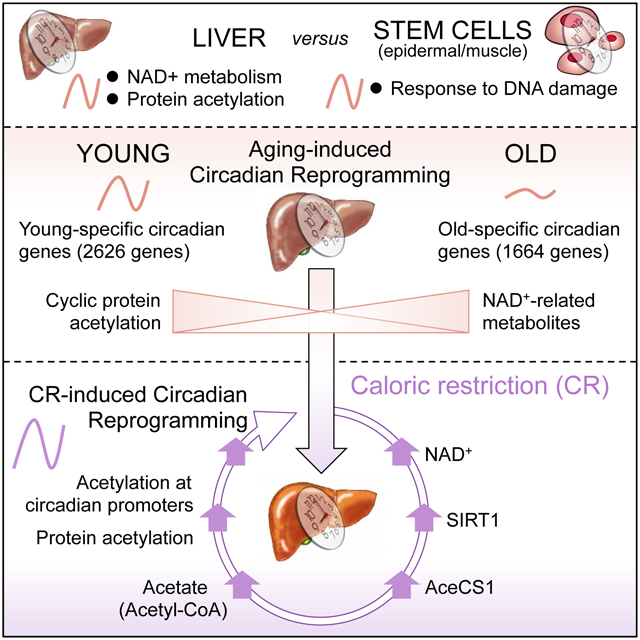
SUMMARY
The process of aging and circadian rhythms are intimately intertwined, but how peripheral clocks involved in metabolic homeostasis contribute to aging remains unknown. Importantly, caloric restriction (CR) extends lifespan in several organisms and rewires circadian metabolism. Using young versus old mice, fed ad libitum or under CR, we reveal reprogramming of the circadian transcriptome in the liver. These age-dependent changes occur in a highly tissue-specific manner, as demonstrated by comparing circadian gene expression in the liver versus epidermal and skeletal muscle stem cells. Moreover, de novo oscillating genes under CR show an enrichment in SIRT1 targets in the liver. This is accompanied by distinct circadian hepatic signatures in NAD+-related metabolites and cyclic global protein acetylation. Strikingly, this oscillation in acetylation is absent in old mice while CR robustly rescues global protein acetylation. Our findings indicate that the clock operates at the crossroad between protein acetylation, liver metabolism and aging.
Graphical Abstract
Etoc
Aging reprograms the circadian transcriptome in a highly tissue-specific manner
INTRODUCTION
It is commonly assumed that interplay exists between the circadian clock and the aging process and, more specifically, that changes in central rhythmic behavior are associated with age-dependent decline (Kondratova and Kondratov, 2012). In addition to a largely heterogeneous and anecdotal literature, it has been shown that neuronal activity rhythms display an age-dependent degradation in the suprachiasmatic nucleus (SCN) (Nakamura et al., 2011), which houses the central circadian pacemaker. This age-dependent decline in the central clock disrupts the physiological ability of mice to entrain to external light cues and has been reported to alter rhythmic behavior (Chang and Guarente, 2013; Sellix et al., 2012). On the other hand, a genetic mouse model has shown that deficiency in the core circadian transcription factor aryl hydrocarbon receptor nuclear-translocator-like (ARNTL or BMAL1) results in reduced lifespan and premature aging phenotype in mice (Kondratov et al., 2006). Yet, how aging influences the metabolic functions of peripheral clocks remains virtually unexplored.
Several hallmarks of aging-associated deterioration are linked to disruption of metabolic homeostasis, including deregulated nutrient sensing, mitochondrial dysfunction, reprogramming of cellular signaling pathways and insulin-resistance (Cartee et al., 2016; Fontana and Partridge, 2015; Houtkooper et al., 2012; Imai and Guarente, 2014; Ocampo et al., 2016; Verdin, 2015). Caloric restriction (CR) and fasting regimens have the unique ability to extend lifespan and enhance healthspan in a variety of organisms (Brandhorst et al., 2015; Grandison et al., 2009; Lee et al., 1999; Lin et al., 2002; Longo and Panda, 2016). Moreover, changes in nutritional availability of metabolic cues could be implicated in lifespan extension and attenuation of the aging-related phenotype in a clock-dependent manner (Gill et al., 2015; Patel et al., 2016a; Zwighaft et al., 2015), but the detailed molecular mechanisms remain unclear. Additionally, metabolic cues function as powerful zeitgebers for peripheral circadian clocks, and CR and time-restricted feeding have been linked to reorganizing peripheral circadian gene expression (Asher and Sassone-Corsi, 2015; Gill et al., 2015; Patel et al., 2016b; Stokkan et al., 2001). Importantly, the potential of the circadian clock to be reprogrammed by nutritional challenges has revealed its intrinsic plasticity (Eckel-Mahan et al., 2013). To date, the contribution of peripheral circadian metabolism in mediating the adaptation to macronutrient limitation that modulates lifespan extension is not mechanistically understood (Katewa et al., 2016; Ulgherait et al., 2016).
To unravel the molecular and metabolic pathways that underlie the interplay between circadian rhythms and aging, we have compared young versus old mice, fed ad libitum with either normal chow or under CR, and paralleled the high-throughput transcriptomic analyses of liver versus muscle and epidermal stem cells. Our findings reveal that aging operates by reprogramming the circadian transcriptome. We show that these age-dependent changes in circadian gene expression occur in a highly tissue-specific manner, as demonstrated by a comparative analysis of the liver transcriptional landscape versus epidermal and skeletal muscle stem cells (see accompanying study (Solanas et al.)). Our analysis of the circadian hepatic transcriptome revealed enriched biological pathways centered on protein acetylation and nicotinamide adenine dinucleotide (NAD+) metabolism, thus prompting subsequent examination. Strikingly, global protein acetylation was found to be highly circadian and this rhythmicity was dampened in aged mice. Also, CR resulted in a dramatic rescue of protein acetylation over the day/night cycle. Comparing the CR-dependent circadian transcriptome with our published SIRT1-dependent transcriptome (Masri et al., 2014), we identified an overlapping gene expression signature that linked SIRT1 with CR-mediated effects on circadian reprogramming in the liver. Further analysis by mass spectrometry revealed that CR regulates NAD+/SIRT1, which modulates the circadian acetylation of acetyl-CoA synthetase (AceCS1), a pathway known to control rhythmic nucleocytoplasmic acetyl-CoA production (Hallows et al., 2006; Sahar et al., 2014). These findings reveal age-dependent metabolic changes in the circadian hepatic transcriptome in young and old mice and link CR-dependent reorganization of circadian metabolism in the liver with NAD+ availability, SIRT1 activity and global protein acetylation over the circadian cycle.
RESULTS
Circadian Hepatic Transcriptome Reveals Aging- and CR-Induced Reprogramming
To determine how the hepatic clock regulates rhythmic metabolism during aging, young and old mice were subjected to a 30% calorie reduction or a normal chow feeding for 6-months (see Methods). Mice were sacrificed every 4 hours over the circadian cycle and hepatic transcriptomic analysis was performed. Venn diagrams and heatmaps display circadian genes selected using the nonparametric algorithm JTK_cycle (Hughes et al., 2010) and a p-value cutoff of 0.01 in young normal diet (YND) and old normal diet (OND) fed mice. Transcriptomics revealed 2626 genes (44.8%) exclusively oscillatory in YND and 1664 (28.4%) genes were only rhythmic in OND groups, while 1575 genes (26.9%) were circadian in both groups (Fig. 1A). Comparison of the amplitude of hepatic oscillatory genes in YND versus OND is shown in Fig. 1B. Heatmaps display circadian genes found exclusively in YND (left panel) versus OND (right panel) (Fig. 1C), and common rhythmic genes (Fig. S1A). The expression of core clock genes enriched in both YND and OND was confirmed. The expression of core clock genes and clock-controlled genes (CCGs) as well as the expression of core clock proteins is virtually unaltered when comparing young and old animals fed a normal diet (Fig. 1D, and S2A and D).
Fig. 1. Circadian transcriptome analysis in the liver: effect of aging and caloric restriction.
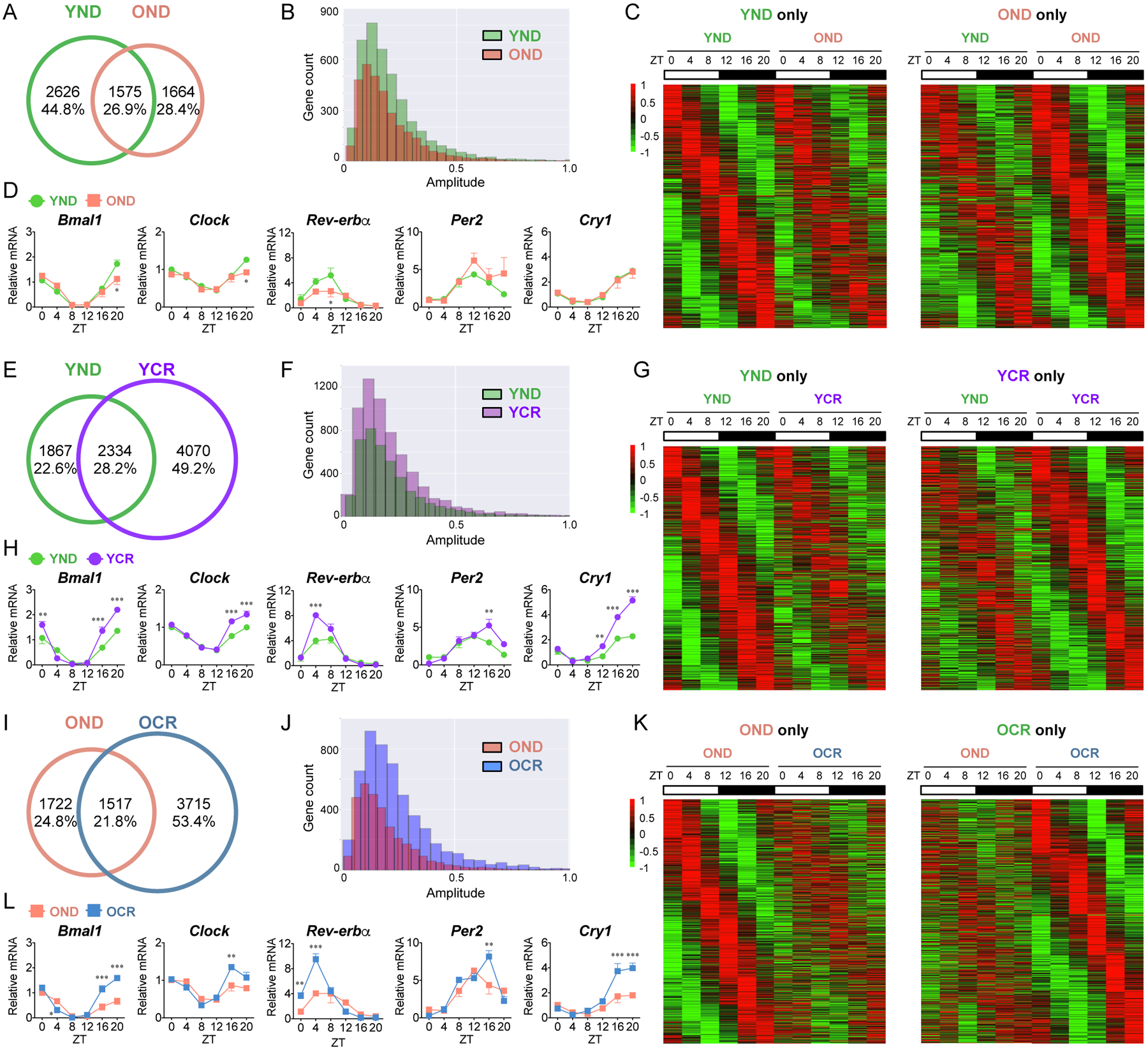
(A) Venn diagram displays the total number (top) and ratio (bottom) of rhythmic genes in the liver from young mice fed a normal diet (YND, green) and old mice fed a normal diet (OND, orange), including common genes. (B) Amplitude plot represents the distribution of oscillatory genes in YND and OND. (C) Heatmaps display rhythmic genes exclusively in YND (left panel) and OND group (right panel). (D) Circadian gene expression of core clock genes, Bmal1, Clock, Rev-erbα, Per2, and Cry1 in YND and OND liver was determined by qPCR. (E) Venn diagram displays hepatic rhythmic genes from YND (green), young mice subjected to CR (YCR, purple), and common genes. (F) Amplitude distribution of rhythmic genes in YND and YCR. (G) Heatmaps display circadian genes found exclusively in YND (left panel) and YCR (right panel). (H) Core clock gene expression in YND and YCR by qPCR. (I) Venn diagram represent circadian genes from OND (orange), old mice subjected to CR (OCR, blue), and common rhythmic genes. (J) Amplitude distribution of circadian genes in OND and OCR. (K) Heatmaps display circadian genes found exclusively in OND (left panel) and OCR (right panel). (L) Determination of core clock gene expression in OND and OCR by qPCR. Data represent mean±SEM and was analyzed by two-way ANOVA using Bonferroni posttest. *, **, and *** indicate P<0.05, P<0.01, and P<0.001, respectively (YND, n=4–5 per time point; OND, n=3–4 per time point; YCR, n=5–6 per time point; OCR, n=3–5 per time point). See also Figure S1, S2, S3, and S4.
