Abstract
Although its diagnosis and treatment have greatly improved in recent decades, cancer remains the major cause of death worldwide. Thus, there is an urgent need to find novel biomarkers and therapeutic targets to improve efficiency of diagnosis and treatment of patients with cancer. Long noncoding RNAs (lncRNAs), a new class of noncoding RNAs (ncRNAs), have been found to play a salient role in human tumorigenesis and progression. Distal-less homeobox 6 antisense RNA 1 (DLX6-AS1) is a novel lncRNA with aberrant expression in various cancers tissues and cell lines compared with nontumor tissues and normal cell lines. Importantly, DLX6-AS1 is closely associated with tumor cell proliferation, apoptosis, invasion, and migration. Patients with high DLX6-AS1 expression often had poorer prognosis than those with low expression. The oncogenicity of DLX6-AS1 mainly (indirectly or indirectly) interacts with targeting genes, and then regulates downstream genes and signaling pathways. Together with the findings of animal model studies, these data suggest that DLX6-AS1 may serve as a feasible predictor or therapeutic target in different cancers. Herein, we summarize the main findings concerning the function and molecular mechanisms of DLX6-AS1 to identify a molecular basis for future clinical application.
Keywords: DLX6-AS1, malignancy, lncRNA, molecular mechanisms, biomarker
Introduction
Several cancers can result in malignant disease marked by unregulated proliferation, migration, invasion, and apoptosis [1-3]. This represents a major public health problem, and thus, studies have focused on malignancies in a variety of cancer types, including breast cancer [4], lung cancer [5], gastric cancer [6], colorectal cancer [7], and hepatocellular carcinoma (HCC) [8]. In 2015, approximately 8.8 million deaths were caused by cancer, a global increase of 17.0% than 2014 [9]. However, treatments remain limited for advanced cancers and effective indicators are still lacking to diagnose cancer at early stages.
In recent years, with technological advances in transcriptome profiling, many noncoding RNAs (ncRNAs) were revealed to be involved in cellular biological processes, as well as several pathological processes, rather than transcriptional noise with no biological function [10-12]. Long noncoding RNAs (lncRNAs), one of the most common types of ncRNAs, are longer than 200 nucleotides but lack protein-coding capacity [13]. Several studies have found that lncRNAs regulate gene expression and that dysfunction is associated with tumorigenesis and progression of human cancers [14-17]. Therefore, researching lncRNAs may provide novel diagnostic biomarkers and therapeutic targets.
Distal-less homeobox 6 antisense RNA 1 (DLX6-AS1) was first identified as an in-trans modifier increasing the activity of the distal-less homeobox 5/6 enhancer [18]. Several studies have demonstrated that DLX6-AS1 expression is increased in tumor tissues, and is significantly associated with unfavorable outcomes in patients [19-21]. Moreover, studies in vivo and in vitro suggest that the knockdown of DLX6-AS1 suppresses tumor growth, migration, and metastasis. In this review, we summarize the current evidence of DLX6-AS1 in human cancers, especially its identification, functions, regulatory mechanisms, and potential clinical application.
Human studies of lncRNA DLX6-AS1
DLX6-AS1 expression in tumor tissues
Many lncRNAs have been found to be aberrantly expressed in cancer, including, PVT1 [22], HORAS5 [23], NEAT1 [24], MNX1-AS1 [25], PCA3 [26], among others. Recently, research groups have explored the expression of DLX6-AS1 in human tissues and its clinical value in various cancers. All studies consistently concluded that DLX6-AS1 is upregulated in patients with cancer. In lung cancer, DLX6-AS1 expression was elevated in lung cancer samples, compared with normal lung samples [20,27-29]. Meanwhile, data regarding the expression levels of DLX6-AS1 were assessed in several other human cancer samples including HCC [30-32], colon cancer [33,34], gastric cancer [35-38], breast cancer [21,39], nasopharyngeal carcinoma (NPC) [40], osteosarcoma [41,42], thyroid cancer [43], bladder cancer [44,45], renal cell carcinoma [46], pancreatic cancer [47], and other cancers [48,49] (Table 1).
Table 1.
Clinical features of DLX6-AS1 in human cancers
| Cancer types | Number (cancer and normal tissues) | Expression | Relationship with clinicopathologic parameters | Property | Reference |
|---|---|---|---|---|---|
| Lung cancer | 51 pairs | upregulated | size of tumor, TNM stage | oncogenic | [20] |
| 48 pairs | upregulated | / | oncogenic | [29] | |
| 72 pairs | upregulated | histological differentiation, TNM stage | oncogenic | [28] | |
| Nasopharyngeal carcinoma (NPC) | 72 pairs | upregulated | / | oncogenic | [40] |
| Hepatocellular carcinoma (HCC) | 48 pairs | upregulated | / | oncogenic | [31] |
| 30 pairs | upregulated | / | oncogenic | [32] | |
| 60 pairs | upregulated | tumor size, TNM stage | oncogenic | [30] | |
| Cervical cancer (CC) | 60 pairs | upregulated | / | oncogenic | [59] |
| Osteosarcoma | 80 pairs | upregulated | clinical stage | oncogenic | [42] |
| Ewing’s sarcoma | 20 pairs | upregulated | / | oncogenic | [41] |
| Laryngeal cancer | 43 pairs | upregulated | primary tumor, tumor stage, lymphatic metastasis, distant metastasis | oncogenic | [62] |
| Thyroid cancer | 60 pairs | upregulated | / | oncogenic | [43] |
| Colorectal Cancer | 76 pairs | upregulated | TNM stage, lymphatic metastasis | oncogenic | [34] |
| 60 pairs | upregulated | T stage, distant metastasis | oncogenic | [33] | |
| Human epithelial ovarian cancer (EOC) | 128 pairs | upregulated | FIGO stage, lymph node metastasis | oncogenic | [50] |
| Ovarian cancer | 58 pairs | upregulated | / | oncogenic | [61] |
| Bladder cancer (BC) | 80 pairs | upregulated | / | oncogenic | [44] |
| 54 pairs | upregulated | TNM stage, lymphatic node metastasis, distant metastasis | oncogenic | [45] | |
| Triple-negative breast cancer (TNBC) | 47 tumor tissues and 28 normal tissues | upregulated | / | oncogenic | [39] |
| Breast cancer | 45 pairs | upregulated | tumor size, lymph node status | oncogenic | [21] |
| Neuroblastoma | 36 tumor tissues and 18 normal tissues | upregulated | TNM stage | oncogenic | [48] |
| 70 pairs | upregulated | distant metastasis | oncogenic | [49] | |
| Renal cell carcinoma | 15 pairs | upregulated | metastasis | oncogenic | [46] |
| Esophageal squamous cell carcinoma (ESCC) | 73 pairs | upregulated | differentiation status, lymph node metastasis, TNM stage | oncogenic | [57] |
| Gastric Cancer | 60 pairs | upregulated | tumor size, lymph node involvement, TNM stage | oncogenic | [35] |
| 56 pairs | upregulated | invasion, distant metastasis | oncogenic | [36] | |
| 62 pairs | upregulated | metastasis, TNM stage | oncogenic | [38] | |
| Pancreatic cancer | 84 pairs | upregulated | tumor size, TNM stage, lymph node metastasis | oncogenic | [47] |
| 60 pairs | upregulated | / | oncogenic | [64] |
Clinical and prognostic values of DLX6-AS1 in various cancers
Notably, Kaplan-Meier analyses indicated that patients could be stratified by DLX6-AS1 expression into low or high expression groups, and those with high expression exhibited worse clinical prognoses. In addition, univariate cox regression analysis and multivariate regression analysis demonstrated that DLX6-AS1 is an independent prognostic indictor for disease-free survival or overall survival (OS) in different cancer types.
