Abstract
Lyme disease is a common tick-borne infection caused by the spirochete Borrelia burgdorferi sensu stricto (s.s.). B. burgdorferi s.s. may utilize chemotaxis, the directional migration towards or away from a chemical stimulus, for transmission, acquisition, and infection. However, the specific signals recognized by the spirochete for these events have not been defined. In this study, we identify an Ixodes scapularis salivary gland protein, Salp12, that is a chemoattractant for the spirochete. We demonstrate that Salp12 is expressed in the I. scapularis salivary glands and midgut and expression is not impacted by B. burgdorferi s.s. infection. Knockdown of Salp12 in the salivary glands or passive immunization against Salp12 reduces acquisition of the spirochete by ticks but acquisition is not completely prevented. Knockdown does not impact transmission of B. burgdorferi s.s. This work suggests a new role for chemotaxis in acquisition of the spirochete and suggests that recognition of Salp12 contributes to this phenomenon.
Keywords: Borrelia burgdorferi sensu stricto, chemotaxis, acquisition, Ixodes scapularis, tick, Lyme disease
Graphical Abstract
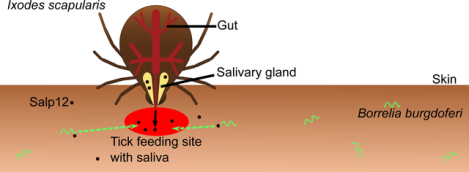
Introduction:
Borrelia burgdorferi sensu stricto (s.s.), the causative agent of Lyme disease, is one of the most prevalent arthropod-transmitted pathogens in the United States and Europe (Kuehn 2013, Hinckley, Connally et al. 2014). There is currently no preventative human vaccine for Lyme disease, and although it is treatable with antibiotics, long lasting symptoms can occur. Additionally, treatment places a significant burden on the medical system (Adrion, Aucott et al. 2015). Therefore, identifying new vaccine and therapeutic targets is warranted, and the elucidation of crucial interaction points between the tick host and B. burgdorferi s.s. has the potential to uncover novel targets for vaccine or therapeutic development.
In the United States, Ixodes scapularis ticks are the most common vector of B. burgdorferi s.s. I. scapularis ticks feed on small mammals and can acquire the spirochete during the first two life stages, the larva and nymph. Infected ticks then transmit the spirochete during subsequent feedings, as a nymph or adult. Both nymphs and adults may feed on human hosts and transmit the spirochete. To complete the infectious cycle, B. burgdorferi s.s. must perform distinct migration patterns through animal and tick tissues for successful transmission, infection, and acquisition. Uninfected ticks acquire B. burgdorferi s.s. when feeding on infected animals, and during acquisition, the spirochetes present in mammalian skin enter the tick midgut as the tick takes a blood meal (Bockenstedt, Gonzalez et al. 2014). The spirochetes are then maintained in the tick gut during molting. During transmission, spirochetes residing in a tick’s midgut migrate to the tick’s salivary glands, where they exit the tick along with saliva (de Silva and Fikrig 1995). After being deposited in mammalian skin, B. burgdorferi s.s. establishes an infection at the bite site and disseminates to other organs (Barthold, Persing et al. 1991, Pahl, Kuhlbrandt et al. 1999). Many steps of these processes are facilitated by or improved by tick proteins. For example, tick protein Salp15 binds to the B. burgdorferi s.s. protein OspC and protects the spirochete from antibody mediated killing(Ramamoorthi, Narasimhan et al. 2005) and knocking down Salp15 or vaccinating animals against Salp15 reduces transmission of the spirochete (Dai, Wang et al. 2009). Additionally, Salp15 is anti-inflammatory and inhibits CD4+ t cells contributing to tick feeding and vector-host-pathogen interactions(Anguita, Ramamoorthi et al. 2002).
Bacteria can perform directed migration patterns through chemotaxis, the directional migration of cells toward or away from a chemical stimulus. Chemotaxis in B. burgdorferi s.s. has been shown to be important for spirochete growth in ticks (Novak, Sekar et al. 2016), transmission from the tick (Sze, Zhang et al. 2012, Novak, Sekar et al. 2016, Xu, Sultan et al. 2017), and infection of mammals (Lin, Gao et al. 2012, Sze, Zhang et al. 2012, Novak, Sekar et al. 2016, Xu, Sultan et al. 2017). Although it has not been directly shown, several lines of evidence suggest that acquisition of the spirochete by the tick host might be facilitated by chemotaxis. When infected and uninfected ticks co-feed on an animal, there is acquisition of the spirochete by nearby ticks prior to disseminated infection (Gern and Rais 1996, Patrican 1997, States, Huang et al. 2017). Additionally, when a nymph feeds on an infected mouse, spirochete levels around the tick bite site decrease concurrent with an increase in spirochetes in the tick (Hodzic, Borjesson et al. 2001). Finally, intravital imaging of spirochetes during tick feeding detected spirochetes migrating into the tick bite site and disappearing (Bockenstedt, Gonzalez et al. 2014). Together these data suggest that spirochetes may migrate toward the tick bite site for acquisition.
Although it is known that chemotaxis is critical for B. burgdorferi s.s., chemoattractants and chemorepellents have not been identified in the infectious cycle. In vitro studies have shown that sugars and amino acids, such as N-Acetylglucosamine and glutamate, are chemoattractants for B. burgdorferi s.s. (Bakker, Li et al. 2007). Additionally, rabbit serum (Shi, Yang et al. 1998) and tick salivary gland extract (SGE, ground filtered salivary glands) (Shih, Chao et al. 2002) have been shown to be chemoattractants for B. burgdorferi s.s. It is likely that spirochete recognition of serum components functions in dissemination of the spirochete during infection. Recognition of tick salivary gland components could facilitate transmission or acquisition of the spirochete by the tick host. In transmission, the spirochete may recognize salivary gland components to migrate from the midgut to the salivary glands. In acquisition, spirochetes in mammalian skin could recognize components of tick saliva to migrate to the tick bite site, where they are ingested by the tick. During feeding, ticks secrete a large quantity of saliva that facilitates the feeding process (Kazimírová and Štibrániová 2013). As tick saliva is made within the salivary glands, SGE and saliva likely contain overlapping repertoires of molecules and therefore chemoattractants.
The specific chemotactic exchanges that occur between the spirochete and the tick are of particular interest because of their potential as therapeutic targets. Previous work has demonstrated that blocking tick-spirochete interactions that function in transmission reduces spirochete burden in the tick and mammal (Ramamoorthi, Narasimhan et al. 2005, Dai, Wang et al. 2009, Zhang, Zhang et al. 2011, Narasimhan, Coumou et al. 2014, Coumou, Narasimhan et al. 2016). Therefore, blocking chemotactic interactions that are critical for transmission or acquisition would likely reduce or prevent these processes. Interactions that are essential for transmission could be targeted in human or companion animal vaccines, as preventing transmission is protective. Interactions that facilitate acquisition could be targeted for environmental control strategies. This study defines a chemoattractant molecule present in tick SGE and saliva and establishes that recognition of this molecule is involved in acquisition in vivo.
Materials and Methods:
Bacterial strains.
B. burgdorferi s.s. strains were obtained from MA Motaleb (Table 1) (Motaleb, Miller et al. 2005, Motaleb, Miller et al. 2007). B. burgdorferi s.s. was grown without aeration in BSK-H (Sigma-Aldrich) at 32°C and were frozen at −80°C in BSK-H supplemented with 20% glycerol. Escherichia coli Top 10 cells (ThermoFisher Scientific) were used for cloning (Table 2) and were grown at 37°C with aeration in Lysogeny Broth supplemented with ampicillin (100 μg/mL).
Table 1.
