Abstract
Background
Angelica L. (family Apiaceae) is an economically important genus comprising ca. One hundred ten species. Angelica species are found on all continents of the Northern Hemisphere, and East Asia hosts the highest number of species. Morphological characters such as fruit anatomy, leaf morphology and subterranean structures of Angelica species show extreme diversity. Consequently, the taxonomic classification of Angelica species is complex and remains controversial, as the classifications proposed by previous studies based on morphological data and molecular data are highly discordant. In addition, the phylogenetic relationships of major clades in the Angelica group, particularly in the Angelica s. s. clade, remain unclear. Chloroplast (cp) genome sequences have been widely used in phylogenetic studies and for evaluating genetic diversity.
Results
In this study, we sequenced and assembled 28 complete cp genomes from 22 species, two varieties and two cultivars of Angelica. Combined with 36 available cp genomes in GenBank from representative clades of the subfamily Apioideae, the characteristics and evolutionary patterns of Angelica cp genomes were studied, and the phylogenetic relationships of Angelica species were resolved. The Angelica cp genomes had the typical quadripartite structure including a pair of inverted repeats (IRs: 5836–34,706 bp) separated by a large single-copy region (LSC: 76,657–103,161 bp) and a small single-copy region (SSC: 17,433–21,794 bp). Extensive expansion and contraction of the IR region were observed among cp genomes of Angelica species, and the pattern of the diversification of cp genomes showed high consistency with the phylogenetic placement of Angelica species. Species of Angelica were grouped into two major clades, with most species grouped in the Angelica group and A. omeiensis and A. sinensis grouped in the Sinodielsia with Ligusticum tenuissimum.
Conclusions
Our results further demonstrate the power of plastid phylogenomics in enhancing the phylogenetic reconstructions of complex genera and provide new insights into plastome evolution across Angelica L.
Keywords: Angelica, Plastome evolution, Phylogenomic, Inverted repeats
Background
The herbaceous perennial genus Angelica L. (family Apiaceae) is a taxonomically complex and controversial group comprising approximately 110 species with extreme polymorphism in leaf morphology, fruit anatomy and subterranean structures [1–3]. Members of Angelica are distributed on all Northern Hemisphere, with the largest number of species (approximately 55) concentrated in East Asia [3–5]. Forty-five Angelica species are distributed in China with 32 of them endemic [3, 6]; some species are extremely rare in the field and are only known from limited specimens [7].
Some of these endemic Angelica species are of great economic value and have been used in traditional Chinese medicines for hundreds of years [3, 8]. Some species of Angelica are official materia medica recorded in Chinese Pharmacopoeia Committee of People’s Republic of China’s 2010, including A. sinensis (Chinese medicine name: Danggui), A. biserrata (Duhuo) and A. dahurica (Baizhi) [7]. Another 15 species of Angelica are also used as herbal medicinal materials in folk remedies (http://frps.eflora.cn).
Previous studies of Angelica systematics have focused on karyotaxonomical analyses [2, 9, 10], pollen morphology [11–13], petiole and fruit anatomy [14], and phytochemistry [15, 16]. Previous molecular phylogenetic analyses of Angelica have exclusively been based on phylogenetic analyses of DNA sequences, especially on the nuclear ribosomal (nr) DNA internal transcribed spacer (ITS) region, and relatively few Chinese representatives of Angelica have been included in analyses [6, 17–20]. Xue et al. (2007) used 44 ITS sequences from species of Angelica sensu stricto (s.s.) and allies from East Asia and proposed that Angelica was polyphyletic. Feng et al. (2009) suggested that Angelica s.s. was monophyletic after including Coelopleurum, Czenaevia, and Ostericum koreanum in analyses but excluding several other species previously recognized in Angelica s.l. Liao et al. (2013) reconstructed the phylogeny of Angelica s.l. and infrageneric relationships in Angelica s.s. with a more extensive sampling of Angelica species from East Asia (including 44 of its approximately 55 known species) and integrated analyses of nrDNA (ITS, ETS), cpDNA (rps16 intron, rps16-trnK intergenic spacer, rpl32-trnL intergenic spacer, and trnL-trnT intergenic spacer), and morphological data. Their analysis suggested that many species of Angelica fell outside of Angelica s.s. and that four species of Angelica occurred outside of the Angelica group. However, the relationships of clades within the Selineae, particularly within the Angelica s.l. group, are still controversial and mostly unresolved.
Chloroplasts are key organelles for photosynthesis and other biochemical pathways in plants [21, 22]. The chloroplast (cp) genome is one of the three DNA genomes (with nuclear and mitochondrial genomes) in plants with a relatively conserved quadripartite circular structure ranging from 115 to 165 kb [23, 24]. Because of their relatively stable genome structure, the complete cp genome sequences have been widely accepted to provide a valuable and informative data source for understanding evolutionary biology and have become a powerful tool for resolving plant phylogenies [24–35].
In this study, we report 28 newly sequenced and complete cp genomes from the genus Angelica (22 species, two varieties and two cultivars) and investigate the structural diversity of cp genomes in Angelica by comparative chloroplast genome analyses. Furthermore, we test the power of complete cp genomes for resolving the phylogeny of the controversial and less well-resolved Angelica group by integrated analyses with another 36 published cp genomes available from NCBI GenBank from representative clades of the Apioideae (subfamily of Apiaceae).
Results
Characteristics of Angelica plastomes
The number of paired-end raw reads obtained by the Illumina HiSeq 4000 system ranged from 8,616,334 to 22,518,619 for the 28 Angelica samples. After mapping the paired-end reads of each Angelica taxon, 52,277 to 1,673,010 reads were extracted, yielding 59× to 1445× chloroplast genome coverage (Table 1). The inverted repeat (IR) junction regions in the assembled chloroplast genomes were further manually checked to avoid potential annotation errors. High-quality chloroplast genome sequences were thus achieved and facilitated for downstream analyses. The 28 Angelica chloroplast genome sequences were deposited in GenBank (accession numbers, MT921958-MT921985).
Table 1.
