (2016). A Functional Switch of NuRD Chromatin Remodeling Complex Subunits Regulates Mouse Cortical Development. Cell reports 17, 1683–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, Haeussler M, Sandoval-Espinosa C, Liu SJ, Velmeshev D, et al. (2017). Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science (New York, NY) 358, 1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. (2008). An integrated genomic analysis of human glioblastoma multiforme. Science (New York, NY) 321, 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathania M, De Jay N, Maestro N, Harutyunyan AS, Nitarska J, Pahlavan P, Henderson S, Mikael LG, Richard-Londt A, Zhang Y, et al. (2017). H3.3(K27M) Cooperates with Trp53 Loss and PDGFRA Gain in Mouse Embryonic Neural Progenitor Cells to Induce Invasive High-Grade Gliomas. Cancer cell 32, 684–700.e689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paugh BS, Zhu X, Qu C, Endersby R, Diaz AK, Zhang J, Bax DA, Carvalho D, Reis RM, Onar-Thomas A, et al. (2013). Novel oncogenic PDGFRA mutations in pediatric high-grade gliomas. Cancer Res 73, 6219–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryniak MA, Potter GB, Rowitch DH, and Rubenstein JL (2007). Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron 55, 417–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, Shuga J, Liu SJ, Oldham MC, Diaz A, et al. (2015). Molecular identity of human outer radial glia during cortical development. Cell 163, 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma Y-A, and Trapnell C (2017a). Single-cell mRNA quantification and differential analysis with Census. Nature methods 14, 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C (2017b). Reversed graph embedding resolves complex single-cell trajectories. Nature methods 14, 979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics (Oxford, England) 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju CS, Spatazza J, Stanco A, Larimer P, Sorrells SF, Kelley KW, Nicholas CR, Paredes MF, Lui JH, Hasenstaub AR, et al. (2018). Secretagogin is Expressed by Developing Neocortical GABAergic Neurons in Humans but not Mice and Increases Neurite Arbor Size and Complexity. Cereb Cortex 28, 1946–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey SA, Knijnenburg TA, Kennedy KA, Zak DE, Gilchrist M, Gold ES, Johnson CD, Lampano AE, Litvak V, and Navarro G (2010). Genome-wide histone acetylation data improve prediction of mammalian transcription factor binding sites. Bioinformatics (Oxford, England) 26, 2071–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, and Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nature protocols 8, 2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, and Lander ES (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmer A, Phan H, Mathieson I, Iqbal Z, Twigg SRF, Wilkie AOM, McVean G, and Lunter G (2014). Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nature genetics 46, 912–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, and Cheetham RK (2012). Strelka: accurate somatic small-variant calling from sequenced tumor–normal sample pairs. Bioinformatics (Oxford, England) 28, 1811–1817. [DOI] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. (2011). Histone methylation by PRC2 is inhibited by active chromatin marks. Molecular cell 42, 330–341. [DOI] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, et al. (2012). Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231. [DOI] [PubMed] [Google Scholar]
- Sergushichev AA (2016). An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv, 060012. [Google Scholar]
- Shim AH-R, Liu H, Focia PJ, Chen X, Lin PC, and He X (2010). Structures of a platelet-derived growth factor/propeptide complex and a platelet-derived growth factor/receptor complex. Proceedings of the National Academy of Sciences 107, 11307–11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi Y, Sato Y, Chiba K, Okuno Y, Nagata Y, Yoshida K, Shiba N, Hayashi Y, Kume H, Homma Y, et al. (2013). An empirical Bayesian framework for somatic mutation detection from cancer genome sequencing data. Nucleic acids research 41, e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidoli S, Bhanu NV, Karch KR, Wang X, and Garcia BA (2016). Complete workflow for analysis of histone post-translational modifications using bottom-up mass spectrometry: from histone extraction to data analysis. JoVE, e54112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm D, Orr BA, Toprak UH, Hovestadt V, Jones DTW, Capper D, Sill M, Buchhalter I, Northcott PA, Leis I, et al. (2016). New Brain Tumor Entities Emerge from Molecular Classification of CNS-PNETs. Cell 164, 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, Pfaff E, Tonjes M, Sill M, Bender S, et al. (2012). Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer cell 22, 425–437. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Kumar SA, Shuai S, Diaz-Navarro A, Gutierrez-Fernandez A, De Antonellis P, Cavalli FM, Juraschka K, Farooq H, and Shibahara I (2019). Recurrent noncoding U1 snRNA mutations drive cryptic splicing in SHH medulloblastoma. Nature 574, 707–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, and Mesirov JP (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in bioinformatics 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tickle T, Tirosh I, Georgescu C, Brown M, and Haas B (2020). inferCNV of the Trinity CTAT Project (Klarman Cell Observatory, Broad Institute of MIT and Harvard, Cambridge, MA, USA.). [Google Scholar]
