Abstract
Eukaryotic membranes can be partitioned into lipid-driven membrane microdomains called lipid rafts, which function to sort lipids and proteins in the plane of the membrane. As protein selectivity underlies all functions of lipid rafts, there has been significant interest in understanding the structural and molecular determinants of raft affinity. Such determinants have been described for lipids and single-spanning transmembrane proteins; however, how multi-pass transmembrane proteins (TMPs) partition between ordered and disordered phases has not been widely explored. Here we used cell-derived Giant Plasma Membrane Vesicles (GPMVs) to systematically measure multi-pass TMP partitioning to ordered membrane domains. Across a set of 24 structurally and functionally diverse multi-pass TMPs, the large majority (92%) had minimal raft affinity. The only exceptions were two myelin-associated four pass TMPs, Myelin and Lymphocyte protein (MAL) and ProteoLipid Protein (PLP). We characterized the potential mechanisms for their exceptional raft affinity and observed that PLP requires cholesterol and sphingolipids for optimal association with ordered membrane domains and that PLP and MAL appear to compete for cholesterol-mediated raft affinity. These observations suggest broad conclusions about the composition of ordered membrane domains in cells and point to previously unrecognized drivers of raft affinity for multi-pass transmembrane proteins.
Keywords: lipid raft, microdomain, plasma membrane, myelin, cholesterol, sphingolipid, MAL, PLP
INTRODUCTION
The PMs of eukaryotic cells are believed to be comprised of a mosaic of compositionally and structurally distinct lateral nanodomains 1. One of the most prominent, but least well understood, examples of such domains are lipid rafts 2, 3. Lipid rafts were originally defined operationally, for example by being resistant to solubilization in non-ionic detergents (i.e. detergent resistant membranes, DRMs 4, 5), or rich in glycolipids, sphingolipids 6, or cholesterol 7. However, the general principle underlying all of these properties is that lipid interactions can enforce lateral membrane organization. Specifically, preferred interactions between sterols, saturated lipids, and sphingolipids form tightly packed (i.e. liquid ordered 8) membrane regions that are capable of selectively sorting lipids and proteins to facilitate cell functions 9. Despite this simple definition and much supporting evidence 10–17, there remains significant controversy about the existence and functionality of raft domains in living cells 3. In large part, the controversies persist because raft domains have not been directly, microscopically observed in living cell PMs, although liquid lipid-driven domains have been imaged directly in yeast vacuoles 17, 18. PM nanodomains may be difficult to image due to the limits on spatial and temporal resolution of currently available technologies. However, other possibilities must be considered, including the dearth of validated, consistent probes for raft domains and indeed even the non-existence of rafts in some, most, or all PMs.
A key piece of supporting evidence for the raft hypothesis is the direct observation of phase separation in isolated plasma membrane vesicles known as Giant Plasma Membrane Vesicles (GPMVs) 19, 20. These vesicles separate into coexisting ordered and disordered lipid phases, like biomimetic model membranes 21, 22, but retain the compositional complexity and high protein content of cell membranes. It remains unresolved why this phase separation is easily observed in isolated PMs, but not in vivo. One possibility, supported by experimental 23–25 and theoretical 26–28 studies is that the cytoskeleton and transmembrane proteins prevent macroscopic phase separation by pinning/scaffolding membrane components. Another possibility is that macroscopic phase separation is manifested in nanodomains 29, due to critical fluctuations 30, 31 or other mechanisms 32, 33. In any case, phase separation in GPMVs provides a practical and tractable model for investigation of raft domains in biological membranes.
The major purpose of membrane domains is to selectively recruit specific proteins. Thus, an essential question is how and why certain proteins are sorted to these domains. For many peripheral proteins, the properties of their lipid-anchor determines their raft residence 34, with saturated lipid or sterol anchors preferring ordered membrane regions 35. For transmembrane proteins, a series of studies in GPMVs has revealed the structural determinants of raft affinity, which largely rely on structural features of the TM α-helix and its interaction with the surrounding bulk membrane environment 36, 37. However, this model is not easily extensible to multi-pass proteins. Firstly, the model was constructed entirely using single-pass proteins, making extrapolation inherently questionable. Moreover, it is difficult to estimate the precise structure of the TMD bundle for multi-pass proteins because it is usually not known how multiple transmembrane helices (TMHs) pack together and which residues face the lipid environment.
Here, we used GPMVs to test the ordered phase affinity of a panel of 24 different multi-pass proteins, spanning from two to twelve TMHs. Consistent with predictions based on single TMH proteins 37, almost all multi-pass proteins were excluded from more ordered raft domains. However, we report the notable exceptions of two 4-TM proteins of similar topologies, MAL (Myelin And Lymphocyte; otherwise known VIP17, Vesicle Integral Protein 17 kDa)38 and PLP (ProteoLipid Protein)39 which have a high affinity for ordered raft phases. Because these proteins do not have the typical features associated with raft affinity, we investigated the origins of their ordered phase preference. Our observations suggest that lipid-protein binding is involved in the raft affinity of these multi-pass TMPs, as depletion of cholesterol and sphingolipids reduced their raft affinity while having no effect on single-pass TMP partitioning. These observations reveal novel determinants of raft affinity for multi-pass TMPs and suggest that specific lipids play essential roles in recruiting specific proteins to raft domains.
MATERIALS AND METHODS
Materials.
The following materials were purchased for this study: Methyl-β-cyclodextrin (MβCD, Sigma); Fast DiO (F-Dio), Fast DiI (F-DiI) and DiD (Life Technologies, Carlsbad, CA); myriocin (Cayman Chemical).
DNA constructs.
All single pass protein constructs were based on the trLAT (LAT) backbone previously described 35–37. The amino acid sequence of WT trLAT is NH2-MEEAILVPCVLGLLLLPILAMLMALCVHCHRLP followed by a short linker (GSGS) and monomeric RFP (mRFP). TMD mutants (LIME, CD4, PAG, LAX, LDLR) were generated by synthesizing the gene of interest (Genscript) and subsequent cloning of the mutant sequence into the LAT construct. Several multi-pass protein constructs were purchased from Addgene: mCherry-CD36-C10 (CD36, 55011), mCherry-Occludin-N-10 (Occludin, 55112), mCherry-CD9–10 (CD9, 55013), mCherry-CD81–10 (CD81, 55012), YFP-CD82 (CD82, 1819), pFCK(1.3)GFP-SynaptophysW (Synaptophysin, 27235), pEGFP-N-Drd1 (DRD1, 104358), Kv7.1/pcDNA3.1 (KCNQ1, 111452), pcDNA3.1(+)mGAT1–0-GFP (GAT1, 41662). Remainder of constructs were kindly gifted: hMOGalpha, hMOGBeta, mMOG, rMOG and Beta2AR (Marcus Reindl, Medical University of Innsbruck); cMAL (Kai Simons, MPI-CBG); hMAL, BENE and MAL2 (Miguel A. Alonso, Universidad Autonoma de Madrid); TRPA1, TRPM8 and TRPV1 (Michael X. Zhu, UTHealth); AC9 (Carmen Dessauer, UTHealth); NKCC1 (Eric Delpire, Vanderbilt University); GPM6a (Michihiro Igarashi, Niigata University). MAL Ct mutants were all based on cVIP17-mRFP and point mutations were performed following the QuickChange II XL Site-directed mutagenesis kit (Agilent).
Cell culture and transfection.
Rat basophilic leukemia (RBL) and HEK-293 (HEK) cells were purchased from ATCC and cultured in medium containing 89% Eagle’s Minimum Essential Medium (EMEM), 10% FCS, and 1% penicillin/streptomycin at 37 °C in humidified 5% CO2. Transfection was done by nucleofection (Amaxa) using the protocols provided with the reagents. 4–6 h after transfection, cells were washed with PBS and then incubated with serum-free medium overnight. To synchronize the cells, 1 h before preparation of GPMV, the cells were given full-serum medium. Cholesterol depletion was accomplished by incubating the cells with 3 mM MβCD for 30 min in MEM before GPMV isolation 40. To inhibit sphingolipid metabolism, the medium was changed six hours after transfection to include 25 μM myriocin and to remove the transfection reagent. GPMVs were isolated after 24 hours of myriocin addition 41.