Bacterial Strains and Plasmidsa
| Bacterial Strain Name | Description |
|---|---|
| B. burgdorferi s.s. B31 | Non-infectious derivative of strain B31A3 |
| B. burgdorferi s.s. B31::ΔcheX | B. burgdorferi s.s. B31 with knockout mutation of the cheX gene (Motaleb, Miller et al. 2005) |
| B. burgdorferi s.s. B31A3 | Infectious strain B31A3 |
| B. burgdorferi s.s. B31::ΔcheX | B. burgdorferi s.s. B31A3 with knockout mutation of the cheX gene (Motaleb et. al, unpublished) |
| E. coli Top 10 | Esherichia coli strain used for cloning |
| Plasmid Name | Description |
| pMT/V5-His A | Plasmid used for cloning |
| pMT/V5-His A::salp12 | pMT/v5-His A containing salp12 |
| pMT/V5-His A::AAEL010228 | pMT/V5-His A containing AAEL010228 |
Table of bacterial strains and plasmids used in this study.
Table 2.
Tick proteins identified by protein fractionation and comparison to tick saliva protein databasea
| Protein IDb | Predicted Function and Transcript Namec | Signal Sequenced |
|---|---|---|
| B7Q407 | Heme lipoprotein, ISCW021710 | Yes |
| B7Q9F1 | Protein disulfide isomerase, ISCW010827 | Yes |
| B7Q406 | Hemelipoglycoprotein, ISCW021709 | Yes |
| B7P4U1 | Protein disulfide isomerase, ISCW016161 | Yes |
| B7PFA2 | Is6 putative, ISCW024249 | Yes |
| B7PEV0 | Chaperonin subunit, ISCW017824 | No |
| B7QI01 | Hsp90 protein, ISCW014265 | No |
| B7QGE3 | Hemelipoglycoprotein, ISCW012424 | No |
| B7QJH6 | Alpha-actinin, ISCW013566 | No |
| B7PAR6 | Heat shock protein, ISCW017456 | No |
| B7PBG2 | Actin putative, ISCW024111 | No |
| B7P1Q2 | Myosin heavy chain, ISCW001340 | No |
| B7PQC5 | 60S acidic ribosomal protein P1, ISCW006538 | No |
| Q8MVB0 | Truncated secreted metalloprotease | No |
| B7P1Z8 | Heat shock protein, ISCW016090 | No |
Table of identified proteins shared in two proteomics analyses of chemoattractant I. scapularis tick protein fractions and a I. scapularis tick saliva protein database (Kim, Tirloni et al. 2016)
ID of protein as found in the VectorBase database
Predicted function based on bioinformatics analysis of VectorBase and the transcript name from the database
Presence (Yes) or absence (No) of a signal sequence as detected by SignalP software
Animals.
Rabbits and mice were housed and handled under the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The animal protocol was approved by the Yale University’s Institutional Animal Care and Use Committee (protocol number 2016–07941, approved December 20, 2016). All animal experiments were performed according to Yale University standards in a biosafety level 2 facility. For all mouse experiments, female, 5–10 weeks of age, C3H/HEJ mice were used (Jackson Laboratory, RRID:IMSR_JAX:000659). For tick saliva collection, New Zealand White rabbits were used (Charles River Laboratories, strain 571).
Ticks.
I. scapularis larvae and adults were acquired from the CDC, and larvae were used to generate nymphs. Briefly, 100 larval ticks were placed on naïve mice and allowed to feed to repletion. For infected nymphs, B. burgdorferi s.s. infected mice were used, see experiments below. Fed larvae were collected and stored until molted, approximately 6 weeks. Ticks were maintained at 23°C with 85% relative humidity with 14-hour light, 10-hour dark photoperiod. To limit variability of ticks, all ticks (i.e. both control and test samples) within a single experiment were from the same egg clutch.
Cell Lines.
Schneider’s Drosophila Line 2 cells purchased from ATCC (CVCL_Z232) were used for protein expression and purification. Cells were grown in Schneider’s medium (Sigma-Aldrich) with 10% FBS at 25°C. For maintenance of plasmids, cells were selected with 200 μg/mL Hygromycin B (Invitrogen). Cells were authenticated by ATCC prior to purchase. Additionally, authentication of the cell line during growth was confirmed by regular morphology checks by microscopy.
Collection of tick products.
Salivary gland extract (SGE) was obtained by feeding nymphs for 48 hours, dissecting, rinsing in PBS three times, and placing the salivary glands in a fresh tube of PBS. Prior to collection, salivary glands were visually inspected for any cross-contaminating midgut tissues. For capillary tube assays, salivary glands from ~40 nymphs were pooled, and for fractionation, salivary glands from ~100 nymphs were pooled. The pooled salivary glands were homogenized using a hand-grinder, filtered through a 0.2-μm filter, and were stored at −80°C until used.
Tick saliva was collected from adult female ticks as previously described (Anguita, Ramamoorthi et al. 2002). Briefly, adult female and male ticks were placed on rabbits and collected after feeding to repletion. Female ticks were immobilized on microscope slides with double-sided tape. Tick mouth parts were placed inside capillary tubes, and 1–3μL of 50 mg/mL pilocarpine was applied. Saliva was collected for 2–3 hours, pooled and stored at −80°C.
Capillary tube chemotaxis assays.
Chemotaxis assays were performed using a capillary tube chemotaxis assay (Motaleb, Miller et al. 2007). B. burgdorferi s.s. cells were grown to logarithmic phase (1×107−8 cells/mL), collected by centrifugation, and re-suspended in motility buffer to a concentration of 1×107 cells/mL. 200 μL of the suspension was added to microcentrifuge tubes and covered with parafilm. Capillary tubes containing chemoattractants or buffer were placed into the suspension and incubated at 32°C for 2 hr. Capillary tubes were removed and the liquid from the tube collected by centrifugation. Bacteria from the capillary tubes were enumerated by extracting DNA using the DNeasy Blood and Tissue Kit (Quiagen) and qPCR. Chemotaxis results are reported as the relative chemotactic response, the number of cells/mL in the test capillary tube divided by the number of cells/mL in the buffer control capillary tube. In each experiment, 3 technical replicates were performed, and the experiment was repeated at least 5 times. For SGE, 50μL of SGE or fractionated SGE (~100 μg/mL protein) was prepared as described above and diluted in 500 μL of motility buffer. For testing purified proteins or bovine serum albumin, 50 μL of 50 μg/mL was diluted in 500 μL of motility buffer. Purified proteins were also tested at half this concentration and showed similar results (data not shown). The empty vector control was purified and diluted in the same ratio as the purified protein. For tick saliva, 40 μL was diluted in 500 μL motility buffer. These solutions were used to fill uncoated Micro-hematocrit Capillary Tubes (ThermoFisher Scientific).
Removing proteins and degrading sugars from SGE.
To remove proteins from SGE, 50 μL of SGE was diluted to 70 μL with PBS and was treated with 30 μL proteinase K (QIAGEN, >600 mAU/mL) (Edwinson, Widmer et al. 2016) at 55°C overnight. The proteinase K was then heat inactivated and the digested solution was rinsed and concentrated to 50 μL using a 3 kDa spin column (Millipore). We confirmed that the heat inactivation conditions were sufficient to remove proteinase K activity. Removal of proteins was confirmed by silver staining of SDS-PAGE gels (Invitrogen). Sugars were degraded using sodium-metaperiodate (Edwinson, Widmer et al. 2016). 50 μL of SGE was diluted to 100 μL with PBS and was treated with 100 μL of 100 mM sodium metaperiodate (pH 7.2) at 4°C overnight. Remaining sodium metaperiodate was inactivated by adding 100 μL of 260mM glycerol. The solution was rinsed and concentrated to 50 μL using a 3 kDa spin column, which also allows for passthrough of small molecules and reacted sodium metaperiodate. Removal of intact sugars was confirmed by blotting (Kropec, Maira-Litran et al. 2005) and phenol-sulfuric acid method (Masuko, Minami et al. 2005). It is important to note that sodium-metaperiodate may also oxidize and therefore degrade other compounds.
