Abstract
Objectives:
Lung cancer (LC) is often accompanied by significant methylation abnormalities. This study aimed to develop a decision tree (DT) accompanied the stature homeobox 2 gene (SHOX2) / prostaglandin E receptor 4 (PTGER4) gene DNA methylation with traditional tumor marker in the differential diagnosis of benign and malignant lung nodule.
Methods:
We performed a study with 104 patients enrolled in the LC group and 36 patients in the benign lung diseases group. All the clinical data of these patients were collected through electronic medical record. Total Methylation (TM) status of both SHOX2 and PTGER4 was defined as methylation levels of SHOX2 plus methylation levels of PTGER4. One-way analysis was used to compare the concentrations of serum samples and t-test was used to compare pairwise mean values between groups. Receiver operating curve (ROC) was used to evaluate the diagnostic value. Furthermore, the strategy was validated in 19 LC patients and 11 patients with benign lung diseases.
Results:
There were significant differences between the concentration of neuron-specific enolase (NSE), carcinoembryonic antigen (CEA), cytokeratin 19 fragments (CYFRA21 -1) and the methylation levels of SHOX2, PTGER4 and TM in lung benign diseases and cancer group. The AUCs of NSE, CEA, CYFRA21 -1, Methylation SHOX2, Methylation PTGER4 and TM were 0.721 (95% CI: 0.627–0.816), 0.753 (95% CI: 0.673–0.833) and 0.778(95% CI: 0.700–0.856), 0.851(0.786-0.916), 0.847(0.780-0.913) and 0.861(0.800-0.922) respectively. We developed a DT model with TM and CYFRA21 -1 used in this study, and the area under the curve (AUC) of DT was 0.921 and the sensitivity up to 0.856. In the validation cohort, the AUC of SHOX2, PTGER4 and TM was also much higher than traditional serum markers.
Conclusions:
Our results indicated that the DT model calculated from the TM and CYFRA21 -1 can accurately classify LC and benign diseases, which showed better diagnostic performance than traditional serum parameter.
Keywords: lung cancer, diagnosis, stature homeobox 2, total methylation, decision tree
Introduction
As reported from the World Health Organization (WHO), lung cancers are the most common strains of cancer, resulting in about 2.0 million deaths in the year of 2018. In recent years, the emergence of low-dose CT (LDCT) screening in high-risk populations significantly increase the early detection and diagnose of lung cancers.1,2 However, according to previous publications, there were some drawbacks as follows: the false positive rate (FP rate) is fairly high, the majority of pulmonary nodules detected being benign; less ideal to detect precancerous lesions and early lung cancer; the radiation risk to patients.3,4 Therefore, it is necessary to find a sensitive and safe method for reducing the FP rate for the diagnosis, particular for the differentiation of malignancy from benign nodule.
The epigenetic events occur during carcinogenesis are a source of biomarkers for cancers. Abnormal DNA methylation has been found in various cancers and extensively described as promising biomarkers for the screening, diagnosis and prognosis of cancers, especially in specimens that are amenable for non-invasive sampling.5-7 In recent years, methylation-based biomarkers have been successfully incorporated into commercially available in vitro diagnostic (IVD) devices. Screening for colorectal cancer in a simple blood draw was made possible with real-time polymerase chain reaction (PCR) assessment of methylated Septin9 DNA derived from plasma.8,9 In lung cancers, methylated short stature homeobox 2 gene (SHOX2) DNA and prostaglandin E receptor 4 gene (PTGER4) have been described as valuable biomarkers in several studies.10,11 Elevated SHOX2/PTGER4 methylation was associated with the detection of lung cancers in blood plasma, pleural effusions, and bronchial aspirates.12,13
However, in south China, SHOX2/ PTGER4 DNA methylation were not routinely used in the investigation of a patient with a suspected lung nodule, mainly due to a lack of evidence as to its utility in the population.14,15 Thus, the need for evaluation of these markers in the diagnosis of pulmonary lesion is imperative in the local area.
The purpose of our study was to evaluate the diagnostic performance of SHOX2 and PTGER4 DNA methylation status in lung nodule, as compared with traditional tumor markers. We aimed to ascertain the use of SHOX2/ PTGER4 DNA methylation in the differential diagnosis of benign and malignant lung nodules. Our model offers a sample and useful tool for prognosis, which could be used to avoid the over diagnosis caused by LDCT.