Partitioning measurements in Giant Plasma Membrane Vesicles (GPMVs).
Cell membranes were stained with 5 μg/ml of FAST-DiO, FAST-DiI or DiD (Invitrogen), respectively, green, red or far-red fluorescent lipid dyes that strongly partition to disordered phases 42, 43. Following staining, GPMVs were isolated from transfected RBLs, HEK-293, or HeLa cells as described 19 (cell type had no effect on results). Briefly, GPMV formation was induced by 2 mM N-ethylmaleimide (NEM) in hypotonic buffer containing 100 mM NaCl, 10 mM HEPES, and 2 mM CaCl2, pH 7.4. To quantify protein partitioning, GPMVs were observed on an inverted epifluorescence microscope (Nikon) at 4°C after treatment with 200 μM DCA to stabilize phase separation; this treatment has been previously demonstrated not to affect raft affinity of various proteins 44. The partition coefficient (Kp,raft) for each protein construct was calculated from fluorescence intensity of the construct in the raft and non-raft phase for >10 vesicles/trial (Fig. 1), with multiple independent experiments for each construct. The results are displayed as log (Kp,raft), in analogy with log P values oil-water partitioning, to avoid appearance of compression for values less than 1.
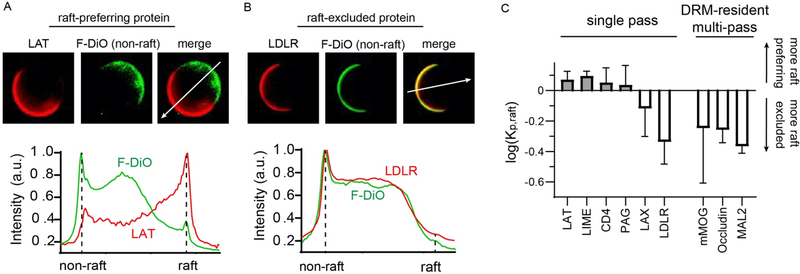
Figure 1 – GPMVs as a biomimetic model of lipid raft affinity for PM proteins.
(A) Example of quantification of raft affinity for a raft-preferring protein. A protein of interest (LAT= TMD of Linker for Activation of T-Cells, red) is expressed in cells, which are stained with a lipid dye with a known phase preference (F-DiO, green, non-raft). The intensity of the protein in the raft versus non-raft phase yields raft partition coefficient (Kp, raft). LAT has Kp, raft > 1, and is thus a raft phase preferring protein. (B) Example of quantification of a non-raft preferring protein. Same principle as panel A, but with the TMD of LDLR, which is largely excluded from the raft phase. (C) Comparison of raft affinity for several single pass proteins and three multi-pass TMPs. Single TMD raft affinity is in good agreement with predictions 37, whereas multi-pass proteins that have been previously associated with rafts have minimal raft phase affinity. Average +/− st. dev. for at least 10 vesicles for each protein, representative of three independent trials.
RESULTS
Quantitative characterization of raft affinity in GPMVs
Membrane domains are functionalized by their selective recruitment of membrane proteins and lipids. However, until recently, the structural principles underlying protein recruitment to ordered membrane domains have not been widely explored. Such investigations are now enabled by direct measurements of raft affinity in GPMVs. GPMVs are intact cell-derived PMs that macroscopically separate into coexisting phases of distinct physical properties and compositions 20. One of these phases exhibits many of the features associated with raft domains in cells: it is cholesterol-dependent 45, more tightly packed 43, 46, less diffusive, enriches sterols, sphingolipids, and saturated lipids and excludes unsaturated lipids 47, recruits GPI-anchored and palmitoylated proteins, and excludes many predicted non-raft proteins 35. Protein partitioning to this “raft phase” in GPMVs has thus been widely validated as a robust tool for quantifying ordered phase affinity in cell-derived membranes 48.
Recent investigations into the raft partitioning of single-pass transmembrane proteins have demonstrated that the length and surface area of membrane-spanning α-helices, together with post-translational palmitoylation, comprise a set of independent, predictive determinants of raft affinity 36, 37. The principle of the assay is demonstrated in Figure 1. Briefly, a protein of interest with a fluorescent tag (e.g. RFP) is expressed in a cultured mammalian cell line (here, RBLs), followed by isolation of GPMVs following standard protocols 19 (see Materials and Methods). Phase separation in these GPMVs is visualized by staining with a marker that partitions strongly to one of the two phases, e.g. FAST-DiO in Fig. 1A–B, which has been shown to strongly label the non-raft phase 42. Some proteins enrich in the opposing (raft) phase, as shown for a widely characterized model transmembrane peptide, the TMD from the linker for activation of T-cells (LAT) (Fig. 1A), while others are almost wholly excluded from the raft domains, as for the TMD of the Low Density Lipoprotein Receptor (LDLR) (Fig. 1B). This behavior can be quantified by fluorescence intensity line scans across the two phases, wherein a ratio between the phases defines the raft partition coefficient (Kp, raft).
We confirmed previously published results that single-pass proteins with relatively long and thin TMHs (e.g. LAT, LIME, CD4, and PAG) partition efficiently to the raft phase, while those with shorter or fatter TMHs (LAX and LDLR) are generally excluded (Fig. 1C). Next, we explored the determinants of raft affinity for proteins with multiple membrane spanning domains. Multi-spanning TMPs would inherently have larger lipid-accessible surface areas than single-pass and would thus be predicted to be excluded from raft phases 37. On the other hand, many multi-pass proteins are found in detergent-resistant fractions (i.e. DRMs), which have been previously used as a proxy for isolated lipid rafts 49–52. Several of these DRM-resident multi-pass proteins were excluded from the raft phase in GPMVs (Fig 1C), leading us to undertake a more systematic analysis of raft affinity across several representative classes of multi-pass TMPs in mammalian PMs.
Almost all multi-pass TM proteins have low raft affinity
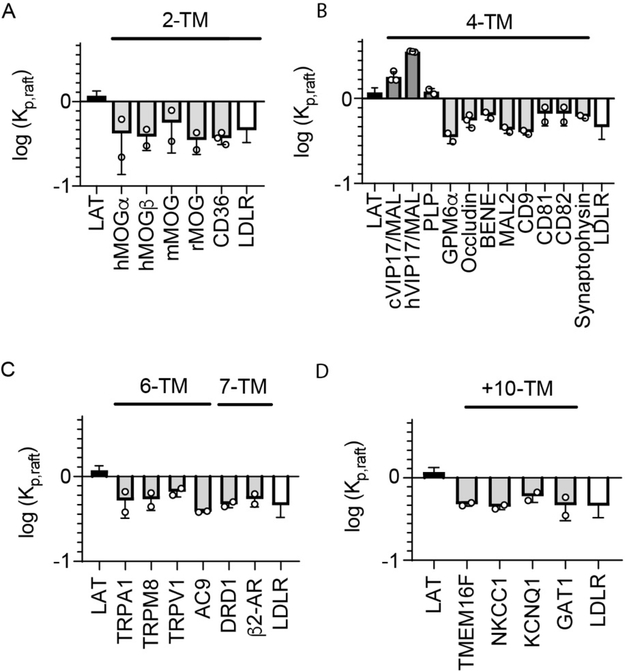
Figure 2 shows the results of raft affinity measurements for proteins with 2, 4, 6, 7, and >10 TMHs, each in comparison to the raft-preferring LAT and non-raft LDLR. The most prominent finding across this set of 24 structurally and functionally diverse multi-pass TMPs is that the vast majority (92%) had minimal raft affinity. This finding is consistent with the predictions from the single-pass model 37, but in many cases inconsistent with previous reports of raft residence for these proteins. Possible reasons for this discrepancy and details of these proteins are expanded upon in Discussion; however, the broad conclusion is clearly that most multi-pass transmembrane proteins are excluded from the raft phase in GPMVs.
Figure 2 – Almost all multi-pass TMPs have low raft affinity in GPMVs.
