Abstract
Acute Lymphoblastic Leukemia (ALL) is the most common type of cancer in children. Polymorphisms that alter the normal function of the microRNAs involved in the development of ALL have been widely investigated, although published data on these polymorphisms in admixed populations are scarce. We investigated the role of 10 polymorphisms in the microRNA and protein-coding genes of the microRNA synthesis complex in susceptibility to pediatric B-cell ALL. The study includes 100 pediatric ALL patients and 180 healthy individuals. The statistical analyses were run in SPSS v.25.0. In the case of the microRNA synthesizing genes, a significant pattern was found in only gene, that is, the rs3805500 polymorphism of DROSHA, in which the homozygous mutant (AA) genotype was associated with a threefold increase in the risk of developing ALL when compared to other genotypes (P=0.004, OR=2.913, CI=1.415-5.998). In the microRNA coding genes, the homozygous mutant rs3746444 genotype of the MIR499A gene was associated with a 17-fold increase in the risk of development of ALL (P<0.001, OR=17.797, CI=5.55-57.016). A protective effect against the development of ALL was also observed in the carriers of the wild homozygous rs2505901 genotype in the MIR938 gene. Our findings highlight the potential of these polymorphisms in the genes involving in the coding of microRNAs for the evaluation of the risk of contracting ALL in the population of the Brazilian Amazon region. These findings contribute to a more complete understanding of the complex etiology of ALL.
Keywords: Acute lymphoblastic leukemia, microRNAs, single nucleotide polymorphism, susceptibility
Introduction
Acute Lymphoblastic Leukemia (ALL) is the most common type of cancer in children and adolescents of up to 19 years of age, representing 28% of all malignancies in this age group and 75% of all cases of leukemia [1-3]. The etiology of ALL is considered to be multifactorial, involving both environmental and genetic factors, which have been investigated extensively. These factors include exposure to carcinogens, as well as chromosomal and molecular alterations [4,5]. In particular, many single nucleotide polymorphisms (SNPs) in key genes of the regulatory pathways have been ascribed a fundamental role in the development of ALL [6].
The investigation of the SNPs found in the microRNA genes has emerged as a promising new field of genomic research. The principal function of these molecules is to regulate gene expression via post-transcriptional silencing, the cleaving of messenger RNAs (mRNA) or inhibiting the initiation of translation through base pairing between the microRNA and its mRNA target [7]. The presence of SNPs in microRNA genes or the protein-coding genes involved in the synthesis of the microRNAs may affect their correct functioning, resulting in impacts on gene regulation processes [8]. Despite the important regulatory potential of these polymorphisms, few studies have focused on the influence of the SNPs located in non-coding genomic regions. In addition, evidence on the occurrence of these polymorphisms in populations with high levels of miscegenation, such as that of the Brazilian Amazon region, is particularly scarce. Given this, the present study investigated the role of 10 polymorphisms of the microRNA and microRNA synthesis genes in the susceptibility of pediatric patients from the Brazilian Amazon region to B-cell ALL.
Material and methods
Patients and controls
The case group of the present study included 100 patients diagnosed with B-cell ALL by immunophenotyping and/or molecular analysis. All these individuals were in treatment at the Otávio Lobo Hospital in Belém, a reference center for the treatment of pediatric cancer in the northern region of Brazil. The patients included in the study were between 1 and 18 years old. Recurrent patients or those with comorbidities were not included in the group. The control group consisted of 180 individuals with no diagnosis for B-cell ALL or any other type of cancer. None of these individuals were related to any of the patients in the case group, and they were all over the peak risk age for the development of ALL.
Ethical aspects
This study was approved by the Research Ethics Committee of the Research Center of Oncology of the Federal University of Pará (CAAE number 11433019.5.0000.5634). All participants (or their legal guardians) signed a term of informed consent authorizing the collection of samples and data.
Selection of the study polymorphisms
The SNPs selected for the present study were chosen due to their known association with susceptibility to ALL and different types of cancer, based on the published data. The references of the journal articles used for the selection of the polymorphisms are listed in the Table S1 [38-48]. A total of 10 polymorphisms were selected, seven in microRNA genes and three in the protein-coding genes essential to the synthesis of microRNAs: rs636832 (AGO1), rs10035440 e rs3805500 (DROSHA), rs213210 and rs107822 (MIR219-1), rs2910164 (MIR146a), rs12894467 (MIR300), rs3746444 (MIR499a), rs4919510 (MIR608), and rs2505901 (MIR938).
Extraction and quantification of the DNA
The DNA was extracted from peripheral blood using a commercial DNA extraction kit (Biopur Mini Spin Plus-250 extraction kit, Biopur, Brazil). The concentration of the genetic material was quantified in a NanoDrop 1000 spectrophotometer (Scientific Term NanoDrop 1000; NanoDrop Technologies Wilmington, DE).
