Abstract
Molecular-targeted therapy has had a significant impact on glioma. Notably, actin-like 6A (ACTL6A) has been indicated to be essential for embryonic development and tumor progression. However, the role of ACTL6A in glioma remains unclear. The present study aimed to investigate the effects of ACTL6A on glioma cell migration and sensitivity to temozolomide (TMZ). The expression levels of ACTL6A were analyzed in patients with glioma, and survival curves were created using data from The Cancer Genome Atlas. U251 and T98G cells were transfected with short hairpin (sh)RNA for use in loss-of-function experiments to investigate the biological function and molecular mechanisms of ACTL6A. Furthermore, an MTT assay was used to assess the effect of ACTL6A on the sensitivity of glioma cells to TMZ. The results demonstrated that ACTL6A was expressed at higher levels in glioma tissues compared with normal brain tissues. Furthermore, high expression of ACTL6A was associated with a poor prognosis. The knockdown of ACTL6A significantly inhibited the migration phenotype in glioma cells and significantly decreased the levels of phosphorylated AKT in glioma cells. The AKT signaling activator SC79 partly attenuated the inhibitory effects of ACTL6A shRNA on glioma cell migration. Additionally, the knockdown of ACTL6A enhanced the sensitivity of glioma cells to TMZ. In conclusion, these results suggest that ACTL6A knockdown inhibited the migration of human glioma cells, at least in part through inactivation of the AKT signaling pathway, and increased the sensitivity of glioma cells to TMZ. Therefore, ACTL6A may be a potential therapeutic target for glioma.
Keywords: actin-like 6A, AKT signaling pathway, glioma, migration, sensitivity
Introduction
Glioblastoma multiforme (GBM), a grade IV glioma, is the most common and aggressive type of primary malignant brain tumor worldwide, and account for ~60 to 70% of malignant gliomas (1). Despite the progress made in the last decade, GBM remains one of the most difficult types of tumors to treat. The median survival rate is only 12-15 months for patients with glioblastomas (2). Thus, investigation of the molecular mechanisms associated with glioma is essential. In recent years, molecular tumour therapy has improved the treatment of a number of different cancer types (3,4). Molecular therapy is expected to attack malignant cells more specifically with fewer side effects than conventional chemotherapy.
Actin-like 6A (ACTL6A), also known as BAF53a, Arp4 or INO80K, is an actin-like protein and chromatin-remodeling factor. Specifically, ACTL6A encodes a subunit of the switch/sucrose non-fermentable complex that has a mainly transcriptional role as a regulator of the function of stem and progenitor cells (5,6). Previous studies have demonstrated that ACTL6A is involved in the differentiation and proliferation of neural progenitor cells (7,8). In addition, other studies have shown that ACTL6A plays an important role in the proliferation of squamous cell carcinoma cells and is associated with epithelial-mesenchymal transition in hematoma (9,10). Although there is evidence to suggest that ACTL6A has an oncogenic role in numerous types of human cancer, the underlying mechanisms of ACTL6A in glioma cell migration remain unknown (11,12).
Temozolomide (TMZ) is a first-line chemotherapeutic drug for GBM. Drug resistance is the predominant obstacle in TMZ therapy. A large number of gliomas are resistant to TMZ, which is detrimental to the effectiveness of clinical treatment (2). Therefore, it is essential to investigate TMZ sensitivity in order to identify how the therapeutic effects of TMZ may be improved. Previous studies have demonstrated that several genes are involved in TMZ sensitivity, including autophagy related 4C cysteine peptidase, N-methylpurine DNA glycosylase, O-6-methylguanine-DNA methyltransferase and STAT3 (13-15). However, whether ACTL6A affects the sensitivity of glioma cells to TMZ is unknown and thus is worthy of investigation.
The aim of the present study was to investigate the potential role of ACTL6A in glioma. In particular, the effect of ACTL6A on cell migration and the underlying mechanisms were evaluated following the knockdown of ACTL6A in two glioma cell lines, T98G and U251. Furthermore, whether the knockdown of ACTL6A sensitizes these cells to TMZ was also analyzed.
Materials and methods
Cell culture
Human glioma cell lines T98G and U251 were obtained from the American Type Culture Collection. The glioma cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.) at 37˚C in a 5% CO2 incubator. Cells in the logarithmic growth phase were used for subsequent experiments.
Cell treatment
The ACTL6A-knockdown and negative control (NC) lentiviruses containing short hairpin (sh)RNA were synthesized by and purchased from Shanghai GeneChem, Co., Ltd. The ACTL6A-knockdown and NC lentiviruses were used to transfect T98G and U251 cells according to the manufacturer's instructions to establish shACTL6A and sh control (shCtrl) groups. Cells transfected with an empty vector were used as controls. The shRNA target sequence for ACTL6A was ACCTTACGTTTCATAGCTTTA and the shRNA NC sequence was TTCTCCGAACGTGTCACGT. The concentration of shACTL6A used was 6x108 TU/ml, and the concentration of shCtrl used was 8x108 TU/ml cells cultured in moderate medium remained in a humid environment with a temperature of 37˚C and 5% CO2, after 72 h, the cells were collected and washed twice with PBS prior to use in further experiments.