Shown are the raft affinity of multi-pass TMPs with varying number of TMHs. Raft affinity is shown in log scale, with 0 representing equal partitioning between the phases and positive and negative values representing enrichment in or depletion from the raft phase, respectively. Included in each panel is the value for LAT (a raft-preferring single pass TMD, black) and LDLR (a non-raft TMD, white). (A) 2-TM proteins. Three homologs of MOG from different species (h:human; m:mouse; r:rat) and both isoforms (alpha and beta) of the human protein are excluded from rafts, as is the unrelated CD36. (B) Two 4-TM proteins are highly raft-preferring, namely MAL and PLP. In contrast, several other members of the MARVEL family (BENE, MAL2, occludin, and synaptophysin) and other tetraspanin family proteins (CD9, CD81, CD82), as well as the neuronal membrane glycoprotein (GPM6a) are almost completely excluded. Two analogs of MAL (c:canine: h:human) were tested and both had significantly greater raft affinity than LAT. (C) All tested 6-TM and 7-TM proteins are raft excluded. Several proteins from the TRP family (TRPA1, TRPM8, TRPV1), Adenylyl Cyclase 9 (AC9), and G Protein-Coupled Receptors (DRD1, β2AR) were not significantly different from LDLR. (D) All tested proteins with 10 or more TMDs - PM scramblase TMEM16F and several transporters (NKCC1, KCNQ1 and GAT1) - were raft excluded. Data points represent the means of individual experiments of >10 vesicles each.
Two 4-TM proteins of similar topology associated with myelin have anomalously high raft affinity
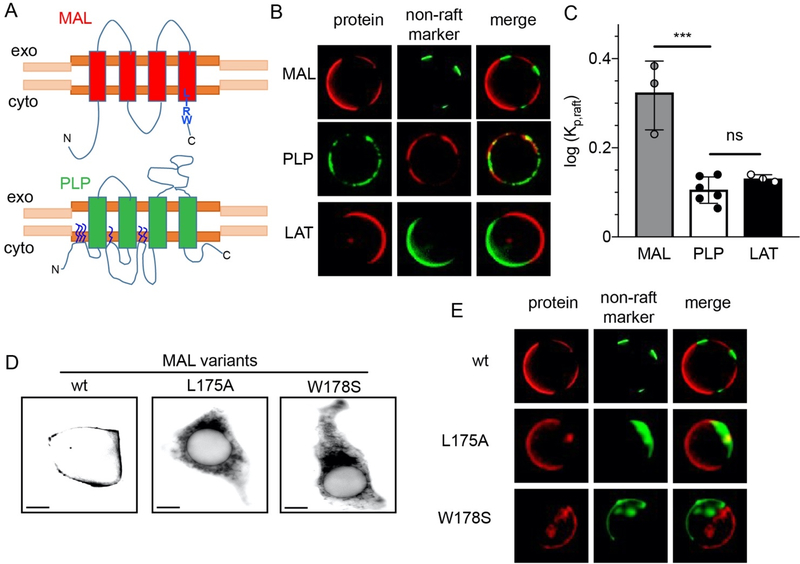
The only exceptions in our dataset to the overall exclusion of multi-pass TM proteins from the raft phase were two 4-TM proteins, MAL and myelin PLP (Fig. 3A–C). As their names imply, both of these proteins are functionally implicated with the structure and physiology of myelin 39, 53. Despite their general topological and functional similarity, a BLAST search revealed no homology between these proteins.
Figure 3 – LIRW peptide is important for MAL ER exit but not raft affinity.
(A) Schematics for MAL (red) with LIRW motif close to the cytosolic C-terminal and PLP (green) with numerous palmitoylation at cytosolic loops. (B) Images of GPMVs containing cMAL-RFP (MAL), PLP-GFP (PLP) and trLAT, with their respective complementary non-raft marker, showing that all three proteins prefer the raft phase. (C) Raft affinity (log Kp, raft) measurements show MAL has higher raft affinity than PLP and LAT, which have similar raft affnity. D) Representative images for HEK-293 cells transfected with cMAL and C-terminal mutants. Wild-type cMAL (wt) accumulates in the PM with some punctate structures while single point mutations of the LIRW motif lead to clear accumulation in the ER. (E) Mutations in LIRW motif do not change lipid raft affinity of the protein. Both mutants, L175A and W178S, showed a retention in the ER, but the minimal amount of protein present in PM still partitioned into the raft phase in GPMVs. Scale bar corresponds to 10 μm. ***p<0.001 by t-test
The strikingly high raft affinity of these two proteins notably diverges with the model for single-pass TMPs, in which lipid-accessible surface area (ASA) and TMD palmitoylation are key drivers of raft affinity. While calculating ASA of a multi-pass TMP cannot be accurately done without a structural model, certainly any 4-TM protein has a larger ASA than any single TMH. Furthermore, several other similar or smaller proteins (e.g. the 2-TM proteins in Fig. 2B) had minimal raft affinity. Additionally, while PLP can be palmitoylated at three sites in the cytoplasmic N-terminus, with these modifications being a major determinant for its distribution to myelin-like membranes 54, MAL is not known to be palmitoylated, and topological modeling revealed no cytoplasmic cysteine residues that would be good candidates for palmitoylation. Thus, despite a relatively large TMD and no palmitoylations, MAL partitions into raft domains in GPMVs with Kp,raft of ~3.5 (Fig. 3C), greater than any previously reported TMPs 36, 37, 55, and similar to the lipid-anchored raft markers like GPI-anchored proteins and cholera toxin 35, 44, 56. Notably, a recent unpublished report also found high raft affinity for another protein with similar topology to MAL and also associated with myelin, Peripheral Myelin Protein 22 (PMP22) 57.
Other MARVEL family members do not partition to raft phase
These surprising observations led us to explore the possible mechanisms for the raft affinity of these 4-TM proteins. MAL is reported to be a member of a protein family named MARVEL (MAL and related proteins for vesicle trafficking and membrane link) 58, 59, which includes occludin, tricellulin, and synaptophysin and has been implicated in mediating membrane apposition 60. We tested the ordered phase affinity of several other MARVEL family members and observed minimal raft preferences for all of them, including two closely MAL-related proteins MAL2 and BENE (also called MAL-like) (Fig. 2B). These results were surprising because of the evolutionary relationship between these proteins and MAL and because MAL2 has been previously implicated in raft-related cellular functions, namely apical transport in polarized cells 52. To ensure consistency of partitioning measurements, we tested a canine homolog of MAL (cMAL) and found that it also partitioned efficiently to raft phases. Thus, it appears that MAL contains unique features that impart very high raft affinity, even compared to closely related proteins.
LIRW peptide is important for ER exit of MAL but not raft affinity
MAL has been previously reported to be a raft-associated protein, with the possible determinants of this behavior sporadically explored. In particular, a short peptide (LIRW, Fig 3A) located at a cytoplasmic interface of the protein with the lipid bilayer 61 has been implicated in MAL recruitment to detergent-resistant membranes, with mutations of this sequence completely abrogating DRM residence. We thus tested whether these mutations also abrogated raft affinity in GPMVs. However, mutations of this C-terminal peptide lead to near absolute ER retention (Fig 3D), consistent with previously reported trafficking disruptions 61, 62. While this ER localization almost completely precluded measurements of these mutants in GPMVs (which are derived from the PM), the few GPMVs with detectable protein signal still showed partitioning preference for the raft phase (Fig 3E). Thus, we conclude that the LIRW motif is important for MAL export from the ER, rather than a bona fide raft partitioning determinant. The absence of the protein from DRMs is likely explained by the trafficking defect.
MAL and PLP compete for raft affinity
The surprising finding that only two multi-pass proteins, MAL and PLP, showed lipid raft affinity and that these possessed some general functional and structural similarities led us to hypothesize that they share characteristics that impart raft affinity. Because both are myelin-related proteins, we tested the raft affinity of other myelin-related proteins, but found that all four isoforms of Myelin Oligodendrocyte Glycoprotein (MOG), a prominent myelin-associated protein 63, were raft-excluded (Fig 2A and B). Similarly, MAL-related proteins MAL2 and BENE were not raft-preferring. Thus, we conclude that myelin-association per se is not a determinant of raft affinity, although the connection to myelin for both raft-preferring proteins found here and another one recently reported 57 is intriguing.