- Velmeshev D, Schirmer L, Jung D, Haeussler M, Perez Y, Mayer S, Bhaduri A, Goyal N, Rowitch DH, and Kriegstein AR (2019). Single-cell genomics identifies cell type-specific molecular changes in autism. Science (New York, NY) 364, 685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Tirosh I, Hebert C, Yizhak K, Neftel C, Filbin MG, Hovestadt V, Escalante LE, Shaw ML, Rodman C, et al. (2017). Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science (New York, NY) 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. (2010). Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer cell 17, 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, McKinsey GL, Pattabiraman K, Silberberg SN, Blow MJ, et al. (2013). A high-resolution enhancer atlas of the developing telencephalon. Cell 152, 895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladoiu MC, El-Hamamy I, Donovan LK, Farooq H, Holgado BL, Sundaravadanam Y, Ramaswamy V, Hendrikse LD, Kumar S, Mack SC, et al. (2019). Childhood cerebellar tumours mirror conserved fetal transcriptional programs. Nature 572, 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltman L, and Van Eck NJ (2013). A smart local moving algorithm for large-scale modularity-based community detection. The European physical journal B 86, 471. [Google Scholar]
- Wang K, Li M, and Hakonarson H (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research 38, e164–e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Li Y, Xi Y, Jiang S, Stratton S, Peng D, Tanaka K, Ren Y, Xia Z, Wu J, et al. (2014). ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature 508, 263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingett S, Ewels P, Furlan-Magaril M, Nagano T, Schoenfelder S, Fraser P, and Andrews S (2015). HiCUP: pipeline for mapping and processing Hi-C data. In F1000Res, pp. 1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, et al. (2012). Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nature genetics 44, 251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Y, Zhu X, Qu C, Chen X, Zhang J, et al. (2014). The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nature genetics 46, 444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younesy H, Möller T, Lorincz MC, Karimi MM, and Jones SJM (2015). VisRseq: R-based visual framework for analysis of sequencing data. BMC bioinformatics 16, S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, and Li W (2008). Model-based analysis of ChIP-Seq (MACS). Genome biology 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shan CM, Wang J, Bao K, Tong L, and Jia S (2017). Molecular basis for the role of oncogenic histone mutations in modulating H3K36 methylation. Sci Rep 7, 43906. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Recurrent somatic mutations in G34R/V HGGs, Related to Figure 1
A. Expression of PDGFRA in HGGs. PDGFRA is significantly upregulated in G34R/V tumours relative to others (adjusted p-value = 0.018, Negative Binomial Wald test). B. Oncoprint depicting recurrently mutated genes (observed in > 3 patients) in G34R/V HGG, grouped by associated gene functions as determined by Gene Ontology. Columns adjacent to gene names represent the number of patients in which mutations were called, in the entire cohort or stratified by PDGFRA mutation status (red = PDGFRAMUT, green = PDGFRAWT). Underlined genes indicate a bias towards PDGFRAWT G34 samples. C. Complex structure diagrams depicting extracellular PDGFRA mutations C235Y/F, Y288C, and K385M. Crystal structures of PDGFRB (PDB: 3MJG) were used to model C235 and Y288 mutations, and VGFR1 (PDB: 5T89) for K385. The C235Y/F mutations destabilize the Ig-like domain, leaving C291 available to form another inter- or intra-molecular disulfide bond, potentially leading to receptor dimerization and activation, analogous to Y288C. Finally, for K385M, in the wildtype protein, the positive charges of two K385 residues at the dimerization interface create an unfavourable repulsive effect. The K385M mutation may promote a small hydrophobic patch adjacent to several salt-bridges, enhancing receptor dimer formation and activation.