Materials and Methods
Subjects
Consecutive patients in the primary cohort, attending the Department of Thoracic Surgery, Jiangmen Hospital from January 2018 to July 2019, were recruited in this study. The inclusion criteria were as follows: (1) patients with histologically diagnosed LC; (2) patients without any treatment before sample collection; (3) patients without other tumors; (4) patients with complete information. Eligible participants were patients with pulmonary nodule detected by low-dose CT screening. Fifty healthy volunteers were also recruited in the same hospital during July 2020. What’s more, patients younger than 18 years or elder than 80 years of age were excluded. Another 30 cases from the First Affiliated Hospital of Sun Yat-sen University (Guangzhou, China) were enrolled into the validation cohorts and used to validate the model. The validation study was conducted from January 2020 to July 2020 with the same inclusion criteria. There were no differences between the primary and validation cohorts in terms of age and sex. The following investigations were carried out in all subjects: physical examination, standard chest roentgenography, low dose CT scan of the chest, upper abdomen and liver sonography. Fiberoptic bronchoscopes or mediastinoscopy was performed when necessary.
Sample Collection
EDTA blood samples (10 mL) were obtained, before the surgery or other invasive operation. Plasma was separated within 2 hours after collection. The plasma supernatant was stored at −80°C until further analysis. Biopsy tissues of the pulmonary lesions were obtained during the operation, fiberoptic bronchoscopes or mediastinoscopy and sent for pathological examination.
Tumor Marker Detection
At the clinical laboratory department, NSE and CEA and were tested using the ARCHITECT NSE and CEA assays (Abbott Diagnostics, Abbott Park, IL, USA) according to the manufacturer’s instructions. Plasma Cyfra21 -1 level was detected with a Cyfra21 -1 test kit (Roche Diagnostics Corp, China) using a Cobas e601 analyzer.
SHOX2/PTGER4 DNA Methylation
Cell-free DNA extraction and bisulfite modification were performed with a commercial kit (TIANGEN Biotech). The concentration of purified DNA was determined by the Qubit dsDNA HS Assay Kit (Life Technologies, Carlsbad, CA). Real-time PCR was performed on ABI Prism 7500 Sequence Detection system (Applied Biosystems, USA) by using a commercial SHOX2/PTGER4 DNA Methylation Detection Kit (Sinomed, Beijing, China). For each sample, a relative methylation value was determined using the ΔΔCT method adapted for DNA methylation analyses as previously described. Percentage methylation was calculated using the following formula: Methylation Sample = 100% *2-ΔΔCT Sample(1).
Diagnosis of Lung Cancers
Tissue specimens were evaluated by experienced pathologists. All histological results were performed blinded to laboratory biomarker detection. Lung cancers were classified according to the WHO histological classification. Staging was carried out according to the Union for International Cancer Control (UICC) tumor node metastasis (TNM) classification at the time of diagnosis.
Statistical Analysis
Statistical analysis was analyzed by R software (Version 3.3.5) with pROC, rPART and Rattle package(3-5). One-way analysis of variance was used to compare the concentrations of serum samples and the 2-sample t-test was used to compare pairwise mean values between groups. Other variables were evaluated by the chi-square test, Fisher’s exact test, and Mann-Whitney U test when appropriate. Receiver operating characteristics (ROC) curve was used to evaluate the diagnostic value. A 2-tailed P value of less than 0.05 was considered statistically significant.
Results
Basic Characteristic
In our study, there were 104 patients enrolled in the lung cancer group and 36 patients enrolled in the benign lung diseases group. All the demographic and clinical characteristics are showed in Table 1. The median age of lung cancer group was 60- year (range: 31-82) and that of benign lung diseases group was 53-year (36-83). There was no significant difference in age distribution between 2 groups (P = 0.362). There were 59 males and 45 females in lung cancer group, while 19 males and 17 females in benign lung diseases group (P = 0.5160.05). There were no significant differences in gender in the 2 groups. The histology of lung cancer included adenocarcinoma (n = 58), squamous cell carcinoma (n =40), adenosquamous carcinoma (n = 2), small cell lung cancer (n = 4), and the benign lung diseases classified into benign sarcoidosis (14), pulmonary tuberculosis(8), organizing pneumonia(10), lymphadenitis(2), hamartoma(2). Meanwhile, in the validation set, there were 19 patients enrolled in the lung cancer group and 11 patients enrolled in the benign lung diseases group. The histology of lung cancer in validation study included adenocarcinoma (n = 14), squamous cell carcinoma (n = 4), lymphoepitheliomatoid carcinoma(1). Among 19 lung cancer patients, 12 patients were at stage Ⅰ, 4 patients were at stage Ⅱ, and 3 patients were at stage Ⅲ.