Post-translational lipidations have been widely implicated as raft-sorting determinants 34, but MAL is not known to be lipidated, whereas several of the raft multi-pass proteins in Figure 2 (CD9, CD81, CD82) can be palmitoylated. Thus, palmitoylation is neither necessary nor sufficient for raft affinity of multi-pass proteins, unlike single-pass TMDs.
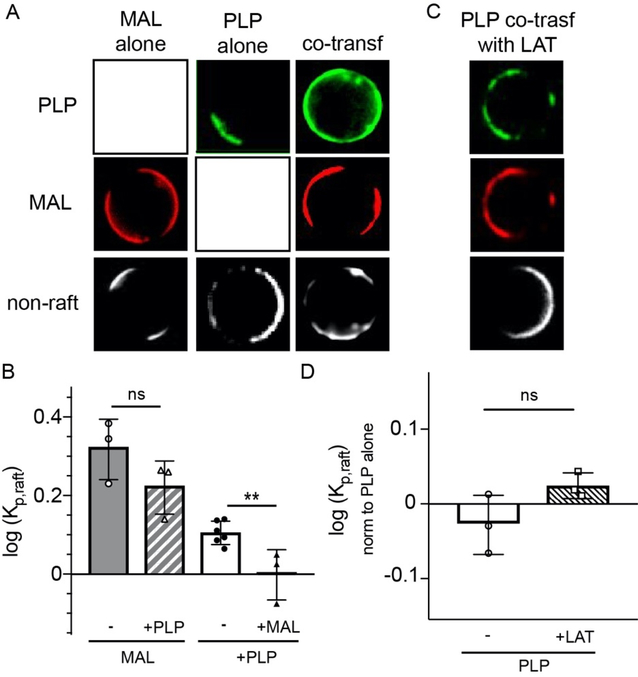
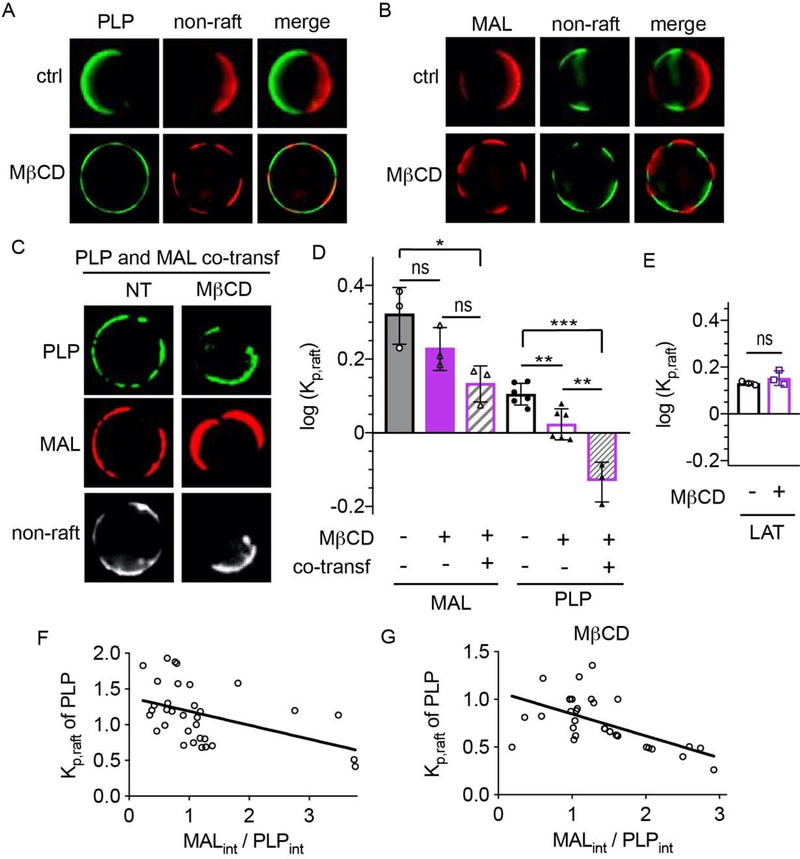
We next hypothesized that both MAL and PLP may acquire raft affinity by binding to a common raft-preferring component (e.g. a protein or lipid with intrinsic ordered phase affinity). To evaluate this possibility, we conducted a ‘competition’ experiment by co-transfecting the two proteins and measuring the effect of co-transfection on raft affinity. The rationale behind this experiment was that if both proteins share a saturable binding partner required for their raft residence, then co-transfection should deplete the availability of this component and reduce raft affinity of MAL and/or PLP. This effect was indeed observed for PLP, whose raft residence was significantly reduced by co-transfection with MAL (Fig. 4A&C). This effect was also dose-dependent, as PLP raft affinity was negatively correlated with the relative abundance of MAL to PLP (quantified as MAL/PLP fluorescence intensity, Fig 5E). MAL raft affinity was also somewhat, though not significantly, reduced by co-transfection. Importantly, PLP raft affinity was not reduced by co-transfection with LAT, which resides in rafts solely due to the physical properties of its TMD, rather than binding to a separate component (Fig. 4C–D).
Figure 4 – MAL displaces PLP from raft phase.
(A) Single transfected PLP and MAL both have preference for raft phase (non-raft labeled with F-DiO, white). Co-transfection reduces PLP raft preference to approximately equal partitioning between phases, but does not significantly affect MAL. (B) PLP partitioning is unaffected by co-transfection with LAT. (C) Raft partitioning of MAL (open symbols) and PLP (closed symbols) as a function of co-transfection (triangles) compared with single transfected (circles). PLP raft affinity is significantly reduced by co-transfection with MAL, (D) but not by the co-transfection with LAT. Partitioning in panel D shown as normalized by PLP alone. Bars represent average +/− s.e.m.; circles represent means of individual experiments (>10 vesicles each). **p<0.01 by t-test.
Figure 5 – MAL and PLP may compete for cholesterol.
(A) Cholesterol depletion does not significantly affect MAL partitioning, which still enriches in raft phase upon cholesterol depletion with MβCD. (B) PLP partitioning is reduced by cholesterol depletion, becoming approximately homogeneously distributed between phases. (C) Cholesterol depletion severely reduces PLP raft affinity when co-expressed with MAL. Under this condition, PLP is excluded from rafts domains and co-enriches with the non-raft marker (F-DiD: white). (D) Quantifications of all conditions. (E) LAT partitioning is not affected by MβCD treatment. Bars represent average +/− s.e.m.; circles represent means of individual experiments (>10 vesicles each). *p<0.1, **p<0.01, ***p<0.001 by t-test. (F) PLP partitioning negatively correlates with the abundance of MAL. (i.e. relative intensity of MAL to PLP). (F) The effect is enhanced by cholesterol depletion with MβCD.
Cholesterol is important for PLP raft affinity
The most prominent components of lipid rafts are sphingolipids and cholesterol 64. Thus, we hypothesized that these lipids may be responsible for recruiting MAL and/or PLP to raft domains. To test this possibility, we assayed whether depletion of cholesterol or sphingolipids had an effect on raft affinity of the multi-pass TMPs. To deplete cholesterol, cells were treated prior to GPMV isolation with methyl-β-cyclodextrin (MβCD), a common method for acute depletion of cholesterol 65. Importantly, phase separation into raft and non-raft domains was still observable after MβCD treatment (Fig. 5A–C), consistent with previous reports and explainable by the fact that only a fraction of PM cholesterol is removed by this treatment 40, 66. MβCD treatment significantly reduced the raft affinity of PLP (Fig. 5A&D), with a smaller and not significant effect on MAL (Fig. 5B&D). As a control, we tested the cholesterol sensitivity of raft partitioning for the single-pass LAT, whose raft affinity was not reduced by MβCD (Fig. 5E). The effect of cholesterol depletion on PLP partitioning was further exacerbated by co-transfection with MAL, with PLP becoming quite depleted from the raft phase under the co-transfected+MβCD condition (Fig. 5C–D). Again, this effect was dependent on MAL expression, with the correlation between PLP partitioning and MAL-to-PLP abundance strongly exacerbated by cholesterol depletion (Fig. 5G). No trend was observed for MAL partitioning as a function of PLP levels (not shown). These observations suggest that MAL competes with PLP for cholesterol binding, which appears to be required for PLP to partition to the raft phase. This hypothesis is consistent with the previously reported interaction of PLP with cholesterol, which has been implicated in PLP transport to myelin in neurons 67. It is important to note that MβCD-mediated cholesterol extraction likely has side-effects and therefore direct binding of cholesterol to these proteins and its effect on raft affinity is a provisional conclusion that will require confirmation by direct experiments and/or computational modeling.