In human epithelial ovarian cancer, overexpression of DLX6-AS1 was closely correlated to lymph node metastasis, tumor stage, and poor prognosis. Further, univariate cox regression analysis and multivariate regression analysis verified that DLX6-AS1 is an independent risk factor in patients with epithelial ovarian cancer [50]. In patients with bladder cancer, 45 samples were collected and classified according to the DLX6-AS1 expression. And the subgroup with elevated DLX6-AS1 expression had shorter OS time (P<0.05) and was remarkably related with the TNM stage (P=0.006), lymphatic metastasis (P=0.038), and tumor size (P=0.033) [22]. A similar conclusion was made in patients with gastric cancer. The findings indicated that DLX6-AS1 may be a promising biomarker because its expression was increased in late-stage gastric cancer samples compared with early-stage samples. In addition, upregulated DLX6-AS1 was correlated with distant metastasis, lymph node metastasis, advanced clinical stage, and poor prognosis [36,38]. Furthermore, accumulating evidence has demonstrated that the level of DLX6-AS1 in NSCLC samples is positively related to TNM stage and tumor size [20,27].
Collectively, these results imply that DLX6-AS1 is an oncogene whose expression is markedly related with tumor progression, which may be helpful to predict the prognosis in various cancers.
Expression and function of lncRNA DLX6-AS1 in different cell lines
Many studies have explored the expression of lncRNA DLX6-AS1 in different types of cell lines. These studies have found that silencing or forced overexpression of DLX6-AS1 affected cell proliferation, migration, invasion, and apoptosis in various cancers. Specifically, expression of this lncRNA affects numerous interconnected signaling pathways, including the mTOR, Wnt/β-catenin, TGF-β, and PI3K/AKT pathways. In the following sections, we summarize the role of DLX6-AS1 in various cancer cell lines.
Lung cancer
Lung cancer is now the most common malignancy and the leading cause of tumor-related death worldwide [51,52]. LncRNAs are novel regulatory molecules in lung cancer development and progression [53-56]. A growing number of findings have shown that DLX6-AS1 affects biological processes of the tumor cell. Sun et al. revealed that DLX6-AS1 is highly expressed in NSCLC cells and its downregulation suppresses NSCLC cell clone formation, migration, and invasion through the miR-27b-3p/GSPT1 axis [20]. Consistent with these results, Huang et al. demonstrated that silencing DLX6-AS1 decreased NSCLC cell proliferation, migration, invasion, and pro-apoptosis through regulating expression of miR-144 and PRR11 genes. In addition, Zhang et al. suggested that DLX6-AS1 expression is increased in NSCLC cell lines and that DLX6-AS1 knockdown suppresses cell proliferation and migration [27].
Hepatocellular carcinoma
Studies have reported that DLX6-AS1 is elevated in HCC cell lines compared with a normal human liver cell line. Downregulated DLX6-AS1 suppresses HCC cell proliferation, migration, and invasion. Moreover, bioinformatics analysis revealed miR-203a potentially targets DLX6-AS1 3’UTR, and miR-203a targets MMP-2 during tumorigenesis [30]. Li et al. demonstrated that DLX6-AS1 could regulate miR-424-5p, and oncogene WEE1 (G2 checkpoint kinase) expression is the target of miR-422-5p and associated with DLX6-AS1 expression [32]. Furthermore, in liver cancer stem cells, silencing lncRNA DLX6-AS1 impaired the stem cell properties of liver cancer stem cells by limiting the methylation of CADM1 promoter and activating the STAT3 signaling pathway [31].
Gastric cancer
In gastric cancer cells, functional analysis had showed that highly expressed DLX6-AS1 is obviously related with the enhancement of cell proliferation, migration, invasion, EMT, and apoptotic induction. Regarding the molecular mechanism, Qian et al. verified that DLX6-AS1 knockdown inhibits aerobic glycolysis but promotes mitochondrial respiration in gastric cancer cells by targeting miR-4290 and 3-phosphoinositide-dependent protein kinase 1 [35]. Liang et al. demonstrated that DLX6-AS1 serves as a competing endogenous RNA through targeting miR-204-5P and increasing OCT1 expression [36]. Meanwhile, Wu et al. reported that DLX6-AS1 regulates gastric cancer cell progression through the DLX6-AS1/FUS/MAP4K1 axis [37].