Figure S2. G34R/V uniquely exhibits a transcriptome and epigenome program specific to the forebrain interneuron lineage, Related to Figure 2 and Tables S2 and S3
A. Gene set enrichment analysis (GSEA) plots for the most enriched or depleted signature of each cell type. Enrichments are computed for G34R/V compared to IDH1 HGGs. NES: normalized enrichment score. B. Heatmap of expression levels of genes driving the enrichment of interneuron gene signature, or the depletion of the oligodendrocyte signature, in G34R/V tumours compared to other HGG subgroups. Expression is z-scored across samples. C. Heatmap of GSEA NES scores of forebrain astrocyte signatures in G34R/V tumours compared to the HGNET-BCOR tumours (non-gliomas). D. Heatmap of GSEA NES scores of 8-10 week old mouse ventricular/sub-ventricular zone (V-SVZ) cell type signatures in G34R/V compared to IDH1, K27M and WT HGG subgroups, as in (A). Scores are shown for all signatures significantly enriched (adjusted p-value < 0.01) in G34R/V vs. IDH1. aNSC: actively dividing neural stem cells, TAC: transit amplifying cells, NB: neuroblasts, OPC: oligodendrocyte progenitor cells, COP: committed oligodendrocyte precursors. E. Expression levels of the human radial glia marker MOXD1 and markers for cortical inhibitory and excitatory neurons in G34R/V, other HGG subgroups, and HGNET-BCOR. For genes upregulated in G34R/V, adjusted p-values (Negative Binomial Wald test) for comparison of G34R/V to other entities are indicated in parentheses. F. Gene ontology analysis of differentially expressed genes that showed significant H3K27ac or H3K27me3 enrichment in G34R/V tumours (z-score > 0.5). G. Core transcription factors (inferred by H3K27ac enrichment) activated in G34R/V compared to other HGGs, non-glioma brain tumours, and normal brain. Indicated TFs from cluster 2 lack activation signals specifically in G34R/V HGGs. Bolded genes indicate transcription factors annotated as necessary for oligodendrocyte differentiation (FDR = 0.0001) based on Gene ontology enrichment analysis.
Figure S3. G34R/V promotes aberrant H3K27me3 retention at terminal neuronal genes, Related to Figure 3 and Tables S2 and S3
A. Quantification of H3K27 and H3K36 methylation states by histone mass spectrometry on the mutant H3.3G34R or H3.3G34V histone normalized by H3.3 WT abundance in PS10-801 (left, n = 3), HSJD-GBM002 (middle, n = 1) or KNS-42 (right, n = 6) cell lines respectively. * denotes p-value < 0.05 (paired t-test). B. Volcano plot depicting differential H3K27me3 enrichment between G34R/V and non-G34R/V cortical HGGs at all annotated promoters. Blue data points indicate G34R/V-specific H3K27me3+ genes (z-score > 0.5, p-value < 0.05) involved in spontaneous synaptic transmission (FDR = 0.0273), calcium ion-regulated exocytosis of neurotransmitter (FDR = 0.000269), and regulation of GABAergic synaptic transmission (FDR = 0.00332). C. Volcano plot depicting differential H3K27ac enrichment between G34R/V and non-G34R/V cortical HGGs at all annotated promoters. Red data points indicate G34R/V-specific H3K27ac+ genes (z-score > 0.5, p-value < 0.05) involved in neuron fate commitment (FDR = 0.00243). D. Targeted deep sequencing validation of CRISPR-mediated editing of H3F3A G34R in HSJD-GBM002 clones. E. Volcano plot depicting transcriptomic change in CRISPR HSJD-GBM002 clones. Genes significantly up-regulated (log2FC > 1, p-adj < 0.05) in G34R unedited clones are highlighted in teal, whereas genes significantly up-regulated upon G34R removal are highlighted in navy (log2FC > 1, p-adj < 0.05). F. Volcano plot depicting H3K36me3 change at all annotated genic promoters in CRISPR HSJD-GBM002 clones (grey points), highlighting differentially expressed genes from RNA-seq (from panel E, teal = up-regulated in G34R, navy = up-regulated in edited). G. Volcano plot depicting H3K27me3 change at all annotated genic promoters in CRISPR HSJD-GBM002 clones (grey points), highlighting differentially expressed genes from RNA-seq (from panel E, teal = up-regulated in G34R, navy = up-regulated in edited).
