Abstract
Over 70% of breast cancers express the estrogen receptor (ER) and depend on ER activity for survival and proliferation. While hormone therapies that target receptor activity are initially effective, patients invariably develop resistance which is often associated with activation of the phosphoinositide 3-kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR) pathway. While the mechanism by which estrogen regulates proliferation is not fully understood, one gene target of ER, growth regulation by estrogen in breast cancer 1 (GREB1), is required for hormone-dependent proliferation. However, the molecular function by which GREB1 regulates proliferation is unknown. Herein, we validate that knockdown of GREB1 results in growth arrest and that exogenous GREB1 expression initiates senescence, suggesting that an optimal level of GREB1 expression is necessary for proliferation of breast cancer cells. Under both of these conditions, GREB1 is able to regulate signaling through the PI3K/Akt/mTOR pathway. GREB1 acts intrinsically through PI3K to regulate phosphatidylinositol (3,4,5)-triphosphate levels and Akt activity. Critically, growth suppression of estrogen-dependent breast cancer cells by GREB1 knockdown is rescued by expression of constitutively activated Akt. Together, these data identify a novel molecular function by which GREB1 regulates breast cancer proliferation through Akt activation and provides a mechanistic link between estrogen signaling and the PI3K pathway.
While GREB1 has been known to regulate estrogen-dependent breast cancer proliferation, the molecular function of this protein has remained elusive. Herein, we show that GREB1 regulates the PI3K/Akt pathway to control breast cancer proliferation.
Introduction
Breast cancer is the most frequently diagnosed malignancy in women (1). Over 70% of breast cancer patients are diagnosed with the estrogen receptor-positive (ER+) subtype, which is characterized by the expression of the transcription factor ER and dependence on ER activity for tumor cell growth and survival (2–6). Patients diagnosed with the ER+ subtype of breast cancer are typically prescribed endocrine therapies that target ER activity (2,3,6). However, resistance to endocrine therapies invariably occurs, leading to reactivation of the ER, expression of ER-target genes, and ultimately, patient relapse (2,3,6). Treatment options for patients that are resistant to endocrine therapies are limited, highlighting the need for innovative therapies that target downstream of the ER (2,3,6).
Crosstalk between the ER and the PI3K/Akt/ mammalian target of rapamycin (mTOR) pathway has been implicated in ER+ breast cancer progression and resistance to endocrine therapies (7–10). The PIK3CA gene, which encodes the catalytic subunit of the PI3K enzyme, is the most commonly mutated gene in ER+ breast cancer patients with over 40% incidence (11). This mutation is speculated to be a causal event in breast cancer progression, suggesting there is some need to upregulate this pathway in the development of ER+ breast cancer (11). Studies have indicated the ER can interact with PI3K to regulate its kinase activity in an estrogen-dependent manner (12). Further, inhibition of the PI3K/Akt/mTOR pathway increases ER expression and sensitivity of breast cancer cells to tamoxifen treatment, suggesting that activation of this pathway is associated with resistance to endocrine therapies through downregulation of ER (13). Together, these data indicate interdependence between ER and PI3K/Akt/mTOR signaling, however, the molecular basis and clinical relevance for this cooperation remains unclear.
Herein, we show that the ER gene target, growth regulation by estrogen in breast cancer 1 (GREB1), is a regulator of the PI3K/Akt/mTOR pathway, linking ER activation to this critical signaling pathway. Expression of GREB1 has been highly correlated to ER-positivity in breast cancer cell lines and patient samples (14–17). Previous studies have shown that knockdown of GREB1 results in significantly reduced proliferation and colony formation of ER+ breast cancer cell lines indicating a required role for GREB1 in regulation of estrogen-dependent proliferation (16–18). However, it appears that an optimal level of GREB1 expression is necessary for proliferation of breast cancer cell lines as exogenous expression of GREB1 inhibits growth (18). Interestingly, growth repression by exogenous expression of GREB1 was also observed in ER-negative cell lines, indicating the ability of GREB1 to regulate proliferation of breast cancer cells is independent of ER activity (18). Despite the clear association of GREB1 and proliferation of ER+ breast cancer, the molecular function of the protein and the mechanism by which it regulates proliferation remain largely unknown. In this study, we characterize a novel mechanism by which GREB1 regulates proliferation of ER+ breast cancer cell lines through activation of Akt.
Materials and methods
Cell lines and reagents
MCF7, T47D, ZR-75-1 and HEK-293T cells were obtained in January 2013 and ZR-75-1 were obtained in September 2015. These cell lines were validated using Short Tandem Repeat analysis by the Genomics Core in the Research Technology Support Facility (Michigan State University, East Lansing, MI 48824) in May 2017. HCC1500 cells were purchased from the American Type Culture Collection in March 2018. Cells were maintained as described previously (18). For experiments with epidermal growth factor (EGF) stimulation, cells were cultured in serum-free media for 16 h before being stimulated with 1 ng/ml recombinant human EGF (Thermo) for the indicated time. For all other assays, cells were cultured in Dulbecco’s modified Eagle’s medium plus fetal bovine serum, containing endogenous estrogens. The inhibitor GDC-0941 was obtained from Cayman Chemicals and were used at the indicated concentration for 24 h prior to harvest of the cells.
Plasmids
3XFLAG PCDNA and H2BGFP have been described previously (18–20). GIPZ lentiviral nonspecific shRNA (# RHS4346) and lentiviral GREB1-targeted shRNA plasmids (V2LHS_139192 and V3LHS_372339) were obtained from Open Biosystems. MISSION shRNA constructs targeted to PIK3CA (TRCN0000196 582, TRCN0000195 203, TRCN0000010 406), PTEN (TRCN0000002 745, TRCN0000002 747, TRCN0000002 749) and PDK1 (TRCN0000001 476, TRCN0000039 778, TRCN0000039 782, TRCN0000010 413) were purchased from Sigma–Aldrich. Myristoylated (Myr) AKT1 from pBabe-Puro-Myr-Flag-AKT1 (21) was cloned into a pLenti-hygro backbone to create pLenti hygro Myr FLAG AKT1 (CA AKT) using standard Gibson cloning (NEB).