Table 1.
Characteristics of all Subjects.
| SCLC(n = 104) | Benign lung disease(n = 36) | Normal(n = 50) | P value | ||
|---|---|---|---|---|---|
| Gender | |||||
| Male | 59(56.7%) | 19(52.8%) | 31(62.0%) | 0.682a | |
| Female | 45(43.3%) | 17(47.2) | 19(19.0%) | ||
| Age, years | |||||
| Median | 62 | 58 | 42 | 0.316b | |
| Range | 31-82 | 36-83 | 20-76 | ||
| Smoking | |||||
| Yes | 60(57.6%) | 20(48.4%) | 11(22.0%) | <0.001a | |
| Never | 44(42.3%) | 16(51.6%) | 39(78.0%) | ||
| Stage | |||||
| Ⅰ | 48 | ||||
| Ⅱ | 15 | ||||
| Ⅲ | 20 | ||||
| Ⅳ | 21 | ||||
| Histology | |||||
| Adenocarcinoma | 53 | ||||
| Squamous cell carcinoma | 15 | ||||
| Adenosquamous carcinoma | 1 | ||||
| Small cell carcinoma | 3 | ||||
| Benign sarcoidosis | 13 | ||||
| Pulmonary tuberculosis | 8 | ||||
| Organizing pneumonia | 10 | ||||
| Lymphadenitis | 1 | ||||
| Hamartoma | 1 | ||||
| Serum markers | |||||
| NSE(pmol/L) | 15.83 ± 12.29 | 10.90 ± 2.66 | 11.80 ± 3.33 | <0.001c | |
| CEA(μg/L) | 24.82 ± 92.08 | 2.08 ± 0.96 | 2.46 ± 1.57 | <0.001c | |
| Cyfra 21-1(ng/ml) | 13.36 ± 64.16 | 2.17 ± 0.92 | 2.21 ± 0.86 | <0.001c | |
| Methylation S2(%) | 57.67 ± 159.17 | 0.30 ± 0.98 | 0.12 ± 0.29 | <0.001d | |
| Methylation P4(%) | 9.95 ± 24.92 | 0.05 ± 0.13 | 0.004 ± 0.02 | <0.001d | |
| Total Methylation(%) | 67.62 ± 168.37 | 0.34 ± 1.12 | 0.13 ± 0.29 | <0.001d |
aanalyzed by Chi Squared test.
banalyzed by Mann–Whitney U test.
canalyzed by One-way ANOVA.
dWilcoxon Rank-Sum test.
NSE, CEA, CYFRA21 -1 and the methylation levels in lung cancer and benign diseases
Total Methylation (TM) status of both SHOX2 and PTGER4 was defined as methylation levels of SHOX2 plus methylation levels of PTGER4. There were significant differences between the concentration of NSE (10.90 ± 2.66 VS. 15.83 ± 12.29, P < 0.001), CEA (2.08 ± 0.96 VS. 24.82 ± 92.08, P < 0.001), CYFRA 21-1(2.17 ± 0.92 VS. 13.36 ± 64.16, P = 0.032) and the methylation levels of SHOX2(0.30 ± 0.98 VS. 57.67 ± 159.17, P < 0.001), PTGER4(0.05 ± 0.13 VS. 9.95 ± 24.92, P < 0.001) and TM (0.34 ± 1.12 VS. 67.62 ± 168.37, P < 0.001) in lung benign diseases and cancer group.