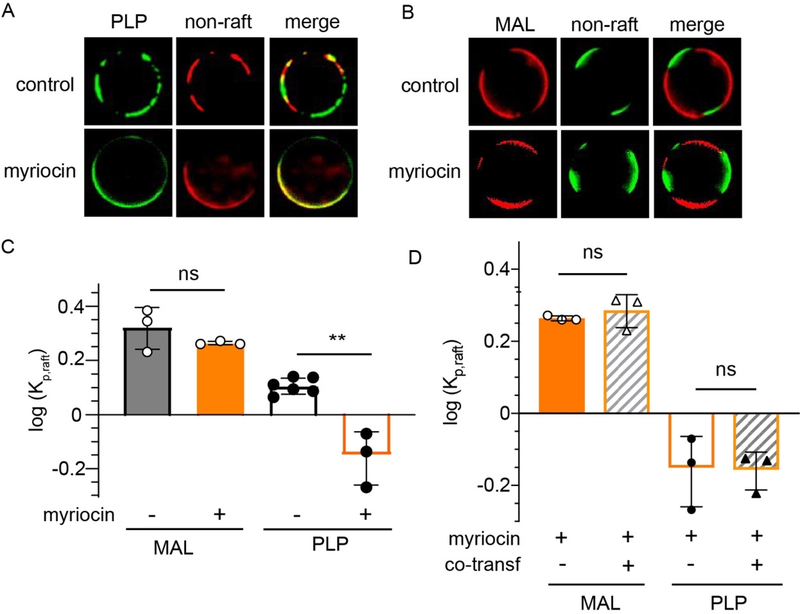
PLP, but not MAL, requires sphingolipids for raft residence
To evaluate whether sphingolipids could similarly promote PLP raft affinity, we depleted sphingolipids in cells using myriocin, a potent inhibitor of serine palmitoyltransferase, which catalyzes the first step in de novo sphingolipid synthesis 68, 69. Myriocin treatment significantly inhibited the raft affinity of PLP (Fig 6A&C), with a subtle and not significant effect on MAL partitioning (Fig 6B–C). Co-transfection had no additive effect in the presence of myriocin (Fig 6D). These observations suggest that sphingolipid interactions are important for PLP raft affinity. This effect is likely mediated by glycosylated sphingolipids (GSLs), as it was previously reported that galactosyl transferase is essential for PLP to associate with DRMs 70.
Figure 6 – Sphingolipids are required for PLP raft enrichment.
(A) MAL raft affinity is not significantly affected by sphingolipid depletion. (B) In contrast, PLP enriched in non-raft phrase upon myriocin treatment. (C) Quantification of myriocin-dependent partitioning. (D) Myriocin effect on PLP is not affected by co-transfection with MAL. Bars represent average +/− s.e.m.; circles represent means of individual experiments (>10 vesicles each). **p<0.01.
In summary, we conclude from these observations that PLP uses both cholesterol and sphingolipids for its recruitment to raft domains. It appears to compete with MAL for the free cholesterol available in the PM, with MAL likely having greater affinity to cholesterol. Finally, the raft affinity of MAL was not strongly dependent on either sphingolipids or cholesterol abundance, revealing a yet-unidentified mechanism of raft affinity.
Discussion
Minimal raft affinity of multi-pass proteins.
The major finding of our study is that the large majority of multi-spanning TMPs are excluded from raft phases in GPMVs. This observation is particularly striking because the 24 proteins in our study were not chosen randomly, but rather many of them had been previously implicated in raft-associated functions or shown to be in DRMs (biochemical preparation enriching for raft components). Included among these proteins are glycosylated and palmitoylated proteins, features that are thought to confer raft affinity 35, 71. Fig 2A shows that several 2-TM glycoproteins Myelin-Oligodendrocyte Glycoprotein (MOG) and the Platelet Glycoprotein (CD36) were raft-excluded. These were tested because glycosylations were implicated in raft affinity for single-pass proteins including P-selectin Glycoprotein Ligand-1 (PSGL1), Leukosialin (CD43), and CD44 Antigen (CD44) 72. All 6-TM and 7-TM receptors were completely excluded from the raft phase (Fig 2C). We tested several members of the TRP family receptors, a large group of calcium-selective ion channels that are likely palmitoylated to regulate their function 73. Again, the fact that these proteins may be palmitoylated was insufficient for their raft affinity, as was also the case for the G Protein-Coupled Receptors DRD1 and β2-AR, which are believed to be palmitoylated in their C-termini 74, 75. It is important to note that the presence of documented palmitoylation sites does not imply that these proteins are necessarily palmitoylated in any given context.
We also analyzed several proteins with more than 10 TM helices, and again all were excluded from the raft phase. Among these was the scramblase TMEM16F, involved in the externalization of phosphatidylserine in the cell PM. The other 10+ TM proteins tested (Solute Carrier Family 12 Member 2 (SLC12A2 or NKCC1), Potassium Voltage-gated Channel Subfamily KQT Member 1 (KCNQ1), and Sodium-and Chloride-dependent GABA Transporter 1 (SLC6A1 or GAT)) also included several other proteins belonging to the sodium transporter family. Finally, we found that neither myelin residence nor 4-TM domains were sufficient to explain raft affinity. The myelin point was discussed above for MOG and MAL2. We also tested several known ‘tetraspanin’ proteins (CD9, CD81, and CD82), a family with specific characteristics including 2 extracellular loops, one short and one large, and highly conserved cysteine-rich domains that are likely palmitoylated. In our experiments, neither CD9 76, 77, CD81 78, nor CD82 79 were raft preferring.
These experiments show a broad survey of partitioning for multi-pass TMPs. A related but separate point of interest is the behavior of protein complexes, from homodimers to more complex multi-subunit assemblies 80. Is raft affinity of such complexes predictable from the raft affinities of individual components or are there non-ideal effects arising from interactions? Such questions remain to largely unexplored.
Cholesterol as a regulator of raft affinity
Our measurements implicate cholesterol as a potential regulator of partitioning for PLP, and to a lesser extent MAL. This inference is based on the observations that these proteins appear to compete for a saturable component and that the partitioning of PLP is strongly sensitive to cholesterol depletion. A role for cholesterol in PLP’s raft affinity would also be consistent with previously suggested cholesterol interaction of PLP via its Cholesterol Recognition Amino acid Consensus (CRAC) motif in the cytosolic interface of its TMD2 and a Sterol Sensing Domain (SSD) in the extracellular interface of its TMD3 81. None of these motifs have been reported in MAL and our bioinformatics analysis also did not identify any such motifs. Obviously, the known cholesterol-binding modes are not exhaustive, and it is possible that MAL (and PLP) interacts with lipids through as-yet-undefined mechanisms.
Cholesterol is highly abundant in mammalian PMs (up to 40 mol% 82), thus it may be surprising that co-transfection of two putatively cholesterol-interacting proteins could deplete a sufficient fraction of cholesterol to effect such large changes on raft affinity. Even in the case of MBCD depletion, there are almost certainly more remaining cholesterol molecules than PLP and MAL combined, regardless of the extent of overexpression. A possible answer to this seeming paradox comes from the extensive literature that reveals that a relatively small fraction of the cholesterol present in the PM is actually available for bioactivity, known as the active cholesterol hypothesis 83. While most of the cholesterol in the PM is “hidden” by its interactions with membrane phospholipids, some small “active” fraction is available for interaction with proteins and it is presumably this small fraction that can be saturated.
