Abstract
Brain tumors are the most common solid tumors of childhood, and the genetic drivers and optimal therapeutic strategies for many of the different subtypes remain unknown. Here we identify that bithalamic gliomas harbor frequent mutations in the EGFR oncogene, only rare histone H3 mutation (in contrast to their unilateral counterparts), and a distinct genome-wide DNA methylation profile compared to all other glioma subtypes studied to date. These EGFR mutations are either small in-frame insertions within exon 20 (intracellular tyrosine kinase domain) or missense mutations within exon 7 (extracellular ligand-binding domain) that occur in the absence of accompanying gene amplification. We find these EGFR mutations are oncogenic in primary astrocyte models and confer sensitivity to specific tyrosine kinase inhibitors dependent on location within the kinase domain or extracellular domain. We initiated treatment with targeted kinase inhibitors in four children whose tumors harbor EGFR mutations with encouraging results. This study identifies a promising genomically-tailored therapeutic strategy for bithalamic gliomas, a lethal and genetically distinct brain tumor of childhood.
Keywords: bithalamic glioma, diffuse midline glioma, EGFR, histone H3, pediatric cancer, molecular neuropathology, tyrosine kinase inhibitor, afatinib, osimertinib, erlotinib, trametinib
INTRODUCTION
Central nervous system (CNS) tumors are the most common solid tumors of childhood and are the leading cause of cancer mortality in children 0–14 years of age [23]. Lethal high-grade brain tumors of childhood include medulloblastoma, atypical teratoid/rhabdoid tumor, ependymoma, and diffuse gliomas. The genetic drivers and optimal therapeutic strategy for many of the different subtypes of pediatric CNS tumors remain unknown.
Diffuse gliomas are malignant tumors of the CNS that cannot be cured by resection alone due to their infiltrative growth. Genomic studies over the past two decades have identified that there are several different molecular subtypes of diffuse gliomas that differ according to anatomic location (e.g. cerebral hemispheres versus midline structures), patient age (e.g. pediatric versus adult), and cell lineage (e.g. astrocytic versus oligodendroglial). For example, glioblastomas in the cerebral hemispheres of older adults are characterized by frequent TERT promoter mutation, EGFR amplification with accompanying mutation or rearrangement, PTEN inactivation, and CDKN2A homozygous deletion [5]. In contrast, diffuse lower-grade gliomas in the cerebral hemispheres of young adults are characterized by IDH1 or IDH2 mutation, with accompanying TP53 and ATRX mutations in astrocytic tumors versus accompanying TERT promoter, CIC, and FUBP1 mutations in oligodendroglial tumors [8]. Unlike diffuse gliomas within the cerebral hemispheres, diffuse gliomas arising within midline structures of the CNS (e.g. thalamus, brainstem, and spinal cord) are characterized by a recurrent lysine to methionine mutation at codon 27 (p.K27M) in the H3F3A or HIST1H3B genes that encode the histone H3 variants H3.3 and H3.1, respectively [18, 36, 42, 43]. The genetic differences in the diffuse glioma subtypes reflect distinct cells of origin and underlying molecular pathogenesis, which correlate with distinct clinical outcomes [10].
A unique and poorly characterized subtype of diffuse glioma involves the bilateral thalami at time of initial presentation, principally affecting young children. These bithalamic diffuse gliomas are not amenable to surgical resection and have a uniformly poor outcome despite radiation and conventional cytotoxic chemotherapy [4, 6, 11, 13, 15, 20, 22, 25, 28, 30, 31, 35, 38, 45]. Though diffuse midline gliomas with unilateral thalamic involvement frequently harbor histone H3 K27M mutation, bithalamic gliomas in children often lack this defining mutation [6]. Genome-wide DNA methylation profiling has also revealed that bithalamic gliomas have a distinct epigenome compared to their unilateral counterparts [6]. As such, a better understanding of the molecular pathogenesis of bithalamic gliomas is desperately needed to enable the implementation of new effective targeted therapies for affected children.
Herein, we performed comprehensive genomic and epigenomic analysis on a cohort of children with bithalamic gliomas. We identified that these tumors harbor frequent mutations in the EGFR oncogene in the absence of accompanying gene amplification, most frequently small in-frame insertions within exon 20 encoding the tyrosine kinase domain. We assessed therapeutic efficacy of a panel of EGFR kinase inhibitors in isogenic primary astrocyte models carrying EGFR mutations within the kinase domain or extracellular domain. We also initiated treatment with targeted kinase inhibitors in four children whose tumors harbor EGFR mutations with encouraging results to date. This study provides the foundation for a precision medicine treatment approach to bithalamic gliomas, a lethal and genetically distinct brain tumor of childhood.
METHODS
Study population and tumor specimens
This study was approved by the Committee on Human Research of the University of California, San Francisco, with a waiver of patient consent. Stereotactic biopsies and genomic testing for seven of the children with bithalamic gliomas were performed as part of standard prospective clinical management for pediatric neuro-oncology patients at UCSF Medical Center, whereas genomic testing was performed on a retrospective research basis for six children. Four of these retrospective patients (annotated in Supplementary Table 1 [Online supplement 1]) were previously reported in part, including histone H3 K27M mutation status and DNA methylation profiling [6]. Imaging features of the thirteen patients were reviewed by an expert neuroradiologist (J.V.M.). Pathologic review of all tumor samples was performed by two expert neuropathologists (A.P. and D.A.S.).
Immunohistochemistry
Immunohistochemistry was performed on whole formalin-fixed, paraffin-embedded tissue sections using the following antibodies: histone H3 K27M mutant protein (RevMAb Biosciences, cat # 31-1175-00, rabbit monoclonal clone RM192, 1:600 dilution), histone H3 lysine 27 trimethylated protein (Cell Signaling, cat #9733, rabbit monoclonal clone C36B11, 1:50 dilution), and EGFR (Ventana, cat # 790–4347, rabbit monoclonal clone 5B7, undiluted). Immunostaining for histone H3 K27M mutant protein and EGFR protein was performed in a Ventana BenchMark Ultra automated stainer using CC1 antigen retrieval. Immunostaining for histone H3 lysine 27 trimethylated protein was performed in a Leica Bond-Max automated stainer using ER2 antigen retrieval. Diaminobenzidine was used as the chromogen, followed by hematoxylin counterstain. Histone H3 lysine 27 trimethylation (H3K27me3) was scored as either intact, lost in a subset of the tumor cells (<75%), lost in the majority of tumor cells (>75%), or lost in all tumor cells (100%).
Targeted next-generation DNA sequencing and mutational analysis
Genomic DNA was extracted from formalin-fixed, paraffin-embedded blocks of tumor tissue from seven children with bithalamic diffuse gliomas using the QIAamp DNA FFPE Tissue Kit (Qiagen). Genomic DNA was also extracted from a peripheral blood sample for patient #10 using the QIAamp DNA Blood Midi Kit (Qiagen). Capture-based next-generation DNA sequencing was performed using an assay that targets all coding exons of 479 cancer-related genes, select introns and upstream regulatory regions of 47 genes to enable detection of structural variants including gene fusions, and DNA segments at regular intervals along each chromosome to enable genome-wide copy number and zygosity analysis, with a total sequencing footprint of 2.8 Mb (UCSF500 Cancer Panel; Supplementary Table 2 [Online supplement 1]; ref. 17). Multiplex library preparation was performed using the KAPA Hyper Prep Kit (Roche) according to the manufacturer’s specifications using 250 ng of sample DNA. Hybrid capture of pooled libraries was performed using a custom oligonucleotide library (Nimblegen SeqCap EZ Choice). Captured libraries were sequenced as paired-end 100 bp reads on an Illumina HiSeq 2500 instrument. Sequence reads were mapped to the reference human genome build GRCh37 (hg19) using the Burrows-Wheeler aligner (BWA). Recalibration and deduplication of reads was performed using the Genome Analysis Toolkit (GATK). Coverage and sequencing statistics (Supplementary Table 3 [Online supplement 1]) were determined using Picard CalculateHsMetrics and Picard CollectInsertSizeMetrics. While the mean on-target sequencing depth was high for most samples (>300x), three samples (patients #2, #4, and #9) had low mean on-target sequencing depth (between 17–26x) due to limited tumor tissue quantity that may have limited the sensitivity of detection for genetic alterations. Single nucleotide variant and insertion/deletion mutation calling was performed with FreeBayes and PinDel. Structural variant calling was performed with Delly. Variant annotation was performed with Annovar. Single nucleotide variants, insertions/deletions, and structural variants were visualized and verified using Integrative Genome Viewer. Genome-wide copy number analysis based on on-target and off-target reads was performed by CNVkit and visualized using Nexus Copy Number (Biodiscovery).
EGFR exon 20 Sanger sequencing
PCR amplification of exon 20 of the EGFR gene was performed on genomic DNA from the bithalamic glioma of patient #7, as well as medulloblastoma from an unrelated patient, using the MyFi DNA Polymerase Kit (Bioline). PCR products were treated with ExoSAP (Affymetrix) and then sequenced using BigDye terminator chemistry (Applied Biosystems) following standard techniques. Primer sequences used for amplification and sequencing of EGFR exon 20 were as follows:
EGFR ex20 Fwd: 5′-CATGCGTCTTCACCTGGAA-3′
EGFR ex20 Rev: 5′-CTGCGTGATGAGCTGCAC-3′.