Bioinformatics analysis
The Cancer Cell Line Encyclopedia database (CCLE; https://portals.broadinstitute.org/ccle) was used to analyze the transcriptomic expression levels of the ACTL6A gene in multiple glioma cell lines. Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn) is an interactive web server that enables users to perform expression analyses, including survival and differential analyses, at the subtype level using data from The Cancer Genome Atlas and Genotype-Tissue Expression portal. In the present study, GEPIA was used to examine the differential expression of the ACTL6A gene between different types of cancer, including glioma, and healthy tissue. A Kaplan-Meier plot was also created using GEPIA to calculate the overall survival (OS) rates of patients with glioma, and analyzed using the log-rank test. The Cox proportional hazard ratio was also calculated. The thresholds for the high/low ACTL6A expression level cohorts were adjusted according to the median value.
Patients
A total of 27 patients with glioma were enrolled in the present study. The clinicopathological features of the patients are described in Table I. All patients had received surgery without chemotherapy or radiotherapy at the First Affiliated Hospital of Nanchang University (Nanchang, China) between March 2013 and August 2014. Tumor biopsies were collected prior to neoadjuvant chemotherapy or wide resection of the tumor and were fresh-frozen and stored at -80˚C. A total of 5 normal control brain tissues were also obtained from patients with cerebral trauma. The procedures were approved by the Ethics Committee of the First Affiliated Hospital of Nanchang University (approval no. 2010-015). Written informed consent was obtained from every participant.
Table I.
Patient characteristics and clinical features for the different types of brain tissue.
| Tissue | ||
|---|---|---|
| Variable | Glioma (n=27) | Normal (n=5) |
| Mean age (years) | ||
| Female | 50.83±8.21 | 44.67±7.37 |
| Male | 45.13±13.53 | 42.50±3.54 |
| Sex (n) | ||
| Female | 12.00 | 3.00 |
| Male | 15.00 | 5.00 |
| Tumor grade | ||
| Low grades I–II | 9.00 | 0.00 |
| High grades III–IV | 16.00 | 0.00 |
Western blot analysis
Total protein was extracted from the cells using RIPA buffer (cat. no. R0278; Sigma-Aldrich; Merck KGaA) which contained 2% phosphatase inhibitor and 1% PMSF. A total of 100 µl protein buffer was harvested from each well of a 6-well plate. The protein concentration was quantified by the BCA method and the mass of protein loaded per lane was ~30 µg. Protein samples were separated by 10% SDS-PAGE and transferred to PVDF membranes. The membranes were blocked in 5% skimmed milk with Tris-buffered saline with 10% Tween-20 (TBST) for 1 h at room temperature, and then incubated with primary antibodies overnight at 4˚C. The next day, the membranes were incubated with secondary antibodies for 1 h at room temperature. After extensive washing using TBST, the blots were visualized using Amersham Hyperfilm ECL (Cytiva). The following primary antibodies were used: Anti-ACTL6A (cat. no. abs133368; absin; dilution 1:3,000), anti-AKT (cat. no. 10176-2-AP; ProteinTech Group, Inc.; dilution 1:2,000), anti-Ser473-phosphorylated (p)-AKT (cat. no. 4060T; Cell Signaling Technology, Inc.; dilution 1:2,000), anti-E-cadherin (cat. no. 3195T; Cell Signaling Technology, Inc,.; dilution 1:6,000), anti-vimentin (cat. no. 5741T; Cell Signaling Technology, Inc.; dilution 1:3,000), anti-PI3K (cat. no. abs123958; Cell Signaling Technology, Inc.; dilution 1:5,000), anti-p-PI3K (cat. no. 17366S; absin; dilution 1:1,000), and anti-β-actin (abs118937, absin, dilution 1:5,000). Horseradish peroxidase-conjugated anti-human IgG was used as the secondary antibody (cat. no. 7074P2; Cell Signaling Technology, Inc.; dilution 1:10,000). Image Lab V3.0 (Bio-Rad Laboratories, Inc.) was used for densitometry.
Reverse transcription-quantitative PCR (RT-qPCR)
The total RNA was extracted from patient tissue using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). A total of 2 µg RNA was converted into cDNA using a Thermo Scientific RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.). qPCR was then performed using Maxima SYBR Green qPCR Master Mix (DBI Bioscience) in an ABI PRISM 7500 Fast Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The PCR conditions were as follows: 95˚C for 5 min, and then 40 cycles of amplification for 30 sec at 95˚C, 45 sec at 60˚C and 45 sec at 72˚C. The primers, purchased from Guangzhou RiboBio Co. Ltd., were as follows: ACTL6A, forward: TCAGAGGCACCGTGGAATAC; reverse: GACATAGCCATCGTGGACTG; GAPDH, forward: TGACTTCAACAGCGACACCCA; reverse: CACCCTGTTGCTGTAGCCAAA. The gene expression was quantified using the 2-ΔΔCq method (16).