Immunoblot analysis and antibodies
Cell lysates were prepared, subjected to immunoblot analysis and visualized on a LI-COR Odyssey system as described previously (18,22). Immunoblots were probed with the following antibodies: GREB1 (abcam; ab72999 or CST; P76195 Clone 9C1), β-actin [Cell Signaling Technologies (CST); 3700], phosphor-p38 (CST; 9211S), p38 (CST; 9212), phosphor-MEK1/2 (CST; 9121), MEK1/2 (CST; 9122), phosphor-ERK1/2 (CST; 4370); ERK1/2 (CST; 4695), phospho-MKK3/6 (CST; 12280), phospho-MSK1 (CST; 9595), phospho-ATF2 (CST; 5112), phospho-HSP27 (CST; 9709), phospho-MAPKAPK2 (CST; 3007), phospho-PTEN (CST; 9551), PTEN (CST; 9556), phospho-PDK1 (CST; 3438), PDK1 (CST; 5662), phospho-Akt Thr308 (CST; 13038), phospho-Akt Ser473 (CST; 9271S), Akt (CST; 2920S), PP2A (CST; 2041T), phospho-glycogen synthase kinase 3β (CST; 5558), mTOR (CST; 2983), Rictor (CST; 2114), Raptor (CST; 2280) and p110α (CST; 4249T). Densitometry analysis was performed using ImageJ. Statistical significance was determined using a two-way analysis of variance with Fisher’s least significant difference test.
Adenovirus
GREB1 adenovirus was purified as described previously (18). Ad5-CMV-eGFP adenovirus (Baylor College of Medicine Vector Development Labs, Houston, TX 77030) was used as a control. The MOI for experiments was determined by monitoring green fluorescent protein (GFP) expression in cells such that the minimum of adenovirus was transduced to generate greater than 95% GFP positivity. For proliferation under varying levels of GREB1 expression, the multiplicity of infection (MOI) was adjusted from 66 to 200% of this preidentified level.
Alamar blue assay
Cells were treated with 0.04 g/l resazurin sodium salt in phosphate-buffered saline (PBS) at 37°C for 1 h. Fluorescence was measured on a BioTek Synergy microplate reader using a 540/35 excitation filter and a 590/20 emission filter. Data are depicted as mean fluorescence normalized to day 0 ± SD for each condition from three biological replicates. Statistical significance for alamar blue assays was determined using either a two-tailed Student’s t-test (exogenous GREB1 expression) or a one-way analysis of variance with post hoc Tukey’s honestly significant difference test (shRNA experiments).
SA-β-gal staining
Cells transduced with GFP or GREB1 adenovirus were plated on poly-l-lysine-coated coverslips. Cells were fixed and stained for SA-β-gal activity using the Senescence β-Galactosidase Staining Kit (CST #9860) as described previously (23). Total and SA-β-gal-positive cells were blindly scored and significance determined by two-tailed Student’s t-test.
Conditioned media assay
MCF7 cells were transduced with either GFP or GREB1 adenovirus. After 24 h, transduced cells were washed in PBS and fresh media added. The following day, media was collected from the transduced cells and centrifuged at 500g for 5 min to pellet any cellular debris. Target cells were washed twice with PBS prior to the addition of conditioned media. After 24 h, all cells were harvested by scraping in cold PBS containing 10 nM calyculin A (Cell Signaling).
Coculture assay
MCF7 cells were transduced with adenovirus expressing either GFP or GREB1 (both adenovirus vectors express GFP). The following day, transduced cells were trypsinized and replated at a 1:1 ratio with untransduced MCF7 cells. Cells were harvested after 24 h by trypsinization and washed twice with cold PBS containing 10 nM calyculin A (Cell Signaling). Cells were sorted from both the adGFP and adGREB1 cocultures using a Becton Dickinson FACSAria II cell sorter into GFP-positive (transduced) and GFP-negative (untransduced) populations. Cell lysates were prepared from the sorted populations and immunoblot analysis was performed as described above.
Immunofluorescence microscopy
Cells were plated on poly-l-lysine-coated coverslips. Following treatment, cells were fixed in 4% methanol-free formaldehyde (Thermo) diluted in PBS for 15 min at room temperature. Cells were then washed three times in PBS before permeabilization with 0.5% saponin, 1% bovine serum albumin PBS solution at room temperature for 15 min. The cells were labeled with the indicated primary antibodies for 2 h at room temperature in a humidified chamber. Coverslips were then washed three times in PBS before incubation with secondary antibodies (Alexa Fluor 555 goat anti-mouse and Alexa Fluor 555 goat anti-rabbit; Invitrogen) at room temperature for 1 h in a humidified chamber, protected from light. Coverslips were mounted on microscope slides with VECTASHIELD Hard Set Mounting Medium with 4′,6-diamidino-2-phenylindole (Vector Laboratories). Images were obtained using a spinning disk confocal microscope (Ultra-VIEW VoX CSU-X1 system; Perkin Elmer) and analyzed using Velocity (Perkin Elmer). Images shown are representative of three biological replicates.
PIP3 quantification
MCF7 cells were transduced with either GFP or GREB1 adenovirus. 24 h after transduction, cells were placed in serum-free, phenol-red-free Dulbecco’s modified Eagle’s medium for 16 h. Lipids were extracted and phosphatidylinositol (3,4,5)-triphosphate (PIP3) levels measured using the PIP3 Mass ELISA kit (Echelon K-2500s) according to the manufacturer’s instructions.
Results
GREB1 initiates cellular senescence
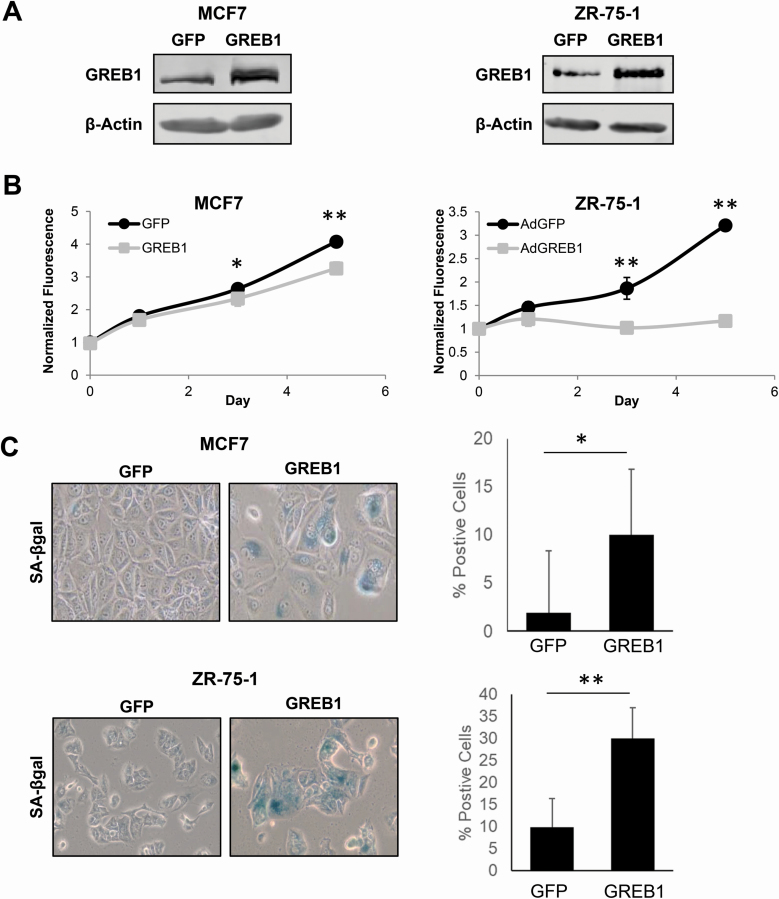
Our previous work has suggested that an optimal level of GREB1 expression is necessary for proliferation of breast cancer cell lines. This work demonstrated that both GREB1 knockdown (18) and exogenous expression of GREB1 results in growth arrest (Figure 1A and B, Supplementary Figure 1, available at Carcinogenesis Online) (18). We previously reported that exogenous expression of GREB1 did not induce apoptosis (18); thus, we investigated the ability of GREB1 overexpression to initiate cellular senescence. Two ER+ breast cancer cell lines, MCF7 and ZR-75-1 cells, were transduced with adenovirus expressing either GFP or GREB1. Following 7 days of exogenous GREB1 expression, the cells were fixed and stained for SA-β-galactosidase activity, a marker of cellular senescence (23,24). Compared with GFP control cells, cells overexpressing GREB1 had a large, flattened morphology and characteristic blue staining associated with SA-β-galactosidase activity (Figure 1C). Quantification revealed a significant increase in the percentage of stained cells when GREB1 was overexpressed compared with GFP control in both MCF7 and ZR-75-1 cells (Figure 1C). These data suggest that exogenous GREB1 expression is able to induce cellular senescence to inhibit proliferation of breast cancer cell lines.