Genome-wide DNA methylation profiling
Genomic DNA from ten of the bithalamic gliomas was bisulfite converted using the EZ DNA Methylation kit following the manufacturer’s recommended protocol (Zymo Research). Bisulfite converted DNA was then amplified, fragmented, and hybridized to Infinium EPIC 850k Human DNA Methylation BeadChips or Infinium 450k Human DNA Methylation BeadChips following the manufacturer’s recommended protocol (Illumina). The approximately 450,000 overlapping CpG sites between the 850K and 450K BeadChips were used in the analysis. Methylation data were preprocessed using the minfi package (v.1.28.4) in R Bioconductor (version 3.5.3) [3]. The detection p-value for each sample was computed. All samples had detection p values less than 0.002. Additional quality control was performed by calculating the median log (base2) intensities for methylated and unmethylated signals for each array. All samples had unmethylated and methylated median intensity values above 10 that were used for analysis. Beta density plots for all samples before and after normalization were also examined. Functional normalization with NOOB background correction and dye-bias normalization was performed [12, 39]. Probe filtering was performed after normalization. Specifically, probes located on sex chromosomes, containing nucleotide polymorphisms (dbSNP132 Common) within five base pairs of and including the targeted CpG site, or mapping to multiple sites on hg19 (allowing for one mismatch), as well as cross reactive probes were removed from analysis. Unsupervised analysis of methylation data from our cohort of 10 pediatric bithalamic gliomas alongside 616 glioma reference samples from GSE90496 (Supplementary Table 9 [Online supplement 1]) or 2,801 reference samples spanning 82 CNS tumor entities and 9 control tissues from GSE90496 (Supplementary Table 8 [Online supplement 1]) was performed using hierarchical clustering and t-distributed stochastic neighbor embedding (t-SNE) using Rtsne (v.0.15). For the t-SNE plots shown in Fig. 3 and Supplementary Fig. 7 [Online supplement 2], the 32,000 most variably methylated CpG probes, as measured by the standard deviation of the probe-level beta values across samples, was selected for analysis. For the hierarchical clustering plot shown in Supplementary Fig. 8 [Online supplement 2], the 5,000 most variably methylated CpG probes was selected for analysis. Pearson correlation was calculated as the distance measured between samples.
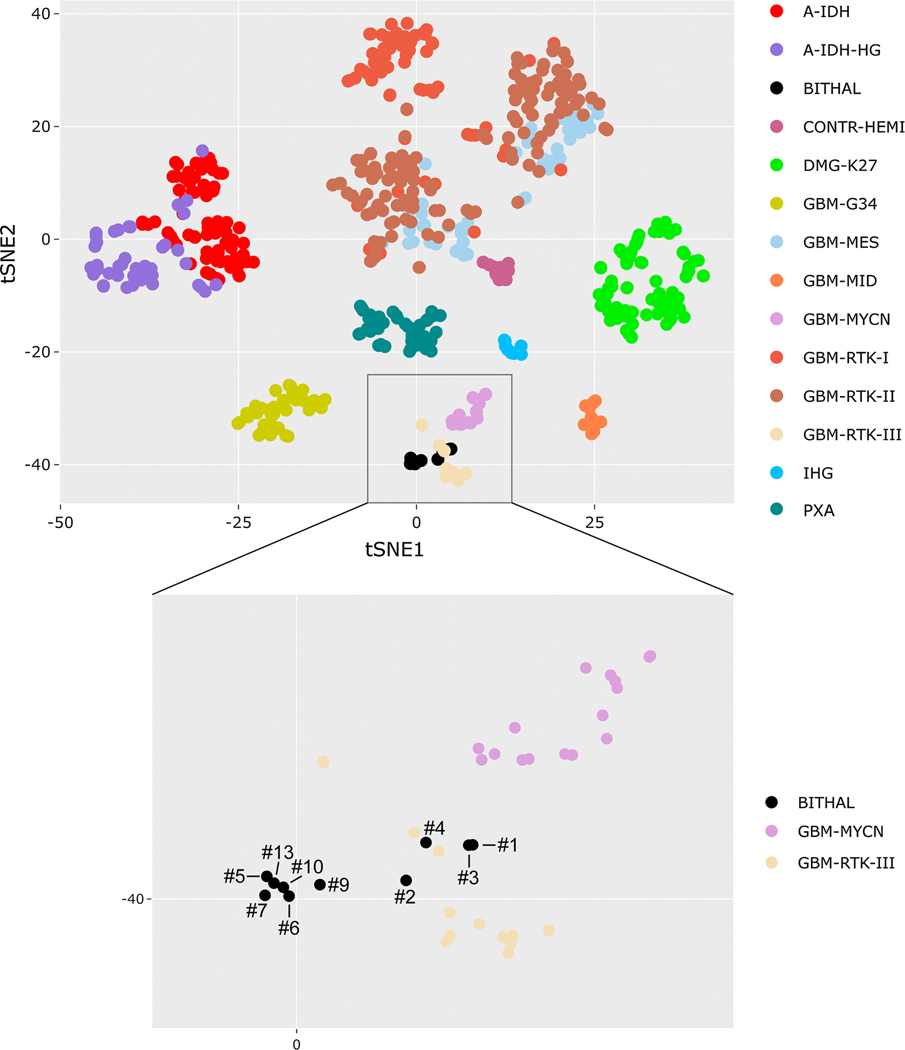
Fig. 3.
Epigenomic analysis reveals that pediatric bithalamic gliomas are a distinct glioma entity. Genome-wide DNA methylation profiles from ten bithalamic gliomas were clustered together with 616 glioma reference samples (Supplementary Table 9 [Online supplement 1]) by t-SNE analysis. Bithalamic gliomas (BITHAL) form a cluster that is distinct from all glioma entities that have been studied to date, albeit with some limited overlap with the “Glioblastoma, IDH-wildtype, RTK III subclass”, which is a poorly characterized subgroup of glioblastomas in the cerebral hemispheres of children [9]. The bithalamic gliomas are entirely distinct from their unilateral thalamic counterparts belonging to the methylation class “Diffuse midline glioma, H3 K27M-mutant” (DMG-K27), and are also distinct from the “Glioblastoma, IDH-wildtype, midline subclass” (GBM-MID) that is comprised of glioblastomas located in midline structures lacking H3 K27M mutation [9]. A-IDH, IDH-mutant astrocytoma. CONTR-HEMI, normal cerebral hemisphere. GBM-G34, diffuse hemispheric glioma with H3 G34 mutation. IHG, infantile hemispheric glioma. PXA, pleomorphic xanthoastrocytoma.
EGFR cDNA expression vector construction and site-directed mutagenesis
The pBabe-EGFR human wildtype cDNA expression vector was obtained from Addgene (plasmid #11011). p.A289T (exon 7, extracellular domain) and p.A767delinsASVG (exon 20, intracellular kinase domain) mutations were engineered into the pBabe-EGFR construct by site-directed mutagenesis using the QuikChange II XL kit (Stratagene) as directed by the manufacturer. The entire coding sequence of all expression vectors was verified by Sanger sequencing. Primer sequences used for the mutagenesis reactions were as follows: A289T Fwd:
5′- CAAATACAGCTTTGGTACCACCTGCGTGAAGAAG −3′
A289T Rev: 5′- CTTCTTCACGCAGGTGGTACCAAAGCTGTATTTG −3′
A767delinsASVG Fwd 5′- CGATGAAGCCTACGTGATGGCCAGCGTGGGCAGCGTGGACAACCCCCACG −3′
A767delinsASVG Rev: 5′- CGTGGGGGTTGTCCACGCTGCCCACGCTGGCCATCACGTAGGCTTCATCG −3′.
Immortalized human astrocytes and cell culture
SVG-p12 immortalized human fetal astrocytes were obtained directly from ATCC. Normal human astrocytes immortalized by retroviral transduction of hTERT, E6, and E7 proteins were generously provided by Dr. Russ Pieper (University of California, San Francisco). Immortalized human astrocyte cultures were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum at 37°C in 5% CO2.
Lentiviral production and infection
Empty pBabe vector or pBabe-EGFR wildtype and mutant expression vectors were co-transfected into 293T cells with pVSV-G and pHR-CMV8.2deltaR helper plasmids using Fugene 6 (Roche) as described by the manufacturer. Virus-containing conditioned medium was harvested 48 hours after transfection, filtered, and used to infect recipient cells in the presence of 5 ug/mL polybrene.
Western blot
Primary antibodies used were EGFR (Cell Signaling, cat #4267, clone D38B1, 1:500 dilution), phosphop-44/42 ERK Thr202/Tyr204 (Cell Signaling, cat #4370, clone D13.14.4E, 1:250 dilution), phospho-Akt Ser473 (Cell Signaling, cat #4060, clone D9E, 1:250 dilution), and β-actin (Sigma, cat# A5441, clone AC-15, 1:5,000 dilution). Total cell lysates were extracted from immortalized human astrocytes in RIPA buffer containing protease and phosphatase inhibitor cocktail (Pierce, cat# 88668), resolved by SDS-PAGE, and immunoblotted following standard biochemical techniques.
Soft agar colony forming assay
EGFR wildtype and mutant expressing lentivirus was added to exponentially growing immortalized human astrocytes in the presence of 5 ug/mL polybrene. At 24 hours following infection, 20,000 cells were suspended in 0.3% agar in DMEM + 10% fetal bovine serum and cast onto a bottom layer of 0.6% agar in DMEM + 10% fetal bovine serum in 6-well tissue culture plates. Soft agar plates were incubated in a humidified chamber at 37°C containing 5% CO2 for 21 days. Colonies were imaged and counted on a GelCount Colony Counter (Oxford Optronix). 12 replicates from two independent experiments were performed for each experimental condition.
