Abstract
This study aimed to investigate factors affecting coronavirus disease 2019 (COVID-19) progression, also to explore the clinical features and prognosis of nervous system symptom (NSS) involved COVID-19 patients. 417 COVID-19 patients were analyzed in this retrospective study, and they were clinically classified as severe patients and non-severe patients. According to NSS involved status, COVID-19 patients were further divided into NSS patients and non-NSS patients. Elderly cases, males, common comorbidities, NSS, respiratory/cardiovascular/gastrointestinal symptoms, bilateral lesion, multifocal lesion, bacterial infection, bacterial&fungal infection were more common in severe patients compared to non-severe patients. Meanwhile, severe COVID-19 patients showed increased baseline APTT, TT, D-dimer, CRP, ESR, CK-MB, creatine kinase, AST, ALT, creatinine, but decreased baseline platelet level, lymphocyte, albumin, GFR compared to non-severe patients. Notably, the continuous differences of lymphocyte, D-dimer, CRP, AST, ALT, albumin, GFR between severe patients and non-severe patients during treatment were observed. Age, NSS, bacterial & fungal infection, CRP and creatinine were further identified as independent risk factors for severe COVID-19, which could predict severe COVID-19 with area under curve of 0.861. Furthermore, severe patients presented with worse prognosis. Regrading NSS patients, they were related to older age, surgery history, diabetes comorbidities, respiratory/cardiovascular/gastrointestinal symptoms, bilateral lesion, multifocal lesion, bacterial infection, bacterial&fungal infection and more dysregulated laboratory indexes compared to non-NSS patients. Besides, NSS patients were correlated with poor prognosis to some extent. More intensive attention should be paid to COVID-19 patients with severe-disease risk factors and those with NSS involvement, in case of rapid deterioration.
Keywords: Coronavirus disease 2019, disease severity, nervous system symptom, disease progression predicting model, laboratory indexes, prognosis
Introduction
The coronaviruses are a class of viruses that have crossed species barriers to become human pathogens; there are seven identified human coronaviruses derived from animal reservoirs (such as bats, mice or even domestic animals) [3]. Most human coronaviruses have been reported to correlate with mild illness (e.g., common cold), whereas severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) may result in severe respiratory tract infections, acute respiratory distress syndrome (ARDS) and even death [1]. Currently, the ongoing outbreak caused by a novel viral pneumonia named coronavirus disease 2019 (COVID-19) in humans has raised acute and grave global concern; this outbreak has been declared a global public health emergency by the World Health Organization [2]. The causative pathogen of COVID-19 has been identified as a novel β-coronavirus, named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), by the International Committee on Taxonomy of Viruses [3]. SARS-CoV-2 is one of the members of the coronavirus family; it has a round, elliptic or pleomorphic shape with a diameter of 60-140 nm [4]. From the existing data, the genetic signature of SARS-CoV-2 is notably different from that of severe SARS-CoV and MERS-CoV, but it presents more than 85% homology with bat-SL-CoVZC45 [4]. In addition, the reproduction number (R0) or transmission rate of SARS-CoV-is 2.24-3.58; by contrast, the R0 of 2009 influenza virus A antigen (H1N1) seasonal influenza is 1.46-1.48 [1,5]. Accordingly, the spread rate of novel SARS-CoV-2 is relatively rapid.
COVID-19 is characterized by rapid spread with major transmission through respiratory droplets as well as fomites, and its incubation period ranges from 2 to 14 days [4,6]. According to the World Health Organization COVID-19 situation report, there have been a total of 4,735,622 laboratory-confirmed infections and 316,289 deaths attributed to COVID-19 worldwide, with 216 countries, areas or territories affected (updated to 19 May 2020) [2]. Regarding clinical symptoms, COVID-19 patients usually present with respiratory system symptoms (including fever, dry cough, fatigue, and abnormal chest computed tomography (CT) findings in the form of pulmonary ground glass opacity changes), while some patients may develop severe pneumonia, ARDS, septic shock, multiple organ failure, or even death [4].
From the existing data, COVID-19 is characterized by rapid progression, and some common patients are prone to progress rapidly to severe illness, critical illness or even death [4]. Therefore, exploration of risk factors predicting COVID-19 progression is necessary, though the relevant information is largely unknown. In addition, SARS-CoV-2 has been confirmed to invade the human body through the angiotensin converting enzyme 2 (ACE2) receptor, and because the ACE2 receptor is widely present in various human organs, SARS-CoV-2 may damage the respiratory system, nervous system, digestive system and other human systems [7,8]. However, most reports focus on describing the clinical characteristics of the acute respiratory system effects in COVID-19 patients, whereas limited information regarding nervous system symptoms (NSSs) in COVID-19 patients is found. Herein, the aim of this study was to investigate factors affecting COVID-19 progression, establish a predictive model for severe COVID-19, and explore the clinical features and prognosis of COVID-19 patients with NSS.
Methods
Patients
A total of 417 COVID-19 patients treated in the Third People’s Hospital of Shenzhen (Second Affiliated Hospital of Southern University of Science and Technology) from January 11, 2020, to February 27, 2020, were analyzed in this retrospective study. All patients met the diagnostic criteria issued in the 7th version of the guidelines on the Diagnosis and Treatment of COVID-19 by the National Health Commission of China (available at: http://www.nhc.gov.cn/). The nasal-pharyngeal swab specimens of all patients were collected and transported to the Shenzhen Centers for Disease Control through a biosafety transport box. SARS-CoV-2 nucleic acid detection by real-time reverse transcriptase polymerase chain reaction (RT-PCR) was carried out in the third-level Laboratory of Biosafety Protection (P3 laboratory) using a nucleic acid detection kit (Shanghai Berger Medical Technology Co., Ltd., Shanghai, China). All 417 patients were confirmed as having COVID-19 by positive results for SARS-CoV-2 nucleic acid detection. This study was performed in accordance with the regulations issued by the National Health Commission of China and the Declaration of Helsinki and was approved by the Ethics Committee of the Third People’s Hospital of Shenzhen. Verbal or written informed consent was collected from patients or their relatives.
Data collection
Clinical data of COVID-19 patients, which mainly included age, sex, blood type, history of surgery, comorbidities (hypertension, obesity, diabetes, cardiovascular disease, hepatitis B, hyperlipidemia, arteriosclerosis, syphilis), respiratory system symptoms (fever, cough, expectoration, throat pain, nasal obstruction, fear of cold), NSS (hypodynamia, muscle aches, headache, dizziness, drowsiness, coma, paralysis, hyposmia, hypogeusia, lalopathy), cardiovascular system symptoms (chest distress, shortness of breath, chest pain, palpitation, arrhythmia), gastrointestinal symptoms (diarrhea, abdominal distension, nausea, poor appetite, emesis, gastralgia, abdominal pain), lesion location, type of lesion, pathogenic microbiology, and laboratory indexes (routine blood, coagulation, inflammation indicators related to serum enzymes, enzyme index, liver function index and renal function index), were collected from the Electronic Medical Record System (EMRS) of the hospital. All medical records were documented by specially trained physicians, and the collection of clinical data required for this study was completed by three trained neurologists. In particular, the NSS were recorded after consensus was unanimously confirmed by the three neurologists. In addition, patients’ clinical outcomes up to February 27, 2020, which included the time to nucleic acid negative and prognostic outcomes (cured, improved, aggravated, dead), were collected from the Electronic Medical Record System (EMRS) of the hospital.