Statistics of NGS sequencing of 28 Angelica samples
| ID | Species | Raw reads no. | Mapped reads No. | Chloroplast genome coverage (x) |
|---|---|---|---|---|
| DG001 | A. morii | 11,122,925 | 522,264 | 546 |
| DG002 | A. tianmuensis | 22,518,619 | 1,673,010 | 1445 |
| DG003 | A. cartilaginomarginata var. foliata | 14,742,849 | 195,610 | 204 |
| DG004 | A. biserrata | 19,320,868 | 129,002 | 136 |
| DG005 | A. polymorpha | 13,445,469 | 258,026 | 290 |
| DG006 | A. megaphylla | 9,985,839 | 133,105 | 141 |
| DG007 | A. valida | 9,519,329 | 52,277 | 59 |
| DG008 | A. decursiva | 14,307,154 | 419,462 | 430 |
| DG009 | A. kangdingensis | 14,576,047 | 176,603 | 199 |
| DG010 | A. apaensis | 11,056,070 | 964,507 | 1096 |
| DG011 | A. maowenensis | 12,236,395 | 186,811 | 211 |
| DG012 | A. pseudoselinum | 11,687,001 | 624,354 | 706 |
| DG013 | A. laxifoliata | 11,817,482 | 216,804 | 245 |
| DG014 | A. omeiensis | 18,910,215 | 1,243,559 | 1134 |
| DG015 | A. tsinlingensis | 14,571,967 | 62,281 | 70 |
| DG016 | A. dahurica var. formosana | 11,086,333 | 55,945 | 62 |
| DG017 | A. dahurica cv. ‘Qibaizhi’ | 10,575,208 | 500,975 | 569 |
| DG018 | A. porphyrocaulis | 8,616,334 | 300,612 | 345 |
| DG019 | A. dahurica cv. ‘Qibaizhi’ | 9,073,284 | 231,034 | 266 |
| DG020 | A. nitida | 9,991,134 | 70,752 | 80 |
| DG023 | A. cartilaginomarginata | 14,135,856 | 145,751 | 163 |
| DG025 | A. anomala | 10,508,302 | 277,618 | 313 |
| DG026 | A. dahurica cv. ‘Hangbaizhi’ | 12,266,363 | 296,498 | 336 |
| DG027 | A. dahurica cv. ‘Hangbaizhi’ | 9,171,688 | 990,934 | 1132 |
| DG028 | A. sinensis | 9,212,560 | 929,902 | 947 |
| DG029 | A. sinensis | 10,265,910 | 1,084,379 | 1188 |
| HG021 | A. dahurica | 17,918,362 | 359,326 | 407 |
| HG022 | A. gigas | 14,989,366 | 86,751 | 96 |
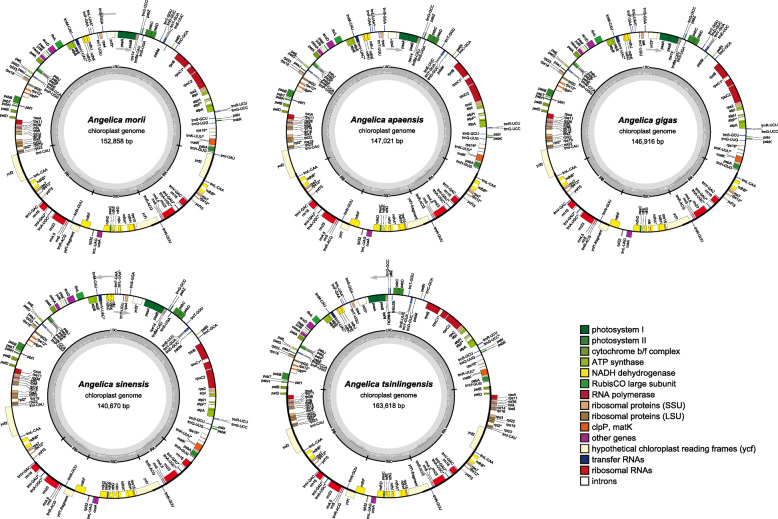
The length of the complete chloroplast genome ranged from 140,670 bp (A. sinensis) to 163,618 bp (A. tsinlingensis) among the 33 cp genomes from 27 Angelica species (varieties or cultivars). All of the cp genomes possessed the typical quadripartite structure of angiosperms, including a pair of inverted repeat regions (IRs: 5836–34,706 bp) separated by a large single-copy region (LSC: 76,657–103,161 bp) and a small single-copy region (SSC: 17,433–21,794 bp) (Fig. 1; Table 2). The average GC content was 37.5%, which was virtually identical among the 33 complete Angelica cp genomes. The total number of genes ranged from 121 (A. sinensis) to 144 (A. tsinlingensis) in these 33 complete Angelica cp genomes. After removing the duplicated genes in IR regions, the 33 Angelica cp genomes harbored 113 to 114 different genes, including 80 protein-coding and 4 rRNA genes shared by all cp genomes (Table 1). While most cp genomes contained 29 tRNA genes, seven cp genomes contained one more tRNA gene (trnG-UCC or trnG-GCC) (Additional file 1: Table S1). The organization, gene order and GC content of cp genomes in Angelica were highly identical and similar to those of other higher plants (Fig. 1).
Fig. 1.
Gene map of five Angelica chloroplast genomes. The genes transcribed in the clockwise and counterclockwise directions are plotted inside and outside the circle, respectively. Different colors indicate genes belonging to different functional groups. The small single-copy (SSC) and large single-copy (LSC) regions are separated by the region of inverted repeats (IRa and IRb) indicated with the thick lines
Table 2.