Genotyping of polymorphisms
The 10 polymorphisms were genotyped by allelic discrimination using the TaqMan OpenArray Genotyping technology with a set of 32 customized assays, which were run in a QuantStudio™ 12K Flex Real-Time PCR system (Applied BiosystEM, Life Technologies, Carlsbad, USA), according to the manufacturer’s protocol. This method is based on real-time Polymerase Chain Reaction (qPCR). The quality of the readings of the genotypes and other data were analyzed in the TaqMan Genotyper software.
Quality control
To ensure an adequate level of accuracy, polymorphisms were only included in the present study if they satisfied three criteria: (i) MAF≥1%; (ii) genotyping rate ≥80%, and (iii) were in Hardy-Weinberg (HWE). The HWE was performed using the Arlequin software (v.3.5.1.2). The significance of the HWE test was adjusted for multiple comparisons by the Bonferroni method (P≤0.001), and the HWE values are shown in the Table S2. As all 10 of the polymorphisms selected for the present study satisfied these criteria, they were all included in the association analyses (Table S2).
Fusion analysis by extraction of the RNA and the reverse transcriptase-polymerase chain reaction (RT-PCR)
For the cytogenetic analysis of the BCR-ABL, ETV6-RUNX1, MLL-AF4, SIL-TAL and E2A-PBX1 fusions, blood samples were collected from 84 patients via venipuncture and stored in EDTA. Ficoll Histopaque® (Sigma-Aldrich, USA) was added to the samples for the separation of the lymphocytes, following the manufacturer’s instructions. The ARNsy MiniKit (Qiagen, USA) and High Capacity cDNA Reverse Transcription kits (Applied Biosystems, USA) were used to extract RNA and convert cDNA, respectively, both according to the manufacturers’ protocols. A multiplex RT-PCR reaction was run using The Master Mix kit (Promega, USA) with the primers designed specifically for the five fusions mentioned above, also following the manufacturer’s instructions. The primers used for RT-PCR are listed in Table 1.
Table 1.
Primers used for RT-PCR in the chromosomal fusion analysis
| Cromossomic Fusion | Genes | Primer (5’-3’) |
|---|---|---|
| t(1;19)(q23;p13) | E2A | CTACTCCCCGGATCACTCAA |
| PBX1 | AGGCTTCATTCTGTGGCAGT | |
| t(4;11)(q21;q23) | MLL | CGCCCAAGTATCCCTGTAAA |
| AF4 | GAGCATGGATGACGTTCCTT | |
| t(9;22)(q34;q11) | BCR | TCGCAGAACTCGCAACAGT |
| ABL | ACACCATTCCCCATTGTGAT | |
| t(12;21)(p13;q22) | TEL | TCTCTCATCGGGAAGACCTG |
| AML1 | TGCGGTAGCATTTCTCAGC | |
| del(1)(p32;p32) | SIL | TCCTACCCTGCAAACAGACC |
| TAL1 | AGGCGGAGGATCTCATTCTT |
Analysis of genomic ancestry
The genomic ancestry of the participants was analyzed following Santos et al. (2010) and Ramos et al. (2016), using a set of 61 Ancestry Informative Markers (AIMs). The individual and global proportions of European, Amerindian, and African genetic ancestry were estimated using STRUCTURE v.2.3.4 [9-11].
Statistical analyses
The Chi-square test was applied for the pairwise comparisons of the categorical variables between the case and control groups, while the quantitative variables were compared using Student’s t. The multivariate analyses considered the sex of the participants and Amerindian ancestry as confounding variables. The Mann-Whitney test was used to compare the estimates of genetic ancestry between the groups. All statistical analyses were run in SPSS v.25.0 (SPSS, Chicago, IL, USA), considering a P≤0.05 significance level.
Results
Significant differences were found between the case (ALL) and control group ages, sex ratio, and genomic ancestry (Table 2). The mean age of the case group was significantly lower than that of the control group (P<0.001) and the sex ratios also varied significantly between groups (P<0.001), with a predominance of males in the case group and of females in the control group.
Table 2.
Demographic parameters for the case (patients with B-cell ALL) and control groups analyzed in the present study
| Variable | Case (100) | Control (180) | p-value |
|---|---|---|---|
| Sex (M/F) | 60/40 | 53/127 | <0.001a |
| Age* | 5.53 ± 3.991 | 65.97 ± 16.021 | <0.001b |
| Genetic ancestry * | |||
| European | 0.429 ± 0.133 | 0.454 ± 0.170 | 0.189c |
| African | 0.203 ± 0.089 | 0.241 ± 0.138 | 0.213c |
| Amerindian | 0.361 ± 0.154 | 0.304 ± 0.149 | 0.004c |
Chi-square;
Studenta0 t;
Mann-Whitney9trU.
Mean ± eStandard Deviation.
The genomic makeup of the case group was dominated by European ancestry (43%), followed by the Amerindian (36%) and African (20%) components. While the control group presented a similar makeup (European-45%, Amerindian-30%, African-24%), its Amerindian ancestry was significantly lower (P=0.004) than that of the case group. Given the observed variation, both sex and Amerindian ancestry were controlled for in the analyses of the potential association between the polymorphisms and susceptibility to ALL.