Other potential modulators of raft affinity
The reduction in PLP raft affinity by myriocin treatment reveals that lipid raft affinity of some proteins could be affected by their binding sphingolipids. Protein motifs that bind sphingolipids 84 or glycolipids 85, 86 have been extensively reported and some of these may have a significant role in conferring raft affinity. MAL itself has been shown to bind sulfatide 87, a glycosphingolipid enriched in myelin sheath. Thus, the interaction of MAL and/or PLP with glycolipids may be involved in their unique raft affinity.
Why is partitioning in GPMVs so different from DRMs?
The fact that so many proteins previously associated with rafts show minimal ordered phase affinity in GPMVs demands critical examination. It should be noted that almost all previous such studies have relied on detergent resistance to infer raft residence. The many caveats associated with detergent resistance have been discussed at length 88, 89 and will not be recapitulated here, except to point out that DRM compositions are rather unpredictable and highly dependent on specific experimental conditions, and that protein appearance in detergent resistant fractions cannot be used as bona fide evidence of raft affinity. However, pointing out the issues with DRMs should not be construed as absolute validation of GPMVs as a system for evaluating raft affinity. Many essential factors of live cell membranes are perturbed in GPMVs, including lipid asymmetry, out-of-equilibrium energy consumption, assembled cytoskeleton, and protein-protein interactions that are disrupted by the isolation procedure, among others previously discussed 48. Any of these factors may be necessary for a given protein to partition to raft domains in vivo. Furthermore, the specific relationships between phase separation in GPMVs and the nanodomains presumed to be present in living membranes remain incompletely understood 29. Ultimately, low raft affinity in GPMVs seems to be the default condition for many proteins and the very large majority of multi-pass TMPs. However, whether this accurately represents their in-situ association with ordered domains remains to be resolved.
Acknowledgements
Funding for this work was provided by the NIH/National Institute of General Medical Sciences (GM114282, GM124072, GM120351, GM134949), the Volkswagen Foundation (grant 93091), and the Human Frontiers Science Program (RGP0059/2019). All authors have no competing interests.
REFERENCES
- 1.Garcia-Parajo MF; Cambi A; Torreno-Pina JA; Thompson N; Jacobson K, Nanoclustering as a dominant feature of plasma membrane organization. J Cell Sci 2014, 127 (Pt 23), 4995–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sezgin E; Levental I; Mayor S; Eggeling C, The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat Rev Mol Cell Biol 2017, 18 (6), 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levental I; Levental KR; Heberle FA, Lipid rafts: controversies resolved, mysteries remain. Trends in cell biology 2020, 30 (5), 341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuck S; Honsho M; Ekroos K; Shevchenko A; Simons K, Resistance of cell membranes to different detergents. PNAS 2003, 100 (10), 5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lingwood D; Simons K, Detergent resistance as a tool in membrane research. Nature Protocols 2007, 2 (9), 2159–2165. [DOI] [PubMed] [Google Scholar]
- 6.Prinetti A; Chigorno V; Tettamanti G; Sonnino S, Sphingolipid-enriched Membrane Domains from Rat Cerebellar Granule Cells Differentiated in Culture. J Biol Chem 2000, 275 (16), 11658–11665. [DOI] [PubMed] [Google Scholar]
- 7.Epand RM, Proteins and cholesterol-rich domains. Biochimica et Biophysica Acta (BBA) - Biomembranes 2008, 1778 (7), 1576–1582. [DOI] [PubMed] [Google Scholar]
- 8.Simons K; Vaz WL, Model systems, lipid rafts, and cell membranes. Annu Rev Biophys Biomol Struct 2004, 33, 269–95. [DOI] [PubMed] [Google Scholar]
- 9.Simons K; Ikonen E, Functional rafts in cell membranes. Nature 1997, 387 (6633), 569–572. [DOI] [PubMed] [Google Scholar]
- 10.Raghupathy R; Anilkumar AA; Polley A; Singh PP; Yadav M; Johnson C; Suryawanshi S; Saikam V; Sawant SD; Panda A; Guo Z; Vishwakarma RA; Rao M; Mayor S, Transbilayer lipid interactions mediate nanoclustering of lipid-anchored proteins. Cell 2015, 161 (3), 581–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Owen DM; Williamson DJ; Magenau A; Gaus K, Sub-resolution lipid domains exist in the plasma membrane and regulate protein diffusion and distribution. Nature Communications 2012, 3 (1), 1256. [DOI] [PubMed] [Google Scholar]
- 12.Kinoshita M; Suzuki KGN; Matsumori N; Takada M; Ano H; Morigaki K; Abe M; Makino A; Kobayashi T; Hirosawa KM; Fujiwara TK; Kusumi A; Murata M, Raft-based sphingomyelin interactions revealed by new fluorescent sphingomyelin analogs. J Cell Biol 2017, 216 (4), 1183–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komura N; Suzuki KGN; Ando H; Konishi M; Koikeda M; Imamura A; Chadda R; Fujiwara TK; Tsuboi H; Sheng R; Cho W; Furukawa K; Furukawa K; Yamauchi Y; Ishida H; Kusumi A; Kiso M, Raft-based interactions of gangliosides with a GPI-anchored receptor. Nature Chemical Biology 2016, 12 (6), 402–410. [DOI] [PubMed] [Google Scholar]
- 14.Stone MB; Shelby SA; Núñez MF; Wisser K; Veatch SL, Protein sorting by lipid phase-like domains supports emergent signaling function in B lymphocyte plasma membranes. Elife 2017, 6, e19891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerl MJ; Sampaio JL; Urban S; Kalvodova L; Verbavatz J-M; Binnington B; Lindemann D; Lingwood CA; Shevchenko A; Schroeder C; Simons K, Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J Cell Biol 2012, 196 (2), 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingólfsson HI; Melo MN; van Eerden FJ; Arnarez C; Lopez CA; Wassenaar TA; Periole X; de Vries AH; Tieleman DP; Marrink SJ, Lipid Organization of the Plasma Membrane. Journal of the American Chemical Society 2014, 136 (41), 14554–14559. [DOI] [PubMed] [Google Scholar]
- 17.Toulmay A; Prinz WA, Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J Cell Biol 2013, 202 (1), 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rayermann SP; Rayermann GE; Cornell CE; Merz AJ; Keller SL, Hallmarks of reversible separation of living, unperturbed cell membranes into two liquid phases. Biophys J 2017, 113 (11), 2425–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sezgin E; Kaiser HJ; Baumgart T; Schwille P; Simons K; Levental I, Elucidating membrane structure and protein behavior using giant plasma membrane vesicles. Nat Protoc 2012, 7 (6), 1042–51. [DOI] [PubMed] [Google Scholar]
- 20.Baumgart T; Hammond AT; Sengupta P; Hess ST; Holowka DA; Baird BA; Webb WW, Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proceedings of the National Academy of Sciences of the United States of America 2007, 104 (9), 3165–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veatch SL; Keller SL, Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol. Biophysical journal 2003, 85 (5), 3074–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heberle FA; Feigenson GW, Phase separation in lipid membranes. Cold Spring Harbor Perspectives in Biology 2011, 3 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honigmann A; Sadeghi S; Keller J; Hell SW; Eggeling C; Vink R , A lipid bound actin meshwork organizes liquid phase separation in model membranes. Elife 2014, 3, e01671–e01671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arumugam S; Petrov EP; Schwille P, Cytoskeletal pinning controls phase separation in multicomponent lipid membranes. Biophysical journal 2015, 108 (5), 1104–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogel SK; Greiss F; Khmelinskaia A; Schwille P, Control of lipid domain organization by a biomimetic contractile actomyosin cortex. Elife 2017, 6, e24350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Machta BB; Papanikolaou S; Sethna JP; Veatch SL, Minimal model of plasma membrane heterogeneity requires coupling cortical actin to criticality. Biophysical journal 2011, 100 (7), 1668–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yethiraj A; Weisshaar JC, Why are lipid rafts not observed in vivo? Biophysical journal 2007, 93 (9), 3113–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin X; Gorfe AA; Levental I, Protein Partitioning into Ordered Membrane Domains: Insights from Simulations. Biophysical journal 2018, 114 (8), 1936–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li G; Wang Q; Kakuda S; London E, Nanodomains can persist at physiologic temperature in plasma membrane vesicles and be modulated by altering cell lipids. J Lipid Res 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veatch SL; Cicuta P; Sengupta P; Honerkamp-Smith A; Holowka D; Baird B, Critical fluctuations in plasma membrane vesicles. ACS Chem Biol 2008, 3 (5), 287–93. [DOI] [PubMed] [Google Scholar]
