Abstract
Background
Non-small cell lung cancer is a common type of lung cancer. Piperlongumine (PL), which is extracted from the roots of piperaceae plant, long pepper, and peppercorn, is an alkaloid amide that inhibits tumor growth and metastasis. However, whether it affects lung cancer cells remains unclear.
Methods
We assessed the effects of PL on the proliferation and apoptosis of A549 and H1299 NSCLC cell lines.
Results
PL was mildly toxic to normal human bronchial epithelial cells and significantly suppressed growth and facilitated apoptosis of A549 and H1299 cells. It also upregulated microRNA (miR)-34b-3p and downregulated the transforming growth factor beta type I receptor (TGFBR1). The dual-luciferase reporter assay showed that TGFBR1 is a target gene of miR-34b-3p. Silencing of miR-34b-3p or overexpression of TGFBR1 partially attenuated the effects of PL on A549 and H1299 cells.
Conclusions
PL inhibits proliferation and induces apoptosis of A549 and H1299 cells by upregulating miR-34b-3p and modulating TGFBR1 signaling pathway.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12906-020-03123-y.
Keywords: Piperlongumine, miR-34b-3p, Non-small-cell lung cancer, TGFBR1
Background
Lung cancer (LC) is a major contributing factor to the cancer-related death worldwide and has a high incidence and mortality rate [1]. The two main types of LC are small cell lung cancer and non-small cell lung cancer (NSCLC). Despite continuous improvements of medical treatment and technology, the vast majority of NSCLC patients are diagnosed with late-stage disease and the 5-year survival rate is low [2]. At present, LC treatment has rapidly developed, but there are many side effects and the prognosis still remains poor [3]. Therefore, there is an urgent need to identify new targets and methods for early diagnosis in order to improve the 5-year survival rate of patients.
MicroRNAs (miRNAs) are non-coding RNAs that regulate the expression of several genes [4, 5] through complementary binding to the 3′ untranslated region (UTR) of target gene mRNAs [6, 7]. It is reported that miRNAs participate in cancer progression via regulation of cell cycle, cell proliferation, and cell invasion etc. [8, 9]. For example, miR-221 and miR-222 inhibits the growth of LC [10, 11], and miR-493 promotes the sensitivity of cancer cells to cisplatin [12]. The abnormal expression of miRNAs in LC (high or low) may lead to tumor heterogeneity and tumor-initiating cell behaviors [13].
Piperlongumine (PL) exists naturally in nature and plays an anticancer role in a variety of tumors [14]. It has been shown to inhibit cell growth and induce apoptosis of cancer cells, such as breast cancer [15], human melanoma [16], head and neck cancer [17], human prostate cancer [18], pancreatic cancer [19], gastric cancer [20], and NSCLC [21]. Furthermore, Lad [22, 23] and Mizuno et al [24] found that PL is involved in the occurrence and development of multiple cancers through regulating different signaling pathways. Mizuno et al [24] reported that miR-34b-3p can inhibit the occurrence and development of LC.
In this study, we performed a miRNA microarray assay in NSCLC cells and found the differential expression of miR-34b-3p after PL treatment. In addition, we also assessed the effects of PL on the proliferation and apoptosis of A549 and H1299 NSCLC cell lines.
Methods
Cells and reagents
The NSCLC cell lines A549, H1299, H520, and SPC-A-1 were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). A549 and H1299 cells were transfected with lentivirus containing miR-34b-3p or NC fragment to obtain stable expression of miR-34b-3p/NC (Hanbio, Shanghai, China). The PL was purchased from Sigma (St. Louis, MO, USA), dissolved in dimethyl sulfoxide (DMSO) (Sigma) and stored at 4 °C for subsequent experiments.
RNA extraction and qPCR
Total RNA was extracted from cells using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Complementary DNA (cDNA) was generated using the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA, USA). miR-34b-3p (primer sequences: F- 5′-CGGCGAATCACTAACTCCACT-3′ and R-5′-GTGCAGGGTCCGAGGT-3′) and TGFBR1 (primer sequences: F-5′-GAACTGTTTTGATTGGCATC-3′ and R-5′-AAGAAGGGACCTACACTATTT-3′) mRNA expression was determined by qPCR. U6 small nuclear RNA (primer sequences: F-5′- CGCTTCGGCAGCACATATAC-3′ and R-5′- TTCACGAATTTGCGTGTCAT-3′) and GAPDH (primer sequences: F-5′-TGCCCAGAACATCATCCCT-3′ and R-5′-TGAAGTCGCAGGAGACAACC-3′) were used as the internal reference for miR-34b-3p and TGFBR1, respectively.
miRNA microarray
A549 cells were divided into two groups: PL treatment (10 μM PL) group and control group (0.1% DMSO). Then the miRNA microarray assay was conducted by the Shanghai Bohao Biotechnology Co., Ltd. (Shanghai, China).
Cell transfection
Transient transfection was conducted using Lipofectamine 2000 reagent (Invitrogen) following the manufacturer’s instructions. Cells were collected for subsequent experiments at different time points after transfection.
CCK-8 assay
A549 and H1299 cells were seeded into 96-well plates. After treatment for 0, 24, 48, 72 h, 10 μL CCK-8 reagent (Dojindo, Kumamoto, Japan) was added to each well and incubated for another 1–2 h followed by measuring the optical density values to assess cell growth.
Cell apoptosis assay
Apoptotic cell numbers were detected by Annexin V-FITC/PI Apoptosis Detection Kit (Solarbio, Beijing, China) in accordance with the kit instructions. Annexin V-FITC and PI were added to the binding buffer and mixed well followed by being added into cells. After 30 min incubation at room temperature under dark, a flow cytometer (BD Biosciences, San Diego, CA, USA) was used to measure cell apoptosis.
