Abstract
Background
Use of immunohistochemistry-based surrogates of molecular breast cancer subtypes is common in research and clinical practice, but information on their comparative validity and prognostic capacity is scarce.
Methods
Data from 2 PAM50-subtyped Swedish breast cancer cohorts were used: Stockholm tamoxifen trial–3 with 561 patients diagnosed 1976-1990 and Clinseq with 237 patients diagnosed 2005-2012. We evaluated 3 surrogate classifications; the immunohistochemistry-3 surrogate classifier based on estrogen receptor, progesterone receptor, and HER2 and the St. Gallen and Prolif surrogate classifiers also including Ki-67. Accuracy, kappa, sensitivity, and specificity were computed as compared with PAM50. Alluvial diagrams of misclassification patterns were plotted. Distant recurrence-free survival was assessed using Kaplan-Meier plots, and tamoxifen treatment benefit for luminal subtypes was modeled using flexible parametric survival models.
Results
The concordance with PAM50 ranged from poor to moderate (kappa = 0.36-0.57, accuracy = 0.54-0.75), with best performance for the Prolif surrogate classification in both cohorts. Good concordance was only achieved when luminal subgroups were collapsed (kappa = 0.71-0.69, accuracy = 0.90-0.91). The St. Gallen surrogate classification misclassified luminal A into luminal B; the reverse pattern was seen with the others. In distant recurrence-free survival, surrogates were more similar to each other than PAM50. The difference in tamoxifen treatment benefit between luminal A and B for PAM50 was not replicated with any surrogate classifier.
Conclusions
All surrogate classifiers had limited ability to distinguish between PAM50 luminal A and B, but patterns of misclassifications differed. PAM50 subtyping appeared to yield larger separation of survival between luminal subtypes than any of the surrogate classifications.
Since their discovery in 2000 (1,2), gene expression–based molecular subtypes of breast cancer have been independently confirmed (3-5) and widely accepted (6-8). To facilitate clinical implementation, gene expression assays mimicking the original subtyping algorithm have been developed, with perhaps the most well-known and well-spread being the PAM50 assay (9). Despite this, the availability of large datasets with gene expression–based subtyping appears limited because most epidemiological studies rely on immunohistochemistry (IHC) surrogate subtypes for studying the intrinsic subtypes (10-18). A 3-marker panel of estrogen receptor (ER), progesterone receptor (PR), and HER2 staining has been frequently used (10,11,15); other surrogates used include the proliferation marker Ki-67 (12-14,16), and/or basal-like markers cytokeratin 5 and 6 (17,18).
Although multiple surrogate classifiers exist, validation studies are scarce even on a single-surrogate basis (19,20). A limitation in choosing surrogate based on said studies (19-21) is that they were evaluated using different metrics and populations, when comparisons of surrogates are ideally performed using within-study comparisons (22). It was only this year within-study comparison of surrogates became available (23). The authors studied luminal tumors, observed low concordance, and requested more knowledge on prognostic implications (23). It is important to continue evaluating concordance considering all molecular subtypes, further assess how misclassification patterns may vary between surrogate classifiers, and to what extent any differences are robust to variability in staining protocols, given the known issues of reproducibility in staining of Ki-67. With the aim to characterize their respective strengths and weaknesses, we examined the concordance of 3 distinct IHC surrogates with PAM50 subtypes to quantify how well these surrogates mimic PAM50. We performed our analysis in 2 datasets representing 2 common scenarios in epidemiological research: one where information is based on centralized staining of IHC markers and one with IHC information collected from original pathology records. We also evaluated whether the luminal A and B subtypes, as defined by the surrogates, mirrored PAM50 luminal subtypes in tamoxifen treatment response and distant recurrence-free survival during 15 years of follow-up.
Methods
Study Samples
For this study, tumor material from the Stockholm tamoxifen trial (STO-3) and the Clinseq study was used. Ethical approval was granted for each study, and all participants provided informed consent for participation.
The STO-3 trial was originally carried out to evaluate the survival benefit of tamoxifen treatment and enrolled lymph-node negative, postmenopausal patients with tumors no more than 30 mm diagnosed in Stockholm during the period 1976-1990 (24). Briefly, participants were randomized to either 2 years of adjuvant tamoxifen or no adjuvant treatment. In 2014, formalin-fixed paraffin-embedded tumor blocks were sectioned for all tumors with enough material available for analysis (n = 727) and stained for ER, PR, HER2, and Ki-67 at University of California Davis Medical Center (Supplementary Methods, available online). Of the 727 tumors, 652 passed the RNA quality checks. PAM50 subtypes were assigned using microarray gene expression data and the PAM50 classifier (Supplementary Methods, available online). For the present study, normal-like tumors and tumors missing ER, PR, HER2, and/or Ki-67 data were excluded, resulting in a final number of 561 analysis samples.
The Clinseq study was designed to evaluate the clinical use of sequencing in breast cancer and consisted of 307 sequenced breast cancer cases from the Libro-1 (25) and KARMA study (26,27) diagnosed in Stockholm, Sweden, between 2001 and 2012. For this study, only the tumors diagnosed in 2005 and onward were included to ensure Ki-67 availability (n = 258). Molecular subtyping was performed in 2013 using RNA-sequencing data, assigning PAM50 subtypes using a nearest shrunken centroid classifier based on the PAM50 gene set (28) (Supplementary Methods, available online). ER, PR, HER2, and Ki-67 were collected in 2015 from pathology reports of the initial surgery. Complete information on all markers was obtained for 92% of the cases, resulting in a final 237 samples. Scoring of IHC markers for STO-3 and Clinseq is described in the Supplementary Methods (available online).