Kinase inhibitor sensitivity assay
The CellTiter-Glo assay (Promega) was used to assess cell survival following treatment with a panel of small molecule kinase inhibitors on two immortalized human astrocyte cell lines. Following lentiviral transduction with empty vector, wildtype EGFR, or mutant EGFR isoforms, 200 exponentially growing cells were seeded per well in clear-bottom white polystyrene 96-well microplates (Corning 3903). At 24 hours after seeding, cells were treated with DMSO vehicle or the small molecule kinase inhibitors at the indicated concentrations. At 4 days following treatment, an equal volume of CellTiter-Glo One Solution reagent was added to each of the wells, followed by shaking for five minutes at 120 rpm at room temperature. Luminescence was measured using the SpectraMax M5 spectrophotometer (Molecular Devices). All drugs were obtained from Selleck Chemicals and dissolved in DMSO. The following agents were used: erlotinib (cat # S1023), gefitinib (cat # S1025), lapatinib (cat # S2111), poziotinib (cat # S7358), afatinib (cat # S1011), osimertinib (cat # S7297), neratinib (cat # S2150), and trametinib (cat # S2673). 9 replicates from 3 independent experiments were performed for each experimental condition in the SVG-p12 cells, and 6 replicates from 2 independent experiments were performed for each experimental condition in the iNHA cells. Percent survival was assessed as the mean luminescence of the experimental condition relative to the mean luminescence of the cells treated with DMSO vehicle.
Quantification and statistical analysis
Statistical analysis was performed using GraphPad Prism. All error bars shown in colony forming assay and kinase inhibitor sensitivity assay plots represent standard error of the mean. The Kaplan-Meier survival analysis shown in Fig. 5b and 5c includes 41 children with bithalamic gliomas, comprised of the 13 patients from this study combined with all historical cases of biopsy-proven bithalamic diffuse gliomas in children reported in the literature (see collated clinical data in Supplementary Table 11 [Online supplement 1]). This survival analysis encompasses all reported cases that could be identified of diffuse gliomas of the bilateral thalami in children with histology reported as diffuse astrocytoma, anaplastic astrocytoma, or glioblastoma (excludes pilocytic astrocytoma and astrocytomas with KIAA1549-BRAF fusion, as these do not represent true bithalamic diffuse gliomas). The Kaplan-Meier survival analysis shown in Fig. 5d is limited to the 11 children from this study cohort with bithalamic gliomas harboring EGFR mutations and is stratified by those whose therapy included a small molecule EGFR or MEK kinase inhibitor versus those children whose treatment did not include a small molecule tyrosine kinase inhibitor. p value was calculated by Log-rank (Mantel-Cox) test.
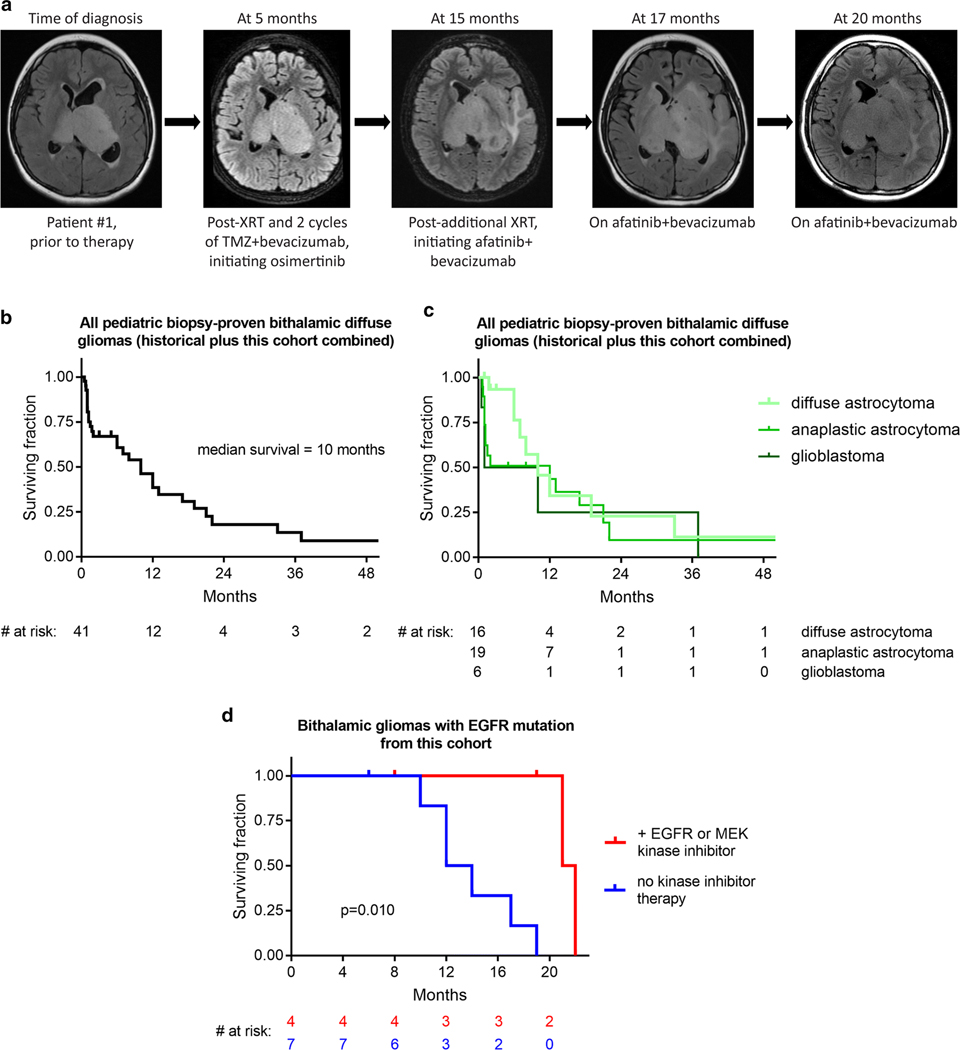
Fig. 5.
Small molecule tyrosine kinase inhibition may extend survival for children with bithalamic gliomas. a, Imaging showing disease course for patient #1 whose therapy included both osimertinib and afatinib. Images are from axial T2/FLAIR sequences taken at different stages during treatment. b, Kaplan-Meier survival analysis of 41 children with bithalamic gliomas, including the 13 children from this study combined with all historical cases of biopsy-proven bithalamic diffuse gliomas reported in the literature (see Supplementary Table 11 [Online supplement 1]). c, Kaplan-Meier survival analysis of these 41 children with bithalamic gliomas stratified based on histologic classification as diffuse astrocytoma, anaplastic astrocytoma, or glioblastoma. d, Kaplan-Meier survival analysis of the 11 children with bithalamic gliomas harboring EGFR mutations from this study cohort stratified by those whose therapy included a small molecule EGFR or MEK kinase inhibitor (n=4) versus those children whose treatment did not include a small molecule tyrosine kinase inhibitor (n=7). p value was calculated by Log-rank (Mantel-Cox) test.
Availability of Data and Material
Targeted DNA sequencing data from this study are available from the European Genome-Phenome Archive (EGA) repository under accession number EGAS00001004033 (https://ega-archive.org/). DNA methylation array data from this study are available from the Gene Expression Omnibus (GEO) repository under accession number GSE140124 (https://www.ncbi.nlm.nih.gov/geo/).
RESULTS
Genomic Landscape of Pediatric Bithalamic Gliomas
In order to investigate the molecular pathogenesis of bithalamic gliomas, we performed targeted next-generation sequencing and genome-wide copy number analysis on tumor samples from thirteen children. The ten male and three female patients, ages 2–16 years, all had diffuse gliomas involving the bilateral thalami at time of initial presentation (Fig. 1a, Table 1, and Supplementary Table 1 [Online supplement 1]). Imaging revealed bulky expansion of the bilateral thalami that was T2 and FLAIR hyperintense, and either lacked enhancement (eleven cases) or had minimal enhancement (two cases) following contrast administration (Supplementary Fig. 1 [Online supplement 2] and Supplementary Table 4 [Online supplement 1]). Infiltration into the basal ganglia, brainstem, or insular cortex was often seen at time of initial imaging. Only one patient (#12) was identified to have disseminated disease throughout the ventricular system and along the spinal cord at time of presentation. All tumors were unresectable, with only a stereotactic biopsy (ten cases) or limited subtotal resection (three cases) performed for diagnosis and molecular profiling. Histologic diagnosis was diffuse astrocytoma for three tumors, anaplastic astrocytoma for nine tumors, and glioblastoma for one tumor (Supplementary Fig. 2 [Online supplement 2] and Supplementary Table 5 [Online supplement 1]). All were diffusely infiltrating astrocytic neoplasms with variable cellularity and mitotic activity, which uniformly lacked both necrosis and microvascular proliferation except for patient #6 whose tumor had foci of necrosis.