COVID-19 classification
According to the 7th version of the guidelines on the Diagnosis and Treatment of COVID-19 by the National Health Commission of China, COVID-19 was clinically classified as follows: (i) Mild type: the clinical symptoms were mild with no abnormal radiological findings; (ii) Common type: fever and respiratory symptoms were present and pneumonia was detected on chest computed tomography; (iii) Severe type: one of the following conditions were met: (a) respiratory distress, respiratory rate ≥30 per min; (b) oxygen saturation on quiescent condition ≤93%; (c) partial pressure of oxygen in arterial blood/fraction of inspired oxygen (PaO2/FiO2)≤300 mmHg (1 mmHg=0.133 kPa); (iv) Critical type: one of the following conditions were met: (a) respiratory failure occurred and mechanical ventilation was required; (b) shock occurred; (c) patients had other organ dysfunction needing intensive care unit monitoring and treatment. In the analysis of this study, mild and common COVID-19 were defined as non-severe COVID-19, and severe and critical COVID-19 were defined as severe COVID-19.
Statistical analysis
SAS 9.4 (SAS Institute, Inc., Cary, North Carolina, USA) was used for statistical analysis. Continuous variables were displayed as the mean with standard deviation (SD) or median with interquartile range (IQR); categorical variables were expressed as count and percentage. Comparisons of continuous variables between two groups were determined by Student’s t test or Wilcoxon rank sum test; comparisons of categorical variables between two groups were determined by chi-square test or Fisher’s exact test. Forward stepwise multivariate logistic regression analysis was carried out to screen the variables independently related to severe COVID-19, while only variables with a P value <0.05 in the univariate analysis were included. Then, a disease progression prediction model was constructed using the independent variables. The performance of the disease progression prediction model was further evaluated by receiver operating characteristic (ROC) curve analysis and derived area under the curve (AUC). A P value <0.05 was considered statistically significant.
Results
Comparison of clinical characteristics between severe patients and non-severe patients
Compared to non-severe patients, severe patients were more likely to be older aged (P<0.001) and males (P<0.001). In addition, hypertension comorbidity (P<0.001), fat comorbidity (P=0.001), diabetes comorbidity (P=0.001), respiratory system symptoms (P=0.001), NSS (P<0.001), cardiovascular system symptoms (P<0.001), gastrointestinal symptoms (P<0.001), bilateral lesions (P=0.001) and multifocal lesions (P=0.005) were more common in severe patients than in non-severe patients. However, there was no difference in duration from onset to fever (P=0.070), blood type (P=0.886) or history of surgery (P=0.193) between severe patients and non-severe patients (all P>0.05). Detailed information about the clinical characteristics is shown in Table 1.
Table 1.
Clinical characteristics of COVID-19 patients
| Characteristics | Total (N=417) | Disease severity | P value | |
|---|---|---|---|---|
|
| ||||
| Severe (n=81) | Non-severe (n=336) | |||
| Age (years), M ± SD | 45.2±17.6 | 56.3±12.5 | 42.6±17.6 | <0.001 |
| Gender, No. (%) | <0.001 | |||
| Male | 198 (47.5) | 53 (65.4) | 145 (43.2) | |
| Female | 219 (52.5) | 28 (34.6) | 191 (56.8) | |
| Duration from onset to fever (days), median (IQR) | 1.0 (1.0-2.0) | 1.0 (1.0-3.0) | 1.0 (1.0-2.0) | 0.070 |
| Blood type, No. (%) | 0.886 | |||
| A+ | 79 (18.9) | 17 (21.0) | 62 (18.5) | |
| AB+ | 43 (10.3) | 8 (9.9) | 35 (10.4) | |
| B+ | 85 (20.4) | 19 (23.4) | 66 (19.6) | |
| O+ | 78 (18.7) | 14 (17.3) | 64 (19.1) | |
| Unknown | 132 (31.7) | 23 (28.4) | 109 (32.4) | |
| History of surgery, No. (%) | 63 (15.1) | 16 (19.8) | 47 (14.0) | 0.193 |
| Comorbidities, No. (%) | ||||
| Hypertension | 74 (17.8) | 27 (33.3) | 47 (14.0) | <0.001 |
| Fat | 44 (10.6) | 17 (21.0) | 27 (8.0) | 0.001 |
| Diabetes | 27 (6.5) | 12 (14.8) | 15 (4.5) | 0.001 |
| Cardiovascular disease | 12 (2.9) | 5 (6.2) | 7 (2.1) | 0.062 |
| Hepatitis B | 12 (2.9) | 2 (2.5) | 10 (3.0) | 0.806 |
| Hyperlipidemia | 6 (1.4) | 1 (1.2) | 5 (1.5) | 0.863 |
| Arteriosclerosis | 5 (1.2) | 3 (3.7) | 2 (0.6) | 0.053 |
| Syphilis | 1 (0.2) | 0 (0.0) | 1 (0.3) | 1.000 |
| Respiratory system symptom, No. (%) | ||||
| Any | 361 (86.6) | 79 (97.5) | 282 (83.9) | 0.001 |
| Fever | 310 (74.3) | 77 (95.1) | 233 (69.3) | <0.001 |
| Cough | 215 (51.6) | 58 (71.6) | 157 (46.7) | <0.001 |
| Expectoration | 116 (27.8) | 33 (40.7) | 83 (24.7) | 0.004 |
| Throat pain | 71 (17.0) | 20 (24.7) | 51 (15.2) | 0.041 |
| Nasal obstruction | 43 (10.3) | 8 (9.9) | 35 (10.4) | 0.886 |
| Fear of cold | 34 (8.2) | 12 (14.8) | 22 (6.5) | 0.015 |
| NSS, No. (%) | ||||
| Any | 122 (29.3) | 40 (49.4) | 82 (24.4) | <0.001 |
| Hypodynamia | 57 (13.7) | 21 (25.9) | 36 (10.7) | <0.001 |
| Muscle aches | 43 (10.3) | 15 (18.5) | 28 (8.3) | 0.007 |
| Headache | 39 (9.4) | 3 (3.7) | 36 (10.7) | 0.052 |
| Dizziness | 33 (7.9) | 11 (13.6) | 22 (6.5) | 0.035 |
| Drowsiness | 4 (1.0) | 3 (3.7) | 1 (0.3) | 0.024 |
| Coma | 2 (0.5) | 2 (2.5) | 0 (0.0) | 0.037 |
| Paralysis | 1 (0.2) | 1 (1.2) | 0 (0.0) | 0.194 |
| Hyposmia | 1 (0.2) | 0 (0.0) | 1 (0.3) | 0.623 |
| Hypogeusia | 1 (0.2) | 0 (0.0) | 1 (0.3) | 0.623 |
| Lalopathy | 1 (0.2) | 1 (1.2) | 0 (0.0) | 0.194 |
| Cardiovascular system symptom, No. (%) | ||||
| Any | 60 (14.4) | 22 (27.2) | 38 (11.3) | <0.001 |
| Chest distress | 37 (8.9) | 13 (16.0) | 24 (7.1) | 0.011 |
| Shortness of breath | 17 (4.1) | 9 (11.1) | 8 (2.4) | 0.002 |
| Chest pain | 14 (3.4) | 4 (4.9) | 10 (3.0) | 0.488 |
| Palpitation | 3 (0.7) | 0 (0.0) | 3 (0.9) | 0.393 |
| Arrhythmia | 3 (0.7) | 2 (2.5) | 1 (0.3) | 0.098 |
| Gastrointestinal symptom, No. (%) | ||||
| Any | 53 (12.7) | 21 (25.9) | 32 (9.5) | <0.001 |
| Diarrhea | 36 (8.6) | 13 (16.0) | 23 (6.8) | 0.008 |
| Abdominal pain | 11 (2.6) | 4 (4.9) | 7 (2.1) | 0.236 |
| Nausea | 10 (2.4) | 4 (4.9) | 6 (1.8) | 0.108 |
| Poor appetite | 7 (1.7) | 1 (1.2) | 6 (1.8) | 0.729 |
| Emesis | 5 (1.2) | 2 (2.5) | 3 (0.9) | 0.250 |
| Gastralgia | 5 (1.2) | 2 (2.5) | 3 (0.9) | 0.250 |
| Abdominal distension | 4 (1.0) | 4 (4.9) | 0 (0.0) | 0.001 |
| Lesion location, No. (%) | 0.001 | |||
| Left | 28 (6.7) | 0 (0.0) | 28 (8.3) | |
| Right | 45 (10.8) | 2 (2.5) | 43 (12.8) | |
| Bilateral | 305 (73.1) | 73 (90.1) | 232 (69.1) | |
| Unknown | 39 (9.4) | 6 (7.4) | 33 (9.8) | |
| Type of lesion, No. (%) | 0.005 | |||
| Unifocal lesion | 46 (11.0) | 1 (1.2) | 45 (13.4) | |
| Multifocal lesion | 329 (78.9) | 73 (90.1) | 256 (76.2) | |
| Unknown | 42 (10.1) | 7 (8.7) | 35 (10.4) | |
Comparison was determined by Student’s t test, Wilcoxon rank sum test, Chi-square test or Fisher’s exact test. COVID-19, coronavirus disease 2019; M ± SD, mean ± standard deviation; IQR, interquartile range; NSS, nervous system symptom.