To determine if CR can reorganize circadian gene expression in the liver, CR was applied to young (YCR) and old (OCR) mice and the circadian hepatic transcriptomes were compared to young and old mice fed a normal diet. Transcriptomic analysis identified 1867 genes (22.6%) in YND and 4070 genes (49.2%) in YCR to be circadian based on a 0.01 p-value cutoff criteria, and 2334 genes (28.2%) were found to be commonly rhythmic in both groups (Fig. 1E). The amplitude of hepatic rhythmic genes is enhanced in YCR mice, as compared to YND (Fig. 1F). Heatmaps display circadian genes found exclusively in YND (left panel) versus YCR (right panel) (Fig. 1G), as well as common circadian genes (Fig. S1B). Though core clock and CCGs expression profiles are rhythmic in both YND and YCR groups, CR enhances the expression of clock genes (Fig. 1H and S2B). The dynamic response in oscillating genes found in YCR demonstrates the sensitivity of the circadian hepatic clock to macronutrient limitation.
Similarly, old mice were subjected to CR and transcriptomic analysis was performed. 1722 genes (24.8%) were found to be circadian in OND, 3715 genes (53.4%) were oscillatory in OCR group, and 1517 genes (21.8%) were commonly rhythmic in both groups (Fig. 1I). Similar to YCR, the amplitude of hepatic rhythmic genes is enhanced in OCR mice, as compared to OND (Fig. 1J). Heatmaps display circadian genes found exclusively in OND (left panel) versus OCR (right panel) (Fig. 1K), as well as common circadian genes (Fig. S1C). A significant increase in core clock and CCGs expression profiles was found in OCR versus OND fed mice (Fig. 1L and S2C). Overall, the effect of CR in both young and old mice is evident as a significant portion of the circadian hepatic genome is rhythmic in response to nutrient deprivation.
Gene ontology (GO) analysis of the oscillatory transcriptome shows that sterol metabolic process, mitogen-activated protein kinase (MAPK) cascade, toll-like receptor (TLR) signaling and glycerolipid metabolism are highly enriched In the YND group (Fig. 2A). Both YND and OND categories are enriched in histone/chromatin modifications, while OND is specifically enriched in protein deubiquitination and cell cycle G1/S transition biological pathways (Fig. 2A). Moreover, as compared to YND, the YCR transcriptome shows highly significant biological function enrichment in genes involved in RNA processing, chromatin modification and peptidyl-lysine modification (Fig. 2B). Similar to the YCR group, GO pathway analysis does reveal that the OCR category of genes is highly enriched in chromatin modification, RNA processing, autophagy and ribosome biogenesis (Fig. 2C). Common genes in YND, OND, YCR and OCR groups contain an enrichment in circadian regulation of gene expression and NAD+ metabolism or biosynthesis (Fig. S1, D–F). Moreover, a comparison between the YCR and OCR groups revealed a 45.8% overlap of 3653 common genes among these two categories (Fig. S3), including consistence in the expression of core clock gene between YCR and OCR groups (Fig. S3E).
Fig. 2. GO ontology (GO) analysis of rhythmic genes from the circadian hepatic transcriptome.
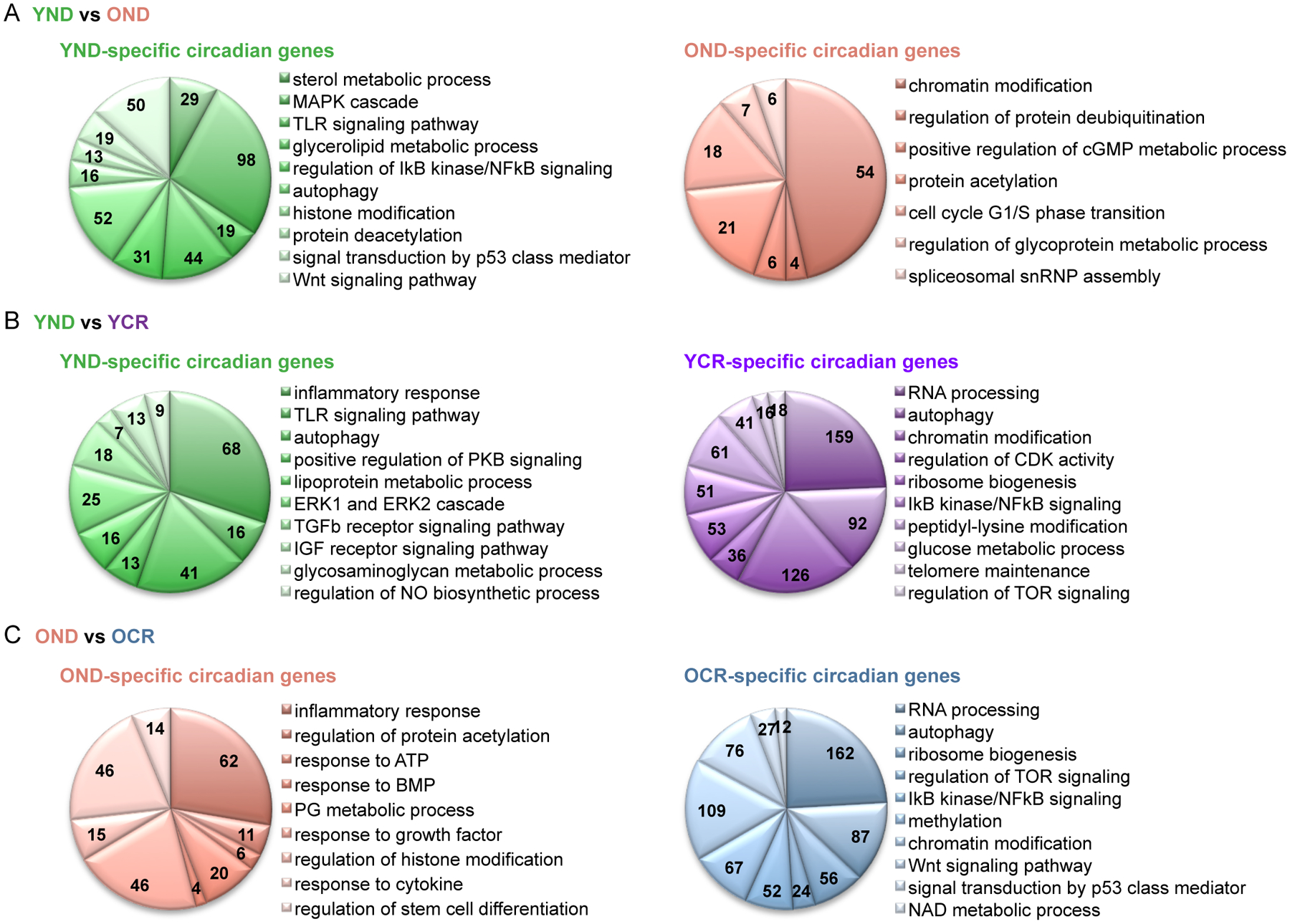
(A) GO analysis of genes exclusively circadian in YND (left, green) and OND (right, orange). Numbers within the pie charts indicate number of circadian genes identified within each biological process, using a p-value cutoff of 0.01. (B) GO analysis of genes exclusively circadian in YND (left, green) versus YCR (right, purple). (C) GO analysis of genes exclusively circadian in OND (left, orange) versus OCR (right, blue). 3–6 biological replicates per time point were subjected to the hepatic transcriptomic analysis. See also Figure S1, S3, and S4.
The circadian hepatic transcriptome and resulting GO analysis reveal a large set of genes with dampened oscillation by aging whereas some genes gain oscillation by CR (Fig. S2, E–G). Remarkably, circadian genes involved in RNA processing and autophagy are highly enriched both in YCR and OCR groups (Fig. 2, B and C, and Data Table S1). Therefore, the question arises: does CR reorganize the circadian transcriptome in the liver? We compared the circadian transcriptome between YND and OCR revealing only 24% overlap of 1824 common genes in the liver (Fig. S4). However, GO analysis uncovered that some of the circadian biological signatures of CR such as autophagy, RNA processing, and target of rapamycin (TOR) signaling are identified within “common” circadian signatures between YND and OCR (Fig. S4D). These observations show that CR reprograms the circadian hepatic transcriptome.
Distinct Phase Signatures of Aging and CR in the Liver as compared to Stem Cells
Additionally, the hepatic transcriptome was analyzed for phase of circadian gene expression, revealing that two sets of circadian genes peaking around zeitgeber time (ZT) 0–6 (phase 1) and ZT 12–18 (phase 2) in both YND and OND liver (Fig. 3A). Interestingly, new peaks in phase of gene expression are evident in YCR and OCR groups, which is likely reflective of feeding anticipatory behavior in calorically restricted mice (Fig. 3A). In support of this notion, the phase of circadian genes is indeed advanced by CR (Fig. 3B). Comparative analysis of phase 3 (ZT 8–16) rhythmic genes of YCR/OCR shows they are enriched in phase 2 (ZT12–18) of YND/OND and advanced in ZT (Fig. 3B). Similarly, circadian genes of YCR/OCR at phase 4 (ZT 20–2) are enriched in phase 1 (ZT 0–6) of YND/OND (Fig. 3B). This data indicates that CR functions to broaden the phase of circadian gene expression over the day/night cycle. In addition to liver, epidermal (EpSCs) and skeletal muscle stem cells (MsSCs) were harvested from young and old mice fed normal diet or CR (see accompanying study (Solanas et al.)). Tissue-specific comparison of phase of gene expression was performed using genes selected to be circadian by JTK_cycle (p-value cutoff of 0.01). Remarkably, similar phase profiles were observed in MsSCs and liver, likely reflective of the metabolic and nutrient responsive nature of these cells (Fig. 3C). Also, EpSCs were altered in phase of gene expression in old mice, but in mice subjected to CR this phase was reverted to a ‘young’ profile, consisting of two sets of genes peaking in the day and the night, respectively (Fig. 3C). The phase profiles suggest that mice fed a CR diet display a reorganization of the transcriptome, and this phase distribution is reflective of tissue-specific function.
Fig. 3. Distinct phase signature of circadian hepatic genes.
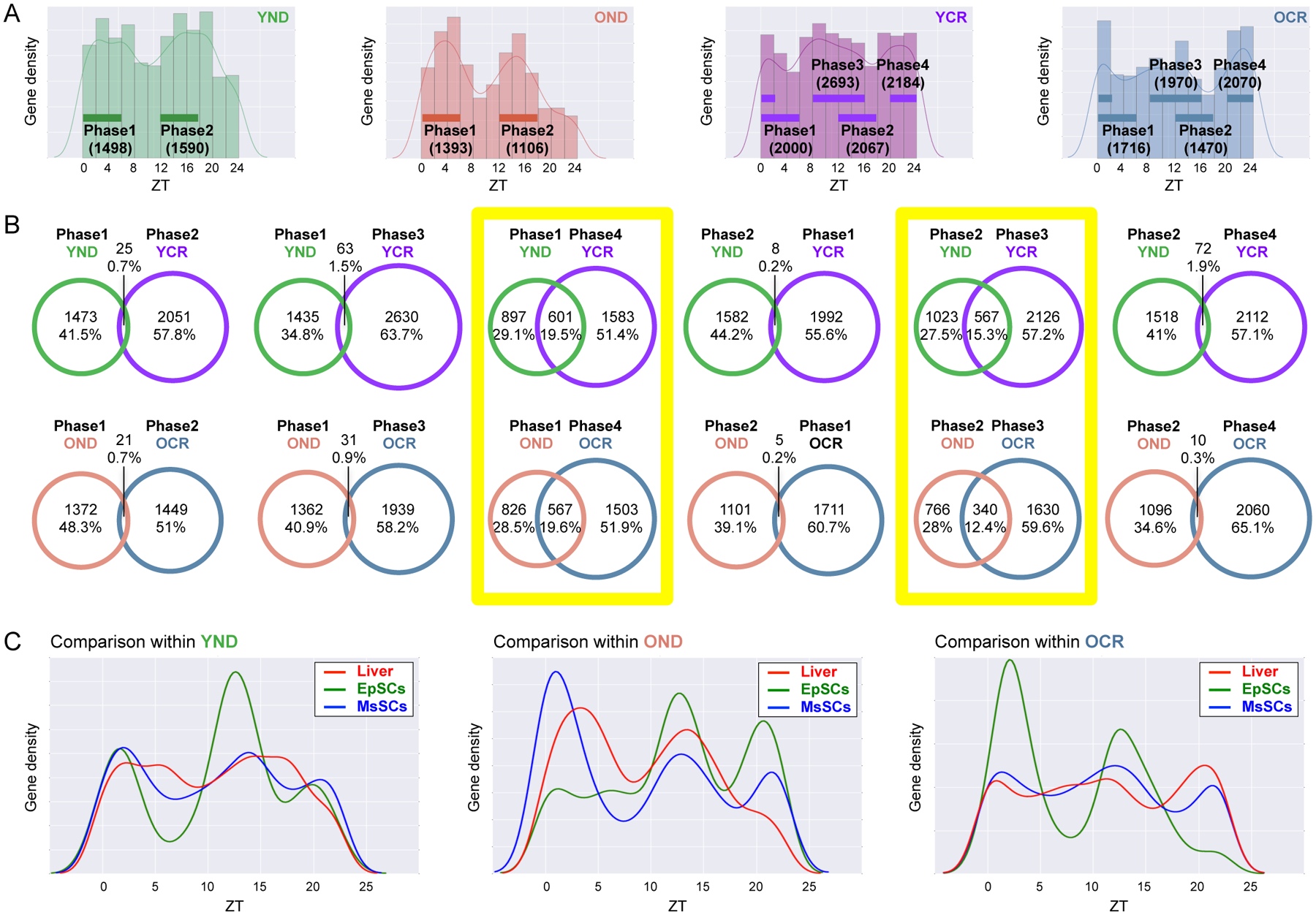
(A) Phase analysis of YND (green), OND (orange), YCR (purple), and OCR (blue) liver-specific oscillating genes selected to be circadian by JTK_cycle, based on a p-value cutoff of 0.01. Phase 1, 2, 3 and 4 indicate ZT 0–6, ZT 12–18, ZT 8–16, and ZT 20–2, respectively. (B) Venn diagrams display the total number (top) and ratio (bottom) of phase-specific rhythmic genes in YND, OND, YCR, and OCR. Phase 1, 2, 3 and 4 correspond to ZT 0–6, ZT 12–18, ZT 8–16, and ZT 20–2, respectively. Yellow squares highlight significant overlap between YND versus YCR, and between OND versus OCR. (C) Tissue-specific phase comparison of gene expression was performed using JTK_cycle, based on p-value cutoff of 0.01. Red lines, green lines, and blue lines indicate the phase of rhythmic genes in the liver, EpSCs, and MsSCs, respectively. 3–6 biological replicates per time point per tissue were subjected to the transcriptomic analysis.