Wound-healing assay
The transfected cells were seeded in 6-well plates containing DMEM with 10% FBS. When cultured to 90% confluence, the cells were starved for 24 h in serum-free medium. A sterile 10-µl pipette tip was used to create wounds in the cell layer. After washing away the dislodged cell fragments using PBS, the cells were cultured in 2% FBS medium. The areas of the wounds were assessed under an Zeiss AG fluorescence inverted microscope at x5 magnification after 24 h. ImageJ 1.8.0 was then used to quantify the results (National Institutes of Health).
Single cell tracking assay
The Operetta® High Content Imaging System (PerkinElmer, Inc.) combines fully automated high-throughput fluorescence microscopy with multi-parameter quantitative image analysis, which enables the effects of drugs on cell properties, such as migration, to be evaluated (10,11). The transfected cells were seeded at a density of 2,000 cells/well in a 96-well plate and cultured in an Operetta High Content Imaging System at 37˚C with 5% CO2 for 20 h. Digital phase contrast images were acquired every 1 h for 20 h, and the mean square displacements of cells in the two groups were compared using Harmony® 3.5 High Content Imaging and Analysis Software (PerkinElmer, Inc.).
MTT assay
U251 and T98G cells were seeded in 96-well plates at a density of 5x103 cells/well and cultured in 5% CO2 at 37˚C. TMZ (Selleck Chemicals) and SC79 were dissolved in dimethyl sulfoxide (Sigma Aldrich; Merck KGaA). The cells were cultured for 12 h prior to the knockdown of ACTL6A, after 48 h transfection, cell were treated with TMZ (0, 100, 500 and 1,000 µM) for 12 h. Similarly, after 48 h of transfection, cells were treated with 5 µM of SC79 for 12 h. A total of 100 µl culture medium (DMEM; Thermo Fisher Scientific, Inc.) containing 20 µl MTT was added into each well and the 96-well plates were placed in an incubator at 37˚C for 4 h. After 4 h, 150 µl/well formanzan solution (Nanjing KeyGen Biotech Co., Ltd.) was used to dissolve the blue-purple crystals. The OD value of each well was detected at the absorbance of 490 nm.
Statistical analysis
Each experiment was repeated three times. Data are presented as the mean ± SD unless otherwise indicated and were compared using Student's t-test or one-way analysis of variance (ANOVA) followed by Bonferroni post hoc tests. The mean square displacement data were analyzed using two-way ANOVA followed by Bonferroni post hoc tests. All analyses were performed using GraphPad Prism (version 7.00; GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
ACTL6A expression is upregulated in glioma
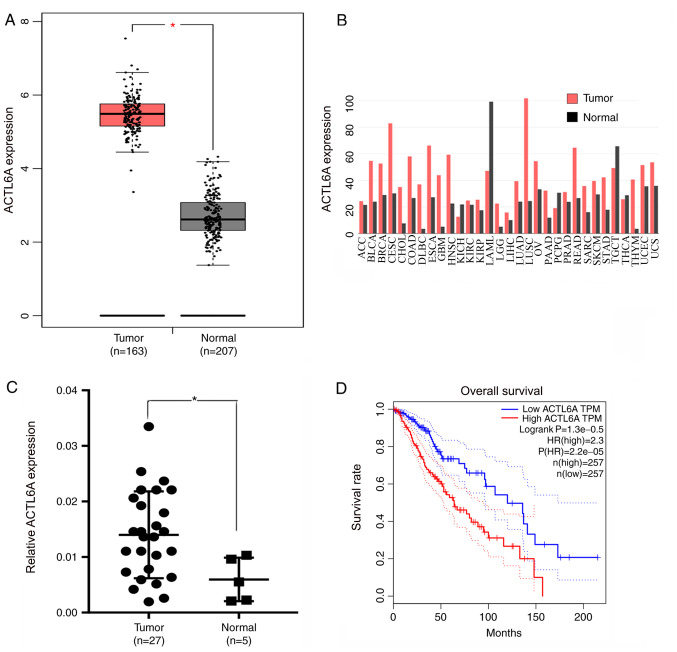
The differences in ACTL6A expression between glioma and normal tissue were analyzed using the GEPIA database. The results revealed a significant difference in the expression levels of ACTL6A between glioma and normal tissue; the ACTL6A expression levels in glioma were significantly higher compared with those in normal tissue (Fig. 1A). The expression levels of ACTL6A in different types of cancer were also analyzed using GEPIA. The results reveal that compared with normal samples, ACTL6A is highly expressed in multiple cancer types, including in glioma, bladder urothelial carcinoma and breast carcinoma. However, ACTL6A expression is low in some cancers, such as in acute myeloid leukemia and testicular germ cell tumors (Fig. 1B). A total of 27 glioma samples and 5 normal control brain tissue samples were obtained from patients at the First Affiliated Hospital of Nanchang University, and RT-qPCR was used to measure the expression levels of ACTL6A. The results revealed that ACTL6A was expressed at significantly higher levels in glioma compared with normal brain tissue (Fig. 1C). The OS curves for glioma generated using GEPIA indicated that high expression of ACTL6A was associated with a poor prognosis, while low expression of ACTL6A was associated with an improved prognosis (Fig. 1D). These results indicate that ACTL6A expression is upregulated in glioma tissue, and that ACTL6A may play a significant role in the development and progression of glioma.