Comparison of pathogenic microbiology between severe patients and non-severe patients
Compared to non-severe patients, bacterial infections (P=0.005) and bacterial and fungal infections (P<0.001) were more common in severe patients. However, no differences in fungal, influenza B virus, respiratory syncytial virus, Mycoplasma pneumonia, Epstein-Barr virus, herpes simplex virus, rubella virus or cytomegalovirus infections between severe patients and non-severe patients was discovered (all P>0.05) (Table 2).
Table 2.
Pathogenic microbiology of COVID-19 patients
| Items | Total (N=417) | Disease severity | P value | |
|---|---|---|---|---|
|
| ||||
| Severe (n=81) | Non-severe (n=336) | |||
| Bacterial infection, No. (%) | 21 (5.0) | 9 (11.1) | 12 (3.6) | 0.005 |
| Fungal infection, No. (%) | 1 (0.2) | 1 (1.2) | 0 (0.0) | 0.194 |
| Bacterial and fungal infection, No. (%) | 18 (4.3) | 16 (19.8) | 2 (0.6) | <0.001 |
| Influenza B virus, No. (%) | 3 (0.7) | 0 (0.0) | 3 (0.9) | 0.393 |
| Respiratory syncytial virus, No. (%) | 4 (1.0) | 0 (0.0) | 4 (1.2) | 0.420 |
| Mycoplasma pneumonia, No. (%) | ||||
| IgG positive | 34 (8.2) | 4 (4.9) | 30 (8.9) | 0.239 |
| IgM positive | 7 (1.7) | 0 (0.0) | 7 (2.1) | 0.354 |
| Epstein-Barr virus, No. (%) | ||||
| IgG positive | 113 (27.1) | 17 (21.0) | 96 (28.6) | 0.168 |
| IgM positive | 12 (2.9) | 5 (6.2) | 7 (2.1) | 0.062 |
| Herpes simplex virus, No. (%) | ||||
| IgG positive | 13 (3.1) | 2 (2.5) | 11 (3.3) | 0.708 |
| IgM positive | 18 (4.3) | 2 (2.5) | 16 (4.8) | 0.545 |
| Rubella virus, No. (%) | ||||
| IgG positive | 16 (3.8) | 2 (2.5) | 14 (4.2) | 0.475 |
| IgM positive | 1 (0.2) | 0 (0.0) | 1 (0.3) | 0.623 |
| Cytomegalovirus, No. (%) | ||||
| IgG positive | 17 (4.1) | 3 (3.7) | 14 (4.2) | 0.850 |
| IgM positive | 0 (0.0) | 0 (0.0) | 0 (0.0) | - |
Comparison was determined by Chi-square test or Fisher’s exact test. COVID-19, coronavirus disease 2019; IgG, immunoglobulin G; IgM, immunoglobulin M.
Comparison of laboratory indexes between severe patients and non-severe patients
Compared to non-severe patients, severe patients presented with increased baseline APTT (P<0.001), TT (P=0.011), D-dimer (P<0.001), CRP (P<0.001), ESR (P=0.014), CK-MB (P=0.004), creatine kinase (P=0.001), AST (P<0.001), ALT (P<0.001) and creatinine (P=0.001) but decreased baseline platelet level (P<0.001), lymphocytes (P<0.001), albumin (P<0.001) and GFR (P<0.001). However, there were no differences in other laboratory indexes at baseline between severe patients and non-severe patients (all P>0.05) (Table 3).
Table 3.
