We read with interest that Sun et al. (2020) identified GGC repeat expansion in the 5′ region of the NOTCH2NLC gene in Chinese patients with essential tremor (ET). Recently, two independent research teams have shown that this novel repeat expansion is the causative mutation for neuronal intranuclear inclusion disease (NIID) (Ishiura et al., 2019; Sone et al., 2019). NIID is a progressive neurodegenerative disease with the pathological hallmarks of eosinophilic ubiquitin-positive and p62-positive intranuclear inclusions in both central and peripheral nervous systems, and other tissue organs (Lindenberg et al., 1968; Liu et al., 2008; Sone et al., 2016). Patients can present with either dementia-dominant or weakness-dominant subtypes (Sone et al., 2016); a Parkinson disease phenocopy has also been recently described (Deng et al., 2019). Tremor is present in approximately one-third of these reported cohorts carrying the GGC repeat expansion (Deng et al., 2019; Okubo et al., 2019; Sone et al., 2019; Tian et al., 2019), although it invariably manifests with other neurological features such as leukoencephalopathy and cognitive impairment.
In contrast to NIID, ET is a clinical syndrome defined as an isolated tremor syndrome of bilateral upper limb action tremor of at least 3 years’ duration (Bhatia et al., 2018). A new addition to the classification is essential tremor plus (ET-plus) (Bhatia et al., 2018): ET with additional neurological signs of uncertain significance such as impaired tandem gait, ambiguous dystonic posturing or memory impairment. There is no definitive radiological or pathological marker for the ET syndromes; and only a handful of genetic variants are reported in single families and none has been reproducible (Hopfner and Helmich, 2018). The finding of 11/197 (5.58%) Chinese ET pedigrees carrying the GGC repeat expansion suggests that this mutation may play a significant role in the genetics of ET (Sun et al., 2020). Given all the patients with the GGC repeat expansion in NOTCH2NLC reported in the literatures are East Asians, we are interested to establish the prevalence of this mutation in a European ET cohort.
We analysed 111 index ET patients of European descent who were recruited from the National Hospital for Neurology and Neurosurgery (NHNN). All patients were clinically assessed by neurologists who have interests in neurogenetics and movement disorders. The distinction between ET and ET-plus was made via retrospective review of patients’ clinical records. The study was approved by the joint ethics committee of UCL institute of Neurology and NHNN, UK (UCLH: 04/N034). To test for the presence of GGC repeat expansion at NOTCH2NLC, we carried out repeat primed PCR (RP-PCR) on all patients’ genomic DNA based on published protocol (Ishiura et al., 2019), followed by fragment length analysis on an ABI 3730xl DNA analyser with a GeneScan 500 LIZ Size Standard (Thermo Fisher Scientific) and GeneMapper software (version 5.0, Scientific) (Supplementary Table 1 and Supplementary Fig. 1A and C). To avoid false negative results, we repeated the RP-PCR in duplicate with two different positive controls. In patients without the expansion, we estimated their GGC repeat sizes by amplifying the genomic DNA region containing the repeat using PCR primers specific for NOTCH2NLC (Ishiura et al., 2019) (Supplementary Fig. 1B and D).