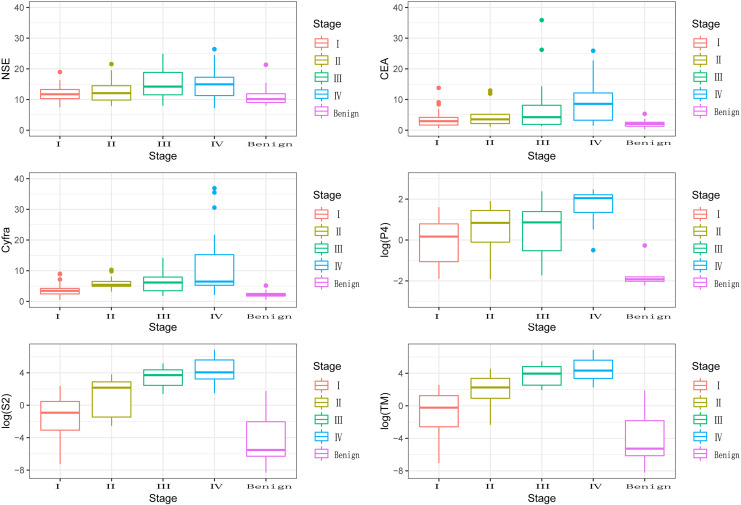
Furthermore, the methylation levels of SHOX2, PTGER4 and TM in different stages of lung cancer (I, II, III, IV) were analyzed. The methylation level of PTGER4 was 2.41 ± 5.27, 13.25 ± 17.11, 54.43 ± 50.36 and 219.13 ± 302.61 respectively (P < 0.001). The methylation level of SHOX2 was 1.18 ± 16.05, 7.51 ± 15.61, 25.96 ± 47.67 and 16.64 ± 18.38 respectively (P < 0.001). The TM were 3.60 ± 8.25, 20.76 ± 26.54, 80.39 ± 75.78 and 235.77 ± 313.85 respectively (P < 0.001). The methylation levels of SHOX2 and TM were significantly associated with the tumor stage (Figure 1).
Figure 1.
Detection of the concentration of NSE, CEA, CYFRA21 -1 and the methylation levels of SHOX2 and PTGER4 genes in different groups.
Diagnostic Values of SHOX2, PTGER4, NSE, CEA and CYFRA 21 -1 levels in the Detection of Lung Cancer
To evaluate the diagnostic value of SHOX2 and PTGER4 methylation levels and compare with traditional markers, we took area under curve (AUC) into consideration. As shown in Table 2 and Figure 2, the AUCs of NSE, CEA, CYFRA21 -1, Methylation SHOX2, Methylation PTGER4 and TM were 0.721 (95% CI: 0.627–0.816), 0.753 (95% CI: 0.673–0.833) and 0.778(95% CI: 0.700–0.856), 0.851(95% CI: 0.786-0.916), 0.847(95% CI: 0.780-0.913) and 0.861(95% CI: 0.800-0.922) respectively. The AUC of SHOX2, PTGER4 and TM was much higher than traditional serum markers (P = 0.039, DeLong’s method was used for AUC comparison). Cut-off value of our study was defined as the maximum of Youden index. At the level of Cut-off, both SHOX2 and PTGER4 were a highly specific marker for the detection of malignant nodule (0.971, 0.971), however, the sensitivity was poor (0.538, 0.654).
Table 2.
The Diagnostic Performance of Different Markers.
| AUC(95%CI) | Cut-off | Sensitivity | Specificity | NPV | PPV | |
|---|---|---|---|---|---|---|
| NSE(pmol/L) | 0.7214(0.6274-0.8155) | 13.770 | 0.404 | 0.857 | 0.326 | 0.894 |
| CEA(μg/L) | 0.7527(0.6723-0.8332) | 3.020 | 0.596 | 0.857 | 0.417 | 0.925 |
| Cyfra 21-1(ng/ml) | 0.7782(0.7002-0.8561) | 3.255 | 0.606 | 0.943 | 0.446 | 0.969 |
| Methylation SHOX2 (%) | 0.8514( 0.7864-0.9164) | 1.031 | 0.606 | 0.971 | 0.453 | 0.984 |
| Methylation PTGER4(%) | 0.8466(0.7801-0.913) | 0.481 | 0.538 | 0.971 | 0.415 | 0.982 |
| Total Methylation(%) | 0.8613(0.8002-0.9223) | 1.567 | 0.654 | 0.971 | 0.486 | 0.986 |
| Decision Tree | 0.921(0.8476-0.951) | 1 | 0.856 | 1.000 | 0.822 | 1.000 |
Figure 2.