Unique Signatures in Circadian NAD+ Metabolism and Protein Acetylation in the Liver
To further compare the tissue-specific circadian signature, we carried out GO analysis. The Venn diagram reveals overlapping genes among the three cell types, including 74 genes in common that comprise the tissue-independent circadian genes in liver, EpSCs and MsSCs from YND mice (Fig. 4A). The EpSCs transcriptome from YND mice reveals a highly significant enrichment in genes involved in response to DNA damage, DNA replication, and DNA repair as well as skin epidermal development (data shown in accompanying study (Solanas et al.). These data were further validated (Solanas et al.) to reveal a circadian reprogramming that occurs in OND versus YND, whereby aged EpSCs conversely lose oscillations of epidermal homeostasis genes while gaining oscillation in genes mediating a stress response to DNA damage and inflammation. Similarly, MsSCs from YND mice display a unique biological function signature that involves genes implicated in skeletal muscle development, glucose homeostasis, and response to insulin (see accompanying study (Solanas et al.)). Comparing both EpSCs and MsSCs from YND mice, a unique stem cell gene signature is uncovered, which includes cellular response to DNA damage stimulus, DNA replication, DNA repair and regulation of protein deacetylation (Fig. 4A). In contrast, a comparison of stem cells versus liver reveals a unique set of genes specifically enriched in the liver implicated in sterol metabolism, fatty acid biosynthetic process, NAD+ biosynthetic process, cellular ketone metabolism, histone H3 acetylation and peptidyl-lysine acetylation (Fig. 4A). Not surprisingly, genes involved in rhythmic processes are common in liver, EpSCs and MsSCs and are listed as tissue-independent circadian genes (Fig. 4A). Lastly, comparing the numbers of circadian genes identified in the tissue-independent group, we observe a robust induction of rhythmic genes in YCR and to a lesser extent in the OCR group (Fig. 4B). Similarly, GO analysis shows that a variety of biological processes are enriched exclusively in CR-fed mice. Among these, histone acetylation and peptidyl-lysine acetylation are prominent (Fig. S5A). These findings indicate the profound and tissue-specific effect of CR on reprogramming the circadian transcriptome in liver, EpSCs and MsSCs. This reorganization involves a robust induction of de novo oscillating genes as a compensatory response to nutritional deprivation.
Fig. 4. Tissue-specific circadian signatures identified by transcriptomic analysis.
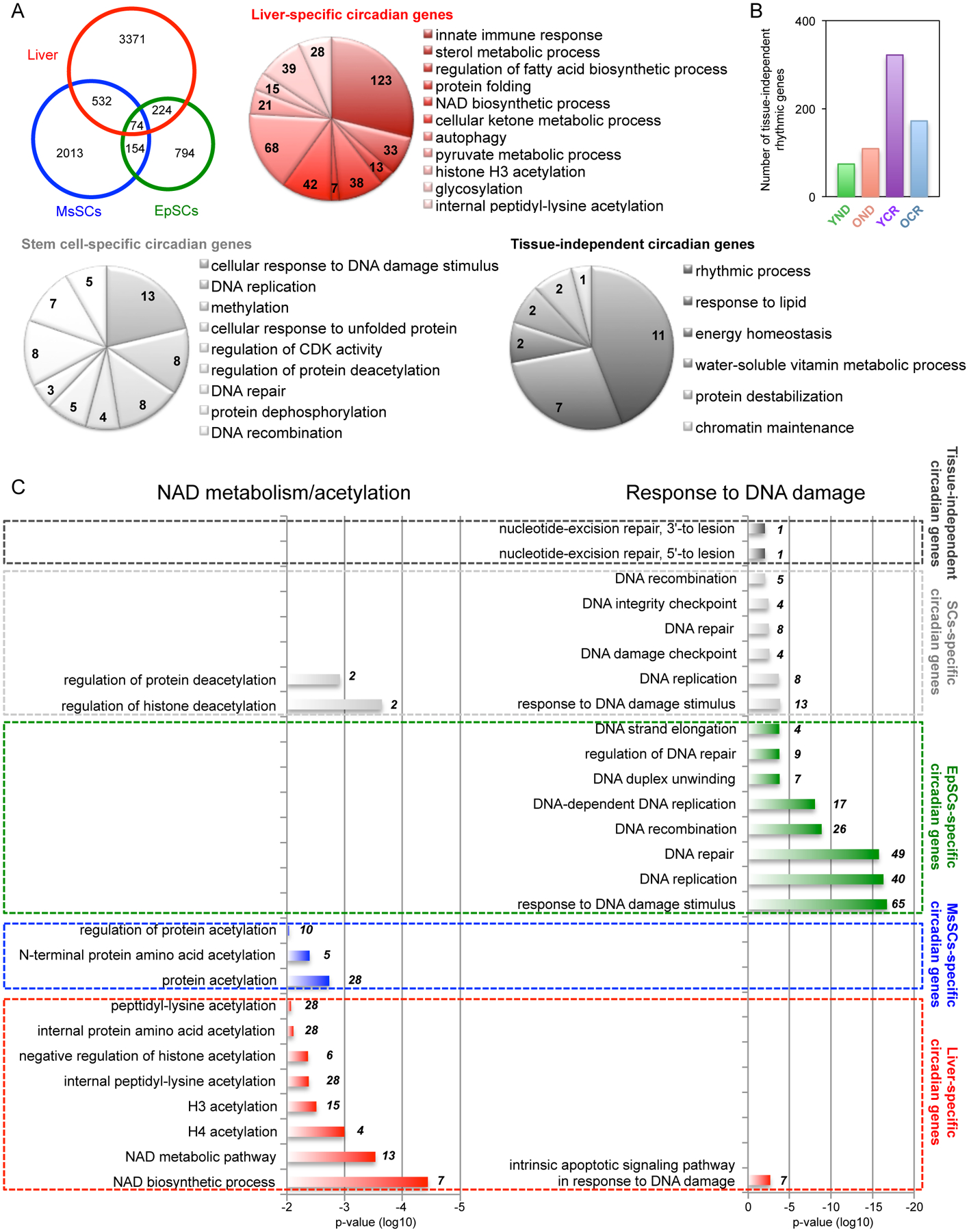
(A) Venn diagram displays the number of genes selected to be circadian by JTK_cycle, based on p-value cutoff of 0.01 in each tissue/cell including liver (red), EpSCs (green), and MsSCs (blue). Pie charts display GO terms of genes exclusively circadian in liver (red). Common circadian genes in EpSCs and MsSCs are indicated (light gray) versus rhythmic genes in all three tissues/cells (dark gray). (B) Histogram represents the number of tissue-independent common circadian genes in each group. (C) Histogram represents the significance of each biological process and numbers within the histogram indicate number of circadian genes identified within each biological process. GO terms regarding NAD metabolism/acetylation processes specifically identified by liver-specific circadian genes (left), while GO terms regarding DNA damage responses specifically identified by EpSCs-specific circadian genes (right). Top eight GO terms regarding DNA damage responses identified by EpScs-specific circadian genes were listed. p-value cutoff of 0.01 was used. 3–6 biological replicates per time point per tissue were subjected to the transcriptomic analysis. See also Figure S5.
The circadian hepatic transcriptome illustrated the relevance of lysine acetylation (Fig. 2 and 4A). Control of acetylation is involved in the aging process as well as CR nutritional challenge (Peleg et al., 2016; Verdin and Ott, 2015). This notion was further strengthened by the remarkable significance found for all metabolic pathways implicated in NAD+ metabolism and protein acetylation in a comparison with the EpSCs and MsSCs transcriptome (Fig. 4C). Indeed, these metabolic pathways are not enriched in EpSCs whereas are present in MsSCs, pointing to metabolic properties as elements of distinction among SCs. Remarkably, the EpSCs transcriptome identified response to DNA damage and DNA repair as an EpSCs- and stem cells-dependent distinct circadian biological signatures (Fig. 4C). These observations indicate that circadian reprogramming of physiological homeostasis due to aging or CR is tissue-specific.
CR Rescues the Aging-Dependent Decline in Global Protein Acetylation in the Liver
Based on circadian transcriptomic evidence that NAD+ metabolism and protein acetylation identify significant liver-specific metabolic pathways, we analyzed liver protein lysates with a pan-acetyl-lysine antibody over the circadian cycle. A striking circadian oscillation was observed in global protein acetylation in YND mice, with a peak at ZT8 (Fig. 5A). This finding is in keeping with previous results by mass spectrometry showing that global protein acetylation is rhythmic and dependent on an intact circadian clock (Masri et al., 2013). Notably, this cycle in protein acetylation was completely absent in the aging OND mice (Fig. 5A and S6A). The age-dependent rhythm of protein acetylation prompted us to investigate the effect of CR. In this respect it is notable that CR results in increased mitochondrial protein acetylation in liver (Hebert et al., 2013). Importantly, under both YCR and OCR, an extensive global protein hyperacetylation was observed in comparison to normal diet-fed mice (Fig. 5B and S6A).
Fig. 5. Hepatic cyclic protein acetylation and protein hyperacetylation under CR.