Esophageal squamous cell carcinoma
The role and potential mechanism of DLX6-AS1 in squamous cell carcinoma has rarely been assessed. The expression of DLX6-AS1 was upregulated in esophageal squamous cell carcinoma cell lines, as shown by qRT-PCR [57]. Functional analysis revealed that inhibiting DLX6-AS1 significantly attenuates cell proliferation, migration, and invasion, yet enhances apoptosis [57,58].
Colorectal cancer
Some research groups have reported that DLX6-AS1 is obviously increased in colorectal cancer cells relative to normal human colorectal epithelial cell lines and that silencing DLX6-AS1 can block the malignant characteristics of colorectal cancer cells. Mechanistically, Kong et al. showed that DLX6-AS1 targets miR-26a and miR-26a upregulates the expression of EZH2 [34]. Additional evidence indicated that DLX6-AS1 promotes colorectal cancer cell malignant characteristics through the PI3K/AKT pathway [33].
Breast cancer
In breast cancer cells, the DLX6 expression was enhanced in cell lines. DLX6-AS1 knockdown increased cell apoptosis and suppressed cell proliferation and EMT. Moreover, dual luciferase reporter and RNA pull-down assays revealed that DLX6-AS1 interacts with miR-199b-5p modulating the expression of paxillin [39]. Meanwhile, another study found that DLX6-AS1 promotes breast cancer malignant characteristics through the miR-505-3p/RUNX2 axis [21].
Other cancers
In cervical cancer cells, elevated DLX6-AS1 expression is significantly related with cell proliferation, EMT, migration, and anti-apoptosis [59]. One study reported that DLX6-AS1 increases the cellular process via sponging miR-16-5p, which targets the downstream gene ARPP19 [60]. Meanwhile, some evidence was uncovered that DLX6-AS1 is much higher in epithelial ovarian cancer cells, and downregulation of this lncRNA weakened cells’ malignant characteristics via targeting miR-613 or regulating the Notch signaling pathway [50,61].
Studies have showed a promotive role for DLX6-AS1 in nasopharyngeal carcinoma [40], osteosarcoma [42], thyroid cancer [43], bladder cancer [45], renal cell carcinoma [46], pancreatic cancer [47], and other cancers. Table 2 summarizes the evidence concerning its expression, functional analyses, and underlying mechanism of DLX6-AS1 in diverse malignant cancers.
Table 2.
Expression pattern and function of DLX6-AS1 in cell lines and animal models
| Cancer types | Assessed cancer cell lines | Expression | Related genes and pathways | Biological significance | Reference |
|---|---|---|---|---|---|
| Lung cancer | CALU3, CALU6, A549, H1229 | up | miR-27b-3p, GSPT1 | proliferation, migration, invasion | [20] |
| NSCLC A549, H1299, 95D | up | / | proliferation, migration | [27] | |
| H1975, A549 | up | miR-144, PRR11 | proliferation, migration, invasion, anti-apoptosis | [29] | |
| A549, H1650 | up | / | / | [28] | |
| Nasopharyngeal carcinoma (NPC) | S26, CNE-1, CNE-2, HONE-1, 5-8F | up | miR-199a-5p, HIF-1α | proliferation, migration, invasion | [40] |
| Hepatocellular carcinoma (HCC) | SMMC-7721, HCCLM3, Hep3B, HepG2, Huh7 | up | CADM1, STAT3 signaling pathway | self-renewal, proliferation, proliferation | [31] |
| MHCC97L, HCCLM3, SK-HEP-1, Hep3B, Huh7 | up | miR-424-5p, WEE1 | proliferation, migration, invasion | [32] | |
| HepG2, HCCLM3 | up | miR-203a, MMP-2 | proliferation, migration, invasion | [30] | |
| Cervical cancer (CC) | SiHa, HeLa, C-33A, CaSki | up | miR-16-5p, ARPP19 | proliferation, migration, EMT, anti-apoptosis | [60] |
| HeLa, SiHa, C4-1, C-33a | up | FUS | proliferation, invasion | [59] | |
| Osteosarcoma | MG63 and U2OS | up | miR-129-5p, DLK1, Wnt signaling | stemness | [42] |
| Ewing’s sarcoma | SK-ES-1, A673, RD-ES, MSCs | up | miR-124-3p, CDK4 | proliferation, anti-apoptosis | [41] |
| Laryngeal cancer | HEp-2, Tu-177 | up | miR-26a, TRPC3 axis | proliferation, mitochondrial metabolism | [62] |
| Thyroid cancer | TPC-1, K1, SW579 | up | UPF1 | migration, invasion | [43] |
| Colorectal Cancer | DLD-1, HCT-116, HT-29, SW480, SW620 | up | miR-26a, EZH2 | proliferation, cell cycle, migration, invasion | [34] |
| HCT116, HT-29, SW480 | up | PI3K, AKT, mTOR pathway | proliferation, invasion, migration, apoptosis | [33] | |
| Epithelial ovarian cancer (EOC) | IOSE80, HEY, SKOV3, OVCAR-3 | up | Notch signaling pathway | proliferation, migration, invasion, cell cycle, anti-apoptosis | [50] |
| Ovarian cancer | A2780, SKOV3, OVCAR-3 | up | miR-613 | cell migration, invasion | [60] |
| Bladder cancer | T24, SW780 | up | miR-223, HSP90B1 | proliferation, invasion | [44] |
| 5637, J82 and T24 | up | Wnt/β-catenin signaling pathway | proliferation, invasion, migration, EMT | [45] | |
| Triple-negative breast cancer (TNBC) | HCC1599, MDA-MB-231, HCC1806, HS578 T | up | miR-199b-5p, paxillin | proliferation, EMT, anti-apoptosis | [39] |
| Breast cancer | MDA-MB-231, MCF-7, MDA-MB-468, T47D, BT-474 | up | miR-505-3p, RUNX2 | proliferation, invasion, migration, anti-apoptosis | [21] |
| Glioma carcinogenesis | U251, U87MG, T98G, SHG44 | up | miR-197-5p, E2F1 | proliferation, invasion | [63] |