Comparison of the chloroplast genome features of Angelica species
| ID | Species | CP genome type | Genome size (bp) | LSC size (bp) | SSC size (bp) | IR size (bp) | Genome GC | Total genes | Protein coding genes | rRNA genes | tRNA genes | Total genes uniq |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DG001 | A. morii | I | 152,858 | 86,914 | 17,524 | 24,210 | 37.5% | 129 | 85 | 8 | 36 | 113 |
| DG025 | A. anomala | II | 147,145 | 93,695 | 17,820 | 17,815 | 37.5% | 127 | 84 | 8 | 35 | 113 |
| DG010 | A. apaensis | II | 147,021 | 93,693 | 17,770 | 17,779 | 37.5% | 127 | 84 | 8 | 35 | 113 |
| DG004 | A. biserrata | II | 146,677 | 93,217 | 17,500 | 17,980 | 37.5% | 128 | 84 | 8 | 36 | 114 |
| DG023 | A. cartilaginomarginata | II | 146,583 | 94,185 | 17,806 | 17,296 | 37.5% | 128 | 84 | 8 | 36 | 114 |
| DG003 | A. cartilaginomarginata var. foliata | II | 147,017 | 93,622 | 17,777 | 17,809 | 37.5% | 127 | 84 | 8 | 35 | 113 |
| DG017 | A. dahurica cv. ‘Qibaizhi’ | II | 146,815 | 93,547 | 17,690 | 17,789 | 37.5% | 127 | 84 | 8 | 35 | 113 |
| DG019 | A. dahurica cv. ‘Qibaizhi’ | II | 146,811 | 93,547 | 17,630 | 17,817 | 37.5% | 127 | 84 | 8 | 35 | 113 |
| KT963037 | A. dahurica | II | 146,918 | 93,605 | 17,677 | 17,818 | 37.5% | 128 | 85 | 8 | 35 | 113 |
| DG026 | A. dahurica cv. ‘Hangbaizhi’ | II | 146,810 | 93,546 | 17,630 | 17,817 | 37.5% | 127 | 84 | 8 | 35 | 113 |
| DG027 | A. dahurica cv. ‘Hangbaizhi’ | II | 146,835 | 93,571 | 17,630 | 17,817 | 37.5% | 127 | 84 | 8 | 35 | 113 |
| HG021 | A. dahurica | II | 147,477 | 93,584 | 18,259 | 17,817 | 37.5% | 128 | 84 | 8 | 36 | 114 |
| DG016 | A. dahurica var. formosana | II | 147,097 | 93,322 | 17,625 | 18,075 | 37.5% | 127 | 84 | 8 | 35 | 113 |
| DG008 | A. decursiva | II | 146,158 | 92,655 | 17,537 | 17,983 | 37.6% | 128 | 84 | 8 | 36 | 114 |
| DG009 | A. kangdingensis | II | 146,529 | 93,853 | 17,530 | 17,573 | 37.5% | 128 | 84 | 8 | 36 | 114 |
| DG013 | A. laxifoliata | II | 146,682 | 93,964 | 17,532 | 17,593 | 37.5% | 127 | 84 | 8 | 35 | 113 |
| DG011 | A. maowenensis | II | 146,882 | 94,105 | 17,433 | 17,672 | 37.4% | 127 | 84 | 8 | 35 | 113 |
| DG006 | A. megaphylla | II | 146,724 | 92,350 | 17,672 | 18,351 | 37.5% | 127 | 84 | 8 | 35 | 113 |
| DG020 | A. nitida | II | 146,789 | 93,081 | 17,506 | 18,101 | 37.4% | 127 | 84 | 8 | 35 | 113 |
| MF594405 | A. nitida | II | 146,512 | 103,161 | 21,794 | 5836 | 37.5% | 127 | 84 | 8 | 35 | 113 |
| DG014 | A. omeiensis | II | 147,814 | 93,787 | 17,635 | 18,196 | 37.6% | 128 | 85 | 8 | 35 | 113 |
| DG005 | A. polymorpha | II | 146,982 | 93,442 | 17,754 | 17,893 | 37.6% | 127 | 84 | 8 | 35 | 113 |
| DG018 | A. porphyrocaulis | II | 146,859 | 93,543 | 17,682 | 17,817 | 37.5% | 128 | 84 | 8 | 36 | 114 |
| DG012 | A. pseudoselinum | II | 148,942 | 91,035 | 17,527 | 20,190 | 37.5% | 128 | 85 | 8 | 35 | 113 |
| DG002 | A. tianmuensis | II | 147,308 | 93,239 | 17,637 | 18,216 | 37.5% | 127 | 84 | 8 | 35 | 113 |
| DG007 | A. valida | II | 146,833 | 94,103 | 17,530 | 17,600 | 37.5% | 127 | 84 | 8 | 35 | 113 |
| KT963036 | A. acutiloba | II | 147,074 | 93,368 | 17,574 | 18,066 | 37.5% | 128 | 85 | 8 | 35 | 113 |
| KX352468 | A. acutiloba | II | 147,074 | 93,368 | 17,574 | 18,066 | 37.5% | 128 | 85 | 8 | 35 | 113 |
| HG022 | A. gigas | III | 146,916 | 93,163 | 17,579 | 18,087 | 37.6% | 127 | 84 | 8 | 35 | 113 |
| KT963038 | A. gigas | III | 146,916 | 93,119 | 17,583 | 18,107 | 37.6% | 128 | 85 | 8 | 35 | 113 |
| DG028 | A. sinensis | IV | 140,694 | 101,695 | 17,591 | 10,704 | 37.4% | 121 | 80 | 8 | 33 | 113 |
| DG029 | A. sinensis | IV | 140,670 | 101,680 | 17,578 | 10,706 | 37.5% | 121 | 80 | 8 | 33 | 113 |
| DG015 | A. tsinlingensis | V | 163,618 | 76,657 | 17,549 | 34,706 | 37.5% | 144 | 99 | 8 | 37 | 114 |
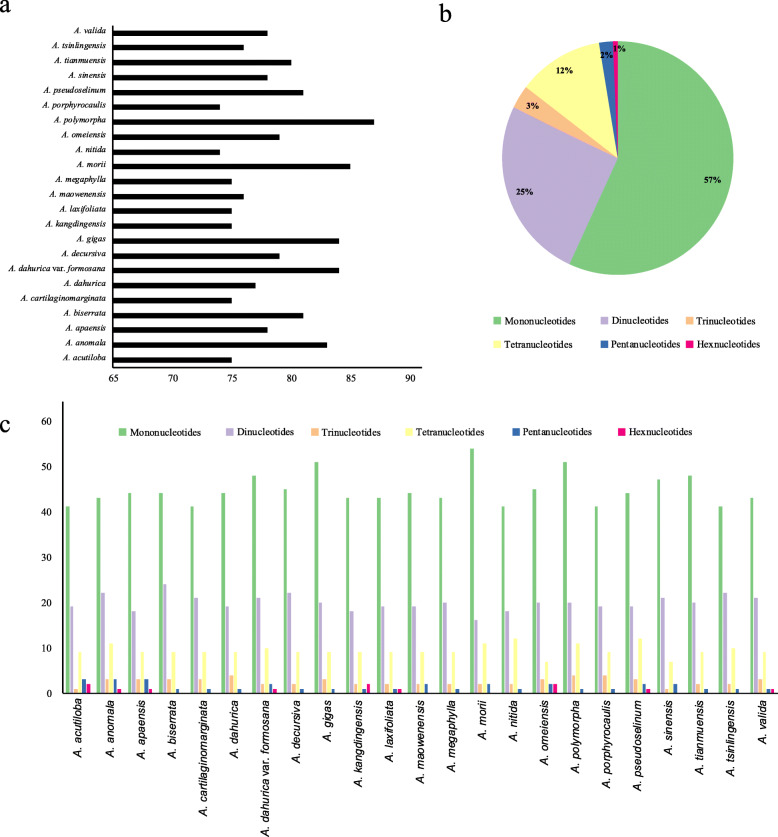
The number of simple sequence repeats (SSRs) ranged from 68 (A. nitida) to 87 (A. polymorpha) among the 33 Angelica cp genomes (Fig. 2a). Most of the SSRs were mono-nucleotide repeats (58%), while di-nucleotide, tri-nucleotide, tetra-nucleotide, penta-nucleotide and hex-nucleotide SSRs made up 24, 4, 11, 2 and 1% of all SSRs, respectively (Fig. 2b). The mono-nucleotide repeat number with the highest variability, ranged from 38 (A. nitida) to 54 (A. morii), while the number of other repeat types did not significantly differ among the 33 Angelica cp genomes (Additional file 2: Table S2, Fig. 2c).
Fig. 2.