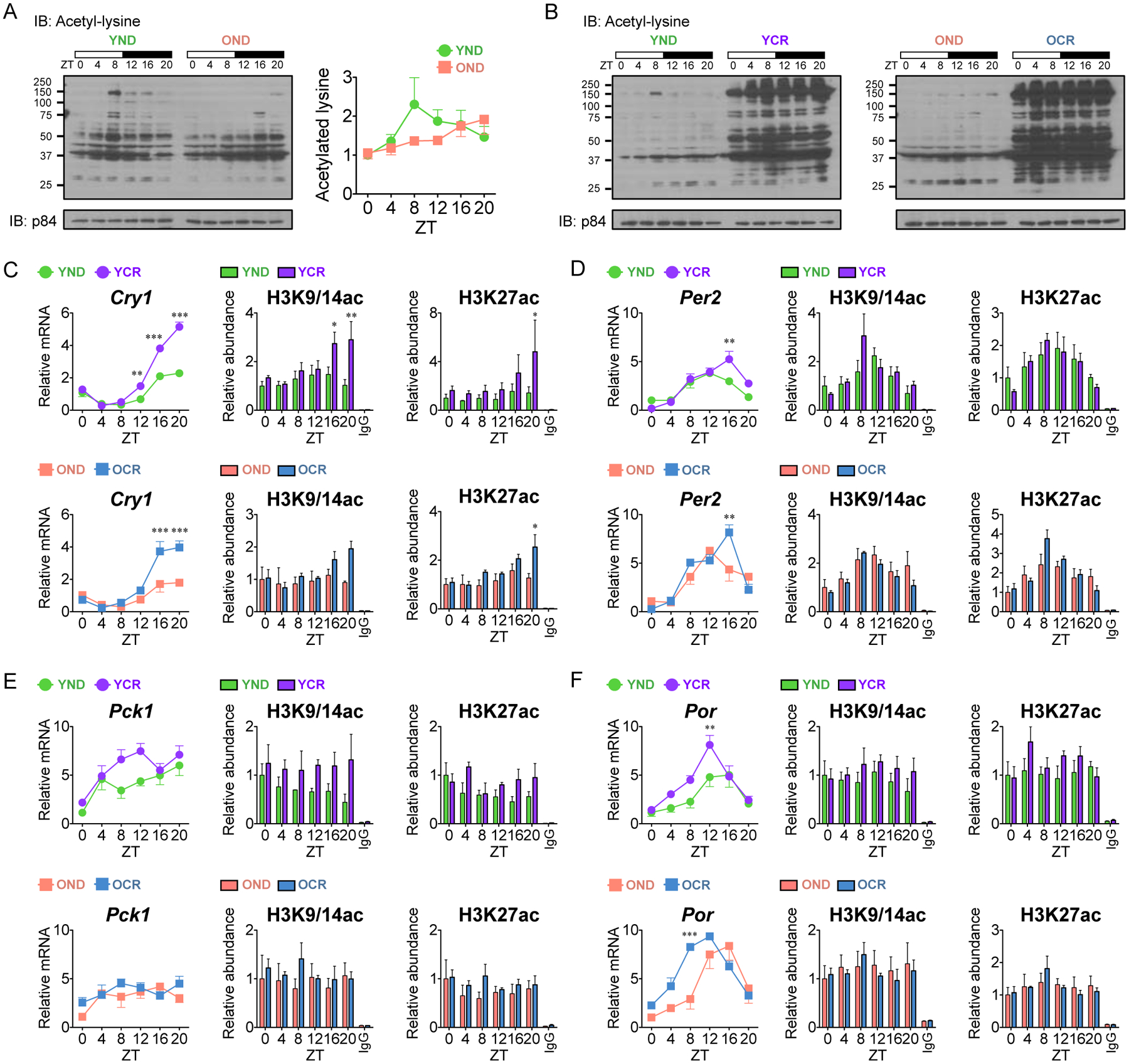
(A and B) Circadian lysates from livers of YND, OND, YCR, and OCR were probed with a pan acetylated lysine antibody by western blot and quantified using three independent samples. p84 was used as a loading control. (C-F) Abundance of acetylated H3K9/K14 and H3K27 was determined by ChIP analysis using YND (green) versus YCR (purple) liver and OND (orange) versus OCR (blue) liver at Cry1 (C), Per2 (D), Pck1 (E), and Por (F) promoters (YND, n=4–5 per time point; OND, n=3–4 per time point; YCR, n=5–6 per time point; OCR, n=3–5 per time point). IgG was used as a negative control. Corresponding gene expression as determined by qPCR is shown on the left. Data represent mean±SEM and was analyzed by two-way ANOVA using Bonferroni posttest. *, **, and *** indicate P<0.05, P<0.01, and P<0.001, respectively. See also Figure S6 and S7.
Accumulation of Histone Acetylation at Circadian Hepatic Promoters by CR
Next, we sought to establish whether the effect of CR on global protein hyperacetylation would extend to histone acetylation and corresponding gene expression. Using EnrichR (http://amp.pharm.mssm.edu/Enrichr/), a comprehensive gene set enrichment analysis web tool, a comparative analysis was performed with our circadian CR transcriptome and published chromatin immunoprecipitation (ChIP)-sequencing datasets for several histone modifications (Chen et al., 2013; Kuleshov et al., 2016). A significant enrichment of H3K9ac and H3K27ac was specifically identified in the YCR/OCR versus YND/OND specific gene lists (Fig. S7). Additionally, we observe an induction of amplitude of clock genes in YCR and OCR versus normal diet-fed mice (Fig. 1, H and L and S2, B and C), which would suggest changes in histone acetylation marks linked to active transcription. Additionally, the levels of hepatic β-hydroxybutyrate are dampened at ZT 4 in the liver from calorie-restricted diet fed mice (Fig. S6B).
Indeed, a significant accumulation of histone H3K9/K14ac and H3K27ac that perfectly matches the gene expression profile was found at the cryptochrome1 (Cry1) gene transcriptional start site (TSS) in YCR versus YND and OCR versus OND (Fig. 5C). A similar trend was also observed at the period2 (Per2) TSS with an increase in H3K9/K14ac and H3K27ac levels in response to CR in YCR and OCR, respectively (Fig. 5D). Similarly, the CR-dependent transcriptome in YCR and OCR mice reveals circadian transcriptional reprogramming of large numbers of genes that become newly oscillatory in response to nutrient deprivation (Fig. 1, E–L). In keeping with this concept, a number of rhythmic metabolic genes are enhanced in gene expression by CR. ChIP analysis revealed that an accumulation of H3K9/K14ac and H3K27ac was seen at the phosphoenolpyruvate carboxykinase1 (Pck1) and cytochrome P450 oxidoreductase (Por) promoters in YCR versus YND, a similarly observed trend that was less pronounced in old mice under CR (Fig 5, E and F). Thus, CR triggers an extensive reorganization of protein acetylation that includes histone marks linked to activation of gene expression.
Liver-Specific Elevation of NAD+ and Related Metabolites
NAD+ and its biosynthetic pathway through the enzyme nicotinamide phosphoribosyltransferase (NAMPT) is a critical regulator of aging, and rescuing the levels of NAD+ is reported to modulate lifespan and/or healthspan extension (Belenky et al., 2007b; Gomes et al., 2013; Mouchiroud et al., 2013; Stein and Imai, 2014; Zhang et al., 2016). Also, CR has been previously shown to be dependent on the biosynthesis of NAD+ and the effects of CR may be tied to the cellular NAD+ (oxidized)/NADH (reduced) ratio and/or the level of NAD+ (Lin et al., 2000; Lin et al., 2004; Lin et al., 2002). Several NAD+-dependent deacetylases are critically involved in facilitating the effects of nutrient deprivation, namely the sirtuins SIRT1 (Chen et al., 2005; Cohen et al., 2004; Rodgers et al., 2005) and mitochondrial SIRT3 (Hebert et al., 2013; Hirschey et al., 2010; Shimazu et al., 2010; Someya et al., 2010). Importantly, an interplay exist between aging, nutritional challenge and circadian metabolism as illustrated by the role played by mammalian SIRTs in circadian control of both transcription and metabolism in the liver (Asher et al., 2008; Masri et al., 2014; Masri and Sassone-Corsi, 2014; Nakahata et al., 2008), but also the cyclic availability of NAD+ (Nakahata et al., 2009; Peek et al., 2013; Ramsey et al., 2009) and acetyl-CoA (Sahar et al., 2014).
We therefore quantitatively measured the hepatic NAD+ metabolome to determine if aging or CR alters the circadian control of NAD+ metabolism. Mass spectrometry analysis of NAD+-related metabolites is displayed as a heatmap and reveals dynamic changes of each metabolite in YND, YCR, OND, and OCR (Fig. 6A). Levels of NAD+ are elevated in the liver from calorie-restricted animals compared to normal diet-fed animals (Fig. 6B). Similarly, a significant increase of nicotinamide adenine dinucleotide phosphate (NADP+) levels in CR group versus ND group was found at both ZT 0 and ZT 12 (Fig. 6B). Upregulation of the levels of nicotinamide riboside (NR) and nicotinamide mononucleotide (NMN) under CR at ZT12 results in robust diurnal changes of these metabolites in YCR and OCR (Fig. 6B). This increase in NR and NMN levels suggests that CR leads to activation of NAD+ turnover through NMN to circulate NR (Bogan and Brenner, 2008). Levels of the metabolites produced by NAD+-consuming enzymes, nicotinamide (NAM) and ADP ribose (ADPR) (Belenky et al., 2007a), are elevated in OND liver harvested at ZT 0 (Fig. 6B). Finally, CR suppresses the levels of nicotinic acid adenine dinucleotide (NAAD) (Fig. 6B), a metabolite that serves as a biomarker of NAD+ reserves (Trammell et al., 2016) and an intermediate of the NAD+ de novo synthesis pathway. These findings suggest that aging increases NAD+ turnover while CR sharpens circadian control of NAD+ metabolism, thereby boosting NAD+, NADP+ and potentially the circulation of NR. These results reveal the dependence of the NAD+ salvage pathway under CR rather than de novo NAD+ synthesis and show that aging and CR reorganize the circadian NAD+ salvage pathway (Fig. 6C).
Fig. 6. Elevation of hepatic NAD+ and related metabolites by CR.
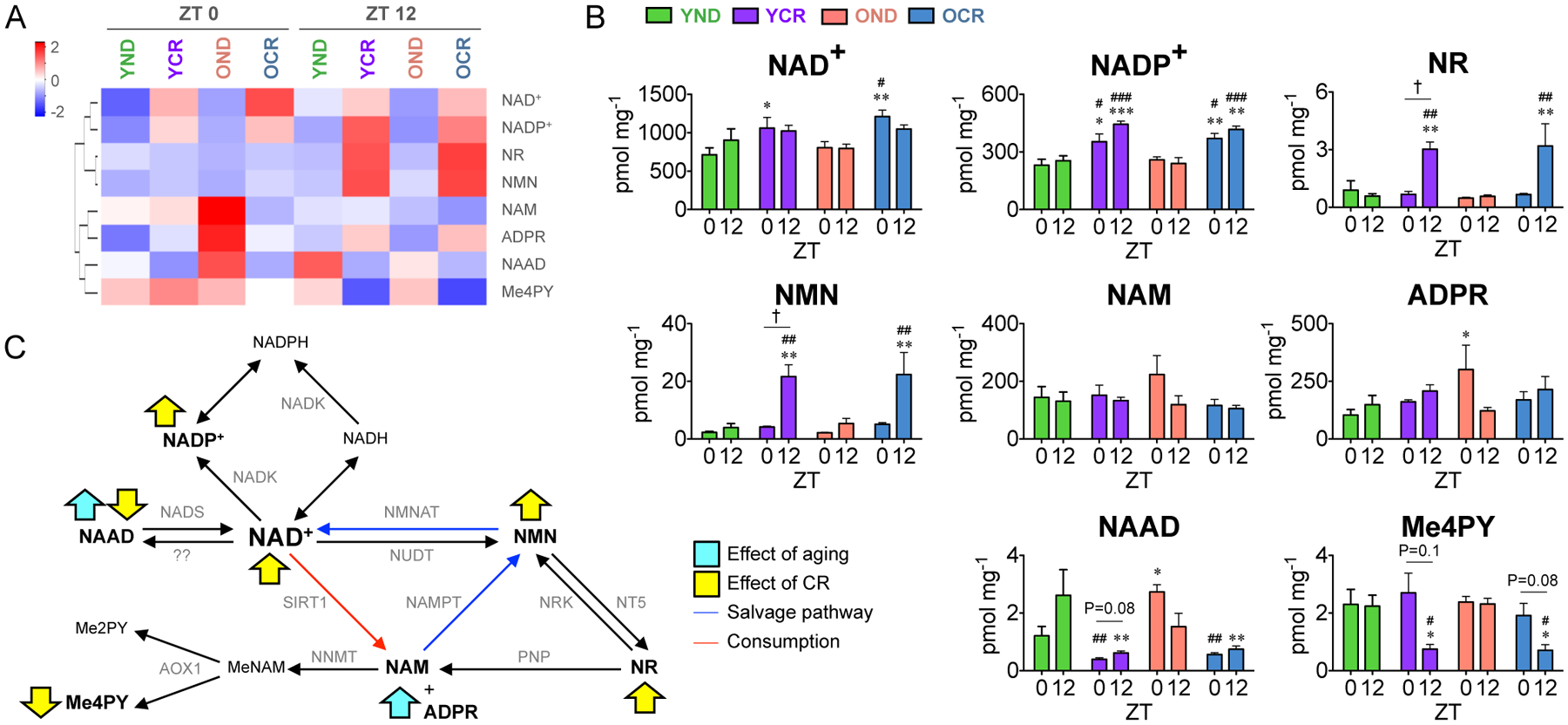
(A) Heatmap displays the levels of NAD+ and related metabolites in YND (n=3 per time point), YCR (n=3 per time point), OND (n=3 per time point), and OCR (n=3 per time point) groups at ZT 0 (left) and ZT 12 (right). (B) Histograms show the concentrations of NAD+ metabolites (NAD+, NADP+, NR, NMN, NAM, ADRP, NAAD, and Me4PY). (C) Schematic summary of the impact of aging (blue arrows) and CR (yellow arrows) on the NAD+ salvage pathway. Data represents mean±SEM from three independent samples and was analyzed by two-way ANOVA using Bonferroni posttest. *, **, and *** indicate P<0.05, P<0.01, and P<0.001 versus YND at corresponding time points, respectively. #, ##, and ### indicate P<0.05, P<0.01, and P<0.001 versus OND at corresponding time points, respectively. † indicates P<0.05 between ZT 0 and ZT 12, respectively. Abbreviations: AOX1, aldehyde oxidase1; Me2PY, N-methyl-2-pyridone-5-carboxamide; Me4PY, N-methyl-4-pyridone-5-carboxamide; MeNAM, methylnicotinamide; NAAD, nicotinic acid adenine dinucleotide; NAD+/NADH, oxidized/reduced nicotinamide adenine dinucleotide; NADK, nicotinamide adenine dinucleotide kinase; NADP+/NADPH, oxidized/reduced nicotinamide adenine dinucleotide phosphate; NADS, nicotinamide adenine dinucleotide synthase; NAM, nicotinamide; NAMPT, nicotinamide phosphoribosyltransferase; NMN, nicotinamide mononucleotide; NMNAT, nicotinamide/nicotinic acid mononucleotide adenylyltransferase; NNMT, NAM N-methyltransferase; NR, nicotinamide riboside; NRK, nicotinamide riboside kinase; NT5, 5-nucleotidase; NUDT, nudix hydrolase; PNP, purine nucleoside phosphorylase; SIRT1, sirtuin1.