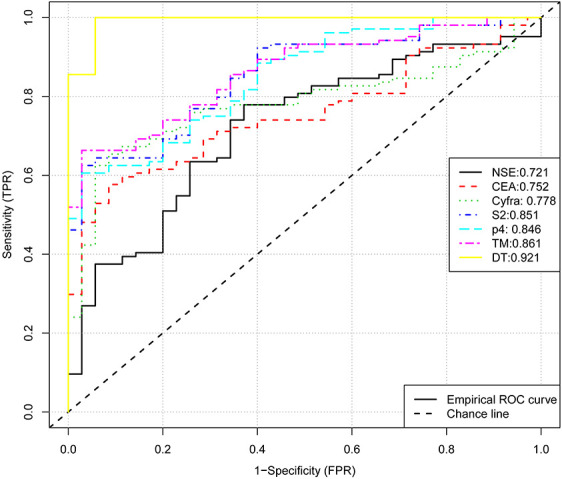
ROC curve for diagnostic value of TM, SHOX2, PTGER4, NSE, CEA, CYFRA 21 -1 and DT.
Classification of Samples Using the Decision Tree Model
Since the sensitivity is critical for the screening of malignant nodule, we try to combine these markers to generate more sensitive diagnostic values. To generate simple splitting criteria for classifying samples into lung cancer and benign diseases, we performed a decision tree(DT) analysis. By using the rPART package, we created a decision tree with all serum makers and methylation markers and only 2 parameters (TM and CYFRA21 -1) remained in this model and the rules were shown in Figure 3. The classification rules were: (1) TM >1.4 was classed as malignant (2) TM <1.4 but CYFRA >3.3 was classed as malignant. The maximum accuracy achieved by the classifier was 85.6%. And the AUC of the decision tree was 0.921 and the sensitivity up to 85.6% (Table 2). Our results indicated that the decision tree model calculated from the 2 parameters can accurately classify lung cancer and benign diseases (Figure 2), which showed better diagnostic performance than a single parameter.
Figure 3.
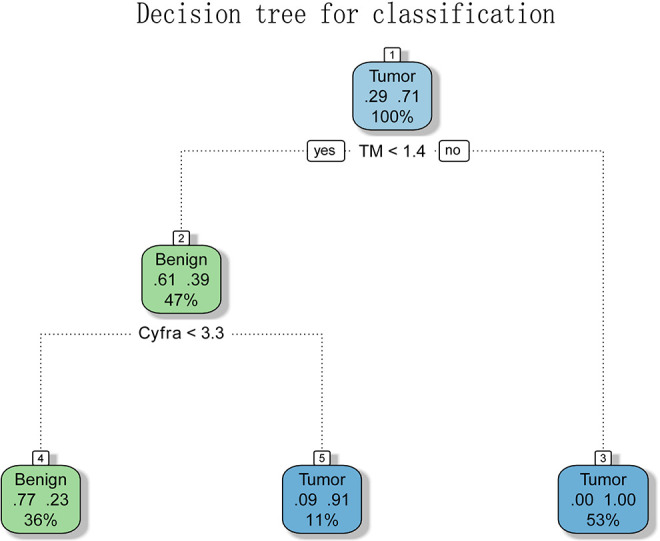
A decision tree to classify samples into the 3 groups. A decision tree constructed using the 2 parameters is shown. The formula in the decision tree split criteria for samples. The classification rules were: (1) TM >1.4 was classed as malignant (2) TM <1.4 but CYFRA >3.3 was classed as malignant.
Validation of the Decision Tree Model
We also tested the accuracy of the model by AUC in externa validation cohort. The discrimination of the model was measured by AUC. In the validation cohort, the AUCs of NSE, CEA, CYFRA21 -1, Methylation SHOX2, Methylation PTGER4 and TM were 0.579((95% CI: 0.360-0.798), 0.617((95% CI: 0.407-0.827), 0.538((95% CI: 0.325-0.751), 0.804((95% CI: 0.644-0.964), 0.617((95% CI: 0.402-0.832), 0.804((95% CI:0.644-0.964) and 0.744((95% CI: 0.8476-0.951) respectively. The AUC of SHOX2, PTGER4 and TM was also much higher than traditional serum markers. The DT model showed better diagnostic performance than single parameter (Table 3).
Table 3.