The distribution of the B-cell ALL subtypes, based on the occurrence of the chromosomal fusions analyzed here, is shown in Figure 1. The most frequent fusions were E2A-PBX1 (21.42%) and BCR-ABL (14.30%), followed by ETV6-RUNX1 (7.14%), MLL-AF4 (2.38%), and SIL-TAL (1.19%).
Figure 1.
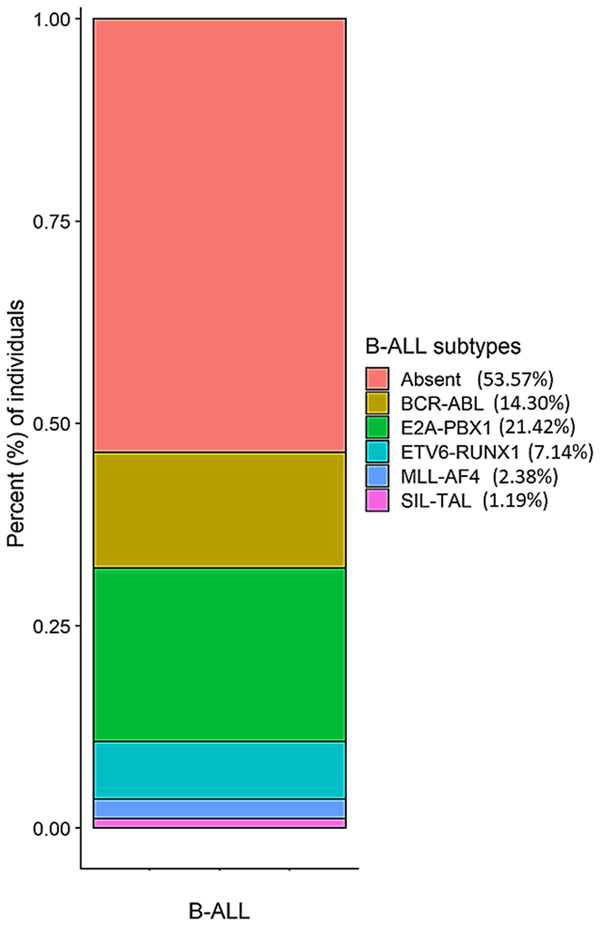
Distribution of the B-cell ALL subtypes in the study patients of the case group.
Three SNPs were investigated further in the case of the genes involved in the synthesis of microRNAs, and a significant pattern was observed in the case of the recessive model (AA vs. GG + GA) of the rs3805500 variant of the DROSHA gene (Table 3). The mutant homozygote (AA) genotype of this gene was associated with a threefold increase in the risk of developing ALL (P=0.004, OR=2.913, CI=1.415-5.998).
Table 3.
Distribution of the alleles and genotypes of the polymorphisms investigated in the B-cell ALL patients, in comparison with control individuals
| Genotype ID | Case (%) | Control (%) | p | Model | OR (95% CI) |
|---|---|---|---|---|---|
| AGO1_rs636832 | 75 | 150 | |||
| AA | 8 (10.7) | 14 (9.3) | |||
| AG | 34 (45.3) | 67 (44.7) | AA + AG vs. GG | ||
| GG | 33 (44) | 69 (46) | 0.163 | Recessive | 1.687 (0.809-3.520) |
| Allele A | 0.333 | 0.316 | |||
| Allele G | 0.666 | 0.683 | |||
| DROSHA_rs10035440 | 80 | 148 | |||
| TT | 60 (75) | 98 (66.2) | |||
| TC | 17 (21.3) | 44 (29.7) | TT + TC vs. CC | ||
| CC | 3 (3.8) | 6 (4.1) | 0.434 | Recessive | 2.082 (0.331-13.085) |
| Allele T | 0.856 | 0.810 | |||
| Allele C | 0.143 | 0.189 | |||
| DROSHA_rs3805500 | 74 | 159 | |||
| GG | 11 (14.9) | 50 (31.4) | |||
| GA | 26 (35.1) | 51 (32.1) | GG + GA vs. AA | ||
| AA | 37 (50) | 58 (36.5) | 0.004 | Recessive | 2.913 (1.415-5.998) |
| Allele G | 0.324 | 0.474 | |||
| Allele A | 0.675 | 0.525 | |||
| MIR219-1_rs213210 | 82 | 166 | AA vs. AG + GG | ||
| AA | 64 (78) | 134 (80.7) | 0.813 | Dominant | 0.908 (0.409-2.015) |
| AG | 16 (19.5) | 19 (11.4) | |||
| GG | 2 (2.4) | 13 (7.8) | |||
| Allele A | 0.878 | 0.864 | |||
| Allele G | 0.121 | 0.135 | |||
| MIR219-1_rs107822 | 73 | 152 | CC vs. CT + TT | ||
| CC | 36 (49.3) | 85 (55.9) | 0.831 | Dominant | 0.929 (0.471-1.832) |
| CT | 30 (41.1) | 53 (34.9) | |||
| TT | 7 (9.6) | 14 (9.2) | |||
| Allele C | 0.698 | 0.733 | |||
| Allele T | 0.301 | 0.266 | |||
| MIR146A_rs2910164 | 81 | 160 | GG vs. GC + CC | ||
| GG | 47 (58) | 68 (42.5) | 0.091 | Dominant | 1.777 (0.912-3.462) |
| GC | 24 (29.6) | 74 (46.3) | |||
| CC | 10 (12.3) | 18 (11.3) | |||
| Allele G | 0.728 | 0.656 | |||
| Allele C | 0.271 | 0.343 | |||
| MIR300_rs12894467 | 76 | 175 | TT vs. TC + CC | ||
| TT | 22 (28.9) | 75 (42.9) | 0.062 | Dominant | 0.513 (0.254-1.035) |
| TC | 42 (55.3) | 80 (45.7) | |||
| CC | 12 (15.8) | 20 (11.4) | |||
| Allele T | 0.565 | 0.657 | |||
| Allele C | 0.474 | 0.342 | |||
| MIR499_rs3746444 | 75 | 167 | |||
| AA | 5 (6.7) | 126 (75.4) | |||
| AG | 43 (57.3) | 36 (21.6) | AA + AG vs. GG | ||
| GG | 27 (36) | 5 (3.0) | <0.001 | Recessive | 17.797 (5.55-57.016) |
| Allele A | 0.353 | 0.862 | |||
| Allele G | 0.646 | 0.137 | |||