Caspase-3/7 activity assay
Caspase-3/7 activity was measured using the Apo-ONE Homogeneous Caspase-3/7 Assay Kit (Promega, Madison, WI, USA). The fluorescence (relative fluorescence units) of each well was measured using a spectrofluorometer (Thermo Fisher Scientific).
Transwell assay
A549 and H1299 cells were cultured in 6-well plates. After treatment with PL or transfection for 24 h, 5 × 104 cells were seeded in the top compartment of 8 μm pores transwell culture inserts (Corning Life Sciences, Corning, NY, USA) in the presence of serum-free medium, and the experiment was performed following the instructions. Three independent experiments were performed.
Xenograft mouse model
Female BALB/c nude mice of 4–6 weeks old (n = 12) were purchased from the Beijing Vital River Laboratory Animal Technology Center (Beijing, China). Three groups of A549 cells (miR-34b-3p, NC, and non-infected) were subcutaneously injected into the dorsal scapular region of the mice at a density of 2 × 106. After tumors were visible (about 1 week), the tumor-bearing nude mice were intraperitoneally injected with PL at a dose of 5 mg/kg (dissolved in 0.01% DMSO) twice per week for 3 weeks in the PL group with non-infected A549 cells, and the other two groups were injected with the same amount of 0.9% saline. The tumor volumes were monitored every week after tumor cell injection. After 4 weeks, the nude mice were sacrificed by Automated CO2 Delivery System. In brief, After the animals were put into the Automated CO2 Delivery System, the CO2 was infused into the box at the rate of 10–30% of the volume of the box per minute. When the animals were determined to be immobile, not breathing and pupil dilation (about 5 min), the CO2 was turned off and observed for 2 min to determine the death of the animal. The tumors were stripped and weighed, and immunohistochemistry (ki-67) and the TUNEL assay were performed. The study protocols were approved by the Animal Experimental Ethics Committee of the First Affiliated Hospital of Zhengzhou University.
Immunohistochemistry
The processed paraffin sections were incubated with primary antibody against Ki-67 (1:50 dilution, AF1738; Beyotime, Beijing, China) followed by incubation with secondary antibodies according to the instructions.
TUNEL assay
The TUNEL Assay Kit (C1088; Beyotime) was purchased, the experiment was conducted according to the manufacturer’s instructions, and the TUNEL staining was observed under a fluorescence microscope.
Luciferase reporter assay
The wild-type (Wt) TGFBR1 3′-UTR and mutant type (Mt) TGFBR1 3′-UTR fragments were amplified by overlapping PCR. Then the two fragments were inserted into the pmirGLO promoter vector (Promega) to generate recombinant Wt TGFBR1 and Mt TGFBR1 plasmids, respectively. 293 T cells were seeded in 24-well plates and co-transfected with recombinant plasmids (Wt TGFBR1 or Mt TGFBR1) and miRNAs (miR-34b-3p mimics or miR-NC). Luciferase activities in each group were measured by a luminescence microplate reader (Berthold, Bad Wildbad, Germany).
Western blotting
Western blotting was performed to detect protein expression after transfection. A549 and H1299 cells were lysed with RIPA lysis buffer (Beyotime), and total protein concentrations were detected by BCA Protein Assay Kit (Beyotime) followed by separation on 10% SDS-PAGE for western blot through incubation of the membrane with polyclonal rabbit anti-human TGFBR1 primary antibody (1: 2000 dilution) (Proteintech, WuHan, Hubei, China) at 4 °C overnight and subsequent incubation with HRP-conjugated secondary antibody (1: 5000 dilution) for 1 h at room temperature (Proteintech).
Statistical analysis
SPSS 21.0 software was used for processing data. Each experiment was repeated three times. Data with normal distribution were presented as mean ± standard deviation. One-way analysis of variance was used to compare the difference among three or more groups; Student’s t-test was used to compare difference between two independent groups. P < 0.05 was considered to be statistically significant.
Results
PL upregulates miR-34b-3p expression in LC cell lines
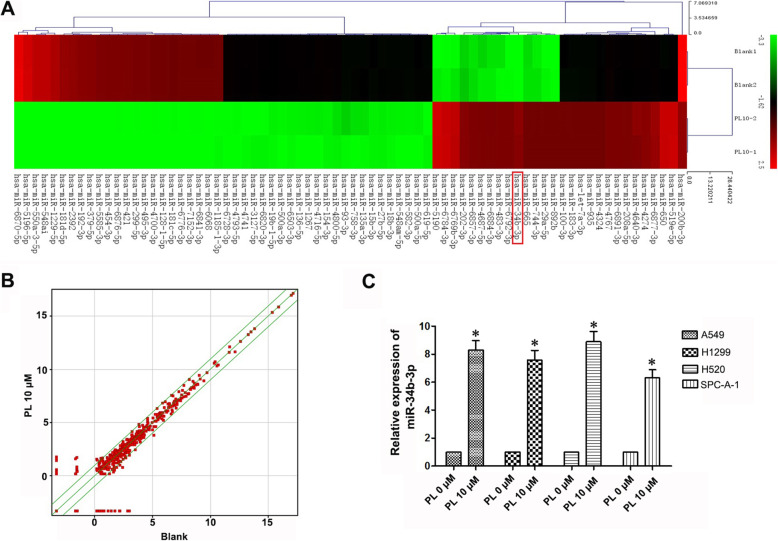
To study the anti-tumor mechanism of PL, miRNA microarray analysis was performed in A549 cells after treatment with PL. We found that there were 74 differentially expressed miRNAs with more than 3-fold changes, of which 27 were upregulated and 47 were downregulated. In addition, miR-34b-3p showed a 14-fold change increase in the 10 μM PL group compared with the 0 μM PL group (Table 1, Fig. 1a, b). To further confirm these results, miR-34b-3p expression was detected by quantitative PCR (qPCR) after total RNA extraction from PL-treated A549, H1299, H520, and SPC-A-1 NSCLC cells. Compared with cells in the 0 μM PL group, miR-34b-3p expression was consistently upregulated after 10 μM PL treatment in NSCLC cell lines (P < 0.05; Fig. 1c). These results showed that PL treatment increased the expression of miR-34b-3p.