CR Activates NAD+-Dependent SIRT1 leading to Circadian Hepatic Acetyl-CoA Metabolism
Given the changes in NAD+ related metabolites under CR, we sought to determine if SIRT1 is activated and thereby identify its downstream targets. We subsequently compared the SIRT1-dependent circadian transcriptome (Masri et al., 2014) with the YND, YCR, OND and OCR datasets from the present study. A 5-fold enrichment in circadian SIRT1-target genes was found in CR groups, as the number of overlapping genes increased from 84 to 435 and 87 to 426 in YCR and OCR, respectively, versus normal diet-fed mice (Fig. 7A and Data Table S2). The expression of Nampt, a clock-controlled gene that encodes the rate-limiting enzyme of the NAD+ salvage pathway (Nakahata et al., 2009; Ramsey et al., 2009), was found to be elevated in mice subjected to CR (Fig. 7B) (Lin et al., 2000; Lin et al., 2004; Lin et al., 2002). The enrichment of SIRT1-dependent targets, especially at ZT 8, indicates an activation of SIRT1 under CR, which also correlates with the increased expression of Nampt at ZT 8 (Fig. 7B). As an additional readout of SIRT1 activity, acetylation at lysine 537 of BMAL1 has been previously demonstrated as a SIRT1-specific deacetylation site (Hirayama et al., 2007). As expected, CR suppresses the acetylation of BMAL1 (Fig. 7C), which supports our hypothesis that CR activates SIRT1.
Fig. 7. CR activates SIRT1 and modulates circadian acetyl CoA metabolic pathways.
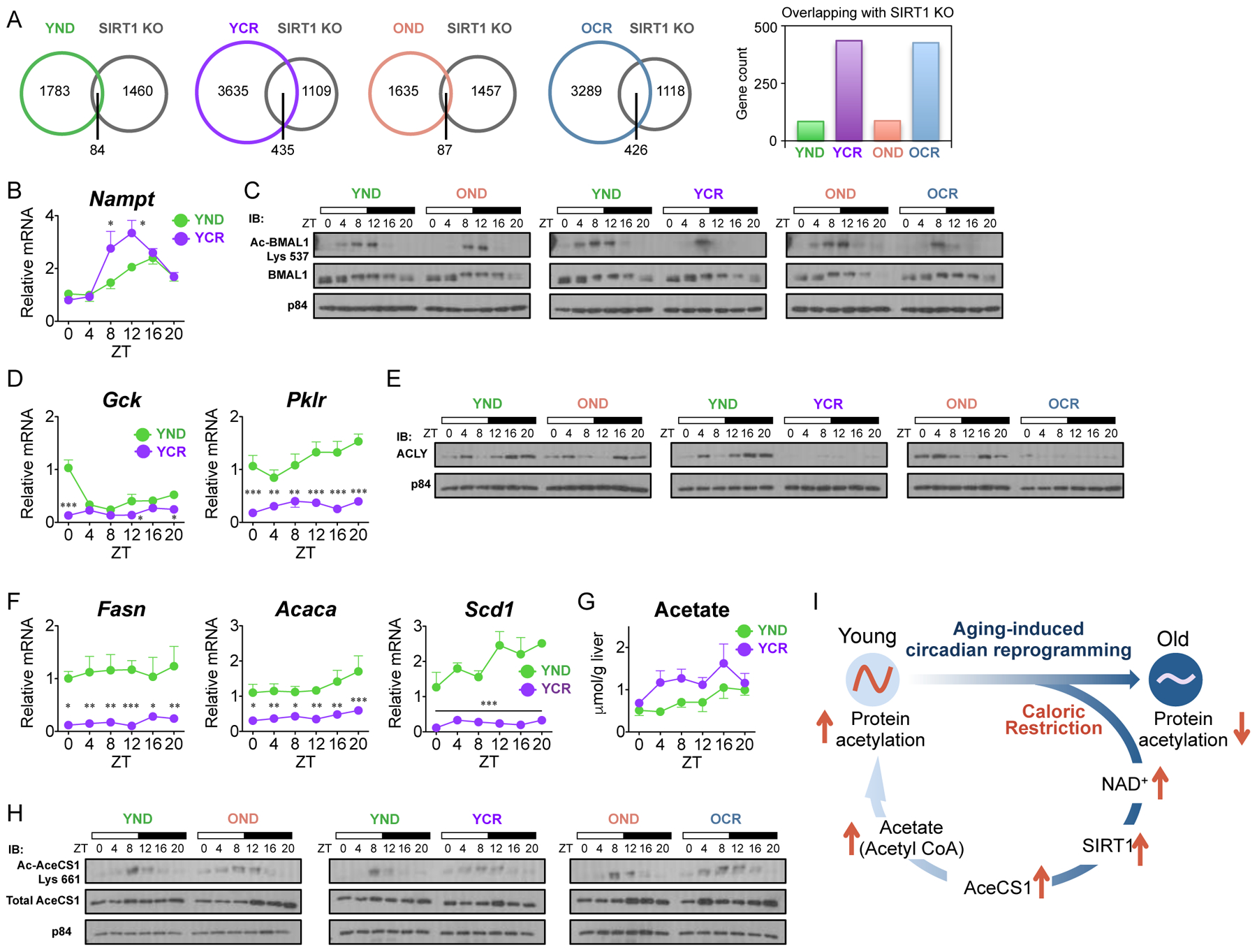
(A) Venn diagrams display the overlap between hepatic SIRT1-dependent circadian genes (Masri et al., 2014) and rhythmic genes in YND (green), OND (orange), YCR (purple), and OCR (blue). Histogram displays the number of overlapping circadian genes in each group. JTK_cycle was used for the selection of circadian genes, based on p-value cutoff of 0.01, for both transcriptome datasets. (B) Circadian gene expression of Nampt, the rate-limiting enzyme of the NAD+ salvage pathway. (C) Acetylated BMAL1 from liver whole cell extracts was determined by western blot. p84 was used as a loading control. (D) Gene expression of glycolytic enzymes, Gck and Pklr, compared between YND (green) and YCR (purple). (E) ACLY protein levels in liver nuclear extracts were determined by western blot, compared to the p84 loading control. (F) Gene expression profiles of SREBP-dependent lipogenic target genes, Fasn, Acaca, and Scd1. (G) Cellular acetate concentrations in liver as measured by colorimetric assay. (H) Circadian acetylation of AceCS1 levels as determined by western blot using liver nuclear extracts from YND, OND, YCR, and OCR. p84 was used as a loading control. (I) Scheme represents the circadian reorganization of CR-dependent metabolism which may rescue age-associated circadian decline in the liver. Western blot images represent a minimum of three independent experiments. Data represent mean±SEM and was analyzed by two-way ANOVA using Bonferroni posttest. *, **, and *** indicate P<0.05, P<0.01, and P<0.001, respectively (YND, n=4–5 per time point; OND, n=3–4 per time point; YCR, n=5–6 per time point; OCR, n=3–5 per time point).
We further explored the role of the hepatic metabolic clock in a CR-dependent model of aging. Glycolysis and the tricarboxylic acid (TCA) cycle are responsible for the production of metabolites such as acetyl-CoA, which potentially supplies the increased demand of acetyl-CoA required for protein hyperacetylation seen in CR conditions (Hagopian et al., 2003; Pietrocola et al., 2015). Yet, given that nutrients are limiting under CR, the expression of glucokinase (Gck) and liver pyruvate kinase (Pklr) are flat in YCR versus YND (Fig. 7D). This corresponds with a drastic loss of protein expression of ATP-citrate lyase (ACLY) in YCR and OCR mice. As ACLY is responsible for conversion of citrate to acetyl-CoA (Fig. 7E), we reasoned that the cellular pool of acetyl-CoA required for the increased global acetylation under CR is not dependent on this pathway. Moreover, Acly is a transcriptional target of sterol regulatory element-binding protein (SREBP) and this pathway is responsible for de novo lipid biosynthesis that utilizes acetyl-CoA as a building block (Horton et al., 2003; Sato et al., 2000). The SREBP pathway controlling lipid metabolism is shut down under CR, as shown by the gene expression profiles of fatty acid synthase (Fasn), acetyl-CoA carboxylase α (Acaca) and stearoyl-CoA desaturase1 (Scd1) in YCR versus YND (Fig. 7F).
Importantly, SIRT1 deacetylates and controls cytoplasmic AceCS1 (Hallows et al., 2006), a regulatory pathway that was found to be circadian and drives cyclic pools of acetate-derived acetyl-CoA (Sahar et al., 2014). Indeed, elevation of hepatic acetate levels was found in YCR versus YND (Fig. 7G) Acetylation of nuclear AceCS1 is dynamically circadian with a sharp peak at ZT 8/12 in normal diet (Fig. 7H). Notably, CR broadens the acetylation profile of AceCS1 over several hours of the circadian cycle in YCR and OCR (Fig. 7H). As the ACLY-centered pathway is impaired in CR (Fig. 7E), and since both acetate (Fig. 7G) and acetyl-coA levels increase under CR in the liver (Hagopian et al., 2003; Pietrocola et al., 2015; Seufert et al., 1974), we hypothesize that the change in circadian acetylation of AceCS1 could be linked to acetyl-CoA and global protein hyperacetylation. While this hypothesis needs confirmation, it is noteworthy that the profile of AceCS1 acetylation under CR extends broadly over several circadian time points (Fig. 7H), in concert with the global protein hyperacetylation observed at all ZTs under CR (Fig. 5B). Although a causal relationship is not directly assessable, this regulatory pathway might also explain the phase of CR-dependent genes that is widely distributed over the circadian cycle (Fig. 3A), paralleling promoter-specific histone hyperacetylation of core clock and metabolic gene targets (Fig. 5, C–F).
DISCUSSION
How aging influences circadian rhythms and how the clock contributes to the aging process have been unresolved questions (Orozco-Solis and Sassone-Corsi, 2014). While some evidence suggests that the central clock mechanism in the brain is associated with an age-dependent decline (Chang and Guarente, 2013; Nakamura et al., 2011; Sellix et al., 2012), the specific cellular pathways implicated have been elusive. Moreover, whether aging influences various clocks in a given organism by operating through different mechanisms is unknown. This question is particularly relevant when considering the central contribution of a circadian pacemaker in governing peripheral metabolic tissues and thereby controlling homeostasis. During aging the homeostatic balance changes and it has been suggested that re-synchronization of metabolic and physiological functions by the circadian clock may slow down the aging process. Indeed, peripheral metabolic clocks are particularly responsive to changes in nutritional availability (Damiola et al., 2000; Gill et al., 2015; Stokkan et al., 2001), and given the role of CR in lifespan extension (Lee et al., 1999; Lin et al., 2002), the question arises as to the contribution of the molecular clockwork in mediating the effects of CR on lifespan.
A question that remains unanswered is whether the interplay between the circadian clock and the aging process influences various tissues in different manners. In addition to the relationship between the central pacemaker in the SCN and peripheral oscillators (Reppert and Weaver, 2001; Welsh et al., 2010), evidence of communication between peripheral clocks is accumulating (Masri et al., 2016). Thus, it is conceivable that altered communication within a network of cell-autonomous clocks could contribute to the aging process. In this context it is notable that circulating serum factors appear to be able to alter cellular clock properties. Serum factors from aged subjects, while not influencing the molecular machinery of peripheral circadian clocks, induce age-related circadian dysfunctions (Pagani et al., 2011). These notions underlie the remarkable plasticity of the circadian clock and its link with the aging process, but do not provide a molecular interpretation of how the two processes are intertwined. Our findings, together with the accompanying study (Solanas et al.), reveal the molecular framework that underlies the above-mentioned plasticity and underscore its cellular specificity.
We have shown that the circadian profiles of all core clock genes are unaltered during the aging process in the liver as well as in EpSCs and MsSCs, stressing that the clock mechanism is intact in the peripheral tissues of aged mice. On the other hand, we show that remarkable circadian changes take place during aging and CR in the liver, where a reorganized metabolic clock defines a remodeled homeostasis. This reorganization was highlighted by the tissue comparative circadian transcriptome and underscores the importance of NAD+ metabolism and protein acetylation as liver-specific circadian metabolic processes. This finding parallels a rhythmic global protein acetylation that appears as a hallmark of the liver clock (Fig. 4). Remarkably, this oscillatory acetylation signature is drastically dampened in old mice whereas CR serves to rescue the age-dependent loss of global protein acetylation. These changes in protein acetylation are likely due to improved NAD+ availability, enhanced SIRT1 activity, and increased levels of acetate and acetyl-coA (Fig. 7I) (Hagopian et al., 2003; Pietrocola et al., 2015; Seufert et al., 1974). We have shown that control of protein acetylation is mediated at least by SIRT1 and the dynamic acetylation of AceCS1. Our results centrally place the circadian NAD+ salvage pathway in mediating the beneficial effects of CR and reveal changes in rhythmic protein acetylation that implicate metabolic cues along the process of aging (Fig. 7I). Strikingly, CR sharpened circadian oscillations while simultaneously boosting hepatic NAD+, NADP+ and the generation and potential circulation of NR (Fig. 6C).