Laboratory indexes of COVID-19 patients
| Items | Total (N=417) | Disease severity | P value | |
|---|---|---|---|---|
|
| ||||
| Severe (n=81) | Non-severe (n=336) | |||
| Blood routine index, median (IQR) | ||||
| Red blood cell (*1012/L) | 4.6 (4.3-5.1) | 4.6 (4.3-4.9) | 4.6 (4.3-5.1) | 0.958 |
| White blood cell (*109/L) | 4.5 (3.6-5.8) | 4.6 (3.8-6.3) | 4.5 (3.6-5.6) | 0.534 |
| Platelet (*109/L) | 180.0 (145.0-223.0) | 149.0 (129.0-184.0) | 188.0 (150.8-230.3) | <0.001 |
| Lymphocyte (*109/L) | 1.3 (1.0-1.8) | 1.1 (0.8-1.3) | 1.4 (1.0-1.9) | <0.001 |
| Hemoglobin (g/L) | 137.0 (127.0-146.0) | 139.0 (130.0-151.0) | 137.0 (126.0-146.0) | 0.266 |
| Coagulation function index, median (IQR) | ||||
| APTT (s) | 35.7 (32.7-38.5) | 37.1 (35.1-41.4) | 34.9 (31.9-38.2) | <0.001 |
| TT (s) | 15.7 (15.2-16.4) | 15.9 (15.4-16.9) | 15.7 (15.2-16.3) | 0.011 |
| D-dimer (μg/L) | 0.4 (0.3-0.5) | 0.5 (0.4-0.7) | 0.3 (0.2-0.5) | <0.001 |
| Inflammation-related indicators, median (IQR) | ||||
| CRP (mg/L) | 11.7 (3.4-26.8) | 29.3 (12.8-53.0) | 8.1 (2.7-21.3) | <0.001 |
| ESR (mm/h) | 28.0 (14.0-48.8) | 36.0 (20.8-52.0) | 25.0 (13.0-46.3) | 0.014 |
| Serum enzyme index, median (IQR) | ||||
| CK-MB (U/L) | 0.9 (0.6-1.1) | 1.0 (0.9-1.4) | 0.8 (0.6-1.1) | 0.004 |
| Creatine kinase (U/L) | 66.5 (50.0-108.0) | 102.0 (65.5-356.1) | 65.0 (49.0-94.8) | 0.001 |
| Liver function index, median (IQR) | ||||
| AST (μ/L) | 26.0 (21.0-36.1) | 36.1 (26.0-51.9) | 25.0 (20.0-33.1) | <0.001 |
| ALT (μ/L) | 21.0 (15.0-32.0) | 27.1 (19.2-40.0) | 20.0 (13.8-28.8) | <0.001 |
| Albumin (g/L) | 43.1 (41.0-45.3) | 41.0 (38.0-43.9) | 43.5 (41.4-45.6) | <0.001 |
| Total protein (g/L) | 70.4 (66.4-74.4) | 69.1 (65.4-73.2) | 70.8 (66.7-74.7) | 0.174 |
| Total bilirubin (μmol/L) | 9.5 (7.8-13.2) | 9.5 (8.1-12.6) | 9.5 (7.7-13.3) | 0.848 |
| Renal function index, median (IQR) | ||||
| Creatinine (μmol/L) | 61.0 (51.0-74.0) | 66.0 (57.0-94.8) | 59.0 (50.0-72.0) | 0.001 |
| GFR (mL/min) | 106.8 (94.6-117.5) | 92.3 (75.0-104.0) | 108.7 (97.5-119.3) | <0.001 |
Comparison was determined by Wilcoxon rank sum test. COVID-19, coronavirus disease 2019; IQR, interquartile range; APTT, activated partial thromboplastin time; TT, thrombin time; CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; CK-MB, creatine kinase MB fraction; AST, aspartate aminotransferase; ALT, alanine transaminase; GFR, Glomerular filtration rate.
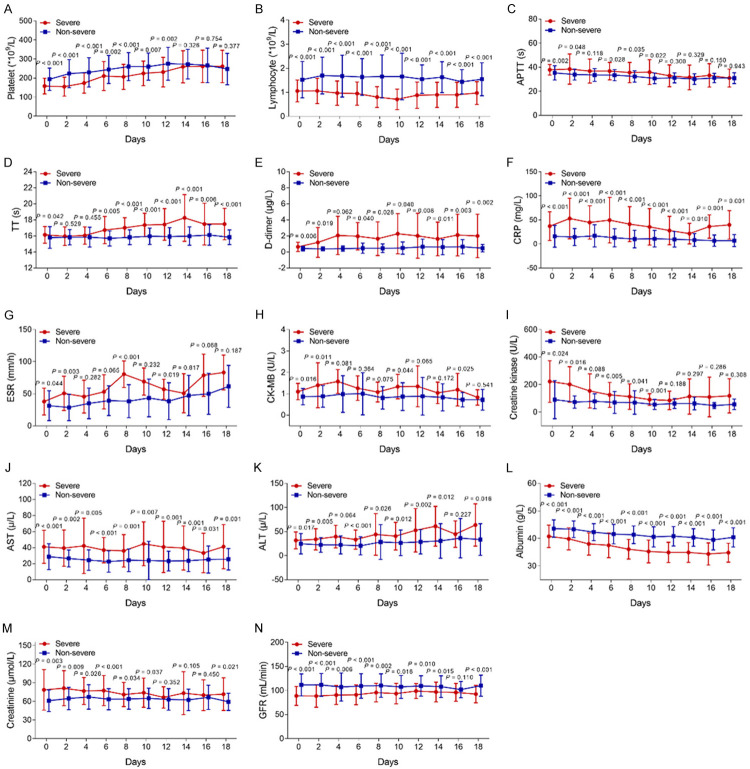
We further evaluated the trends of laboratory indexes over time in severe patients and non-severe patients, and we found that the differences in platelets (Figure 1A), APTT (Figure 1C), ESR (Figure 1G), CK-MB (Figure 1H), creatine kinase (Figure 1I) and creatine (Figure 1M) gradually decreased over time and eventually became consistent between severe patients and non-severe patients. In addition, there were continuously obvious differences in lymphocytes (Figure 1B), D-dimer (Figure 1E), CRP (Figure 1F), AST (Figure 1J), ALT (Figure 1K), albumin (All P<0.001) (Figure 1L) and GFR (Figure 1N) between the two groups. Meanwhile, the difference in TT (Figure 1D) was increased between the two groups over time.
Figure 1.
The longitudinal changes of major laboratory indexes between severe patients and non-severe patients. Comparison of platelet (A), lymphocyte (B), APTT (C), TT (D), D-dimer (E), CRP (F), ESR (G), CK-MB (H), Creatine kinase (I), AST (J), ALT (K), albumin (L), Creatine (M) and GFR (N) between severe patients and non-severe patients at different time points. APTT, activated partial thromboplastin time; TT, thrombin time; CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; CK-MB, creatine kinase MB fraction; AST, aspartate aminotransferase; ALT, alanine transaminase; GFR, Glomerular filtration rate.
Construction of the disease progression prediction model
Forward stepwise multivariate logistic regression model analysis revealed that age (P=0.002), any NSS (P=0.009), bacterial and fungal infection (P=0.022), CRP (P=0.019) and creatinine (P=0.018) were factors independently associated with severe COVID-19 (Table 4). In addition, we further built a disease progression prediction model based on these independent factors, and we found that this disease progression prediction model had good value for predicting severe COVID-19 risk with an AUC of 0.861 (95% CI: 0.799-0.922) (Figure 2).
Table 4.
Forward stepwise multivariate logistic regression model analysis of independent factors related to severe COVID-19
| Items | Multivariate logistic regression model | |||
|---|---|---|---|---|
|
| ||||
| P value | OR | 95% CI | ||
|
| ||||
| Lower | Higher | |||
| Age | 0.002 | 1.063 | 1.023 | 1.106 |
| Any NSS | 0.009 | 3.517 | 1.366 | 9.057 |
| Bacterial and fungal infection | 0.022 | 24.034 | 1.573 | 367.202 |
| CRP | 0.019 | 1.022 | 1.004 | 1.040 |
| Creatinine | 0.018 | 1.035 | 1.006 | 1.065 |
The factors with P value <0.05 in univariate analysis (Tables 1, 2 and 3) were included in the forward stepwise multivariate logistic regression model. The model was as follows: P=exp [-8.004 + 0.061*(age) + 1.258*(any nervous system symptom) + 3.179*(bacterial and fungal infection) + 0.022*(CRP) + 0.034*(creatinine)]/1+ exp [-8.004 + 0.061*(age) + 1.258*(any nervous system symptom) + 3.179*(bacterial and fungal infection) + 0.022*(CRP) + 0.034*(creatinine)]. Goodness of fit: -2ln(R) =117.127, Nagelkerke R2 =0.457. COVID-19, coronavirus disease 2019; OR, odds ratio; CI, confidence interval; CRP, C-reactive protein; NSS, nervous system symptom.
Figure 2.