| Neuroblastoma | NB-1643, NB-1691, SK-N-AS, IMR-32, SH-SY5Y, SK-N-SH | up | miR107, BDNF | growth, invasion, metastasis, differentiation | [48] |
| SK-N-SH, SH-SY5Y, SK-N-AS, SK-N-BE | up | miR-497-5p, YAP1 | proliferation, migration, invasion ability, EMT | [49] | |
| Renal cell carcinoma | A498, ACHN, Caki-1, Caki-2, 786-O, G401 | up | miR-26a, PTEN | proliferation, cell cycle, anti-apoptosis | [46] |
| Esophageal squamous cell carcinoma (ESCC) | Eca109, Ec9706, TE-1, TE-10, TE-11, KYSE-520 | up | / | proliferation, invasion | [58] |
| EC109, KYSE30 | up | proliferation, anti-apoptosis, invasion | [57] | ||
| Gastric Cancer | HGC-27, MGC803, SGC7901, MKN45 | up | miR-4290, PDK1 | anti-apoptosis, proliferation, glucose metabolism | [35] |
| MKN-7, MKN-28, MGC-803, HGC-27, MKN-45, AGS, SGC-7901 | up | miR-204-5p, OCT1 | proliferation, migration, invasion, EMT | [36] | |
| AGS, HGC-27, SGC-7901, BGC-823 | up | FUS, MAP4K1 | proliferation, migration, EMT | [37] | |
| HGC27, BGC823, SGC7901, AGS | up | / | proliferation, cell cycle, migration, invasion, EMT | [38] | |
| Pancreatic cancer | CAPAN-1, BxPC-3, SW 1990, PANC-1 | up | miR181b | proliferation, migration, invasion | [47] |
| Panc-1, Bxpc-3, AsPC-1, Capan-1, CFPAC-1, MIA PaCa-2 | up | miR-497-5p, FZD4, FZD6, Wnt/β-catenin | proliferation, invasion, migration, anti-apoptosis | [64] |
Studies of lncRNA DLX6-AS1 in animal models
Studies have explored the effects of DLX6-AS1 knockdown or overexpression in tumor-bearing nude mice. Regarding NSCLC, nude mice were injected with NSCLC cells transfected with shDLX6-AS1 or shNC. The results showed that tumors with shDLX6-AS1 obviously decreased than those with shNC (P<0.001). In addition, the Ki67 and GSPT1 expression of tumor tissues were lower in the shDlX6-AS1 subgroup than in the control group [20]. Regarding HCC, Sun et al. found that DLX6-AS1 knockdown mice had remarkably reduced tumor size after being subcutaneously implanted with HCC cells, including HCCLM3 cells and HepG2 cells [30]. Moreover, a signal study investigated the effect of DLX6-AS1 in laryngeal tumor-bearing mice model, silencing DLX6-AS1 promoted mitochondrial metabolism but inhibited the IRPC3 expression [62]. Researchers also concluded that DLX6-AS1 was a tumor-promoting lncRNA in breast cancer [39], glioma carcinogenesis [63], and pancreatic cancer [47,64] using xenografted nude mice.
All evidence indicates that DLX6-AS1 facilitates disease progression but silencing DLX6-AS1 can suppress tumor growth in several cancers. Table 3 summarizes the studies of silencing or overexpressing DLX1-AS1 in animal models.
Table 3.
Assessed function of DXL6-AS1 in animal models
| Cancer | Model type | Know-down DXL6-AS | Reference |
|---|---|---|---|
| Lung cancer | male BALB/c nude mice | suppressed tumor growth | [20] |
| male BALB/c nude mice | suppressed tumor growth | [29] | |
| Hepatocellular carcinoma (HCC) | NOD-SCID mice | suppressed tumor growth | [31] |
| male BALB/c nude mice | suppressed tumor growth | [30] | |
| Cervical cancer (CC) | BALB/c nude mice | suppressed tumor growth | [60] |
| NOD/SCID mice | suppressed tumor growth and lung metastatic | [59] | |
| Osteosarcoma | male BALB/c nude mice | suppressed tumor formation | [42] |
| Laryngeal cancer | male BALB/c nude mice | suppressed tumor growth | [62] |
| Thyroid cancer | NOD/SCID mice | suppressed lung metastatic | [43] |
| Bladder cancer | BALB/c nude mice | suppressed tumor growth | [44] |
| Triple-negative breast cancer (TNBC) | BALB/c nude | suppressed tumor growth | [39] |
| Glioma carcinogenesis | male nude mic | suppressed tumor growth | [63] |
| Neuroblastoma | male BALB/c athymic nude mice | suppressed tumor growth | [48] |
| female nude BALB/c mice | suppressed tumor growth | [49] | |
| Pancreatic cancer | female BABL/c athymic nude mice | suppressed tumor growth and metastasis | [47] |
| BALB/c nude mice | suppressed tumor growth and metastasis | [64] |
Conclusions
As a novel class of ncRNAs, lncRNAs have attracted much attention about their crucial regulation function, such as affecting cancer cell proliferation, apoptosis, invasion, migration, and drug sensitivity [65]. LncRNA DLX6-AS1, a tumor-promoting gene, has pleiotropic effects in several cancers. Notably, the expression levels of DLX6-AS1 in tumor samples were remarkably elevated than normal samples, as assessed by qRT-PCR and immunohistochemical analysis. This observation emphasized that aberrant expression of DLX6-AS1 does not depend on the tissue type. Through further analysis of the relationship within the expression level of DLX6-AS1 and clinicopathological characters in patients with different kinds of cancers, we found that DLX6-AS1 is strongly connected with advanced TNM stage. Together with Kaplan-Meier analyses, univariate cox regression analysis, and multivariate regression analysis, we concluded that this lncRNA is an independent risk factor and may be a promising biomarker for more than a dozen human cancers, including NSCLC, HCC, nasopharyngeal carcinoma, osteosarcoma, renal cell carcinoma, and cancers of the thyroid, bladder, colon, stomach, breast, and pancreas.