Protein fractionation and identification.
SGE was fractionated using anion exchange medium pressure chromatography (NGC, Bio-rad). SGE was diluted in 10mM NaCl, pH 7.0 for column binding. Proteins were salted-off the column using 1M NaCL, pH 7.0, and 3 mL fractions were collected. Fractions were concentrated using a 3 kDa spin column and were re-suspended in PBS for chemotaxis assays and mass spectrometry analysis.
Proteins from fractionated pools processed for bottom-up proteomics using high resolution LC-MS/MS of a Waters NanoACQUITY UPLC coupled to a Q-Exactive Plus mass spectrometer. Mass spectral data were analyzed with Mascot Search Engine (Matrix Science, Boston, MA) against the tick protein database (VectorBase). Significant protein hits were those with a Mascot expectation value of 0.05 (equivalent to a 95% confidence identification) (https://www.ncbi.nlm.nih.gov/BLAST/tutorial/Altschul-1.html) and represented by at least 2 unique peptides. These analyses resulted in 86 and 168 significant protein identifications, within two biological replicate fractionated pools. The intersection (70 shared proteins) of these two replicates were compared to a database of adult tick saliva proteins (Kim, Tirloni et al. 2016) to obtain 15 putative targets. These targets were assessed for secretion signals, resulting in 5 possible proteins of interest. One protein was selected for additional analysis based on our ability to clone and express the intact protein.
Bioinformatic analysis of Salp12.
The protein sequences were analyzed using blastp (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) to identify homologs, InterPro (https://www.ebi.ac.uk/interpro/) (Finn, Attwood et al. 2017) to identify domains, and SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/) (Petersen, Brunak et al. 2011) to identify signal sequences. Alignment of Salp12 to human co-lipase was done using MUSCLE (https://www.ebi.ac.uk/Tools/msa/muscle/).
Protein Purification.
For cloning of tick proteins, cDNA from ticks fed for 48 hours was produced as described above. The transcript was cloned into pMT/V5-His A using primers specific to the gene (Table 4). The resulting plasmid (Table 1) was transfected into Drosophila S2 cells using Effectene (Quiagen). For expression, cells were grown in Insect Express medium (Lonza) and induced by addition of copper sulfate (500 μM). The cell supernatant was collected after 7 days and the protein was purified using a Talon column (Clontech). The eluted protein was concentrated using a 3 kDa filter (Millipore) and checked by Western blot with anti-V5 (Thermo Fisher Scientific) and anti-His (Thermo Fisher Scientific) antibodies. Mosquito protein AAEL010228 was produced and purified from S2 cells similarly (Table 1, S5). Mosquito RNA was extracted from salivary glands of Aedes aegypti mosquitoes and used to produce cDNA for cloning. Gels of the purification can be seen in Figure S3. Gels were run with the Precision Plus Protein Dual Color Standards (Bio-Rad), and stained with either Simply Blue Safe Stain (Thermo Fisher Scientific) or SilverQuest Silver Staining (Invitrogen).
Table 4.
Primers
| Primer Namea | Sequenceb |
|---|---|
| Salp12_cloning_F | GAT AT CGACATGGCCGCGTGGCCGGATGACGAT AA |
| Salp12_cloning_R | CTCGAGGTCGTCCTTATCTTCGTCGGTGTTT |
| 010228_F | CTCGCTCGGGAGATCTAGGCCAACTCCCGAAGACGATGGTGGTACCA |
| 010228_R | GCCCTCTAGACTCGAGTGATCTATTGGAAGCAACGCAAGACG |
| Salp12_qPCR_F | AAGGTTCTCGTAGTCCTCTACTT |
| Salp12_qPCR_R | TGGCCACTTGCTGTATGTTAT |
| tick_actin_qPCR_F | GGCGACGTAGCAG |
| tick_actin_qPCR_R | GGTATCGTGCTCGACTC |
| mouse_actin_qPCR_F | AGCGGGAAATCGTGCGTG |
| mouse_actin_qPCR_R | CAGGGTACATGGTGGTGCC |
| flaB_qPCR_F | TTCAATCAGGTAACGGCACA |
| flaB_qPCR_F | GACGCTTGAGACCCTGAAAG |
| dsRNA_GFP_F | TAATACGACTCACTATAGGGGCGACGTAAACGGCCACAAGTT |
| dsRNA_GFP_R | TAATACGACTCACTATAGGGAGAGGGTCTTGTAGTTGCCGTC |
| dsRNA_Salp12_F | TAATACGACTCACTATAGGGAGACAGGACCACGAGGACAGG |
| dsRNA_Salp12_R | TAATACGACTCACTATAGGGAGATTAACGTGACACTGGGCAGG |
Descriptive name of the primer that includes the gene name and use.
Nucleic acid sequence of the primer.
Tick expression studies.
For gene expression studies, ticks were placed on mice and allowed to feed for 24, 48, or 72 hours. Ticks were then removed and dissected to collect salivary glands and midguts. To prevent cross contamination of the salivary glands and midguts, the organs were visually inspected for inappropriate tissues and rinsed three times prior to collection. Organs from 2–3 ticks were pooled for RNA extraction using the RNeasy Kit (Quiagen), reverse transcribed using iScript (Bio-rad), and used for qPCR.
Immunofluorescence assay.
IFA was performed similarly to previously described methods (Ramamoorthi, Narasimhan et al. 2005, Narasimhan, Coumou et al. 2014). B. burgdorferi s.s. cultures were incubated with glass coverslips for 3 days under normal growth conditions in a 24-well tissue culture plate. Coverslips were removed and airdried, and spirochetes were fixed using 4% paraformaldehyde. Coverslips were blocked with 1% bovine serum albumin and were incubated with 50 μg of Salp12 overnight at 4°C. Control coverslips for detecting Salp12 alone were incubated with Salp12 overnight at 4°C and then blocked. Salp12 was detected using a mouse anti-V5 antibody (Invitrogen, 36–7500) and a goat anti-mouse antibody conjugated to Alexa Fluor 555 (Life Technologies, A-21147). B. burgdorferi s.s. was detected using a BacTrace® anti-Borrelia antibody conjugated to FITC (Kirkegaard and Perry Laboratories, 5330–0064). Coverslips were mounted using Prolong® Gold anti-fade reagent (Invitrogen) and were viewed at 20x magnification using an Axiovert 200M microscope (ZEISS). Imaging acquisition and processing was performed using ZEN software (ZEISS).
RNAi silencing of Salp12.
Knockdown of Salp12 and subsequent transmission and acquisition studies were performed as previously described (Pedra, Narasimhan et al. 2006). Briefly, dsRNA was amplified using primers specific to the sequence (Table 4) and cloned into the T7 script II vector (Fire, Xu et al. 1998). Complimentary dsRNA was synthesized in vitro using the Megascript RNAi kit (Ambion). I. scapularis nymphs, clean or B. burgdorferi s.s. infected, were injected in the body with ds salp12 or ds gfp control (6 nl, 3×1012 molecules) (Schuijt, Coumou et al. 2011). To determine knockdown efficiency, ticks were fed on mice for 24 hours, and samples were collected and analyzed as described above.
Passive immunization.
Antibodies against Salp12 and ovalbumin were produced in New Zealand white rabbits at Cocalico Biologicals, Inc. Rabbits, 4–6 weeks old, were immunized subcutaneously with 30 μg of protein in complete Freud’s adjuvant and boosted twice, every 3 weeks. Test bleeds were collected from the ear vein at 2 weeks and after the final boost. After the final boost, rabbits were euthanized and the serum was collected by cardiac puncture. Recognition of purified Salp12 or ovalbumin was performed by Western blot and ELISA (data not shown). To passively immunize mice, 200 μL of whole serum injected intraperitoneally 24 hours prior to tick placement and at the time of tick placement.
Acquisition and transmission studies.
