Abstract
Traditional methods of cancer treatment are usually based on the morphological and histological diagnosis of tumors, and they are not optimized according to the specific situation. Precision medicine adjusts the existing treatment regimen based on the patient’s genomic information to make it most suitable for patients. Detection of genetic mutations in tumors is the basis of precise cancer medicine. Through the analysis of genetic mutations in patients with cancer, we can tailor the treatment plan for each patient with cancer to maximize the curative effect, minimize damage to healthy tissues, and optimize resources. In recent years, next-generation sequencing technology has developed rapidly and has become the core technology of precise targeted therapy and immunotherapy for cancer. From early cancer screening to treatment guidance for patients with advanced cancer, liquid biopsy is increasingly used in cancer management. This is as a result of the development of better noninvasive, repeatable, sensitive, and accurate tools used in early screening, diagnosis, evaluation, and monitoring of patients. Cell-free DNA, which is a new noninvasive molecular pathological detection method, often carries tumor-specific gene changes. It plays an important role in optimizing treatment and evaluating the efficacy of different treatment options in clinical trials, and it has broad clinical applications.
Keywords: cancer, NGS, cfDNA, targeted therapy, immunotherapy
Tumorigenesis is characterized by uncontrolled cell growth leading to cancer.1-3 Normal cells can become cancerous due to genetic mutations and epigenetic modifications.4-6 Patients with different cancer have different genetic mutations and epigenetic modifications, which increase the complexity and heterogeneity of tumors.7,8 In addition, intratumoral heterogeneity increases over the course of disease development, making the treatment of tumors particularly challenging.9-12
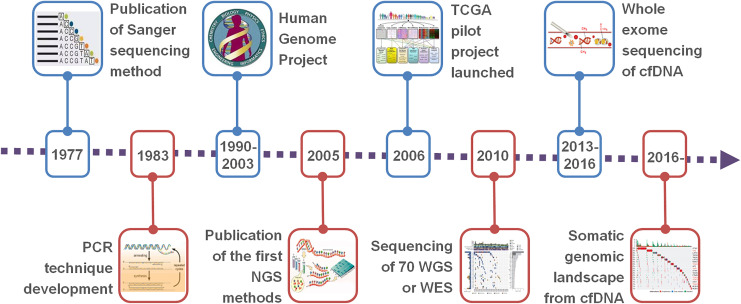
The development of next-generation sequencing (NGS) technology and bioinformatics has decoded a large number of cancer genome data, which has also promoted the development of targeted therapy and immunotherapy, especially for invasive cancer types that do not respond to traditional treatment options13-15 (Figure 1). Through the detection of cell-free DNA (cfDNA) genetic information in cancer cells, immune cells, or liquid biopsy samples by NGS, we can access key genetic mutations in patients with cancer. Assessment of these mutations can help inform whether patients will respond to targeted therapy or immunotherapy. Furthermore, new treatment options can be developed by creating new drugs to target these mutations.16-18 Immune drugs induce the body’s immune system to attack and treat tumors and develop more effective immunological checkpoint inhibitors or Chimeric Antigen Receptor T-Cell Immunotherapy(CAR-T) or cancer vaccines.19-21
Figure 1.
Time line of major achievements in sequencing technologies.
Precision Medicine and Tumor Gene Detection
Precision medicine is a new term, replacing individualized medicine. There are many overlaps between precision medicine and individualized medicine.22-24 However, precision medicine mainly refers to adjusting the existing treatment plan according to the patient’s genomic information, making it most suitable for patients, while individualized medicine is often based on creating new treatment methods or plans based on the patient’s genomic information.25-27 This process is lengthy and may not benefit patients with cancer. For this reason, the National Research Council, which is responsible for the management of science, technology, engineering, and mathematics in the United States, recommended replacing “individualized medicine” with “precision medicine” in 2011.28 In early 2015, President Barack Obama launched the Precision Medicine program. At the same time, at the end of 2015, China also launched the “Precision Medicine” program and included it in the 13th Five-Year Plan of China.29
As it pertains to precision medicine for cancer treatment, detection of genetic aberrations is the basis of developing effective treatment options.30-32 Through the analysis of genetic mutations in patients with cancer, we can tailor the treatment plan for each patient with cancer to maximize the curative effect, minimize the damage, and optimize the resources.33-36
Next-Generation Sequencing and Precise Targeting Therapy for Tumors
Next-generation sequencing, also known as large-scale parallel sequencing, can simultaneously sequence millions or even billions of DNA molecules, achieving the goal of large-scale, high-throughput sequencing.37-40 This makes NGS a revolutionary progress following Sanger sequencing (first-generation sequencing).8,41,42 In recent years, NGS technology has been developed and applied rapidly, especially in cancer gene detection. In the United States, medical NGS services (FMI and MSK) from commercial companies and academic institutions were approved by Food and Drug Administration (FDA) in 2017, suggesting that NGS has been formally applied in clinical practice.43 At the same time, the approval process for various medical NGS services is also accelerating. In 2017, the National Cancer Institute launched a National Cancer Precision Medical Survey, which found that 75.6% of oncologists in the United States are using NGS gene detection technology to guide cancer treatment.44
From 2018 to 2019, 5 cancer NGS products were approved by National Medical Products Administration, suggesting that NGS has officially entered the stage of clinical application in China.45-47 Currently, the FDA has approved concomitant diagnostic technologies in oncology, such as immunohistochemistry (IHC), in situ hybridization (fluorescence in situ hybridization/chromogenic in situ hybridization), real-time fluorescence quantitative polymerase chain reaction, and NGS.48 Excluding IHC, the other 3 are gene detection techniques; among them, NGS has become the core technology of precise targeted therapy and immunotherapy for cancer.49-52
The NGS mutation profiles obtained from different tumors have led to the emergence of targeted therapies for tumors, which include identifying mutations in signaling pathways and blocking them with existing or newly developed drugs.53-56 The detected mutations are classified as driver mutations if they are critical to the maintenance of tumors and passenger mutations if they have no definite role in the maintenance of tumors.57-60 This classification therapy led to the development of imatinib, a constitutive inhibitor of BCR-ABL kinase, for the treatment of leukemia.61-63
At present, targeted therapeutic drugs include small molecule inhibitors and macromolecular monoclonal antibody drugs. For drugs with definite targets, it is necessary to use gene detection before they can be used (Figure 2).