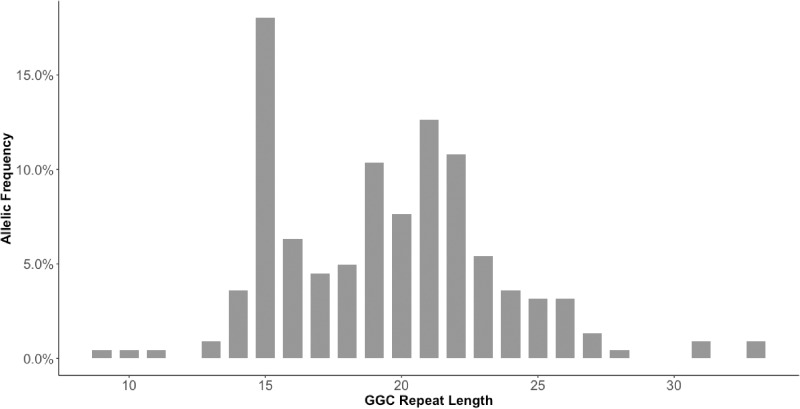
In our cohort of 111 index European patients with ET, 35 patients had a family history of autosomal dominant inheritance. The ‘Pure’ ET subgroup comprised 74 patients, and the ET-plus subgroup comprised 37 patients. In the latter subgroup, 62% had ambiguous dystonic posturing, 32% had impaired tandem gait, and 5% had equivocal parkinsonism. Table 1 outlines the demographic data of the cohort. In our screening using RP-PCR, we did not identify any patient carrying the GGC repeat expansion in NOTCH2NLC. The repeat sizes of our patients ranged from 9 to 33, with an average of 19.44 ± 4.02 (Fig. 1).
Table 1.
Demographic data of European ET cohort screened
| All probands | ET | ET-plus | |
|---|---|---|---|
| n | 111 | 74 | 37 |
| Sex, female, n (%) | 55 (49.5) | 42 (56.8) | 13 (35.1) |
| Age at onset, years, mean (range) | 42.4 (10–70) | 45.5 (15–52) | 36 (13–70) |
| Tremor | |||
| Upper limbs, n (%) | 111 (100) | 74 (100) | 37 (100) |
| Lower limbs, n (%) | 12 (10.8) | 7 (9.5) | 5 (13.5) |
| Head, n (%) | 62 (55.9) | 35 (47.3) | 27 (73) |
| Voice, n (%) | 21 (18.9) | 15 (20.3) | 6 (16.2) |
| Ataxia, n (%)a | 12 (10.8) | 0 | 12 (32.4) |
| Parkinsonism, n (%)a | 2 (1.8) | 0 | 2 (5.4) |
| Dystonia, n (%)a | 22 (19.8) | 0 | 22 (59.5) |
| Memory impairment, n (%)a | 0 | 0 | 0 |
| Family history, n (%) | 33 (29.7) | 23 (31.1) | 10 (27) |
Additional signs of unknown significance.
Figure 1.
Distribution of the GGC repeat length of NOTCH2NLC in the 111 patients with ET from the UK.
The absence of a positive screening result in our cohort infers that this mutation is unlikely a major contributor to the genetic aetiology of ET in the Europeans. The ethnic difference in mutation prevalence may be a result of a founder effect in East Asian populations. Another example of this is dentatorubral-pallidoluysian atrophy, a CAG repeat expansion disorder, which has a relatively high prevalence in Japan, but is rare elsewhere (Le Ber et al., 2003). The GGC repeat sizes of our cohort are similar to those in the Chinese ET patients without expanded GGC repeats. The weaknesses of our study include: (i) our cohort size may not be sufficiently large, especially for familial ET patients; and (ii) we retrospectively classified patients into the ET and ET-plus subgroups. However, we would still expect to identify patients carrying the GGC repeat expansion in NOTCH2NLC if the prevalence of this mutation is similar in Chinese and European cohorts of ET patients. Further studies with larger cohorts of European patients will help us to better define the role of this mutation for ET, NIID and other movement disorders outside of Japan and China.
Data availability
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
This study is made possible by the generous participation of the patients. We are grateful to Professor Shoji Tsuji from Department of Neurology of the University of Tokyo providing us with genomic DNA of the positive controls.
Funding
W.Y.Y. receives a PhD studentship from Ataxia UK and Rosetree Trust. E.O.C. receives a studentship from the brain research UK. Z.C. is supported by a clinical fellowship from the Leonard Wolfson Foundation. We are grateful to the Medical Research Council (MRC), The Wellcome Trust Synaptopathies award, MRC Centre grant (G0601943), Ataxia UK, The Rosetrees Trust, Brain Research UK, UCL ODA/LMIC award, The MSA Trust, MDUK, The Muscular Dystrophy Association (MDA). This research was also supported by the UCL/UCLH National Institute for Health Research University College London Hospitals Biomedical Research Centre.