Figure S4. Introduction of G34R in murine embryonic forebrain induces H3K27me3 epigenomic reprogramming, Related to Figure 3
A. Volcano plot depicting differential promoter H3K27me3 enrichment between G34R and non-G34R sorted cells 72-hours post-electroporation (left) or >1-year post-electroporation (right). Genic promoters with significantly increased H3K27me3 enrichment in G34R cells are indicated in red. B-C. Genome browser snapshot of ChIP-seq profiles in IUE-derived neural precursor cells showing H3K27me3 profiles associated with G34R-introduction. Some genic promoters, such as Foxp2 (B) and Jph4 (C) show striking H3K27me3 enrichment uniquely in G34R-electroporated cells. EV: empty vector. WT: wildtype H3.3. APP: Atrx shRNA, sgRNA for Trp53, PDGFRAWT. AP: Atrx shRNA, sgRNA for Trp53.
Figure S5. Active chromatin conformation promotes GSX2-PDGFRA co-expression in G34R/V HGGs, Related to Figure 4 and Table S4
A. Normalized expression of PDGFRA and GSX2 in bulk RNA-seq of G34R/V tumours (circles) and cell lines (triangles). Pearson correlation=0.69, R2=0.48, p-value=0.0002. B. PDGFRA, GSX2, and DLX1/2 expression in cell populations from scRNAseq atlas of the developing (5-37 PCW) human telencephalon. For each cell population (tree leaves), mean expression and proportion of cells expressing the gene are indicated. Dendrograms were constructed based on pairwise Spearman correlations of gene expression. MGE: medial ganglionic eminence, OPC: oligodendrocyte progenitor cells. *: GSX2 is not detected in this dataset due to low coverage. C. Top, H3K27ac profiles at the PDGFRA-GSX2-hs687 locus in individual G34R/V (n = 7), IDH1 (n = 5), SETD2 (n = 5), WT (n = 3), and K27M (n = 4) tumours. PDGFRAMUT samples are distinguished by darker teal. Note that PDGFRA is amplified in one K27M sample (first yellow track). Bottom, H3K27me3 profiles at the PDGFRA-GSX2 locus in individual G34R/V tumours (n=5), IDH1 (n=5), SETD2 (n=5), WT (n=3), and K27M (n=2) tumours. D. Hi-C heatmaps from a G34R, H3/IDH WT and K27M glioma cell lines depicting sub-TAD structure at the PDGFRA-GSX2-hs687 locus. Small black triangle illustrates a TAD between PDGFRA and hs687 enriched for contacts in G34R/V lines. Large dashed triangle demarcates the TAD formed by contact to a known distal insulator. Heatmap represents the log2 ratios of observed interactions relative to expected interactions across the PDGFRA-hs687 locus at a 5kb resolution (red = positive, blue = negative). E. Manhattan plot depicting H3K27ac enhancer activation difference between PDGFRAMUT and PDGFRAWT G34R/V HGGs (z-score > 0 indicate H3K27ac enriched in PDGFRAMUT) at far distal regions of PDGFRA-GSX2 locus. Black points depict comparison across different patients (n=5 PDGFRAMUT and n=2 PDGFRAWT), and teal points depict recurrence (PDGFRAMUT) vs. primary (PDGFRAWT) comparison in the same patient P-1190. F. Hi-C heatmaps from two G34R/V cell lines, HSJD-GBM002 (PDGFRAMUT) and KNS-42 (PDGFRAWT), depicting TAD structure at the PDGFRA-GSX2-hs687 locus. Black rectangles illustrate long-distance interactions between PDGFRA