Figure 1.
Exogenous GREB1 initiates cellular senescence. MCF7 or ZR-75-1 cells were transduced with adenovirus expressing GFP or GREB1. (A) Immunoblot depicting the relative overexpression of GREB1 in cells transduced with GREB1 adenovirus compared with GFP control. (B) Proliferation of transduced cells was measured by alamar blue assay. Data are plotted as mean fluorescence normalized to day 0 ± SD; n = 3 for each cell line. *P ≤ 0.05, **P ≤ 0.005. (C) Cells were fixed and stained for SA-β-galactosidase activity 7 days post-transduction. Data are plotted as percent of SA-β-galactosidase stained cells ± SD. *P ≤ 0.05, **P ≤ 0.01.
Exogenous GREB1 expression induces hyperactivation of the PI3K/Akt/mTOR pathway
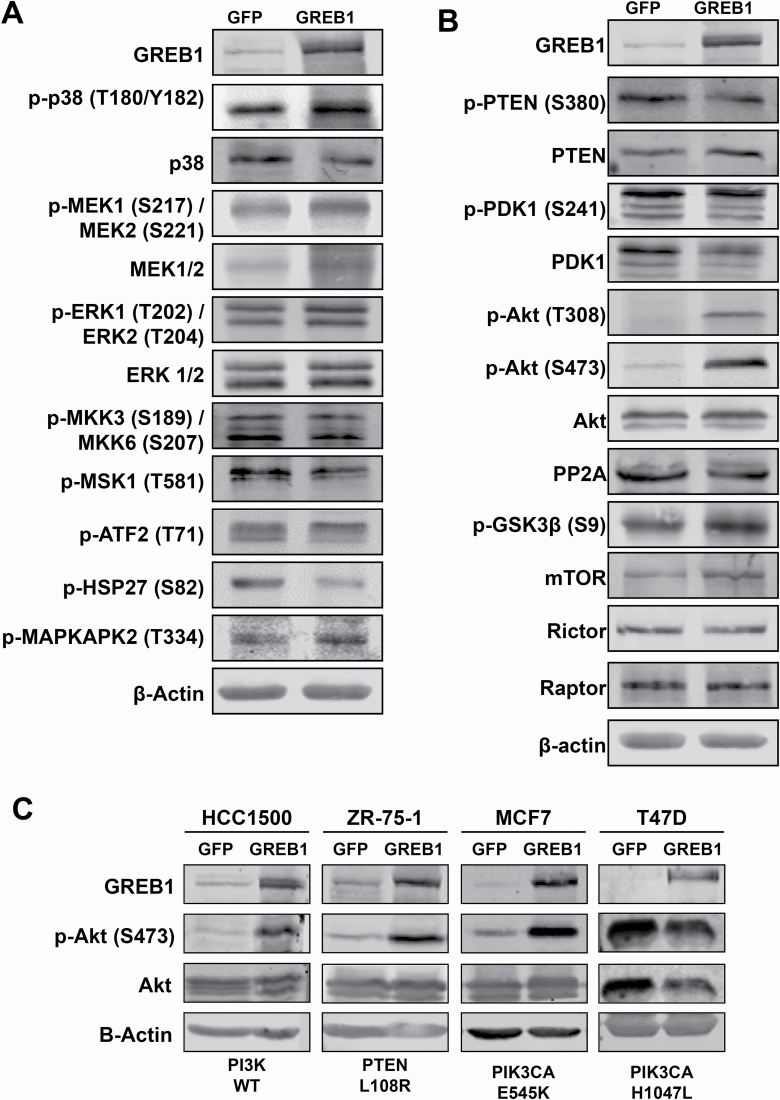
In order to delineate the mechanism by which GREB1 regulates proliferation, we chose to focus our attention on two signaling pathways thought to play a critical role in regulating both senescence and proliferation phenotypes: the p38 mitogen-activated protein kinase (MAPK) pathway and the PI3K/Akt/mTOR pathway (25–27). MCF7 cells were transduced with adenovirus expressing GFP or GREB1 and after 48 h, cell lysates were analyzed by immunoblot for activation of various nodes in the p38 MAPK pathway (Figure 2A) and the PI3K/Akt/mTOR pathway (Figure 2B). Our data indicate that exogenous GREB1 expression induces an increase in the activation and phosphorylation of p38, however, the activation and/or expression of upstream regulators of p38 (MEK1/2, ERK1/2 and MKK3/6) and downstream effectors (MSK1, ATF2, HSP27 and MAPKAPK2) were largely unaffected (Figure 2A). Analysis of the PI3K/Akt/mTOR pathway revealed hyperactivation of Akt, as well as increased phosphorylation of glycogen synthase kinase 3β, a downstream effector of Akt, when GREB1 was exogenously expressed in MCF7 cells (Figure 2B). As the PI3K/Akt/mTOR pathway is frequently altered in breast cancer, we analyzed the effect of GREB1 overexpression on Akt activation in a panel of ER+ breast cancer cell lines with wildtype or differing alterations to the PI3K/Akt/mTOR pathway (28). While MCF7, ZR-75-1 and T47D cells harbor mutations in this pathway, each cell line has varying levels of Akt activation with MCF7 cells demonstrating the least constitutive activity (Supplementary Figure 2A, available at Carcinogenesis Online). In cells lines without constitutive maximal activity of this pathway, GREB1 induces a strong activation of Akt (Figure 2C). In contrast, T47D cells, which have a PIK3CA mutation resulting in constitutively hyperactive Akt activity (Supplementary Figure 2A, available at Carcinogenesis Online), are unaffected by GREB1 expression (Figure 2C).