Table 1.
Summary of differentially expressed miRNAs by piperlongumine treatment using microarray
| Name | miRNA location | Fold change | Regulation |
|---|---|---|---|
| hsa-miR-5190 | chr18 | 33.42283 | up |
| hsa-miR-6784-3p | chr17 | 31.10156 | up |
| hsa-miR-6769b-3p | chr1 | 26.50146 | up |
| hsa-miR-202-3p | chr10 | 14.70523 | up |
| hsa-miR-34b-3p | chr11 | 14.6738 | up |
| hsa-miR-6857-3p | chrX | 13.53686 | up |
| hsa-miR-6792-3p | chr19 | 13.4376 | up |
| hsa-miR-483-3p | chr11 | 13.22746 | up |
| hsa-miR-4687-5p | chr11 | 13.18184 | up |
| hsa-miR-6884-3p | chr17 | 12.94226 | up |
| hsa-miR-892b | chrX | 11.49669 | up |
| hsa-miR-744-3p | chr17 | 11.49669 | up |
| hsa-miR-665 | chr14 | 11.49669 | up |
| hsa-miR-29a-5p | chr7 | 11.49669 | up |
| hsa-miR-519e-5p | chr19 | 9.546338 | up |
| hsa-miR-650 | chr22 | 9.533337 | up |
| hsa-miR-6877-3p | chr9 | 5.105282 | up |
| hsa-miR-4274 | chr4 | 4.374106 | up |
| hsa-miR-4640-3p | chr6 | 3.953356 | up |
| hsa-miR-6891-3p | chr6 | 3.834103 | up |
| hsa-miR-208a-5p | chr14 | 3.791038 | up |
| hsa-miR-4767 | chrX | 3.689143 | up |
| hsa-miR-935 | chr19 | 3.635668 | up |
| hsa-miR-183-3p | chr7 | 3.635668 | up |
| hsa-miR-100-3p | chr11 | 3.635668 | up |
| hsa-let-7a-3p | chr9 | 3.635668 | up |
| hsa-miR-4324 | chr19 | 3.577973 | up |
| hsa-miR-6870-5p | chr20 | 42.62034 | down |
| hsa-miR-5196-5p | chr19 | 33.30788 | down |
| hsa-miR-550a-3-5p | chr7 | 26.47759 | down |
| hsa-miR-548ai | chr6 | 26.33067 | down |
| hsa-miR-1229-5p | chr5 | 22.00426 | down |
| hsa-miR-181d-5p | chr19 | 17.0129 | down |
| hsa-miR-2392 | chr14 | 15.64695 | down |
| hsa-miR-192-3p | chr11 | 14.80487 | down |
| hsa-miR-379-5p | chr14 | 14.55127 | down |
| hsa-miR-5585-3p | chr1 | 13.2484 | down |
| hsa-miR-454-3p | chr17 | 13.15125 | down |
| hsa-miR-6876-5p | chr8 | 13.04917 | down |
| hsa-miR-421 | chrX | 12.66647 | down |
| hsa-miR-299-5p | chr14 | 12.45378 | down |
| hsa-miR-495-3p | chr14 | 12.42371 | down |
| hsa-miR-4700-3p | chr12 | 11.56176 | down |
| hsa-miR-128-1-5p | chr2 | 11.53145 | down |
| hsa-miR-6776-3p | chr17 | 11.18739 | down |
| hsa-miR-7152-3p | chr10 | 11.18294 | down |
| hsa-miR-1185-1-3p | chr14 | 11.16815 | down |
| hsa-miR-6841-3p | chr8 | 11.15 | down |
| hsa-miR-6068 | chr1 | 11.07605 | down |
| hsa-miR-181c-5p | chr19 | 11.07164 | down |
| hsa-miR-6728-3p | chr1 | 3.467114 | down |
| hsa-miR-4716-5p | chr15 | 3.448778 | down |
| hsa-miR-154-3p | chr14 | 3.44251 | down |
| hsa-miR-4800-5p | chr4 | 3.403111 | down |
| hsa-miR-3127-5p | chr2 | 3.401971 | down |
| hsa-miR-136-5p | chr14 | 3.384909 | down |
| hsa-miR-4741 | chr18 | 3.381469 | down |
| hsa-miR-4793-5p | chr3 | 3.371059 | down |
| hsa-miR-6503-3p | chr11 | 3.367946 | down |
| hsa-miR-500a-3p | chrX | 3.352311 | down |
| hsa-miR-6820-3p | chr22 | 3.339673 | down |
| hsa-miR-1267 | chr13 | 3.338454 | down |
| hsa-miR-19b-1-5p | chr13 | 3.284702 | down |
| hsa-miR-500a-5p | chrX | 3.183913 | down |
| hsa-miR-200b-3p | chr1 | 3.150891 | down |
| hsa-miR-93-3p | chr7 | 3.094811 | down |
| hsa-miR-758-3p | chr14 | 3.094811 | down |
| hsa-miR-619-5p | chr12 | 3.094811 | down |
| hsa-miR-582-3p | chr5 | 3.094811 | down |
| hsa-miR-548am-5p | chrX | 3.094811 | down |
| hsa-miR-27b-5p | chr9 | 3.094811 | down |
| hsa-miR-18b-3p | chrX | 3.094811 | down |
| hsa-miR-15b-3p | chr3 | 3.094811 | down |
| hsa-miR-135a-3p | chr3 | 3.094811 | down |
Fig. 1.
