Abstract
Objectives
To analyse the diagnostic performance of eosinopenia, alone or combined with polymorphonuclear neutrophils (PMN) and/or lymphocytes, as a marker of active COVID-19 in patients hospitalized for suspicion of SARS-CoV-2 infection.
Methods
A prospective observational study including patients hospitalized for suspicion of COVID-19 in a COVID unit was performed from 20th March to 5th April 2020, in Perpignan, France. Patients for which there was a doubt upon diagnosis, who were recently under oral corticosteroids, had myeloid malignancy or human immunodeficient virus infection were excluded. SARS-CoV-2 detection was performed using an RT-PCR assay, from nasopharyngeal swab specimens. Complete blood count were performed for all patients.
Results
One-hundred and twenty-one patient were included: 57 patients were diagnosed with COVID-19, 64 patients were not. Eosinophil count was lower in the COVID-19 group (median: 0/μL versus 70/μL, p < 0.0001). To diagnose COVID-19, eosinopenia had a sensitivity of 89.5% and a specificity of 78.1% while lymphopenia's were 73.7% and 62.5% respectively. Using area under curve (AUC) of receiving operating characteristics (ROC) curves, eosinophil's optimal cut-off level was 10/μL, sensitivity and specificity were 86%, and 79.7% respectively. Regarding the eosinophil/PMN ratio, the optimal cut-off level was 3.344, sensitivity and specificity were 87.7% and 73.4% respectively. The AUC of lymphocyte/PMN ratio was significantly lower than eosinophil/PMN ratio's (0.621 versus 0.846, p = 0.0003).
Conclusion
Eosinopenia – <10/μL – and eosinophil/PMN ratio are useful, low-cost, reproducible tools to help diagnose COVID-19, during an epidemic period, in a population of hospitalized patients admitted for suspicion of COVID-19.
Keywords: COVID-19, SARS-CoV-2, Eosinopenia, Marker
Introduction
Since December 2019, starting in Wuhan, China, a novel coronavirus - severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) - has rapidly spread worldwide, causing a new pandemic viral pneumonia: coronavirus disease 2019 (COVID-19).1 The SARS-CoV-2 reached Europe in January 2020 and the first three cases in France were confirmed on 24th January 2020 2. Diagnosis of COVID-19 is confirmed by SARS-CoV-2 detection, performed by real-time reverse transcriptase-polymerase chain reaction (RT-PCR) assays, from respiratory track swab specimens (mostly nasopharyngeal). In France, hospital or common practice laboratories had to face a shortage of diagnostic tests on a national scale, so tests were prioritized for patients with risk factors or serious illness.
In addition, the RT-PCR assay for SARS-CoV-2 detection seems to have a limited sensitivity (59–89%).3, 4, 5 Chest computed tomography (CT) is another primary diagnostic tool but can be source of false positive results.5 Lymphopenia is a simple biological tool parameter associated to COVID-19 and is more common in severe cases 6 but not specific for COVID-19.8
In our centre, one male patient was highly suspected of COVID-19 despite the fact that he had a negative result of RT-PCR. He was part of a confirmed cluster of COVID-19 cases, had typical ground glass pulmonary opacities on chest CT and got worse despite antibiotic therapy during 72 h. His complete blood count (CBC) at admission included normal polymorphonuclear neutrophil (PMN) count, eosinopenia and lymphopenia.
Eosinopenia as an indirect argument for infection has been well described in previous publications.9 More recently, it has been studied as an important tool to distinguish bacterial infection from other causes of acute inflammatory syndromes, when associated to leukocytosis,10 , 11 without additional cost or extra-time. Recent data also showed that eosinopenia may be a potential indicator for COVID-19 diagnosis 6 without leukocytosis.
The aim of our study was to evaluate the diagnostic performance of eosinopenia, alone or combined with PMN and/or lymphocytes, as a marker of active COVID-19 in patients hospitalized for suspicion of SARS-CoV-2 infection.
Methods
Population
We performed an observational prospective study in our Internal Medicine department, converted into a COVID-19 unit, in Perpignan's Hospital, France, from March 20th to April 5th, 2020. This study included all consecutive hospitalized COVID-19 patients.
All patients included were 18 or older, addressed because of COVID-19 suspicion based on a clinical diagnosis score, with or without confirmed diagnosis, from the emergency room. The clinical diagnosis score was the following: unexplained fever >38 °C: 2 points, recent/recently worsened cough: 2 points, recent/recently worsened dyspnoea: 1 point, odynophagia OR anosmia OR dysgeusia: 1 point, flu-like syndrome: 1 point, diarrhea >65 years-old: 1 point, contact with proven case: 1 point.