| MIR608_rs4919510 | 81 | 161 | GG vs. GC + CC | ||
| GG | 5 (6.2) | 18 (11.2) | 0.325 | Dominant | 0.566 (0.182-1.760) |
| GC | 29 (35.8) | 58 (36) | |||
| CC | 47 (58) | 85 (52.8) | |||
| Allele G | 0.240 | 0.291 | |||
| Allele C | 0.759 | 0.708 | |||
| MIR938_rs2505901 | 76 | 150 | CC vs. CT + TT | ||
| CC | 15 (19.7) | 53 (35.3) | 0.013 | Dominant | 0.359 (0.160-0.805) |
| CT | 38 (50) | 46 (37.7) | |||
| TT | 23 (30.3) | 51 (34) | |||
| Allele C | 0.447 | 0.506 | |||
| Allele T | 0.552 | 0.493 |
OR: Odds Ratio; CI: Confidence interval. The p values refer to the logistic regression adjusted for sex and Amerindian ancestry.
Seven variants of the microRNA genes were analyzed, of which, two were related significantly to the susceptibility of the carrier to ALL (Table 3)-MIR499A (rs3746444) and MIR938 (rs2505901). The mutant homozygote rs3746444 (GG) genotype was associated with a 17-fold increase in the risk of developing ALL (P<0.001, OR=17.797, CI=5.55-57.016). By contrast, the wild homozygous rs2505901 (CC) genotype was associated with a lower risk for the development of ALL, apparently conferring a protective effect (P=0.013, OR=0.359, CI=0.160-0.805).
Discussion
The population of Brazil is one of the world’s most genetically heterogeneous, and the pediatric ALL patients analyzed in the present study had a high level of admixture (42% European, 36% Amerindian, and 20% African). This is a very distinct genetic background in comparison with that of the other ethnic populations in which the association between SNPs in the microRNA genes have typically been evaluated [12].
We identified three variants with the apparent potential to influence the risk of developing ALL - the rs3805500 variant of the DROSHA gene, rs3746444 (MIR499A gene), and rs250590 (MIR938 gene). The differential expression of these genes and their apparent role in the development of cancer has been demonstrated in a number of previous studies [13-15]. However, a number of both internal and external environmental factors may affect the levels of gene expression, which impedes the full understanding of the role of these variants in the susceptibility of the individual to ALL, which reinforces the importance of evaluating germline polymorphisms [16,17].
The DROSHA gene is a key component of the synthesis of microRNAs. This gene encodes a type III RNase, which is essential for the maturation of pri-miRNAs and pre-miRNAs [18]. We found that the homozygous mutant (AA) rs3805500 genotype is associated with a threefold increase of the development of ALL. This is the first study to report the role of this polymorphism as a risk factor of ALL, although the rs3805500 mutant has already been identified as a factor in the susceptibility of individuals to other types of leukemia [19,20]. A GWAS study [19] found that the rs3805500 mutant A allele, in a haplotype with two other polymorphisms, was associated with an increased risk of Chronic Lymphocytic Leukemia (CLL), which is consistent with our finding that the rs3805500 mutant A allele contributes to an increased susceptibility to leukemia.