Cyclic global protein acetylation appears to be age-dependent and controlled by nutritional cues. These concepts implicate a precisely timed activation of both histone acetyltransferases (HATs) and histone deacetylases (HDACs) in regulating histone and non-histone protein acetylation over the circadian cycle in response to aging and CR. As an additional level of control, the availability of metabolites such as NAD+ and acetyl-CoA suggest an intimate link between nutritional cues and regulatory pathways that ultimately result in controlled chromatin remodeling and regulation of gene expression.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Paolo Sassone-Corsi (psc@uci.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C57B6/J mice were bred and aged at the animal facilities of the Barcelona Science Park, Spain, in accordance with the Spanish and European Union regulations. Animal care and use was approved by the Government of Catalonia, Spain, in accordance with applicable legislation. Both male and female mice were used for the experiment. Mice were maintained on a 12 hr light/dark cycle.
METHODS DETAILS
Animal Experiments
Mice of the two different age groups were fed either a control diet (Harlan TD.120685, Harlan Industries) ad libitum or a calorie-restricted diet (Harlan TD.120686, Harlan Industries) with a 30% food reduction as compared to the ad libitum control diet group, according to previously published work (Cao et al., 2001) with a slight modification. The control diet was composed of 18.8 %/w protein, 7.3 %/w fatty acids and 55.1 %/w carbohydrates, with 3.6 Kcal/g, whereas the calorie-restricted diet was composed of 32.9 %/w protein, 12.7 %/w fatty acids and 31.9 %/w carbohydrates, with 3.7 Kcal/g. The calorie-restricted group mice were adapted to the regimen for 3-weeks, receiving 10 % less calories every week until reaching the final 30 % restriction. The calorie-restricted group received food once a day at zeitgeber time (ZT) 12. Animals subjected to caloric restriction were housed individually to prevent food competition. All animals were weighed every 2 weeks. At the start of the adaptation period, the age of young and old animals was 19–29 weeks and 55–69 weeks, respectively. After 25-weeks of this feeding regimen, livers were collected every 4 hr over the circadian cycle and immediately flash frozen in liquid nitrogen for subsequent processing.
Microarrays Analyses
Microarrays were performed at the IRB Barcelona Functional Genomics Core Facility, IRB Barcelona, Spain. Total RNA was extracted from mouse liver using Trizol reagent (Life Technologies). cDNA library preparation and amplification were performed from 25 ng of total RNA using WTA2 (Sigma-Aldrich), with 17 cycles of amplification. cDNA was purified using Purelink (Invitrogen). cDNA (8 ug) was then fragmented by DNaseI and biotinylated by terminal transferase obtained from GeneChip Human Mapping 10K 2.0 Assay Kit (Affymetrix). Hybridization mixtures were prepared according to the Gene Atlas protocol (Affymetrix). Each sample target was hybridized to Mouse Genome 430 PM array. After hybridization for 16 h at 45 °C, samples were washed and stained in the GeneAtlas Fluidics Station (Affymetrix). Arrays were scanned in a GeneAtlas Imaging Station (Affymetrix). All processing was performed according to manufacturer’s recommendations. CEL files were generated from DAT files using Affymetrix Command Console software.
Processing of microarray samples was carried out separately for each dataset using R (http://www.R-project.org/) and Bioconductor (Gentleman et al., 2004; Irizarry et al., 2003). Probesets were annotated using information available on the Affymetrix webpage (http://www.affymetrix.com/analysis/index.affx). Prior to downstream analysis, expression values were corrected for amplification and scanning batches using a liner model in which gender, time point, diet, age, and the pairwise interaction terms between the three last were included as covariates. Microarrays were analyzed at IRB Barcelona Biostatistics/Bioinformatics Core Facility.
Gene Ontology (GO) Analysis
GO analysis was performed using Genomatix (http://www/genomatix.de/), using the Gene Ranker package, based on a p-value cutoff of 0.01.
Real-Time Quantitative Polymerase Chain Reaction (Real-Time qPCR)
Using Trizol reagent, total RNA was extracted from liver isolated from mice at ZT 0, 4, 8, 12,16, and 20. One ug of total RNA was reverse transcribed to cDNA using iScript cDNA Synthesis Kit (Bio-Rad Laboratories). cDNA was applied to real-time qPCR using iQ SYBR Green Supermix (Bio-Rad Laboratories). Gene expression was normalized to 18S rRNA. Primers used for qPCR are listed in Table S1.
Chromatin Immunoprecipitation (ChIP)
ChIP assays were carried out as described previously (Masri et al., 2014). Briefly, liver nuclei were sonicated to fragment DNA to ~0.5 kb using Qsonica Q125 Sonicator (Qsonica). Sheared DNA was pre-cleared for 2 hr at 4 °C with protein G sepharose beads (Sigma-Aldrich) and immunoprecipitated with antibodies, including anti-H3K27ac (Abcam, ab4729), and anti-H3K9/K14ac (Diagenode, C15410200), overnight at 4°C. Protein G beads were added and rocked for 3 hr at 4 °C. The beads were washed and subjected to reverse-crosslink in elution buffer overnight at 65 °C. DNA was purified by phenol/chloroform and eluted in water for qPCR. Primers for ChIP are listed in Table S2. Anti-rabbit IgG antibody (Santa Cruz, sc-2027) was used as a negative control.
Western Blot
Liver whole cell lysates and nuclear extracts were prepared as described previously (Masri et al., 2014). Generally, 20 ug of liver whole cell lysates or 5 ug of liver nuclear extracts was loaded on 6–10% polyacrylamide gels. Antibodies used for western blots are as follows: anti-BMAL1 (Abcam, ab93806), anti-REV-ERBα, anti-acetylated lysine, anti-ACLY, anti-AceCS1 (Cell signaling, 13418, 9441, 4332 and 3658, respectively), anti-CRY1 (Bethyl Laboratories, A302–614), anti-PER2 (Alpha diagnostic, PER21-A), anti-acetylated BMAL1 (Millipore, AB15396), and anti-p84 (Genetex, 5E10). Anti-acetylated AceCS1 was kindly provided from the laboratory of Dr. John Denu (University of Wisconsin) as described previously (Hallows et al., 2006).
Comparative Analysis of Transcriptome Data among Epidermal (EpSCs), Skeletal Muscle Stem Cell (MsSCs) and Liver
Tissue-specific comparison of phase of gene expression was carried out using genes selected to be significantly circadian (p-value cutoff of 0.01) in each tissue/cell from young and old mice fed normal diet or under caloric restriction (see accompanying study (Solanas et al.)). Hepatic microarray dataset in this study was also crossed with stem cells microarray datasets used in the accompanying study (GEO accession number is GSE84580 (Solanas et al.)).
NAD+-Targeted Quantitative Metabolomics
NAD+ metabolome was performed as described previously (Trammell and Brenner, 2013; Trammell et al., 2016). Mouse liver was pulverized using a Bessman pulverizer (Spectrum Laboratories, Rancho Dominguez, CA). For analysis of NR, Nam and Me4PY (group A analytes), samples were spiked with [18O1]-NR, [18O1]-NAM and [D3]-Me4PY (internal standard (IS) A). For analysis of NAD+, NADP+, NMN, NAAD and ADPR (group B analytes), samples were dosed with 13C-yeast extract (IS B) as described (Trammell and Brenner, 2013). Before extraction, IS A and IS B were added to aliquots for quantification of group A and B analytes, respectively. Samples were extracted by addition of 0.4 ml of buffered ethanol at 80 °C. Samples were vortexed vigorously until thawed, sonicated in a bath sonicator (10 s followed by 15 s on ice, repeated twice), then placed into a Thermomixer (Eppendorf) set to 55 °C and shaken at 1,050 rpm for 5 min. After centrifugation, the supernatants were transferred to fresh tubes and dried via speed vacuum. Before LC-MS analysis, samples were resuspended in 60 ul of 10 mM ammonium acetate (>99% pure) in LC-MS-grade water.
Separation and quantitation of analytes were performed with an Acquity LC interfaced with a TQD mass spectrometer (Waters) operated in positive ion multiple reaction monitoring mode as described (Trammell and Brenner, 2013) with minor modifications. Group A analytes were analyzed using an acid separation, whereas group B analytes were analyzed using an alkaline separation on a 2.1 X 100 mm Hypercarb column (Thermo Fisher Scientific) (Trammell and Brenner, 2013). Metabolites were quantified by dividing their peak areas by IS peak areas and comparing the ratio to a standard curve in 10 mM ammonium acetate. Metabolites were normalized to wet liver weights.
Measurement of Hepatic β-Hydroxybutyrate Concentration
Liver β-hydroxybutyrate levels were determined by using a β-hydroxybutyrate assay kit (Abcam). Briefly, 20 mg of frozen liver tissue was homogenized with a Dounce homogenizer. After collecting supernatant, samples were deproteonized using ice cold perchloric acid and then neutralized with KOH. The deproteonized samples were subjected to enzymatic reaction and measured at OD450 nm using a microplate reader.
Measurement of Hepatic Acetate Concentration
Liver acetate levels were quantified by using an acetate colorimetric assay kit (BioVision) according to the manufacturer’s protocol. Ten mg of frozen liver was homogenized and the supernatant was collected after centrifugation. The samples were incubated with enzyme and substrate at room temperature and measured at OD450 nm using a microplate reader.
Enrichment Analysis with SIRT1-Dependent Transcriptome Dataset
Each YND, OND, YCR, and OCR microarray dataset in this study was crossed with published microarray dataset that includes genes with more robust circadian expression when SIRT1 is disrupted (GEO accession number is GSE57830, (Masri et al., 2014)).
Enrichment Analysis with Hepatic Histone Modification ChIP-Sequence Dataset
EnrichR (http://amp.pharm.mssm.edu/Enrichr/), a comprehensive gene set enrichment analysis web tool (Chen et al., 2013; Kuleshov et al., 2016), was used for a comparative enrichment analysis between the circadian hepatic transcriptome in this study and ChIP-sequencing datasets in the liver for several histone modifications. Adjusted p-values extending to 0.05 were allowed for enriched histone marks.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification
Western blots for pan acetylated lysine antibody were quantified using ImageJ (NIH).
Statistical Analysis
3–6 biological replicates were used for the animal experiment. Data were analyzed by two-way ANOVA using Bonferroni posttest (Prism 5.0). All of the data are presented as the mean±SEM and represent a minimum of three independent experiments. For the analysis of circadian genes, the nonparametric test JTK_cycle was used incorporating a window of 20–28 hr for the determination of circadian periodicity (Hughes et al., 2010), including amplitude and phase analysis. A gene was considered circadian based on a p-value cutoff of 0.01.
DATA AND SOFTWARE AVAILABILITY
The GEO accession number for the microarray data reported in this paper is GSE93903.
Supplementary Material
Data Table S1. Lists of circadian genes identified within top two circadian biological processes, “RNA processing” and “autophagy”, exclusively in YCR and OCR (Excel file, related to Fig. 2).
Data Table S2. Lists of overlapping genes with SIRT1-dependent genes (Excel file, related to Fig. 7).
Fig. S1. Heatmaps and gene ontology (GO) analysis of common circadian genes from the hepatic transcriptome (Related to Fig. 1 and 2). Heatmaps display common circadian genes in YND versus OND (A), YND versus YCR (B), and OND versus OCR (C). GO analysis of common circadian genes within the comparison between YND versus OND (D), YND versus YCR (E), and OND versus OCR (F). Numbers within the pie charts indicate number of circadian genes identified within each biological process, using a p-value cutoff of 0.01. 3–6 biological replicates per time point were subjected to the hepatic transcriptomic analysis.
Fig. S2. Circadian gene and protein expression of core clock genes in livers from YND, OND, YCR and OCR, and circadian hepatic gene expression shown as age- and diet-specific circadian genes (Related to Fig. 1). Circadian gene expression of Dbp, Per1, Per3, Rev-erbβ, Rora, and Rorc was compared between YND versus OND (A), YND versus YCR (B), and OND versus OCR (C). (D) Circadian protein expression of BMAL1, REV-ERBα, CRY1, and PER2 in the whole cell extracts from YND, OND, YCR, and OCR livers was determined by western blot. p84 was used as a loading control. Images represent a minimum of three independent experiments. Age- and diet-dependent rhythmic genes, cyclin-dependent kinase inhibitor 1a (Cdkn1a), heat shock protein 90aa1 (Hsp90aa1), Hspa4, interleukin 6 receptor, alpha (Il6ra), S-adenosylhomocysteine hydrolase (Ahcy), DNA methyltransferase 3b (Dnmt3b), pyruvate dehydrogenase kinase 4 (Pdk4), and Sirt1 were compared between YND versus OND (E), YND versus YCR (F), and OND versus OCR (G). Data represent mean±SEM and were analyzed by two-way ANOVA using Bonferroni posttest. *, **, and *** indicate P<0.05, P<0.01, and P<0.001, respectively (YND, n=4–5 per time point; OND, n=3–4 per time point; YCR, n=5–6 per time point; OCR, n=3–5 per time point).