Briefly, cutoff for ER- and PR-positive tumors was 10%. HER2 status had been determined differently in the 2 original studies. In the STO-3 cohort, HER2 was positive if IHC staining was 3+; 0-2+ was coded as negative. In Clinseq, HER2 status followed Swedish pathology guidelines at the time, using fluorescence in situ hybridization (FISH) to assign status of 2+ tumors. Ki-67 was counted using whole slide in STO-3, whereas hotspot counting was used clinically during the Clinseq period.
Surrogate Classifications
We evaluated 3 surrogate classifiers, denoted hereafter as IHC3, St. Gallen, and Prolif. The surrogates are described in Table 1. In brief, IHC3 refers to a surrogate subtyping based on ER, PR, and HER2 staining alone, not uncommon in the literature (10,11,15,18). The St. Gallen 2013 surrogate subtypes (additionally including Ki-67) were originally created to identify ER-positive, HER2-negative tumors that may benefit from chemotherapy (12). In 2015, we developed the Prolif surrogate (29) with the aim to identify a simplistic IHC surrogate classifier that would still capture the signature among genes differentially expressed between luminal types A and B (1,2,4). For St. Gallen and Prolif, 3 Ki-67 cutoffs were investigated: 14%, 20%, and a “free” cutoff tailored to each dataset (as described in the Supplementary Methods, available online). The cutoff at 14% was recommended for optimal classification of luminal B (12), and the others were recommended by the St. Gallen 2013 consensus statement (7). The “free” Ki-67 cutoff for the Prolif surrogate was 20% in STO-3 and 40% in Clinseq, whereas the “free” cutoff for the St. Gallen surrogate was 45% and 40%, respectively (Supplementary Methods, available online).
Table 1.
Definitions of the IHC surrogate classifiers used
| Surrogate | Luminal A–like | Luminal B–like | HER2 enriched | Basal-like | Unclassifiable |
|---|---|---|---|---|---|
| IHC3 | ER+/PR+/HER2- | ER+/PR+/HER2+ | ER-/PR-/HER2+ | ER-/PR-/HER2- | None |
| St. Gallen | ER+/PR ≥ 20a/HER2-/Ki-67lowb | ER+/PR-/HER2- or ER+/HER2+ or ER+/HER2-/Ki-67highb | ER-/PR-/HER2+ | ER-/PR-/HER2- | ER-/PR+ |
| Prolif | ER+/Ki-67lowc | ER+/Ki-67highc | ER-/PR-/HER2+ | ER-/PR-/HER2- | ER-/PR+ |
The cutoff for PR in Clinseq was kept at 10%, because too few cases with continuous PR staining were available. ER = estrogen receptor; IHC = immunohistochemistry; PR = progesterone receptor.
The cutoff for KI67 high/low for St. Gallen14 was 14%, and for St. Gallen 20 20%. In the St. Gallenfree, the cutoff was 45% in the STO-3 and 40% in Clinseq.
The cutoff for KI67 high/low for the Prolif14 was 14% and for Prolif 20 20%. In the Prolif free, the cutoff was 20% in the STO-3 and 40% in Clinseq.
Statistical Analysis
Accuracy and Cohen unweighted kappa with 95% confidence intervals (CIs) were calculated for assessment of overall concordance with PAM50 subtypes. Accuracy is a measure of percentage agreement, whereas kappa estimates the excess in agreement taking into account the possibility of the agreement occurring by chance. Sensitivity and specificity were calculated separately by subtype. To visually compare the patterns of reclassification of samples going from PAM50 to each surrogate, alluvial flow diagrams were plotted. The calculations were repeated as a 3-class problem, after collapsing luminal subtypes.
Receiver operating characteristics (ROC) plots were constructed to assess the balance between sensitivity and specificity for luminal A vs B comparisons. For this purpose, data were restricted to luminal subtypes as defined by PAM50, and sensitivity and specificity were calculated as a 2-class problem.
Survival Analysis
Distant recurrence-free survival was assessed in the STO-3 cohort. Individuals were followed until the date of distant recurrence in the quality register, date of death, emigration from Sweden, or end of follow-up at 15 years after diagnosis, whichever came first. Follow-up was achieved by linkages to the Swedish Cause of Death Register (30), Total Population Register (31) and the nationwide breast cancer quality register (32) using the Swedish unique personal identity number (33).
Five- and 10-year survival proportions with 95% confidence interval were calculated for each subtype. To visually assess differences between surrogate classifiers and PAM50 in survival over time, Kaplan-Meier curves of distant recurrence-free survival for ER-positive luminal tumors were plotted for luminal A and B for each surrogate and overlaid on top of the equivalent curves obtained using PAM50.
To compare treatment response prediction of tamoxifen using surrogate classifiers and PAM50, hazard ratios (HRs) of distant recurrence were estimated at 5 and 10 years after diagnosis. In this analysis, data was restricted to ER-positive luminal samples. The hazards of the event were modeled using flexible parametric survival models (34), allowing for time-varying effects. Two degrees of freedom were chosen for the cubic spline function of the baseline hazard, after assessing model fit by Akaike information criterion when modeling PAM50 subtypes. Two covariates were allowed to vary with time: treatment effect of tamoxifen and the effect of subtype, using 1 degree of freedom for each. Underlying time scale was time since diagnosis, adjusted for age and calendar effects.
All statistical analyses were performed in R v3.2.2 and v3.5.2 (35). R package caret (36) was used to define the surrogates and model Ki-67 cutoffs, and the pROC package (37) was used to derive ROC plots. Alluvial diagrams were plotted using the alluvial package (38). The rstpm2 (39) and survival (40) packages were used to model survival. The survminer package (41) was used to draw survival plots.