Figure 2.
GREB1 modulates PI3K/Akt pathway signaling. (A) MCF7 cells were transduced with adenovirus expressing GFP or GREB1. Cell lysates were harvested 2 days post-transduction and analyzed by immunoblot for indicated proteins in the p38 MAPK pathway. (B) MCF7 cells were transduced with adenovirus expressing GFP or GREB1. Cell lysates were harvested 2 days post-transduction and analyzed by immunoblot for indicated proteins in the PI3K/Akt/mTOR pathway. (C) HCC1500, ZR-75-1, MCF7 and T47D cells were transduced with adenovirus expressing GFP or GREB1. Cell lysates were harvested 2 days post-transduction and analyzed by immunoblot for indicated proteins.
GREB1-induced hyperactivation of Akt is PI3K dependent
Akt requires phosphorylation at two sites, Thr308 and Ser473, by PDK1 and mTORC2, respectively, for maximal activation (29–31). Both phosphorylation events are dependent on PI3K and occur downstream of PI3K conversion of PIP2 to PIP3 (29,31). Thus, we sought to determine if GREB1 was acting to regulate Akt activation upstream or downstream of PI3K. To this end, we targeted PI3K activity with the pharmaceutical inhibitor GDC0941. MCF7 cells were simultaneously treated with dimethyl sulfoxide or GDC0941 and transduced with adenovirus expressing GFP or GREB1. After 24 h, cell lysates were harvested and activation of Akt at Thr308 and Ser473 were evaluated by immunoblot analysis. The expected hyperactivation of Akt was observed in dimethyl sulfoxide-treated cells that were transduced with GREB1 adenovirus (Figure 3A). PI3K inhibition by GDC0941 demonstrated a decrease in basal Akt activation in the control GFP-transduced cells (Figure 3A). A significant decrease in GREB1-mediated phosphorylation of Akt at both Ser473 and Thr308 was observed upon PI3K inhibition in comparison to dimethyl sulfoxide control (Figure 3A). These data suggest that GREB1 is activating the canonical PI3K/Akt signaling axis.
Figure 3.
GREB1-induced hyperactivation of Akt is PI3K dependent. (A) MCF7 cells were transduced with adenovirus expressing GFP or GREB1 and treated with dimethyl sulfoxide (DMSO) or 40 nM GDC0941 simultaneously. Cell lysates were harvested 24 h post-transduction and immunoblot analysis was performed with the indicated antibodies. Immunoblot shown is representative of at least four biological replicates. Histograms depict average phospho-Akt levels normalized to total Akt relative to GFP-overexpressing, DMSO-treated cells ± SD. *P ≤ 0.05 comparison to GFP DMSO; **P ≤ 0.01 comparison to GFP DMSO; ΔΔ P ≤ 0.01 comparison to GREB1 DMSO; ++P ≤ 0.01 comparison to GFP GDC0941. (B) MCF7 cells were transduced with lentivirus targeted to a nonspecific control (shNS), PIK3CA, PTEN or PDK1. Following selection, cells were transduced with GFP or GREB1 adenovirus. Cell lysates were harvested 24 h post adenovirus transduction. Following sodium dodecyl sulfate–polyacrylamide gel electrophoresis, immunoblot analysis was performed with indicated antibodies. Immunoblot shown is representative of at least four biological replicates. Histograms depict average phospho-Akt levels normalized to total Akt relative to GFP-overexpressing, shNS cells ± SD. *P ≤ 0.05 comparison to shNS GFP, **P ≤ 0.01 comparison to shNS GFP, +P ≤ 0.05 comparison to shNS GREB1, Δ P ≤ 0.05 comparison within same shRNA between GFP and GREB1.
To confirm this result and further probe other nodes in the pathway, MCF7 cells were transduced with lentivirus expressing shRNA targeted to a nonspecific control (shNS), PIK3CA, PTEN or PDK1. Following selection, cells were transduced with adenovirus expressing GFP or GREB1. After 24 h, cell lysates were harvested and analyzed via immunoblot. Control cells transduced with nonspecific shRNA demonstrated the expected significant increase in phosphorylation of Akt (Thr308 and Ser473) when GREB1 was exogenously expressed (Figure 3B). Knockdown of PIK3CA expression reduced Akt activation in control cells and significantly impaired Akt activation in GREB1-expressing cells (Figure 3B). Knockdown of PTEN had no effect on GREB1-induced hyperactivation of Akt at Thr308 compared with the nonspecific (NS) control, suggesting the mechanism of GREB1 action is not through phosphatase inhibition (Figure 3B). Further, GREB1-induced hyperactivation of Akt at Thr308 was reduced by knockdown of PDK1, the primary kinase for this site (29–32), to the point at which there was no significant difference between the level of activation of Akt at Thr308 between GFP- and GREB1-overexpressing cells (Figure 3B). Knockdown of all PI3K pathway components diminished phosphorylation of the downstream target, glycogen synthase kinase 3β (Figure 3B, Supplementary Figure 2B, available at Carcinogenesis Online). These data further demonstrate that GREB1-induced hyperactivation of Akt is dependent on signaling through the canonical PI3K pathway.
GREB1 activates Akt through intracellular mechanisms
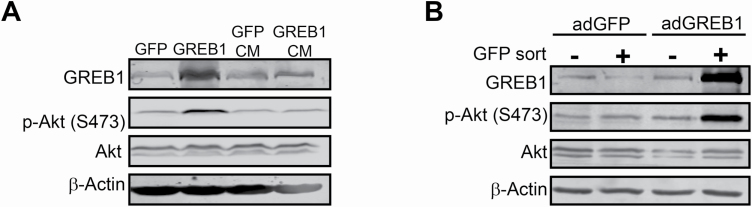
Canonical activation of PI3K and Akt occurs through activation of receptor tyrosine kinases (RTK) or G-protein-coupled receptors by external stimuli (29,33). We first sought to determine if GREB1-mediated Akt regulation is dependent upon induction and secretion of a signaling molecule that activates the PI3K/Akt/mTOR pathway. To this end, MCF7 cells were transduced with adenovirus expressing GFP or GREB1. Media from transduced cells was transferred to untransduced MCF7 cells. After 24 h, cell lysates were harvested and activation of Akt was analyzed by immunoblot. As expected, exogenous expression of GREB1-induced hyperactivation of Akt at Ser473 when compared with GFP-transduced cells (Figure 4A). However, conditioned media was unable to induce hyperactivation of Akt in untransduced cells (Figure 4A).