Fig. S3. Comparison of the circadian hepatic transcriptome between YCR and OCR (Related to Fig. 1 and 2). (A) Venn diagram displays the total number (top) and ratio (bottom) of rhythmic genes in the liver from YCR (purple, n=5–6 per time point) and OCR (blue, n=3–5 per time point), including common genes. (B) Amplitude plot represents the distribution of hepatic oscillatory genes in YCR and OCR. (C) Heatmaps display rhythmic genes exclusively in YCR (top left panel), OCR (top right panel), and in common (bottom panel). (D) GO analysis of genes exclusively circadian in YCR (top, purple), OCR (middle, blue), and common circadian genes (bottom, gray). Numbers within the pie charts indicate number of circadian genes identified within each biological process, based on a p-value cutoff of 0.01. (E) Determination of core clock gene expression in YCR and OCR by qPCR. Data represent mean±SEM and were analyzed by two-way ANOVA using Bonferroni posttest. *, **, and *** indicate P<0.05, P<0.01, and P<0.001, respectively.
Fig. S4. Comparison of the circadian hepatic transcriptome between YND and OCR (Related to Fig. 1 and 2). (A) Venn diagram displays the total number (top) and ratio (bottom) of rhythmic genes in the liver from YND (green, n=4–5 per time point) and OCR (blue, n=3–5 per time point), including common genes. (B) Amplitude plot exhibits the distribution of hepatic oscillatory genes in YND and OCR. (C) Heatmaps display genes exclusively circadian in YND (top left panel), OCR (top right panel), and in common (bottom panel). (D) GO analysis of genes exclusively circadian in YND (top, green), OCR (middle, blue), and common circadian genes (bottom, gray). Numbers within the pie charts indicate number of circadian genes identified within each biological process, based on a p-value cutoff of 0.01.
Fig. S5. Unique circadian signatures in NAD+ metabolism and protein acetylation in the liver versus stem cells (Related to Fig. 4). GO analysis of genes commonly circadian in all three tissues (liver, EpSCs, and MsSCs) harvested from YND (green), OND (orange), YCR (purple), and OCR (blue) animals. Numbers within the pie charts indicate number of circadian genes identified within each biological process. p-value cutoff of 0.01 was used. 3–6 biological replicates per time point per tissue were subjected to the transcriptomic analysis.
Fig. S6. Hepatic protein hyperacetylation under CR (Related to Fig. 5). (A) Circadian lysates from livers of YND, OND, YCR, and OCR were probed with a pan acetylated lysine antibody by western blot and quantified using three independent samples. The far right panel displays a low exposure image with a parallel comparison of YCR and OCR liver lysates. p84 was used as a loading control. (B) Hepatic β-hydroxybutyrate concentration was determined among YND, OND, YCR, and OCR groups by colorimetric assay. Data represent Means±SEM from three independent samples and was analyzed by two-way ANOVA using Bonferroni posttest. *, **, and *** indicate P<0.05, P<0.01, and P<0.001, respectively.
Fig. S7. Comprehensive analysis of histone modifications enriched at circadian promoters in YND, OND, YCR, and OCR (Related to Fig. 5). A comparative analysis was performed with the hepatic circadian transcriptome and ChIP-sequencing datasets for several histone modifications using EnrichR (http://amp.pharm.mssm.edu/Enrichr/). Adjusted p-values extending to 0.05 were allowed for enriched histone marks listed in each histogram.
Table S1. Primers used for qPCR (Related to Fig. 1, 5, 7, S2 and S3, and STAR Methods).
Table S2. Primers used for ChIP-qPCR (Related to Fig. 5 and STAR Methods).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-BMAL121 | Abcam | Cat# ab93806 |
| Anti-H3K27ac | Abcam | Cat# ab4729 |
| Anti-H3K9/K14ac | Diagenode | Cat# C15410200 |
| Anti-rabbit IgG | Santa Cruz Biotechnology | Cat# sc-2027 |
| Anti-acetylated BMAL1 | EMD Millipore | Cat# AB15396 |
| Anti-REV-ERBα | Cell Signaling Technology | Cat# 13418 |
| Anti-acetylated lysine | Cell Signaling Technology | Cat# 9441 |
| Anti-ACLY | Cell Signaling Technology | Cat# 4332 |
| Anti-AceCS1 | Cell Signaling Technology | Cat# 3658 |
| Anti-CRY1 | Bethyl Laboratories | Cat# A302–614 |
| Anti-PER2 | Alpha diagnostic | Cat# PER21-A |
| Anti-p84 | Genetex | Cat# 5E10 |
| Anti-acetylated AceCS1 | Denu lab (University of Wisconsin) | (Hallows et al., 2006) |
| Anti-Mouse IgG, HRP conjugate | EMD Millipore | Cat# AP160P |
| Anti-Rabbit IgG, HRP-linked | Cell Signaling Technology | Cat# 7074 |
| Chemicals and Reagent | ||
| TRIzol | ThermoFisher Scientific | Cat# 15596026 |
| WTA2 | Sigma-Aldrich | Cat# WTA2 |
| Purelink | ThermoFisher Scientific | Cat# K182002 |
| GeneChip Human Mapping 10K 2.0 Assay Kit | Affymetrix | Cat# 511060 |
| iScript Reverse Transcription Supermix | Bio-Rad Laboratories | Cat# 1708840 |
| iQ SYBR Green Supermix | Bio-Rad Laboratories | Cat# 1708880 |
| Protein G Sepharose | Sigma-Aldrich | Cat# P3296 |
| Protein Assay Dye Reagent | Bio-Rad Laboratories | Cat# 500–0006 |
| Nitrocellulose Membrane, 0.45 μm | Bio-Rad Laboratories | Cat# 1620115 |
| Immobilon Western Chemiluminescent HRP substrate | EMD Millipore | Cat# WBKLS0500 |
| DSG (disuccinimidyl glutarate) | ThermoFisher Scientific | Cat# 20593 |
| cOmplete, EDT-free Protease Inhibitor Cocktail | Roche | Cat# 11873580001 |
| Critical Commercial Assays | ||
| β-Hydroxybutyrate Assay Kit | Abcam | Cat# ab83390 |
| Acetate Colorimetric Assay Kit | BioVision | Cat# K658–100 |
| Deposited Data | ||
| Transcriptomics data | This paper | GEO# GSE93903 |
| Transcriptomics data | Accompanying paper by (Solanas et al.) | GEO# GSE84580 |
| Transcriptomics data | (Masri et al., 2014) | GEO# GSE57830 |
| Experimental Models: Organisms/Strains | ||
| C57B6/J mice | Benitah lab (IRB Barcelona) | N/A |
| Oligonucleotides | ||
| See Table S1 for qPCR primer list | N/A | N/A |
| See Table S2 for ChIP-qPCR primer list | N/A | N/A |
| Software and Algorithms | ||
| Prism 5.0 | GraphPad Software | http://www.graphpad.com |
| JTK_cycle | Hughes lab (Washington University School of Medicine) | www.openwetware.org/wiki/HughesLab:JTK_Cycle |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| EnrichR | (Chen et al., 2013; Kuleshov et al., 2016) | http://amp.pharm.mssm.edu/Enrichr/ |
| CircadiOmics | (Patel et al., 2012) | circadiomics.igb.uci.edu |
| Bioconductor | (Gentleman et al., 2004) | https://www.bioconductor.org/ |
| Other (Research diet) | ||
| Calorie-restricted diet | Harlan laboratories | Cat# 120685 |
| Control diet | Harlan laboratories | Cat# 120686 |
Highlights.
Aging reprograms clockwork with distinct modalities in the liver versus stem cells
Liver circadian genomic signatures of aging are reverted by caloric restriction (CR)
Cyclic protein acetylation is lost in old mice while CR results in hyperacetylation
CR reorganizes circadian metabolic pathway linked to NAD+-SIRT1-AceCS1 in the liver
ACKNOWLEDGMENTS
We thank all members of the Sassone-Corsi laboratory for help and critical discussions. S. S. is supported by the Della Martin Foundation; L. B. by Italian Association for Cancer Research; this work was possible through support by the National Institutes of Health, INSERM (Institut National de la Sante et Recherche Medicale, France), the Roy J. Carver Trust, and a Novo Nordisk Foundation Challenge Grant. Research for this project in the lab of S.A.B. was supported by the European Research Council (ERC), the Spanish Ministry of Economy and Development, and the Institute for Research in Biomedicine (IRB-Barcelona). F.O.P. is supported by a La Caixa International PhD fellowship. IRB Barcelona is the recipient of a Severo Ochoa Award of Excellence from MINECO (Government of Spain).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, and Schibler U (2008). SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134, 317–328. [DOI] [PubMed] [Google Scholar]
- Asher G, and Sassone-Corsi P (2015). Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell 161, 84–92. [DOI] [PubMed] [Google Scholar]
- Belenky P, Bogan KL, and Brenner C (2007a). NAD+ metabolism in health and disease. Trends Biochem Sci 32, 12–19. [DOI] [PubMed] [Google Scholar]
- Belenky P, Racette FG, Bogan KL, McClure JM, Smith JS, and Brenner C (2007b). Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+. Cell 129, 473–484. [DOI] [PubMed] [Google Scholar]
- Bogan KL, and Brenner C (2008). Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu Rev Nutr 28, 115–130. [DOI] [PubMed] [Google Scholar]
- Brandhorst S, Choi IY, Wei M, Cheng CW, Sedrakyan S, Navarrete G, Dubeau L, Yap LP, Park R, Vinciguerra M, et al. (2015). A Periodic Diet that Mimics Fasting Promotes Multi-System Regeneration, Enhanced Cognitive Performance, and Healthspan. Cell Metab 22, 86–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao SX, Dhahbi JM, Mote PL, and Spindler SR (2001). Genomic profiling of short- and long-term caloric restriction effects in the liver of aging mice. Proc Natl Acad Sci U S A 98, 10630–10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartee GD, Hepple RT, Bamman MM, and Zierath JR (2016). Exercise Promotes Healthy Aging of Skeletal Muscle. Cell Metab 23, 1034–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, and Guarente L (2013). SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell 153, 1448–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Steele AD, Lindquist S, and Guarente L (2005). Increase in activity during calorie restriction requires Sirt1. Science 310, 1641. [DOI] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, and Ma’ayan A (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, and Sinclair DA (2004). Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 305, 390–392. [DOI] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, and Schibler U (2000). Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14, 2950–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, and Sassone-Corsi P (2013). Reprogramming of the Circadian Clock by Nutritional Challenge. Cell 155, 1464–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, and Partridge L (2015). Promoting health and longevity through diet: from model organisms to humans. Cell 161, 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S, Le HD, Melkani GC, and Panda S (2015). Time-restricted feeding attenuates age-related cardiac decline in Drosophila. Science 347, 1265–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, et al. (2013). Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell 155, 1624–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandison RC, Piper MD, and Partridge L (2009). Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature 462, 1061–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagopian K, Ramsey JJ, and Weindruch R (2003). Caloric restriction increases gluconeogenic and transaminase enzyme activities in mouse liver. Exp Gerontol 38, 267–278. [DOI] [PubMed] [Google Scholar]
- Hallows WC, Lee S, and Denu JM (2006). Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A 103, 10230–10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, et al. (2013). Calorie Restriction and SIRT3 Trigger Global Reprogramming of the Mitochondrial Protein Acetylome. Mol Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, and Sassone-Corsi P (2007). CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature 450, 1086–1090. [DOI] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, et al. (2010). SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464, 121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, and Goldstein JL (2003). Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci U S A 100, 12027–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Pirinen E, and Auwerx J (2012). Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol 13, 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, and Kornacker K (2010). JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms 25, 372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, and Guarente L (2014). NAD+ and sirtuins in aging and disease. Trends Cell Biol 24, 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, and Speed TP (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264. [DOI] [PubMed] [Google Scholar]
- Katewa SD, Akagi K, Bose N, Rakshit K, Camarella T, Zheng X, Hall D, Davis S, Nelson CS, Brem RB, et al. (2016). Peripheral Circadian Clocks Mediate Dietary Restriction-Dependent Changes in Lifespan and Fat Metabolism in Drosophila. Cell Metab 23, 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, and Antoch MP (2006). Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev 20, 1868–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratova AA, and Kondratov RV (2012). The circadian clock and pathology of the ageing brain. Nat Rev Neurosci 13, 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44, W90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Klopp RG, Weindruch R, and Prolla TA (1999). Gene expression profile of aging and its retardation by caloric restriction. Science 285, 1390–1393. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, and Guarente L (2000). Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289, 2126–2128. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Ford E, Haigis M, Liszt G, and Guarente L (2004). Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev 18, 12–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, and Guarente L (2002). Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 418, 344–348. [DOI] [PubMed] [Google Scholar]