Figure 2.
Epigenetic drugs for cancer therapy. Epigenetic drugs being studied for human cancer are listed.
Next-Generation Sequencing and Immunotherapy for Tumors
In 2018, a study in Nature Medicine showed that when a patient with metastatic breast cancer did not respond to several kinds of chemotherapy and had a life expectancy of only a few months, somatic cell mutations were detected by NGS technology, and immunotherapy was administered to completely eliminate the tumor.64 Thus, the genomic information of tumors detected by NGS can identify patients who may respond to immunotherapy, use immunodrugs to induce the body’s immune system to attack and treat tumors, or develop more effective immune checkpoint inhibitors or CAR-T or cancer vaccines.65-67
Under normal physiological conditions, the immune system recognizes and eliminates mutant cells.68 However, tumors occur when cancer cells escape the immune system by creating an immunosuppressive environment.69-72 Therefore, the focus of recent research has shifted from targeted therapy to immunotherapy, hoping to be used to treat more patients with cancer. This is as a result of immune escape being common for all tumors, and restoring the immune system can help destroy tumors.
Immunotherapy is not equally effective for all types of tumors, and the efficacy varies from patient to patient.73-77 The possible reasons are the heterogeneity of T cells and tumor cells and their complex interactions in the tumor microenvironment.78-81 Immunogenomics is a relatively new field of cancer research. The detection and analysis of whole-genome sequencing (WGS), whole-exome sequencing (WES), and RNA sequencing (RNA-Seq) on T cells and tumor cells by NGS technology can obtain genome maps of tumors and immune cells, which can help to customize treatment schemes for specific characteristics of tumors and increase the possibility of success.82-84 At the same time, NGS technology can be used to evaluate the changes in biomarkers of immunological checkpoint inhibitors, such as tumor mutational burden (TMB), microsatellite instability, and PD-L1 amplification and other therapeutic effects, drug resistance, and genetic mutations related to hyperprogression.64,85-87
In cancer vaccines, the immune system is stimulated to produce antibodies.88 In adoptive T-cell therapy, T cells are isolated from the body, stimulated and amplified in vitro, and then infused back into the patient.89-91 Genetic modification of T cells (CAR-T cells) by chimeric antigen receptors can improve the immune response of T cells.92 Detection and analysis of WGS, WES, and RNA-seq in T cells and tumor cells by NGS technology will help to improve the design of CAR-T cells and the selection of new antigens. Tumor cells secrete and express new antigens on the surface of cells to escape recognition of T cells.93-96 Patient-derived T cells can grow in vitro and can be stimulated with these new antigens to elicit a strong T-cell response.97 To further enhance the ability of T cells to recognize tumors, CAR-encoded DNA was introduced into T cells (CAR-T cell therapy).98 Therefore, once T cells increase, they will be transferred back to the patient, where they can now recognize tumor antigens, thereby improving the effectiveness of inducing cancer cell death and clearance. Detection and analysis of WGS, WES, and RNA-seq in T cells and tumor cells by NGS technology will help to improve the design of CAR-T cells and the selection of new antigens.
The progress of NGS technology and bioinformatics is expected to improve the recognition of new antigens and the effectiveness of cancer vaccines.99-102 Single-cell genomics will be particularly helpful in revealing the expression, mutation of tumor genes, and the heterogeneity of new immune cells in the same tumor, which can be used to develop cancer vaccines targeting different clonal populations in tumors.103-106 Therefore, NGS technology (WGS, WES, RNA-seq, ChIP-seq, NGS panel, etc) has become the core development and application technology of precise targeted therapy and immunotherapy for cancer. It can help us better understand tumors, tumor microenvironment, and T cells and then provide personalized treatment programs for patients with cancer.
Next-Generation Sequencing and cfDNA Detection
“Liquid biopsy” is often used to analyze cfDNA in plasma and other body fluids (such as pleural effusion, ascites, and cerebrospinal fluid), as well as circulating tumor cells (CTCs) and other nucleic acids (such as RNA and microRNA) in blood.107-109 Circulating tumor cells in blood exist at very low concentrations, usually less than 10 CTCs per milliliter of blood, even in patients with metastatic disease.110 This low concentration characteristic greatly limits the diagnostic and analytical potential of CTCs. Compared with CTC, the proportion of cfDNA contributed by cancer cells was significantly higher.111 In advanced patients with hepatocellular carcinoma (HCC), DNA fragments carrying cancer-specific mutations account for more than 50% of cfDNA.105 It is also because the proportion of cancer-derived DNA in cfDNA is higher than that of CTCs in nucleated blood cells, and therefore, the analysis of cfDNA is more widely used in cancer management than that of CTCs.
From early cancer screening to treatment guidance for patients with advanced cancer, liquid biopsy is increasingly used in cancer management. Liquid biopsy may overcome the limitations of tumor markers (mainly proteins or glycoproteins) in conventional tissue samples.112-115 Cancer-related mutations, including single-nucleotide mutations, copy number changes, methylation changes, and DNA fragmentation patterns have been detected in cfDNA of patients with various cancer by NGS technology (Figure 3).74,116-118
Figure 3.
Time line of the main important discoveries of circulating tumor DNA.