Figure 4.
GREB1 activates Akt through intracellular mechanisms. (A) Conditioned media from MCF7 cells transduced with GFP or GREB1 adenovirus was added to untransduced cells. Cell lysates from transduced cells (GFP or GREB1) and untransduced cells cultured in conditioned media (GFP CM or GREB1 CM) were harvested 24 h later. Lysates were analyzed by immunoblot for indicated proteins. (B) MCF7 cells were transduced with adenovirus expressing GFP or GREB1. Transduced cells were then cultured with untransduced cells at a 1:1 ratio for 24 h. Cells were harvested and sorted for GFP. Cell lysates were analyzed via immunoblot for expression of the indicated proteins.
Alternatively, exogenous GREB1 may induce the expression of a membrane-bound signaling molecule that could activate the PI3K/Akt/mTOR pathway. To investigate this possibility, MCF7 cells were transduced with adenovirus expressing GFP or GREB1 and then cocultured at a 1:1 ratio with untransduced MCF7 cells. After 24 h, the cells were harvested and GFP-positive, adenovirus-transduced cells (GFP or GREB1), were sorted from GFP-negative, untransduced cells. All populations were then analyzed for activation of Akt by immunoblot. Both GFP-transduced cells and cells cocultured with the GFP-transduced cells had similar levels of Akt activation (Figure 4B). Exogenous expression of GREB1 induced the expected hyperactivation of Akt at Ser473 within the transduced cells; however, the untransduced coculture cells did not demonstrate Akt hyperactivation (Figure 4B). Together, these data clearly demonstrate that GREB1 regulates Akt activation through in an intracellular mechanism that does not require extracellular activation of RTKs.
Exogenous GREB1 promotes recruitment of Akt to the plasma membrane
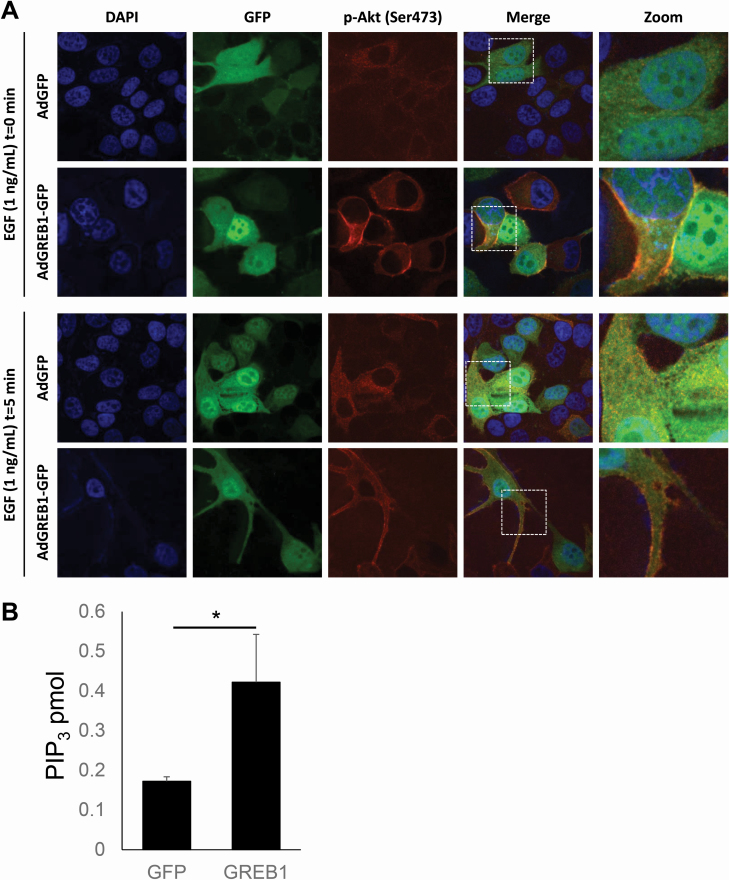
Akt is typically activated via recruitment to the plasma membrane by interaction with PIP3, however, it is believed that there are other pools of activated Akt on endomembrane surfaces and within the nucleus (29). To determine the localization of activated Akt induced by exogenous GREB1 expression, MCF7 cells were transduced with either GFP or GREB1 adenovirus. Following transduction, the cells were serum-starved for 16 h to reduce basal Akt activation before stimulation of the pathway with EGF. Cells were then fixed and stained with 4′,6-diamidino-2-phenylindole and the indicated Akt antibodies. As both adenoviral vectors expressed GFP, we focused our imaging on transduced cell populations. In serum-starved, GFP-transduced cells, staining for activated Akt was minimal and staining for total Akt resulted in diffuse staining throughout the cytoplasm (Figure 5A, Supplementary Figure 3A and B, available at Carcinogenesis Online). In contrast, cells transduced with GREB1 adenovirus under serum-starved conditions had distinct staining for activated and total Akt, primarily localized to the plasma membrane (Figure 5A, Supplementary Figure 3A and B, available at Carcinogenesis Online). When stimulated with EGF for 5 min, both GFP- and GREB1-transduced cells demonstrated activated Akt and total Akt at the plasma membrane (Figure 5A, Supplementary Figure 3A and B, available at Carcinogenesis Online). Activation of Akt and focal localization of total Akt at the plasma membrane was noticeably stronger in GREB1-transduced cells when compared with GFP-transduced cells in the presence of EGF (Figure 5A, Supplementary Figure 3A and B, available at Carcinogenesis Online). These data further suggest that GREB1 may act through PI3K to increase PIP3 levels and resulting in relocalization of Akt to the plasma membrane.
Figure 5.
Exogenous GREB1 promotes recruitment of Akt to the plasma membrane. (A) MCF7 cells were transduced with adenovirus expressing GFP or GREB1. The cells were then cultured in serum-free media for 16 h before being stimulated with 1 ng/ml EGF for 0 or 5 min. Cells were fixed and stained for 4′,6-diamidino-2-phenylindole or p-Akt (Ser473). Immunofluorescence microscopy was used to visualize the activation and localization of Akt. (B) MCF7 cells were transduced with adenovirus to express exogenous GFP or GREB1 and serum starved for 16 h. Lipids were extracted from all samples and levels of PIP3 were measured via enzyme-linked immunosorbent assay. Graphs represent mean PIP3 (pmol) + SD (n = 3). *P ≤ 0.05.