If at least 4 points: COVID-19 was suspected.
All patients had a CBC in the 24 h after their admission in the hospital.
The COVID-19 patients were defined as the following: either they had a positive SARS-CoV-2 detection by RT-PCR, or, in cases in which RT-PCR was negative or not performed, they had clinical, biological and radiological evidence supporting the diagnosis of COVID-19.
The non COVID-19 patients were defined as the following: all had a negative PCR test, and either they had no evidence of infection whatsoever, or they had an alternate proven diagnosis, without any clinical sign nor the evolution suggestive of COVID-19.
All medical files were reviewed by at least two independent physicians.
Patients who were recently under oral corticosteroids, had myeloid malignancy or human immunodeficient virus infection, or for which there was a doubt upon diagnosis were excluded.
Data collection
The data collection was carried out for the specific study question and began on day 1 of the study, following the previous cited observation.
Demographic information, medical history, clinical, laboratory and computed tomographic (CT) scan results were collected in real time, from electronic medical records. The delay between the onset of symptoms to hospital admission was also collected.
All patients were treated according the usual standard of care.
The clinical outcomes of all COVID-19 patients were checked at least four weeks after the last patient was admitted. All deaths, COVID-19 related or not, were accounted.
All patients were given verbal and written information about the collection and analysis of data for future studies. The patients’ consent was reported to the Commission Nationale Informatique et Libertés (CNIL) and the local Ethics Committee gave their approval.
Laboratory testing
SARS-CoV-2 detection was performed in Perpignan's Hospital, using a RT-PCR assay, using nasopharyngeal swab specimens. We used the RealStar®SARS-CoV-2 RT-PCR Kit 1.0∗ (RUO) from Altona Diagnostics and Coronavirus (COVID-19) genesig® Real Time PCR∗ from Primerdesign NOVACYT.
Due to a lack of kits for SARS-CoV-2 detection, and limited intensive care wards on a national scale, the pharyngeal swabs specimens were, as a priority, obtained for all patients who were presumed not to have COVID-19 so as to move them in a non COVID-19 ward. They were also carried out in patients for whom there was a doubt upon diagnosis.
Laboratory blood count collected for all patients included the number of eosinophils (Eo), lymphocytes (Ly), PMN (with calculation of Eo/PMN ratio × 1000 and Ly/PMN ratio × 1000), and platelets, concentration of hemoglobin (Hb), and C-reactive protein (CRP). All laboratory tests were performed in Perpignan's Hospital laboratory using standard protocols and CBC were performed with XN3100 device from SYSMEX®. According to our local laboratory guidelines, eosinopenia was defined when eosinophil count was <300/μL, lymphopenia with a lymphocyte count <1300/μL.
Statistical analysis
Descriptive data were described as frequencies and percentages, using mean with standard deviation values and median with interquartile range values for variables regarding leukocytes. Categorical variables were compared using χ2 test and continuous variables were compared using independent t-test. We evaluated an optimal cut-off level for each continuous variable using Youden's index. Receiver operating characteristics (ROC) curves were calculated using the easyROC tool that implements pROC and Optimal Cutpoints in R. Comparative analysis between areas under curves (AUC) of receiving operating characteristics (ROC) curves were performed using the Delong's method.
Tests with p-value <0.05 were considered statistically significant.
Results
Clinical characteristics
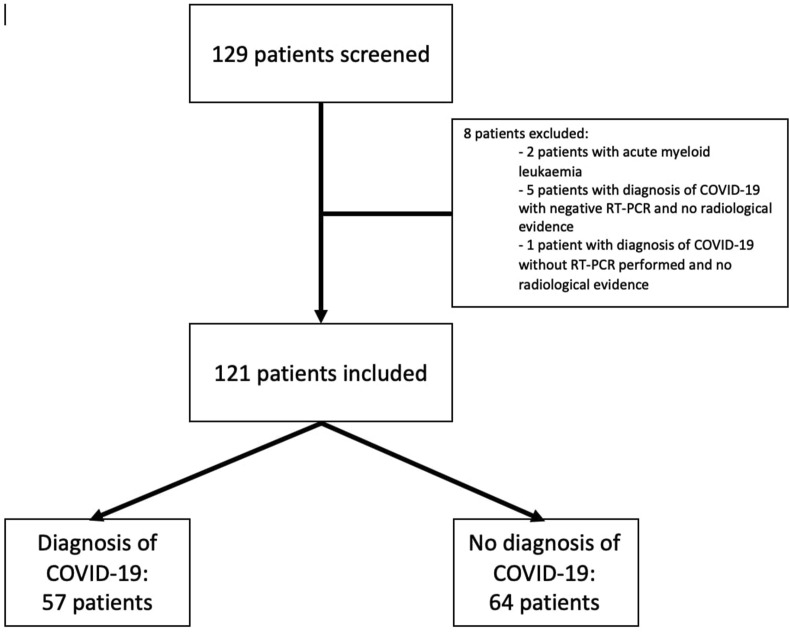
One hundred and twenty-nine patients were screened (Fig. 1 ). Two patients were excluded because they had acute myeloid leukaemia. Six patients were excluded because PCR was negative for 5 of them and not performed for 1, and no radiological evidence supported a COVID-19 diagnosis (4 not performed because considered irrelevant to treatment and 1 considered normal). One patient excluded also had centrilobular emphysema, pulmonary fibrosis, with moderate ground glass opacities, unspecific of COVID-19 in their case.