MiRNA chip and qPCR validation results. a Heat map of miRNA array in A549 cells with or without PL treatment. b Scatter plot of differentially expressed miRNAs with a more than 3-fold change. c Relative expression of miR-34b-3p with or without PL (0, 10 μM) treatment in different LC cells lines by qPCR. *, P < 0.05 compared with the control group
PL treatment and upregulation of miR-34b-3p inhibits proliferation and invasion of A549 and H1299 LC cell lines
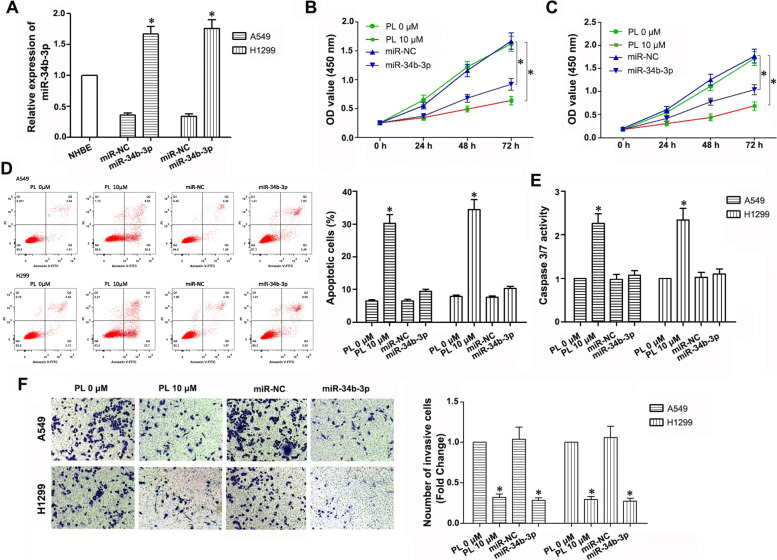
To evaluate the tumor-suppressing effects of miR-34b-3p and PL on LC, we constructed a lentiviral expression vector expressing miR-34b-3p or miR-negative control (NC), and then transfected separately into A549 and H1299 cells. Increased expression of miR-34b-3p after infection of the miR-34b-3p expressing lentivirus was found as demonstrated by qPCR, which indicated successful transfection (Fig. 2a). The CCK-8 assay showed that both upregulation of miR-34b-3p and PL inhibited cell proliferation (P < 0.05; Fig. 2b, c). In addition, a significant apoptosis-promoting effect was found after 10 μM PL treatment (P < 0.05); however, upregulation of miR-34b-3p had little effect on cell apoptosis (P > 0.05; Fig. 2d and e). In the transwell experiments, PL treatment and upregulation of miR-34b-3p effectively decreased the invasion ability of LC cells (P < 0.05; Fig. 2f). Overall, these data indicated that upregulating miR-34b-3p and PL treatment inhibited the proliferation and invasion of A549 and H1299 cells in vitro.
Fig. 2.
Effects of PL treatment and upregulation of miR-34b-3p on the proliferation, invasion, and apoptosis of NSCLC cells in vitro. a Relative expression of miR-34b-3p in normal human bronchial epithelial (NHBE) cells and lentivirus-infected A549 and H1299 cells using qPCR. b, c Optical density value was detected to represent the cell proliferation ability by CCK-8 Kit in the different groups. d, e Cell apoptosis was determined by flow cytometry and the caspase3/7 activity assay. f Cell invasion ability was detected 24 h after PL treatment by the transwell assay. A549 and H1299 were infected with miR-34b-3p lentivirus or treated with PL, then transwell assay was performed to detect invasive cell number. Invasive cells were stained with crystal violet and cell images were captured, then cell number were counted. All of the experiments were repeated three times
miR-34b-3p overexpression and PL treatment inhibits the growth of A549 xenograft tumors in vivo
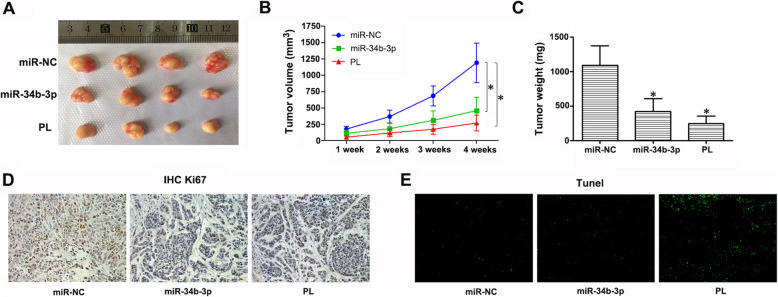
To compare the effects of upregulated miR-34b-3p and PL on the proliferation of tumors in vivo, we performed a nude mouse xenograft experiment. The tumor volume was significantly reduced in the inoculated nude mice of miR-34b-3p group and PL group compared with mice of the miR-NC group after 2, 3, and 4 weeks (P < 0.05; Fig. 3a, b). In addition, the tumor weight in the miR-34b-3p and PL groups showed a significant decrease (P < 0.05; Fig. 3c). Immunohistochemical staining of Ki-67 in the xenograft tumor tissue showed that the positive staining of Ki-67 in the tumor cells was significantly lower in the miR-34b-3p and PL groups than that in the miR-NC group (P < 0.05; Fig. 3d). We also investigated the effect of miR-34b-3p and PL on tumor apoptosis using the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay. As shown in Fig. 3e, the number of apoptotic cells in the PL group was significantly higher than that in the miR-NC and miR-34b-3p group. Taken together, the in vivo results were consistent with in vitro assays indicating that both PL and miR-34b-3p inhibits the proliferation of NSCLC cells and xenograft tumor growth, but only PL promotes tumor cell apoptosis.