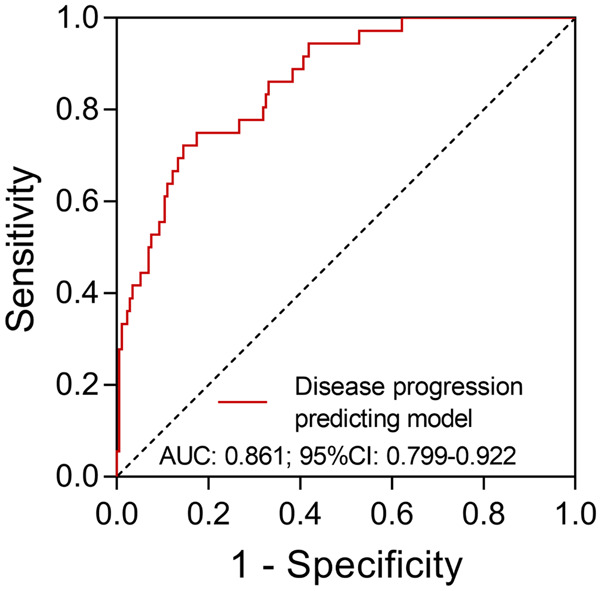
The predictive value of a disease progression predicting model for severe COVID-19. AUC: area under the curve; CI: confidence interval; COVID-19: coronavirus disease 2.
Comparison of outcomes between severe patients and non-severe patients
In all COVID-19 patients, the median time to nucleic acid negative was 15.0 (11.0-22.0) days (Table 5). Regarding the disease condition, 3 (0.7%) patients were cured, 400 (95.9%) patients improved, 11 (2.6%) patients had aggravated disease and 3 (0.07%) patients had died as of the last follow-up date. Compared to non-severe patients, the time to nucleic acid negative was longer in severe patients (P<0.001). Meanwhile, improved outcomes (P<0.001) were decreased, but aggravated outcomes (P<0.001) and death (P=0.007) were increased in severe patients compared to non-severe patients.
Table 5.
Outcomes of COVID-19 patients
| Items | Total (N=417) | Disease severity | P value | |
|---|---|---|---|---|
|
| ||||
| Severe (n=81) | Non-severe (n=336) | |||
| Time to nucleic acid negative (days), median (IQR) | 15.0 (11.0-22.0) | 19.0 (13.0-28.0) | 14.0 (10.0-20.5) | <0.001 |
| Disease condition, No. (%) | ||||
| Cured | 3 (0.7) | 0 (0.0) | 3 (0.9) | 0.393 |
| Improved | 400 (95.9) | 70 (86.5) | 330 (98.2) | <0.001 |
| Aggravated | 11 (2.6) | 8 (9.9) | 3 (0.9) | <0.001 |
| Dead | 3 (0.7) | 3 (3.7) | 0 (0.0) | 0.007 |
Comparison was determined by Wilcoxon rank sum test, Chi-square test or Fisher’s exact test. COVID-19, coronavirus disease 2019; IQR, interquartile range.
Comparison of clinical features between NSS patients and non-NSS patients
According to the findings mentioned above, NSS was discovered as an independent factor associated with severe COVID-19. However, there is little current research focusing on NSS in COVID-19 patients; herein, we explored the clinical features in COVID-19 patients with NSS. In the present study, we divided all COVID-19 patients into NSS patients (N=122) and non-NSS patients (N=295). Then, the following comparison analysis revealed that older age (P<0.001), history of surgery (P=0.048), diabetes comorbidity (P=0.008), respiratory system symptoms (P<0.001), cardiovascular system symptoms (P<0.001), gastrointestinal symptoms (P<0.001), bilateral lesions (P=0.005) and multifocal lesions (P=0.017) were more common in NSS patients than in non-NSS patients (Table 6).
Table 6.
Comparison of clinical characteristics between NSS COVID-19 patients and non-NSS COVID-19 patients
| Characteristics | NSS patients (n=122) | Non-NSS patients (n=295) | P value |
|---|---|---|---|
| Age (years), M ± SD | 50.1±14.7 | 43.2±18.4 | <0.001 |
| Gender, No. (%) | 0.817 | ||
| Male | 59 (48.4) | 139 (47.1) | |
| Female | 63 (51.6) | 156 (52.9) | |
| Duration from onset to fever (days), median (IQR) | 1.0 (1.0-2.8) | 1.0 (1.0-2.0) | 0.608 |
| Blood type, No. (%) | 0.405 | ||
| A+ | 18 (14.8) | 61 (20.7) | |
| AB+ | 21 (17.2) | 64 (21.7) | |
| B+ | 14 (11.5) | 29 (9.8) | |
| O+ | 26 (21.3) | 52 (17.6) | |
| Unknown | 43 (35.2) | 89 (30.2) | |
| History of surgery, No. (%) | 25 (20.5) | 38 (12.9) | 0.048 |
| Comorbidities, No. (%) | |||
| Hypertension | 25 (20.5) | 49 (16.6) | 0.345 |
| Fat | 15 (12.3) | 29 (9.8) | 0.456 |
| Diabetes | 14 (11.5) | 13 (4.4) | 0.008 |
| Cardiovascular disease | 6 (4.9) | 6 (2.0) | 0.118 |
| Hepatitis B | 5 (4.1) | 7 (2.4) | 0.338 |
| Hyperlipidemia | 2 (1.6) | 4 (1.4) | 0.825 |
| Arteriosclerosis | 3 (2.5) | 2 (0.7) | 0.152 |
| Syphilis | 0 (0.0) | 1 (0.3) | 0.520 |
| Respiratory system symptom, No. (%) | |||
| Any | 118 (96.7) | 243 (82.4) | <0.001 |
| Fever | 108 (88.5) | 202 (68.5) | <0.001 |
| Cough | 71 (58.2) | 144 (48.8) | 0.081 |
| Expectoration | 38 (31.1) | 78 (26.4) | 0.329 |
| Throat pain | 27 (22.1) | 44 (14.9) | 0.074 |
| Nasal obstruction | 19 (15.6) | 24 (8.1) | 0.023 |
| Fear of cold | 17 (13.9) | 17 (5.8) | 0.006 |
| Cardiovascular system symptom, No. (%) | |||
| Any | 31 (25.4) | 29 (9.8) | <0.001 |
| Chest distress | 18 (14.8) | 19 (6.4) | 0.007 |
| Shortness of breath | 10 (8.2) | 7 (2.4) | 0.012 |
| Chest pain | 8 (6.6) | 6 (2.0) | 0.032 |
| Palpitation | 2 (1.6) | 1 (0.3) | 0.206 |
| Arrhythmia | 2 (1.6) | 1 (0.3) | 0.206 |
| Gastrointestinal symptom, No. (%) | |||
| Any | 27 (22.1) | 26 (8.8) | <0.001 |
| Diarrhea | 17 (13.9) | 19 (6.4) | 0.020 |
| Abdominal pain | 7 (5.7) | 4 (1.4) | 0.017 |
| Nausea | 7 (5.7) | 3 (1.0) | 0.008 |
| Poor appetite | 4 (3.3) | 3 (1.0) | 0.202 |
| Emesis | 4 (3.3) | 1 (0.3) | 0.027 |
| Gastralgia | 4 (3.3) | 1 (0.3) | 0.027 |
| Abdominal distension | 1 (0.8) | 3 (1.0) | 1.000 |
| Lesion location, No. (%) | 0.005 | ||
| Left | 2 (1.6) | 26 (8.8) | |
| Right | 12 (9.9) | 33 (11.2) | |
| Bilateral | 102 (83.6) | 203 (68.8) | |
| Unknown | 6 (4.9) | 33 (11.2) | |
| Type of lesion, No. (%) | 0.017 | ||
| Unifocal lesion | 7 (5.7) | 39 (13.2) | |
| Multifocal lesion | 107 (87.7) | 222 (75.3) | |
| Unknown | 8 (6.6) | 34 (11.5) |
Comparison was determined by Student’s t test, Wilcoxon rank sum test, Chi-square test or Fisher’s exact test. NSS, nervous system symptom; COVID-19, coronavirus disease 2019; M ± SD, mean ± standard deviation; IQR, interquartile range.