Competing interests
The authors report no competing interests.
References
- Bhatia KP, Bain P, Bajaj N, Elble RJ, Hallett M, Louis ED, et al. Consensus Statement on the classification of tremors. from the task force on tremor of the International Parkinson and Movement Disorder Society. Mov Disord 2018; 33: 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Gu M, Miao Y, Yao S, Zhu M, Fang P, et al. Long-read sequencing identified repeat expansions in the 5’UTR of the NOTCH2NLC gene from Chinese patients with neuronal intranuclear inclusion disease. J Med Genet 2019; 56: 758–64. [DOI] [PubMed] [Google Scholar]
- Hopfner F, Helmich RC. The etiology of essential tremor: genes versus environment. Parkinsonism Relat Disord 2018; 46 (Suppl 1): S92–S6. [DOI] [PubMed] [Google Scholar]
- Ishiura H, Shibata S, Yoshimura J, Suzuki Y, Qu W, Doi K, et al. Noncoding CGG repeat expansions in neuronal intranuclear inclusion disease, oculopharyngodistal myopathy and an overlapping disease. Nat Genet 2019; 51: 1222–32. [DOI] [PubMed] [Google Scholar]
- Le Ber I, Camuzat A, Castelnovo G, Azulay JP, Genton P, Gastaut JL, et al. Prevalence of dentatorubral-pallidoluysian atrophy in a large series of white patients with cerebellar ataxia. Arch Neurol 2003; 60: 1097–9. [DOI] [PubMed] [Google Scholar]
- Lindenberg R, Rubinstein LJ, Herman MM, Haydon GB. A light and electron microscopy study of an unusual widespread nuclear inclusion body disease. A possible residuum of an old herpesvirus infection. Acta Neuropathol 1968; 10: 54–73. [DOI] [PubMed] [Google Scholar]
- Liu Y, Mimuro M, Yoshida M, Hashizume Y, Niwa H, Miyao S, et al. Inclusion-positive cell types in adult-onset intranuclear inclusion body disease: implications for clinical diagnosis. Acta Neuropathol 2008; 116: 615–23. [DOI] [PubMed] [Google Scholar]
- Okubo M, Doi H, Fukai R, Fujita A, Mitsuhashi S, Hashiguchi S, et al. GGC repeat expansion of NOTCH2NLC in adult patients with leukoencephalopathy. Ann Neurol 2019; 86: 962–8. [DOI] [PubMed] [Google Scholar]
- Sone J, Mitsuhashi S, Fujita A, Mizuguchi T, Hamanaka K, Mori K, et al. Long-read sequencing identifies GGC repeat expansions in NOTCH2NLC associated with neuronal intranuclear inclusion disease. Nat Genet 2019; 51: 1215–21. [DOI] [PubMed] [Google Scholar]
- Sone J, Mori K, Inagaki T, Katsumata R, Takagi S, Yokoi S, et al. Clinicopathological features of adult-onset neuronal intranuclear inclusion disease. Brain 2016; 139: 3170–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun QY, Xu Q, Tian Y, Hu ZM, Qin LX, Yang JX, et al. Expansion of GGC repeat in the human-specific NOTCH2NLC gene is associated with essential tremor. Brain 2020; 143: 222–33. [DOI] [PubMed] [Google Scholar]
- Tian Y, Wang JL, Huang W, Zeng S, Jiao B, Liu Z, et al. Expansion of human-specific GGC repeat in neuronal intranuclear inclusion disease-related disorders. Am J Hum Genet 2019; 105: 166–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available from the corresponding author upon reasonable request.