Most cancers are relatively asymptomatic in their early stages.119 As such, most patients are diagnosed with advanced cancers. In this regard, liquid biopsy can be used for cancer surveillance and diagnosis. Detection of genetic mutations by enlarging the target region can improve sensitivity.120 Mutations from tumors can be used to monitor clinical progress and detect residual lesions after treatment.116,121-123 Abnormal methylation signals in cfDNA molecules enable detection of ultra-early tumors, and genome-wide methylation histological analysis of plasma DNA tissue location can be used as a “whole-body molecular imaging” method to identify potential tissue origins of mutations detected in cfDNA.124-127
An important application of cfDNA analysis is to guide treatment decisions, especially in targeted therapy.128-130 For example, liquid biopsy to analyze EGFR mutations has been widely used to guide the use of epidermal growth factor receptor-tyrosine kinase inhibitors.131 Many studies have demonstrated that targeted large-scale parallel sequencing can be used to identify cancer-related driver mutations in the cfDNA of patients with HCC.132-134 Further studies have shown that the mutation characteristics of cfDNA reflect the state of the corresponding tumor tissues.114,135,136 Of course, the potential of cfDNA mutation analysis for cancer treatment management should not be underestimated.137 An NGS-based liquid biopsy provides a noninvasive method for large-scale assessment of mutation profiles in patients with advanced cancer (Figure 4).
Figure 4.
Multitude of factors secreted in the circulatory system with the sustained growth of tumor. A liquid biopsy contains wealth of information relevant to determining tumor status, metastatic potential, and likelihood of relapse. Some of the contributing factors to making such an assessment include circulating tumor cell (CTC) counts, CTC genetic profile and protein expression, levels of circulating tumor DNA/RNA, and the presence or absence of known mutations or epigenetic signatures. A thorough analysis of liquid biopsies from patients with cancer screening for these factors can reveal essential information for personalized care.
Recent studies have shown that TMB can predict the clinical response of patients to immunotherapy in a variety of solid tumors.138-140 Tumor mutational burden detection based on NGS and cfDNA can be used to predict the therapeutic response of patients with non-small-cell lung cancer to the PD-L1 immunosuppressant atezolizumab, and its immunotherapeutic effect is independent of the expression of PD-L1.141-143
Conclusion
Cancer is a relatively heterogeneous disease with multiple causes and carcinogenic driving events. The analysis of cfDNA mutations based on NGS can better characterize patients, and it can be applied to early cancer screening and treatment guidance for patients with advanced cancer. It can optimize patient’s treatment and evaluate different treatment options in drug trials.
Acknowledgments
The authors would like to thank Prof. Pei Yu for data analysis and critical discussion of the manuscript. The authors thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Authors’ Note: T.-M.W., J.-B.L., W.L., and G.-R.W. contributed equally to this work. Y.S.M. and D.F. designed the study. All authors performed the statistical analyses and interpreted the data. D.F. wrote the manuscript. All authors contributed to the final version of the manuscript and approved the final manuscript.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported partly by grants from the National Natural Science Foundation of China (81972214, 81772932, 81472202, 81201535, 81302065, 81671716, 81301993, 81372175 and 81472209), The Fundamental Research Funds for the Central Universities (22120170212 and 22120170117), The Scientific Research Fund Project of Anhui Medical University (2018xkj058), Shanghai Natural Science Foundation (12ZR1436000), Shanghai Municipal Commission of Health and Family Planning (201540228), Special Funding Fund for Clinical Research of Wu Jieping Medical Foundation (320.6750.14326), Nantong Science and Technology Project (YYZ15026), The Peak of Six personnel Foundation in Jiangsu Province (WSW-009), The Fifth Phase of 333 Talents Engineering Science and Technology Project of Jiangsu Province (2017205), and Jiangsu Province Science Foundation for Youths (BK2012101).
ORCID iD: Da Fu  https://orcid.org/0000-0002-0878-2575
https://orcid.org/0000-0002-0878-2575
References
- 1.Laissue P. The forkhead-box family of transcription factors: key molecular players in colorectal cancer pathogenesis. Mol Cancer. 2019;18(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cristóbal I, Sanz-Alvarez M, Torrejón B, et al. Potential therapeutic impact of miR-145 deregulation in colorectal cancer. Mol Ther. 2018;26(6):1399–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su R, Cao S, Ma J, et al. Knockdown of SOX2OT inhibits the malignant biological behaviors of glioblastoma stem cells via up-regulating the expression of miR-194-5p and miR-12. Mol Cancer. 2018;16(1):171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aupy P, Echevarría L, Relizani K, et al. Identifying and avoiding tcDNA-ASO sequence-specific toxicity for the development of DMD Exon 51 skipping therapy. Mol Ther Nucleic Acids. 2019;19:371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santos JM, Cervera-Carrascon V, Havunen R, et al. Adenovirus coding for interleukin-2 and tumor necrosis Factor alpha replaces lymphodepleting chemotherapy in adoptive T Cell therapy. Mol Ther. 2018;26(9):2243–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang KW, Reebye V, Czysz K, et al. Liver activation of hepatocellular nuclear factor-4α by small activating RNA rescues dyslipidemia and improves metabolic profile. Mol Ther Nucleic Acids. 2019;19:361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu Y, Wang C, Becker SA, et al. miR-145 antagonizes snai1-mediated stemness and radiation resistance in colorectal cancer. Mol Ther. 2018;26(3):744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai X, Ni J, Beretov J, Graham P, Li Y. Cancer stem cell in breast cancer therapeutic resistance. Cancer Treat Rev. 2018;69:152–163. [DOI] [PubMed] [Google Scholar]