Comparison of pathogenic microbiology between NSS patients and non-NSS patients
Compared to non-NSS patients, more NSS patients had bacterial infections (P=0.004) and bacterial and fungal infections (P=0.048). However, no differences in any other pathogenic microbiologic characteristics were discovered between NSS patients and non-NSS patients (all P>0.05) (Table 7).
Table 7.
Comparison of pathogenic microbiology between NSS COVID-19 patients and non-NSS COVID-19 patients
| Items | NSS patients (n=122) | Non-NSS patients (n=295) | P value |
|---|---|---|---|
| Bacterial infection, No. (%) | 12 (9.8) | 9 (3.1) | 0.004 |
| Fungal infection, No. (%) | 1 (0.8) | 0 (0.0) | 0.293 |
| Bacterial and fungal infection, No. (%) | 9 (7.4) | 9 (3.1) | 0.048 |
| Influenza B virus, No. (%) | 0 (0.0) | 3 (1.0) | 0.559 |
| Respiratory syncytial virus, No. (%) | 0 (0.0) | 4 (1.4) | 0.326 |
| Mycoplasma pneumonia, No. (%) | |||
| IgG positive | 10 (8.2) | 24 (8.1) | 0.983 |
| IgM positive | 0 (0.0) | 7 (2.4) | 0.112 |
| Epstein-Barr virus, No. (%) | |||
| IgG positive | 38 (31.1) | 75 (25.4) | 0.276 |
| IgM positive | 4 (3.3) | 8 (2.7) | 0.753 |
| Herpes simplex virus, No. (%) | |||
| IgG positive | 4 (3.3) | 9 (3.1) | 0.903 |
| IgM positive | 2 (1.6) | 16 (5.4) | 0.084 |
| Rubella virus, No. (%) | |||
| IgG positive | 3 (2.5) | 13 (4.4) | 0.416 |
| IgM positive | 0 (0.0) | 1 (0.3) | 0.520 |
| Cytomegalovirus, No. (%) | |||
| IgG positive | 5 (4.1) | 12 (4.1) | 1.000 |
| IgM positive | 0 (0.0) | 0 (0.0) | - |
Comparison was determined by Chi-square test or Fisher’s exact test. NSS, nervous system symptom; COVID-19, coronavirus disease 2019; IgG, immunoglobulin G; IgM, immunoglobulin M.
Comparison of laboratory indexes between NSS patients and non-NSS patients
Compared to non-NSS patients, NSS patients presented with decreased platelets (P<0.001), lymphocytes (P=0.025) and GFR (P=0.007) but increased APTT (P=0.004), CRP (P=0.002) and ESR (P=0.008). However, there was no difference in other laboratory indexes between NSS patients and non-NSS patients (all P>0.05) (Table 8).
Table 8.
Comparison of laboratory indexes between NSS COVID-19 patients and non-NSS COVID-19 patients
| Items | NSS patients (n=122) | Non-NSS patients (n=295) | P value |
|---|---|---|---|
| Blood routine index, median (IQR) | |||
| Red blood cell (*1012/L) | 4.6 (4.2-5.0) | 4.6 (4.3-5.1) | 0.317 |
| White blood cell (*109/L) | 4.7 (3.9-5.8) | 4.5 (3.6-5.8) | 0.347 |
| Platelet (*109/L) | 164.0 (139.0-195.3) | 188.0 (150.0-233.5) | <0.001 |
| Lymphocyte (*109/L) | 1.2 (1.0-1.5) | 1.3 (1.0-1.9) | 0.025 |
| Hemoglobin (g/L) | 137.5 (125.0-146.0) | 137.0 (127.0-147.0) | 0.719 |
| Coagulation function index, median (IQR) | |||
| APTT (s) | 36.6 (34.1-39.1) | 35.0 (31.8-38.2) | 0.004 |
| TT (s) | 15.7 (15.1-16.3) | 15.7 (15.2-16.5) | 0.512 |
| D-dimer (μg/L) | 0.4 (0.3-0.6) | 0.4 (0.2-0.5) | 0.234 |
| Inflammation-related indicators, median (IQR) | |||
| CRP (mg/L) | 14.8 (6.3-34.3) | 8.3 (2.7-23.9) | 0.002 |
| ESR (mm/h) | 32.5 (17.3-57.8) | 25.0 (13.0-44.0) | 0.008 |
| Serum enzyme index, median (IQR) | |||
| CK-MB (U/L) | 0.9 (0.6-1.1) | 0.9 (0.7-1.1) | 0.480 |
| Creatine kinase (U/L) | 65.0 (49.5-109.5) | 67.0 (50.0-103.5) | 0.789 |
| Liver function index, median (IQR) | |||
| AST (μ/L) | 26.4 (21.0-39.2) | 26.0 (20.0-35.0) | 0.284 |
| ALT (μ/L) | 24.0 (16.0-33.2) | 20.0 (14.1-31.8) | 0.210 |
| Albumin (g/L) | 43.1 (39.9-44.9) | 43.1 (41.3-45.6) | 0.231 |
| Total protein (g/L) | 70.4 (67.5-73.8) | 70.4 (66.1-75.0) | 0.908 |
| Total bilirubin (μmol/L) | 9.6 (7.9-13.4) | 9.4 (7.6-13.0) | 0.560 |
| Renal function index, median (IQR) | |||
| Creatinine (μmol/L) | 64.0 (53.0-80.0) | 60.0 (50.0-72.0) | 0.054 |
| GFR (mL/min) | 103.2 (90.6-112.1) | 108.6 (96.5-119.4) | 0.007 |
Comparison was determined by Wilcoxon rank sum test. NSS, nervous system symptom; COVID-19, coronavirus disease 2019; IQR, interquartile range; APTT, activated partial thromboplastin time; TT, thrombin time; CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; CK-MB, creatine kinase MB fraction; AST, aspartate aminotransferase; ALT, alanine transaminase; GFR, Glomerular filtration rate.
Comparison of outcomes between NSS patients and non-NSS patients
Compared to non-NSS patients, the median time to nucleic acid negative was longer in NSS patients (P=0.037). However, no difference was found in the cure rate (P=1.000), improved outcomes (P=0.283), aggravated outcomes (P=0.231) or deaths (P=1.000) between NSS patients and non-NSS patients (Table 9).
Table 9.