Figure 1.
ACTL6A expression is upregulated in glioma. (A) Data from GEPIA showing the expression levels of ACTL6A in glioma samples compared with normal brain samples. *P<0.05. (B) Data from GEPIA showing the differential expression of ACTL6 in different types of cancer. (C) Expression of ACTL6A in human glioma and normal brain tissue samples as measured by reverse transcription-quantitative PCR. *P<0.05. (D) Survival curves showing overall survival in ACTL6A high and low expression groups. ACTL6A, actin-like 6A; GEPIA, Gene Expression Profiling Interactive Analysis.
ACTL6A promotes glioma cell migration in vitro
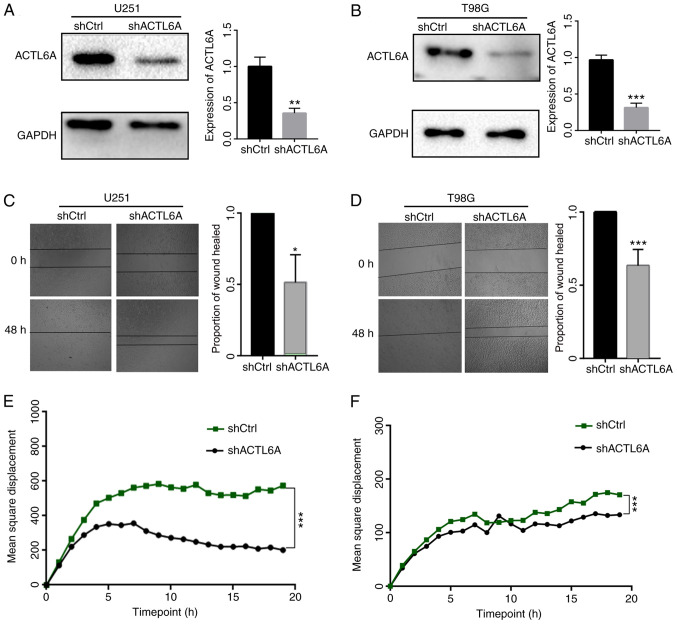
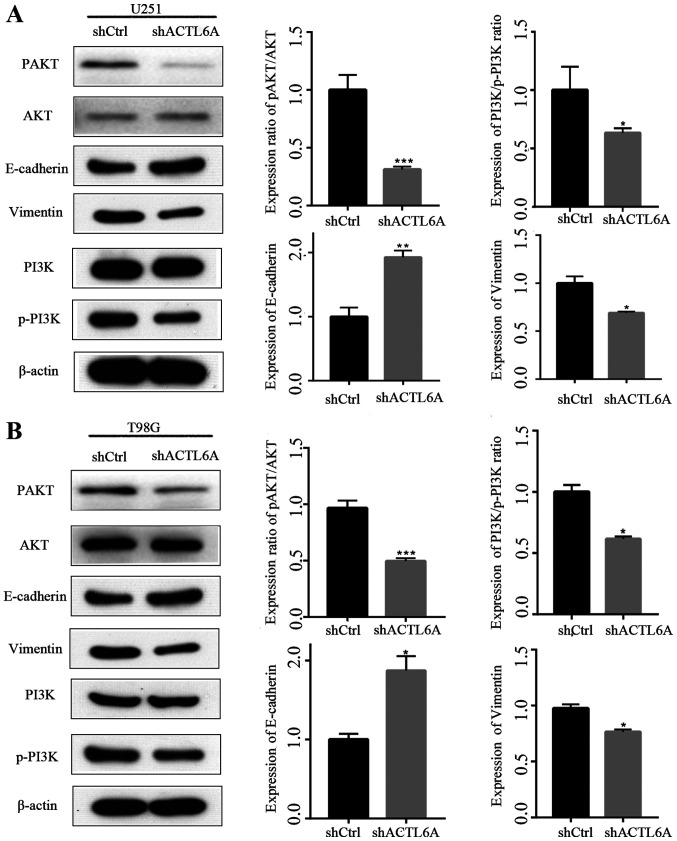
ACTL6A knockdown U251 and T98G cell lines were constructed to investigate the functional role of ACTL6A in glioma cells in vitro. First, the efficiency of ACTL6A knockdown in U251 and T98G cells was determined by comparison with NC transfected controls, and the results revealed that transfection with ACTL6A shRNA significantly reduced the expression of ACTL6A (Fig. 2A and B). Next, a wound-healing assay was performed to detect the migration ability of the glioma cells. The results revealed that the knockdown of ACTL6A significantly reduced the wound-healing rate compared with that of the shCtrl group (Fig. 2C and D). These results indicate that the knockdown of ACTL6A inhibited the migration capacity of the glioma cells. In addition, the 20-h mean square displacements of the glioma cells were analyzed. The results confirm that the cells in which ACTL6A was knocked down migrated more slowly compared with those in the shCtrl group (Fig. 2E and F). The expression levels of proteins associated with migration, namely E-cadherin and vimentin, were also detected and the results revealed that the expression of E-cadherin was upregulated while that of vimentin was downregulated in the ACTL6A knockdown groups compared with the respective shCtrl groups, which further indicate that ACTL6A knockdown inhibited cell migration (Fig. 3). These results suggest that ACTL6A promotes the migration capacity of glioma cells.