Fig. 3.
Effects of PL and miR-34b-3p on A549 xenograft mice. a, b The tumor volumes were measured once a week after transplantation, and the tumor nodules were stripped 4 weeks after transplantation after which the final volumes were measured. c The stripped tumor nodules were weighed. d Immunohistochemical staining of Ki-67. e Results of the TUNEL assay in three groups
miR-34b-3p targets TGFBR1
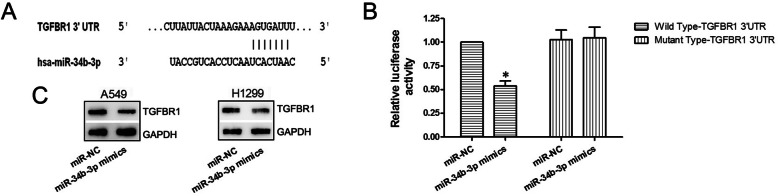
The result of bioinformatics analysis through TargetScan indicated that the TGFBR1 mRNA shared a complementary region with miR-34b-3p, and thus may be a potential target of miR-34b-3p (Fig. 4a). We verified this through dual-luciferase reporter assays and found that in the miR-34b-3p mimics and wild-type pmirGLO co-transfection group, the fluorescence activity was significantly reduced compared to the other three groups (P < 0.05; Fig. 4b), suggesting that miR-34b-3p bound to the TGFBR1 mRNA 3’UTR and decreased firefly luciferase activity. Moreover, Western blotting showed an apparent decrease in TGFBR1 protein expression after miR-34b-3p mimics transfection (P < 0.05; Fig. 4c). Taken together, these data showed that TGFBR1 is a target gene of miR-34b-3p.
Fig. 4.
TGFBR1 is a target of miR-34b-3p. a TargetScan bioinformatics analysis results. b Relative luciferase activity results of the dual-luciferase reporter assay. c Relative protein expression level of TGFBR1 after a 48 h transfection of miR-NC or miR-34b-3p mimics by western blotting. GAPDH served as the internal reference
Overexpression of TGFBR1 attenuates the inhibitory effects of PL on the proliferation and migration of A549 and H1299 cells
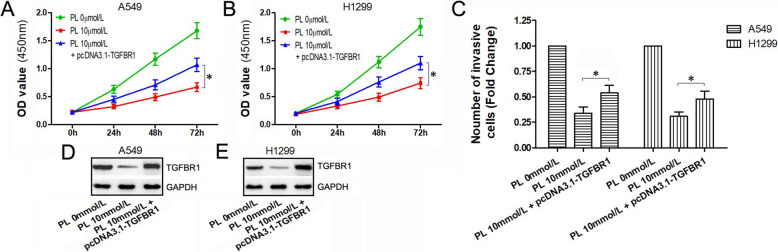
To further demonstrate the role of PL/miR-34b-3p/TGFBR1 axis in LC cells, we constructed an expression vector of TGFBR1 lacking the 3 ‘UTR (named pcDNA3.1-TGFBR1) which was transfected into PL-treated A549 and H1299 cells (combination group). The CCK-8 assay showed that compared to cells with PL treatment alone, the combination group showed few inhibitory effects on both A549 and H1299 cells (Fig. 5a, b), indicating that overexpression of TGFBR1 partially restored the inhibitory effects on PL-induced cell proliferation. In addition, the migration effects of PL on tumor cells were attenuated after TGFBR1 overexpression (Fig. 5c). Western blot analysis showed that PL treatment alone significantly decreased TGFBR1 protein expression which was rarely affected in the combination group (Fig. 5d, e).
Fig. 5.
Overexpression of TGFBR1 lacking 3′ UTR may restore the effects of PL. a, b Cell growth curve and optical density value were obtained from the CCK-8 assay in cells treated with PL alone, PL combined with TGFBR1 lacking 3′ UTR, and blank control. c Cell migration ability was evaluated by the transwell assay in cells treated with PL alone, PL combined with TGFBR1 lacking 3′ UTR, and blank control. d, e Relative protein expression level of TGFBR1 was detected by western blotting after 48 h of PL treatment or TGFBR1 transfection
Discussion
There is evidence that PL has anti-tumor effects in the progression of LC through various pathways, including the Akt [25] and nuclear factor-kappa B (NF-κB) signaling pathways [26]. PL suppresses NF-κB signaling pathway by directly binding to the DNA binding site of NF-κB p50 subunit and decreasing nuclear translocation of NF-κB p65 subunit, and inhibits Akt signaling pathway by suppressing phosphorylation of Akt in lung tumor cells. Moreover, PL can regulate expressions of c-Myc-regulated miRNAs (miR-27a, miR-20a, and miR-17) by decreasing c-Myc via an epigenetic pathway [14]. Here, we also supposed that PL may upregulate miR-34b-3p by some transcription factors induced transcriptional regulation. However, the related mechanisms are quite complicated and involve several pathways, thus needing further exploration. If the anti-tumor effects of PL are confirmed in the clinic, it will certainly be beneficial to patients. In this study, we showed that PL inhibited the proliferation of NSCLC cell lines (A549 and H1299), upregulated the expression of miR-34b-3p, and reduced the expression of TGFBR1. It has been reported lower miR-34b-3p level in cancers, such as LC, pancreatic cancer, and prostate cancer [14, 27, 28]. miR-34b-3p plays a role in tumor inhibition, such as pancreatic cancer [29], oral cancer [30], bladder cancer [31], cholangiocarcinoma [32], breast cancer [22], cervical cancer [33], ovarian cancer [23], and colorectal cancer [34]. In LC studies, the abnormal expression of miR-34b-3p in H1299 and A549 cells was shown to represses cell proliferation, cell cycle progression, and promote cell apoptosis by targeting cyclin-dependent kinase 4 [35]. Consistently, our study found that PL/miR-34b-3p/TGFBR1 axis influenced cell growth, invasion, and apoptosis of A549 and H1299 cells. TGFBR1 is a component of the transforming growth factor beta (TGF-β)/SMAD signaling pathway, which can regulate cell growth, differentiation, and migration [36]. TGF-β (tumor promoter or inhibitor) plays a key role in tumorigenesis [37]. TGFBR1 participates in cellular processes, differentiation, and apoptosis [38, 39]. The activation or overexpression of TGFBR1 is found in various types of cancer. For example, it can enhance the migration and invasion of breast cancer cells and promote the invasion and metastasis of colorectal cancer [39–42]. Furthermore, TGFBR1 affects tumor invasion and metastasis through the epithelial-mesenchymal pathway [43]. Together, these results indicate that TGFBR1 inhibition may be a novel therapeutic target for tumor treatment, consistent with previous results showing that downregulation of TGFBR1 suppresses cell proliferation, migration, and invasion in NSCLC [44]. In this study, the expression of TGFBR1 was affected after PL treatment and TGFBR1 knockdown inhibited cell proliferation and promoted cell apoptosis, consistent with PL-induced upregulation of miR-34b-3p expression. The overexpression of TGFBR1 repressed the proliferation and invasion of A549 and H1299 cells, suggesting that PL affected growth and apoptosis of NSCLC cells, and upregulated miR-34b-3p by targeting TGFBR1.