- 31.Levental I; Veatch SL, The continuing mystery of lipid rafts. J Mol Biol 2016, 428 (24), 4749–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heberle FA; Petruzielo RS; Pan J; Drazba P; Kucerka N; Standaert RF; Feigenson GW; Katsaras J, Bilayer thickness mismatch controls domain size in model membranes. J Am Chem Soc 2013, 135 (18), 6853–9. [DOI] [PubMed] [Google Scholar]
- 33.Goh SL; Amazon JJ; Feigenson GW, Toward a better raft model: modulated phases in the four-component bilayer, DSPC/DOPC/POPC/CHOL. Biophys J 2013, 104 (4), 853–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levental I; Grzybek M; Simons K, Greasing their way: lipid modifications determine protein association with membrane rafts. Biochemistry 2010, 49 (30), 6305–16. [DOI] [PubMed] [Google Scholar]
- 35.Levental I; Lingwood D; Grzybek M; Coskun U; Simons K, Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proceedings of the National Academy of Sciences of the United States of America 2010, 107 (51), 22050–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diaz-Rohrer BB; Levental KR; Simons K; Levental I, Membrane raft association is a determinant of plasma membrane localization. Proceedings of the National Academy of Sciences of the United States of America 2014, 111 (23), 8500–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lorent JH; Diaz-Rohrer B; Lin X; Spring K; Gorfe AA; Levental KR; Levental I, Structural determinants and functional consequences of protein affinity for membrane rafts. Nat Commun 2017, 8 (1), 1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheong KH; Zacchetti D; Schneeberger EE; Simons K, VIP17/MAL, a lipid raft-associated protein, is involved in apical transport in MDCK cells. Proceedings of the National Academy of Sciences of the United States of America 1999, 96 (11), 6241–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weimbs T; Stoffel W, Proteolipid protein (PLP) of CNS myelin: positions of free, disulfide-bonded, and fatty acid thioester-linked cysteine residues and implications for the membrane topology of PLP. Biochemistry 1992, 31 (49), 12289–96. [DOI] [PubMed] [Google Scholar]
- 40.Levental KR; Lorent JH; Lin X; Skinkle AD; Surma MA; Stockenbojer EA; Gorfe AA; Levental I, Polyunsaturated lipids regulate membrane domain stability by tuning membrane order. Biophys J 2016, 110(8), 1800–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levental KR; Surma MA; Skinkle AD; Lorent JH; Zhou Y; Klose C; Chang JT; Hancock JF; Levental I, omega-3 polyunsaturated fatty acids direct differentiation of the membrane phenotype in mesenchymal stem cells to potentiate osteogenesis. Sci Adv 2017, 3 (11), eaao1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levental KR; Levental I, Isolation of giant plasma membrane vesicles for evaluation of plasma membrane structure and protein partitioning. Methods in molecular biology 2015, 1232, 65–77. [DOI] [PubMed] [Google Scholar]
- 43.Levental I; Grzybek M; Simons K, Raft domains of variable properties and compositions in plasma membrane vesicles. Proceedings of the National Academy of Sciences of the United States of America 2011, 108 (28), 11411–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Y; Maxwell KN; Sezgin E; Lu M; Liang H; Hancock JF; Dial EJ; Lichtenberger LM; Levental I, Bile acids modulate signaling by functional perturbation of plasma membrane domains. J. Biol. Chem. 2013, 288 (50), 35660–35670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levental I; Byfield FJ; Chowdhury P; Gai F; Baumgart T; Janmey PA, Cholesterol-dependent phase separation in cell-derived giant plasma-membrane vesicles. Biochem J 2009, 424 (2), 163–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaiser HJ; Lingwood D; Levental I; Sampaio JL; Kalvodova L; Rajendran L; Simons K, Order of lipid phases in model and plasma membranes. Proceedings of the National Academy of Sciences of the United States of America 2009, 106 (39), 16645–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sezgin E; Levental I; Grzybek M; Schwarzmann G; Mueller V; Honigmann A; Belov VN; Eggeling C; Coskun U; Simons K; Schwille P, Partitioning, diffusion, and ligand binding of raft lipid analogs in model and cellular plasma membranes. Biochimica et biophysica acta 2012, 1818 (7), 1777–84. [DOI] [PubMed] [Google Scholar]
- 48.Levental KR; Levental I, Giant plasma membrane vesicles: models for understanding membrane organization. Current topics in membranes 2015, 75, 25–57. [DOI] [PubMed] [Google Scholar]
- 49.Marta CB; Montano MB; Taylor CM; Taylor AL; Bansal R; Pfeiffer SE, Signaling Cascades Activated upon Antibody Cross-linking of Myelin Oligodendrocyte Glycoprotein: POTENTIAL IMPLICATIONS FOR MULTIPLE SCLEROSIS. Journal of Biological Chemistry 2005, 280 (10), 8985–8993. [DOI] [PubMed] [Google Scholar]
- 50.Boyanapalli M; Kottis V; Lahoud O; Bamri-Ezzine S; Braun PE; Mikol DD, Oligodendrocyte-myelin glycoprotein is present in lipid rafts and caveolin-1-enriched membranes. Glia 2005, 52 (3), 219–227. [DOI] [PubMed] [Google Scholar]
- 51.Nusrat A; Parkos CA; Verkade P; Foley CS; Liang TW; Innis-Whitehouse W; Eastburn KK; Madara JL, Tight junctions are membrane microdomains. Journal of Cell Science 2000, 113 (10), 1771. [DOI] [PubMed] [Google Scholar]
- 52.de Marco MC; Martín-Belmonte F; Kremer L; Albar JP; Correas I; Vaerman JP; Marazuela M; Byrne JA; Alonso MA, MAL2, a novel raft protein of the MAL family, is an essential component of the machinery for transcytosis in hepatoma HepG2 cells. J Cell Biol 2002, 159 (1), 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frank M, MAL, a proteolipid in glycosphingolipid enriched domains: functional implications in myelin and beyond. Prog Neurobiol 2000, 60 (6), 531–44. [DOI] [PubMed] [Google Scholar]
- 54.Schneider A; Länder H; Schulz G; Wolburg H; Nave K-A; Schulz JB; Simons M, Palmitoylation is a sorting determinant for transport to the myelin membrane. J Cell Sci 2005, 118 (11), 2415. [DOI] [PubMed] [Google Scholar]
- 55.Sengupta P; Hammond A; Holowka D; Baird B, Structural determinants for partitioning of lipids and proteins between coexisting fluid phases in giant plasma membrane vesicles. Biochimica et biophysica acta 2008, 1778 (1), 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson SA; Stinson BM; Go MS; Carmona LM; Reminick JI; Fang X; Baumgart T, Temperature-dependent phase behavior and protein partitioning in giant plasma membrane vesicles. Biochimica et biophysica acta 2010, 1798 (7), 1427–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marinko JT; Li GC; Kenworthy AK; Sanders CR, Peripheral Myelin Protein 22 preferentially partitions into ordered phase membrane domains. bioRxiv 2020, 2020.01.28.923771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yaffe Y; Shepshelovitch J; Nevo-Yassaf I; Yeheskel A; Shmerling H; Kwiatek JM; Gaus K; Pasmanik-Chor M; Hirschberg K, The MARVEL transmembrane motif of occludin mediates oligomerization and targeting to the basolateral surface in epithelia. J Cell Sci 2012, 125 (Pt 15), 3545–56. [DOI] [PubMed] [Google Scholar]