Comparison of simple sequence repeats among 33 Angelica chloroplast genomes. a Number of SSRs detected in 33 Angelica chloroplast genomes; b Frequencies of identified SSRs in different repeat types; c Number of SSRs in different repeat types in 33 Angelica chloroplast genomes
Expansion and contraction of the IR region
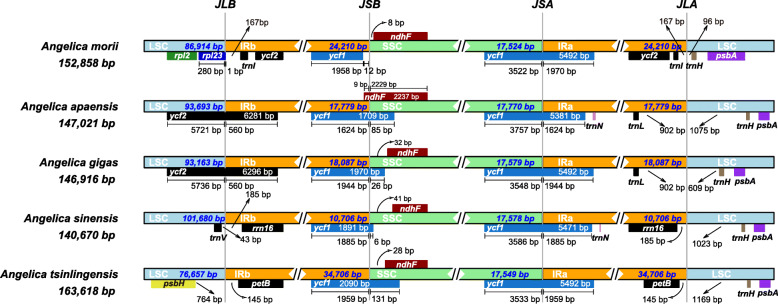
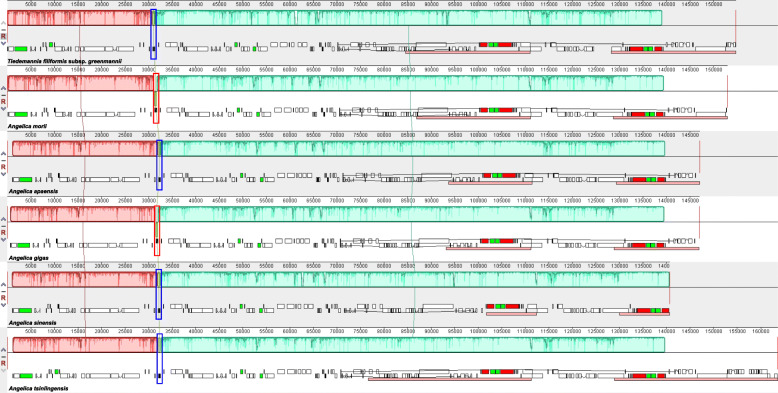
Although cp genomes are highly conserved in genomic structure and size, the change in the size of the IR/SC junction caused by the expansion and contraction of the IR/SC boundary regions has been considered a primary mechanism for creating length variation in cp genomes of higher plants [26, 36–38]. Extensive expansion and contraction of the IR region were observed among the 33 Angelica cp genomes examined in this study and could be classified into five different types based on the characteristics in the IR/SC junction region and with/without inversion. The IR region of A. morii expanded and contained a duplicate copy of the ycf2 gene (Type I); in most (25/33) Angelica cp genomes, the junction site of IR/SSC was located in the ycf1 gene, and the junction site of IR/LSC was located between genes of trnL and trnH (Type II) (Fig. 3). An inversion of approximately 490 bp in the trnY-trnD-trnE gene was observed in the cp genome of A. gigas (Type III) and in A. moii (Fig. 4). Significant contraction of the IR region was detected in A. sinensis (10,706 bp) and ended with the rrn16 gene in the IR region (Type IV); the largest expansion of the IR region was observed in A. tsinlingensis and ended with the petB gene in the IR region (Type V) (Fig. 3).
Fig. 3.
Comparison of the borders of LSC, SSC, and IR regions of chloroplast genomes in six Angelica species
Fig. 4.
MAUVE alignment of chloroplast genomes in six Angelica species. The chloroplast genome of Tiedemannia filiformis subsp. greenmannii is shown at the top as the reference genome. Within each of the alignments, local collinear blocks are represented by blocks of the same color connected by lines
Phylogenetic analysis
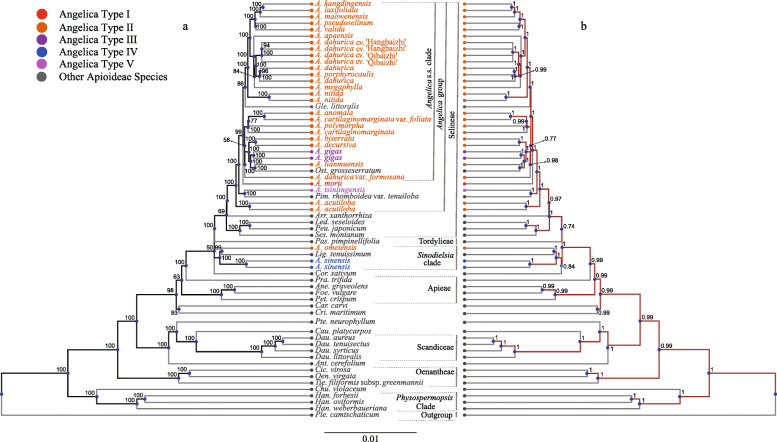
The ML and Bayesian trees yielded highly similar topologies. Members of Angelica fell primarily into two major lineages: (1) the Angelica group occurring in tribe Selineae (BS = 100, PP = 1), and (2) the Sinodielsia clade (BS = 100, PP = 1) (Fig. 5). The names of major clades determined by previous studies are followed [1, 39–41]. The Angelica group made up most of the Angelica accessions (30/33), and 26 Angelica accessions formed the well-supported Angelica s.s. clade (BS = 99, PP = 1) which also included Glehnia littoralis and Ostericum grosseserratum (Fig. 5). Within the Angelica s.s. clade, four major lineages were recovered (A. kangdingensis to A. valida, A. apaensis to A. megaphylla, A. anomala to A. cartilaginomarginata, and A. biserrata to Ost. grosseserratum). The support value of the placement of clade A. anomala to A. dahurica var. formosana was relatively low (BS = 58, PP = 0.77). The littoral Angelica species A. morii, which inhabits the East Asian littoral regions or islands, and A. tsinlingensis, which is clearly different from members of the Angelica s.s. clade by its thin-winged dorsal ribs and triple vittae in the furrow [1], were placed outside of the Angelica s.s. clade based on the molecular findings (Fig. 5). A. acutiloba is also isolated from the Angelica s.s. clade and occupies an early diverging branch of the Angelica group (Fig. 5). The Sinodielsia clade consisted of A. omeiensis, A. sinensis and Ligusticum tenuissimum. Most clades in the Angelica group received high BS/PP support with the exception of the clade that included A. anomala to A. dahurica var. formosana (BS = 58, PP = 0.77) (Fig. 5). Most accessions of A. dahurica (A. dahurica, A. dahurica cv. hangbaizhi and A. dahurica cv. xingan) were placed in a well-supported clade that also included A. porphyrocaulis, with the exception of A. dahurica var. formosana, which was placed in a relatively distant clade that included A. anomala to A. tianmuensis and Ostericum grosseserratum (Fig. 5). Clades of non-Angelica species were generally consistent with those inferred by previous studies [1, 6, 40, 42, 43].