For acquisition studies, mice were infected subcutaneously with 1×104−5 spirochetes and used for studies 14–21 days post infection. Uninfected and injected ticks were placed and allowed to feed to repletion. Ticks were assessed for time to repletion and weight. Ticks were dissected to collect salivary glands and midguts. RNA was collected as described above and used for qPCR. For time course experiments, ticks were removed at 24 hours post placement or collected after repletion and incubated for 7 days in normal storage conditions. Ticks were dissected to collect salivary glands and midguts for RNA extraction and qPCR.
For transmission studies, 5 B. burgdorferi s.s. infected and injected ticks were placed on each mouse (5 total per experimental condition) and allowed to feed to repletion. Fed ticks were assessed for time to repletion and weight. To monitor infection, skin samples were collected at 3, 5, 7, and 14 days. At 21 days, mice were euthanized, and skin, bladder, heart, and joints were collected. B. burgdorferi s.s. burden in mouse tissues was determined by DNA extraction using the DNeasy Blood and Tissue Kit (Quiagen) and qPCR.
Absolute quantification qPCR.
All qPCR results were obtained by absolute quantification qPCR. The qPCR reactions used 1 μL of template DNA with primers specific to the gene (Table 4) in SYBR Green (Invitrogen). Quantification of copy number was performed using a standard curve for each gene.
Statistics.
All details regarding biological and technical replicates can be found in the figure legends. Details regarding the statistical tests used for each experiment can be found in both the results section and figure legends. Bar graphs and dot plots show the mean and SEM. Significance was defined as p<0.05, and specific p-values can be found in both the text and figures. To assess distribution of the data, all data sets were analyzed in both the variable and outcome using the Shapiro-Wilk normality test performed in PRISM. Data sets with normal distribution were assessed by ANOVA in R. Data sets with non-normal distribution were assessed by Mann-Whitney tests, performed in PRISM. Significance of proteomics hits were assessed using Mascot Search Engine criteria.
Data Availability.
The mass spectrometry proteomic data are available via the ProteomeXchange Consortium and the PRIDE partner repository (http://www.proteomexchange.org/) under the identifier PXD011273.
Results:
Proteins in tick salivary glands and saliva are chemoattractants for B. burgdorferi s.s.
To assess which components of tick salivary glands are chemoattractants for the spirochete, we removed specific compounds from SGE and tested these solutions in in vitro assays. Small molecules, such as sugar monomers, dipeptides, or ions, are common chemoattractants for bacteria (Porter, Wadhams et al. 2011). Therefore, we tested whether small molecules in SGE are chemoattractants by assessing B. burgdorferi s.s. chemotaxis towards small molecule (<3 kDa) or large molecule (>3 kDa) fractions (Fig. 1A). We examined chemotaxis in vitro using a capillary tube assay with our putative chemoattractants relative to a buffer control (Bakker, Li et al. 2007, Motaleb, Miller et al. 2007). Bacterial numbers were enumerated using absolute quantification qPCR, and data are shown as the relative chemotactic response (number of cells in the chemoattractant tube divided by the number of cells in the buffer control tube). Assays were performed with a wildtype, non-infectious isolate, B31A, and a chemotaxis deficient mutant, B31A ΔcheX (Table 1) (Motaleb, Miller et al. 2005), to confirm that migration toward putative chemoattracts was due to chemotaxis. Wildtype B. burgdorferi s.s. but not the ΔcheX mutant migrated toward the large molecule fraction of SGE (ANOVA, df=3, p=0.02), indicating small molecules within SGE are not the chemoattractant detected in the in vitro assay.
Figure 1. I. scapularis salivary gland and saliva proteins are chemoattractants for B. burgdorferi.
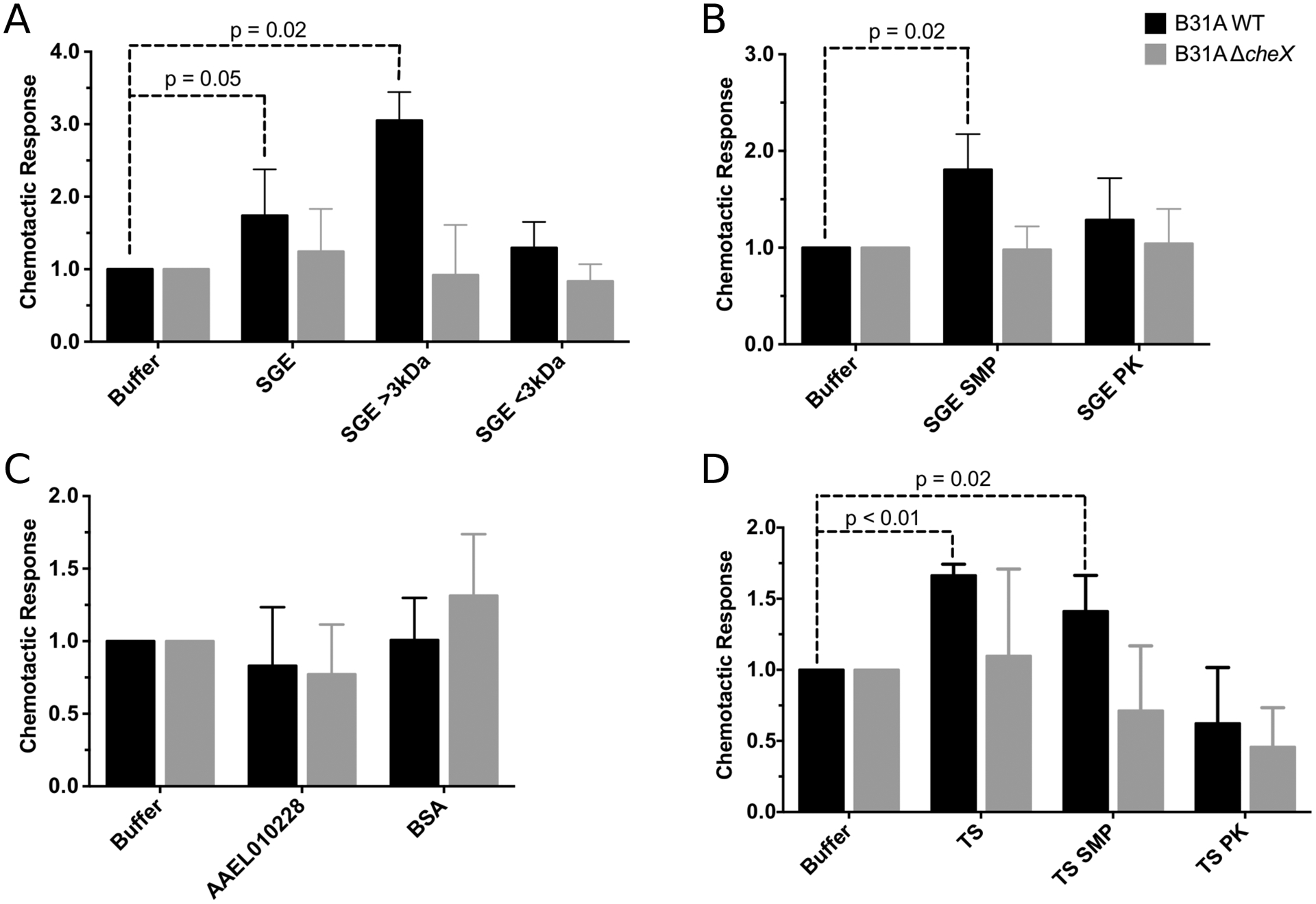
The wild type (WT, black bars) and chemotaxis mutant (ΔcheX, grey bars) of the B. burgdorferi isolate B31A were tested for in vitro chemotaxis towards potential chemoattractants. Data is shown as the relative chemotactic response (number of cells responding to the test sample divided by the number of cells responding to the buffer control). Chemotaxis was tested for I. scapularis salivary gland extract (SGE) (A, B), non-tick proteins (C) bovine serum albumin (BSA) and mosquito protein (AAEL010228), and I. scapularis tick saliva (TS) (D). SGE was separated into fractions greater than and less than 3 kDa (A). SGE and TS were also treated with sodium-metaperiodate (SMP) to remove sugars and proteinase K (PK) to remove proteins (B, D) to assess of which molecules are chemoattractants. Data are the average of 5 biological replicates each containing 3 technical replicates. Significance was tested using ANOVA.