- 9.Abenza EG, Molero SI, Moreno DG, et al. Zebrafish modeling reveals that SPINT1 regulates the aggressiveness of skin cutaneous melanoma and its crosstalk with tumor immune microenvironment. J Exp Clin Cancer Res. 2019;38(1):405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Xu D, Yao J, et al. Inhibition of miR-155-5p exerts anti-fibrotic effects in silicotic mice by regulating meprin α. Mol Ther Nucleic Acids. 2019;19:350–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russell SJ. For the success of oncolytic viruses: single cycle cures or repeat treatments? (One cycle should be enough). Mol Ther. 2018;26(8):1876–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gokita K, Inoue J, Ishihara H, Kojima K, Inazawa J. Therapeutic potential of LNP-mediated delivery of mir-634 for cancer therapy. Mol Ther Nucleic Acids. 2019;19:330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li XN, Wang ZJ, Ye CX, et al. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res. 2018;37(1):325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen XJ, Zhang ZC, Wang XY, et al. The circular RNome of developmental retina in mice. Mol Ther Nucleic Acids. 2019;19:339–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kowalsky SJ, Liu Z, Feist M, et al. Superagonist IL-15-armed oncolytic virus elicits potent antitumor immunity and therapy that are enhanced with pd-1 blockade. Mol Ther. 2018;26(10):2476–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu H, Wang C, Song H, Xu Y, Ji G. RNA-Seq profiling of circular RNAs in human colorectal cancer liver metastasis and the potential biomarkers. Mol Cancer. 2019;18(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toiyama Y, Okugawa Y, Fleshman J, Richard Boland C, Goel A. MicroRNAs as potential liquid biopsy biomarkers in colorectal cancer: a systematic review. Biochim Biophys Acta Rev Cancer. 2018;1870(2):274–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng HC, Lin JY, Hsu SH, et al. Low folate metabolic stress reprograms DNA methylation-activated sonic hedgehog signaling to mediate cancer stem cell-like signatures and invasive tumour stage-specific malignancy of human colorectal cancers. Int J Cancer. 2017;141(12):2537–2550. [DOI] [PubMed] [Google Scholar]
- 19.Fletcher R, Wang YJ, Schoen RE, et al. Colorectal cancer prevention: immune modulation taking the stage. Biochim Biophys Acta Rev Cancer. 2018;1869(2):138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rezvani K, Rouce R, Liu E, Shpall E. Engineering natural killer cells for cancer immunotherapy. Mol Ther. 2017;25(8):1769–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng S, Deng Q, Zhang Y, et al. Non-platelet-derived CXCL4 differentially regulates cytotoxic and regulatory T cells through CXCR3 to suppress the immune response to colon cancer. Cancer Lett. 2019;443:1–12. [DOI] [PubMed] [Google Scholar]
- 22.Ibarrola-Villava M, Cervantes A, Bardelli A. Preclinical models for precision oncology. Biochim Biophys Acta Rev Cancer. 2018;1870(2):239–246. [DOI] [PubMed] [Google Scholar]
- 23.Arcis MS, Bargiela A, Furling D, Artero R. miR-7 restores phenotypes in myotonic dystrophy muscle cells by repressing hyperactivated autophagy. Mol Ther Nucleic Acids. 2019;19:278–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naumenko V, Van S, Dastidar H, et al. Visualizing oncolytic virus-host interactions in live mice using intravital microscopy. Mol Ther Oncolytics. 2018;10:14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhartiya D, Patel H, Ganguly R, et al. Novel insights into adult and cancer stem cell biology. Stem Cells Dev. 2018;27(22):1527–1539. [DOI] [PubMed] [Google Scholar]
- 26.Hou Z, Guo K, Sun X, et al. TRIB2 functions as novel oncogene in colorectal cancer by blocking cellular senescence through AP4/p21 signaling. Mol Cancer. 2018;17(1):172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma YS, Huang T, Zhong XM, et al. Proteogenomic characterization and comprehensive integrative genomic analysis of human colorectal cancer liver metastasis. Mol Cancer. 2018;17(1):139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carrasco-Ramiro F, Pastor RP, Aguado B. Human genomics projects and precision medicine. Gene Ther. 2017;24(9):551–561. [DOI] [PubMed] [Google Scholar]
- 29.Chen W, Zheng R, Zhang S, et al. Cancer incidence and mortality in China, 2013. Cancer Lett. 2017;401:63–71. [DOI] [PubMed] [Google Scholar]
- 30.Segal M, Biscans A, Gilles ME, et al. Hydrophobically modified let-7b miRNA enhances biodistribution to NSCLC and downregulates HMGA2 In vivo. Mol Ther Nucleic Acids. 2019;19:267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng F, Chen D, Swartz MC, Sun H. A pilot study of a culturally tailored lifestyle intervention for Chinese American cancer survivors. Cancer Control. 2019;26(1):1073274819895489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beug ST, Pichette SJ, St-Jean M, et al. Combination of IAP antagonists and TNF-α-armed oncolytic viruses induce tumor vascular shutdown and tumor regression. Mol Ther Oncolytics. 2018;10:28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smekalova EM, Gerashchenko MV, O’Connor PBF, et al. In vivo RNAi-mediated eif3 m knockdown affects ribosome biogenesis and transcription but has limited impact on mRNA-specific translation. Mol Ther Nucleic Acids. 2019;19:252–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosales Gerpe MC, van Vloten JP, Santry LA, et al. Use of precision-cut lung slices as an ex vivo tool for evaluating viruses and viral vectors for gene and oncolytic therapy. Mol Ther Methods Clin Dev. 2018;10:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim MK, Shin SJ, Lee HM, et al. Mycoplasma infection promotes tumor progression via interaction of the mycoplasmal protein p37 and epithelial cell adhesion molecule in hepatocellular carcinoma. Cancer Lett. 2019;454:44–52. [DOI] [PubMed] [Google Scholar]