Conclusions
PL upregulates the expression of miR-34b-3p in NSCLC cell lines and inhibit cell proliferation and migration through TGFBR1 signaling pathway.
Supplementary Information
Additional file 1. Fig. 4C-A549-GAPDH: The original Fig. 4C GAPDH image of WB in A549 cells. Fig. 4C-A549-GAPDH-L: The original Fig. 4C GAPDH image with text labels in A549 cells. Fig. 4C-A549-TGFBR1: The original Fig. 4C TGFBR1 image of WB in A549 cells. Fig. 4C-A549-TGFBR1-L: The original Fig. 4C TGFBR1 image with text labels in A549 cells. Fig. 4C-H1299-GAPDH: The original Fig. 4C GAPDH image of WB in H1299 cells. C- Fig. 4H1299-GAPDH-L: The original Fig. 4C GAPDH image with text labels in H1299 cells. C- Fig. 4H1299-TGFR1: The original Fig. 4C TGFR1 image of WB in H1299 cells. C- Fig. 4H1299-TGFR1-L: The original Fig. 4C TGFR1 image with text labels in H1299 cells. Fig. 5 DE-GAPDH: The original GAPDH image in Figure 5D and 5E. Fig. 5 DE-GAPDH-L: The original GAPDH image with text labels in Figure 5D and 5E. Fig. 5 DE-TGFBR1: The original TGFBR1 image in Figure 5D and 5E. Figure 5 DE-TGFBR1-L: The original TGFBR1 image with text labels in Figure 5D and 5E.
Acknowledgements
Not applicable
Data availability statements
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Authors’ contributions
XHL and GJZ designed this study. XHL and CYX wrote this manuscript. XHL and ZXX performed the experiments. CYL and RY collected the data. CYX, FRZ and ZXX analyzed the data. XHL and GJZ interpreted the data. GJZ reviewed this manuscript. All authors have reviewed and approved the final version of this manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (grant no.81874042).
Availability of data and materials
Not applicable
Ethics approval and consent to participate
This study was approved by the Life science ethics review committee of Zhengzhou University.
Consent for publication
N/A
Competing interests
The authors declare that they have no conflicts of interest to report regarding the present study.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman PC, Mauer AM, Vokes EE. Lung cancer. Lancet. 2000;355(9202):479–485. doi: 10.1016/S0140-6736(00)82038-3. [DOI] [PubMed] [Google Scholar]
- 3.Higgins KA, Pillai RN, Chen Z, Tian S, Zhang C, Patel P, et al. Concomitant chemotherapy and radiotherapy with SBRT boost for unresectable stage III non–small cell lung cancer: a phase I study. J Thorac Oncol. 2017;12:1687–1695. doi: 10.1016/j.jtho.2017.07.036. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 5.Stadhouders R, Vidal E, Serra F, Di Stefano B, Le Dily F, Quilez J, et al. Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat Genet. 2018;50:238. doi: 10.1038/s41588-017-0030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223(2):102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Šulc M, Marín RM, Robins HS, Vaníček J. PACCMIT/PACCMIT-CDS: identifying microRNA targets in 3′ UTRs and coding sequences. Nucleic Acids Res. 2015;43(W1):W474–W479. doi: 10.1093/nar/gkv457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest. 2017;127(3):761–771. doi: 10.1172/JCI84424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Zeng H, Li H, Chen T, Wang L, Zhang K, et al. MicroRNA-101 inhibits growth, proliferation and migration and induces apoptosis of breast cancer cells by targeting sex-determining region Y-box 2. Cell Physiol Biochem. 2017;43(2):717–732. doi: 10.1159/000481445. [DOI] [PubMed] [Google Scholar]
- 10.Yamashita R, Sato M, Kakumu T, Hase T, Yogo N, Maruyama E, et al. Growth inhibitory effects of miR-221 and miR-222 in non-small cell lung cancer cells. Cancer Med. 2015;4(4):551–564. doi: 10.1002/cam4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filipska M, Skrzypski M, Czetyrbok K, Stokowy T, Stasiłojć G, Supernat A, et al. MiR-192 and miR-662 enhance chemoresistance and invasiveness of squamous cell lung carcinoma. Lung Cancer. 2018;118:111–118. doi: 10.1016/j.lungcan.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Gu Y, Zhang Z, Yin J, Ye J, Song Y, Liu H, et al. Epigenetic silencing of miR-493 increases the resistance to cisplatin in lung cancer by targeting tongue cancer resistance-related protein 1 (TCRP1) J Exp Clin Cancer Res. 2017;36(1):114. doi: 10.1186/s13046-017-0582-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang WC, Chin TM, Yang H, Nga ME, Lunny DP, Lim EK, et al. Tumour-initiating cell-specific miR-1246 and miR-1290 expression converge to promote non-small cell lung cancer progression. Nat Commun. 