We sought to directly test if GREB1 expression influenced the conversion of PIP2 to PIP3. MCF7 cells were transduced with GFP and GREB1 adenovirus and then placed in serum-free media as in the experiments described above for Figure 5A. Following serum starvation for 16 h, lipids were extracted and PIP3 levels measured by enzyme-linked immunosorbent assay. Expression of exogenous GREB1 induced a significant increase in PIP3 levels, further indicating that GREB1 augments PI3K activity.
Previous studies have suggested that GREB1 is primarily localized to the nucleus in patient samples and breast cancer cell lines (15,16), thus, it remained unclear how GREB1 was able to regulate signaling through a primarily cytoplasmic pathway. Interestingly, we discovered that under serum-starved conditions, endogenous GREB1 is diffuse throughout the cytoplasm and nucleus in MCF7 cells, but upon stimulation with EGF, the vast majority of GREB1 rapidly relocalizes to the cytoplasm (Supplementary Figure 4A, available at Carcinogenesis Online). As this is contradictory to previously published reports, we performed nuclear/cytoplasmic fractionation to verify cytoplasmic expression of GREB1. Under normal growth conditions (i.e. media containing fetal bovine serum), GREB1 is primarily located within the cytoplasm of MCF7 cells (Supplementary Figure 4B and C, available at Carcinogenesis Online). Thus, in response to growth factor activation, GREB1 localizes to the cytoplasm wherein it can modulate the PI3K pathway.
GREB1 regulates breast cancer proliferation through activation of the PI3K/Akt/mTOR pathway
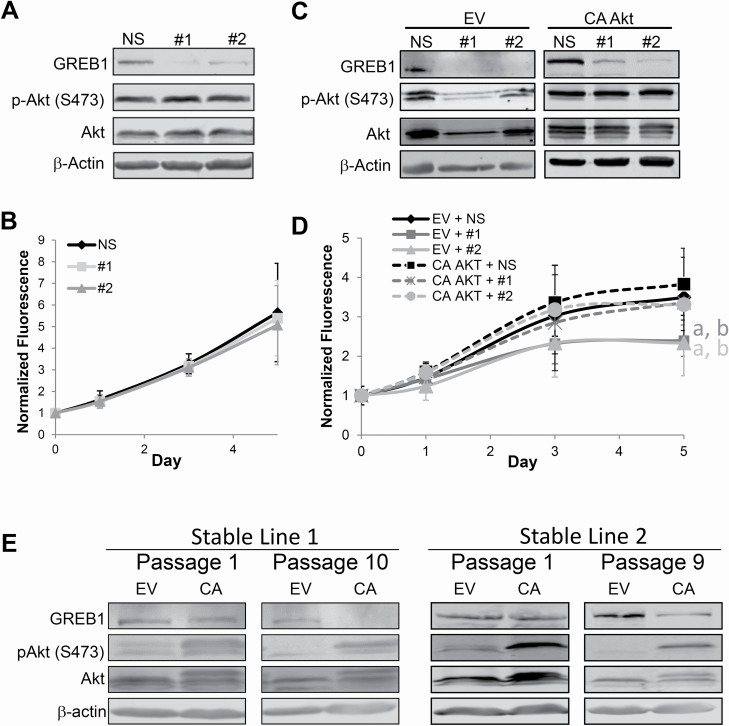
In order to determine if GREB1-mediated Akt activation is imperative for proliferation of ER+ breast cancer cells, we tested whether constitutively activated Akt can rescue proliferation loss in GREB1-depleted cells. Thus, we made use of T47D cells which are ER+ and GREB1-expressing, but harbor a PI3KCAH1047L mutation rendering this pathway constitutively active and unresponsive to typical PI3K/Akt/mTOR-activating stimuli, including GREB1 exogenous expression (Figure 2C and Supplementary Figure 2A, available at Carcinogenesis Online). T47D cells were transduced with shRNA targeting a NS control or shRNA targeting GREB1 (#1 or #2). Immunoblot analysis confirmed knockdown of GREB1, as well as hyperactivation of Akt in T47D cells (Figure 6A). Proliferation of these cells was then monitored via alamar blue assay. GREB1 knockdown had no effect on the proliferation of T47D cells (Figure 6B), suggesting constitutive Akt activation abrogates the need for GREB1 expression.
Figure 6.
GREB1 regulates breast cancer proliferation through activation of the PI3K/Akt pathway. (A) T47D cells were transduced with lentivirus expressing NS shRNA or shRNA targeted to GREB1 (#1 or #2). Immunoblot depicting the expression of indicated proteins. (B) Proliferation was measured via alamar blue assay. Data are plotted as mean fluorescence normalized to day 0 ± SD; n = 3. (C) MCF7 cells were transduced with lentivirus expressing empty vector (EV) or myristoylated Akt (CA AKT) and either NS shRNA NS or shRNA targeted to GREB1 (#1 or #2). Representative immunoblot depicting expression of indicated proteins. (D) Proliferation was measured via alamar blue assay. Data are plotted as mean fluorescence normalized to day 0 ± SD; n = 3; aP < 0.05 versus EV + NS; bP < 0.05 versus CA AKT + NS. (E) Following transduction with control (EV) or constitutively active Akt (CA) expressing lentivirus, MCF7 stable lines were generated by placing cells on selection. Lysates from different passages were probed for GREB1 expression and Akt activation. Two distinct stable lines are depicted (left and right panels, respectively).
The proliferation of ER+ and GREB1-expressing MCF7 cells has previously been shown to be dependent on expression of GREB1 (16–18). While MCF7 cells also harbor a mutation in PIK3CAE545K, which is thought to be an activating mutation (34), these cells are still responsive to typical PI3K/Akt/mTOR-activating stimuli (Supplementary Figure 2, available at Carcinogenesis Online). Thus, we sought to determine if constitutively activated Akt (myristoylated Akt) would rescue proliferation in GREB1-depleted MCF7 cells. MCF7 cells were transduced with empty vector lentivirus or lentivirus expressing constitutively activated Akt (CA AKT) in combination with lentivirus expressing shRNA targeted to a NS control or to GREB1 (#1 or #2). Knockdown of endogenous GREB1 resulted in decreased Akt activation in cells cotransduced with GREB1-targeted shRNA and empty vector lentivirus (Figure 6C) as well as parental cells infected with control lentivirus (Supplementary Figure 5, available at Carcinogenesis Online). The knockdown of GREB1 significantly impaired the growth of MCF7 cells cotransduced with empty vector lentivirus (Figure 6D). However, expression of constitutively active Akt (Figure 6C), rescues the proliferation phenotype caused by GREB1 knockdown to that of control transduced cells (Figure 6D). Together, these data demonstrate that the primary mechanism by which GREB1 drives estrogen-dependent proliferation is through modulation of Akt activity. Interestingly, stable expression of constitutively active Akt results in long-term silencing of GREB1 (Figure 6E), suggesting a feedback loop and potentially explaining the observation that GREB1 expression is reduced in hormone-refractory disease (16).