Because sugars, such as N-acetyl glucosamine, are chemoattractants for B. burgdorferi s.s. (Bakker, Li et al. 2007), we next assessed if sugars and carbohydrates present in SGE are chemoattractants for B. burgdorferi s.s.. Sugars in SGE were degraded using a sodium-metaperiodate treatment (Edwinson, Widmer et al. 2016), and the absence of intact sugar molecules was confirmed by blotting (Kropec, Maira-Litran et al. 2005) and the phenol-sulfuric acid method (Masuko, Minami et al. 2005) (Fig. S1 A, B). B. burgdorferi s.s. migration toward sodium-metaperiodate treated SGE was statistically significant (ANOVA, df=2, p=0.02), indicating sugars in SGE are not a significant chemoattractant as measured by the in vitro assay (Fig. 1B).
Another major component of a tick salivary gland is protein (Chmelař, Kotál et al. 2016). We therefore tested whether proteins in SGE are chemoattractants by removing proteins using proteinase K (Edwinson, Widmer et al. 2016) and testing the chemotactic response (Fig. 1B). Removal of proteins was confirmed by silver staining of protein gels (Invitrogen) (Fig. S1C). B. burgdorferi s.s. did not migrate toward the proteinase K treated SGE, demonstrating that this treatment removed the chemoattractant present in SGE. Therefore, proteins are the chemoattractant molecules present SGE. Because amino acids, such as glutamate (Bakker, Li et al. 2007), are known to be chemoattractants for B. burgdorferi s.s., we assessed if this interaction is specific to proteins within SGE. We tested migration of B. burgdorferi s.s. toward proteins not expected to function as chemoattractants, mosquito salivary protein AAEL010228 (Conway, Londono-Renteria et al. 2016) and bovine serum albumin (Fig. 1C). These proteins were not chemoattractants for B. burgdorferi s.s., indicating that the chemotactic response of B. burgdorferi s.s. is specific to proteins in tick SGE.
As tick saliva and tick salivary glands overlap in proteins and sugars, we assessed if proteins in tick saliva are chemoattractants for B. burgdorferi s.s. Similarly, we found that B. burgdorferi s.s. migrated toward tick saliva and that proteins, but not sugars, within saliva are chemoattractants (Fig. 1D, ANOVA, df=3, p<0.02).
Tick protein Salp12 is an attractant.
To determine which proteins in tick SGE and saliva are chemoattractants for B. burgdorferi s.s., we fractionated the proteins and tested the fractions in an in vitro capillary tube chemotaxis assay. Because tick saliva can only be collected in small amounts and at low concentrations, we utilized SGE for these experiments. Using anion exchange medium pressure chromatography, we fractionated SGE into 4 fractions (Fig. S2). B. burgdorferi s.s. seemed migrated toward one of these fractions, Fraction 3, more consistently than other fractions and had the largest difference between wildtype and ΔcheX mutant B. burgdorferi s.s. (Fig. 2). This was not a statistically significant difference.
Figure 2. Identification of chemoattractant fractions.
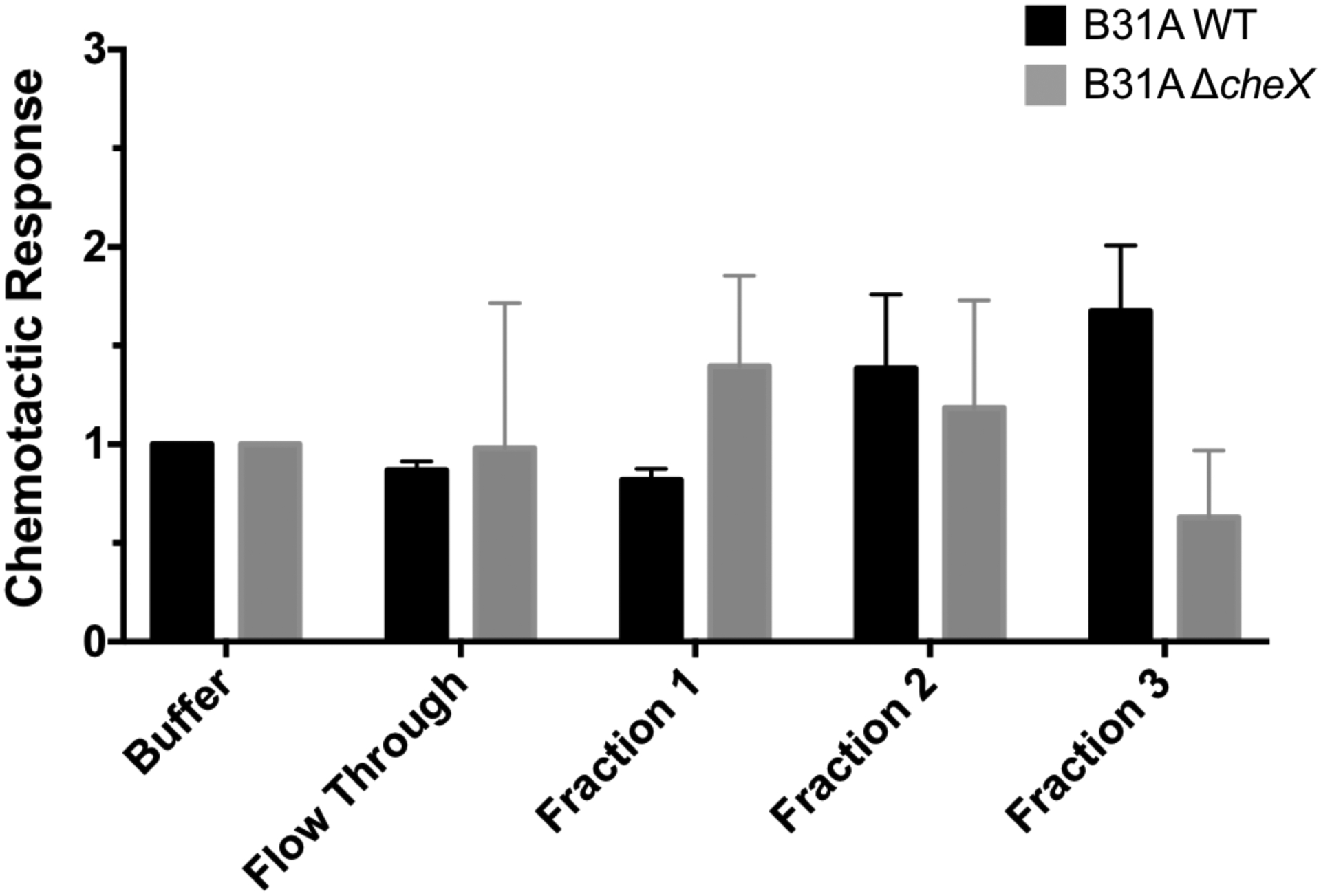
I. scapularis salivary gland proteins were fractionated using anion exchange medium pressure liquid chromatography. Fractions were tested using the in vitro chemotaxis assay for the B. burgdorferi isolate B31A, wild type (WT, black bars) and chemotaxis mutant (ΔcheX, grey bars). Data are the average of 2 biological replicates each containing 3 technical replicates, and statistical significance was not identified using ANOVA.