Figure 2.
ACTL6A affects the migration of glioma cells. Following transfection with shACTL6A or shCtrl, the expression of ACTL6A was detected in (A) U251 and (B) T98G cells using western blotting. Cell migration was determined by wound-healing assays in the transfected (C) U251 and (D) T98G cells from 0 to 48 h. The area of wound healing represents the rate of cell migration. A high-content imaging and analysis instrument was used to determine the mean square displacements of transfected at x20 magnification (E) U251 and (F) T98G cells. Cell migration was measured over a period of 20 h. ACTL6A, actin-like 6A; sh, short hairpin; Ctrl, control. *P<0.05, **P<0.01, ***P<0.0001 vs. shCtrl.
Figure 3.
Effects of ACTL6A on AKT signaling protein expression. (A) U251 and (B) T98G cells were transfected with shACTL6A or shCtrl and the levels of p-AKT, AKT, p-PI3K, PI3K, E-cadherin and Vimentin were detected by western blotting. The results reveal that the PI3K/AKT signaling pathway was inhibited after the knockdown of ACTL6A. ACTL6A, actin-like 6A; sh, short hairpin; Ctrl, control; p, phosphorylated. . *P<0.05, **P<0.01 and ***P<0.0001 vs. shCtrl.
Knockdown of ACTL6A inhibits the PI3K/AKT signaling pathway
Numerous studies have confirmed that the PI3K/AKT signaling pathway plays an important role in the regulation of cell proliferation (17,18). However, whether ACTL6A affects the PI3K/AKT pathway remains unknown. Western blotting analysis revealed that the knockdown of ACTL6A significantly decreased p-AKT/AKT and p-PI3K/PI3K ratios compared with those in the shCtrl group, while the levels of total AKT and total PI3K were not changed (Fig. 3). These results suggest that ACTL6A may regulate biological functions in glioma via the PI3K/AKT signaling pathway.
PI3K/AKT agonist reverses the effect of ACTL6A knockdown
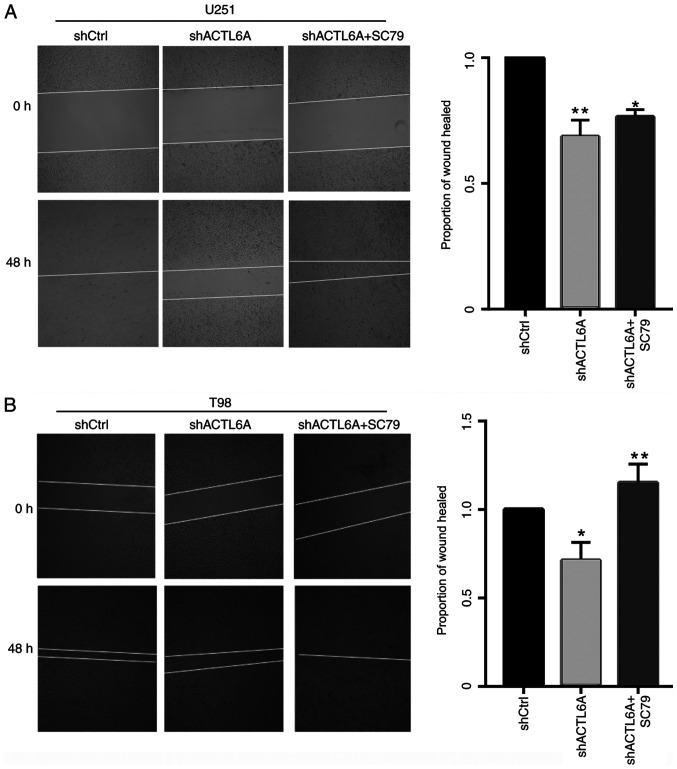
In order to further investigate the role and mechanism of ACTL6A in cell migration, U251 and T98G cells were treated with the AKT pathway agonist SC79 or DMSO solvent after transfection. Following treatment, the wound-healing assay was used to detect cell migration, and the results showed that the migration ability of the U251 and T98G cells, which was significantly decreased following transfection with sh-ACTL6A was recovered by SC79 (Fig. 4). These results supports the hypothesis that silencing ACTL6A inhibits cell migration through deactivation of the AKT pathway.
Figure 4.
Activation of AKT reverses the effect of ACTL6A knockdown on cell migration. A wound-healing assay was used to demonstrate the restorative effect of the AKT agonist SC79 on the migration of (A) U251 and (B) T98G cells following ACTL6A knockdown. sh, short hairpin; ACTL6A, actin-like 6A; Ctrl, control. The areas of the wounds were assessed at 5x magnification. *P<0.05, **P<0.01 vs. shCtrl.