Figure 1.
Flowchart of the study population at admission
A total of 121 patients were included: 57 patients diagnosed with COVID-19 and 64 patients without COVID-19. About half (51%) patients in the COVID-19 group were male, and 36% of them in the non COVID-19 group (Table 1 ), with no significant difference (p = 0.098).
Table 1.
Diagnosis in non COVID-19 patients.
| Bacterial infection (n = 25) | Viral infection (n = 9) | Cardiopulmonary affections (n = 15) | Miscellaneous (n = 5) | No infection/no disease (n = 10) |
|---|---|---|---|---|
| Respiratory tract infection (n = 14) | Bronchiolitis (n = 2) | Acute pulmonary oedema (n = 9) | Anaemia (n = 1) | |
| Urinary tract infection (n = 2) | Bronchitis (n = 1) | Pulmonary embolism (n = 2) | Cancer (n = 1) | |
| Liver and digestive tract infection (n = 2) | Common cold (n = 6) | Emphysema (n = 1) | Palliative care - terminal kidney disease (n = 1) | |
| Bacteraemia (n = 2) | Asthma (n = 3) | Vasovagal syncope (n = 1) | ||
| Osteo-articular infection (n = 1) | Gastric reflux and orthostatic hypotension (n = 1) | |||
| Cellulitis (n = 3) | ||||
| Dialysis catheter infection (n = 1) |
About half patients (47%) in the COVID-19 group had fever >38.3 °C at admission, significantly higher than in the non COVID-19 group (30%) (p = 0.046). It was also the case for fatigue (54% versus 34%, p = 0.027). Respiratory symptoms (dyspnoea, cough, chest tightness) and ear-nose-throat symptoms (anosmia or dysgeusia) were significantly more present in the COVID-19 group (93% versus 75%, p = 0.008; 23% versus 2%, p < 0.0001 respectively).
There was no significant difference between the two groups regarding comorbidities, except for obesity (44% in the COVID-19 group versus 23% in the non COVID-19 group, p = 0.017).
Diagnosis in non-COVID patients
All non COVID-19 patients had a negative PCR; 10 (15.6%) of them had no sign of infection or of any active disease, and 54 (84.4%) of them had an alternate diagnosis (Table 1). Twenty-four patients (37.5%) had a bacterial infection, 13 of which a bacterial pneumonia resolved with antibiotics. Ten patients (15.7%) had typical signs of benign undocumented viral infection not related to SARS-CoV-2 virus which resolved spontaneously (bronchitis, bronchiolitis). Fifteen patients (23.4%) suffered from a cardiorespiratory affection (emphysema, asthma, acute pulmonary oedema, pulmonary embolism). The five remaining patients (7.8%) had other conditions (anaemia, cancer, palliative care due to kidney failure, vasovagal syncope, and gastric reflux).
Leukocytes and COVID-19
Laboratory blood count results are summarized in Table 2 . Compared to the non COVID-19 group, median values of eosinophil count was significantly lower in the COVID-19 group (0/μL versus 70/μL, p < 0.0001) as well as lymphocyte count (1000/μL versus 1580/μL, p = 0.001). Median values of neutrophil count, Eo/PMN ratio and CRP were also significantly lower in the COVID-19 group.
Table 2.