Specifically, this lncRNA was increased in tumor cells. Furthermore, using gene knockdown and overexpression technology, it was demonstrated that this lncRNA participates in diverse cancer biological processes, such as metastasis, and apoptosis. In terms of molecular mechanisms, the oncogenic function of DLX6-AS1 is complicated due to its ability to regulate the target multiple genes expression and interlinking signaling pathways related with various cancers (Figure 1). Nude mice were subcutaneously implanted with tumor cells transfected with shDLX6-AS1, overexpressed DLX6-AS1, or shNC to assess the function of DLX6-AS1 in vivo. Taken together, current evidence indicates that DLX6-AS1 is an oncogene and interfering with this lncRNA can impair the tumor growth of several cancers (Figure 2).
Figure 1.
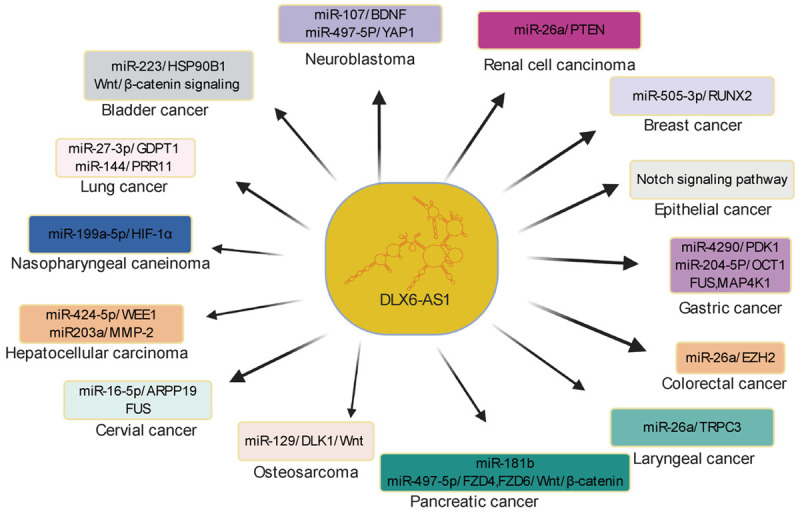
Schematic representation showing interaction between DLX6-AS1 and target miRNA or/and interlinking signaling pathways in various cancers.
Figure 2.
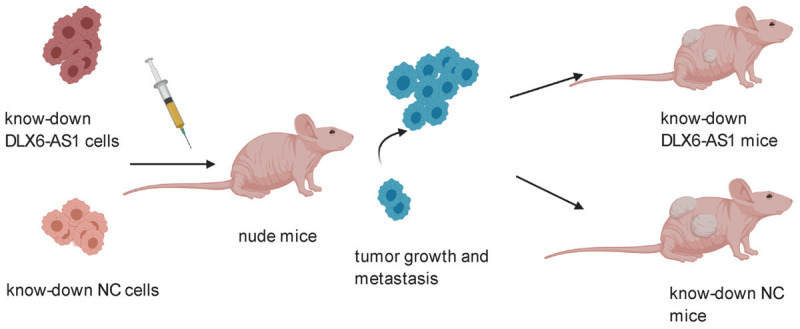
Knock-down DLX6-AS1 suppress the rumor growth in animal models.
The diversity of involved miRNAs, target genes, and signaling pathways even within one cancer type suggests the complexity of human cancer. Based on the literature presented herein, the findings uncover a critical role for DLX6-AS1 in cancers. Importantly, the central role of DXL6-AS1 is an essential shared character in almost all cancer types and indicates to the enormous possibility of DXL6-AS1 as a target in cancer therapy. Considering the complicated mechanism of DXL6-AS1 promote tumor development, targeting this gene may also induce toxicity to normal human tissues. The detailed regulatory mechanisms including upstream and downstream molecules remain to be systematically researched. Extensive research concluded that the expression level of DXL6-AS1 could serve as a prognostic indicator because of its high expression in tumor tissues related with poorer prognosis and clinicopathological parameters, but the expression level of DXL6-AS1 in easily obtained human samples (for example, plasma) have not been clearly explored. Thus, ongoing efforts to explore the expression level of this lncRNA in human samples through the method of liquid biopsies and to clarify the underlying mechanisms promise that DXL6-AS1 will ultimately reach the clinic. We are optimistic that this review will contribute to better understanding of DLX6-AS1 and its relationship with a variety of cancers to act as a stepping stone for future clinical application.
Acknowledgements
This study is funded by the National Natural Science Foundation of China (81790631, 81570512 and 81672422), the National Key Research and Development Program of China (2018YFC2000500), the Natural Science Foundation of Zhejiang Province in China (LQ19H030007), Zhejiang Province Health Department Program (2021KY657), and Dr. Xue was supported by Zhejiang University Academic Award for Outstanding Doctoral Candidates (2020055).
Disclosure of conflict of interest
None.