Proteomic analysis of Fraction 3 revealed 70 significant hits shared by two replicate protein fractions (Table S1). As this is a large number of hits, we narrowed this list by comparing the significant hits to a database of proteins known to be in tick saliva (Kim, Tirloni et al. 2016). Because tick saliva is produced in the salivary glands and also contains chemoattractant proteins, it is likely that the chemoattractants identified in tick saliva and SGE are overlapping. However, this comparison is more likely to identify proteins involved in acquisition, as tick saliva is encountered by the spirochete at the tick bite site during tick feeding. During transmission, spirochetes exit the tick with the tick saliva but there is not an obvious gradient of tick saliva that would be encountered in this process. This comparison yielded 15 proteins (Table 2). We then prioritized this list by significance of the hit and the presence of a secretion signal sequence to yield 5 candidates. The presence of a secretion signal sequence indicates that the protein could be secreted outside of the salivary gland cells at higher concentrations, which may make the protein better signal for chemotaxis. Due to its small size and our ability to purify it, we selected the protein from transcript ISCW024249 for testing. B. burgdorferi s.s. migration toward the recombinant protein was significant, indicating this protein is a chemoattractant for the spirochete (ANOVA, df=3, p=0.04, Fig. 3A). The spirochete failed to migrate toward purified empty vector, containing only the protein tag. Additionally, specificity of this interaction is further supported by the previously mentioned lack of migration toward an irrelevant protein, AAEL010228, produced in the same manner (Fig. 1C).
Figure 3. The I. scapularis protein from isolate ISCW024249 is a chemoattractant for B. burgdorferi.

The recombinant I. scapularis protein from transcript ISCW024249 and an empty vector control were tested in the in vitro chemotaxis assay with the B. burgdorferi isolate B31A (A), wild type (WT, black bars) and chemotaxis mutant (ΔcheX, grey bars). The protein was also tested with the infectious B. burgdorferi isolate B31A3 (B). Significance was tested using ANOVA.
The B31A strains used in our in vitro experiments are noninfectious, laboratory adapted strains that genetically differ from infectious isolates. Therefore, it is possible that the chemotactic response of infectious isolates might differ. To assess if the protein from transcript ISCW024249 is also a chemoattractant for infectious B. burgdorferi s.s., we tested the protein in the capillary tube chemotaxis assay using an infectious background, B31A3 and B31A3 ΔcheX (Table 1) (Fig. 3B). As expected, wild type B. burgdorferi s.s. B31A3 migrated toward the purified protein from transcript ISCW024249, but the cheX mutant did not (ANOVA, df=2, p=0.03). This demonstrates that the protein is a chemoattractant for an infectious isolate of B. burgdorferi s.s. and may contribute to the in vivo infectious cycle of the spirochete.
The protein encoded by transcript ISCW024249 is a small, 12.18 kDa protein, not previously assessed for function (Fig. S3). Analysis of the protein by blastp yielded proteins of undefined function and human colipase-like proteins (Table 3). Human colipase is a small, 10kDa, cofactor that functions to aid pancreatic lipase in the digestion and absorption of lipids and lipid soluble nutrients (Lowe 1997). However, further analysis of the ISCW024249 protein by InterPro did not reveal any predicted functions or known domains, including colipase domains, and alignment of the ISCW024249 protein to known colipases did not yield significant homology (data not shown). Therefore, we hypothesize that this protein is sufficiently different that it is likely to have a distinct function from human colipase. As there is not significant homology of this protein to proteins of known function, we will name the protein according to the nomenclature used for other I. scapularis salivary proteins (Salp), and throughout this manuscript the protein will be termed Salp12. This protein does not share homology with other previously identified Salp proteins (Das, Banerjee et al. 2001).
Table 3.
BLAST hits for the protein from transcript ISCW024249a
| Accession Numberb | Predicted Functionc | Percent Identityd | E-valuee | Bit Scoref |
|---|---|---|---|---|
| AAY66501.1 | putative secreted salivary gland protein [Ixodes scapularis] | 47.8% | 5.1E-10 | 62.4 |
| CAX51408.1 | hypothetical protein [Opisthacanthus cayaporum] | 30.6% | 6.7E-09 | 59.3 |
| EEC14111.1 | secreted salivary gland protein, putative [Ixodes scapularis] | 40.6% | 3.9E-06 | 52 |
| KPM07042.1 | hypothetical protein QR98_0055240 [Sarcoptes scabiel] | 34.6% | 2.1E-04 | 47.8 |
| XP_023241637.1 | colipase-like [Centruroides sculpturatus] uncharacterized protein LOC106813686 | 31.1% | 2.9E-04 | 47 |
| XP_014673379.1 | [Priapulus caudatus] uncharacterized protein LOC111634649 | 33.3% | 3.8E-04 | 47.4 |
| XP_023235249.1 | [Centruroides sculpturatus] | 33.3% | 4.9E-04 | 46.6 |
| OTF74413.1 | hypothetical protein BLA29_000290 [Euroglyphus maynei] | 32.3% | 2.0E-03 | 45.4 |
| EEC14112.1 | secreted protein, putative [Ixodes scapularis] | 31.4% | 5.0E-03 | 43.5 |
| XP_020024523.1 | colipase-like [Castor canadensis] | 36.5% | 7.0E-03 | 43.1 |
Table of the top ten BLASTP hits for the protein of transcript ISCW024249
Accession number of protein as found in GenBank
Predicted function based on the submission to GenBank. The species that the hit came from is found in the brackets.
Percent identity of the protein hit to the protein of transcript ISCW024249
E-value of the match between the hit and the protein of transcript ISCW024249
Bit score of the match between the hit and the protein of transcript ISCW024249
As other tick salivary gland proteins have been shown to bind B. burgdorferi s.s., we assessed if we could detect binding between Salp12 and the spirochete in an in vitro immunofluorescence assay (IFA) similar to IFAs previously used for Salp15 (Ramamoorthi, Narasimhan et al. 2005) and Ixofin3D (Narasimhan, Coumou et al. 2014). In this experiment, in vitro grown B. burgdorferi s.s. B31A was incubated with Salp12 and protein localization was detected through fluorescent antibodies (Fig. S4). We were unable to detect any binding between the spirochete and Salp12. Although some Salp proteins bind B. burgdorferi s.s., it is not surprising that binding was not detected between Salp12 and the spirochete. In vivo chemotactic interactions are transient in nature, and therefore, interactions between B. burgdorferi s.s. and Salp12 may not be stable enough to be identified by an IFA. It is also possible that Salp12 interacts with secreted spirochete proteins that are then able to serve as chemoattractants or that the protein is cleaved by proteases prior to binding that prevent visualization.
Salp12 is expressed in the midguts and salivary glands of I. scapularis ticks.
To gain a better understanding of Salp12 expression and its role within the tick, we assessed salp12 mRNA expression during the course of feeding in I. scapularis nymphs. salp12 was detectable in both salivary glands and midguts throughout feeding, and expression in tick midguts was significantly higher than expression in tick salivary glands at every time point (Fig. 4A) (Mann-Whitney, df=2, p=0.002). To determine whether the expression of salp12 is altered by the presence of B. burgdorferi s.s., we assessed salp12 expression in infected and uninfected nymphs. These data show no difference in salp12 expression between uninfected and B. burgdorferi s.s. infected salivary glands (Fig. 4B) or midguts (Fig. 4C).
Figure 4. I. scapularis protein Salp12 is expressed throughout tick feeding in both the salivary glands and midgut.

I. scapularis ticks were collected at 24, 48, or 72 hours of feeding and assessed for expression of Salp12 relative to tick actin in their salivary glands (SG, circles) or midguts (MG, squares) using absolute quantification qPCR. Expression levels were compared between tick organs (A). Transcript expression was also compared for SG (B) and MG (C) between uninfected (black) and B. burgdorferi infected (grey) samples. Each panel shows a representative experiment, one of two biological replicates. For each experiment at least five ticks were collected per time point. Significance was tested using a Mann-Whitney test.
Salp12 contributes to B. burgdorferi s.s. acquisition.