Baseline characteristics of patients.
| Patients diagnosed with COVID-19 n = 57 (%) | Patients undiagnosed with COVID-19 n = 64 (%) | p | |
|---|---|---|---|
| Male | 29 (51) | 23 (36) | 0.098 |
| Age, years (median ± standard deviation) | 60 ± 18 | 69 ± 24 | 0.508 |
| Onset of symptom to hospital admission, days (median) | 7 | 3 | 0.358 |
| Fever >38.3 °C | 27 (47) | 19 (30) | 0.046 |
| Fatigue | 31 (54) | 22 (34) | 0.027 |
| Respiratory symptoms | 53 (93) | 48 (75) | 0.008 |
| ENT symptoms | 13 (23) | 1 (2) | <0.0001 |
| Digestive symptoms | 21 (37) | 15 (37) | 0.107 |
| Comorbidities | |||
| Asthma | 6 (11) | 8 (13) | 0.735 |
| COPD | 3 (5) | 7 (11) | 0.258 |
| Diabetes mellitus | 21 (37) | 15 (23) | 0.107 |
| Heart disease | 16 (28) | 20 (31) | 0.703 |
| Chronic kidney disease | 6 (11) | 6 (9) | 0.832 |
| Active neoplasia | 2 (4) | 2 (3) | 0.906 |
| Obesity | 25 (44) | 15 (23) | 0.017 |
| Cirrhosis | 0 (0) | 1 (2) | 0.343 |
| Immunosuppressive treatment | 3 (5) | 0 (0) | 0.063 |
| Laboratory findings | |||
| Polymorphonuclear neutrophil count (G/L) | |||
| Mean (standard deviation) | 5.891 ± 2.51 | 7.361 ± 4.29 | 0.025 |
| Median (interquartile range) | 6.02 (3.49–7.37) | 5.95 (4.20–8.91) | |
| Increased >10 G/L | 6 (11) | 15 (23) | 0.061 |
| Eosinophil count (G/L) | |||
| Mean (standard deviation) | 0.014 ± 0.037 | 0.123 ± 0.149 | |
| Median (interquartile range) | 0.00 (0.00–0.01) | 0.07 (0.02–0.16) | <0.0001 |
| Decreased <0.03 G/L | 51 (89) | 14 (22) | <0.0001 |
| Lymphocyte count (G/L) | |||
| Mean (standard deviation) | 1.075 ± 0.541 | 1.909 ± 1.700 | 0.001 |
| Median (interquartile range) | 1.00 (0.73–1.32) | 1.58 (1.2–2.16) | |
| Decreased <1.3 G/L | 42 (74) | 24 (38) | <0.0001 |
| Eo/PMN × 1000 ratio | |||
| Mean (standard deviation) | 2.493 ± 6.698 | 24.274 ± 29.354 | <0.0001 |
| Median (interquartile range) | 0 (0–1.538) | 13.021 (2.899–29.851) | |
| Ly/PMN × 1000 ratio | |||
| Mean (standard deviation) | 220.346 ± 137.169 | 357.344 ± 312.837 | 0.003 |
| Median (interquartile range) | 180.505 (114.650–309.572) | 282.427 (125.960–445.238) | |
| Hb (g/dl, median (interquartile range)) | 13.1 (11.4–14.6) | 12.9 (12.0–14.0) | 0.481 |
| Platelets (G/L, median (interquartile range)) | 238 (175–329) | 268 (220–323) | 0.348 |
| CRP (mg/L, median (interquartile range)) | 69.8 (14.7–120.5) | 14.7 (2.9–65.2) | 0.001 |
ENT: ear-nose-throat; COPD: chronic obstructive pulmonary disease; PMN: polymorphonuclear neutrophil; Eo: eosinophil; Ly: lymphocyte; Hb: haemoglobin; CRP: C-reactive protein.
As an indirect marker of COVID-19, eosinopenia alone had a sensitivity (Se) of 89.5% and a specificity (Sp) of 78.1% (Table 3 ), whereas lymphopenia's were 73.7% and 62.5% respectively. Each variable combined with a neutrophil count <10,000/μL had their Sp increased (92.2% and 75% respectively) at the expense of their Se (78.9% and 68.4% respectively). Specificity of eosinopenia and lymphopenia combined (84.4%) was also increased when associated to a neutrophil count <10,000/μL (93.8%) at the expense of their Se (66.7% versus 61.4% respectively). ROC curves (Fig. 2 ) were made to establish Se, Sp, PPV and NPV of the following variables for the diagnosis of COVID-19 (Table 4 ) compared to the non COVID-19 group: CRP, eosinophil count, lymphocyte count, Eo/PMN ratio × 1000 and Ly/PMN ratio × 1000.
