Chronic kidney disease (CKD) presents unique challenges in our efforts to identify preventative, diagnostic, and therapeutic strategies aimed at reducing the burden of cardiovascular disease. Not only do patients with CKD disproportionately suffer from cardiovascular disease, but traditional risk factors alone are not sufficient to explain their increased cardiovascular risk. 1 Changing this paradigm presents several challenges for translational and precision medicine. Chief among these challenges is the fact that the repertoire of circulating factors available to be measured in cardiovascular risk studies is influenced directly and indirectly by both glomerular and tubular function. 2 The complexity of this challenge cannot be underestimated, because CKD is not a single disease, but rather a constellation of renal diseases capable of arising from multiple causes with varying degrees of tubular and glomerular involvement. Thus, the impact of kidney function on biomarkers of cardiovascular risk must be assessed and validated. Ideally, unique cardiovascular risk biomarkers will be defined across the spectrum of CKD and inform preventative, diagnostic, and therapeutic strategies designed to significantly improve cardiovascular outcomes in individuals burdened with this disease.
See Article by Yang et al.
In this issue of the Journal of the American Heart Association (JAHA), Yang and colleagues3 sought to address these challenges through a novel approach. These investigators performed high‐throughput proteomic analyses using modified aptamers to identify the importance of kidney function on the circulating proteome of patients with stable coronary artery disease from the Heart and Soul Study, an observational cohort study. 3 Aptamers are short single‐stranded oligonucleotides, capable of folding into secondary structures to achieve various complex molecular forms that bind with high affinity and specificity to proteins, peptides, and even small molecules, such as uremic toxins (Figure). High‐throughput proteomics routinely uses mass spectrometry and antibody‐based technologies. Despite advancements of mass spectrometry in clinical proteomics, its power can decrease with the increasing complexity of the biological samples. Moreover, compared with antibodies, aptamer generation is significantly easier, is cheaper, has less batch‐to‐batch variation, and offers a wide target range. 4 These features make aptamers a promising tool for diagnostic applications as well as for purification of target molecules from biological samples. In the current study, Yang and colleagues3 used a modified aptamer‐based proteomics platform that uses “slow off‐rate modified DNA aptamers” in conjunction with high‐affinity protein capture reagents to quantify 1054 plasma proteins in these patients. Their approach has been used in several studies, including validation of a 9‐protein–based risk score to improve prediction of cardiovascular outcomes in patients with stable coronary heart disease. 5
Figure 1.
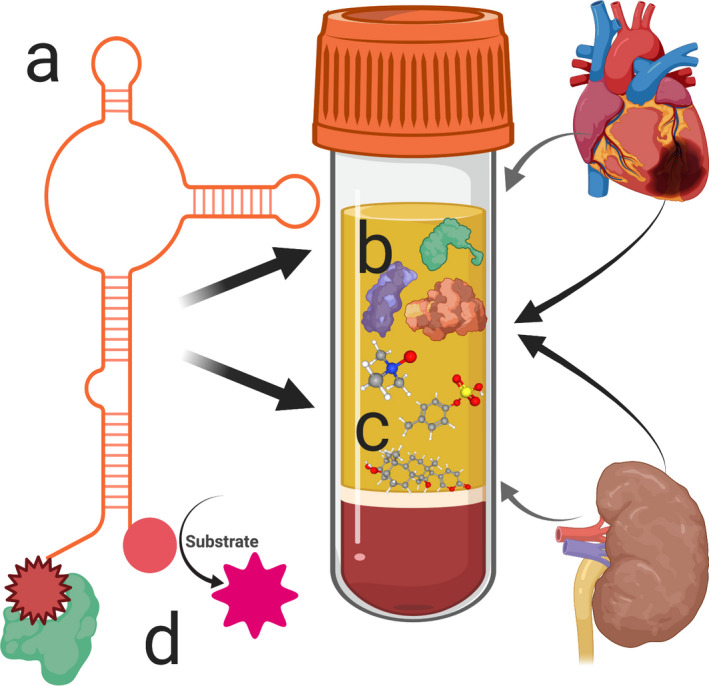
Schematic of aptamer‐based detection of cardiovascular risk markers in chronic kidney disease. Aptamers are short single‐stranded oligonucleotides, capable of folding into secondary structures to achieve various complex molecular forms (A). They are screened and selected from large random libraries for their ability to bind with high affinity and specificity to proteins (B), peptides, and even small molecules, such as uremic toxins (C). Their affinity, specificity, stability, and binding kinetics for these targets can be enhanced through modifications, including the addition of functional groups, such as proteins or protein‐like moieties, at the 5′ or 3′ ends (D). Thus, modified aptamer technology may significantly enhance our ability to probe cardiovascular risk markers beyond standard proteomics in diseases such as chronic kidney disease.