Fig. 5.
Phylogenetic trees derived from analyses of chloroplast genomes from 33 Angelica and 31 other representative Apioideae species. a Majority-rule consensus tree derived from maximum likelihood (ML) analysis. Numbers at each node are bootstrap values calculated from 2000 replicates. b Majority-rule consensus tree derived from Bayesian (BI) analysis. Numbers at each node are posterior probability estimates from 2 × 5,000,000 MCMC generations with sampling every 1000 generations. Different colors are used to indicate different types of chloroplast genomes based on characteristics of the LSC, SSC, and IR boundary regions
Discussion
Expansion and contraction of the IR region in Angelica Plastomes
In this study, we sequenced 28 chloroplast genomes of Angelica using the Illumina HiSeq-4000 platform and performed comparative analyses of these genomes with five other published chloroplast genomes of the same genus available from GenBank. The chloroplast genomes of Angelica had a typical quadripartite structure of higher plants, were conserved in gene order and content and consisted of 113 to 114 different genes. The cp genomes among Angelica species were similar in GC content, but the GC contents in LSC and SSC regions were significantly lower than those in the IR region because of the inclusion of eight rRNA genes with high GC contents in the IR region. The IR region is considered the most conserved region of the chloroplast genome [44].
The primary causes of differences in the lengths of chloroplast genomes are considered to be the expansion and contraction in IR, LSC, and SSC regions, which are relative common during evolution [45].
The lengths of cp genomes varied between 140,670 base pairs (A. sinensis) to 163,618 base pairs (A. tsinlingensis). Shrinkage, expansion, or loss of the IR region has been proposed to be one of the main reasons explaining the change in the size of cp genomes [46]. Large-scale expansion and contraction of the IR region were reported in Apiaceae; indeed the frequency and large size of JLB shifts documented in Apioideae cp genomes are unprecedented among the angiosperms [47]. In our study, extensive expansion and contraction of the IR region were detected in the Angelica species, with five types of changes in the IR and boundary between IR and SSC or LSC of the chloroplast genomes discovered (Fig. 3). Most Angelica species (25/33) had Type II cp genomes with the junction site of IR/SSC located in the ycf1 gene and the junction site of IR/LSC located between the genes trnL and trnH and were significantly clustered in the Angelica s.s. group, with the exception of A. omeiensis, which was grouped in the Sinodielsia clade. The expansion of the IR region results in the inclusion of extra genes in this region; for example, expansion of this region in the littoral species A. morii resulted in the inclusion of a duplicated copy of the ycf2 gene (Type I). The largest expansion of the IR region was observed in the cp genome of A. tsinlingensis (34,706 bp) and ended with the petB gene in the IR region. A significant contraction of the IR region was observed in A. sinensis (10,704 bp). The patterns of variation observed were generally consistent with the groups of Angelica species recovered in the phylogenetic analyses, reflecting the high diversification of species and cp genomes in this controversial genus (Fig. 5).
Phylogenetics of the genus Angelica
With the use of the whole cp genome sequence from 33 Angelica species and another 31 representative species of Apioideae, a highly consistent topology was recovered by ML and Bayesian analyses (Fig. 5). The allocation of the main clades of Apioideae (e.g., Oenantheae, Scandiceae, Apieae, Tordylieae, and Selineae) were consistent with those inferred by previous studies [18, 40, 42, 43]. Species of Angelica were not grouped in a monophyletic clade but distributed in four clades, with most Angelica species grouped in a well-supported clade (the Angelica group), supporting the phylogenetic topologies of previous studies [1, 41]. This group also consisted of species from the genus Glehnia (Gle. littoralis), Ostericum (Ost. grosseserratum) and Pimpinella (Pim. rhomboidei var. tenuiloba).
The Angelica s.s. clade in Liao et al. (2013) primarily contained East Asian Angelica species and species from Ostericum (Ost. Koreanum, Ost. huangdongensis) and Czemaevia (Cze. Laevigata var. larvigata) but excluded species from Glehnia (Gle. littoralis var. littoralis, Gle. var. leiocarpa). Based on whole cp genome data, Gle. littoralis was grouped within the Angelica s.s. clade with relatively high support (BS = 86, PP = 1). A. anomala was previously grouped with Ostericum grosseserratum and species from Peucedanum within the Acronema clade [6, 43] based on nrITS sequences but was then placed into the Angelica s.s clade when both nrITS, nrETS, cpDNA and morphological characters were used [1]. In the study, A. anomala was grouped with A. cartilaginomarginata, A. cartilaginomarginata var. foliata, and A. polymorpha in a clade within the Angelica s.s. clade. The allocation of A. morii and A. tsinglingensis within the Angelica group but outside the Angelica s.s. clade was also consistent with previous studies and was supported by studies of morphological characters (e.g., dorsal ribs and triple vittae in each furrow) [1]. Because of its unusual fruit characteristics, the taxonomic position of A. acutiloba has been controversial for many years. A. acutiloba was previously placed within the Angelica s.s. clade based on nrDNA ITS sequences [6] and was then placed outside the Angelica s.s. clade with data from nrDNA, cpDNA, and morphological characters [1]. In our study, A. acutiloba was also isolated from the Angelica s.s. clade and occupies an early diverging branch of the Angelica group. A. ameiensis was previously grouped with A. apaensis and A. nitida in a clade within the Angelica s.s. clade based on nrDNA ITS and cpDNA sequences [1, 6] but was grouped with A. sinensis and Ligusticum tenuissimum in the Sinodielsia clade with high support (BS = 100, PP = 1) by whole cp genome data from this study.
This study reports the results of a comparative analysis of 33 Angelica cp genomes and found extensive expansion and contraction in the IR region among species of Angelica. The changes in cp genomes can be classified into five types that are consistent with the general phylogenetic placement of these Angelica species. The relationships of Angelica species examined here are were clear, and the lineages within the Angelica group were classified with a better resolution compared with previous studies. We suggest that the results of our study facilitate our understanding of the evolutionary history of Angelica species; nevertheless, more extensive cp genome sampling (e.g., A. roseana, A. ampla, A. hirsutiflora, and A. oncosepala) may be necessary to further characterize the relationships between Angelica species. These findings also provide an informative and valuable genetic source for Angelica germplasm resources to aid species identification and future taxonomic reconstructions of Angelica.
Conclusions
Our analyses not only reveal extensive expansion and contraction of the IR region among cp genomes of Angelica species, but also show the power of plastome for resolving relationships in currently less-resolved and controversial groups. The variation patterns of IR region can be classified into five different types and are generally consistent with the groups of Angelica species in phylogenetic analyses. The relationships of Angelica species investigated here are mainly clearly classified and the lineages within the Angelica group are classified with a better resolution than previous studies, which we believe will facilitate the understanding of the evolutionary history of Angelica species, yet more extensive cp genome sampling may be necessary to further illustrate the relationships of species in Angelica.