- 59.Raleigh DR; Marchiando AM; Zhang Y; Shen L; Sasaki H; Wang Y; Long M; Turner JR, Tight junction-associated MARVEL proteins marveld3, tricellulin, and occludin have distinct but overlapping functions. Mol Biol Cell 2010, 21 (7), 1200–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanchez-Pulido L; Martin-Belmonte F; Valencia A; Alonso MA, MARVEL: a conserved domain involved in membrane apposition events. Trends Biochem Sci 2002, 27 (12), 599–601. [DOI] [PubMed] [Google Scholar]
- 61.Puertollano R; Alonso MA, A Short Peptide Motif at the Carboxyl Terminus Is Required for Incorporation of the Integral Membrane MAL Protein to Glycolipid-enriched Membranes. Journal of Biological Chemistry 1998, 273 (21), 12740–12745. [DOI] [PubMed] [Google Scholar]
- 62.Puertollano R; Alonso MA, Substitution of the two carboxyl-terminal serines by alanine causes retention of MAL, a component of the apical sorting machinery, in the endoplasmic reticulum. Biochem Biophys Res Commun 1999, 260 (1), 188–92. [DOI] [PubMed] [Google Scholar]
- 63.Peschl P; Bradl M; Hoftberger R; Berger T; Reindl M, Myelin Oligodendrocyte Glycoprotein: Deciphering a Target in Inflammatory Demyelinating Diseases. Front Immunol 2017, 8, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heberle FA; Doktorova M; Scott HL; Skinkle A; Waxham MN; Levental I, Direct label-free imaging of nanodomains in biomimetic and biological membranes by cryogenic electron microscopy. bioRxiv 2020, 2020.02.05.935551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mahammad S; Parmryd I, Cholesterol Depletion Using Methyl-β-cyclodextrin In Methods in Membrane Lipids, Owen DM, Ed. Springer New York: New York, NY, 2015; pp 91–102. [DOI] [PubMed] [Google Scholar]
- 66.Mahammad S; Dinic J; Adler J; Parmryd I, Limited cholesterol depletion causes aggregation of plasma membrane lipid rafts inducing T cell activation. Biochimica et biophysica acta 2010, 1801 (6), 625–34. [DOI] [PubMed] [Google Scholar]
- 67.Simons M; Krämer EM; Thiele C; Stoffel W; Trotter J, Assembly of myelin by association of proteolipid protein with cholesterol- and galactosylceramide-rich membrane domains. J Cell Biol 2000, 151 (1), 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miyake Y; Kozutsumi Y; Nakamura S; Fujita T; Kawasaki T, Serine Palmitoyltransferase Is the Primary Target of a Sphingosine-like Immunosuppressant, ISP-1/Myriocin. Biochemical and Biophysical Research Communications 1995, 211 (2), 396–403. [DOI] [PubMed] [Google Scholar]
- 69.Osuchowski MF; Johnson VJ; He Q; Sharma RP, Myriocin, a serine palmitoyltransferase inhibitor, alters regional brain neurotransmitter levels without concurrent inhibition of the brain sphingolipid biosynthesis in mice. Toxicology Letters 2004, 147 (1), 87–94. [DOI] [PubMed] [Google Scholar]
- 70.Fitzner D; Schneider A; Kippert A; Mobius W; Willig KI; Hell SW; Bunt G; Gaus K; Simons M, Myelin basic protein-dependent plasma membrane reorganization in the formation of myelin. EMBO J 2006, 25 (21), 5037–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Benting JH; Rietveld AG; Simons K, N-Glycans mediate the apical sorting of a GPI-anchored, raft-associated protein in Madin-Darby canine kidney cells. J Cell Biol 1999, 146 (2), 313–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shao B; Yago T; Setiadi H; Wang Y; Mehta-D’souza P; Fu J; Crocker PR; Rodgers W; Xia L; McEver RP, O-glycans direct selectin ligands to lipid rafts on leukocytes. Proceedings of the National Academy of Sciences 2015, 112 (28), 8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Han H; Yi F, New insights into TRP channels: Interaction with pattern recognition receptors. Channels (Austin) 2014, 8 (1), 13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McGraw DW; Liggett SB, Molecular mechanisms of beta2-adrenergic receptor function and regulation. Proc Am Thorac Soc 2005, 2 (4), 292–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arango-Lievano M; Sensoy O; Borie A; Corbani M; Guillon G; Sokoloff P; Weinstein H; Jeanneteau F, A GIPC1-Palmitate Switch Modulates Dopamine Drd3 Receptor Trafficking and Signaling. Mol Cell Biol 2016, 36 (6), 1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Andreu Z; Yáñez-Mó M, Tetraspanins in Extracellular Vesicle Formation and Function. Frontiers in Immunology 2014, 5 (442). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang XH; Kovalenko OV; Kolesnikova TV; Andzelm MM; Rubinstein E; Strominger JL; Hemler ME, Contrasting Effects of EWI Proteins, Integrins, and Protein Palmitoylation on Cell Surface CD9 Organization. Journal of Biological Chemistry 2006, 281 (18), 12976–12985. [DOI] [PubMed] [Google Scholar]
- 78.Zhu Y-Z; Luo Y; Cao M-M; Liu Y; Liu X-Q; Wang W; Wu D-G; Guan M; Xu Q-Q; Ren H; Zhao P; Qi Z-T, Significance of palmitoylation of CD81 on its association with tetraspanin-enriched microdomains and mediating hepatitis C virus cell entry. Virology 2012, 429 (2), 112–123. [DOI] [PubMed] [Google Scholar]
- 79.Zhou B; Liu L; Reddivari M; Zhang XA, The Palmitoylation of Metastasis Suppressor KAI1/CD82 Is Important for Its Motility- and Invasiveness-Inhibitory Activity. Cancer Research 2004, 64 (20), 7455. [DOI] [PubMed] [Google Scholar]
- 80.Beck-Garcia K; Beck-Garcia E; Bohler S; Zorzin C; Sezgin E; Levental I; Alarcon B; Schamel WW, Nanoclusters of the resting T cell antigen receptor (TCR) localize to non-raft domains. Biochimica et biophysica acta 2015, 1853 (4), 802–9. [DOI] [PubMed] [Google Scholar]
- 81.Werner HB; Krämer-Albers E-M; Strenzke N; Saher G; Tenzer S; Ohno-Iwashita Y; De Monasterio-Schrader P; Möbius W; Moser T; Griffiths IR; Nave K-A, A critical role for the cholesterol-associated proteolipids PLP and M6B in myelination of the central nervous system. Glia 2013, 61 (4), 567–586. [DOI] [PubMed] [Google Scholar]
- 82.Lorent JH; Levental KR; Ganesan L; Rivera-Longsworth G; Sezgin E; Doktorova M; Lyman E; Levental I, Plasma membranes are asymmetric in lipid unsaturation, packing and protein shape. Nat Chem Biol 2020, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Steck TL; Lange Y, Cell cholesterol homeostasis: mediation by active cholesterol. Trends Cell Biol 2010, 20 (11), 680–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Björkholm P; Ernst AM; Hacke M; Wieland F; Brügger B; von Heijne G, Identification of novel sphingolipid-binding motifs in mammalian membrane proteins. Biochimica et Biophysica Acta (BBA) - Biomembranes 2014, 1838 (8), 2066–2070. [DOI] [PubMed] [Google Scholar]
- 85.Fantini J; Garmy N; Yahi N, Prediction of Glycolipid-Binding Domains from the Amino Acid Sequence of Lipid Raft-Associated Proteins: Application to HpaA, a Protein Involved in the Adhesion of Helicobacter pylori to Gastrointestinal Cells. Biochemistry 2006, 45 (36), 10957–10962. [DOI] [PubMed] [Google Scholar]
- 86.Sasaki T, Glycolipid-binding proteins. Chem Phys Lipids 1985, 38 (1), 63–77. [DOI] [PubMed] [Google Scholar]
- 87.Frank M; van der Haar ME; Schaeren-Wiemers N; Schwab ME, rMAL is a glycosphingolipid-associated protein of myelin and apical membranes of epithelial cells in kidney and stomach. J Neurosci 1998, 18 (13), 4901–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lichtenberg D; Goni FM; Heerklotz H, Detergent-resistant membranes should not be identified with membrane rafts. Trends Biochem Sci 2005, 30 (8), 430. [DOI] [PubMed] [Google Scholar]
- 89.Brown DA, Lipid rafts, detergent-resistant membranes, and raft targeting signals. Physiology 2006, 21, 430–9. [DOI] [PubMed] [Google Scholar]