Polymorphisms in the DROSHA gene may influence shared pathways in the development of both ALL and CLL [19]. The rs3805500 mutant is also in linkage disequilibrium with the rs640831 variant of the same gene, which is related to a reduction in the expression of the DROSHA mRNA, and an alteration in the maturation of pri-miRNAs and pre-miRNAs, a condition linked to the progression of a number of different types of cancer [21-25].
Another variant analyzed here is the rs3746444, which is located in the seed region of the MIR-499a-3p gene, and may impede the microRNA from binding to its targets [12,26]. Our results indicate an association between the homozygous mutant genotype (GG) and a 17-fold increase in the risk of developing ALL. Other studies have reported an association between the rs3746444 polymorphism and susceptibility to a number of malignant neoplasia, although these findings have been controversial [27-30]. The homozygous mutant (GG) rs3746444 genotype has been related to the risk of gastric and lung cancer in Asian populations, but not in Caucasians [29,30]. The contradictory nature of these results is probably due to the distinct genetic backgrounds of the different types of cancer and interethnic differences between the Asian and Caucasian populations [12,29,30]. In the ALL, few studies have investigated the role of rs3746444 polymorphism in its development, which were carried out in homogeneous Caucasian populations [12,26]. Hasani et al. (2014) showed no association of this variant with the risk of ALL [12], while Gutierrez-Camino (2014), described a protective effect of the G allele on the risk of ALL [26]. Further studies are needed to better elucidate the role of this variant in the ALL development. Furthermore, many findings indicate the potential influence of ethnicity in genetic associations, and reinforce the need for the analysis of ancestry to guarantee more conclusive studies.
The MIR938 gene is responsible for the regulatory pathways of the genes related to cell survival and apoptosis [31]. Variants of the SNP type present in the MIR938 gene have been associated with modifications in its biogenesis and stability [32,33]. We investigated the rs2505901 variant, which corresponds to a C>T nucleotide swap in the intronic region of the MIR938 gene, and found that the wild homozygous (CC) genotype of this variant is associated with a 33% decrease in the risk of development of ALL. A decrease in the risk of gastric cancer was also observed in wild homozygous genotype carriers [34], which corroborates the protective effect of the rs2505901 variant. It is important to note here that only a few studies have investigated the role of this polymorphism in the development of cancer, in general, and that the present study is the first to evaluate its role in ALL.
Chromosome abnormalities have an important role in the predisposition, prognosis, and treatment of ALL [35]. In the present study, a majority (53.57%) of the patients did not present any of the chromosomal translocations investigated. Even so, 45.24% of the translocations observed were of high or intermediate risk (BCR-ABL, E2A-PBX1, ETV6-RUNX1, MLL-AF4) [36,37].
Conclusions
Overall, then, the present study provides convincing evidence of the influence of the rs3805500 (DROSHA), rs3746444 (MIR499), and rs2505901 (MIR938) variants on the susceptibility of the Brazilian Amazon population to ALL. From a methodological viewpoint, the use of SNPs as biomarkers is more practicable than the analysis of mRNA expression, given that germline variants can be investigated through the analysis of peripheral blood, requiring less costly laboratory procedures. Our findings are fundamental to the better understanding of the susceptibility of the population of the Brazilian Amazon region to ALL. This is an important advance, given the unique, ethnically diverse background of this population, which is quite distinct from the more homogeneous populations, which have been the focus of most previous ALL research.
Acknowledgements
We thank Antonio A.C. Modesto and R.B. Andrade, for technical assistance and we also thank all research participants, their families and the clinical staff for their skilled assistance. This research was supported by the National Council for Scientific and Technological Development (CNPq) and Pro-rector of Research and Post-graduation (Propesp) of Federal University of Pará.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ward E, DeSantis C, Robbins A, Kohler B, Jemal A. Childhood and adolescent cancer statistics, 2014. CA Cancer J Clin. 2014;64:83–103. doi: 10.3322/caac.21219. [DOI] [PubMed] [Google Scholar]