To assess if B. burgdorferi s.s. chemotaxis towards Salp12 is involved in acquisition, we performed RNAi knockdown experiments with I. scapularis nymphs (Fig. 5). Ticks received thoracic injection of dsRNA to knockdown expression in the salivary glands, and ticks were injected with ds salp12 for knockdown or ds gfp as a control. RNAi knockdown of salp12 was not complete and resulted in a 50% decrease of transcript levels in the salivary glands at 24 hours of feeding (Fig. 5A) (ANOVA, df=1, p<0.05). Uninfected nymphs were injected and fed to repletion on B. burgdorferi s.s. B31A3 infected mice. We first assessed if knockdown of salp12 impacted tick feeding, which could alter acquisition. There were no significant differences in tick feeding, as measured by engorgement weight (Fig. 5B) and time to repletion (Fig. 5C). B. burgdorferi s.s. burdens in replete ticks injected with ds salp12 trended lower than ds gfp infected ticks, but this was not statistically significant (Fig. 5D, Mann-Whitney test, df=1, p=0.15).
Figure 5. Knockdown of Salp12 expression decreases B. burgdorferi acquisition by I. scapularis ticks.

Protein expression was knocked down in I. scapularis salivary glands using RNAi with ds salp12 (white) or control dsRNA, ds gfp (black), which was measured in I. scapularis salivary glands (SG) and midgut (MG) (A). Knock down I. scapularis ticks were fed on B. burgdorferi infected mice to assess acquisition of the spirochete. The impact of salp12 knockdown on tick feeding was measured by engorgement weight (B) and time to repletion (C). B. burgdorferi burden in replete tick midguts (D) was determined using absolute quantification qPCR of flaB relative to tick actin. Analysis of acquisition during the course of tick feeding (E) was determined by assess B. burgdorferi burden in the midgut at 24 hours and at 7 days post detachment. Data (A-D) is the average of 2 biological replicates, and panel E shows the average of 3 biological replicates. For B and C, statistical significance was tested using ANOVA, and for A, D, and E, statistical significance was tested using a Mann-Whitney test.
There are two distinct steps for B. burgdorferi s.s. acquisition. Spirochetes enter the tick midgut during tick feeding and then colonize the midgut to be maintained during molting. Therefore, to gain a more nuanced view of the acquisition process, we assessed the impact of Salp12 on spirochete entry (after 24 hours of tick attachment) and on colonization (7 days post detachment) in I. scapularis nymphs (Fig. 5E). The 24 hour timepoint was chosen because Salp12 is most highly expressed at 24 hours (Fig. 4A) and would therefore likely have the largest impact at this time. Spirochete entry into the ds salp12 injected ticks trended lower than in ds gfp injected ticks at 24 hours, but this was not statistically significant (Mann-Whitney test). By seven days post detachment, the spirochete burden in the midgut was the same between control and ds salp12 injected ticks. The trend in the data suggests that Salp12 contributes to spirochete entry but does not impact maintenance or growth of the spirochete in the tick midgut after entry.
To circumvent the incomplete knockdown of Salp12 by RNAi, we utilized passive immunization to block Salp12-spirochete interactions and assessed acquisition I. scapularis nymphs (Fig. 6). Antibodies against Salp12 or control antibodies against ovalbumin were transferred into mice infected with B. burgdorferi s.s. B31A3, and acquisition was assessed. Ticks fed on mice immunized against Salp12 had a decrease in spirochete entry at 24 hours (Mann-Whitney test, df=2, p<0.03). There was also a decrease in spirochete burden at 7 days post detachment (not statistically significant, Mann-Whitney test), indicating that colonization of the spirochete is affected. Similar to the RNAi experiments, these data suggest that Salp12 contributes to spirochete entry during tick feeding but blocking Salp12 interactions with the spirochete does not completely prevent acquisition.
Figure 6. Passive immunization against Salp12 decreases B. burgdorferi acquisition of by I. scapularis ticks.
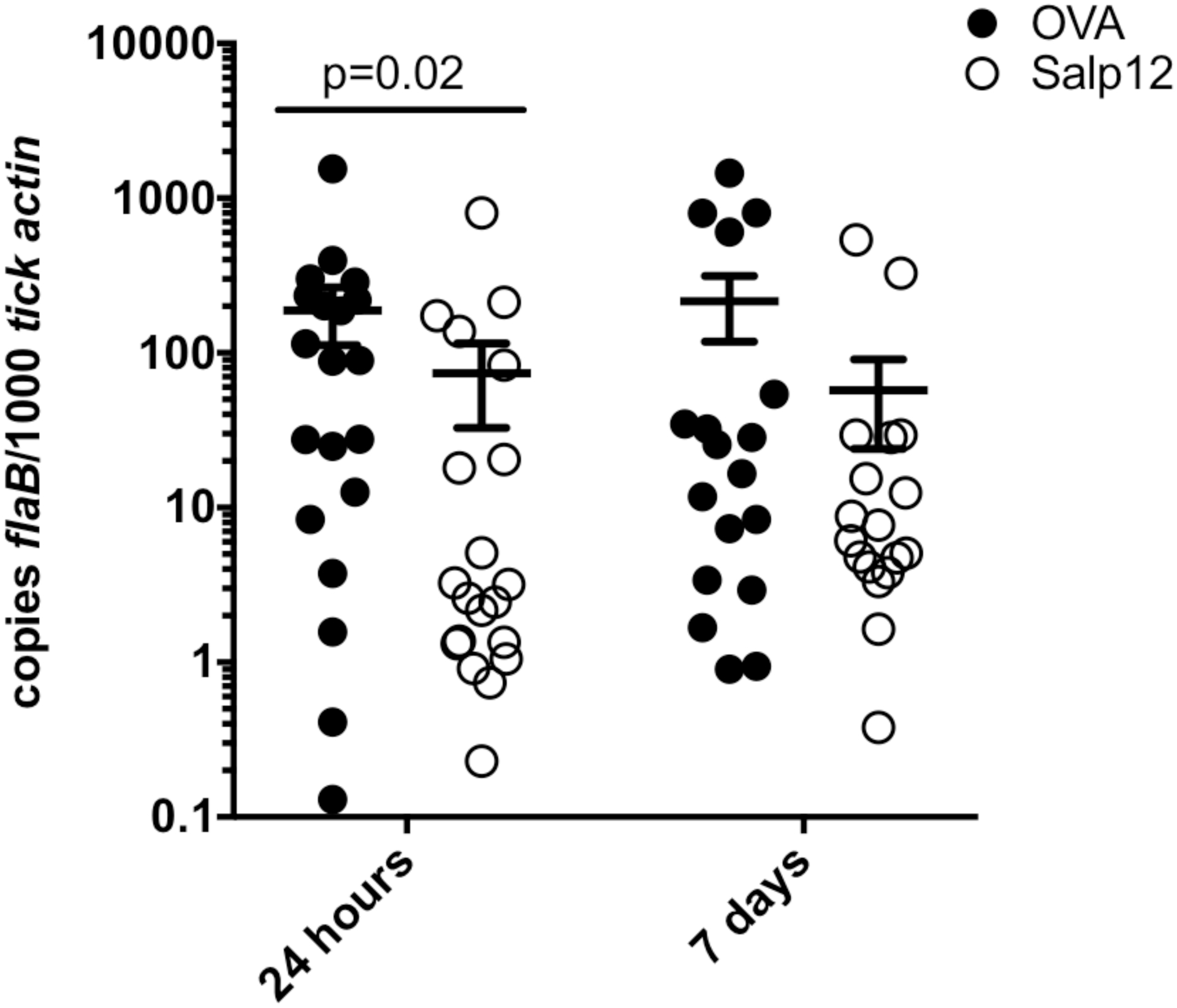
Mice infected with B. burgdorferi were passively immunized with rabbit antibodies against Salp12 (black circles) or control antibodies against ovalbumin (OVA, white circles). Uninfected I. scapularis ticks were fed on B. burgdorferi infected mice and removed at 24 hours or allowed to feed to repletion and incubated for 7 days. B. burgdorferi burden in tick midguts was measured by absolute quantification qPCR of flaB relative to tick actin. The graph shows an average of two biological replicates. Statistical significance was assessed using a Mann-Whitney test.