- Longo VD, and Panda S (2016). Fasting, Circadian Rhythms, and Time-Restricted Feeding in Healthy Lifespan. Cell Metab 23, 1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Papagiannakopoulos T, Kinouchi K, Liu Y, Cervantes M, Baldi P, Jacks T, and Sassone-Corsi P (2016). Lung Adenocarcinoma Distally Rewires Hepatic Circadian Homeostasis. Cell 165, 896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Patel VR, Eckel-Mahan KL, Peleg S, Forne I, Ladurner AG, Baldi P, Imhof A, and Sassone-Corsi P (2013). Circadian acetylome reveals regulation of mitochondrial metabolic pathways. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Rigor P, Cervantes M, Ceglia N, Sebastian C, Xiao C, Roqueta-Rivera M, Deng C, Osborne TF, Mostoslavsky R, et al. (2014). Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 158, 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, and Sassone-Corsi P (2014). Sirtuins and the circadian clock: bridging chromatin and metabolism. Science signaling 7, re6. [DOI] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Canto C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, et al. (2013). The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell 154, 430–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, and Sassone-Corsi P (2008). The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, and Sassone-Corsi P (2009). Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324, 654–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TJ, Nakamura W, Yamazaki S, Kudo T, Cutler T, Colwell CS, and Block GD (2011). Age-related decline in circadian output. J Neurosci 31, 10201–10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo A, Reddy P, and Izpisua Belmonte JC (2016). Anti-Aging Strategies Based on Cellular Reprogramming. Trends Mol Med 22, 725–738. [DOI] [PubMed] [Google Scholar]
- Orozco-Solis R, and Sassone-Corsi P (2014). Circadian clock: linking epigenetics to aging. Curr Opin Genet Dev 26, 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani L, Schmitt K, Meier F, Izakovic J, Roemer K, Viola A, Cajochen C, Wirz-Justice A, Brown SA, and Eckert A (2011). Serum factors in older individuals change cellular clock properties. Proc Natl Acad Sci U S A 108, 7218–7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SA, Chaudhari A, Gupta R, Velingkaar N, and Kondratov RV (2016a). Circadian clocks govern calorie restriction-mediated life span extension through BMAL1- and IGF-1-dependent mechanisms. FASEB J 30, 1634–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SA, Velingkaar N, Makwana K, Chaudhari A, and Kondratov R (2016b). Calorie restriction regulates circadian clock gene expression through BMAL1 dependent and independent mechanisms. Scientific reports 6, 25970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VR, Eckel-Mahan K, Sassone-Corsi P, and Baldi P (2012). CircadiOmics: integrating circadian genomics, transcriptomics, proteomics and metabolomics. Nat Methods 9, 772–773. [DOI] [PubMed] [Google Scholar]
- Peek C, Affinati A, Ramsey K, Kuo H, Yu W, Sena L, Ilkayeva O, Marcheva B, Kobayashi Y, Omura C, et al. (2013). Circadian Clock NAD+ Cycle Drives Mitochondrial Oxidative Metabolism in Mice. Science 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg S, Feller C, Ladurner AG, and Imhof A (2016). The Metabolic Impact on Histone Acetylation and Transcription in Ageing. Trends Biochem Sci 41, 700–711. [DOI] [PubMed] [Google Scholar]
- Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F, and Kroemer G (2015). Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab 21, 805–821. [DOI] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, et al. (2009). Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324, 651–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM, and Weaver DR (2001). Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol 63, 647–676. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, and Puigserver P (2005). Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 434, 113–118. [DOI] [PubMed] [Google Scholar]
- Sahar S, Masubuchi S, Eckel-Mahan K, Vollmer S, Galla L, Ceglia N, Masri S, Barth TK, Grimaldi B, Oluyemi O, et al. (2014). Circadian Control of Fatty Acid Elongation by SIRT1-mediated Deacetylation of Acetyl-CoA Synthetase 1. J Biol Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato R, Okamoto A, Inoue J, Miyamoto W, Sakai Y, Emoto N, Shimano H, and Maeda M (2000). Transcriptional regulation of the ATP citrate-lyase gene by sterol regulatory element-binding proteins. J Biol Chem 275, 12497–12502. [DOI] [PubMed] [Google Scholar]
- Sellix MT, Evans JA, Leise TL, Castanon-Cervantes O, Hill DD, DeLisser P, Block GD, Menaker M, and Davidson AJ (2012). Aging differentially affects the re-entrainment response of central and peripheral circadian oscillators. J Neurosci 32, 16193–16202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert CD, Graf M, Janson G, Kuhn A, and Soling HD (1974). Formation of free acetate by isolated perfused livers from normal, starved and diabetic rats. Biochem Biophys Res Commun 57, 901–909. [DOI] [PubMed] [Google Scholar]
- Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, et al. (2010). SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab 12, 654–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solanas G, Oliveira Peixoto F, Perdiguero E, Jardí M, Ruiz-Bonilla V, Datta D, Symeonidi A, Castellanos A, Welz P, Martín Caballero J, et al. Adult stem cells undergo circadian reprogramming during ageing. [Google Scholar]
- Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, and Prolla TA (2010). Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell 143, 802–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LR, and Imai S (2014). Specific ablation of Nampt in adult neural stem cells recapitulates their functional defects during aging. EMBO J 33, 1321–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, and Menaker M (2001). Entrainment of the circadian clock in the liver by feeding. Science 291, 490–493. [DOI] [PubMed] [Google Scholar]
- Trammell SA, and Brenner C (2013). Targeted, LCMS-based Metabolomics for Quantitative Measurement of NAD(+) Metabolites. Comput Struct Biotechnol J 4, e201301012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trammell SA, Schmidt MS, Weidemann BJ, Redpath P, Jaksch F, Dellinger RW, Li Z, Abel ED, Migaud ME, and Brenner C (2016). Nicotinamide riboside is uniquely and orally bioavailable in mice and humans. Nat Commun 7, 12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulgherait M, Chen A, Oliva MK, Kim HX, Canman JC, Ja WW, and Shirasu-Hiza M (2016). Dietary Restriction Extends the Lifespan of Circadian Mutants tim and per. Cell Metab. Verdin, E. (2015). NAD(+) in aging, metabolism, and neurodegeneration. Science 350, 1208–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E, and Ott M (2015). 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Biol 16, 258–264. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Takahashi JS, and Kay SA (2010). Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol 72, 551–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ryu D, Wu Y, Gariani K, Wang X, Luan P, D’Amico D, Ropelle ER, Lutolf MP, Aebersold R, et al. (2016). NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 352, 1436–1443. [DOI] [PubMed] [Google Scholar]
- Zwighaft Z, Aviram R, Shalev M, Rousso-Noori L, Kraut-Cohen J, Golik M, Brandis A, Reinke H, Aharoni A, Kahana C, et al. (2015). Circadian Clock Control by Polyamine Levels through a Mechanism that Declines with Age. Cell Metab 22, 874–885. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Table S1. Lists of circadian genes identified within top two circadian biological processes, “RNA processing” and “autophagy”, exclusively in YCR and OCR (Excel file, related to Fig. 2).
Data Table S2. Lists of overlapping genes with SIRT1-dependent genes (Excel file, related to Fig. 7).
Fig. S1. Heatmaps and gene ontology (GO) analysis of common circadian genes from the hepatic transcriptome (Related to Fig. 1 and 2). Heatmaps display common circadian genes in YND versus OND (A), YND versus YCR (B), and OND versus OCR (C). GO analysis of common circadian genes within the comparison between YND versus OND (D), YND versus YCR (E), and OND versus OCR (F). Numbers within the pie charts indicate number of circadian genes identified within each biological process, using a p-value cutoff of 0.01. 3–6 biological replicates per time point were subjected to the hepatic transcriptomic analysis.
Fig. S2. Circadian gene and protein expression of core clock genes in livers from YND, OND, YCR and OCR, and circadian hepatic gene expression shown as age- and diet-specific circadian genes (Related to Fig. 1). Circadian gene expression of Dbp, Per1, Per3, Rev-erbβ, Rora, and Rorc was compared between YND versus OND (A), YND versus YCR (B), and OND versus OCR (C). (D) Circadian protein expression of BMAL1, REV-ERBα, CRY1, and PER2 in the whole cell extracts from YND, OND, YCR, and OCR livers was determined by western blot. p84 was used as a loading control. Images represent a minimum of three independent experiments. Age- and diet-dependent rhythmic genes, cyclin-dependent kinase inhibitor 1a (Cdkn1a), heat shock protein 90aa1 (Hsp90aa1), Hspa4, interleukin 6 receptor, alpha (Il6ra), S-adenosylhomocysteine hydrolase (Ahcy), DNA methyltransferase 3b (Dnmt3b), pyruvate dehydrogenase kinase 4 (Pdk4), and Sirt1 were compared between YND versus OND (E), YND versus YCR (F), and OND versus OCR (G). Data represent mean±SEM and were analyzed by two-way ANOVA using Bonferroni posttest. *, **, and *** indicate P<0.05, P<0.01, and P<0.001, respectively (YND, n=4–5 per time point; OND, n=3–4 per time point; YCR, n=5–6 per time point; OCR, n=3–5 per time point).
Fig. S3. Comparison of the circadian hepatic transcriptome between YCR and OCR (Related to Fig. 1 and 2). (A) Venn diagram displays the total number (top) and ratio (bottom) of rhythmic genes in the liver from YCR (purple, n=5–6 per time point) and OCR (blue, n=3–5 per time point), including common genes. (B) Amplitude plot represents the distribution of hepatic oscillatory genes in YCR and OCR. (C) Heatmaps display rhythmic genes exclusively in YCR (top left panel), OCR (top right panel), and in common (bottom panel). (D) GO analysis of genes exclusively circadian in YCR (top, purple), OCR (middle, blue), and common circadian genes (bottom, gray). Numbers within the pie charts indicate number of circadian genes identified within each biological process, based on a p-value cutoff of 0.01. (E) Determination of core clock gene expression in YCR and OCR by qPCR. Data represent mean±SEM and were analyzed by two-way ANOVA using Bonferroni posttest. *, **, and *** indicate P<0.05, P<0.01, and P<0.001, respectively.
Fig. S4. Comparison of the circadian hepatic transcriptome between YND and OCR (Related to Fig. 1 and 2). (A) Venn diagram displays the total number (top) and ratio (bottom) of rhythmic genes in the liver from YND (green, n=4–5 per time point) and OCR (blue, n=3–5 per time point), including common genes. (B) Amplitude plot exhibits the distribution of hepatic oscillatory genes in YND and OCR. (C) Heatmaps display genes exclusively circadian in YND (top left panel), OCR (top right panel), and in common (bottom panel). (D) GO analysis of genes exclusively circadian in YND (top, green), OCR (middle, blue), and common circadian genes (bottom, gray). Numbers within the pie charts indicate number of circadian genes identified within each biological process, based on a p-value cutoff of 0.01.
Fig. S5. Unique circadian signatures in NAD+ metabolism and protein acetylation in the liver versus stem cells (Related to Fig. 4). GO analysis of genes commonly circadian in all three tissues (liver, EpSCs, and MsSCs) harvested from YND (green), OND (orange), YCR (purple), and OCR (blue) animals. Numbers within the pie charts indicate number of circadian genes identified within each biological process. p-value cutoff of 0.01 was used. 3–6 biological replicates per time point per tissue were subjected to the transcriptomic analysis.
Fig. S6. Hepatic protein hyperacetylation under CR (Related to Fig. 5). (A) Circadian lysates from livers of YND, OND, YCR, and OCR were probed with a pan acetylated lysine antibody by western blot and quantified using three independent samples. The far right panel displays a low exposure image with a parallel comparison of YCR and OCR liver lysates. p84 was used as a loading control. (B) Hepatic β-hydroxybutyrate concentration was determined among YND, OND, YCR, and OCR groups by colorimetric assay. Data represent Means±SEM from three independent samples and was analyzed by two-way ANOVA using Bonferroni posttest. *, **, and *** indicate P<0.05, P<0.01, and P<0.001, respectively.
Fig. S7. Comprehensive analysis of histone modifications enriched at circadian promoters in YND, OND, YCR, and OCR (Related to Fig. 5). A comparative analysis was performed with the hepatic circadian transcriptome and ChIP-sequencing datasets for several histone modifications using EnrichR (http://amp.pharm.mssm.edu/Enrichr/). Adjusted p-values extending to 0.05 were allowed for enriched histone marks listed in each histogram.
Table S1. Primers used for qPCR (Related to Fig. 1, 5, 7, S2 and S3, and STAR Methods).
Table S2. Primers used for ChIP-qPCR (Related to Fig. 5 and STAR Methods).
Data Availability Statement
The GEO accession number for the microarray data reported in this paper is GSE93903.