- 3.Linet MS, Brown LM, Mbulaiteye SM, Check D, Ostroumova E, Landgren A, Devesa SS. International long-term trends and recent patterns in the incidence of leukemias and lymphomas among children and adolescents ages 0-19 years. Int J Cancer. 2016;138:1862–1874. doi: 10.1002/ijc.29924. [DOI] [PubMed] [Google Scholar]
- 4.Vijayakrishnan J, Studd J, Broderick P, Kinnersley B, Holroyd A, Law PJ, Kumar R, Allan JM, Harrison CJ, Moorman AV, Vora A, Roman E, Rachakonda S, Kinsey SE, Sheridan E, Thompson PD, Irving JA, Koehler R, Hoffmann P, Nöthen MM, Heilmann-Heimbach S, Jöckel KH, Easton DF, Pharaoh PDP, Dunning AM, Peto J, Canzian F, Swerdlow A, Eeles RA, Kote-Jarai Z, Muir K, Pashayan N PRACTICAL Consortium. Greaves M, Zimmerman M, Bartram CR, Schrappe M, Stanulla M, Hemminki K, Houlston RS. Genome-wide association study identifies susceptibility loci for B-cell childhood acute lymphoblastic leukemia. Nat Commun. 2018;9:1340. doi: 10.1038/s41467-018-03178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.International Agency for Research on Cancer Iyon. IARC monographs on the evaluation of carcinogenic risks to humans: outdoor air pollution. 109th edition. Lyon: International Agency for Research on Cancer; 2015. [Google Scholar]
- 6.Brisson GD, Alves LR, Pombo-de-Oliveira MS. Genetic susceptibility in childhood acute leukaemias: a systematic review. Ecancermedicalscience. 2015;9:539. doi: 10.3332/ecancer.2015.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drobna M, Szarzyńska-Zawadzka B, Dawidowska M. T-cell acute lymphoblastic leukemia from miRNA perspective: basic concepts, experimental approaches, and potential biomarkers. Blood Rev. 2018;32:457–472. doi: 10.1016/j.blre.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Santos NP, Ribeiro-Rodrigues EM, Ribeiro-Dos-Santos AK, Pereira R, Gusmão L, Amorim A, Guerreiro JF, Zago MA, Matte C, Hutz MH, Santos SE. Assessing individual interethnic admixture and population substructure using a 48-insertion-deletion (INSEL) ancestry-informative marker (AIM) panel. Hum Mutat. 2010;31:184–190. doi: 10.1002/humu.21159. [DOI] [PubMed] [Google Scholar]
- 10.Ramos BR, D’Elia MP, Amador MA, Santos NP, Santos SE, da Cruz Castelli E, Witkin SS, Miot HA, Miot LD, da Silva MG. Neither self-reported ethnicity nor declared family origin are reliable indicators of genomic ancestry. Genetica. 2016;144:259–265. doi: 10.1007/s10709-016-9894-1. [DOI] [PubMed] [Google Scholar]
- 11.Pritchard JK, Donnelly P. Case-control studies of association in structured or admixed populations. Theor Popul Biol. 2001;60:227–237. doi: 10.1006/tpbi.2001.1543. [DOI] [PubMed] [Google Scholar]
- 12.Hasani SS, Hashemi M, Eskandari-Nasab E, Naderi M, Omrani M, Sheybani-Nasab M. A functional polymorphism in the miR-146a gene is associated with the risk of childhood acute lymphoblastic leukemia: a preliminary report. Tumor Biol. 2014;35:219–225. doi: 10.1007/s13277-013-1027-1. [DOI] [PubMed] [Google Scholar]
- 13.Poursadegh Zonouzi AA, Shekari M, Nejatizadeh A, Shakerizadeh S, Fardmanesh H, Poursadegh Zonouzi A, Rahmati-Yamchi M, Tozihi M. Impaired expression of Drosha in breast cancer. Breast Dis. 2017;37:55–62. doi: 10.3233/BD-170274. [DOI] [PubMed] [Google Scholar]
- 14.Wang BQ, Yang B, Yang HC, Wang JY, Hu S, Gao YS, Bu XY. MicroRNA-499a decelerates glioma cell proliferation while accelerating apoptosis through the suppression of Notch1 and the MAPK signaling pathway. Brain Res Bull. 2018;142:96–106. doi: 10.1016/j.brainresbull.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Ahmed FE, Ahmed NC, Vos PW, Bonnerup C, Atkins JN, Casey M, Nuovo GJ, Naziri W, Wiley JE, Mota H, Allison RR. Diagnostic MicroRNA markers to screen for sporadic human colon cancer in stool: I. Proof of principle. Cancer Genomics Proteomics. 2013;10:93–113. [PubMed] [Google Scholar]
- 16.Lobo I. Environmental influences on gene expression | learn science at scitable. Nat Educ. 2008;1:39. [Google Scholar]