The current study reveals a significant correlation between estimated glomerular filtration rate (eGFR) and alterations in 709 of the 1054 circulating proteins measured, where reduced eGFR associated with increased circulating protein levels. As renal function declines, the kidney's glomerular barrier changes, and its ability to filter water‐soluble molecules <50 kDa also decreases, resulting in the accumulation of blood proteins. Furthermore, impaired renal function can result in altered protein synthesis as well as the loss of renal parenchyma cells, consequently causing significant alterations in the circulating proteome. 6 The situation is compounded by common comorbidities, such as hypertension, which can amplify multiorgan injury. These findings are supported by proteomic analysis of the discovery cohort from the PIVUS (Prospective Investigation of the Vasculature in Uppsala Seniors) study, which identified >2 dozen unique plasma proteins that were significantly associated with eGFR decline. 7 Interestingly, proteomic analysis of the replication cohort from the ULSAM (Uppsala Longitudinal Study of Adult Men) validated that 20 of these proteins were also found to be significantly associated with eGFR decline. 7
The current study further demonstrates that these eGFR associated changes in the circulating proteome alter proteins with critical functions implicated in cardiovascular and renal diseases, such as angiogenesis, blood pressure, and fibrosis, thus providing potential mechanistic links to the cardiovascular events reported in patients with CKD. In fact, many of the measured proteins were cytokines, chemokines, adipokines, growth factors, and hormones that mediate crucial biological functions and play essential roles in mediating prevalent cardiovascular events in CKD. Consistent with these findings, other recent reports have demonstrated that glomerular filtration rate–mediated changes in the proteome are linked to known processes involved in CKD complications, such as inflammation, complement activation, heart failure, and vascular damage. 8 More important, the impact of reduced eGFR on the circulating proteome was supported by the authors' comparison of their results from the Heart and Soul Study cohort with those of the Lund CKD cohort in Sweden. 9 Of the top 100 proteins, 98 were common between the 2 cohorts, demonstrating wide agreement and important validation for these findings.
The present study further determined the extent to which a previously validated set of 196 cardiovascular biomarkers related to kidney function by adjusting the hazard ratios of these biomarkers for eGFR. The number of prognostic biomarkers decreased from 196 to 87 proteins after correcting for eGFR. This highly suggests the importance of kidney function in cardiovascular risk information contained in circulating proteins as well as recognizes a new set of biomarkers that may better predict cardiovascular risk independently of kidney function.
To identify biomarkers that are prognostic of cardiovascular risk in patients with CKD compared with non‐CKD patients, Yang and coworkers3 divided the Heart and Soul Study cohort according to eGFR. They determined that of the 1054 proteins quantified, 84 predicted cardiovascular risk in patients with normal renal function and 21 predicted cardiovascular risk especially in patients with CKD. These 21 proteins are known to be involved in various pathological functions, including inflammation, tissue repair, malignancy, and cell signaling with extracellular matrix communication. Interestingly, 8 of the 21 cardiovascular risk proteins predicted were exclusive to the CKD group and were absent in the patients with normal renal function. One of the predicted proteins, tumor necrosis factor receptor superfamily member 1A, is a well‐known predictor of cardiovascular outcomes in patients with CKD. 10 Consistent with the previous studies, the 2 other predicted proteins, including PEBP‐1 (phosphatidylethanolamine‐binding protein 1) and platelet‐activating factor acetylhydrolase IB subunit β, are involved in heart failure and atherosclerosis, respectively. 11 , 12 Future studies of these predicted cardiovascular biomarkers are required to better understand their potential role in mediating the cardiovascular disease associated with CKD.