Table 3.
Diagnostic performances of categorical studied variables alone and combined.
| Sensitivity [IC 95%] | Specificity [IC 95%] | Positive predictive value [IC 95%] | Negative predictive value [IC 95%] | |
|---|---|---|---|---|
| Eosinopenia (<0.03 G/L) | 89.5% [78.4–95.4] | 78.1% [66.4–86.6] | 78.5% [68.5–88.5] | 89.3% [81.2–97.4] |
| Eosinopenia (<0.03 G/L) + PMN < 10 G/L | 78.9% [66.5–87.6] | 92.2% [82.5–96.9] | 90% [81.7–98.3] | 83.1% [74.4–91.8] |
| Eosinopenia (<0.03 G/L) + lymphopenia (<1.3 G/L) | 66.7% [53.7–77.5] | 84.4% [73.3–91.4] | 79.2% [67.7–90.7] | 74% [63.9–84.0] |
| Eosinopenia (<0.03 G/L) + lymphopenia (<1.3 G/L) + PMN < 10 G/L | 61.4% [48.4–72.9] | 93.8% [84.4–97.9] | 89.7% [80.2–99.3] | 73.2% [63.6–82.9] |
| Lymphopenia (<1.3 G/L) | 73.7% [60.9–83.4] | 62.5% [50.2–73.3] | 63.6% [52.0–75.2] | 72.7% [61.0–84.5] |
| Lymphopenia (<1.3 G/L) + PMN < 10 G/L | 68.4% [55.4–79.0] | 75% [63.0–84.0] | 70.9% [58.9–82.9] | 72.7% [62.0–83.5] |
PMN: polymorphonuclear neutrophil.
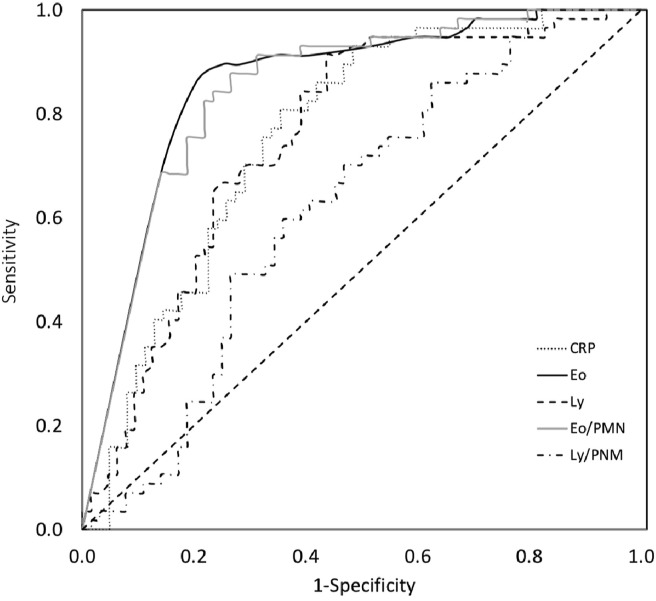
Figure 2.
Receiver-operating characteristics (ROC) curves in the total cohort for various biomarker cutoff levels. Areas under the ROC curves: CRP, 0.759 (95% confidence interval (CI), 0.671–0.846); Eo, 0.852 (95% CI, 0.784–0.920); Ly (95% CI, 0.667–0.842); Eo/PMN ratio × 1000, 0.846 (95% CI, 0.778–0.914); Ly/PMN ratio × 1000, 0.621 (95% CI, 0.521–0.721). Statistical difference between areas under the ROC curves of Eo and Ly counts using Delong's method: p-value=0.0845. Statistical difference between areas under the ROC curves of Eo/PMN ratio × 1000 and Ly/PMN ratio × 1000 using Delong's method: p-value=0.0003. CRP: C-reactive protein; Eo: eosinophil; Ly: lymphocyte; PMN: polymorphonuclear neutrophil.
Table 4.