- 17.Goldinger A, Shakhbazov K, Henders AK, McRae AF, Montgomery GW, Powell JE. Seasonal effects on gene expression. PLoS One. 2015;10:e0126995. doi: 10.1371/journal.pone.0126995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phua SL, Sivakamasundari V, Shao Y, Cai X, Zhang LF, Lufkin T, Featherstone M. Nuclear accumulation of an uncapped RNA produced by Drosha cleavage of a transcript encoding miR-10b and Hoxd4. PLoS One. 2011;6:e25689. doi: 10.1371/journal.pone.0025689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin-Guerrero I, Gutierrez-Camino A, Lopez-Lopez E, Bilbao-Aldaiturriaga N, Pombar-Gomez M, Ardanaz M, Garcia-Orad A. Genetic variants in MiRNA processing genes and Pre-MiRNAs are associated with the risk of chronic lymphocytic leukemia. PLoS One. 2015;10:e0118905. doi: 10.1371/journal.pone.0118905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hashemi M, Hasani SS, Naderi M. DROSHA rs642321 polymorphism influence susceptibility to childhood acute lymphoblastic leukemia: a preliminary report. Indian J Med Paediatr Oncol. 2017;38:416–419. doi: 10.4103/ijmpo.ijmpo_4_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.López-López E, Gutiérrez-Camino Á, Piñán MÁ, Sánchez-Toledo J, Uriz JJ, Ballesteros J, García-Miguel P, Navajas A, García-Orad Á. Pharmacogenetics of MicroRNAs and MicroRNAs biogenesis machinery in pediatric acute lymphoblastic leukemia. PLoS One. 2014;9:e91261. doi: 10.1371/journal.pone.0091261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mi S, Lu J, Sun M, Li Z, Zhang H, Neilly MB, Wang Y, Qian Z, Jin J, Zhang Y, Bohlander SK, Le Beau MM, Larson RA, Golub TR, Rowley JD, Chen J. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc Natl Acad Sci U S A. 2007;104:19971–19976. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugito N, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Kurehara H, Ando T, Mori R, Takashima N, Ogawa R, Fujii Y. RNASEN regulates cell proliferation and affects survival in esophageal cancer patients. Clin Cancer Res. 2006;12:7322–7328. doi: 10.1158/1078-0432.CCR-06-0515. [DOI] [PubMed] [Google Scholar]
- 24.Muralidhar B, Goldstein LD, Ng G, Winder DM, Palmer RD, Gooding EL, Barbosa-Morais NL, Mukherjee G, Thorne NP, Roberts I, Pett MR, Coleman N. Global microRNA profiles in cervical squamous cell carcinoma depend on Drosha expression levels. J Pathol. 2007;212:368–377. doi: 10.1002/path.2179. [DOI] [PubMed] [Google Scholar]
- 25.Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick AM, Deavers MT, Mourad-Zeidan A, Wang H, Mueller P, Lenburg ME, Gray JW, Mok S, Birrer MJ, Lopez-Berestein G, Coleman RL, Bar-Eli M, Sood AK. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gutierrez-Camino A, Lopez-Lopez E, Martin-Guerrero I, Piñan MA, Garcia-Miguel P, Sanchez-Toledo J, Carbone Bañeres A, Uriz J, Navajas A, Garcia-Orad A. Noncoding RNA-related polymorphisms in pediatric acute lymphoblastic leukemia susceptibility. Pediatr Res. 2014;75:767–773. doi: 10.1038/pr.2014.43. [DOI] [PubMed] [Google Scholar]
- 27.Zhang LH, Hao BB, Zhang CY, Dai XZ, Zhang F. Contributions of polymorphisms in mir146a, mir196a, and mir499 to the development of hepatocellular carcinoma. Genet Mol Res. 2016;9:15. doi: 10.4238/gmr.15038582. [DOI] [PubMed] [Google Scholar]
- 28.Nouri R, Ghorbian S. Association of single nucleotide polymorphism in hsa-miR-499 and hsa-miR-196a2 with the risk of prostate cancer. Int Urol Nephrol. 2019;51:811–816. doi: 10.1007/s11255-019-02099-0. [DOI] [PubMed] [Google Scholar]
- 29.Yang X, Li X, Zhou B. A meta-analysis of miR-499 rs3746444 polymorphism for cancer risk of different systems: evidence from 65 case-control studies. Front Physiol. 2018;9:737. doi: 10.3389/fphys.2018.00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang F, Sun G, Zou Y, Li Y, Hao L, Pan F. Association of microRNA-499 rs3746444 polymorphism with cancer risk: evidence from 7188 cases and 8548 controls. PLoS One. 2012;7:e45042. doi: 10.1371/journal.pone.0045042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Y, Jia Z, Cao D, Wang C, Wu X, You L, Wen S, Pan Y, Cao X, Jiang J. Predictive value of miR-219-1, miR-938, miR-34b/c, and miR-218 polymorphisms for gastric cancer susceptibility and prognosis. Dis Markers. 2017;2017:4731891. doi: 10.1155/2017/4731891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torruella-Loran I, Ramirez Viña MK, Zapata-Contreras D, Muñoz X, Garcia-Ramallo E, Bonet C, Gonzalez CA, Sala N, Espinosa-Parrilla Y. rs12416605:C>T in miR938 associates with gastric cancer through affecting the regulation of the CXCL12 chemokine gene. Mol Genet Genomic Med. 2019;7:832. doi: 10.1002/mgg3.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim WH, Min KT, Jeon YJ, Kwon C Il, Ko KH, Park PW, Hong SP, Rim KS, Kwon SW, Hwang SG, Kim NK. Association study of microRNA polymorphisms with hepatocellular carcinoma in Korean population. Gene. 2012;504:92–97. doi: 10.1016/j.gene.