Compassion of outcomes between NSS COVID-19 patients and non-NSS COVID-19 patients
| Items | NSS patients (n=122) | Non-NSS patients (n=295) | P value |
|---|---|---|---|
| Time to nucleic acid negative (days), median (IQR) | 16.0 (11.3-24.0) | 14.0 (10.0-21.0) | 0.037 |
| Disease condition, No. (%) | |||
| Cured | 1 (0.8) | 2 (0.7) | 1.000 |
| Improved | 115 (94.3) | 285 (96.6) | 0.283 |
| Aggravated | 5 (4.1) | 6 (2.0) | 0.231 |
| Dead | 1 (0.8) | 2 (0.7) | 1.000 |
Comparison was determined by Wilcoxon rank sum test, Chi-square test or Fisher’s exact test. NSS, nervous system symptom; COVID-19, coronavirus disease 2019; IQR, interquartile range.
Discussion
Several studies have addressed the risk factors for COVID-19 progression. For instance, a previous study recruiting a total of 174 COVID-19 patients revealed that diabetes is a risk factor for rapid progression and poor prognosis of COVID-19 [9]. Furthermore, an interesting study with 323 COVID-19 patients discovered hypersensitive troponin I as a novel risk factor for severe COVID-19 [10]. Although these previous studies paid attention to the risk factors for disease progression of COVID-19, the sample size was relatively small in these previous studies, which may have resulted in statistical power. Meanwhile, the number of factors included in the risk analyses was relatively low in these previous studies, which indicates a potential increased risk for missing important relevant factors. Herein, our study, which had a relatively larger sample size (417 COVID-19 patients) included more candidate risk factors for severe COVID-19, and we found that age, NSS, bacterial and fungal infection, CRP and creatinine were independent factors for predicting severe COVID-19. The possible explanations are as follows (1) Regarding age, older patients were characterized by poor physical state and decreased immune function that led them to become vulnerable to infections and have a harder time defending against SARS-CoV-2, thereby making progression to severe disease or even death more likely; hence, older age could predict severe COVID-19 [11]. (2) Regarding NSS, SARS-CoV-2, as a virus with high sequence homology with SARS-CoV and MERS-CoV, might exert an effect on neuroinvasiveness and enter the central nervous system via the synaptic transmission pathway [12,13]. Hence, many COVID-19 patients, particularly severe patients, showed NSS. In addition, some COVID-19 patients presented with hyposphraesia or hypogeusia as the first clinical symptoms (SARS-CoV-2 might first invade the peripheral nerves, such as the olfactory nerve, and then travel retrograde along the olfactory bulb into the cranium to affect COVID-19 patients’ NSS). (3) Bacterial and fungal infection could directly affect the disease progression in COVID-19 patients, though we also found another interesting result that more non-severe patients had a history of Mycoplasma pneumonia, Epstein-Barr virus and Herpes simplex virus infection compared to severe patients; thus, it was very likely that previous infection with these viruses could induce human memory T cell response, and the response of memory T cells to SARS-CoV-2 was induced again when faced with SARS-CoV-2, thereby helping COVID-19 patients with their defense capability to some extent [14-17]. (4) Regarding CRP, SARS-CoV-2 could induce an inflammatory storm and then cause myocardial cell injury; thus, proinflammatory cytokine-induced reactions in severe patients might be more intense. Thus, increased CRP and creatinine could predict severe COVID-19. (5) Regarding creatinine, severe COVID-19 was more likely to cause worse renal dysfunction; thus, severe patients presented with higher levels of creatinine. In addition, we further constructed a disease progression prediction model, using these independent factors, by forward stepwise multivariate logistic analysis; this model presented a good value for predicting severe COVID-19 risk with an AUC of 0.861. These findings provide good evidence to remind clinicians to focus on potential risk factors for severe COVID-19 to prevent COVID-19 progression.
In the existing data, many studies focused on the descriptions of baseline laboratory indexes in COVID-19 patients, whereas little is known about the change in major laboratory indexes over time in these patients. To the best of our knowledge, this study was the first to explore the trend in major laboratory indexes over time in severe patients and non-severe patients. Our results showed that there were continuously obvious differences in lymphocytes, D-dimer, CRP, AST, ALT, albumin and GFR between severe patients and non-severe patients, which meant that severe SARS-CoV-2 infection could more obviously induce the release of inflammatory cytokines and immune complexes to worsen multiple system dysfunction (including renal dysfunction, liver dysfunction, coagulation dysfunction and hematopoietic dysfunction). In addition, we also found that the differences in platelets, APTT, ESR, CK-MB, creatine kinase and creatine gradually decreased over time and eventually became consistent between severe patients and non-severe patients, which may have been caused by the fact that after confirmation of COVID-19, most patients may have received relevant antiviral therapies and symptomatic therapies, and their multiple system dysfunction improved as a result. Hence, some laboratory indexes were normal in these COVID-19 patients.
In terms of the prognosis of COVID-19 patients, some studies have been carried out. For instance, there were 965 deaths (2.2%) reported a recent study of 44 672 confirmed COVID-19 cases up to February 11, 2020 (both adults and children) [18]. Another report disclosed that the mortality rate of COVID-19 patients was 1.37% (15/1099) [19]. In line with the previous data, our findings showed that the median time to nucleic acid negative was 15.0 (11.0-22.0) days, and 0.7% of patients were cured, but 0.7% of patients died. In addition, we found that severe COVID-19 patients had a longer time to nucleic acid negative and more aggravated outcomes and deaths than non-severe patients, suggesting a worse prognosis in severe COVID-19 patients, which might be caused by the fact that severe COVID-19 patients were more likely to have worse multiple organ dysfunction, which is directly related to poor prognosis. Taken together, these findings also indicate the importance of the disease progression prediction model (abovementioned), which may be helpful preventing severe progression and improve prognosis in COVID-19 patients.
Currently, most reports focus on the acute respiratory system of COVID-19 patients, while little attention has been paid to other systems in COVID-19 patients. Based on the results mentioned above, we found an interesting and important result that NSS was more common in severe patients, and it was observed to be an independent risk factor for severe COVID-19, suggesting that NSS patients were more likely to deteriorate into severe COVID-19 patients. Thus, further exploring the clinical characteristics of NSS patients may provide assistance in preventing disease deterioration and improving prognosis in COVID-19 patients. In this study, we investigated the clinical features in COVID-19 patients with NSS and found that older age, history of surgery, diabetes comorbidities, respiratory system symptoms, cardiovascular system symptoms, gastrointestinal symptoms, bilateral lesion, multifocal lesion, bacterial infection, bacterial and fungal infection and worse laboratory indexes were more common in NSS patients. These relevant factors were similar to the factors related to severe progression in COVID-19 patients (mentioned above). The possible reasons were as follows. (1) Age: Older patients more frequently had neurodegeneration and decreased immune function. Thus, they were more likely to present with nervous dysfunction when faced with SARS-CoV-2 infection [11]. (2) Diabetes comorbidities: Diabetes is widely considered a chronic, low-grade inflammatory disease that could cause peripheral neuropathy; thus, diabetes is more closely related to NSS in COVID-19 patients [9]. (3) Respiratory/cardiovascular/gastrointestinal symptoms: The occurrence of NSS in COVID-19 patients indicates nervous system impairment, which could directly affect neuro-regulation in multiple systems and then impact the function of multiple system function; thus, NSS is closely related to respiratory/cardiovascular/gastrointestinal symptoms in COVID-19 patients. Meanwhile, NSS was found to be an independent risk factor for severe COVID-19 (mentioned above) and severe patients showed more obvious respiratory/cardiovascular/gastrointestinal symptoms; thus, NSS might be correlated with respiratory/cardiovascular/gastrointestinal symptoms in COVID-19 patients. (4) Bilateral lesions and multifocal lesions: Bilateral lesions and multifocal lesions might indicate severe SARS-CoV-2 infection, which is prone to enter the CNS through the hematogenous or retrograde neuronal route, thereby causing NSS in COVID-19 patients [20]. Regarding the prognosis of NSS patients, the results showed a longer time to nucleic acid negative in NSS patients, which indicated a worse prognosis in NSS patients. Our findings might provide good evidence to remind clinicians to focus on NSS patients to prevent disease progression and improve prognosis.