promoter and potential enhancers, enriched for contacts in HSJD-GBM002 vs. KNS-42. Heatmap represents the normalized observed interactions across the locus at a 5kb resolution (red = high interaction, white = low interaction). G. Hi-C heatmaps from murine E13.5 cortex and ganglionic eminences (GE) depicting sub-TAD structure at the Pdgfra-Gsx2-hs687 locus. The dashed square represents the interaction between GSX2 promoter and hs687. Heatmap represents the normalized observed interactions across the locus at a 10kb resolution (red = high interaction, white = low interaction).
Figure S6. G34 HGGs are devoid of the oligodendroglial lineage, and co-option of PDGFRAMUT promotes neuronal-to-astrocytic transition, Related to Figure 5 and Table S5
A. Bubble plot representing the composition of cell types in each tumour. Circle size represents the proportion of each cell type per tumour, and circle colour represents the total number of malignant cells within the tumour. B. UMAP embedding of IDH1, K27M, H3/IDH1-WT HGG (pediatric and adult) scRNA-seq datasets, colour coded by patient of origin (top) and cell type projection (bottom). Oligodendrocytes (in green) are clearly detected in all non G34R/V HGG entities. C. Mean expression of foetal interneuron and astrocyte gene signatures in individual cells from the normal human telencephalon. Cells of the interneuron lineage, as well as radial glial cells and astrocytes were included.
Figure S7. G34R/V mutations are dispensable for tumour maintenance, Related to Figure 6
A. Hematoxylin & eosin (H&E) and immunohistochemical analysis of representative KNS-42 parental and edited clone-derived PDOX tumours using a G34V-specific antibody. B. H&E and immunohistochemical analysis of representative HSJD-GBM002 parental and edited clone-derived PDOX tumours using a G34R-specific antibody.
Table S1. PDGFRA mutations in G34R/V, K27M, IDH1/SETD2, and WT HGG subgroups, Related to Figure 1
Table S2. Analysis of bulk RNA-seq profiles in G34R/V HGGs, Related to Figures 2, 3, S2 and S3
Table S3. Analysis of H3K27me3 and H3K27ac ChIP-seq profiles in G34R/V HGGs, Related to Figures 2, 3, S2 and S3
Table S4. Analysis of Hi-C profiles in G34R/V and other HGG cell lines, and E13.5 murine cortex and GE samples, Related to Figure 4, and S5
Table S6. G34R/V primary/recurrence mutation comparison for P-1190, P-3200, and P-1978, Related to Figure 6
Table S7. Extended information on in-house G34R/V HGG samples, G34R/V cell lines and murine IUE samples characterized by pathology, transcriptomics, and epigenomics, Related to STAR Methods
Data Availability Statement
ChIP-seq, RNA-seq, Hi-C sequencing data for cell lines and murine samples (Table S7) have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE146731. WES, bulk RNA-seq, ChIP-seq, and scRNA-seq sequencing data for human tumours (Table S7) have been deposited in the European Genome-phenome Archive (EGA) under accession number EGAS00001004301. Bulk RNA-seq sequencing data for non-G34R/V tumour entities have been previously deposited in EGA under dataset accession number EGAD00001005131.