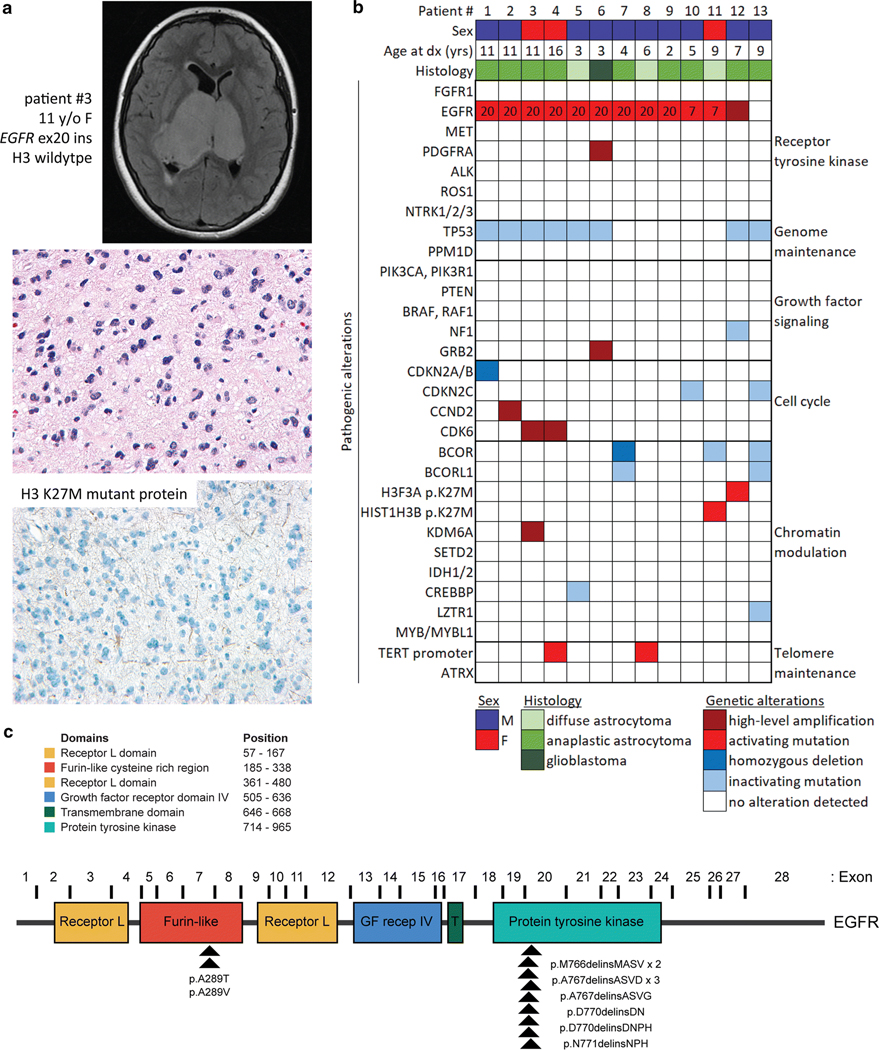
Fig. 1.
Genetic landscape of pediatric bithalamic gliomas. a, Axial T2-weighted fluid-attenuated inversion recovery (FLAIR) MR image from a 11-year-old girl (patient #3) showing an expansile mass involving the bilateral thalami (top). Histology from the stereotactic biopsy revealing a diffuse astrocytic neoplasm with pleomorphic nuclei and occasional mitotic figures (H&E stain, 400x magnification, middle). Negative immunohistochemical stain for histone H3 K27M mutant protein (400x magnification, bottom). b, Oncoprint table of the clinical features and likely pathogenic alterations identified in biopsies from thirteen children with bithalamic gliomas. Exon number where the EGFR mutations are located is annotated. c, Diagram of human EGFR protein with the location of the recurrent exon 20 small in-frame insertions within the intracellular tyrosine kinase domain and p.A289T/V missense mutations within the extracellular ligand-binding domain. UniProt P00533, RefSeq NM_005228.
Table 1.
Clinical data for the cohort of thirteen children with bithalamic gliomas.
| Patient | Age (yrs) | Sex | Surgical procedure | Histology | Genetic alterations | Radiation therapy | Initial chemotherapy | Additional radiation | Additional chemotherapy | Outcome | Follow-up (months) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| #1 | 11 | M | biopsy | anaplastic astrocytoma | EGFR ex20 ins, TP53 fs, CDKN2A/B del | localized 59 Gy | temozolomide, bevacizumab, osimertinib | 30 Gy | bevacizumab, afatinib | died of disease | 22 |
| #2 | 11 | M | biopsy | anaplastic astrocytoma | EGFR ex20 ins, TP53 mis, CCND2 amp | localized 54.4 Gy | temozolomide, valproic acid | none | etoposide | died of disease | 14 |
| #3 | 11 | F | biopsy | anaplastic astrocytoma | EGFR ex20 ins, TP53 mis, CDK6 amp, KDM6A amp | localized 59.4 Gy | temozolmide, CCNU | none | trametinib, ribociclib | died of disease | 21 |
| #4 | 16 | F | biopsy | anaplastic astrocytoma | EGFR ex20 ins, TP53 fs, CDK6 amp, TERT promoter | localized 54 Gy | temozolomide, valproic acid | none | none | died of disease | 12 |
| #5 | 3 | M | biopsy | diffuse astrocytoma | EGFR ex20 ins, TP53 indel, CREBBP fs | localized 54 Gy | temozolmide, osimertinib | none | bevacizumab, CCNU | alive with progressive disease | 8 |
| #6 | 3 | M | STR | glioblastoma | EGFR ex20 ins, PDGFRA amp, TP53 mis, GRB2 amp | localized 54 Gy | methotrexate, vincristine, cisplatin, cyclophosphamide | none | none | died of disease | 12 |
| #7 | 4 | M | STR | anaplastic astrocytoma | EGFR ex20 ins, BCOR del, BCORL1 non | localized 54 Gy | temozolomide, carboplatin, vincristine | none | bevacizumab, enzastaurin, CCNU | died of disease | 17 |
| #8 | 6 | M | biopsy | diffuse astrocytoma | EGFR ex20 ins, TERT promoter | declined | carboplatin, vincristine | none | thioguanine, procarbazine, CCNU, vincristine | died of disease | 19 |
| #9 | 2 | M | biopsy | anaplastic astrocytoma | EGFR ex20 ins | localized 46 Gy | methotrexate, vincristine, cisplatin, cyclophosphamide | none | bevacizumab, topotecan, vorinostat | died of disease | 10 |
| #10 | 5 | M | biopsy | anaplastic astrocytoma | EGFR ex7 mis, CDKN2C indel | localized 59 Gy | temozolomide, afatinib | 30 Gy | bevacizumab, CCNU, osimertinib | alive with progressive disease | 19 |
| #11 | 9 | F | biopsy | diffuse astrocytoma | EGFR ex7 mis, HIST1H3B K27M, BCORL1 fs | localized 54 Gy | ONC201 | none | none | alive with stable disease | 6 |
| #12 | 7 | M | biopsy | anaplastic astrocytoma | EGFR amp, TP53 mis, NF1 non+fs, H3F3A K27M | craniospinal 36 Gy + 4 Gy thalamic boost | temozolomide, bevacizumab, cetuximab | none | none | died of disease | 2 |
| #13 | 9 | M | STR | anaplastic astrocytoma | TP53 mis, CDKN2C indel, BCOR fs, BCORL1 non, LZTR1 fs | localized 59 Gy | bevacizumab | none | none | died of disease | 13 |
STR, subtotal resection. ins, small in-frame insertion. fs, frameshift mutation. del, homozygous deletion. mis, missense mutation. amp, focal amplification. indel, small in-frame insertion or deletion, nonsense mutation.
EGFR and MEK small molecule tyrosine kinase inhibitors are in bold.
Eleven tumors (85%) harbored mutations in the EGFR oncogene in the absence of accompanying EGFR gene amplification (Fig. 1b and Supplementary Table 6 [Online supplement 1]). Nine of these EGFR mutations were small in-frame insertions within exon 20 that encodes a portion of the intracellular tyrosine kinase domain (Fig. 1c and Supplementary Figs. 3 and 4 [Online supplement 2]), where mutations are common in lung adenocarcinoma [2, 24, 44] and fibrous hamartoma of infancy [26] but have not been previously identified in gliomas. The other two cases had EGFR mutations (p.A289T or p.A289V) located in exon 7 within the extracellular ligand-binding domain (Fig. 1c and Supplementary Fig. 5 [Online supplement 2]). While two of the bithalamic gliomas harbored p.A289T/V that is a mutational hotspot in the extracellular domain recurrently seen in glioblastomas in adults, the exon 20 insertions identified in nine of the bithalamic gliomas were not seen in any of 326 pediatric high-grade and diffuse intrinsic pontine gliomas from a recent meta-analysis [18], nor the greater than 1,100 adult diffuse gliomas studied to date by The Cancer Genome Atlas [10].
Only two of the thirteen bithalamic gliomas lacked identifiable EGFR mutations, one of which harbored focal high-level amplification of a wildtype EGFR allele (patient #12). The one bithalamic glioma lacking EGFR mutation or amplification in this cohort (patient #13) was both radiographically and histologically distinct compared to the other twelve tumors (Supplementary Fig. 2 [Online supplement 2] and Supplementary Tables 4 and 5 [Online supplement 1]). A more heterogeneous appearance with both enhancement and restricted diffusion were seen on MR imaging, whereas higher cell density, calcifications, and more severe pleomorphism were seen on histology compared to the other twelve tumors.
Whereas histone H3 K27M mutation is frequent in diffuse gliomas with unilateral involvement of the thalamus at time of presentation, we find that bithalamic gliomas in children rarely harbor H3 K27M mutation (only two of thirteen cases, 15%). Instead, we find that they often harbor alterations in other epigenetic regulatory genes. One tumor harbored focal high-level amplification of KDM6A, which encodes a lysine demethylase enzyme with specificity for lysine 27 of histone H3. Three tumors harbored homozygous deletions or truncating mutations in the BCOR and BCORL1 transcriptional co-repressor genes (two with dual inactivation of both genes), one tumor harbored a truncating mutation in the CREBBP histone acetyltransferase gene, and one harbored a truncating mutation in the LZTR1 transcriptional regulatory gene (Fig. 1b). Thus, while bithalamic gliomas in children often lack the histone H3 K27M mutation that is frequent in their unilateral counterparts, they often harbor alterations in other epigenetic regulators. Immunohistochemical staining for histone H3 lysine 27 trimethylation (H3K27me3) revealed loss in eight of the nine evaluated tumors (89%, Supplementary Fig. 2 [Online supplement 2] and Supplementary Table 5 [Online supplement 1]), despite only one of these harboring HIST1H3B p.K27M mutation, indicating alternative mechanisms in bithalamic gliomas of blocking this histone tail modification critical for glial cell differentiation.