- 36.Wambalaba FW, Son B, Wambalaba AE, Nyong’o D, Nyong’o A. prevalence and capacity of cancer diagnostics and treatment: a demand and supply survey of health-care facilities in Kenya. Cancer Control. 2019;26(1):10732.74819886930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee WR, Lin YK, Alalaiwe A, et al. Fractional laser-mediated siRNA delivery for mitigating psoriasis-like lesions via IL-6 silencing. Mol Ther Nucleic Acids. 2019;19:240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim YJ, Baek DS, Lee S, et al. Dual-targeting of EGFR and neuropilin-1 attenuates resistance to EGFR-targeted antibody therapy in KRAS-mutant non-small cell lung cancer. Cancer Lett. 2019;466:23–34. [DOI] [PubMed] [Google Scholar]
- 39.Huff AL, Wongthida P, Kottke T, et al. APOBEC3 mediates resistance to oncolytic viral therapy. Mol Ther Oncolytics. 2018;11:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pedersen L, Hagedorn P, Vikeså J, et al. Targeting repeated regions unique to a gene is an effective strategy for discovering potent and efficacious antisense oligonucleotides. Mol Ther Nucleic Acids. 2019;19:124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nimmakayala RK, Batra SK, Ponnusamy MP. Unraveling the journey of cancer stem cells from origin to metastasis. Biochim Biophys Acta Rev Cancer. 2019;1871(1):50–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu F, Liu JB, Wu ZJ, et al. Tumor suppressive microRNA-124a inhibits stemness and enhances gefitinib sensitivity of non-small cell lung cancer cells by targeting ubiquitin-specific protease 1. Cancer Lett. 2018;427:74–84. [DOI] [PubMed] [Google Scholar]
- 43.Lapin V, Mighion LC, da Silva CP, et al. Regulating whole exome sequencing as a diagnostic test. Hum Genet. 2016;135(6):655–673. [DOI] [PubMed] [Google Scholar]
- 44.Vetsch J, Wakefield CE, Techakesari P, et al. Healthcare professionals’ attitudes toward cancer precision medicine: a systematic review. Semin Oncol. 2019;46(3):291–303. [DOI] [PubMed] [Google Scholar]
- 45.Pan Y, Zhang Y, Ye T, et al. Detection of novel NRG1, EGFR and MET fusions in lung adenocarcinomas in the Chinese population. J Thorac Oncol. 2019;14(11):2003–2008. [DOI] [PubMed] [Google Scholar]
- 46.Hoshikawa N, Sakai A, Takai S, Suzuki H. Targeting extracellular miR-21-TLR7 signaling provides long-lasting analgesia in osteoarthritis. Mol Ther Nucleic Acids. 2019;19:199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moreno R, Fajardo CA, Farrera-Sal M, et al. Enhanced antitumor efficacy of oncolytic adenovirus-loaded menstrual blood-derived mesenchymal stem cells in combination with peripheral blood mononuclear cells. Mol Cancer Ther. 2019;18(1):127–138. [DOI] [PubMed] [Google Scholar]
- 48.Dellis A, Zagouri F, Liontos M, et al. Management of advanced prostate cancer: a systematic review of existing guidelines and recommendations. Cancer Treat Rev. 2019;73:54–61. [DOI] [PubMed] [Google Scholar]
- 49.Zamay TN, Zamay GS, Shnayder NA, et al. Nucleic acid aptamers for molecular therapy of epilepsy and blood-brain barrier damages. Mol Ther Nucleic Acids. 2019;19:157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giulietti MV, Vespa A, Ottaviani M, et al. Personality (at intrapsychic and interpersonal level) associated with quality of life in patients with cancer (lung and colon). Cancer Control. 2019;26(1):1073274819880560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rovira-Rigau M, Raimondi G, Marín MÁ, et al. Bioselection reveals miR-99b and miR-485 as enhancers of adenoviral oncolysis In pancreatic cancer. Mol Ther. 2019;27(1):230–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karali M, Guadagnino I, Marrocco E, et al. AAV-miR-204 protects from retinal degeneration by attenuation of microglia activation and photoreceptor cell death. Mol Ther Nucleic Acids. 2019;19:144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oshima G, Guo N, He C, et al. In vivo delivery and therapeutic effects of a microRNA on colorectal liver metastases. Mol Ther. 2018;25(7):1588–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kennaway DJ. Melatonin-deficient balb/c mice and their use in cancer research. Cancer Control. 2019;26(1):1073274819886825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cuomo O, Cepparulo P, Anzilotti S, et al. Anti-miR-223-5p ameliorates ischemic damage and improves neurological function by preventing NCKX2 downregulation after ischemia in rats. Mol Ther Nucleic Acids. 2019;18:1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phelps MP, Yang H, Patel S, et al. Oncolytic virus-mediated RAS targeting in rhabdomyosarcoma. Mol Ther Oncolytics. 2018;11:52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ryu J, Kwon DH, Choe N, et al. Characterization of circular RNAs in vascular smooth muscle cells with vascular calcification. Mol Ther Nucleic Acids. 2019;19:31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fogliatto FS, Anzanello MJ, Soares F, Brust-Renck PG. Decision support for breast cancer detection: classification improvement through feature selection. Cancer Control. 2019;26(1):1073274819876598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Denton NL, Chen CY, Hutzen B, et al. Myelolytic treatments enhance oncolytic herpes virotherapy in models of Ewing sarcoma by modulating the immune microenvironment. Mol Ther Oncolytics. 2018;11:62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Catuogno S, Di Martino MT, Nuzzo S, et al. An anti-BCMA RNA aptamer for miRNA intracellular delivery. Mol Ther Nucleic Acids. 2019;18:981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Monteleone F, Taverna S, Alessandro R, Fontana S. SWATH-MS based quantitative proteomics analysis reveals that curcumin alters the metabolic enzyme profile of CML cells by affecting the activity of miR-22/IPO7/HIF-1α axis. J Exp Clin Cancer Res. 2018;37(1):170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Havunen R, Santos JM, Sorsa S, et al. Abscopal effect in non-injected tumors achieved with cytokine-armed oncolytic adenovirus. Mol Ther Oncolytics. 2018;11:109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Delviks-Frankenberry KA, Ackerman D, Timberlake ND, et al. Development of lentiviral vectors for HIV-1 gene therapy with Vif-resistant APOBEC3G. Mol Ther Nucleic Acids. 2019;18:1023–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wei F, Zhang T, Deng SC, et al. PD-L1 promotes colorectal cancer stem cell expansion by activating HMGA1-dependent signaling pathways. Cancer Lett. 2019;450:1–13. [DOI] [PubMed] [Google Scholar]