The Diagnostic Performance of Different Markers in Validation Set.
| AUC(95%CI) | Cut-off | Sensitivity | Specificity | NPV | PPV | |
|---|---|---|---|---|---|---|
| NSE(pmol/L) | 0.579(0.360-0.798) | 14.76 | 0.579 | 0.727 | 0.633 | 0.680 |
| CEA(μg/L) | 0.617(0.407-0.827) | 2.33 | 0.684 | 0.636 | 0.668 | 0.653 |
| Cyfra 21-1(ng/ml) | 0.538(0.325-0.751) | 2.42 | 0.368 | 0.818 | 0.564 | 0.669 |
| Methylation SHOX2 (%) | 0.804( 0.644-0.964) | 0.206 | 0.632 | 0.909 | 0.712 | 0.874 |
| Methylation PTGER4(%) | 0.617(0.402-0.832) | 0.001 | 0.526 | 0.727 | 0.605 | 0.658 |
| Total Methylation(%) | 0.804(0.644-0.964) | 0.203 | 0.684 | 0.919 | 0.744 | 0.894 |
| Decision Tree | 0.849(0.8476-0.951) | 1 | 0.785 | 0.909 | 0.783 | 0.864 |
Discussions
Lung cancer is the most common malignant tumor in China, and it is the leading cause of mortality of all cancers.16 Studies show that the 5-year survival rate of early lung cancer is about 80%, which is much higher than that of advanced lung cancer.17,18 In the diagnosis of early lung cancer, LDCT has a high false-positive rate while serum biomarkers have low sensitivity, such as NSE, CEA and CYFRA 21 -1. Therefore, effective and rapid diagnosis of early lung cancer is of great significance.
DNA methylation plays a key role in regulating gene expression, development and cells differentiation. It is well known that the occurrence and development of tumors are often accompanied with significant abnormal methylation, which results in abnormal gene expression, thus affecting the activity of downstream signaling pathways.19,20 Many studies reported that abnormal DNA methylation correlates with tumor aggressiveness, tumor stage, and prognosis.21-23 Aberrant DNA methylations in plasma are emerging biomarkers for liquid biopsy. In this study, we evaluated whether the detection of SHOX2 and PTGER4 methylation levels in serum would benefit the differential diagnosis of lung nodule found in LDCT screening.
During January 2018 to July 2019, 139 subjects in Jiangmen Hospital were recruited in our study. We evaluated the levels of methylated SHOX2 and PTGER4 and 3 common serum tumor markers in lung cancer. The results showed that methylation levels of SHOX2 and PTGER4 were significantly associated with the tumor stage, patients with advanced-stage have high methylation levels (Figure 1). Then, the ROC curve was used to estimate the diagnostic efficiency of all the parameters, the AUC of SHOX2, PTGER4 and TM (0.851, 0.847 and 0.861) was much higher than traditional serum markers (P < 0.05), but with low sensitivity (0.606, 0.538 and 0.654). Furthermore, we created a decision tree with TM and CYFRA21 -1, and found that the AUC of the decision tree was 0.921 and the sensitivity increased to 0.856, which showed better diagnostic performance than a single parameter. Also, in the validation set, the AUC of SHOX2, PTGER4, TM and DT model was also much higher than traditional serum markers.
In our study, the correlation between the levels of aberrant methylated SHOX2 and PTGER4 and the progression of lung cancer has been confirmed by both previous research,24 which operates as a suitable biomarker at a high specificity in diagnosing lung cancer patients, whereas the sensitivity is low. The possible reasons might be as follows: (1) the amount of ctDNA in peripheral blood is limited in the early stage of lung cancer; (2) the molecular biotechnology needs to be improved in some fields, such as ctDNA enrichment; (3) the loss of DNA during modification and amplification. Therefore, DNA methylation may be preferentially employed as a diagnostic test with traditional biomarkers.
This study indicates that a combination of TM and CYFRA21 -1 is promising for the screening of lung cancer and lower the false positive of LDCT with the AUC was 0.921 and the sensitivity up to 85.6%. With the development of the tumor, more malignant tumor cells necrosis and lysis, resulting in the release of more methylation genes in cells.25 Gene methylation is well acknowledged as a feature of transcribed genes, and gene methylation level in tumor have been observed in various studies.26 The biological relevance of SHOX2 and PTGER4 expression during carcinogenesis and tumor progression, however, is only poorly understood. SHOX2 involvement in epithelial-mesenchymal transition (EMT) has been reported in many tumors, such as breast cancer and esophageal squamous cell carcinoma. EMT is a process by which epithelial cells lose their cell-cell adhesion and cell polarity and gain migratory and invasive properties, ultimately leading to the initiation of metastasis in cancer progression.27-29 This might explain SHOX2 could induce proliferation, invasion, and metastasis. PTGER4 play the role in tumor proliferation through activating the phosphorylation of glycogen synthase kinase-3 (GSK3).30,31
In the validation study, the diagnostic performance of our DT model is inferior than that in the primary study. We consider that this is due to the high proportion of patients with early stage in the validation study. However, the sensitivity and specificity of DT model was still much higher than traditional markers. These data suggested that our strategy is promising for the identification of lung cancer patients correctly.