2016;7:11702. doi: 10.1038/ncomms11702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karki K, Hedrick E, Kasiappan R, Jin UH, Safe S. Piperlongumine induces reactive oxygen species (ROS)-dependent downregulation of specificity protein transcription factors. Cancer Prev Res. 2017;10(8):467–477. doi: 10.1158/1940-6207.CAPR-17-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee HN, Jin HO, Park JA, Kim JH, Kim JY, Kim B, et al. Heme oxygenase-1 determines the differential response of breast cancer and normal cells to piperlongumine. Mol Cell. 2015;38(4):327. doi: 10.14348/molcells.2015.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song X, Gao T, Lei Q, Zhang L, Yao Y, Xiong J. Piperlongumine induces apoptosis in human melanoma cells via reactive oxygen species mediated mitochondria disruption. Nutr Cancer. 2018;70(3):502–511. doi: 10.1080/01635581.2018.1445769. [DOI] [PubMed] [Google Scholar]
- 17.Roh JL, Kim EH, Park JY, Kim JW, Kwon M, Lee BH. Piperlongumine selectively kills cancer cells and increases cisplatin antitumor activity in head and neck cancer. Oncotarget. 2014;5(19):9227. doi: 10.18632/oncotarget.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piska K, Koczurkiewicz P, Wnuk D, Karnas E, Bucki A, Wójcik-Pszczoła K, et al. Synergistic anticancer activity of doxorubicin and piperlongumine on DU-145 prostate cancer cells–the involvement of carbonyl reductase 1 inhibition. Chem Biol Interact. 2019;300:40–48. doi: 10.1016/j.cbi.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Dhillon H, Mamidi S, McClean P, Reindl KM. Transcriptome analysis of piperlongumine-treated human pancreatic cancer cells reveals involvement of oxidative stress and endoplasmic reticulum stress pathways. J Med Food. 2016;19(6):578–585. doi: 10.1089/jmf.2015.0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou P, Xia Y, Ji J, Zhang J, Chen X, Rajamanickam V, et al. Piperlongumine as a direct TrxR1 inhibitor with suppressive activity against gastric cancer. Cancer Lett. 2016;375(1):114–126. doi: 10.1016/j.canlet.2016.02.058. [DOI] [PubMed] [Google Scholar]
- 21.Zhou L, Li M, Yu X, Gao F, Li W. Repression of hexokinases II-mediated glycolysis contributes to Piperlongumine-induced tumor suppression in non-small cell lung cancer cells. Int J Biol Sci. 2019;15(4):826. doi: 10.7150/ijbs.31749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thongsom S, Suginta W, Lee KJ, Choe H, Talabnin C. Piperlongumine induces G2/M phase arrest and apoptosis in cholangiocarcinoma cells through the ROS-JNK-ERK signaling pathway. Apoptosis. 2017;22(11):1473–1484. doi: 10.1007/s10495-017-1422-y. [DOI] [PubMed] [Google Scholar]
- 23.Lad NP, Kulkarni S, Sharma R, Mascarenhas M, Kulkarni MR, Pandit SS. Piperlongumine derived cyclic sulfonamides (sultams): synthesis and in vitro exploration for therapeutic potential against HeLa cancer cell lines. Eur J Med Chem. 2017;126:870–878. doi: 10.1016/j.ejmech.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 24.Mizuno K, Mataki H, Arai T, Kamikawaji K, Kumamoto T, Hiraki T, et al. The microRNA expression signature of small cell lung cancer: tumor suppressors of miR-27a-5p and miR-34b-3p and their targeted oncogenes. J Hum Genet. 2017;62(7):671. doi: 10.1038/jhg.2017.27. [DOI] [PubMed] [Google Scholar]
- 25.Seok JS, Jeong CH, Petriello MC, Seo HG, Yoo H, Hong K, et al. Piperlongumine decreases cell proliferation and the expression of cell cycle-associated proteins by inhibiting Akt pathway in human lung cancer cells. Food Chem Toxicol. 2018;111:9–18. doi: 10.1016/j.fct.2017.10.058. [DOI] [PubMed] [Google Scholar]
- 26.Zheng J, Son DJ, Gu SM, Ham YW, Lee HP, Kim WJ, et al. Piperlongumine inhibits lung tumor growth via inhibition of nuclear factor kappa B signaling pathway. Sci Rep. 2016;6:26357. doi: 10.1038/srep26357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daugaard I, Knudsen A, Kjeldsen TE, Hager H, Hansen LL. The association between miR-34 dysregulation and distant metastases formation in lung adenocarcinoma. Exp Mol Pathol. 2017;102(3):484–491. doi: 10.1016/j.yexmp.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 28.Liu C, Cheng H, Shi S, Cui X, Yang J, Chen L, et al. MicroRNA-34b inhibits pancreatic cancer metastasis through repressing Smad3. Curr Mol Med. 2013;13(4):467–478. doi: 10.2174/1566524011313040001. [DOI] [PubMed] [Google Scholar]