Additionally, inactivating mutations of the TP53 tumor suppressor gene were present in eight cases (62%), and alterations of cell cycle regulators were also frequent (38%). Two tumors harbored mutations in CDKN2C, two harbored focal high-level amplification of CDK6, one harbored focal high-level amplification of CCND2, and one harbored focal homozygous deletion of CDKN2A/B (Fig. 1b, Supplementary Fig. 6 [Online supplement 2], and Supplementary Table 7 [Online supplement 1]). Only two cases harbored hotspot mutations in the promoter region of the TERT gene, and none of the other cases were found to harbor alterations associated with telomere maintenance (e.g. ATRX mutation/deletion or TERT amplification or promoter rearrangement). Except for one tumor that harbored focal PDGFRA amplification in addition to EGFR exon 20 insertion (patient #6), this cohort of bithalamic gliomas lacked fusions, amplifications, or mutations in other receptor tyrosine kinase genes known to be common in pediatric gliomas (e.g. MET, FGFR1, ALK, ROS1, and NTRK1-3). Notably, mutations or rearrangements involving BRAF, RAF1, MYB, and MYBL1 that are common in pediatric low-grade gliomas were also not seen in any of the tumors. Additionally, mutations in components of the PI3-kinase pathway (PIK3CA, PIK3R1, and PTEN) that are common in both pediatric and adult gliomas were absent in this cohort of bithalamic gliomas. The somatic mutation burden was very low in each of the tumors (less than 3 somatic mutations per Mb). Two of the tumors (#5 and #8) demonstrated flat copy number profiles, while the other cases demonstrated several chromosomal gains and losses. Recurrent cytogenetic alterations included trisomy 1q present in seven tumors (54%), trisomy 7 in six tumors (46%), trisomy 5 or 5p in four tumors (31%), and monosomy 17p in four tumors (31%, Supplementary Table 7 [Online supplement 1] and representative genome-wide copy number plots shown in Supplementary Fig. 6 [Online supplement 2]). Four of the six tumors with trisomy 7 were those harboring EGFR mutations that was likely resulting in extra copies of the mutant allele in tumor cells.
Multi-Region Sequencing of Bithalamic Gliomas Confirms Clonality of EGFR Mutations
The variant allele frequency for the EGFR mutations identified in the eleven bithalamic gliomas ranged from 17% to 61% (Supplementary Table 6 [Online supplement 1]). Cases with the highest EGFR variant allele frequencies had genomic DNA isolated from areas histologically visualized to contain a high tumor cell content (and included those harboring trisomy 7), whereas cases with the lowest mutant allele frequencies had genomic DNA that was isolated from tumors containing more sparse infiltrating tumor nuclei. Therefore, these data suggest that EGFR mutation is a clonal heterozygous alteration in bithalamic gliomas, indicating that it is probably an early or initiating event in tumorigenesis. To further assess this hypothesis, we performed multi-region sequencing analysis on two of the bithalamic gliomas harboring EGFR mutations.
Sequencing of a stereotactic biopsy at time of initial presentation for patient #10 revealed EGFR p.A289T mutation, CDKN2C p.D95del mutation, and gains of chromosomes 1q, 5, 7, and distal Xq. This child was treated with 59 Gy of intensity modulated radiation therapy, followed by a combination of temozolomide plus afatinib, a small molecule kinase inhibitor with specificity for EGFR. Surveillance imaging revealed slow disease progression over the course of 14 months, with increasing disease burden extending into the bilateral basal ganglia and insular cortices (Fig. 2a). A repeat stereotactic biopsy from the right temporal lobe was then performed for clinical trial eligibility purposes. Sequencing analysis performed on this biopsy revealed the identical EGFR p.A289T mutation and gains of chromosomes 1q, 5, and 7; however, the CDKN2C mutation and gain of distal chromosome Xq seen in the initial biopsy from the right thalamus were not present.
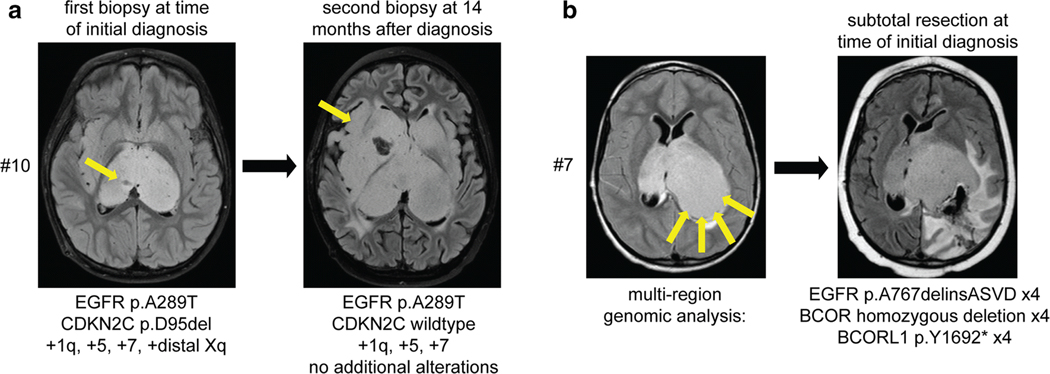
Fig. 2.
Multi-region sequencing of pediatric bithalamic gliomas reveals that EGFR mutations are clonal and likely an early or initiating event during tumorigenesis. a, Sequencing of a stereotactic biopsy from the right thalamus at time of initial presentation for patient #10 revealed EGFR p.A289T mutation, CDKN2C p.D95del mutation, and gains of chromosomes 1q, 5, 7, and distal Xq. At 14 months following radiation and chemotherapy, a repeat stereotactic biopsy from the right temporal lobe was performed with sequencing analysis revealing the identical EGFR p.A289T mutation and gains of chromosomes 1q, 5, and 7. b, Imaging at time of initial presentation for patient #7 revealed substantial involvement of the bilateral thalami but with more bulky disease burden in the left thalamus. A limited subtotal resection from the posterior aspect of the left thalamus was performed, with sequencing analysis performed on four independent areas of tumor from this resection specimen. The identical EGFR p.A767delinsASVD insertion within exon 20, focal homozygous deletion of BCOR, and nonsense mutation in BCORL1 was present in each of the four tumor regions.
Brain imaging at time of initial presentation for patient #7 revealed substantial involvement of the bilateral thalami, but was asymmetric with more bulky disease burden in the left thalamus. This child underwent a limited subtotal resection from the posterior aspect of the left thalamus, and sequencing analysis was performed on four independent areas of tumor from this resection specimen (Fig. 2b). The identical EGFR p.A767delinsASVD insertion within exon 20 was present in each of the four tumor regions, as was focal homozygous deletion of BCOR and a truncating nonsense mutation in BCORL1. Together, the findings from multi-region sequencing of these two patients further support that EGFR mutation is a clonal alteration in bithalamic gliomas, thereby indicating it is likely an early or initiating event in tumorigenesis.
Epigenomic Analysis Reveals that Bithalamic Gliomas are a Distinct Glioma Entity
We next studied the genome-wide DNA methylation profiles from ten of the thirteen bithalamic gliomas using Infinium Methylation 850k or 450k BeadChip arrays (Illumina). Analysis of DNA methylation profiles was performed by t-SNE plotting (Fig. 3 and Supplementary Fig. 7 [Online supplement 2]) and unsupervised hierarchical clustering (Supplementary Fig. 8 [Online supplement 2]) against 616 glioma reference samples or 2,801 reference samples spanning 82 CNS tumor entities and 9 control tissues (Supplementary Tables 8 and 9 [Online supplement 1]). Additionally, random forest classification using the DKFZ MolecularNeuropathology.org online classifier tool (version 11b4) was performed (Supplementary Table 10 [Online supplement 1]). This analysis demonstrated that bithalamic gliomas harbor DNA methylation patterns distinct from all glioma and other brain tumor entities that have been studied to date, albeit with some limited overlap with the “RTK III subclass” of the methylation class “Glioblastoma, IDH-wildtype”, which is a poorly characterized subgroup of IDH-wildtype glioblastomas in the cerebral hemispheres of children [9]. Notably, the bithalamic gliomas were entirely distinct from their unilateral counterparts belonging to the methylation class “Diffuse midline glioma, H3 K27M-mutant”, and were also distinct from the “Glioblastoma, IDH-wildtype, midline subclass” that is comprised of glioblastomas located in midline structures lacking H3 K27M mutation but frequently harboring FGFR1 mutations [9]. Unfortunately, no remaining tumor tissue was available for methylation analysis from the two bithalamic gliomas in this cohort which harbored histone H3 K27M mutations (patient #11 with dual EGFR p.A289T and HIST1H3B p.K27M mutations and patient #12 with dual EGFR amplification and H3F3A p.K27M mutation).