Knockdown of ACTL6A contributes to the sensitivity of glioma cells to TMZ
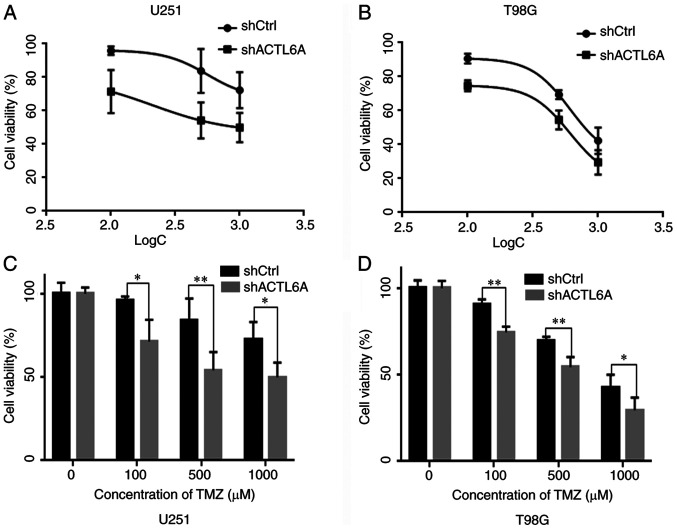
In order to determine whether ACTL6A affects the sensitivity of U251 and T98G cells to TMZ-induced cell death, the viability of U251 and T98G cells following treatment with TMZ was tested using an MTT assay. The results revealed that as the concentration of TMZ increased, cell viability decreased. The cell viability of the ACTL6A group was significantly lower than that of the shCtrl group at all concentrations tested, suggesting that the sensitivity to TMZ was greater in the shACTL6A group compared with the shCtrl group (Fig. 5).
Figure 5.
Knockdown of ACTL6A enhances the sensitivity of glioma cells to TMZ. (A) U251 and (B) T98G cells transfected with shACTL6A or shCtrl were treated with TMZ at different concentrations, and the cell survival rate was measured using an MTT assay. Cell viability was assayed in the (C) U251 and (D) T98G cells by MTT assay and compared between the shACTL6A and shCtrl groups following treatment with different concentrations of TMZ for 24 h. *P<0.05, **P<0.01 as indicated. ACTL6A, actin-like 6A; TMZ, temozolomide; sh, short hairpin; Ctrl, control; logC, log TMZ concentration.
Discussion
Glioma-related literature and cell databases were consulted to assist the selection of cell lines for inclusion in the present study. The CCLE database revealed that the expression of ACTL6A was similar in different glioma cell lines. The current study observed that the long synapse of the U87 cell line renders it unsuitable for cell scratch experiments, and the proliferation rate of HS683 cells is rapid, which would adversely affect the cell migration experiment. Therefore, U251 cells and T98G cell lines were selected for investigation in the present study. The results demonstrated that ACTL6A is highly expressed in glioma cells, and its expression is associated with tumor migration. In vitro assays indicated that the knockdown of ACTL6A inhibited cell migration via reduction of the phosphorylation of AKT at Ser473, and the decrease in cell migration induced by ACTL6A knockdown was reversed by SC79, an activator of the AKT pathway. Furthermore, MTT assay results revealed that ACTL6A knockdown enhanced the sensitivity of the glioma cells to TMZ.
Previous studies have demonstrated that ACTL6A is involved in the migration of certain types of cancer cells, such as epidermal squamous cell carcinoma and colorectal carcinoma cells (19,20). Furthermore, ACTL6A has been shown to be upregulated in glioma cells and associated with the progression of glioma (11). The results of present study are consistent with these previous findings. They revealed that the knockdown of ACTL6A inhibited the migration ability of glioma cells, and demonstrated that the ability to migrate was restored following treatment with an AKT agonist.
AKT is a major cellular kinase involved in cell proliferation, apoptosis, invasion and migration (21). Numerous studies have shown that AKT activation increases the migratory or invasive activity of various types of cells (22-25). The AKT pathway is regulated by multiple genes, including insulin-like growth factor 1 receptor and mTOR, and contributes to wound healing (26-28). SC79, an AKT pathway agonist, reversed the wound healing by activating the AKT pathway in glioma (29,30). The results of the present study suggest that ACTL6A induces the phosphorylation of AKT at Ser473, and stimulates the migration of human glioma cells, at least in part through activation of the AKT signaling pathway.
TMZ is an imidazotetrazine derivative and a prodrug, the active metabolite of which can also be derived from the alkylating agent dacarbazine. TMZ is often used as a standard treatment for glioma. It causes the death of cancer cells via its ability to methylate or alkylate DNA at the O6 or N7 sites of guanine residues (31,32). However, tumor cells can acquire the ability to repair DNA damage, which may greatly reduce the efficacy of TMZ. Therefore, to improve the effectiveness of TMZ, it is necessary to further investigate the mechanism by which TMZ inhibits tumor progression. The present study revealed that ACTL6A knockdown increased the sensitivity of glioma cells to TMZ. Future studies are required to assess whether the enhancement of TMZ sensitivity by ACTL6A knockdown is associated with DNA damage repair or other mechanisms.