- 65.Van Minh H, Van Thuan T, Shu XO. Scientific evidence for cancer control in Vietnam. Cancer Control. 2019;26(1):1073274819866450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mulholland EJ, Green WP, Buckley NE, McCarthy HO. Exploring the potential of microRNA Let-7c as a therapeutic for prostate cancer. Mol Ther Nucleic Acids. 2019;18:927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kemler I, Ennis MK, Neuhauser CM, Dingli D. In vivo imaging of oncolytic measles virus propagation with single-cell resolution. Mol Ther Oncolytics. 2018;12:68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Choi HR, Song IA, Oh TK. Association of opioid use in the week before death among patients with advanced lung cancer having sepsis. Cancer Control. 2019;26(1):1073274819871326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cultrara CN, Shah S, Antuono G, et al. Size matters: arginine-derived peptides targeting the PSMA receptor can efficiently complex but not transfect siRNA. Mol Ther Nucleic Acids. 2019;18:863–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tam BY, Chiu K, Chung H, et al. The CLK inhibitor SM08502 induces anti-tumor activity and reduces Wnt pathway gene expression in gastrointestinal cancer models. Cancer Lett. 2019;S0304–3835(19):30473–30482. [DOI] [PubMed] [Google Scholar]
- 71.Mooney R, Majid AA, Covello JB, et al. Enhanced delivery of oncolytic adenovirus by neural stem cells for treatment of metastatic ovarian cancer. Mol Ther Oncolytics. 2018;12:79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Del Corpo O, Goguen RP, Malard CMG, et al. A U1i RNA that enhances HIV-1 RNA splicing with an elongated recognition domain is an optimal candidate for combination HIV-1 gene therapy. Mol Ther Nucleic Acids. 2019;18:815–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Durham NM, Mulgrew K, McGlinchey K, et al. Oncolytic VSV primes differential responses to immuno-oncology therapy. Mol Ther. 2017;25(8):1917–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rajaraman S, Canjuga D, Ghosh M, et al. Measles virus-based treatments trigger a pro-inflammatory cascade and a distinctive immunopeptidome in glioblastoma. Mol Ther Oncolytics. 2018;12:147–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fadaka AO, Pretorius A, Klein A. Biomarkers for stratification in colorectal cancer: microRNAs. Cancer Control. 2019;26(1):1073274819862784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taschauer A, Polzer W, Alioglu F, et al. Peptide-targeted polyplexes for aerosol-mediated gene delivery to CD49f-overexpressing tumor lesions in lung. Mol Ther Nucleic Acids. 2019;18:774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hotani T, Mizuguchi H, Sakurai F. Systemically administered reovirus-induced ownregulation of hypoxia inducible factor-1α in subcutaneous tumors. Mol Ther Oncolytics. 2018;12:162–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gasparello J, Lomazzi M, Papi C, et al. Efficient delivery of MicroRNA and antimiRNA molecules using an argininocalix[4]arene macrocycle. Mol Ther Nucleic Acids. 2019;18:748–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reda M, Ngamcherdtrakul W, Gu S, et al. PLK1 and EGFR targeted nanoparticle as a radiation sensitizer for non-small cell lung cancer. Cancer Lett. 2019;467:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pearl TM, Markert JM, Cassady KA, Ghonime MG. Oncolytic virus-based cytokine expression to improve immune activity in brain and solid tumors. Mol Ther Oncolytics. 2019;13:14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lindel F, Dodt CR, Weidner N, et al. TraFo-CRISPR: enhanced genome engineering by transient foamy virus vector-mediated delivery of CRISPR/Cas9 components. Mol Ther Nucleic Acids. 2019;18:708–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mishchenko T, Mitroshina E, Balalaeva I, et al. An emerging role for nanomaterials in increasing immunogenicity of cancer cell death. Biochim Biophys Acta Rev Cancer. 2019;1871(1):99–108. [DOI] [PubMed] [Google Scholar]
- 83.Belhadj S, Moutinho C, Mur P, et al. Germline variation in O6-methylguanine-DNA methyltransferase (MGMT) as cause of hereditary colorectal cancer. Cancer Lett. 2019;447:86–92. [DOI] [PubMed] [Google Scholar]
- 84.Tzchori I, Falah M, Shteynberg D, et al. Improved patency of ePTFE grafts as a hemodialysis access site by seeding autologous endothelial cells expressing fibulin-5 and VEGF. Mol Ther. 2018;26(7):1660–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yue B, Liu C, Sun H, et al. A positive feed-forward loop between LncRNA-CYTOR and Wnt/β-catenin signaling promotes metastasis of colon cancer. Mol Ther. 2018;26(5):1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jiang X, Wang J, Deng X, et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer. 2019;18(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lewinsky H, Barak AF, Huber V, et al. CD84 regulates PD-1/PD-L1 expression and function in chronic lymphocytic leukemia. J Clin Invest. 2018;128(12):5465–5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dhiman A, Kumar C, Mishra SK, et al. Theranostic application of a novel G-quadruplex-forming DNA aptamer targeting malate synthase of mycobacterium tuberculosis. Mol Ther Nucleic Acids. 2019;18:661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Christian WJ, Vanderford NL, McDowell J, et al. Spatiotemporal analysis of lung cancer histological types in Kentucky, 1995-2014. Cancer Control. 2019;26(1):1073274819845873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Horita K, Kurosaki H, Nakatake M, et al. lncRNA UCA1-mediated Cdc42 signaling promotes oncolytic vaccinia virus cell-to-cell spread in ovarian cancer. Mol Ther Oncolytics. 