Results
Distributions of Immunohistochemistry Markers by PAM50 Subtypes
The proportions of tumor characteristics and subtypes were similar across cohorts, except for a higher frequency of low-grade, luminal A tumors in STO-3, which only contained node-negative samples (Table 2). The degree of overlap in the distributions of Ki-67, PR, and ER between luminal subtypes, and the overlap in Ki-67 between luminal B and HER2-enriched, was substantial in both cohorts (Supplementary Figure 1, available online). Relative to STO-3, the distributions of Ki-67 were shifted to the right in Clinseq and showed better separation between subtypes (Supplementary Figure 1, C and G, available online).
Table 2.
Patient and tumor characteristics in samples from STO-3 and Clinseq included in the studya
| Characteristic | STO-3 | Clinseq |
|---|---|---|
| No. of tumors | 561 | 237 |
| Diagnostic period, y | 1976-1990 | 2005-2012 |
| Age at diagnosis, y | ||
| Range | 45-73 | 28-79 |
| Mean (SD) | 62.01 (5.43) | 58 (11.22) |
| Tumor size ≥20 mm, No. (%) | 134 (24.2) | 153 (64) |
| ER positive, No. (%) | 463 (82.5) | 200 (84) |
| PR positive, No. (%) | 326 (58.1) | 157 (66) |
| HER2 positive, No. (%) | 44 (7.8) | 38 (16) |
| Ki-67 % staining, mean (SD) | 10.75 (13.05) | 30.35 (24.88) |
| Grade, No. (%) | ||
| 1 | 97 (17.5) | 27 (12) |
| 2 | 321 (57.9) | 113 (48) |
| 3 | 136 (24.5) | 94 (40) |
| Node-positive status, No. (%) | 0 (0) | 29 (12) |
| PAM50 subtype, No. (%) | ||
| Luminal A | 322 (57) | 127 (54) |
| Luminal B | 123 (22) | 56 (24) |
| HER2-like | 56 (10) | 31 (13) |
| Basal-like | 60 (11) | 23 (8) |
ER = estrogen receptor; PR = progesterone receptor; STO-3 = Stockholm tamoxifen trial.
Concordance Measurements of Agreement Between PAM50 and Surrogate
Table 3 shows the accuracy and Cohen unweighted kappa for overall classification performance for each surrogate. The concordance with PAM50 ranged across surrogates from poor to moderate (kappa = 0.36-0.57, accuracy = 0.54-0.75) (Table 3). Consistently, at every cutoff of Ki-67, the Prolif surrogate had higher kappa and accuracy than the St. Gallen surrogate, and the IHC3 surrogate had values in line with the St. Gallen. The best concordance was seen for the Prolif surrogate using the “free” Ki-67 cutoff, with a kappa value in STO-3 cohort of 0.48 (95% CI = 0.41 to 0.54) and accuracy of 0.71 (95% CI = 0.67 to 0.75). Similar ranking of surrogates by kappa metric was observed in the Clinseq material, but the values for the metric were higher (Table 3). The impact of choice of Ki-67 cutoff on kappa and accuracy values was negligible in STO-3 data but did impact the degree of concordance in Clinseq. Good concordance was only achieved when luminal subgroups were collapsed (kappa = 0.71-0.69, accuracy = 0.90-0.91) (Table 3).
Table 3.
Accuracy and kappa metrics for each surrogate classifier and dataset
| Cohen kappa unweighted (95% CI) |
Overall accuracy (95% CI) |
|||
|---|---|---|---|---|
| Surrogate | STO-3 | Clinseq | STO-3 | Clinseq |
| IHC3 | 0.41 (0.35 to 0.47) | 0.43 (0.34 to 0.52) | 0.69 (0.65 to 0.73) | 0.68 (0.62 to 0.74) |
| St. Gallen14 | 0.38 (0.32 to 0.45) | 0.36 (0.27 to 0.44) | 0.60 (0.55 to 0.64) | 0.54 (0.47 to 0.60) |
| St. Gallen20 | 0.40 (0.33 to 0.46) | 0.43 (0.34 to 0.51) | 0.61 (0.57 to 0.65) | 0.61 (0.54 to 0.67) |
| St. GallenFree | 0.39 (0.32 to 0.45) | 0.52 (0.43 to 0-61) | 0.62 (0.58 to 0.66) | 0.69 (0.63 to 0.75) |
| Prolif14 | 0.48 (0.41 to 0.54) | 0.42 (0.34 to 0.51) | 0.70 (0.66 to 0.74) | 0.60 (0.53 to 0.66) |
| Prolif20 | 0.48 (0.41 to 0.54) | 0.49 (0.40 to 0.58) | 0.71 (0.67 to 0.75) | 0.67 (0.60 to 0.73) |
| ProlifFree | 0.48 (0.41 to 0.54) | 0.57 (0.48 to 0.66) | 0.71 (0.67 to 0.75) | 0.75 (0.69 to 0.80) |
| 3-class simplea: luminal vs HER2 vs basal-like | 0.71 (0.65 to 0.78) | 0.69 (0.58 to 0.80) | 0.91 (0.88 to 0.93) | 0.90 (0.85 to 0.93) |
3-class simple: collapsing luminal A and B into 1 class. CI = confidence interval; IHC = immunohistochemistry.