References
- 1.Wang J, Sheng Z, Cai Y. Effects of microRNA-513b on cell proliferation, apoptosis, invasion, and migration by targeting HMGB3 through regulation of mTOR signaling pathway in non-small-cell lung cancer. J Cell Physiol. 2019;234:10934–10941. doi: 10.1002/jcp.27921. [DOI] [PubMed] [Google Scholar]
- 2.Ao R, Guan L, Wang Y, Wang JN. Silencing of COL1A2, COL6A3, and THBS2 inhibits gastric cancer cell proliferation, migration, and invasion while promoting apoptosis through the PI3k-Akt signaling pathway. J Cell Biochem. 2018;119:4420–4434. doi: 10.1002/jcb.26524. [DOI] [PubMed] [Google Scholar]
- 3.He YM, Zhang ZL, Liu QY, Xiao YS, Wei L, Xi C, Nan X. Effect of CLIC1 gene silencing on proliferation, migration, invasion and apoptosis of human gallbladder cancer cells. J Cell Mol Med. 2018;22:2569–2579. doi: 10.1111/jcmm.13499. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Vasan N, Toska E, Scaltriti M. Overview of the relevance of PI3K pathway in HR-positive breast cancer. Ann Oncol. 2019;30(Suppl 10):x3–x11. doi: 10.1093/annonc/mdz281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL, Paz-Ares L. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 6.Shah SC, McKinley M, Gupta S, Peek RM Jr, Martinez ME, Gomez SL. Population-based analysis of differences in gastric cancer incidence among races and ethnicities in individuals age 50 years and older. Gastroenterology. 2020;159:1705–1714. e2. doi: 10.1053/j.gastro.2020.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schürch CM, Bhate SS, Barlow GL, Phillips DJ, Noti L, Zlobec I, Chu P, Black S, Demeter J, McIlwain DR, Samusik N, Goltsev Y, Nolan GP. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell. 2020;182:1341–1359. e1319. doi: 10.1016/j.cell.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xue C, Zhao Y, Jiang J, Li L. Expression levels of lncRNAs are prognostic for hepatocellular carcinoma overall survival. Am J Transl Res. 2020;12:1873–1883. [PMC free article] [PubMed] [Google Scholar]
- 9.GBD 2015 Mortality and Causes of Death Collaborators. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1459–1544. doi: 10.1016/S0140-6736(16)31012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 11.Anastasiadou E, Faggioni A, Trivedi P, Slack FJ. The nefarious nexus of noncoding RNAs in cancer. Int J Mol Sci. 2018;19:2072. doi: 10.3390/ijms19072072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei L, Wang X, Lv L, Zheng Y, Zhang N, Yang M. The emerging role of noncoding RNAs in colorectal cancer chemoresistance. Cell Oncol (Dordr) 2019;42:757–768. doi: 10.1007/s13402-019-00466-8. [DOI] [PubMed] [Google Scholar]
- 13.Choudhari R, Sedano MJ, Harrison AL, Subramani R, Lin KY, Ramos EI, Lakshmanaswamy R, Gadad SS. Long noncoding RNAs in cancer: from discovery to therapeutic targets. Adv Clin Chem. 2020;95:105–147. doi: 10.1016/bs.acc.2019.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarfi M, Abbastabar M, Khalili E. Long noncoding RNAs biomarker-based cancer assessment. J Cell Physiol. 2019;234:16971–16986. doi: 10.1002/jcp.28417. [DOI] [PubMed] [Google Scholar]
- 16.Mirhosseini SA, Sarfi M, Samavarchi Tehrani S, Mirazakhani M, Maniati M, Amani J. Modulation of cancer cell signaling by long noncoding RNAs. J Cell Biochem. 2019;120:12224–12246. doi: 10.1002/jcb.28847. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Wang Z, Wu J, Ma R, Feng J. Long noncoding RNAs predict the survival of patients with colorectal cancer as revealed by constructing an endogenous RNA network using bioinformation analysis. Cancer Med. 2019;8:863–873. doi: 10.1002/cam4.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long J, Bai Y, Yang X, Lin J, Yang X, Wang D, He L, Zheng Y, Zhao H. Construction and comprehensive analysis of a ceRNA network to reveal potential prognostic biomarkers for hepatocellular carcinoma. Cancer Cell Int. 2019;19:90. doi: 10.1186/s12935-019-0817-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun W, Zhang L, Yan R, Yang Y, Meng X. LncRNA DLX6-AS1 promotes the proliferation, invasion, and migration of non-small cell lung cancer cells by targeting the miR-27b-3p/GSPT1 axis. Onco Targets Ther. 2019;12:3945–3954. doi: 10.2147/OTT.S196865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao P, Guan H, Dai Z, Ma Y, Zhao Y, Liu D. Long noncoding RNA DLX6-AS1 promotes breast cancer progression via miR-505-3p/RUNX2 axis. Eur J Pharmacol. 2019;865:172778. doi: 10.1016/j.ejphar.2019.172778. [DOI] [PubMed] [Google Scholar]
- 22.Shen SN, Li K, Liu Y, Yang CL, He CY, Wang HR. Down-regulation of long noncoding RNA PVT1 inhibits esophageal carcinoma cell migration and invasion and promotes cell apoptosis via microRNA-145-mediated inhibition of FSCN1. Mol Oncol. 2019;13:2554–2573. doi: 10.1002/1878-0261.12555. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Parolia A, Venalainen E, Xue H, Mather R, Lin D, Wu R, Pucci P, Rogalski J, Evans JR, Feng F, Collins CC, Wang Y, Crea F. The long noncoding RNA HORAS5 mediates castration-resistant prostate cancer survival by activating the androgen receptor transcriptional program. Mol Oncol. 2019;13:1121–1136. doi: 10.1002/1878-0261.12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klec C, Prinz F, Pichler M. Involvement of the long noncoding RNA NEAT1 in carcinogenesis. Mol Oncol. 2019;13:46–60. doi: 10.1002/1878-0261.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H, Han L, Liu Z, Gao N. Long noncoding RNA MNX1-AS1 contributes to lung cancer progression through the miR-527/BRF2 pathway. J Cell Physiol. 2019;234:13843–13850. doi: 10.1002/jcp.28064. [DOI] [PubMed] [Google Scholar]