One of the challenges with biomarker studies based purely on proteomics is that not every marker of risk is reflected in the proteome. Thus, creating aptamers that can capture and detect metabolites and small molecules in addition to circulating proteins would provide a significantly more complete picture. This is particularly true in CKD, where gut metabolites, such as trimethylamine N‐oxide, 13 small molecules, such as cardiotonic steroids, 14 and protein‐bound uremic toxins, such as indoxyl sulfate and p‐cresyl sulfate, 15 have all been linked to cardiovascular risk in the setting of CKD. Thus, extending the modified aptamer approach to capture these important classes of molecules may provide more robust measures of cardiovascular risk as well as present opportunities for therapeutic advances to reduce cardiovascular morbidity and mortality in CKD.
Another limitation of strictly focusing on the circulating proteome is that the functional status of the protein may be at least equally important to its circulating level. In this regard, data from the Cleveland Clinic GeneBank study have demonstrated that activity levels of serum paraoxonase‐1 predict cardiovascular risk in both patients with stable coronary artery disease (after adjustment for renal function) 16 as well as those with CKD. 17 Furthermore, data from the SKS (Salford Kidney Study) demonstrated that the lactonase activity of paraoxonase‐1, but not its circulating protein levels, predicted cardiovascular risk in patients with CKD. 18 In the current study, Yang et al3 identified platelet‐activating factor acetylhydrolase IB subunit β as a unique cardiovascular risk marker in CKD. As the enzymatic activity of platelet‐activating factor acetylhydrolase IB subunit β can be measured, it will be important to assess this in follow‐up studies to provide functional insight into this potentially prognostic enzyme. Of course, the challenge with activity assays is that they are inherently more complex than strict proteomics and perhaps less amenable to high‐throughput approaches. Some groups are solving this challenge by coupling methods that are well adapted to high‐throughput screens, such as liquid chromatography–mass spectrometry, with activity‐based methods, such as those used to interrogate the renin‐angiotensin‐aldosterone system, to form powerful assays that may provide a more complete phenotype of the functional status of the circulating proteome. 19
As with all biomarker studies of cardiovascular risk, the challenge will be validating the identified risk markers through additional clinical‐translational and mechanistic studies. Nevertheless, expansion of aptamer‐based proteomics may be a welcome advancement to help accelerate the discovery and application of preventative, diagnostic, and therapeutic strategies aimed at combatting the significant cardiovascular health disparities that patients with CKD face.
Disclosures
None.
(J Am Heart Assoc. 2020;9:e017427 DOI: 10.1161/JAHA.120.017427.)
The opinions expressed in this article are not necessarily those of the editors or of the American Heart Association.
For Disclosures, see page 3.
References
- 1. Lee AK, Katz R, Jotwani V, Garimella PS, Ambrosius WT, Cheung AK, Gren LH, Neyra JA, Punzi H, Raphael KL, et al. Distinct dimensions of kidney health and risk of cardiovascular disease, heart failure, and mortality. Hypertension. 2019;74:872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dubin RF, Rhee EP. Proteomics and metabolomics in kidney disease, including insights into etiology, treatment, and prevention. Clin J Am Soc Nephrol. 2020;15:404–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang J, Brody EN, Murthy AC, Mehler RE, Weiss SJ, DeLisle RK, Ostroff R, Williams SA, Ganz P. Impact of kidney function on the blood proteome and on protein cardiovascular risk biomarkers in patients with stable coronary heart disease. J Am Heart Assoc. 2020;9:e016463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, Carter J, Dalby AB, Eaton BE, Fitzwater T, et al. Aptamer‐based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5:e15004 DOI:10.1371/journal.pone.0015004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ganz P, Heidecker B, Hveem K, Jonasson C, Kato S, Segal MR, Sterling DG, Williams SA. Development and validation of a protein‐based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. 2016;315:2532–2541. [DOI] [PubMed] [Google Scholar]