EGFR Exon 20 Kinase Domain Insertion Transforms Human Astrocytes
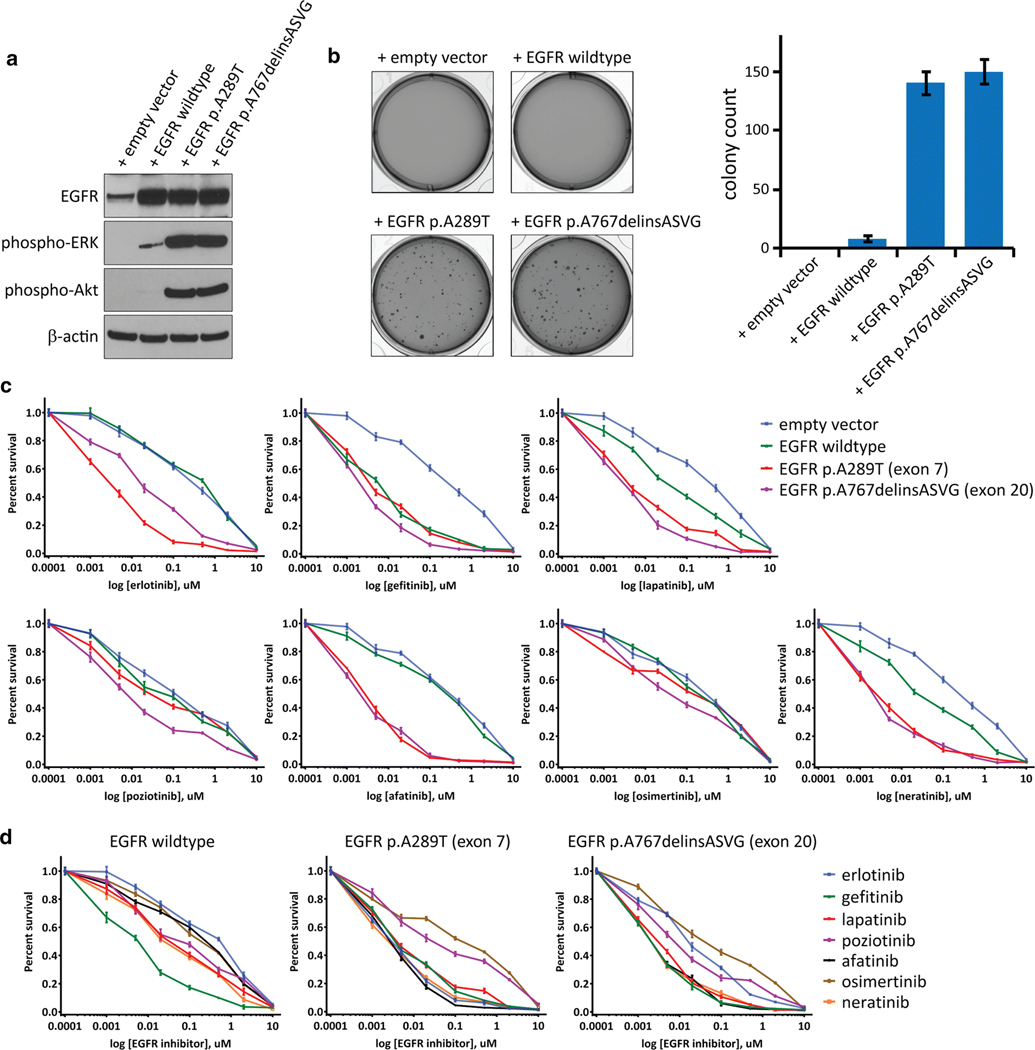
As EGFR exon 20 small in-frame insertions have not been previously identified in gliomas, we examined the effects of introducing one of these insertion mutations in primary human astrocytes. Lentiviral transduction of SVG-p12 cells, an immortalized fetal astrocyte cell line, with empty vector, EGFR wildtype, EGFR p.A289T mutation located within exon 7 in the extracellular ligand-binding domain, and EGFR p.A767delinsASVG mutation located within exon 20 in the intracellular tyrosine kinase domain was performed. Similar levels of ectopic overexpression were observed for the wildtype and mutant EGFR isoforms (Fig. 4a). The exon 20 insertion mutant resulted in increased phosphorylation of ERK1/2 and Akt, similar to the p.A289T mutation (Fig. 4a). Both the exon 20 insertion mutation and the p.A289T extracellular domain mutation conferred anchorage-independent colony formation in soft agar to a similar extent (Fig. 4b). In contrast, SVG-p12 cells transduced with empty vector or wildtype EGFR did not yield appreciable colony formation (Fig. 4b). These data indicate that the exon 20 insertion mutations present in pediatric bithalamic gliomas are oncogenic alterations, equivalent in tumorigenic capacity to the extracellular domain mutations in EGFR that are known to be common in glioblastomas in adults.
Fig. 4.
EGFR mutations in pediatric bithalamic gliomas are oncogenic and confer sensitivity to specific tyrosine kinase inhibitors. a, Western blots on total cell lysate from SVG-p12 immortalized human astrocytes after lentiviral transduction with empty vector, EGFR wildtype, or EGFR mutant isoforms. b, Colony formation in soft agar of SVG-p12 immortalized human astrocytes after lentiviral transduction with empty vector, EGFR wildtype, or EGFR mutant isoforms. Images of representative wells (left) and quantitation of colony number per well (right) are shown. Error bars represent standard error from the mean of twelve replicates from two independent experiments. c, Survival plots of SVG-p12 immortalized human astrocytes transduced with empty vector, EGFR wildtype, and EGFR mutant isoforms followed by treatment with various tyrosine kinase inhibitors at the indicated doses for 72 hours. Each data point is the mean of 9 replicates from three independent experiments, and error bars show standard error of the mean. d, Comparison of kinase inhibitor efficacy in SVG-p12 immortalized human astrocytes expressing EGFR wildtype or mutant isoforms.
Targeted Inhibition of Mutant EGFR Depends on Mutation Location in the Extracellular Domain Versus Intracellular Kinase Domain
In order to guide the precision treatment of children with bithalamic gliomas, we investigated the efficacy of a panel of small molecule tyrosine kinase inhibitors in two isogenic human astrocyte models, SVG-p12 cells and normal human astrocytes immortalized with ectopic expression of TERT and HPV E6 and E7 proteins (iNHAs) that have been previously described [37]. Following lentiviral transduction with empty vector, wildtype EGFR, p.A289T mutant, and p.A767delinsASVG mutant isoforms, cells were treated with the panel of kinase inhibitors. All kinase inhibitors reduced viability of the astrocytes expressing mutant EGFR isoforms compared to empty vector (Fig. 4c and Supplementary Fig. 9 [Online supplement 2]). However, the relative difference in survival between EGFR mutant isoforms, wildtype EGFR, and empty vector were variable among the different compounds tested. Gefitinib indiscriminately reduced survival of cells overexpressing either wildtype or mutant EGFR isoforms in comparison to empty vector. In contrast, afatinib only reduced survival of astrocytes overexpressing mutant EGFR isoforms but not wildtype. Erlotinib reduced survival of astrocytes expressing p.A289T extracellular domain mutation to a greater extent than those expressing p.A767delinsASVG exon 20 kinase domain mutation, whereas poziotinib reduced survival of astrocytes expressing p.A767delinsASVG exon 20 kinase domain mutation to a greater extent than those expressing p.A289T extracellular domain mutation. To ascertain which kinase inhibitor produced the greatest survival reduction in astrocytes overexpressing wildtype and mutant EGFR isoforms, we next compared the relative survival data across the different compounds (Fig. 4d). For astrocytes overexpressing wildtype EGFR, gefitinib produced the greatest reduction in relative survival among the tested inhibitors. For the p.A289T extracellular domain mutation, erlotinib, gefitinib, lapatinib, afatinib, and neratinib produced comparable reductions in relative survival. For the p.A767delinsASVG intracellular kinase domain mutation, gefitinib, lapatinib, afatinib, and neratinib produced comparable reductions in relative survival. Together, these data provide the foundation for a genomically-guided treatment approach using specific kinase inhibitors dependent on the precise EGFR alteration present in a patient’s glioma.
Treatment of Children with Bithalamic Gliomas Harboring EGFR Exon 20 Insertion with Kinase Inhibitors
The treatment regimens and clinical outcomes for the thirteen children with bithalamic gliomas in this study cohort are shown in Table 1 (with more extensive clinical data including radiation method and chemotherapy dosage listed in Supplementary Table 1 [Online supplement 1]). As there are no known effective treatments for bithalamic gliomas, four children in this study cohort were treated with EGFR or MEK small molecule kinase inhibitors. Patient #1 is an 11-year-old boy who was treated with intensity-modulated radiation therapy and then with a combination of osimertinib, temozolomide, and bevacizumab given the finding of EGFR exon 20 insertion. Slow tumor growth over the following 12 months was seen on follow-up imaging, at which point this child was treated with an additional course of radiation therapy and then started on a combination of afatinib and bevacizumab (Fig. 5a). Continued slow tumor progression was observed, and he succumbed to disease at 22 months after initial diagnosis. Patient #3 is an 11-year-old girl who was treated with intensity-modulated radiation therapy followed by temozolomide and then lomustine (CCNU) for progressive disease. A second biopsy was performed for molecular profiling purposes at 9 months after initial diagnosis revealing EGFR exon 20 insertion and CDK6 amplification, for which therapy was transitioned to the MEK inhibitor trametinib in combination with ribociclib (a CDK4/6 small molecule inhibitor). Slow tumor progression was observed over the next several months, and she succumbed to disease at 21 months from initial diagnosis. Patient #5 is a 3-year-old boy who was treated with intensity-modulated proton therapy with concurrent osimertinib and temozolomide, and then was continued on osimertinib and temozolomide given the finding of EGFR exon 20 insertion. Slow tumor progression was observed on follow-up imaging over the next several months. This child is currently alive with progressive disease at 8 months since diagnosis. Patient #10 is a 5-year-old boy who was treated with intensity-modulated radiation therapy, and then with a combination of afatinib and temozolomide given the finding of EGFR p.A289T missense mutation. Slow tumor progression was observed over the next several months. He was treated with an additional course of radiation therapy at 12 months from initial diagnosis and subsequently underwent an additional biopsy for molecular profiling purposes, which demonstrated the identical EGFR p.A289T without any additional pathogenic alterations (Fig. 2a). He remains alive with slowly progressive disease at 19 months since initial diagnosis, currently on a combination of osimertinib, CCNU, and bevacizumab. No significant toxicities associated with the osimertinib, afatinib, or trametinib were experienced by the four children in this cohort.