- 26.Zhang G, He X, Ren C, Lin J, Wang Q. Long noncoding RNA PCA3 regulates prostate cancer through sponging miR-218-5p and modulating high mobility group box 1. J Cell Physiol. 2019;234:13097–13109. doi: 10.1002/jcp.27980. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Guo H, Bao Y, Yu H, Xie D, Wang X. Exosomal long non-coding RNA DLX6-AS1 as a potential diagnostic biomarker for non-small cell lung cancer. Oncol Lett. 2019;18:5197–5204. doi: 10.3892/ol.2019.10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Li P, Zhao W, Yang R, Chen S, Bai Y, Dun S, Chen X, Du Y, Wang Y, Zang W, Zhao G, Zhang G. Expression of long non-coding RNA DLX6-AS1 in lung adenocarcinoma. Cancer Cell Int. 2015;15:48. doi: 10.1186/s12935-015-0201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang Y, Ni R, Wang J, Liu Y. Knockdown of lncRNA DLX6-AS1 inhibits cell proliferation, migration and invasion while promotes apoptosis by downregulating PRR11 expression and upregulating miR-144 in non-small cell lung cancer. Biomed Pharmacother. 2019;109:1851–1859. doi: 10.1016/j.biopha.2018.09.151. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L, He X, Jin T, Gang L, Jin Z. Long non-coding RNA DLX6-AS1 aggravates hepatocellular carcinoma carcinogenesis by modulating miR-203a/MMP-2 pathway. Biomed Pharmacother. 2017;96:884–891. doi: 10.1016/j.biopha.2017.10.056. [DOI] [PubMed] [Google Scholar]
- 31.Wu DM, Zheng ZH, Zhang YB, Fan SH, Zhang ZF, Wang YJ, Zheng YL, Lu J. Down-regulated lncRNA DLX6-AS1 inhibits tumorigenesis through STAT3 signaling pathway by suppressing CADM1 promoter methylation in liver cancer stem cells. J Exp Clin Cancer Res. 2019;38:237. doi: 10.1186/s13046-019-1239-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Li D, Tang X, Li M, Zheng Y. Long noncoding RNA DLX6-AS1 promotes liver cancer by increasing the expression of WEE1 via targeting miR-424-5p. J Cell Biochem. 2019;120:12290–12299. doi: 10.1002/jcb.28493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang JJ, Xu WR, Chen B, Wang YY, Yang N, Wang LJ, Zhang YL. The up-regulated lncRNA DLX6-AS1 in colorectal cancer promotes cell proliferation, invasion and migration via modulating PI3K/AKT/mTOR pathway. Eur Rev Med Pharmacol Sci. 2019;23:8321–8331. doi: 10.26355/eurrev_201910_19143. [DOI] [PubMed] [Google Scholar]
- 34.Kong WQ, Liang JJ, Du J, Ye ZX, Gao P, Liang YL. Long noncoding RNA DLX6-AS1 regulates the growth and aggressiveness of colorectal cancer cells via mediating miR-26a/EZH2 axis. Cancer Biother Radiopharm. 2020 doi: 10.1089/cbr.2020.3589. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 35.Qian Y, Song W, Wu X, Hou G, Wang H, Hang X, Xia T. DLX6 antisense RNA 1 modulates glucose metabolism and cell growth in gastric cancer by targeting microRNA-4290. Dig Dis Sci. 2020 doi: 10.1007/s10620-020-06223-4. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 36.Liang Y, Zhang CD, Zhang C, Dai DQ. DLX6-AS1/miR-204-5p/OCT1 positive feedback loop promotes tumor progression and epithelial-mesenchymal transition in gastric cancer. Gastric Cancer. 2020;23:212–227. doi: 10.1007/s10120-019-01002-1. [DOI] [PubMed] [Google Scholar]
- 37.Wu Q, Ma J, Meng W, Hui P. DLX6-AS1 promotes cell proliferation, migration and EMT of gastric cancer through FUS-regulated MAP4K1. Cancer Biol Ther. 2020;21:17–25. doi: 10.1080/15384047.2019.1647050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu X, Tian Y, Kuang W, Wen S, Guo W. Long non-coding RNA DLX6-AS1 silencing inhibits malignant phenotypes of gastric cancer cells. Exp Ther Med. 2019;17:4715–4722. doi: 10.3892/etm.2019.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du C, Wang Y, Zhang Y, Zhang J, Zhang L, Li J. LncRNA DLX6-AS1 contributes to epithelial-mesenchymal transition and cisplatin resistance in triple-negative breast cancer via modulating mir-199b-5p/paxillin axis. Cell Transplant. 2020;29:963689720929983. doi: 10.1177/0963689720929983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang B, Jia L, Ren H, Jin C, Ren Q, Zhang H, Hu D, Zhang H, Hu L, Xie T. LncRNA DLX6-AS1 increases the expression of HIF-1α and promotes the malignant phenotypes of nasopharyngeal carcinoma cells via targeting miR-199a-5p. Mol Genet Genomic Med. 2020;8:e1017. doi: 10.1002/mgg3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lei X, Yang S, Yang Y, Zhang J, Wang Y, Cao M. Long noncoding RNA DLX6-AS1 targets miR-124-3p/CDK4 to accelerate Ewing’s sarcoma. Am J Transl Res. 2019;11:6569–6576. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Zhang RM, Tang T, Yu HM, Yao XD. LncRNA DLX6-AS1/miR-129-5p/DLK1 axis aggravates stemness of osteosarcoma through Wnt signaling. Biochem Biophys Res Commun. 2018;507:260–266. doi: 10.1016/j.bbrc.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 43.Zhong ZB, Wu YJ, Luo JN, Hu XN, Yuan ZN, Li G, Wang YW, Yao GD, Ge XF. Knockdown of long noncoding RNA DLX6-AS1 inhibits migration and invasion of thyroid cancer cells by upregulating UPF1. Eur Rev Med Pharmacol Sci. 2019;23:10867–10873. doi: 10.26355/eurrev_201912_19790. [DOI] [PubMed] [Google Scholar]