- 6. Defilippi CR, Herzog CA. Interpreting cardiac biomarkers in the setting of chronic kidney disease. Clin Chem. 2017;63:59–65. [DOI] [PubMed] [Google Scholar]
- 7. Carlsson AC, Ingelsson E, Sundström J, Carrero JJ, Gustafsson S, Feldreich T, Stenemo M, Larsson A, Lind L, Ärnlöv J. Use of proteomics to investigate kidney function decline over 5 years. Clin J Am Soc Nephrol. 2017;12:1226–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Glorieux G, Mullen W, Duranton F, Filip S, Gayrard N, Husi H, Schepers E, Neirynck N, Schanstra JP, Jankowski J, et al. New insights in molecular mechanisms involved in chronic kidney disease using high‐resolution plasma proteome analysis. Nephrol Dial Transplant. 2015;30:1842–1852. [DOI] [PubMed] [Google Scholar]
- 9. Christensson A, Ash JA, DeLisle RK, Gaspar FW, Ostroff R, Grubb A, Lindström V, Bruun L, Williams SA. The impact of the glomerular filtration rate on the human plasma proteome. Proteomics Clin Appl. 2018;12:1700067. [DOI] [PubMed] [Google Scholar]
- 10. Carlsson AC, Östgren CJ, Nystrom FH, Länne T, Jennersjö P, Larsson A, Ärnlöv J. Association of soluble tumor necrosis factor receptors 1 and 2 with nephropathy, cardiovascular events, and total mortality in type 2 diabetes. Cardiovasc Diabetol. 2016;15:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schmid E, Neef S, Berlin C, Tomasovic A, Kahlert K, Nordbeck P, Deiss K, Denzinger S, Herrmann S, Wettwer E, et al. Cardiac RKIP induces a beneficial β‐adrenoceptor–dependent positive inotropy. Nat Med. 2015;21:1298. [DOI] [PubMed] [Google Scholar]
- 12. Gonçalves I, Edsfeldt A, Ko NY, Grufman H, Berg K, Björkbacka H, Nitulescu M, Persson A, Nilsson M, Prehn C, et al. Evidence supporting a key role of Lp‐PLA2‐generated lysophosphatidylcholine in human atherosclerotic plaque inflammation. Arterioscler Thromb Vasc Biol. 2012;32:1505–1512. [DOI] [PubMed] [Google Scholar]
- 13. Tang WW, Wang Z, Kennedy DJ, Wu Y, Buffa JA, Agatisa‐Boyle B, Li XS, Levison BS, Hazen SL. Gut microbiota‐dependent trimethylamine N‐oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ Res. 2015;116:448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khalaf FK, Dube P, Mohamed A, Tian J, Malhotra D, Haller ST, Kennedy DJ. Cardiotonic steroids and the sodium trade balance: new insights into trade‐off mechanisms mediated by the Na+/K+‐ATPase. Int J Mol Sci. 2018;19:2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hung SC, Kuo KL, Wu C‐C, Tarng D‐C. Indoxyl sulfate: a novel cardiovascular risk factor in chronic kidney disease. J Am Heart Assoc. 2017;6:e005022 10.1161/JAHA.116.005022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tang WW, Hartiala J, Fan Y, Wu Y, Stewart AF, Erdmann J, Kathiresan S; CARDIoGRAMConsortium , Roberts R, McPherson R, Allayee H, et al. Clinical and genetic association of serum paraoxonase and arylesterase activities with cardiovascular risk. Arterioscler Thromb Vasc Biol. 2012;32:2803–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kennedy DJ, Wilson Tang W, Fan Y, Wu Y, Mann S, Pepoy M, Hazen SL. Diminished antioxidant activity of high‐density lipoprotein–associated proteins in chronic kidney disease. J Am Heart Assoc. 2017;2:e000104 10.1161/JAHA.112.000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mohammed CJ, Xie Y, Brewster PS, Ghosh S, Dube P, Sarsour T, Kleinhenz AL, Crawford EL, Malhotra D, James RW, et al. Circulating lactonase activity but not protein level of PON‐1 predicts adverse outcomes in subjects with chronic kidney disease. J Clin Med. 2019;8:1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pavo N, Wurm R, Goliasch G, Novak JF, Strunk G, Gyöngyösi M, Poglitsch M, Säemann MD, Hülsmann M. Renin‐angiotensin system fingerprints of heart failure with reduced ejection fraction. J Am Coll Cardiol. 2016;68:2912–2914. [DOI] [PubMed] [Google Scholar]