Diagnostic performances of continuous studied variables and cut-off levels.
| AUC [IC 95%] | Cut-off level | Sensitivity [IC 95%] | Specificity [IC 95%] | Positive predictive value [IC 95%] | Negative predictive value [IC 95%] | p-value | |
|---|---|---|---|---|---|---|---|
| CRP (mg/L) | 0.759 [0.671–0.846] | 36 | 80.7% [68.4–89.0] | 64.5% [52.0–75.2] | 67.6% | 78.4% | N/A |
| Eosinophil count (G/L) | 0.852 [0.784–0.920] | 0.010 | 86% [74.3–92.9] | 79.7% [68.1–87.8] | 79% | 86.4% | 0.0845a |
| Lymphocyte count (G/L) | 0.754 [0.667–0.842] | 1.520 | 91.2% [80.5–96.5] | 56.3% [44.1–67.7] | 65% | 87.8% | |
| Eo/PMN ratio × 1000 | 0.846 [0.778–0.914] | 3.344 | 87.7% [76.3–94.1] | 73.4% [61.4–82.7] | 74.6% | 87.0% | 0.0003b |
| Ly/PMN ratio × 1000 | 0.621 [0.521–0.721] | 203.98 | 59.6% [46.7–71.4] | 64.1% [51.8–74.7] | 59.6% | 64.1% |
CRP: C-reactive protein; Eo: eosinophil; PMN: polymorphonuclear neutrophil; Ly: lymphocyte; AUC: area under curve; N/A: not applicable; 95% CI: 95% confidence interval.
Comparison between eosinophil count and lymphocyte count AUC using Delong's method.
Comparison between Eo/PMN ratio × 1000 and Ly/PMN ratio × 1000 AUC using Delong's method.
Lymphocyte count's performance as a diagnostic marker of COVID-19
Regarding lymphocyte count, AUC was 0.754 for an optimal cut-off level of 1520/μL, Se, Sp, positive predictive value (PPV) and negative predictive value (NPV) were 91.2%, 56.3%, 65% and 87.8% respectively. Regarding the Ly/PMN ratio, AUC was 0.621 for an optimal cut-off level of 203.98, Se, Sp, PPV and NPV were 59.6%, 64.1%, 59.6% and 64.1% respectively.
Eosinophil count's performance as a diagnostic marker of COVID-19
Regarding eosinophil count, AUC was 0.852, and for an optimal cut-off level of 10/μL: Se, Sp, PPV, NPV were 86%, 79.7%, 79% and 86.4% respectively. Regarding the Eo/PMN ratio, AUC was 0.846 for an optimal cut-off level of 3.344; Se, Sp, PPV and NPV were 87.7%, 73.4%, 74.6% and 87.0% respectively.
Comparing AUC of ROC curves between eosinophil count and lymphocyte count showed that eosinophil count tended to be a better diagnostic marker than lymphocyte count though the difference is not statistically significant (p = 0.0845). However comparing AUC of ROC curves between Eo/PMN ratio and Ly/PMN ratio showed that diagnostic performance of Eo/PMN ratio was statistically better than Ly/PMN ratio's (p = 0.0003).
COVID-19 suspicion and chest CT compatible
When analysing patients with chest CT compatible with COVID-19 with negative PCR or no performed RT-PCR: 29 patients were diagnosed with COVID-19, 10 patients were not (Table 5 ). The eosinophil count was lower in the COVID-19 group compared to the non COVID-19 group (median 0/μL versus 70/μL, p = 0.001), and eosinopenia was also more frequent in the COVID-19 group (93% versus 30%, p < 0.0001). The Eo/PMN ratio was lower in the COVID-19 group compared to the non COVID-19 group (median 0.0 versus 16.886, p = 0.0002).
Table 5.
Leukocytes and ratio values in patients with chest CT compatible with COVID-19 with negative or not performed RT-PCR.
| Non COVID-19 patients (n = 10) | COVID-19 patients (n = 29) | p | |
|---|---|---|---|
| PMN count (G/L) | |||
| Mean (standard deviation) | 7.491 ± 4.030 | 6.103 ± 2.270 | 0.1185 |
| Median (interquartile range) | 7.160 (3.840–10.170) | 6.170 (4.280–7.560) | |
| Increased >10 G/L | 3 (30) | 3 (10) | 0.137 |
| Eo count (G/L) | |||
| Mean (standard deviation) | 0.109 | 0.011 ± 0.040 | 0.001 |
| Median (interquartile range) | 0.07 (0.02–0.09) | 0 (0–0) | |
| Decreased <0.03 G/L | 3 (30) | 27 (93) | <0.0001 |
| Ly count (G/L) | |||
| Mean (standard deviation) | 1.509 ± 0.609 | 1.013 ± 0.474 | 0.012 |
| Median (interquartile range) | 1.570 (1.200–1.830) | 1.000 (0.73–1.24) | |
| Decreased <1.3 G/L | 5 (50) | 23 (79) | 0.076 |
| Eo/PMN × 1000 ratio | |||
| Mean (standard deviation) | 18.509 ± 18.305 | 2.070 ± 7.109 | 0.0002 |
| Median (interquartile range) | 16.886 (1.536–30.482) | 0 (0–0) | |
| Ly/PMN × 1000 ratio | |||
| Mean (standard deviation) | 312.141 ± 281.586 | 193.929 ± 119.114 | 0.071 |
| Median (interquartile range) | 219.273 (125.196–379.573) | 168.116 (106.154–292.056) | |
PMN: polymorphonuclear neutrophil; Eo: eosinophil; Ly: lymphocyte.