There were some limitations in this experiment. Firstly, the number of samples was not enough, which made the results of the analysis not universal, especially in some histological subtype groups the exact number of patients was insufficient that the diagnostic value may be over-evaluated, so more patients in this group needed to be included in further studies. Secondly, although the number of malignant tumors and benign lung disease patients were matched with sex and age, the number of patients varied greatly. Thirdly, the test fee including TM and CYFRA21-11 might be higher. Especially the number of benign lung disease patients was too small, so the results could not represent all the patients.
Conclusion
The results of this study confirmed the potential value of SHOX2 and PTGER4 methylation markers in lung cancer diagnosis. A decision tree has been developed to predict lung cancer. The predictive accuracy of our DT in this study was higher than current marker alone, and it offers a sample and useful tool for prognosis, which could be used to avoid the over diagnosis caused by LDCT .To generalize the use of this model in other groups, additional validation with data from other institutions is required.
Acknowledgments
We thank all the staff at the department of Laboratory Medicine of The First Affiliated Hospital of Sun Yat-sen University.
Authors Note: Wenhai Huang and Hao Huang contributed equally to this work. Patients are prospectively selected from the EMR(electronic medical record), the data of tumor marker in this study are extracted from the LIS(Laboratory information system).
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethics Statement: This study was approved by the First Affiliated Hospital of Sun Yat-sen University Ethical Committee (Approval Number 2018-45). All patients provided written informed consent prior to enrollment in the study.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Natural Science Foundation of Guangdong province (Number: 2018A0303130246). Guangdong Basic and Applied Basic Research Foundation (2019A1515011663).
ORCID iD: Peisong Chen, PhD  https://orcid.org/0000-0003-0640-3663
https://orcid.org/0000-0003-0640-3663
References
- 1. Zhou M, Wang H, Zeng X, et al. Mortality, morbidity, and risk factors in China and its provinces, 1990-2017: a systematic analysis for the global burden of disease study 2017. Lancet. 2019;394(10204):1145–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. de Koning HJ. Lung cancer screening and its continuous risk assessment. Lancet Oncol. 2017;18(11):1434–1436. [DOI] [PubMed] [Google Scholar]
- 3. Muller DC, Johansson M, Brennan P. Lung cancer risk prediction model incorporating lung function: development and validation in the UK biobank prospective cohort study. J Clin Oncol. 2017;35(8):861–869. [DOI] [PubMed] [Google Scholar]
- 4. Barnett R. Lung cancer. Lancet. 2017;390(10098):928. [DOI] [PubMed] [Google Scholar]
- 5. Thierry AR, Tanos R. Liquid biopsy: a possible approach for cancer screening. Med Sci (Paris). 2018;34(10):824–832. [DOI] [PubMed] [Google Scholar]
- 6. Han X, Wang J, Sun Y. Circulating tumor DNA as biomarkers for cancer detection. Genomics Proteomics Bioinformatics. 2017;15(2):59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rohrich M, Koelsche C, Schrimpf D, et al. Methylation-based classification of benign and malignant peripheral nerve sheath tumors. Acta Neuropathol. 2016;131(6):877–887. [DOI] [PubMed] [Google Scholar]
- 8. Church TR, Wandell M, Lofton-Day C, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63(2):317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Potter NT, Hurban P, White MN, et al. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin Chem. 2014;60(9):1183–1191. [DOI] [PubMed] [Google Scholar]
- 10. Weiss G, Schlegel A, Kottwitz D, Konig T, Tetzner R. Validation of the SHOX2/PTGER4 DNA methylation marker panel for plasma-based discrimination between patients with malignant and nonmalignant lung disease. J Thorac Oncol. 2017;12(1):77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Song L, Yu H, Li Y. Diagnosis of lung cancer by SHOX2 gene methylation assay. Mol Diagn Ther. 2015;19(3):159–167. [DOI] [PubMed] [Google Scholar]