- 29.Fang LL, Sun BF, Huang LR, Yuan HB, Zhang S, Chen J, et al. Potent inhibition of miR-34b on migration and invasion in metastatic prostate cancer cells by regulating the TGF-β pathway. Int J Mol Sci. 2017;18(12):2762. doi: 10.3390/ijms18122762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohammad J, Dhillon H, Chikara S, Sreedasyam A, Chittem K, Orr M, Wilkinson JC, et al. Piperlongumine potentiates the effects of gemcitabine in in vitro and in vivo human pancreatic cancer models. Oncotarget. 2018;9(12):10457. doi: 10.18632/oncotarget.23623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen YJ, Kuo CC, Ting LL, Lu YC, Cheng AJ, Lin YT, et al. Piperlongumine inhibits cancer stem cell properties and regulates multiple malignant phenotypes in oral cancer. Oncol Lett. 2018;15(2):1789–1798. doi: 10.3892/ol.2017.7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu D, Qiu XY, Wu X, Hu DX, Li CY, Yu SB, et al. Piperlongumine suppresses bladder cancer invasion via inhibiting epithelial mesenchymal transition and F-actin reorganization. Biochem Biophys Res Commun. 2017;494(1–2):165–172. doi: 10.1016/j.bbrc.2017.10.061. [DOI] [PubMed] [Google Scholar]
- 33.Jin HO, Park JA, Kim HA, Chang YH, Hong YJ, Park IC, et al. Piperlongumine downregulates the expression of HER family in breast cancer cells. Biochem Biophys Res Commun. 2017;486(4):1083–1089. doi: 10.1016/j.bbrc.2017.03.166. [DOI] [PubMed] [Google Scholar]
- 34.Gong LH, Chen XX, Wang H, Jiang QW, Pan SS, Qiu JG, et al. Piperlongumine induces apoptosis and synergizes with cisplatin or paclitaxel in human ovarian cancer cells. Oxidative Med Cell Longev. 2014;2014. [DOI] [PMC free article] [PubMed]
- 35.Wang H, Jiang H, Corbet C, de Mey S, Law K, Gevaert T, et al. Piperlongumine increases sensitivity of colorectal cancer cells to radiation: involvement of ROS production via dual inhibition of glutathione and thioredoxin systems. Cancer Lett. 2019;450:42–52. doi: 10.1016/j.canlet.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 36.Feng H, Ge F, Du L, Zhang Z, Liu D. MiR-34b-3p represses cell proliferation, cell cycle progression and cell apoptosis in non-small-cell lung cancer (NSCLC) by targeting CDK4. J Cell Mol Med. 2019. [DOI] [PMC free article] [PubMed]
- 37.Akhurst RJ, Hata A. Targeting the TGFβ signalling pathway in disease. Nat Rev Drug Discov. 2012;11(10):790. doi: 10.1038/nrd3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bierie B, Moses HL. Tumour microenvironment: TGFβ: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6(7):506. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- 39.Wang H, Zhang Q, Wang BB, Wu W, Wei J, Li P, et al. miR-22 regulates C2C12 myoblast proliferation and differentiation by targeting TGFBR1. Eur J Cell Biol. 2018;97(4):257–268. doi: 10.1016/j.ejcb.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 40.Cheng R, Dang R, Zhou Y, Ding M, Hua H. MicroRNA-98 inhibits TGF-β1-induced differentiation and collagen production of cardiac fibroblasts by targeting TGFBR1. Hum Cell. 2017;30(3):192–200. doi: 10.1007/s13577-017-0163-0. [DOI] [PubMed] [Google Scholar]
- 41.Rosman DS, Phukan S, Huang CC, Pasche B. TGFBR1* 6A enhances the migration and invasion of MCF-7 breast cancer cells through RhoA activation. Cancer Res. 2008;68(5):1319–1328. doi: 10.1158/0008-5472.CAN-07-5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou R, Huang Y, Cheng B, Wang Y, Xiong B. TGFBR1* 6A is a potential modifier of migration and invasion in colorectal cancer cells. Oncol Lett. 2018;15(3):3971–3976. doi: 10.3892/ol.2018.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z, He J, Gao P, Niu Y, Zhang J, Wang L, et al. miR-769-5p suppressed cell proliferation, migration and invasion by targeting TGFBR1 in non-small cell lung carcinoma. Oncotarget. 2017;8(69):113558. doi: 10.18632/oncotarget.23060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park SJ, Choi YS, Lee S, et al. BIX02189 inhibits TGF-β1-induced lung cancer cell metastasis by directly targeting TGF-β type I receptor. Cancer Lett. 2016;381(2):314–322. doi: 10.1016/j.canlet.2016.08.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Fig. 4C-A549-GAPDH: The original Fig. 4C GAPDH image of WB in A549 cells. Fig. 4C-A549-GAPDH-L: The original Fig. 4C GAPDH image with text labels in A549 cells. Fig. 4C-A549-TGFBR1: The original Fig. 4C TGFBR1 image of WB in A549 cells. Fig. 4C-A549-TGFBR1-L: The original Fig. 4C TGFBR1 image with text labels in A549 cells. Fig. 4C-H1299-GAPDH: The original Fig. 4C GAPDH image of WB in H1299 cells. C- Fig. 4H1299-GAPDH-L: The original Fig. 4C GAPDH image with text labels in H1299 cells. C- Fig. 4H1299-TGFR1: The original Fig. 4C TGFR1 image of WB in H1299 cells. C- Fig. 4H1299-TGFR1-L: The original Fig. 4C TGFR1 image with text labels in H1299 cells. Fig. 5 DE-GAPDH: The original GAPDH image in Figure 5D and 5E. Fig. 5 DE-GAPDH-L: The original GAPDH image with text labels in Figure 5D and 5E. Fig. 5 DE-TGFBR1: The original TGFBR1 image in Figure 5D and 5E. Figure 5 DE-TGFBR1-L: The original TGFBR1 image with text labels in Figure 5D and 5E.
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Not applicable