Methods
Taxon sampling
We sampled 24 species (including two varieties and two cultivars) of Angelica located in 14 provinces, representing approximately 85% species and covering most of the distribution of Angelica in China (http://freps.eflora.cn/). Details of sampling information of the 28 samples collected in this study were shown in Supporting information Additional file 3: Table S3. All the samples were identified by Nian-He Wang (Institute of Botany, Jiangsu Province and Chinese Academy of Sciences) based on the morphological characters and the species were preserved in the herbarium of National Resource Center for Chinese Materia Medica, China Academy of Chinese Medical Sciences. Permission was not necessary for collecting these samples, which have not been included in the list of national key protected plants. The fresh leaves from each accession were immediately dried with silica gel for further DNA extraction.
Plant material and DNA extraction
The Plant Genomic DNA Kit (DP305) from Tiangen Biotech (Beijing) Co., Ltd., China was used to extract total genomic DNA from each sample. Both a NanoDrop spectrophotometer (ND-1000; Thermo Fisher Scientific, USA) and a Qubit 2.0 fluorometer (Invitrogen, Life Technologies) were used to assess the quality and quantity of DNA.
Illumina sequencing
A Covaris S2 was used to fragment total genomic DNA (30–150 ng) to a mean fragment size of 550 bp. The TruSeq DNA Nano Library Prep kit (Illumina) was used for DNA libraries preparation per the manufacturer’s instructions. Libraries were quantified using a KAPA Illumina Library Quantification Kit (KAPA Biosystems) by quantitative polymerase chain reaction, and the pooled libraries were sequenced (2 × 150 bp) using the Illumina HiSeq 4000 platform (Illumina, San Diego, CA).
Chloroplast genome assembly and annotation
The raw sequencing reads were qualitatively assessed and assembled using the GetOrganelle version 1.6.4 [48] with default settings. Manual revision was performed to confirm the ambiguous nucleotides or gaps and the four junction regions between the IRs and SSC/LSC in the chloroplast genome sequences. The annotation of chloroplast genomes was performed using the GeSeq version 1.79 [49]. The annotation results were further manually checked using Geneious version 8.0.2 (http://www.Geneious.com) to avoid potential annotation errors. The gene maps of chloroplast genomes were plotted with OGDRAW version 1.3.1 [50].
Simple sequence repeat analysis
The simple sequence repeat (SSRs or microsatellites) loci in the cp genomes were searched using the Perl script MISA version 2.0 [51]. The minimum numbers (thresholds) of the SSRs for mono-, di-, tri-, tetra-m penta-, and hexa-nucleotides, were 10, 5, 4, 3, 3, and 3 respectively. Manual verifications of the repeats were performed with abundant results removed.
Comparative analysis of cp genomes
The statistics of genome size, GC content, LSC/SSC/IR size and number of genes were summarized using in-house python scripts. Comparative analysis of cp genome structure and gene content was performed using Mauve version 2015-02-13 [52] to locate potential rearrangements (e.g., inversion) and changes in gene order using the cp genome of Tiedemannia filiformis subsp. Greenmannii (GenBank accession: HM596071). The junction sites of LSC-IRa/b and SSC-IRa/b were checked by visualization using IRscope [53].
Phylogenetic analysis
Phylogenetic analysis was conducted using all 33 Angelica cp genomes together with 31 species from major lineages of the subfamily Apoideae (Additional file 4: Table S4). The best-fit substitution models were selected using the PartitionFinder 2 version 2.1.1 [54] for Maximum likelihood (ML) and Bayesian inference (BI). The ML analyses were performed using RAxML-NG version 0.9.0 [55] with the general time-reversible (GTR) + G model, and node support was assessed with 2000 bootstrap replicates. The BI analyses were performed with MrBayes version 3.2.7a [56]. Two chains of 5,000,000 generations were performed for the Markov chain Monte Carlo (MCMC) analysis with trees sampled every 1000 generations. The first 25% of the sampled trees were discarded as burn-in and the remaining trees were used to build a 50% majority-rule consensus tree. Stationarity was considered achieved when the average standard deviation of split frequencies remained below 0.001.
Supplementary Information
Additional file 1: Table S1. Gene content in 64 Apioideae chloroplast genomes.
Additional file 2: Table S2. Number of SSR loci detected in 64 Apoideae samples.
Additional file 3: Table S3. Sampling information of 24 species (including two varietas and two cultivars) of Angelica.
Additional file 4: Table S4. Comparison of the chloroplast genome features of 31 Apoideae species.
Acknowledgements
Not applicable.
Abbreviations
- BI
Bayesian Inference
- GTR
General time reversible
- IR
Inverted repeat
- ITS
Internal transcribed spacer of ribosomal DNA
- LSC
Large single copy
- ML
Maximum Likelihood
- rRNA
Ribosomal RNA
- SSC
Small single copy
- tRNA
Transfer RNA
Authors’ contributions
LH, QY and WD conceived and designed the study. WD performed de novo assembly, genome annotation, phylogenetic and other analyses. QY, XW, ML and YG collected the leaf materials. ML, JS, XW and YW performed the experiments. QY, WD, and JS drafted the manuscript. The authors read and approved the final manuscript.
Funding
This study was funded by the National Key Research and Development Program of China (2017YFC1703700; 2017YFC1703704), the National Natural Science Foundation of China (NSFC: 81891010 and 81891014), Key Project at Central Government Level: The Ability Establishment of Sustainable Use for Valuable Chinese Medicine Resources (2060302) to Q-JY and L-QH. The funding agencies had no role in the design of the experiment, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All sequences used in this study are available from the National Center for Biotechnology Information (NCBI) (see Additional file 3: Table S3 and Additional file 4: Table S4). All raw reads are available in the short sequence archive under accession no. PRJNA684804.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mengli Wang, Xin Wang and Jiahui Sun contributed equally to this work.
Contributor Information
Wenpan Dong, Email: wpdong@bjfu.edu.cn.
Qingjun Yuan, Email: yuanqingjun@icmm.ac.cn.
Luqi Huang, Email: huangluqi01@126.com.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-020-02801-w.