Given that pediatric bithalamic diffuse gliomas appear to represent a distinct tumor entity based on their unique anatomic location, genetic signature, and epigenomic landscape, we sought to define the clinical outcomes for affected children. In order to do this, we collated clinical data from all children with biopsy-proven diffuse gliomas of the bilateral thalami reported in the published literature, including patient age, sex, diagnostic surgical procedure, histologic diagnosis, radiation treatment, chemotherapy, and survival (Supplementary Table 11 [Online supplement 1]). Tumors with a diagnosis of pilocytic astrocytoma or those identified to harbor KIAA1549-BRAF fusion (the defining genetic alteration of pilocytic astrocytoma) were excluded, as these do not represent true bithalamic diffuse gliomas. Kaplan-Meier survival analysis was performed on this historical cohort of pediatric bithalamic gliomas, our study cohort of pediatric bithalamic gliomas, and these two cohorts combined (combined cohorts shown in Fig. 5b). The median survival was 7 months for the historical cohort, 14 months for our study cohort, and 10 months for the combined study cohort. Nearly all children from both cohorts succumbed to disease within 2 years, with only a few children in the historical cohort having longer survival, which may potentially represent inclusion of other tumor types (especially given the absence of detailed molecular characterization in these few tumors from children with long-term survival). Kaplan-Meier survival analysis of the separate and combined study cohorts with stratification based on histologic classification as diffuse astrocytoma vs. anaplastic astrocytoma vs. glioblastoma did not reveal any appreciable differences, thus indicating that histologic grading is not a robust predictor of differential survival in pediatric bithalamic gliomas (Fig. 5c). This is not unexpected given that only small stereotactic biopsies are typically performed in affected children that may preclude accurate histologic grading. Overall, this analysis demonstrates that survival for children with bithalamic gliomas is poor irrespective of histologic grade, with outcomes similar to other diffuse glioma entities with grade IV designations in the 2016 World Health Organization Classification of Tumors of the Central Nervous System such as “diffuse midline glioma, H3 K27M-mutant” and “glioblastoma, IDH-wildtype”.
To evaluate the potential survival benefit conferred by EGFR and MEK kinase inhibitor therapy, we examined clinical outcomes for the eleven children in our study cohort whose bithalamic gliomas harbored EGFR mutation (patients #1–11). We performed Kaplan-Meier survival analysis stratified by those children whose treatment included a small molecule EGFR or MEK kinase inhibitor (n=4) versus those whose treatment did not include a small molecule kinase inhibitor (n=7; Fig. 5d). Median survival was 21.5 months for those children whose treatment included a kinase inhibitor versus 13 months for those children whose treatment did not include a kinase inhibitor (p=0.010 by Log-rank test). While the patient numbers are small, this analysis does suggest a potential survival benefit from the addition of a small molecule tyrosine kinase inhibitor to the treatment regimen for children with bithalamic gliomas harboring EGFR mutation.
DISCUSSION
Brain tumors are the most common solid tumors of childhood. While outcomes are good for some low-grade subtypes such as pilocytic astrocytoma, other subtypes such as diffuse intrinsic pontine glioma and bithalamic glioma are unresectable tumors and usually rapidly lethal. Genomically-tailored therapy has the potential to improve outcomes for these malignant childhood brain tumors.
We have identified that bithalamic gliomas represent a distinct molecular entity characterized by frequent EGFR and TP53 mutations in the absence of EGFR gene amplification and only rare histone H3 p.K27M mutation. This is in contrast to their unilateral thalamic counterparts that lack EGFR mutation, have a high frequency of H3F3A or HIST1H3B p.K27M mutation, along with recurrent accompanying alterations in ATRX, FGFR1, PIK3CA, and ACVR1 that were not present in our cohort of bithalamic gliomas [18, 36, 42, 43]. The EGFR mutations in bithalamic gliomas are most commonly small in-frame insertions within exon 20 in the intracellular tyrosine kinase domain, unlike the missense mutations and intragenic deletions involving exons encoding the extracellular domain that are often present on the amplified EGFR allele in glioblastomas in adults [5]. Apart from bithalamic gliomas, EGFR mutations or amplifications are otherwise rare in pediatric gliomas, being found in less than 4% of pediatric high-grade and diffuse brainstem gliomas according to the largest meta-analysis to date [18]. Notably, while the bithalamic gliomas in this cohort share a similar epigenome that is largely distinct from all other glioma subtypes that have been studied to date, we did observe some overlap with the RTK III methylation subclass of IDH-wildtype glioblastomas. This RTK III methylation subclass is a poorly characterized group of glioblastomas in the cerebral hemispheres of children which have been identified to harbor frequent EGFR amplification in the small number of cases studied to date [9].
Studies have shown that glioblastoma cells from adults harboring EGFR activation via amplification accompanied by intragenic deletion of exons 2–7 (EGFRvIII variant) are inhibited by erlotinib [34]. Unfortunately, clinical trials examining the efficacy of erlotinib in adults with glioblastoma produced largely unsuccessful results [7, 27, 29]. Subsequent investigation suggested potential explanations for the failure of erlotinib in adult glioblastoma. One study found that erlotinib was less efficacious for extracellular domain mutations in adult glioblastomas compared with the kinase domain mutations in lung cancers [40]. Other studies have reported that inactivation of the PTEN tumor suppressor gene, derepression of PDGFRB expression, and dynamic regulation of extrachromosomal mutant EGFR amplicons are also potential resistance mechanisms of glioblastoma to erlotinib [1, 19, 21]. Among EGFR kinase domain mutations in lung cancer, exon 20 insertions have been correlated with worse outcomes and relative resistance to erlotinib [2, 24, 44]. Poziotinib has been identified as a compound with enhanced efficacy in lung carcinomas harboring exon 20 insertions relative to other more common kinase domain mutations (e.g. p.T790M), thought to be attributable to poziotinib’s smaller size and increased flexibility compared to other large, rigid inhibitors [14, 32, 33]. However, the sensitivity of different EGFR mutations to the currently available armamentarium of EGFR kinase inhibitors had not been examined in isogenic glioma model systems.
Given the frequent EGFR mutation in bithalamic gliomas, we speculated that this creates a therapeutic vulnerability that can be targeted with small molecule kinase inhibitors in children affected with this disease. We therefore tested a panel of kinase inhibitors in two isogenic astrocyte cell cultures modeling exon 20 kinase domain mutation (specific to pediatric bithalamic glioma), exon 7 extracellular domain mutation (seen in both pediatric bithalamic glioma and adult glioblastoma), and amplification of wildtype EGFR (specific to adult glioblastoma). We found that these mutant EGFR isoforms or overexpression of wildtype EGFR cause sensitivity to kinase inhibitor treatment. However, the relative efficacy of the different compounds tested was variable dependent on location of the EGFR mutation versus overexpression of wildtype EGFR. For example, gefitinib indiscriminately killed astrocytes overexpressing either wildtype or mutant EGFR isoforms. In contrast, afatinib killed astrocytes expressing mutant EGFR isoforms but not overexpressing wildtype EGFR. This indicates that for glioblastomas in adults harboring amplification of a wildtype EGFR allele, gefitinib is more likely than afatinib to be efficacious. We also observed differential drug sensitivity based on location of the EGFR mutation, with erlotinib being more efficacious for astrocytes expressing extracellular domain mutation and poziotinib being more efficacious for astrocytes expressing exon 20 kinase domain mutation. Among the tested inhibitors, afatinib had the widest therapeutic window between astrocytes expressing either of the mutant isoforms versus astrocytes with wildtype EGFR expression. Together, these data provide the preclinical rationale for an individualized treatment strategy using kinase inhibitors targeting the specific EGFR alteration present in a patient’s glioma. Additionally, as EGFR mutation results in downstream activation of the MAP kinase signaling pathway, MEK inhibition represents another potential therapeutic option. Notably, blood-brain barrier penetration represents an important consideration when selecting small molecule tyrosine kinase inhibitors for use in glioma patients.
In summary, we have identified that bithalamic gliomas in children represent a molecularly distinct brain tumor entity characterized by frequent EGFR mutations and rare histone H3 mutations. These EGFR mutations confer selective vulnerability to EGFR kinase inhibitors. Our initial experience with treatment of four children with bithalamic gliomas harboring EGFR mutations with targeted small molecule kinase inhibitors has shown promising survival benefit for this lethal pediatric brain tumor. Further evaluation of targeted kinase inhibition in a randomized clinical trial setting for children with bithalamic gliomas appears warranted based on this initial study.
Supplementary Material
Acknowledgments
We thank the staff of the UCSF Clinical Cancer Genomics Laboratory for assistance with genetic profiling. B.A. Orr is supported by the National Cancer Institute, National Institutes of Health (P30 CA021765) and the American Lebanese Syrian Associated Charities (ALSAC). D.A. Solomon is supported by the NIH Director’s Early Independence Award from the Office of the Director, National Institutes of Health (DP5 OD021403) and a Developmental Research Program Award from the UCSF Brain Tumor SPORE grant from the National Cancer Institute, National Institutes of Health (P50 CA097257).