The lymphocyte count was significantly lower in the COVID-19 group compared to the non COVID-19 group (median 1000/μL versus 1570/μL, p = 0.012), but lymphopenia was not statistically more frequent in the COVID-19 group (79% versus 50%, p = 0.076). The Ly/PMN ratio was lower in the COVID-19 group compared to the non COVID-19 group, but the difference was also not statistically significant (median 168.116 versus 219.273, p = 0.071).
Clinical outcomes
With a follow-up of one month, one patient died in the COVID-19 group (1.8%) at 97 years and two patients in the non COVID-19 group (3.1%) at 79 and 91 years. The latter died from a bacterial pneumonia associated with Staphyloccoccus hominis bacteraemia and the other one was in palliative care due to a terminal kidney disease who could no longer handle dialysis. All other patients could be discharged.
In the COVID-19 group, 13 patients were transferred to intensive care unit (ICU), including 5 who were intubated (mean duration of intubation in days: 16.3). All patients survived and were successfully transferred to medical units. Comparison of COVID-19 patients who were admitted in ICU with the non-ICU patients did not show any significant difference regarding clinical or biological variables (data not shown).
Discussion
Our results support the usefulness of eosinopenia as a simple biological marker of active COVID-19 among patients hospitalized for suspicion of COVID-19 with a sensitivity of 89.5% during a COVID-19 epidemic period. Useful to distinguish patients from non-infectious conditions (i.e. heart failure), its association with PMN <10 G/L also helps distinction with non SARS-CoV-2 infections, especially bacterial infections, with a good specificity, at the expense of sensitivity, compared to eosinopenia alone. Our findings showed that the optimal cut-off level under which the eosinophil count is useful to diagnose COVID-19 is 10/μL. Despite the fact that PPV and NPV regarding eosinopenia or Eo/PMN ratio were above 70%, conclusions cannot be drawn as they depend on prevalence of COVID-19.
Eosinopenia might not be specific of COVID-19 - even when associated with normal PMN count, but might be useful and should be considered as part of a body of diagnostic evidence during the current epidemic period.
Lymphopenia has been well described as associated with COVID-19, especially in severe cases.6 When comparing predictive value of eosinopenia and lymphopenia, respective diagnostic performance of each biological variable favoured eosinopenia, though the difference was not statistically significant. However, comparing Eo/PMN ratio and Ly/PMN ratio showed that Eo/PMN had significantly better diagnostic performances than Ly/PMN ratio.
Our findings are consistent with those of Zhang et al. who found that eosinopenia, as well as lymphopenia, is an indicator for COVID-19 diagnosis.7 Li et al. also aimed to identify a simple biomarker to facilitate triage for sorting suspected COVID-19 patients, and found an interesting biological tool: the combination of eosinopenia (<200 cell/μL) with elevated CRP.12 Their work differed from ours as it took place in fever clinics and systemically considered RT-PCR negative patients as non-COVID-19 patients. Our work began from the fact that a patient was clinically diagnosed with COVID-19 but with a false negative RT-PCR test, and we aimed to argue eosinopenia was a useful complementary tool to diagnose COVID-19. We also used PMN cells count to be included into the Eo/PMN ratio × 1000.
It should be emphasized that the eosinophil count is easily obtained from CBC with neither additional cost nor additional time, contrary to RT-PCR or chest-CT.
This study is a real-life assessment of a COVID unit managing triage of patients with all suspicions of COVID-19. Goals of such a unit are twofold: taking care of COVID-19 patients after confirming diagnosis and ruling out COVID-19 for uninfected patients so as to transfer them in the appropriate department to avoid nosocomial COVID-19. It also illustrates daily practice difficulties to perform RT-PCR tests for a selected number of patients, accordingly to national instructions. Notably, our results showed that eosinopenia or Eo/PMN ratio - associated with a physical examination - can be a useful tool when patients are addressed because of ground glass opacities on chest CT. One of the strengths of this study is also the prospective character, providing accurate information regarding clinical diagnosis when possible.