Patterns of Misclassification
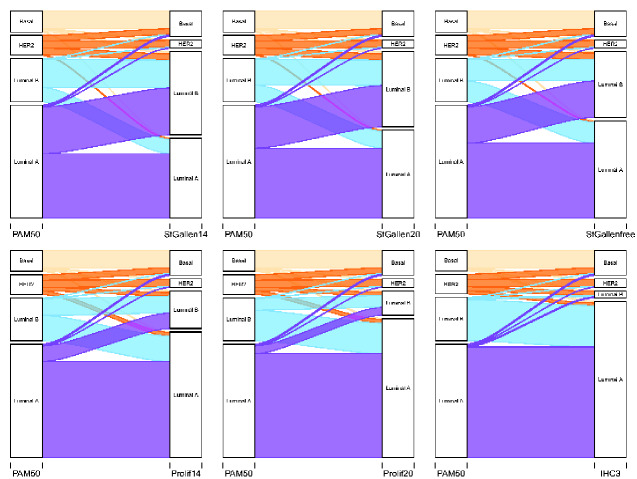
Inspection of the alluvial diagrams for patterns of misclassification showed that all surrogates misclassified approximately half of HER2 as basal-like and luminal B–like but had little misclassification of the basal-like subtype (Figure 1; STO-3; Supplementary Figure 2, available online; Clinseq). The St. Gallen surrogate misclassified luminal A tumors as luminal B–like, and the Prolif surrogate misclassified the luminal B tumors as luminal A–like. The IHC3 surrogate was, however, the most extreme in misclassifying luminal B, with 95% and 82% of the luminal Bs classified as luminal A–like in the respective cohorts (Supplementary Tables 1 and 2, available online).
Figure 1.
Alluvial diagrams showing the flow of tumors reclassified from PAM50 subtypes (right nodes) to surrogate defined subtype (left nodes), for the IHC3, St. Gallen, and Prolif surrogates in STO-3. Upper panel, left to right: St. Gallen14, St. Gallen20, St. GallenFree. Lower panel, left to right: Prolif14, Prolif20/ProlifFree (only labeled as Prolif20), IHC3. IHC = immunohistochemistry; STO-3 = Stockholm tamoxifen trial.
The most balance between sensitivity and specificity for distinguishing between luminal A and B subtypes was seen with the Prolif surrogate, as judged by ROC plots (Figure 2). Sensitivity and specificity for all subtypes are shown in Supplementary Table 3 (available online). The simple 3-class surrogate of “any luminal” vs “HER2-enriched” vs “basal-like” showed good precision for identifying a general luminal subtype, with a sensitivity of 0.98 and specificity of 0.77 in STO-3 and 0.99 and 0.66 in Clinseq.
Figure 2.
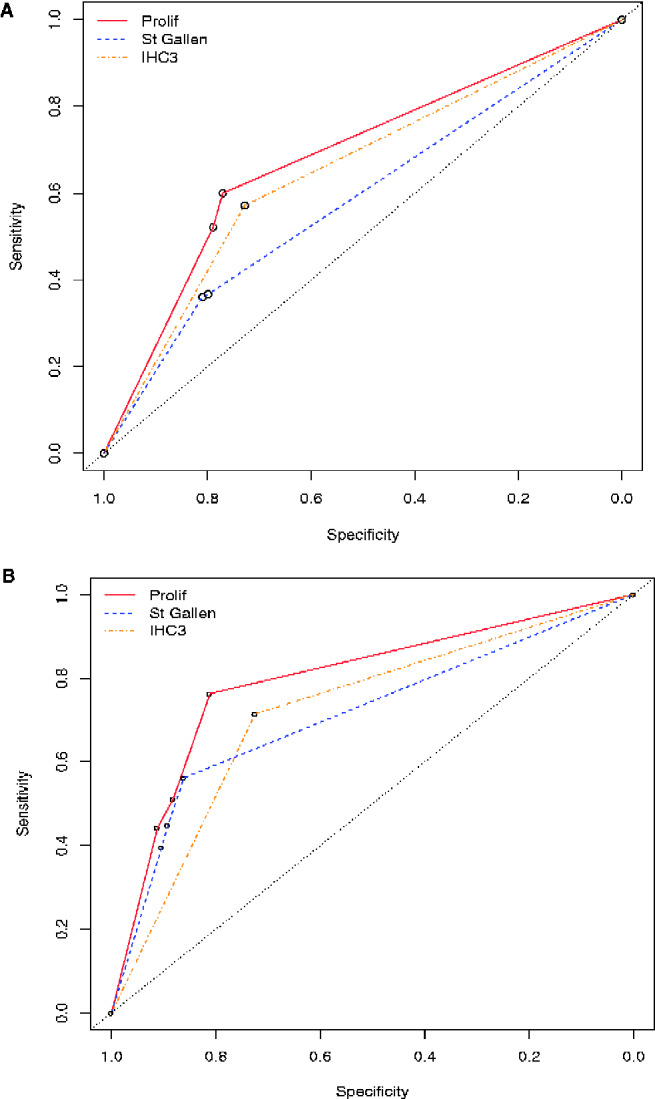
ROC plot, or sensitivity vs 1-specificity, for classifying luminal B in the STO-3 (A) and Clinseq (B) cohorts when restricting to only PAM50 luminal A and B tumors. For St. Gallen and Prolif, the 3 estimates corresponding to 14%, 20%, and “free” Ki-67 cutoff are plotted. For IHC3, there is only 1 estimate to plot. IHC = immunohistochemistry; STO-3 = Stockholm tamoxifen trial.
Distant Recurrence-Free Survival
Survival proportions for luminal A and B at 5 and 10 years only differed with PAM50, for all surrogates confidence intervals overlapped (Table 4).
Table 4.