Discussion
Despite extensive research on hormone signaling in breast cancer, the explicit mechanism by which ER drives proliferation remains largely undefined. In order to delineate this mechanism, concerted efforts have been made to identify ER-target genes involved in estrogen-induced proliferation of breast cancer cells. Several of these studies have identified GREB1 as a gene that is required for estrogen-stimulated proliferation of breast cancer cell lines (14,16,17). Previous studies have suggested that GREB1 regulates proliferation through modulation of ER activity (16). However, our findings show that GREB1 is not a potent regulator of ER activity and has the ability to affect the proliferation of breast cancer cell lines independent of ER expression and action (18). Here, we suggest a novel mechanism by which GREB1 regulates proliferation through fine-tuning of PI3K/Akt/mTOR signaling.
Optimal level of GREB1 expression is necessary for proliferation
Our findings demonstrate that knockdown of GREB1 results in growth arrest and overexpression of GREB1 induces cellular senescence (Figure 1). These data are similar to that of other oncogenes (e.g. Ras) in which activation of the oncogene acts to stimulate proliferation but hyperexpression and activation results in senescence and growth arrest (35). When we probed pathways associated with both proliferation and senescence, we found that overexpression of GREB1 results in hyperactivation of Akt (Figure 2) through the canonical PI3K pathway (Figure 3). Interestingly, constitutive activation of Akt alone did not result in cellular senescence (Figure 6, CA AKT + NS), suggesting GREB1 may regulate additional pathways involved in senescence and growth arrest. Thus, while exogenous expression of GREB1 may not be biologically relevant, it provides us with a tool to investigate the role of GREB1 in regulating cell signaling and proliferation in parallel with loss-of-function assays.
GREB1 regulates proliferation of ER+ breast cancer cells through modulation of Akt activity
Several studies have indicated complex crosstalk between ER signaling and PI3K/Akt/mTOR pathway activation and implications for this crosstalk in resistance to endocrine therapy in breast cancer patients (7–9,36–38). However, no studies have described a comprehensive connection between activation of these signaling pathways and proliferation of breast cancer cells. The difficulty to assess this connection is compounded by the fact that the vast majority of available breast cancer cell lines contain mutations involved in the PI3K/Akt/mTOR pathway (39). Although most ER+ breast cancer cell lines contain mutations within this pathway, the specific mutations have distinctly different effects on the activation of the PI3K/Akt/mTOR pathway. Specifically, breast cancer cell lines harboring PIK3CAH1047R mutations (e.g. T47D) have significantly higher intrinsic PI3K activity compared with breast cancer cell lines harboring PIK3CAE545K mutations (e.g. MCF7), which have subtle effects on activation of the PI3K/Akt/mTOR pathway (11,40). Using this to our advantage we show that in T47D cells, which harbor a constitutively active PI3K/Akt/mTOR pathway, GREB1 is no longer required for proliferation (Figure 6A and B). However, in MCF7 cells, which harbor a PIK3CA mutation but still respond to pathway-activating stimuli, GREB1 is still required but knockdown of GREB1 can be rescued by constitutively active Akt (Figure 6C and D). Together, these data demonstrate that the requirement of GREB1 for hormone-responsive proliferation is dependent upon this novel function to alter Akt activity. Furthermore, the activation of the PI3K/Akt by GREB1 occurs through intracellular mechanisms (Figure 4), highlighting a potentially new mode of modulating this signaling pathway without the need for RTK activation.
GREB1 and endocrine resistance
Despite the clear association between GREB1 and proliferation of breast cancer cells, expression of GREB1 has been correlated to better prognosis in ER+ breast cancer patients (16,41). In a study that included only patients that received adjuvant tamoxifen monotherapy, higher GREB1 expression correlated with both prolonged disease-free survival and sensitivity to tamoxifen treatment (41). Similarly, in an in vitro model of tamoxifen resistance, MCF7 cells that were resistant to tamoxifen treatment had significantly less GREB1 expression compared with the parental line, suggesting GREB1 expression is lost in hormone-refractory breast cancer cells (16). These data have implicated loss of GREB1 as a causal event for therapy resistance. Here, we show that proliferation of breast cancer cells with constitutively active PI3K/Akt/mTOR signaling no longer require GREB1 expression (Figure 6). Constitutive activation of the PI3K/Akt/mTOR pathway is frequently associated with resistance to endocrine therapies and is the basis for numerous clinical trials investigating PI3K/Akt/mTOR pathway inhibitors in endocrine-resistant patient populations (7–10). In patients with hormone-refractory disease with hyperactivation of the PI3K/Akt/mTOR pathway, the pressure to express GREB1 is lost. Thus, decreased GREB1 expression in advanced disease may be the result of constitutive PI3K/Akt/mTOR activity rather than a cause of therapeutic bypass. In support of this notion, stable expression of constitutively active Akt resulted in silencing of GREB1 expression after several passages (Figure 6). These findings warrant further research into the use of GREB1 as a clinical biomarker for treatment selection.
Conclusions
Herein, we identify a novel function of GREB1 by which it modulates cell signaling pathways to influence proliferation. We demonstrate that GREB1 has the ability to influence Akt activation through regulation of PI3K activity and PIP3 conversion. This regulation is dependent upon intracellular mechanisms and not through induced expression of autocrine/paracrine factors that would canonically activate PI3K through RTKs. Critically, GREB1-mediated activation of Akt is essential for estrogen-dependent proliferation, as GREB1 knockdown can be rescued by Akt activation. Together, these identify a novel function of the GREB1 protein that has long been known to influence cancer proliferation.
Supplementary Material
Acknowledgements
We thank Paul Herman, Clarissa Wormsbaecher, Weiwei Liu and Makanko Komara for critical reading of this manuscript prior to submission. We thank Philip Tsichlis for input into the design of this project.
Glossary
Abbreviations
- EGF
epidermal growth factor
- ER
estrogen receptor
- MAPK
mitogen-activated protein kinase
- PBS
phosphate-buffered saline
- RTK
receptor tyrosine kinase
Funding
The work was supported by startup funds from The Ohio State University, P30CA016058 (National Cancer Institute, National Institutes of Health), and a Pelotonia Fellowship to Corinne Haines.
Conflict of Interest Statement: None declared.