Interesting and important findings were observed in this study, while some limitations still existed. (1) All patients were from our hospital only, which might have resulted in selected bias; hence, further multicenter studies are necessary. (2) There was no validation cohort in this study, and further study with a validation cohort is necessary. (3) The sample size in this study was relatively small. Further study with more COVID-19 patients is needed. (4) This study only focused on a review of inpatient medical records from January 11, 2020, to February 27, 2020, while there were still many patients continually receiving treatment (inpatient or community treatment) after that time. Hence, the clinical prognosis summarized in this study was not the final prognosis of the 417 included patients.
In conclusion, age, NSS, bacterial and fungal infection, CRP and creatinine were independent risk factors for severe COVID-19, and the disease progression prediction model using these independent factors had good predictive value for severe COVID-19. In addition, more intensive attention should be paid to COVID-19 patients with NSS to prevent rapid deterioration.
Disclosure of conflict of interest
None.
References
- 1.Segars J, Katler Q, McQueen DB, Kotlyar A, Glenn T, Knight Z, Feinberg EC, Taylor HS, Toner JP, Kawwass JF American Society for Reproductive Medicine Coronavirus/COVID-19 Task Force. Prior and novel coronaviruses, Coronavirus Disease 2019 (COVID-19), and human reproduction: what is known? Fertil Steril. 2020;113:1140–1149. doi: 10.1016/j.fertnstert.2020.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronavirus disease (COVID-19) outbreak situation. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019. Accessed May 20, 2020.
- 3.Liu J, Li S, Liu J, Liang B, Wang X, Wang H, Li W, Tong Q, Yi J, Zhao L, Xiong L, Guo C, Tian J, Luo J, Yao J, Pang R, Shen H, Peng C, Liu T, Zhang Q, Wu J, Xu L, Lu S, Wang B, Weng Z, Han C, Zhu H, Zhou R, Zhou H, Chen X, Ye P, Zhu B, Wang L, Zhou W, He S, He Y, Jie S, Wei P, Zhang J, Lu Y, Wang W, Zhang L, Li L, Zhou F, Wang J, Dittmer U, Lu M, Hu Y, Yang D, Zheng X. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55:102763. doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The guidelines on The Diagnosis and Treatment of COVID-19 (fourth version) 2020. http://www.nhc.gov.cn/. Accessed 20 April 2020.
- 5.Zhao S, Lin Q, Ran J, Musa SS, Yang G, Wang W, Lou Y, Gao D, Yang L, He D, Wang MH. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: a data-driven analysis in the early phase of the outbreak. Int J Infect Dis. 2020;92:214–217. doi: 10.1016/j.ijid.2020.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gemelli Against COVID-19 Geriatrics Team. Landi F, Barillaro C, Bellieni A, Brandi V, Carfi A, Cipriani MC, D’Angelo E, Falsiroli C, Fusco D, Landi G, Liperoti R, Lo Monaco MR, Martone AM, Marzetti E, Pagano FC, Pais C, Russo A, Salini S, Tosasto M, Tummolo AM, Benvenuto F, Bramato G, Catalano L, Ciciarello F, Martis I, Rocchi S, Rota E, Salerno A, Tritto M, Sgadari A, Zuccàla G, Bernabei R. The geriatrician: the frontline specialist in the treatment of COVID-19 patients. J Am Med Dir Assoc. 2020;21:937–938. doi: 10.1016/j.jamda.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann M, Kleine-Weber H, Krüger N, Müller M, Drosten C, Pöhlmann S. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. bioRxiv. 2020 2020.2001.2031.929042. [Google Scholar]
- 8.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo W, Li M, Dong Y, Zhou H, Zhang Z, Tian C, Qin R, Wang H, Shen Y, Du K, Zhao L, Fan H, Luo S, Hu D. Diabetes is a risk factor for the progression and prognosis of COVID-19. Diabetes Metab Res Rev. 2020:e3319. doi: 10.1002/dmrr.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu L, Chen S, Fu Y, Gao Z, Long H, Wang JM, Ren HW, Zuo Y, Li H, Wang J, Xu QB, Yu WX, Liu J, Shao C, Hao JJ, Wang CZ, Ma Y, Wang Z, Yanagihara R, Deng Y. Risk factors associated with clinical outcomes in 323 COVID-19 hospitalized patients in Wuhan, China. Clin Infect Dis. 2020;71:2089–2098. doi: 10.1093/cid/ciaa539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu K, Chen Y, Lin R, Han K. Clinical features of COVID-19 in elderly patients: a comparison with young and middle-aged patients. J Infect. 2020;80:e14–e18. doi: 10.1016/j.jinf.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Netland J, Meyerholz DK, Moore S, Cassell M, Perlman S. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J Virol. 2008;82:7264–7275. doi: 10.1128/JVI.00737-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li K, Wohlford-Lenane C, Perlman S, Zhao J, Jewell AK, Reznikov LR, Gibson-Corley KN, Meyerholz DK, McCray PB Jr. Middle east respiratory syndrome coronavirus causes multiple organ damage and lethal disease in mice transgenic for human dipeptidyl peptidase 4. J Infect Dis. 2016;213:712–722. doi: 10.1093/infdis/jiv499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 15.Benninger-Doring G, Pepperl S, Deml L, Modrow S, Wolf H, Jilg W. Frequency of CD8(+) T lymphocytes specific for lytic and latent antigens of Epstein-Barr virus in healthy virus carriers. Virology. 1999;264:289–297. doi: 10.1006/viro.1999.9996. [DOI] [PubMed] [Google Scholar]
- 16.Hislop AD, Annels NE, Gudgeon NH, Leese AM, Rickinson AB. Epitope-specific evolution of human CD8(+) T cell responses from primary to persistent phases of Epstein-Barr virus infection. J Exp Med. 2002;195:893–905. doi: 10.1084/jem.20011692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janice Oh HL, Ken-En Gan S, Bertoletti A, Tan YJ. Understanding the T cell immune response in SARS coronavirus infection. Emerg Microbes Infect. 2012;1:e23. doi: 10.1038/emi.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Epidemiology Working Group for NCIP Epidemic Response, Chinese Center for Disease Control and Prevention. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41:145–151. doi: 10.3760/cma.j.issn.0254-6450.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS China Medical Treatment Expert Group for Covid-19. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao L, Jin H, Wang M, Hu Y, Chen S, He Q, Chang J, Hong C, Zhou Y, Wang D, Miao X, Li Y, Hu B. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 2020;77:683–690. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]