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 34.Arisawa T, Tahara T, Shiroeda H, Matsue Y, Minato T, Nomura T, Yamada H, Hayashi R, Saito T, Matsunaga K, Fukuyama T, Hayashi N, Otsuka T, Fukumura A, Nakamura M, Shibata T. Genetic polymorphisms of IL17A and pri-microRNA-938, targeting IL17A 3’-UTR, influence susceptibility to gastric cancer. Hum Immunol. 2012;73:747–752. doi: 10.1016/j.humimm.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 35.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354:166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 36.Duque-Afonso J, Lin CH, Han K, Wei MC, Feng J, Kurzer JH, Schneidawind C, Wong SH, Bassik MC, Cleary ML. E2A-PBX1 remodels oncogenic signaling networks in B-cell precursor acute lymphoid leukemia. Cancer Res. 2016;76:6937–6949. doi: 10.1158/0008-5472.CAN-16-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pui CH, Nichols KE, Yang JJ. Somatic and germline genomics in paediatric acute lymphoblastic leukaemia. Nat Rev Clin Oncol. 2019;16:227–240. doi: 10.1038/s41571-018-0136-6. [DOI] [PubMed] [Google Scholar]
- 38.Song X, Zhong H, Wu Q, Wang M, Zhou J, Zhou Y, Lu X, Ying B. Association between SNPs in microRNA machinery genes and gastric cancer susceptibility, invasion, and metastasis in Chinese Han population. Oncotarget. 2017;8:86435–86446. doi: 10.18632/oncotarget.21199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobrijević Z, Matijašević S, Savić-Pavićević D, Brajušković G. Association between genetic variants in genes encoding argonaute proteins and cancer risk: a meta-analysis. Pathol Res Pract. 2020;216:152906. doi: 10.1016/j.prp.2020.152906. [DOI] [PubMed] [Google Scholar]
- 40.Gutiérrez-Malacatt H, Ayala-Sanchez M, Aquino-Ortega X, Dominguez-Rodriguez J, Martinez-Tovar A, Olarte-Carrillo I, Martinez-Hernandez A, C CC, Orozco L, Cordova EJ. The rs61764370 functional variant in the KRAS oncogene is associated with chronic myeloid leukemia risk in women. Asian Pac J Cancer Prev. 2016;17:2265–70. doi: 10.7314/apjcp.2016.17.4.2265. [DOI] [PubMed] [Google Scholar]
- 41.López-López E, Gutiérrez-Camino Á, Piñán MÁ, Sánchez-Toledo J, Uriz JJ, Ballesteros J, García-Miguel P, Navajas A, García-Orad Á. Pharmacogenetics of microRNAs and microRNAs biogenesis machinery in pediatric acute lymphoblastic leukemia. PLoS One. 2014;9:e91261. doi: 10.1371/journal.pone.0091261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng C, Li X, Xia L, Fang X, Quan X, Yin Z, Zhao Y, Zhou B. Polymorphisms of pri-miR-219-1 are associated with the susceptibility and prognosis of non-small cell lung cancer in a Northeast Chinese population. Oncotarget. 2017;8:56533–56541. doi: 10.18632/oncotarget.17035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song X, You W, Zhu J, Cui X, Hu J, Chen Y, Liu W, Wang L, Li S, Wei Y, Yang L, Li F. A genetic variant in miRNA-219-1 is associated with risk of esophageal squamous cell carcinoma in chinese kazakhs. Dis Markers. 2015;2015:541531. doi: 10.1155/2015/541531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pei JS, Chang WS, Hsu PC, Chen CC, Chin YT, Huang TL, Hsu YN, Kuo CC, Wang YC, Tsai CW, Gong CL, Bau DT. Significant association between the miR146a genotypes and susceptibility to childhood acute lymphoblastic leukemia in Taiwan. Cancer Genomics Proteomics. 2020;17:175–180. doi: 10.21873/cgp.20178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Z, Lin Y, Kang S, Xu Q, Xiong W, Cai L, He F. miR-300 rs12894467 polymorphism may be associated with susceptibility to primary lung cancer in the Chinese Han population. Cancer Manag Res. 2018;10:3579–3588. doi: 10.2147/CMAR.S172514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He F, Lin J, Yu T, Zhang X, Liu Z, Xiong W, Cai L. Interaction research on smoking and microRNA genes SNP related to lung cancer in Fujian Han population. Zhonghua Yu Fang Yi Xue Za Zhi. 2016;50:168–74. doi: 10.3760/cma.j.issn.0253-9624.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 47.Dai ZM, Lv JR, Liu K, Lei XM, Li W, Wu G, Liu XH, Zhu YX, Hao Q, Dai ZJ. The role of microRNA-608 polymorphism on the susceptibility and survival of cancer: a meta-analysis. Aging (Albany NY) 2018;10:1402–1414. doi: 10.18632/aging.101476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu H, Zhou Y, Liu Q, Xiao G, Wang B, Li W, Ye D, Yu S. Association of miR-608 rs4919510 polymorphism and cancer risk: a meta-analysis based on 13,664 subjects. Oncotarget. 2017;8:37023–37031. doi: 10.18632/oncotarget.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