References
- 1. Siegel RL et al. (2018) Cancer statistics, 2018. CA Cancer J. Clin., 68, 7–30. [DOI] [PubMed] [Google Scholar]
- 2. Clarke R et al. (2015) Endocrine resistance in breast cancer—an overview and update. Mol. Cell. Endocrinol., 418(Pt 3), 220–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dixon JM. (2014) Endocrine resistance in breast cancer. N. J. Sci., 2014, 1–27. [Google Scholar]
- 4. Libson S et al. (2014) A review of clinical aspects of breast cancer. Int. Rev. Psychiatry, 26, 4–15. [DOI] [PubMed] [Google Scholar]
- 5. Patani N et al. (2014) Understanding response and resistance to oestrogen deprivation in ER-positive breast cancer. Mol. Cell. Endocrinol., 382, 683–694. [DOI] [PubMed] [Google Scholar]
- 6. Ali S et al. (2002) Endocrine-responsive breast cancer and strategies for combating resistance. Nat. Rev. Cancer, 2, 101–112. [DOI] [PubMed] [Google Scholar]
- 7. Campbell RA et al. (2001) Phosphatidylinositol 3-kinase/AKT-mediated activation of estrogen receptor alpha: a new model for anti-estrogen resistance. J. Biol. Chem., 276, 9817–9824. [DOI] [PubMed] [Google Scholar]
- 8. Miller TW et al. (2011) Phosphatidylinositol 3-kinase and antiestrogen resistance in breast cancer. J. Clin. Oncol., 29, 4452–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miller TW et al. (2010) Hyperactivation of phosphatidylinositol-3 kinase promotes escape from hormone dependence in estrogen receptor-positive human breast cancer. J. Clin. Invest., 120, 2406–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miller WR et al. (2009) Gene expression profiles differentiating between breast cancers clinically responsive or resistant to letrozole. J. Clin. Oncol., 27, 1382–1387. [DOI] [PubMed] [Google Scholar]
- 11. Ellis MJ et al. (2013) The genomic landscape of breast cancer as a therapeutic roadmap. Cancer Discov., 3, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Simoncini T et al. (2000) Interaction of oestrogen receptor with the regulatory subunit of phosphatidylinositol-3-OH kinase. Nature, 407, 538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Creighton CJ et al. (2010) Proteomic and transcriptomic profiling reveals a link between the PI3K pathway and lower estrogen-receptor (ER) levels and activity in ER+ breast cancer. Breast Cancer Res., 12, R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ghosh MG et al. (2000) PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer Res., 60, 6367–6375. [PubMed] [Google Scholar]
- 15. Hnatyszyn HJ et al. (2010) Correlation of GREB1 mRNA with protein expression in breast cancer: validation of a novel GREB1 monoclonal antibody. Breast Cancer Res. Treat., 122, 371–380. [DOI] [PubMed] [Google Scholar]
- 16. Mohammed H et al. (2013) Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep., 3, 342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rae JM et al. (2005) GREB 1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res. Treat., 92, 141–149. [DOI] [PubMed] [Google Scholar]
- 18. Haines CN et al. (2018) GREB1 isoforms regulate proliferation independent of ERα co-regulator activities in breast cancer. Endocr. Relat. Cancer, 25, 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Burd CJ et al. (2005) Cyclin D1 binding to the androgen receptor (AR) NH2-terminal domain inhibits activation function 2 association and reveals dual roles for AR corepression. Mol. Endocrinol., 19, 607–620. [DOI] [PubMed] [Google Scholar]
- 20. Lee MS et al. (2015) PI3K/AKT activation induces PTEN ubiquitination and destabilization accelerating tumourigenesis. Nat. Commun., 6, 7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boehm JS et al. (2007) Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell, 129, 1065–1079. [DOI] [PubMed] [Google Scholar]
- 22. Patterson AR et al. (2015) Sustained reprogramming of the estrogen response after chronic exposure to endocrine disruptors. Mol. Endocrinol., 29, 384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guan X et al. (2017) Stromal senescence by prolonged CDK4/6 inhibition potentiates tumor growth. Mol. Cancer Res., 15, 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Debacq-Chainiaux F et al. (2009) Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc., 4, 1798–1806. [DOI] [PubMed] [Google Scholar]
- 25. Xu Y et al. (2014) Emerging roles of the p38 MAPK and PI3K/AKT/mTOR pathways in oncogene-induced senescence. Trends Biochem. Sci., 39, 268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Freund A et al. (2011) p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. EMBO J., 30, 1536–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Courtois-Cox S et al. (2008) Many roads lead to oncogene-induced senescence. Oncogene, 27, 2801–2809. [DOI] [PubMed] [Google Scholar]
- 28. She QB et al. (2008) Breast tumor cells with PI3K mutation or HER2 amplification are selectively addicted to Akt signaling. PLoS One, 3, e3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Manning BD et al. (2017) AKT/PKB signaling: navigating the network. Cell, 169, 381–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Du K et al. (2005) Regulation of the Akt kinase by interacting proteins. Oncogene, 24, 7401–7409. [DOI] [PubMed] [Google Scholar]
- 31. Bellacosa A et al. (1998) Akt activation by growth factors is a multiple-step process: the role of the PH domain. Oncogene, 17, 313–325. [DOI] [PubMed] [Google Scholar]
- 32. Bellacosa A et al. (2005) Activation of AKT kinases in cancer: implications for therapeutic targeting. Adv. Cancer Res., 94, 29–86. [DOI] [PubMed] [Google Scholar]
- 33. Liu P et al. (2009) Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov., 8, 627–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bosch A et al. (2015) PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor-positive breast cancer. Sci. Transl. Med., 7, 283ra51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu XL et al. (2018) Oncogene-induced senescence: a double edged sword in cancer. Acta Pharmacol. Sin., 39, 1553–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bhat-Nakshatri P et al. (2008) AKT alters genome-wide estrogen receptor alpha binding and impacts estrogen signaling in breast cancer. Mol. Cell. Biol., 28, 7487–7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen D et al. (2002) Phosphorylation of human estrogen receptor alpha at serine 118 by two distinct signal transduction pathways revealed by phosphorylation-specific antisera. Oncogene, 21, 4921–4931. [DOI] [PubMed] [Google Scholar]
- 38. Kato S et al. (1995) Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science, 270, 1491–1494. [DOI] [PubMed] [Google Scholar]
- 39. Smith SE et al. (2017) Molecular characterization of breast cancer cell lines through multiple omic approaches. Breast Cancer Res., 19, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jamieson S et al. (2011) A drug targeting only p110α can block phosphoinositide 3-kinase signalling and tumour growth in certain cell types. Biochem. J., 438, 53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu Y et al. (2018) Tamoxifen resistance in breast cancer is regulated by the EZH2-ERα-GREB1 transcriptional axis. Cancer Res., 78, 671–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.