In conclusion, to the best of our knowledge, the present study revealed for the first time that the knockdown of ACTL6A reduced the migration of human glioma cells, at least in part through inactivation of the AKT signaling pathway, and enhanced the sensitivity of glioma cells to TMZ. The results of the present study may provide a new therapeutic strategy for the treatment of GBM.
Acknowledgements
Not applicable.
Funding
The study was funded by the Hunan Provincial Natural Science Foundation of China (grant no. 2018JJ2600) and the Project of Scientific Research Plan of Hunan Provincial Health Commission (grant no. B2017029).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
XP designed the current study and collected data from recruited cases. XC performed the majority of the experiments. ZX analyzed data and performed the statistics. XZ and XC analyzed and wrote the manuscript. DL completed the experiments that were suggested by reviewers. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All patients provided written informed consent for their tissues to be used for clinical research. The present study was approved by the Independent Ethics Committee of the Institute of Clinical Pharmacology, Central South University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet. 2018;392:432–446. doi: 10.1016/S0140-6736(18)30990-5. [DOI] [PubMed] [Google Scholar]
- 2.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 3.Posadas EM, Limvorasak S, Figlin RA. Targeted therapies for renal cell carcinoma. Nat Rev Nephrol. 2017;13:496–511. doi: 10.1038/nrneph.2017.82. [DOI] [PubMed] [Google Scholar]
- 4.Lee YT, Tan YJ, Oon CE. Molecular targeted therapy: Treating cancer with specificity. Eur J Pharmacol. 2018;834:188–196. doi: 10.1016/j.ejphar.2018.07.034. [DOI] [PubMed] [Google Scholar]
- 5.Marom R, Jain M, Burrage LC, Song IW, Graham BH, Brown CW, Stevens SJC, Stegmann APA, Gunter AT, Kaplan JD, et al. Heterozygous variants in ACTL6A, encoding a component of the BAF complex, are associated with intellectual disability. Hum Mutat. 2017;38:1365–1371. doi: 10.1002/humu.23282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sima X, He J, Peng J, Xu Y, Zhang F, Deng L. The genetic alteration spectrum of the SWI/SNF complex: The oncogenic roles of BRD9 and ACTL6A. PLoS One. 2019;14(e0222305) doi: 10.1371/journal.pone.0222305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Son EY, Crabtree GR. The role of BAF (mSWI/SNF) complexes in mammalian neural development. Am J Med Genet C Semin Med Genet. 2014;166C:333–349. doi: 10.1002/ajmg.c.31416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng Z, Yang H, Xiao S. ACTL6A expression promotes invasion, metastasis and epithelial mesenchymal transition of colon cancer. BMC Cancer. 2018;18(1020) doi: 10.1186/s12885-018-4931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao S, Chang RM, Yang MY, Lei X, Liu X, Gao WB, Xiao JL, Yang LY. Actin-like 6A predicts poor prognosis of hepatocellular carcinoma and promotes metastasis and epithelial-mesenchymal transition. Hepatology. 2016;63:1256–1271. doi: 10.1002/hep.28417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng L, Wang X, Liao W, Liu J, Liao Y, He Q. BAF53a is a potential prognostic biomarker and promotes invasion and epithelial-mesenchymal transition of glioma cells. Oncol Rep. 2017;38:3327–3334. doi: 10.3892/or.2017.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji J, Xu R, Zhang X, Han M, Xu Y, Wei Y, Ding K, Wang S, Bin Huang, Chen A, et al. Actin like-6A promotes glioma progression through stabilization of transcriptional regulators YAP/TAZ. Cell Death Dis. 2018;9(517) doi: 10.1038/s41419-018-0548-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee ES, Ko KK, Joe YA, Kang SG, Hong YK. Inhibition of STAT3 reverses drug resistance acquired in temozolomide-resistant human glioma cells. Oncol Lett. 2011;2:115–121. doi: 10.3892/ol.2010.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan Q, Yang XJ, Wang HM, Dong XT, Wang W, Li Y, Li JM. Chemoresistance to temozolomide in human glioma cell line U251 is associated with increased activity of O6-methylguanine-DNA methyltransferase and can be overcome by metronomic temozolomide regimen. Cell Biochem Biophys. 2012;62:185–191. doi: 10.1007/s12013-011-9280-7. [DOI] [PubMed] [Google Scholar]