Footnotes
Disclosure of Potential Conflicts of Interest
S.J. Allen is a current employee of Illumina, Inc. No potential conflicts of interest were disclosed by any of the other authors.
Electronic supplementary material
The online version of this article contains supplementary material, which is available to authorized users:
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
REFERENCES
- 1.Akhavan D, Pourzia AL, Nourian AA, Williams KJ, Nathanson D, Babic I, et al. (2013) De-repression of PDGFRβ transcription promotes acquired resistance to EGFR tyrosine kinase inhibitors in glioblastoma patients. Cancer Discov 3:534–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arcila ME, Nafa K, Chaft JE, Rekhtman N, Lau C, Reva BA, et al. (2013) EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther 12:220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA (2014) Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30:1363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benbir G, Sayilir I, Oz B, et al. (2008) Bilateral thalamic glioma. A case report. J Neurol Sci 25:301–5. [Google Scholar]
- 5.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. (2013) The somatic genomic landscape of glioblastoma. Cell 155:462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broniscer A, Hwang SN, Chamdine O, Lin T, Pounds S, Onar-Thomas A, et al. (2018) Bithalamic gliomas may be molecularly distinct from their unilateral high-grade counterparts. Brain Pathol 28:112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown PD, Krishnan S, Sarkaria JN, Wu W, Jaeckle KA, Uhm JH, et al. (2008) Phase I/II trial of erlotinib and temozolomide with radiation therapy in the treatment of newly diagnosed glioblastoma multiforme: North Central Cancer Treatment Group Study N0177. J Clin Oncol 26:5603–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research Network, Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, et al. (2015) Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372:2481–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, et al. (2018) DNA methylation-based classification of central nervous system tumours. Nature 555:469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, et al. (2016) Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 164:550–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Rocco C, Iannelli A (2002) Bilateral thalamic tumors in children. Childs Nerv Syst 18:440–4. [DOI] [PubMed] [Google Scholar]
- 12.Fortin JP, Labbe A, Lemire M, Zanke BW, Hudson TJ, Fertig EJ, et al. (2014) Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol 15:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gudowius S, Engelbrecht V, Messing-Junger M, Reifenberger G, Gartner J (2002) Diagnostic difficulties in childhood bilateral thalamic astrocytomas. Neuropediatrics 33:331–5. [DOI] [PubMed] [Google Scholar]
- 14.Hirano T, Yasuda H, Tani T, Hamamoto J, Oashi A, Ishioka K, et al. (2015) In vitro modeling to determine mutation specificity of EGFR tyrosine kinase inhibitors against clinically relevant EGFR mutants in non-small-cell lung cancer. Oncotarget 6:38789–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain P, Mohamed A, Sigamani E, Suri V, Mahapatra AK, Kumar A, et al. (2013) Bilateral thalamic lesions in a child. Eur Neurol 70:33–4. [DOI] [PubMed] [Google Scholar]
- 16.Lee JC, Vivanco I, Beroukhim R, Huang JH, Feng WL, DeBiasi RM, et al. (2006) Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med 3:e485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kline CN, Joseph NM, Grenert JP, van Ziffle J, Talevich E, Onodera C, et al. (2017) Targeted next-generation sequencing of pediatric neuro-oncology patients improves diagnosis, identifies pathogenic germline mutations, and directs targeted therapy. Neuro Oncol 19:699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackay A, Burford A, Carvalho D, Izquierdo E, Fazal-Salom J, Taylor KR, et al. (2017) Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell 32:520–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, et al. (2005) Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med 353:2012–24. [DOI] [PubMed] [Google Scholar]
- 20.Messing-Junger AM, Floeth FW, Pauleit D, Reifenberger G, Willing R, Gartner J, et al. (2002) Multimodal target point assessment for stereotactic biopsy in children with diffuse bithalamic astrocytomas. Childs Nerv Syst 18:445–9. [DOI] [PubMed] [Google Scholar]
- 21.Nathanson DA, Gini B, Mottahedeh J, Visnyei K, Koga T, Gomez G, et al. (2014) Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science 343:72–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niu X, Wang T, Yang Y, Gan Y, Li J, Liu Y, et al. (2018) Prognostic factors for the survival outcome of bilateral thalamic glioma: an integrated survival analysis. World Neurosurg 110:e222-e230. [DOI] [PubMed] [Google Scholar]
- 23.Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS (2018) CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro Oncol 20:iv1-iv86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oxnard GR, Lo PC, Nishino M, Dahlberg SE, Lindeman NI, Butaney M, et al. (2013) Natural history and molecular characteristics of lung cancers harboring EGFR exon 20 insertions. J Thorac Oncol 8:179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pandey N, Singh PK, Mahapatra AK, Kakkar A, Sharma BS (2014) Pediatric bilateral large concurrent thalamic glioblastoma: An unusual case report. J Pediatr Neurosci 9:76–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park JY, Cohen C, Lopez D, Ramos E, Wagenfuehr J, Rakheja D (2016) EGFR Exon 20 insertion/duplication mutations characterize fibrous hamartoma of infancy. Am J Surg Pathol 40:1713–8. [DOI] [PubMed] [Google Scholar]
- 27.Peereboom DM, Shepard DR, Ahluwalia MS, Brewer CJ, Agarwal N, Stevens GH, et al. (2010) Phase II trial of erlotinib with temozolomide and radiation in patients with newly diagnosed glioblastoma multiforme. J Neurooncol 98:93–9. [DOI] [PubMed] [Google Scholar]
- 28.Peruzzi L, Iuvone L, Ruggiero A, Colosimo C, Stefanini MC, Riccardi R (2016) Neuropsychological deterioration predicts tumor progression in a young boy with bithalamic glioma. Appl Neuropsychol Child 5:76–81. [DOI] [PubMed] [Google Scholar]
- 29.Prados MD, Chang SM, Butowski N, DeBoer R, Parvataneni R, Carliner H, et al. (2009) Phase II study of erlotinib plus temozolomide during and after radiation therapy in patients with newly diagnosed glioblastoma multiforme or gliosarcoma. J Clin Oncol 27:579–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rafique MZ, Ahmad MN, Yaqoob N, Ahsan H (2007) Diffuse bilateral thalamic astrocytoma. J Coll Physicians Surg Pak 17:170–2. [PubMed] [Google Scholar]
- 31.Rajput DK, Mehrotra A, Srivastav AK, Kumar R, Mahapatra AK (2010) Bilateral thalamic glioma in a 6-year-old child. J Pediatr Neurosci 5:45–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robichaux JP, Elamin YY, Tan Z, Carter BW, Zhang S, Liu S, et al. (2018) Mechanisms and clinical activity of an EGFR and HER2 exon 20-selective kinase inhibitor in non-small cell lung cancer. Nat Med 24:638–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruan Z, Kannan N (2018) Altered conformational landscape and dimerization dependency underpins the activation of EGFR by αC-β4 loop insertion mutations. Proc Natl Acad Sci USA 115:e8162–e8171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarkaria JN, Yang L, Grogan PT, Kitange GJ, Carlson BL, Schroeder MA, et al. (2007) Identification of molecular characteristics correlated with glioblastoma sensitivity to EGFR kinase inhibition through use of an intracranial xenograft test panel. Mol Cancer Ther 6:1167–74. [DOI] [PubMed] [Google Scholar]
- 35.Sharaf AF, Hamouda ES, Teo JG (2016) Bilateral thalamic and right fronto-temporo-parietal gliomas in a 4 years old child diagnosed by magnetic resonance imaging. J Radiol Case Rep 10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solomon DA, Wood MD, Tihan T, Bollen AW, Gupta N, Phillips JJ, et al. (2016) Diffuse midline gliomas with histone H3-K27M mutation: A series of 47 cases assessing the spectrum of morphologic variation and associated genetic alterations. Brain Pathol 26:569–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonoda Y, Ozawa T, Hirose Y, Aldape KD, McMahon M, Berger MS, et al. (2001) Formation of intracranial tumors by genetically modified human astrocytes defines four pathways critical in the development of human anaplastic astrocytoma. Cancer Res 61:4956–60. [PubMed] [Google Scholar]
- 38.Steinbok P, Gopalakrishnan CV, Hengel AR, Vitali AM, Poskitt K, Hawkins C, et al. (2016) Pediatric thalamic tumors in the MRI era: a Canadian perspective. Childs Nerv Syst 32:269–80. [DOI] [PubMed] [Google Scholar]
- 39.Triche TJ, Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD (2013) Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res 41:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vivanco I, Robins HI, Rohle D, Campos C, Grommes C, Nghiemphu PL, et al. (2012) Differential sensitivity of glioma- versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov 2:458–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong AJ, Ruppert JM, Bigner SH, Grzeschik CH, Humphrey PA, Bigner DS, et al. (1992) Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci USA 89:2965–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, et al. (2012) Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 44:251–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Y, et al. (2014) The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet 46:444–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yasuda H, Park E, Yun CH, Sng NJ, Lucena-Araujo AR, Yeo WL, et al. (2013) Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med 5:216ra177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshida M, Fushiki S, Takeuchi Y, Takanashi M, Imamura T, Shikata T, et al. (1998) Diffuse bilateral thalamic astrocytomas as examined serially by MRI. Childs Nerv Syst 14:384–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Targeted DNA sequencing data from this study are available from the European Genome-Phenome Archive (EGA) repository under accession number EGAS00001004033 (https://ega-archive.org/). DNA methylation array data from this study are available from the Gene Expression Omnibus (GEO) repository under accession number GSE140124 (https://www.ncbi.nlm.nih.gov/geo/).