References
- 1.Liao C, Downie SR, Li Q, Yu Y, He X, Zhou B. New insights into the phylogeny of Angelica and its allies (Apiaceae) with emphasis on east Asian species, inferred from nrDNA, cpDNA, and morphological evidence. Syst Bot. 2013;38(1):266–281. [Google Scholar]
- 2.Vasil'eva MG, Pimenov MG. Karyotaxonomical analysis in the genusAngelica (Umbelliferae) Plant Syst Evol. 1991;177(3):117–138. [Google Scholar]
- 3.Meng-lan S, Fa-ting P, Zehui P, Watson MF, Cannon JFM, Holmes-Smith I, et al. In: Apiaceae. Pp. 1–205 in Flora of China, vol. 14. Wu ZY, Raven PH, et al., editors. St. Louis: Missouri Botanical Garden Press and Beijing: Science Press; 2005. [Google Scholar]
- 4.Wen J. Evolution of eastern Asian and eastern north American Disjunct distributions in flowering plants. Annu Rev Ecol Syst. 1999;30(1):421–455. [Google Scholar]
- 5.Liao CY, Downie SR, Yu Y, He XJ. Historical biogeography of the Angelica group (Apiaceae tribe Selineae) inferred from analyses of nrDNA and cpDNA sequences. J Syst Evol. 2012;50(3):206–217. [Google Scholar]
- 6.Feng T, Downie S, Yu Y, Zhang X, Chen W, He X-J, et al. Molecular systematics of Angelica and allied genera (Apiaceae) from the Hengduan Mountains of China based on nrDNA ITS sequences: phylogenetic affinities and biogeographic implications. J Plant Res. 2009;122:403–414. doi: 10.1007/s10265-009-0238-4. [DOI] [PubMed] [Google Scholar]
- 7.Yuan QJ, Zhang B, Jiang D, Zhang WJ, Lin TY, Wang NH, et al. Identification of species and materia medica within Angelica L.(Umbelliferae) based on phylogeny inferred from DNA barcodes. Mol Ecol Resour. 2015;15(2):358–71. [DOI] [PMC free article] [PubMed]
- 8.Shan RH. In: Umbelliferae. Pp. 13–62 in Flora Reipublicae Popularis Sinicae, vol. 55(3) Shan RH, Sheh ML, editors. Beijing: Acade- mia Sinica; 1992. [Google Scholar]
- 9.Ze-Hui P, Xin-Tian L, Meng-Lan S, Lang-Ran X. A study on karyotypes of eight species and geographical distribution of Angelica (Umbelliferae) in Sichuan. Acta Phytotaxonomica Sinica. 1991;29(5):431–438. [Google Scholar]
- 10.Zhang QY, Xing-Jin HE, Zhang YC, Peng L, Ning WU. Study on karyotypes of six species in Angelica from Sichuan,China. Acta Bot Yunnanica. 2005;27(5):539-44.
- 11.Chen WW, He XJ, Zhang XM, Pu JX. Pollen morphology of the genus Angelica from Southwest China and its systematic evolution analysis. Acta Botan Boreali-Occiden Sin. 2007; 27(3):1364-72.
- 12.Lan SM, Su P, Pan ZH. The comparative study of pollen morphology of Angelica L. between East Asia and North America. J Plant Res Environ. 1997;6(1):41-47.
- 13.Meng D. Pollen morphology of the genus Peucedanum from Sichuan and its systematic significance. Acta Bot Boreal Occident Sin. 2004;24(12):2341-45.
- 14.Zhang QY, Xing Jin HE, Zhang YC, Luo P, Ning WU. Anatomical studies on fruits and petioles of 8 species of Angelica L.from Sichuan Province. J Wuhan Botanical Res. 2005;23(6):549-54.
- 15.Chen X, Changqi Y. The generic position of Zihua Qianhu and its comparative taxonomic studies with Korean Danggui. J Nanjing Univ. 1987;23(1):23-31.
- 16.Shneyer VS, Kutyavina NG, Pimenov MG. Systematic relationships within and between Peucedanum and Angelica (Umbelliferae–Peucedaneae) inferred from immunological studies of seed proteins. Plant Syst Evol. 2003;236(3–4):175–94.
- 17.Katz-Downie DS, Valiejo-Roman CM, Terentieva EI, Troitsky AV, Pimenov MG, Lee B, et al. Towards a molecular phylogeny of Apiaceae subfamily Apioideae: additional information from nuclear ribosomal DNA ITS sequences. Plant Syst Evol. 1999;216(3–4):167–195. [Google Scholar]
- 18.Downie S, Katz-Downie D. A molecular phylogeny of apiaceae subfamily apioideae: evidence from nuclear ribosomal DNA internal transcribed spacer sequences. Am J Bot. 1996;83(2):234–51. [PubMed]
- 19.Spalik K, Reduron JP, Downie SR. The phylogenetic position of Peucedanum sensu lato and allied genera and their placement in tribe Selineae (Apiaceae, subfamily Apioideae) Plant Syst Evol. 2004;243(3):189–210. [Google Scholar]
- 20.Xue HJ. Taxonomic study of Angelica from East Asia: Inferences from ITS sequences of nuclear ribosomal DNA. Acta Phytotaxonomica Sinica. 2007;45(6):783–795. [Google Scholar]
- 21.Dong W, Liu J, Yu J, Wang L, Zhou S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS One. 2012;7(4):e35071. doi: 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gurusamy R, Park S. The complete chloroplast genome sequence of ampelopsis: Gene Organization, comparative analysis, and phylogenetic relationships to other angiosperms. Front Plant Sci. 2016;7:341. [DOI] [PMC free article] [PubMed]
- 23.Wang H-X, Liu H, Moore MJ, Landrein S, Liu B, Zhu Z-X, et al. Plastid phylogenomic insights into the evolution of the Caprifoliaceae s.l. (Dipsacales) Mol Phylogenet Evol. 2020;142:106641. doi: 10.1016/j.ympev.2019.106641. [DOI] [PubMed] [Google Scholar]
- 24.Xu C, Dong W, Li W, Lu Y, Xie X, Jin X, et al. Comparative analysis of six Lagerstroemia complete chloroplast genomes. Front Plant Sci. 2017;8:15. [DOI] [PMC free article] [PubMed]
- 25.Dong W, Chao X, Tao C, Lin K, Zhou S. Sequencing angiosperm plastid genomes made easy: a complete set of universal primers and a case study on the phylogeny of Saxifragales. Genome Biol Evol. 2013;5:5. doi: 10.1093/gbe/evt063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong W, Xu C, Li D, Jin X, Li R, Lu Q, et al. Comparative analysis of the complete chloroplast genome sequences in psammophytic Haloxylon species (Amaranthaceae) PeerJ. 2016;4:e2699. doi: 10.7717/peerj.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruhlman T, Lee S-B, Jansen RK, Hostetler JB, Tallon LJ, Town CD, et al. Complete plastid genome sequence of Daucus carota: implications for biotechnology and phylogeny of angiosperms. BMC Genomics. 2006;7(1):222. [DOI] [PMC free article] [PubMed]
- 28.Huang DI, Hefer CA, Kolosova N, Douglas CJ, Cronk QCB. Whole plastome sequencing reveals deep plastid divergence and cytonuclear discordance between closely related balsam poplars, Populus balsamifera and P. trichocarpa (Salicaceae). New Phytol. 2014; 204(3):693-703. [DOI] [PubMed]
- 29.Perdereau A, Klaas M, Barth S, Hodkinson TR. Plastid genome sequencing reveals biogeographical structure and extensive population genetic variation in wild populations of Phalaris arundinacea L. In North-Western Europe. GCB Bioenergy. 2017;9(1):46–56.