- 15.Wen ZP, Zeng WJ, Chen YH, Li H, Wang JY, Cheng Q, Yu J, Zhou HH, Liu ZZ, Xiao J, Chen XP. Knockdown ATG4C inhibits gliomas progression and promotes temozolomide chemosensitivity by suppressing autophagic flux. J Exp Clin Cancer Res. 2019;38(298) doi: 10.1186/s13046-019-1287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Wang S, Zheng Y, He Z, Zhou W, Cheng Y, Zhang C. SH2B1 promotes NSCLC cell proliferation through PI3K/Akt/mTOR signaling cascade. Cancer Cell Int. 2018;18(132) doi: 10.1186/s12935-018-0632-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu XY, Zhang J, Qi YH, Kong M, Liu SA, Hu JJ. Linc-ROR promotes endometrial cell proliferation by activating the PI3K-Akt pathway. Eur Rev Med Pharmacol Sci. 2018;22:2218–2225. doi: 10.26355/eurrev_201804_14807. [DOI] [PubMed] [Google Scholar]
- 19.Zhu B, Ueda A, Song X, Horike SI, Yokota T, Akagi T. Baf53a is involved in survival of mouse ES cells, which can be compensated by Baf53b. Sci Rep. 2017;7(14059) doi: 10.1038/s41598-017-14362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun W, Wang W, Lei J, Li H, Wu Y. Actin-like protein 6A is a novel prognostic indicator promoting invasion and metastasis in osteosarcoma. Oncol Rep. 2017;37:2405–2417. doi: 10.3892/or.2017.5473. [DOI] [PubMed] [Google Scholar]
- 21.Chu N, Salguero AL, Liu AZ, Chen Z, Dempsey DR, Ficarro SB, Alexander WM, Marto JA, Li Y, Amzel LM, et al. Akt Kinase activation mechanisms revealed using protein semisynthesis. Cell. 2018;174:897–907.e14. doi: 10.1016/j.cell.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwiatkowska A, Symons M. Signaling determinants of glioma cell invasion. Adv Exp Med Biol. 2013;986:121–141. doi: 10.1007/978-94-007-4719-7_7. [DOI] [PubMed] [Google Scholar]
- 23.Ao R, Guan L, Wang Y, Wang JN. Silencing of COL1A2, COL6A3, and THBS2 inhibits gastric cancer cell proliferation, migration, and invasion while promoting apoptosis through the PI3k-Akt signaling pathway. J Cell Biochem. 2018;119:4420–4434. doi: 10.1002/jcb.26524. [DOI] [PubMed] [Google Scholar]
- 24.Luo KW, Lung WY, Chun-Xie Luo XL, Huang WR. EGCG inhibited bladder cancer T24 and 5637 cell proliferation and migration via PI3K/AKT pathway. Oncotarget. 2018;9:12261–12272. doi: 10.18632/oncotarget.24301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng X, Tu Z, Xiong M, Tembo K, Zhou L, Liu P, Pan S, Xiong J, Yang X, Leng J, et al. Wnt5a and CCL25 promote adult T-cell acute lymphoblastic leukemia cell migration, invasion and metastasis. Oncotarget. 2017;8:39033–39047. doi: 10.18632/oncotarget.16559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ilowski M, Putz C, Weiss TS, Brand S, Jauch KW, Hengstler JG, Thasler WE. Augmenter of liver regeneration causes different kinetics of ERK1/2 and Akt/PKB phosphorylation than EGF and induces hepatocyte proliferation in an EGF receptor independent and liver specific manner. Biochem Biophys Res Commun. 2010;394:915–920. doi: 10.1016/j.bbrc.2010.03.074. [DOI] [PubMed] [Google Scholar]
- 27.Guo YY, Wu Y, Jia XW, An W. Augmenter of liver regeneration potentiates doxorubicin anticancer efficacy by reducing the expression of ABCB1 and ABCG2 in hepatocellular carcinoma. Lab Invest. 2017;97:1400–1411. doi: 10.1038/labinvest.2017.72. [DOI] [PubMed] [Google Scholar]
- 28.Shang X, Lin K, Yu R, Zhu P, Zhang Y, Wang L, Xu J, Chen K. Resveratrol protects the myocardium in sepsis by activating the phosphatidylinositol 3-Kinases (PI3K)/AKT/mammalian target of rapamycin (mTOR) pathway and inhibiting the nuclear factor-kappaB (NF-kappaB) signaling pathway. Med Sci Monit. 2019;25:9290–9298. doi: 10.12659/MSM.918369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu W, Jing ZT, Wu SX, He Y, Lin YT, Chen WN, Lin XJ, Lin X. A Novel AKT Activator, SC79, prevents acute hepatic failure induced by fas-mediated apoptosis of hepatocytes. Am J Pathol. 2018;188:1171–1182. doi: 10.1016/j.ajpath.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 30.Xu Y, Gao YW, Yang Y. SC79 protects dopaminergic neurons from oxidative stress. Oncotarget. 2018;9:12639–12648. doi: 10.18632/oncotarget.23538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hegi ME, Liu L, Herman JG, Stupp R, Wick W, Weller M, Mehta MP, Gilbert MR. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26:4189–4199. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- 32.Preusser M, Charles JR, Felsberg J, Reifenberger G, Hamou MF, Diserens AC, Stupp R, Gorlia T, Marosi C, Heinzl H, et al. Anti-O6-methylguanine-methyltransferase (MGMT) immunohistochemistry in glioblastoma multiforme: Observer variability and lack of association with patient survival impede its use as clinical biomarker. Brain Pathol. 2008;18:520–532. doi: 10.1111/j.1750-3639.2008.00153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.