Distant metastasis-free survival proportions with 95% confidence intervals for all subtypes, by surrogate classifier, in STO-3a
| Subtyping method and follow-up time point | Luminal A Survival proportion % (95% CI) |
Luminal B Survival proportion % (95% CI) |
HER2 Survival proportion % (95% CI) |
Basal-like Survival proportion % (95% CI) |
|---|---|---|---|---|
| PAM50 | ||||
| 5 y | 95.8 (93.6 to 98.1) | 78.8 (71.8 to 86.6) | 80.1 (70.2 to 91.3) | 81.2 (71.7 to 91.8) |
| 10 y | 90.4 (87.2 to 93.8) | 68.6 (60.6 to 77.7) | 70.2 (59.0 to 83.6) | 73.5 (62.8 to 86.0) |
| IHC3 | ||||
| 5 y | 91.4 (88.8 to 94.1) | 70.0 (52.5 to 93.3) | 86.5 (73.3 to 100.0) | 79.9 (71.0 to 89.9) |
| 10 y | 84.0 (80.6 to 87.6) | 65.0 (47.1 to 89.7) | 70.8 (53.5 to 93.8) | 76.7 (67.3 to 87.4) |
| St. Gallen14 | ||||
| 5 y | 94.1 (91.1 to 97.3) | 86.9 (82.7 to 91.4) | 86.5 (73.3 to 100.0) | 79.9 (71.0 to 89.9) |
| 10 y | 88.3 (84.1 to 92.7) | 78.7 (73.5 to 84.2) | 70.8 (53.5 to 93.8) | 76.7 (67.3 to 87.4) |
| St. Gallen20 | ||||
| 5 y | 93.4 (90.4 to 96.6) | 87.0 (82.5 to 91.7) | 86.5 (73.3 to 100.0) | 79.9 (71.0 to 89.9) |
| 10 y | 88.1 (84.1 to 92.4) | 78.0 (72.5 to 83.9) | 70.8 (53.5 to 93.8) | 76.7 (67.3 to 87.4) |
| St. Gallenfree | ||||
| 5 y | 92.6 (89.5 to 95.8) | 87.4 (82.7 to 92.3) | 86.5 (73.3 to 100.0) | 79.9 (71.0 to 89.9) |
| 10 y | 86.5 (82.4 to 90.8) | 78.9 (73.1 to 85.1) | 70.8 (53.5 to 93.8) | 76.7 (67.3 to 87.4) |
| Prolif14 | ||||
| 5 y | 92.2 (89.4 to 95.1) | 84.6 (78.0 to 91.9) | 86.5 (73.3 to 100.0) | 79.9 (71.0 to 89.9) |
| 10 y | 85.8 (82.2 to 89.6) | 75.2 (67.2 to 84.2) | 70.8 (53.5 to 93.8) | 76.7 (67.3 to 87.4) |
| Prolif20/Proliffree | ||||
| 5 y | 91.4 (88.6 to 94.3) | 85.1 (76.9 to 94.1) | 86.5 (73.3 to 100.0) | 79.9 (71.0 to 89.9) |
| 10 y | 85.3 (81.8 to 89.0) | 72.5 (62.4 to 84.2) | 70.8 (53.5 to 93.8) | 76.7 (67.3 to 87.4) |
CI = confidence interval; IHC = immunohistochemistry; STO-3 = Stockholm tamoxifen trial.
The 10-year survival proportion in STO-3 appeared higher for St. Gallen luminal B–like (St. GallenFree = 78.87, 95% CI = 73.12 to 85.08) than PAM50 luminal B (68.63, 95% CI = 60.60 to 77.73) (Table 4). For IHC3, the 10-year survival appeared worse for the luminal A–like than PAM50 luminal A (IHC3 = 84.04, 95% CI = 80.60 to 87.63; PAM50 = 90.41, 95% CI = 87.15 to 93.80). Both of these patterns were also observed for the Prolif surrogate, but survival for luminal B–like was closer to PAM50 luminal B than those obtained with St. Gallen (Table 4). Survival at 10 years was close to 0.70 for both surrogate HER2 and PAM50. For basal-like, 10-year survival was very similar using the surrogate definition (76.71, 95% CI = 67.31 to 87.42) and PAM50 (73.52, 95% CI = 62.83 to 86.03) (Table 4).
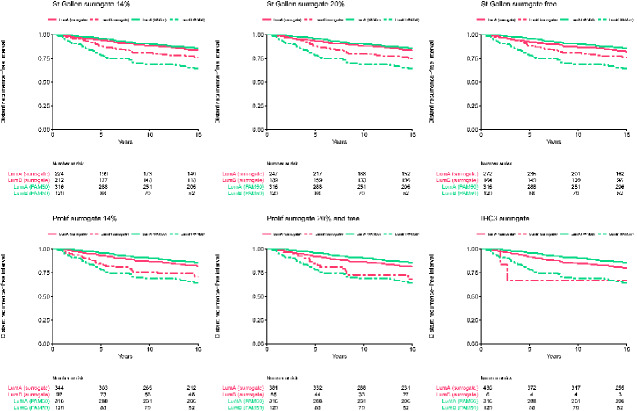
Kaplan-Meier curves of distant recurrence-free survival for ER-positive luminal tumors visually showed that maximum separation of survival estimates between luminal A and B was seen with PAM50; the St. Gallen and Prolif surrogates showed survival curves for luminal A– and B–like that were closer to each other. IHC3 showed more resemblance to PAM50, but the very few observations for luminal B hampers interpretation (Figure 3).
Figure 3.
Kaplan-Meier plots and risk tables of 15-year distant recurrence-free survival for luminal A-like and luminal B-like, defined by each surrogate. Upper panel, left to right: St. Gallen14, St. Gallen20, St. GallenFree. Lower panel, left to right: Prolif14, Prolif20/ProlifFree, IHC3. Plots are overlaid on top of the survival curves for PAM50 luminal A (lumA) and luminal B (lumB). IHC = immunohistochemistry.