2019;13:35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.van Ingen E, Foks AC, Kröner MJ, et al. Antisense oligonucleotide inhibition of microRNA-494 halts atherosclerotic plaque progression and promotes plaque stabilization. Mol Ther Nucleic Acids. 2019;18:638–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fan P, Zhao J, Meng Z, et al. Overexpressed histone acetyltransferase 1 regulates cancer immunity by increasing programmed death-ligand 1 expression in pancreatic cancer. J Exp Clin Cancer Res. 2019;38(1):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Steponaviciene L, Briediene R, Vanseviciute R, Smailyte G. Trends in breast cancer incidence and stage distribution before and during the introduction of the mammography screening program in Lithuania. Cancer Control. 2019;26(1):1073274818821096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Barthélémy F, Wang RT, Hsu C, et al. Targeting RyR activity boosts antisense exon 44 and 45 skipping in human DMD skeletal or cardiac muscle culture models. Mol Ther Nucleic Acids. 2019;18:580–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Foloppe J, Kempf J, Futin N, et al. The enhanced tumor specificity of TG6002, an armed oncolytic vaccinia virus deleted in two genes involved in nucleotide metabolism. Mol Ther Oncolytics. 2019;14:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Drozd M, Delhaye S, Maurin T, et al. Reduction of Fmr1 mRNA levels rescues pathological features in cortical neurons in a model of FXTAS. Mol Ther Nucleic Acids. 2019;18:546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sengupta S, Katz SC, Sengupta S, Sampath P. Glycogen synthase kinase 3 inhibition lowers PD-1 expression, promotes long-term survival and memory generation in antigen-specific CAR-T cells. Cancer Lett. 2018;433:131–139. [DOI] [PubMed] [Google Scholar]
- 98.Duong MT, Collinson-Pautz MR, Morschl E, et al. Two-dimensional regulation of CAR-T cell therapy with orthogonal switches. Mol Ther Oncolytics. 2018;12:124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.John S, Chen H, Deng M, et al. A novel anti-LILRB4 CAR-T cell for the treatment of monocytic AML. Mol Ther. 2018;26(10):2487–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Araújo D, Azevedo NM, Barbosa A, et al. Application of 2’-OMethylRNA’ antisense oligomer to control candida albICANS EFG1 virulence determinant. Mol Ther Nucleic Acids. 2019;18:508–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jennings VA, Scott GB, Rose AMS, et al. Potentiating oncolytic virus-induced immune-mediated tumor cell killing using histone deacetylase inhibition. Mol Ther. 2019;27(6):1139–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhao B, Baloch Z, Ma Y, et al. Identification of potential key genes and pathways in early-onset colorectal cancer through bioinformatics analysis. Cancer Control. 2019;26(1):1073274819831260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kocher T, Wagner RN, Klausegger A, et al. Improved double-Nicking strategies for COL7A1-editing by homologous recombination. Mol Ther Nucleic Acids. 2019;18:496–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ylösmäki E, Malorzo C, Capasso C, et al. Personalized cancer vaccine platform for clinically relevant oncolytic enveloped viruses. Mol Ther. 2018;26(9):2315–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Warner SG, Kim SI, Chaurasiya S, et al. A novel chimeric poxvirus encoding hNIS Is tumor-tropic, imageable, and synergistic with radioiodine to sustain colon cancer regression. Mol Ther Oncolytics. 2019;13:82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bremer J, van der Heijden EH, Eichhorn DS, et al. Natural exon skipping sets the stage for exon skipping as therapy for dystrophic epidermolysis bullosa. Mol Ther Nucleic Acids. 2019;18:465–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Herrera M, Llorens C, Rodríguez M, et al. Differential distribution and enrichment of non-coding RNAs in exosomes from normal and cancer-associated fibroblasts in colorectal cancer. Mol Cancer. 2018;17(1):114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.El Enein MA, Grainger DW, Kili S. Registry contributions to strengthen cell and gene therapeutic evidence. Mol Ther. 2018;26(5):1172–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Deng S, Zhang G, Kuai J, et al. Lentinan inhibits tumor angiogenesis via interferon γ and in a T cell independent manner. J Exp Clin Cancer Res. 2018;37(1):260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Matsuyama T, Ishikawa T, Takahashi N, et al. Transcriptomic expression profiling identifies ITGBL, an epithelial to mesenchymal transition (EMT)-associated gene, is a promising recurrence prediction biomarker in colorectal cancer. Mol Cancer. 2019;18(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zainfeld D, Goldkorn A. Liquid biopsy in prostate cancer: circulating tumor cells and beyond. Cancer Treat Res. 2018;175:87–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li J, Han X, Yu X, et al. Clinical applications of liquid biopsy as prognostic and predictive biomarkers in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. J Exp Clin Cancer Res. 2018;37(1):213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ma YS, Lv ZW, Yu F, et al. MiRNA-302a/d inhibits the self-renewal capability and cell cycle entry of liver cancer stem cells by targeting the E2F7/AKT axis. J Exp Clin Cancer Res. 2018;37(1):252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fergusson DA, Wesch NL, Leung GJ, et al. Assessing the completeness of reporting in preclinical oncolytic virus therapy studies. Mol Ther Oncolytics. 2019;14:179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim SH, Lee S, Lee H, et al. AAVR-displaying interfaces: serotype-independent adeno-associated virus capture and local delivery systems. Mol Ther Nucleic Acids. 2019;18:432–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Del Papa J, Petryk J, Bell JC, Parks RJ. An oncolytic adenovirus vector expressing p14 FAST protein induces widespread syncytium formation and reduces tumor growth rate in vivo. Mol Ther Oncolytics. 2019;14:107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wu TH, Hsiue EH, Yang JC. Opportunities of circulating tumor DNA in lung cancer. Cancer Treat Rev. 2019;78:31–41. [DOI] [PubMed] [Google Scholar]