- 44.Fang C, Xu L, He W, Dai J, Sun F. Long noncoding RNA DLX6-AS1 promotes cell growth and invasiveness in bladder cancer via modulating the miR-223-HSP90B1 axis. Cell Cycle. 2019;18:3288–3299. doi: 10.1080/15384101.2019.1673633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo J, Chen Z, Jiang H, Yu Z, Peng J, Xie J, Li Z, Wu W, Cheng Z, Xiao K. The lncRNA DLX6-AS1 promoted cell proliferation, invasion, migration and epithelial-to-mesenchymal transition in bladder cancer via modulating Wnt/β-catenin signaling pathway. Cancer Cell Int. 2019;19:312. doi: 10.1186/s12935-019-1010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng X, Hu Z, Ke X, Tang H, Wu B, Wei X, Liu Z. Long noncoding RNA DLX6-AS1 promotes renal cell carcinoma progression via miR-26a/PTEN axis. Cell Cycle. 2017;16:2212–2219. doi: 10.1080/15384101.2017.1361072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.An Y, Chen XM, Yang Y, Mo F, Jiang Y, Sun DL, Cai HH. LncRNA DLX6-AS1 promoted cancer cell proliferation and invasion by attenuating the endogenous function of miR-181b in pancreatic cancer. Cancer Cell Int. 2018;18:143. doi: 10.1186/s12935-018-0643-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang HY, Xing MQ, Guo J, Zhao JC, Chen X, Jiang Z, Zhang H, Dong Q. Long noncoding RNA DLX6-AS1 promotes neuroblastoma progression by regulating miR-107/BDNF pathway. Cancer Cell Int. 2019;19:313. doi: 10.1186/s12935-019-0968-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li C, Wang S, Yang C. Long non-coding RNA DLX6-AS1 regulates neuroblastoma progression by targeting YAP1 via miR-497-5p. Life Sci. 2020;252:117657. doi: 10.1016/j.lfs.2020.117657. [DOI] [PubMed] [Google Scholar]
- 50.Zhao J, Liu HR. Down-regulation of long noncoding RNA DLX6-AS1 defines good prognosis and inhibits proliferation and metastasis in human epithelial ovarian cancer cells via notch signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:3243–3252. doi: 10.26355/eurrev_201904_17684. [DOI] [PubMed] [Google Scholar]
- 51.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 52.Arbour KC, Riely GJ. Systemic therapy for locally advanced and metastatic non-small cell lung cancer: a review. JAMA. 2019;322:764–774. doi: 10.1001/jama.2019.11058. [DOI] [PubMed] [Google Scholar]
- 53.Sun CC, Zhu W, Li SJ, Hu W, Zhang J, Zhuo Y, Zhang H, Wang J, Zhang Y, Huang SX, He QQ, Li DJ. FOXC1-mediated LINC00301 facilitates tumor progression and triggers an immune-suppressing microenvironment in non-small cell lung cancer by regulating the HIF1α pathway. Genome Med. 2020;12:77. doi: 10.1186/s13073-020-00773-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gandhi M, Groß M, Holler JM, Coggins SA, Patil N, Leupold JH, Munschauer M, Schenone M, Hartigan CR, Allgayer H, Kim B, Diederichs S. The lncRNA lincNMR regulates nucleotide metabolism via a YBX1 - RRM2 axis in cancer. Nat Commun. 2020;11:3214. doi: 10.1038/s41467-020-17007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen J, Liu A, Wang Z, Wang B, Chai X, Lu W, Cao T, Li R, Wu M, Lu Z, Pang W, Xiao L, Chen X, Zheng Y, Chen Q, Zeng J, Li J, Zhang X, Ren D, Huang Y. LINC00173. v1 promotes angiogenesis and progression of lung squamous cell carcinoma by sponging miR-511-5p to regulate VEGFA expression. Mol Cancer. 2020;19:98. doi: 10.1186/s12943-020-01217-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Z, Chen X, Lu B, Gu Y, Chen Q, Lei T, Nie F, Gu J, Huang J, Wei C, Sun M, Wang Z. Up-regulated LINC01234 promotes non-small-cell lung cancer cell metastasis by activating VAV3 and repressing BTG2 expression. J Hematol Oncol. 2020;13:7. doi: 10.1186/s13045-019-0842-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang M, Li Y, Yang Y, Liu X, Zang M, Li Y, Yang K, Yang W, Zhang S. Long non-coding RNA DLX6-AS1 is associated with malignant progression and promotes proliferation and invasion in esophageal squamous cell carcinoma. Mol Med Rep. 2019;19:1942–1950. doi: 10.3892/mmr.2018.9786. [DOI] [PubMed] [Google Scholar]
- 58.Tian W, Jiang C, Huang Z, Xu D, Zheng S. Comprehensive analysis of dysregulated lncRNAs, miRNAs and mRNAs with associated ceRNA network in esophageal squamous cell carcinoma. Gene. 2019;696:206–218. doi: 10.1016/j.gene.2019.02.051. [DOI] [PubMed] [Google Scholar]
- 59.Tian Y, Wang YR, Jia SH. Knockdown of long noncoding RNA DLX6-AS1 inhibits cell proliferation and invasion of cervical cancer cells by downregulating FUS. Eur Rev Med Pharmacol Sci. 2019;23:7307–7313. doi: 10.26355/eurrev_201909_18836. [DOI] [PubMed] [Google Scholar]
- 60.Xie F, Xie G, Sun Q. Long noncoding RNA DLX6-AS1 promotes the progression in cervical cancer by targeting miR-16-5p/ARPP19 axis. Cancer Biother Radiopharm. 2020;35:129–136. doi: 10.1089/cbr.2019.2960. [DOI] [PubMed] [Google Scholar]
- 61.You Q, Shi HY, Gong CF, Tian XY, Li S. Long non-coding RNA DLX6-AS1 acts as an oncogene by targeting miR-613 in ovarian cancer. Eur Rev Med Pharmacol Sci. 2019;23:6429–6435. doi: 10.26355/eurrev_201908_18524. [DOI] [PubMed] [Google Scholar]
- 62.Liu Y, Liu X, Zhang X, Deng J, Zhang J, Xing H. lncRNA DLX6-AS1 promotes proliferation of laryngeal cancer cells by targeting the miR-26a/TRPC3 pathway. Cancer Manag Res. 2020;12:2685–2695. doi: 10.2147/CMAR.S237181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li X, Zhang H, Wu X. Long noncoding RNA DLX6-AS1 accelerates the glioma carcinogenesis by competing endogenous sponging miR-197-5p to relieve E2F1. Gene. 2019;686:1–7. doi: 10.1016/j.gene.2018.10.065. [DOI] [PubMed] [Google Scholar]
- 64.Yang J, Ye Z, Mei D, Gu H, Zhang J. Long noncoding RNA DLX6-AS1 promotes tumorigenesis by modulating miR-497-5p/FZD4/FZD6/Wnt/β-catenin pathway in pancreatic cancer. Cancer Manag Res. 2019;11:4209–4221. doi: 10.2147/CMAR.S194453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]