Time-Varying Analysis of Long-Term Tamoxifen Treatment Benefit
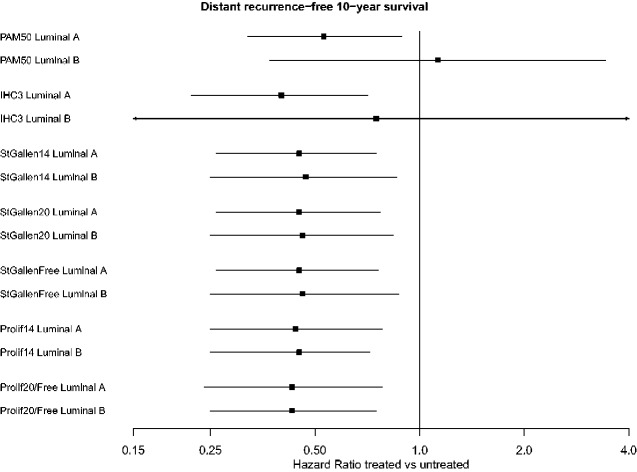
The hazard ratios at 10 years after diagnosis ranged narrowly from 0.40 to 0.45 across surrogates for tamoxifen-treated vs untreated luminal A–like and 0.43 to 0.47 for luminal B–like, except for IHC3 that had more similar estimates to PAM50 (Figure 4; Supplementary Table 4, available online). However, the estimates for IHC3 had huge uncertainty. Within surrogate classifiers, the luminal A and B hazard ratio estimates were near identical (St. Gallen, IHC3) or identical (Prolif) to each other. It was only when using PAM50 that there was any discernable difference between hazard ratio estimates for luminal subtypes, with a treatment benefit for luminal A 10 years after diagnosis (10 year HR = 0.53, 95% CI = 0.32 to 0.89) but no indication of treatment benefit for luminal B (10 year HR = 1.13, 95% CI = 0.37 to 3.43, confidence intervals including 1.00) (Figure 4).
Figure 4.
Forest plot showing hazard ratios (HR) of distant recurrence-free survival estimated at 10 years after diagnosis, comparing tamoxifen-treated to untreated estrogen receptor–positive luminal tumors (according to each classifier) in STO-3. Modeled using flexible parametric survival models using 15 years of follow-up. STO-3 = Stockholm tamoxifen trial.
Discussion
In this study, the ability of 3 separate IHC surrogate classifiers to classify breast cancer cases into molecular subtypes was assessed in 2 independent datasets: one with centrally stained IHC and one with IHC performed across several clinical pathology labs. We found that irrespective of cohort, all the surrogates showed poor to moderate agreement with the PAM50 classifier as judged by overall kappa values, with best performance for the proliferation-based surrogate. Alluvial diagrams and sensitivity-specificity measures illustrated that the main limitation of the surrogates was in separating luminal A and B, as well as luminal B and HER2-enriched, from each other. This comes as no surprise when one considers the overlap of distributions of IHC marker staining between these groups. Prolif was the best performing surrogate in this study, in line with observations of Lundgren and colleagues (23), who found that a grade-based surrogate classifier outperformed St. Gallen in a cohort of ER-positive and HER2-negative tumors. Interestingly, both surrogates that showed the best in-study concordance relied either entirely (Prolif) or in part (Grade-based) on measures of proliferation, and neither relies on PR negative or HER2 status for distinguishing luminal A from luminal B. Our concordance metrics cannot be directly compared because Lundgren colleagues only studied luminal tumors, but we observed the same pattern of misclassification of luminal A tumors as luminal B–like as they did (23).
We could conclude that IHC surrogates and PAM50 subtyping were different when considering crude survival proportions for all 4 subtypes, and no surrogate had as clear a separation of survival between the luminal groups as the PAM50. The misclassification patterns observed in the alluvial diagrams were mirrored in the Kaplan-Meier plots and survival proportions. The low specificity for luminal B (specifically mixed up with misclassified luminal A tumors) in St. Gallen translated into a better survival for St. Gallen luminal B–like than PAM50 luminal B. For IHC3, the very low numbers of luminal B–like tumors make any conclusions difficult to draw, but the luminal A–like group for IHC3 had worse survival than PAM50 luminal A, owning to the severe misclassification of luminal B into this group. These patterns were also true for the Prolif surrogate, albeit to a lesser extent. It should be stressed that, at present, it is not clear if IHC subtypes or PAM50 subtypes provide the most accurate prognostic information.
The time-varying patterns of tamoxifen-treatment benefit differs between luminal A and B tumors (42). We therefore investigated whether this was captured using any of the IHC surrogates, but the answer appeared to be no. There was hardly any difference in hazard ratios within surrogates between the effect for luminal A and B–like tumors. In addition, the hazard ratios of tamoxifen treatment benefit were very similar across all surrogates.
The St. Gallen surrogate was designed to identify ER-positive patients who may benefit from chemotherapy (7,13). We did not have the data to investigate chemotherapy response; however, the misclassification patterns might suggest how the surrogate classifiers could differ in this aspect. Assuming that mainly luminal B benefits from chemotherapy, undertreatment would be minimized (at the cost of potential overtreatment) using the St. Gallen surrogate, because the sensitivity for luminal B was highest with St. Gallen. Using the IHC3 surrogate could instead lead to undertreatment, because it misclassified almost all luminal B tumors as luminal A–like.
For observational studies where the goal is to be able to separate between the luminal subtypes, the optimal choice of surrogate may vary with the study objective. However, in studies that need not distinguish between luminal A and B, a 3-class surrogate of luminal (ER-positive) vs HER2 and basal-like subtypes should be recommended as the first choice.
We performed an independent assessment of 3 IHC surrogate classifiers in the same datasets, including one with markers stained in one study site with high standardization, considered optimal for assessments of validity and concordance (22). The high coverage of the Swedish registers enabled virtually complete follow-up for our survival analysis. The survival estimates are from a historic cohort and do not reflect current treatment options (such as anti-HER2 therapies) and clinical management.
Our analysis is based on the Swedish cutoff for ER positivity at 10%. Because tumors with ER 1%-9% are similar to ER-negative tumors (43,44), concordance may be lower in settings where cutoff for ER is at 1%. However, the fraction of borderline ER-positive tumors is probably too low to influence concordance greatly.