- 12. Fleischhacker M, Dietrich D, Liebenberg V, Field JK, Schmidt B. The role of DNA methylation as biomarkers in the clinical management of lung cancer. Expert Rev Respir Med. 2013;7(5):363–383. [DOI] [PubMed] [Google Scholar]
- 13. Dietrich D, Kneip C, Raji O, et al. Performance evaluation of the DNA methylation biomarker SHOX2 for the aid in diagnosis of lung cancer based on the analysis of bronchial aspirates. Int J Oncol. 2012;40(3):825–832. [DOI] [PubMed] [Google Scholar]
- 14. Ni S, Ye M, Huang T. Short stature homeobox 2 methylation as a potential noninvasive biomarker in bronchial aspirates for lung cancer diagnosis. Oncotarget. 2017;8(37):61253–61263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao QT, Guo T, Wang HE, et al. Diagnostic value of SHOX2 DNA methylation in lung cancer: a meta-analysis. Onco Targets Ther. 2015;8:3433–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shinagare AB, Guo M, Hatabu H, et al. Incidence of pulmonary embolism in oncologic outpatients at a tertiary cancer center. Cancer-Am Cancer Soc. 2011;117(16):3860–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Da CD, Parrish JW, Singh NK, Hsia DW. Lung cancer: advances and insights in diagnosis, treatment, and palliation. Am J Respir Crit Care Med. 2018;198(9):667–669. [DOI] [PubMed] [Google Scholar]
- 18. Brody H. Lung cancer. Nature. 2014;513:S1. [DOI] [PubMed] [Google Scholar]
- 19. Goldstein M, Kastan MB. The DNA damage response: implications for tumor responses to radiation and chemotherapy. Annu Rev Med. 2015;66:129–143. [DOI] [PubMed] [Google Scholar]
- 20. Esteller M. Aberrant DNA methylation as a cancer-inducing mechanism. Annu Rev Pharmacol Toxicol. 2005;45:629–656. [DOI] [PubMed] [Google Scholar]
- 21. Yang Z, Qi W, Sun L, Zhou H, Zhou B, Hu Y. DNA methylation analysis of selected genes for the detection of early-stage lung cancer using circulating cell-free DNA. Adv Clin Exp Med. 2019;28(3):355–360. [DOI] [PubMed] [Google Scholar]
- 22. Ma K, Cao B, Guo M. The detective, prognostic, and predictive value of DNA methylation in human esophageal squamous cell carcinoma. Clin Epigenetics. 2016;8(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheng J, Wei D, Ji Y, et al. Integrative analysis of DNA methylation and gene expression reveals hepatocellular carcinoma-specific diagnostic biomarkers. Genome Med. 2018;10(1):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schneider KU, Dietrich D, Fleischhacker M, et al. Correlation of SHOX2 gene amplification and DNA methylation in lung cancer tumors. BMC Cancer. 2011;11(1):102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bartosch C, Lopes JM, Jeronimo C. Epigenetics in endometrial carcinogenesis—part 1: DNA methylation. Epigenomics-Uk. 2017;9(5):737–755. [DOI] [PubMed] [Google Scholar]
- 26. Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. [DOI] [PubMed] [Google Scholar]
- 27. Branchi V, Schaefer P, Semaan A, et al. Promoter hypermethylation of SHOX2 and SEPT9 is a potential biomarker for minimally invasive diagnosis in adenocarcinomas of the biliary tract. Clin Epigenetics. 2016;8(1):133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang T, Zhang H, Cai SY, et al. Elevated SHOX2 expression is associated with tumor recurrence of hepatocellular carcinoma. Ann Surg Oncol. 2013;20(3):S644–S649. [DOI] [PubMed] [Google Scholar]
- 29. Mari-Alexandre J, Diaz-Lagares A, Villalba M, et al. Translating cancer epigenomics into the clinic: focus on lung cancer. Transl Res. 2017;189:76–92. [DOI] [PubMed] [Google Scholar]
- 30. Heinrichs S, Hess T, Becker J, et al. Evidence for PTGER4, PSCA, and MBOAT7 as risk genes for gastric cancer on the genome and transcriptome level. Cancer Med. 2018;7(10):5057–5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fujino H, West KA, Regan JW. Phosphorylation of glycogen synthase kinase-3 and stimulation of T-cell factor signaling following activation of EP2 and EP4 prostanoid receptors by prostaglandin E2. J Biol Chem. 2002;277(4):2614–2619. [DOI] [PubMed] [Google Scholar]