- 30.Spooner DM, Ruess H, Iorizzo M, Senalik D, Simon P. Entire plastid phylogeny of the carrot genus (Daucus, Apiaceae): concordance with nuclear data and mitochondrial and nuclear DNA insertions to the plastid. Am J Bot. 2017;104(2):296–312. [DOI] [PubMed]
- 31.Wang H, Liu H, Moore M, Landrein S, Liu B, Zhu Z, et al. Plastid phylogenomic insights into the evolution of the Caprifoliaceae s.l.(Dipsacales) Mol Phylogenet Evol. 2019;142:106641. doi: 10.1016/j.ympev.2019.106641. [DOI] [PubMed] [Google Scholar]
- 32.Yao G, Jin J, Li H-T, Yang J-B, Mandala V, Croley M, et al. Plastid phylogenomic insights into the evolution of Caryophyllales. Mol Phylogenet Evol. 2019;134:74-86. [DOI] [PubMed]
- 33.Liu D, Tu X-D, Zhao Z, Zeng M-Y, Zhang S, Ma L, et al. Plastid phylogenomic data yield new and robust insights into the phylogeny of Cleisostoma–Gastrochilus clades (Orchidaceae, Aeridinae) Mol Phylogenet Evol. 2020;145:106729. doi: 10.1016/j.ympev.2019.106729. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, Wei R, Xiang Q-P. Plastome-based phylogenomics resolves the placement of the sanguinolenta group in the spikemoss of lycophyte (Selaginellaceae). Mol Phylogenet Evol. 2020;106788. [DOI] [PubMed]
- 35.Zhang R, Wang Y-H, Jin J, Moore M, Zhang S-D, Chen S-Y, et al. Exploration of plastid Phylogenomic conflict yields new insights into the deep relationships of Leguminosae. Syst Biol. 2020;69(4):613-22. [DOI] [PMC free article] [PubMed]
- 36.Kim KJ, Lee HL. Widespread occurrence of small inversions in the chloroplast genomes of land plants. Mol Cell. 2005;19(1):104–113. [PubMed] [Google Scholar]
- 37.Sajjad A, Khan AL, Khan AR, Muhammad W, Kang SM, Khan MA, et al. Complete chloroplast genome of Nicotiana otophora and its comparison with related species. Front Plant Sci. 2016;7:843. [DOI] [PMC free article] [PubMed]
- 38.Yang Y, Zhou T, Duan D, Yang J, Feng L, Zhao G. Comparative analysis of the complete chloroplast genomes of five Quercus species. Front Plant Sci. 2016;07:959. [DOI] [PMC free article] [PubMed]
- 39.Downie SR, Katz-Downie DS, Watson MF. A phylogeny of the flowering plant family Apiaceae based on chloroplast DNA rpl16 and rpoC1 intron sequences: towards a suprageneric classification of subfamily Apioideae. Am J Bot. 2000;87(2):273–292. [PubMed] [Google Scholar]
- 40.Zhou J, Gong X, Downie SR, Peng H. Towards a more robust molecular phylogeny of Chinese Apiaceae subfamily Apioideae: additional evidence from nrDNA ITS and cpDNA intron (rpl16 and rps16) sequences. Mol Phylogenet Evol. 2009;53(1):56–68. doi: 10.1016/j.ympev.2009.05.029. [DOI] [PubMed] [Google Scholar]
- 41.Downie SR, Spalik K, Katz-Downie DS, Reduron J-P. Major clades within Apiaceae subfamily Apioideae as inferred by phylogenetic analysis of nrDNA ITS sequences. Plant Diversity Evol. 2010;128(1–2):111–136. [Google Scholar]
- 42.Gong X, Liu ZW, Downie SR, Peng H, Zhou J. A molecular phylogeny of Chinese Apiaceae subfamily Apioideae inferred from nuclear ribosomal DNA internal transcribed spacer sequences. Taxon. 2008;57(2):402-416.
- 43.Downie SR, Spalik K, Katz-Downie DS, Reduron JP. Major clades within Apiaceae subfamily Apioideae as inferred by phylogenetic analysis of nrDNA ITS sequences. Plant Diversity Evol. 2010;128(1):111–136. [Google Scholar]
- 44.Li R, Ma PF, Wen J, Yi TS. Complete sequencing of five Araliaceae chloroplast genomes and the phylogenetic implications. PLoS One. 2013;8(10):e78568. [DOI] [PMC free article] [PubMed]
- 45.Xue S, Shi T, Luo W, Ni X, Iqbal S, Ni Z, et al. Comparative analysis of the complete chloroplast genome among Prunus mume, P. armeniaca, and P. salicina. Horticulture Res. 2019;6(1):89. doi: 10.1038/s41438-019-0171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bock R, Knoop V, editors. Genomics of chloroplasts and mitochondria, Adv. Photosyn Resp, vol. 35. Dordrecht: Springer Netherlands; Dordrecht; 2012. p. 1–458.
- 47.Plunkett GM, Downie SR. Expansion and contraction of the chloroplast inverted repeat in Apiaceae subfamily Apioideae. Syst Bot. 2000;25(4):648–667. [Google Scholar]
- 48.Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, et al. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biology. 2020;21(1):241. [DOI] [PMC free article] [PubMed]
- 49.Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones E, Fischer A, Bock R, et al. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45(1):W6–W11. [DOI] [PMC free article] [PubMed]
- 50.Greiner S, Lehwark P, Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47(W1):W59–W64. doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thiel T, Michalek W, Varshney R, Graner A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.) Theoretical Appl Genet. 2003;106(3):411–422. doi: 10.1007/s00122-002-1031-0. [DOI] [PubMed] [Google Scholar]
- 52.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14(7):1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amiryousefi A, Hyvönen J, Poczai P. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34(17):3030–3031. doi: 10.1093/bioinformatics/bty220. [DOI] [PubMed] [Google Scholar]
- 54.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 2016;34(3):772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 55.Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35(21):4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Gene content in 64 Apioideae chloroplast genomes.
Additional file 2: Table S2. Number of SSR loci detected in 64 Apoideae samples.
Additional file 3: Table S3. Sampling information of 24 species (including two varietas and two cultivars) of Angelica.
Additional file 4: Table S4. Comparison of the chloroplast genome features of 31 Apoideae species.
Data Availability Statement
All sequences used in this study are available from the National Center for Biotechnology Information (NCBI) (see Additional file 3: Table S3 and Additional file 4: Table S4). All raw reads are available in the short sequence archive under accession no. PRJNA684804.