The 2 data sources had distinct strengths and limitations. In STO-3, IHC was centrally assessed, whereas Clinseq had markers retrospectively collected from different clinics. In STO-3, we used the original 20% cutoff for PR for St. Gallen, whereas Clinseq was limited to binary information based on a 10% cutoff clinically implemented at the time. In STO-3, PAM50 subtypes were assigned according to the original algorithm, whereas Clinseq had PAM50 subtypes assigned using a novel RNAseq-based approach. In STO-3, the number of HER2-positive tumors was slightly underestimated as all 2+ tumors were deemed negative in absence of FISH, whereas in Clinseq, 2+ tumors were assigned HER2-status on the basis of FISH.
We were able to show robustness of our findings because the same conclusions were reached in both cohorts, irrespective of the many differences between cohorts. It will be of interest to replicate our findings in other settings.
In conclusion, no surrogate achieved better than moderate overall agreement with the PAM50 subtypes. Their main limitations laid in differentiating luminal A and B from each other and distinguishing HER2 from luminal B. These limitations were also mirrored in the prognosis assessment and treatment response prediction.
Funding
This work was supported by the Swedish Research Council (Grant No. 2018-02547), Swedish Cancer Society (Grant No. 19 0266), and Stockholm County Council (Grant No. 20170088).
Notes
Role of the funder: The funding sources had no involvement in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Disclosures: None of the authors have conflicts of interest or financial disclosures.
Author contributions: KC, JH, AP, PH, and LS designed the study, conceived by JH and AP. LL, KC, and PH provided previously collected data. JH, NY, AP, and AJ performed data analysis. All authors interpreted the analysis. JH drafted the article. All authors provided critical revision of the article and final approval of the version to be published.
Acknowledgments: Parts of the analysis (concordance analysis) have been published in a doctoral thesis from Karolinska Institutet (29).
Data Availability
The data underlying this article cannot be shared publicly due to regulations under the Swedish law. According to the Swedish Ethical Review Act, the General Data Protection Regulation, the Public Access to Information and Secrecy Act, and the Administrative Procedure Act, data can only be made available, after legal review, for researchers who meet the criteria for access to this type of sensitive and confidential data. Requests regarding the data may be made to the senior author.
Supplementary Material
References
- 1. Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98(19):10869-10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100(14):8418-8423. doi:10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yu K, Lee CH, Tan PH, Tan P. Conservation of breast cancer molecular subtypes and transcriptional patterns of tumor progression across distinct ethnic populations. Clin Cancer Res. 2004;10(16):5508-5517. [DOI] [PubMed] [Google Scholar]
- 4. Calza S, Hall P, Auer G, et al. Intrinsic molecular signature of breast cancer in a population-based cohort of 412 patients. Breast Cancer Res. 2006;8(4):R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Van Laere SJ, Van den Eynden GG, Van der Auwera I, et al. Identification of cell-of-origin breast tumor subtypes in inflammatory breast cancer by gene expression profiling. Breast Cancer Res Treat. 2006;95(3):243-255. [DOI] [PubMed] [Google Scholar]
- 6. Mullan PB, Millikan RC. Molecular subtyping of breast cancer: opportunities for new therapeutic approaches. Cell Mol Life Sci. 2007;64(24):3219-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goldhirsch A, Winer EP, Coates AS, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen international expert consensus on the primary therapy of early breast Cancer 2013. Ann Oncol. 2013;24(9):2206-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Prat A, Pineda E, Adamo B, et al. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast. 2015;24(Suppl 2):S26-S35. [DOI] [PubMed] [Google Scholar]
- 9. Parker JS, Mullins M, Cheang MCU, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bandera EV, Chandran U, Hong C-C, et al. Obesity, body fat distribution, and risk of breast cancer subtypes in African American women participating in the AMBER Consortium. Breast Cancer Res Treat. 2015;150(3):655-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Palmer JR, Viscidi E, Troester MA, et al. Parity, lactation, and breast cancer subtypes in African American women: results from the AMBER Consortium. J Natl Cancer Inst. 2014;106(10):dju237. doi:10.1093/jnci/dju237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheang MCU, Chia SK, Voduc D, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101(10):736-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Prat A, Cheang MCU, Martín M, et al. Prognostic significance of progesterone receptor-positive tumor cells within immunohistochemically defined luminal A breast cancer. J Clin Oncol. 2013;31(2):203-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Falck AK, Röme A, Fernö M, et al. St Gallen molecular subtypes in screening‐detected and symptomatic breast cancer in a prospective cohort with long‐term follow‐up. Br J Surg. 2016;103(5):513-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Howlader N, Altekruse SF, Li CI, et al. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J Natl Cancer Inst. 2014;106(5):dju055. doi:10.1093/jnci/dju055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Inwald EC, Koller M, Klinkhammer-Schalke M, et al. 4-IHC classification of breast cancer subtypes in a large cohort of a clinical cancer registry: use in clinical routine for therapeutic decisions and its effect on survival. Breast Cancer Res Treat. 2015;153(3):647-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. O’Brien KM, Cole SR, Engel LS, et al. Breast cancer subtypes and previously established genetic risk factors: a Bayesian approach. Cancer Epidemiol Biomarkers Prev. 2014;23(1):84-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Millikan RC, Newman B, Tse C-K, et al. Epidemiology of basal-like breast cancer. Breast Cancer Res Treat. 2008;109(1):123-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Romero A, Prat A, García-Sáenz JÁ, et al. Assignment of tumor subtype by genomic testing and pathologic-based approximations: implications on patient’s management and therapy selection. Clin Transl Oncol. 2014;16(4):386-394. [DOI] [PubMed] [Google Scholar]
- 20. Jamshidi N, Yamamoto S, Gornbein J, Kuo MD. Receptor-based surrogate subtypes and discrepancies with breast cancer intrinsic subtypes: implications for image biomarker development. Radiology. 2018;289(1):210-217. [DOI] [PubMed] [Google Scholar]
- 21. Allott EH, Cohen SM, Geradts J, et al. Performance of three-biomarker immunohistochemistry for intrinsic breast cancer subtyping in the AMBER consortium. Cancer Epidemiol Biomarkers Prev. 2016;25(3):470-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bossuyt P, Davenport C, Deeks J, Hyde C, Leeflang MS. Chapter 11: Interpreting results and drawing conclusions. Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy Version 0.9; 2013. http://srdta.cochrane.org/. Accessed December 31, 2019.