Nevertheless, our study limitations include the fact that all patients included were hospitalized; therefore our results cannot be extended to ambulatory patients for which a CBC is not constantly performed. They also cannot be extended to patients under corticosteroids, or immunocompromised patients.
Correlation between correction of eosinopenia and clinical improvement, as seen in bacterial infections treated at day 1,13 , 14 has been suspected in our cohort but was not the specific aim of our study.
To summarize, the eosinophil count is a useful, low-cost, reproducible tool to help diagnose COVID-19, in a population of hospitalized patients admitted for suspicion of COVID-19. Eosinopenia – with a cut-off level of 10/μL - or the Eo/PMN ratio can also be of help in patients suspected of COVID-19 with negative PCR and suspicious chest CT.
Funding
No external funding was received.
Declaration of competing interest
None of the authors mentioned above received any financial support or other benefits from commercial sources for the work reported on in the manuscript, or any other financial interests which could create a potential conflict of interest or the appearance of a conflict of interest with regard to the work.
Acknowledgments
We thank Doctor Laurence Sanhes for the advice and help, and Doctor Léa Colombain for her support.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernard Stoecklin S., Rolland P., Silue Y., Mailles A., Campese C., Simondon A. First cases of coronavirus disease 2019 (COVID-19) in France: surveillance, investigations and control measures, January 2020. Euro Surveill Bull Eur Sur Mal Transm Eur Commun Dis Bull. 2020:25. doi: 10.2807/1560-7917.ES.2020.25.6.2000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020:200642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G. Detection of SARS-CoV-2 in different types of clinical specimens. J Am Med Assoc. 2020 May 12;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim H., Hong H., Yoon S.H. Diagnostic performance of CT and reverse transcriptase-polymerase chain reaction for coronavirus disease 2019: a meta-analysis. Radiology. 2020:201343. doi: 10.1148/radiol.2020201343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim E.S., Chin B.S., Kang C.K., Kim N.J., Kang Y.M., Choi J.P. Clinical course and outcomes of patients with severe acute respiratory syndrome coronavirus 2 infection: a preliminary report of the first 28 patients from the Korean cohort study on COVID-19. J Kor Med Sci. 2020;35:e142. doi: 10.3346/jkms.2020.35.e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J.-J., Dong X., Cao Y.-Y., Yuan Y.-D., Yang Y.-B., Yan Y.-Q. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020 Jul;75(7):1730–1741. doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- 8.Warny M., Helby J., Nordestgaard B.G., Birgens H., Bojesen S.E. Lymphopenia and risk of infection and infection-related death in 98,344 individuals from a prospective Danish population-based study. PLoS Med. 2018;15 doi: 10.1371/journal.pmed.1002685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan J.E., Beeson P.B. Experimental observations on the eosinopenia induced by acute infection. Br J Exp Pathol. 1971;52:214–220. [PMC free article] [PubMed] [Google Scholar]
- 10.Gil H., Magy N., Mauny F., Dupond J.-L. [Value of eosinopenia in inflammatory disorders: an “old” marker revisited] Rev Med Interne. 2003;24:431–435. doi: 10.1016/s0248-8663(03)00138-3. [DOI] [PubMed] [Google Scholar]
- 11.Bouldoires B., Gil H., Soumagne T., Humbert S., Meaux Ruault N., Magy Bertrand N. [A predictive bacterial infection score according to eosinophil level: an observational study] Rev Med Interne. 2018;39:10–16. doi: 10.1016/j.revmed.2017.10.425. [DOI] [PubMed] [Google Scholar]
- 12.Li Q., Ding X., Xia G., Chen H.-G., Chen F., Geng Z. Eosinopenia and elevated C-reactive protein facilitate triage of COVID-19 patients in fever clinic: a retrospective case-control study. E Clin Med. 2020;23:100375. doi: 10.1016/j.eclinm.2020.100375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mu T., Yi Z., Wang M., Wang J., Zhang C., Chen H. Expression of eosinophil in peripheral blood of patients with COVID-19 and its clinical significance. J Clin Lab Anal [Internet] 2020 Oct 29 doi: 10.1002/jcla.23620. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7645967/ Online ahead of print, Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davido B., Makhloufi S., Matt M., Calin R., Senard O., Perronne C. Changes in eosinophil count during bacterial infection: revisiting an old marker to assess the efficacy of antimicrobial therapy. Int J Infect Dis IJID Off Publ Int Soc Infect Dis. 2017;61:62–66. doi: 10.1016/j.ijid.2017.06.005. [DOI] [PubMed] [Google Scholar]