- 118.Gao T, Hu Q, Hu X, et al. Novel selective TOPK inhibitor SKLB-C05 inhibits colorectal carcinoma growth and metastasis. Cancer Lett. 2019;445:11–23. [DOI] [PubMed] [Google Scholar]
- 119.Fontana G, Martin HL, Lee JS, et al. Mineral-coated microparticles enhance mRNA-based transfection of human bone marrow cells. Mol Ther Nucleic Acids. 2019;18:455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ye Q, Ling S, Zheng S, Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer. 2019;18(1):114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sivanandam V, LaRocca CJ, Chen NG, Fong Y, Warner SG. Oncolytic viruses and immune checkpoint inhibition: the best of both worlds. Mol Ther Oncolytics. 2019;13:93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lietzén N, Pitkäniemi J, Heinävaara S, Ilmonen P. On exploring hidden structures behind cervical cancer incidence. Cancer Control. 2018;25(1):1073274818801604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Barkowsky G, Lemster AL, Pappesch R, et al. Influence of different cell-penetrating peptides on the antimicrobial efficiency of PNAs in streptococcus pyogenes. Mol Ther Nucleic Acids. 2019;8:444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ma YS, Yu F, Zhong XM, et al. miR-30 family reduction maintains self-renewal and promotes tumorigenesis in NSCLC-initiating cells by targeting oncogene TM4SF. Mol Ther. 2018;26(12):S1525–1538(18):30447–30457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Matés JM, Campos Sandoval JA, Santos Jiménez JL, Márquez J. Dysregulation of glutaminase and glutamine synthetase in cancer. Cancer Lett. 2019;467():29–39. [DOI] [PubMed] [Google Scholar]
- 126.Dutta RK, Chinnapaiyan S, Unwalla H. Aberrant MicroRNAomics in pulmonary complications: implications in lung health and diseases. Mol Ther Nucleic Acids. 2019;18:413–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yuanyue L, Baloch Z, Shanshan L, et al. Cervical cancer, human papillomavirus infection, and vaccine-related knowledge: awareness in Chinese women. Cancer Control. 2018;25(1):1073274818799306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mauseth B, Camilio KA, Shi J, et al. The novel oncolytic compound LTX-401 induces antitumor immune responses in experimental hepatocellular carcinoma. Mol Ther Oncolytics. 2019;14:139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kenneweg F, Bang C, Xiao K, et al. Long noncoding RNA-enriched vesicles secreted by hypoxic cardiomyocytes drive cardiac fibrosis. Mol Ther Nucleic Acids. 2019;18:363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nakatake M, Kurosaki H, Kuwano N, et al. Partial deletion of glycoprotein B5 R enhances vaccinia virus neutralization escape while preserving oncolytic function. Mol Ther Oncolytics. 2019;14:159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kaczor-Urbanowicz KE, Wei F, Rao SL, et al. Clinical validity of saliva and novel technology for cancer detection. Biochim Biophys Acta Rev Cancer. 2019;1872(1):49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Vitiello PP, Cardone C, Martini G, et al. Receptor tyrosine kinase-dependent PI3 K activation is an escape mechanism to vertical suppression of the EGFR/RAS/MAPK pathway in KRAS-mutated human colorectal cancer cell lines. J Exp Clin Cancer Res. 2019;38(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang J, Zhang N, Han Q, et al. Pin1 inhibition reverses the acquired resistance of human hepatocellular carcinoma cells to regorafenib via the Gli1/Snail/E-cadherin pathway. Cancer Lett. 2019;444:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ylä-Herttuala S. The pharmacology of gene therapy. Mol Ther. 2017;25(8):1731–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang L, Liang Y, Li S, et al. The interplay of circulating tumor DNA and chromatin modification, therapeutic resistance, and metastasis. Mol Cancer. 2019;18(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Janssen MJ, Nieskens TTG, Steevels TAM, et al. Therapy with 2’-O-Me phosphorothioate antisense oligonucleotides causes reversible proteinuria by inhibiting renal protein reabsorption. Mol Ther Nucleic Acids. 2019;18:298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nong J, Gong Y, Guan Y, et al. Circulating tumor DNA analysis depicts subclonal architecture and genomic evolution of small cell lung cancer. Nat Commun. 2018;9(1):3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Buono G, Gerratana L, Bulfoni M, et al. Circulating tumor DNA analysis in breast cancer: is it ready for prime-time? Cancer Treat Rev. 2019;73:73–83. [DOI] [PubMed] [Google Scholar]
- 139.Giordano G, Parcesepe P, D’Andrea MR, et al. JAK/Stat5-mediated subtype-specific lymphocyte antigen 6 complex, locus G6D (LY6G6D) expression drives mismatch repair proficient colorectal cancer. J Exp Clin Cancer Res. 2019;38(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sun J, Luo Q, Liu L, Song G. Low-level shear stress promotes migration of liver cancer stem cells via the FAK-ERK1/2 signalling pathway. Cancer Lett. 2018;427:1–8. [DOI] [PubMed] [Google Scholar]
- 141.Yu J, Han Z, Sun Z, Wang Y, Zheng M, Song C. LncRNA SLCO4A1-AS1 facilitates growth and metastasis of colorectal cancer through β-catenin-dependent Wnt pathway. J Exp Clin Cancer Res. 2018;37(1):222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Herkt M, Batkai S, Thum T. Studying interactions between 2’-O-Me-modified inhibitors and microRNAs utilizing microscale thermophoresis. Mol Ther Nucleic Acids. 2019;18:259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yi M, Jiao D, Xu H, et al. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol Cancer. 2018;17(1):129. [DOI] [PMC free article] [PubMed] [Google Scholar]