- 23. Lundgren C, Bendahl P-O, Borg Å, et al. Agreement between molecular subtyping and surrogate subtype classification: a contemporary population-based study of ER-positive/HER2-negative primary breast cancer. Breast Cancer Res Treat. 2019;178(2):459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rutqvist LE, Johansson H; on behalf of the Stockholm Breast Cancer Study Group. Long-term follow-up of the randomized Stockholm trial on adjuvant tamoxifen among postmenopausal patients with early stage breast cancer. Acta Oncol (Madr). 2007;46(2):133-145. [DOI] [PubMed] [Google Scholar]
- 25. Holm J, Li J, Darabi H. Associations of breast cancer risk prediction tools with tumor characteristics and metastasis. J Clin Oncol. 2016;34(3):251-258. doi:10.1200/J Clin Oncol.2015.63.0624 [DOI] [PubMed] [Google Scholar]
- 26. Swedish National Breast Cancer Study. KARMA (Karolinska Mammography Project for Risk Prediction of Breast Cancer). 2011. www.karmastudy.org. Accessed March 3, 2016.
- 27. Gabrielson M, Eriksson M, Hammarström M, et al. Cohort profile: the Karolinska Mammography Project for Risk Prediction of Breast Cancer (KARMA). Int J Epidemiol. 2017;46(6):1740-1741g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rantalainen M, Klevebring D, Lindberg J, et al. Sequencing-based breast cancer diagnostics as an alternative to routine biomarkers. Sci Rep. 2016;6(1):38037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Holm J. Aggressive Breast Cancer: Epidemiological Studies Addressing Disease Heterogeneity Doctoral thesis. Karolinska Institutet; 2018. https://openarchive.ki.se/xmlui/handle/10616/46178. Accessed May 30, 2020.
- 30. Brooke HL, Talbäck M, Hörnblad J, et al. The Swedish cause of death register. Eur J Epidemiol. 2017;32(9):765-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ludvigsson JF, Almqvist C, Bonamy A-KE, et al. Registers of the Swedish total population and their use in medical research. Eur J Epidemiol. 2016;31(2):125-136. [DOI] [PubMed] [Google Scholar]
- 32. Emilsson L, Lindahl B, Köster M, Lambe M, Ludvigsson JF. Review of 103 Swedish healthcare quality registries. J Intern Med. 2015;277(1):94-136. [DOI] [PubMed] [Google Scholar]
- 33. Ludvigsson JF, Otterblad-Olausson P, Pettersson BU, Ekbom A. The Swedish personal identity number: possibilities and pitfalls in healthcare and medical research. Eur J Epidemiol. 2009;24(11):659-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Royston P, Parmar MKB. Flexible parametric proportional-hazards and proportional-odds models for censored survival data, with application to prognostic modelling and estimation of treatment effects. Stat Med. 2002;21(15):2175-2197. [DOI] [PubMed] [Google Scholar]
- 35. R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R foundation for Statistical Computing; 2015. https://www.r-project.org/. Accessed March 30, 2020.
- 36. Max Kuhn and contributors. caret: Classification and Regression Training R package; 2018. https://cran.r-project.org/web/packages/caret/index.html. Accessed March 30, 2020.
- 37. Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bojanowski M, Edwards R. alluvial: R Package for Creating Alluvial Diagrams R package; 2016. https://github.com/mbojan/alluvial. Accessed March 30, 2020.
- 39. Clements M, Liu X-R. rstpm2: Generalized Survival Models R package; 2018. https://cran.r-project.org/web/packages/rstpm2/index.html. Accessed March 30, 2020.
- 40. Therneau T. Survival. A Package for Survival Analysis in S Version 2.38. R Package; 2015. https://CRAN.R-project.org/package=survival. Accessed March 30, 2020.
- 41. Kassambara A, Kosinski M, Biecek P, Fabian S. survminer: Drawing Survival Curves using’ggplot2’. R package. 2018. https://cran.r-project.org/web/packages/survminer/index.html. Accessed March 30, 2020.
- 42. Yu NY, Iftimi A, Yau C, et al. Assessment of long-term distant recurrence-free survival associated with tamoxifen therapy in postmenopausal patients with luminal A or luminal B breast cancer. JAMA Oncol. 2019;5(9):1304-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Iwamoto T, Booser D, Valero V, et al. Estrogen receptor (ER) mRNA and ER-related gene expression in breast cancers that are 1% to 10% ER-positive by immunohistochemistry. J Clin Oncol. 2012;30(7):729-734. [DOI] [PubMed] [Google Scholar]
- 44. Chen T, Zhang N, Moran MS, Su P, Haffty BG, Yang Q. Borderline ER-positive primary breast cancer gains no significant survival benefit from endocrine therapy: a systematic review and meta-analysis. Clin Breast Cancer. 2018;18(1):1-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article cannot be shared publicly due to regulations under the Swedish law. According to the Swedish Ethical Review Act, the General Data Protection Regulation, the Public Access to Information and Secrecy Act, and the Administrative Procedure Act, data can only be made available, after legal review, for researchers who meet the criteria for access to this type of sensitive and confidential data. Requests regarding the data may be made to the senior author.