Chemotaxis Toward Salp12 is not involved in transmission of the spirochete.
Because Salp12 has a role in acquisition and signals for acquisition and transmission are likely different, we hypothesized that Salp12 would not have a role in transmission. To confirm this, we performed transmission studies with RNAi knockdown ticks using thoracic injection. There were no differences in tick feeding in ds salp12 injected ticks compared to control ticks, as measured engorgement weight (Fig. 7A) and time to repletion (Fig. 7B). Replete ticks that were injected with ds salp12 had significantly different burdens of B. burgdorferi s.s. in their midguts at repletion (Fig. 7C, Mann-Whitney test, df=2, p<0.0001). However, this difference was driven by one tick sample, indicating that transmission would likely not differ between ds salp12 injected and control ticks. Differences in B. burgdorferi s.s. burden did not impact the level of transmission to mice, as measured by B. burgdorferi s.s. levels in the skin, heart, bladder, and joints (Fig. 7D, E). These data support that chemotaxis toward salivary gland Salp12 is not involved in transmission.
Figure 7. Knockdown of Salp12 expression does not alter B. burgdorferi transmission.

Expression of Salp12 in I. scapularis salivary glands was knocked down by RNAi with ds salp12 (black), and control ticks were exposed to ds gfp (white). I. scapularis feeding was measured by tick engorgement weight (A) and time to repletion (B), and significance was not detected using ANOVA. B. burgdorferi burden in replete I. scapularis ticks (C) was measured by absolute quantification qPCR of flaB relative to tick actin, and significance was not detected using ANOVA. Transmission from knockdown I. scapularis ticks was measured by qPCR of flaB relative to mouse actin in mouse skin (D) or organs (E), significance was not detected using a Mann-Whitney test. Assessment of B. burgdorferi burden during the course of feeding was measured by absolute quantification qPCR of tick midguts (F) and salivary glands (G), significance was not detected using a Mann-Whitney test. Data for tick feeding and B. burgdorferi burden shows the average of two biological replicates (A-C, F-G). Data for transmission experiments (D-E) is shown by a representative experiment, one of two biological replicates.
As there were differences in the level of spirochetes in replete nymphs, we next assessed if migration of the spirochete during transmission was impacted by knockdown of salp12. During feeding, spirochetes grow within the tick midgut for 36 hours and then migrate to the salivary glands. From the tick salivary glands, spirochetes are able to exit with the tick saliva. Because Salp12 is present in the tick salivary glands and midgut, it is possible that the spirochete is recognizing the protein for egress from the midgut or invasion of the salivary glands. If chemotaxis toward Salp12 is involved in transmission, a decrease in spirochete burden in the salivary glands would be observed. Knockdown experiments showed that levels of B. burgdorferi s.s. in the salivary glands were unchanged (Fig. 7G), indicating that knockdown of salivary gland Salp12 does not impact invasion of the salivary glands by the spirochete or migration through the tick. However, B. burgdorferi s.s. levels in the tick midgut trended higher than in control ticks (not statistically significant, Mann-Whitney test, Fig. 7F), indicating a possible role in egress or growth in the midgut. As spirochetes are still able to migrate to the tick salivary glands and infect mice, chemotaxis toward salivary gland Salp12 does not significantly contribute to transmission of the spirochete.
Discussion:
Our fractionation experiments demonstrate that proteins, and not sugars or other small molecules, are the chemoattractant compounds contributing to B. burgdorferi s.s. migration in the capillary tube chemotaxis assay and that Salp12 is a chemoattractant for B. burgdorferi s.s. However, it is important to note that there is likely a minimum necessary concentration of chemoattractants to identify a response in the capillary tube assay. Therefore, it is possible that other chemoattract compounds may be present in I. scapularis SGE, fractionated SGE, and saliva that were not identified by this in vitro assay. Further analysis of higher concentrations of these fractions or in more sensitive assays may identify other chemoattracts. It is also of note that while B. burgdorferi s.s. migrated toward solutions of purified Salp12 in a reproducible and statistically significant manner, B. burgdorferi s.s. migration toward the SGE fraction containing Salp12 was modest and not statistically significant. This is likely due in part to the fact that a lower concentration of Salp12 would be in the protein fraction compared to the solution of purified Salp12.
Our data demonstrate that Salp12 is sufficient to induce chemotaxis of B. burgdorferi s.s. in vitro, and Salp12 contributes to spirochete entry during acquisition. However, knockdown of Salp12 or passive immunization against Salp12 did not completely prevent acquisition of the spirochete. For the RNAi experiments, this is at least partially due to incomplete knockdown of the protein, as RNAi only reduced transcription by 50%. Continued acquisition of the spirochete could also be explained by the presence of an additional chemoattractant in the tick saliva or a chemoattractant produced by wounding from tick feeding. Moreover, some acquisition likely occurs through passive processes, such as the spirochetes already residing within the tick bite site or migrating through the bite site at random.
Our RNAi and passive immunization studies differed in spirochete burden at 7 days post detachment. Spirochete colonization trended lower in ticks fed on mice passively immunized against Salp12 than on control mice, but this trend was not identified in RNAi experiments. This difference is likely due to passive immunization affecting Salp12 function in the midgut, where RNAi injections only target Salp12 in the salivary glands. In passive immunization, Salp12 antibodies will enter the tick with the blood meal and likely impair midgut Salp12, in addition to Salp12 in the skin. This indicates that midgut Salp12 may influence the midgut environment. It is unlikely that this phenotype is dependent on chemotaxis toward Salp12 because the trend was not observed in RNAi experiments.
As expected, our transmission data support that chemotaxis toward Salp12 does not play a significant role in transmission of the spirochete. However, we identified that Salp12 may affect egress from the tick midgut or growth within the midgut during transmission. As thoracic injection specifically decreases mRNA expression in the tick salivary glands and salp12 levels are unchanged in the midgut, it is unclear how growth or egress are affected by RNAi injections. It is possible that changes in Salp12 expression in the salivary glands could alter global expression of tick proteins, which in turn alters the midgut environment. Together with the findings that Salp12 contributes to spirochete colonization after acquisition and that salp12 is expressed in the midgut, these data suggest that Salp12 likely shapes the midgut environment.
Although the spirochete encounters Salp12 during acquisition and transmission, migration of the spirochete is only altered in acquisition, which highlights that the spirochete is sensing different signals for these processes. This is supported by the fact that although the environment within the tick is similar during acquisition and transmission, spirochetes migrate to the tick salivary glands during transmission but not acquisition. The difference in spirochete response to a similar environment is likely due to divergent gene expression by the spirochete (de Silva and Fikrig 1997).
Identifying a protein as a chemoattractant is quite surprising, as chemoattractants identified for other bacteria are small molecules, such as sugars and short peptides. Additionally, the bacterial receptors for chemotaxis, methyl-accepting chemotaxis proteins (MCPs), have binding pockets that accommodate smaller molecules, and MCPs are localized to the inner-membrane of bacteria. The presence of the bacterial outer-membrane prevents direct interaction of the MCPs with extracellular molecules. The B31A strains used in this study have five predicted MCPs (Fraser, Casjens et al. 1997), all with structural similarity to previously identified MCPs (Fraser, Casjens et al. 1997, Xu, Raddi et al. 2011), and two of these MCPs have been shown to localize to the inner-membrane (Xu, Raddi et al. 2011). Therefore, B. burgdorferi s.s. must utilize a mechanism different from canonical systems for recognition of a large protein. There are two possible hypotheses. One is that the tick protein is autoproteolytic or cleaved by proteases, and the peptides are able to interact with the MCPs directly. A second possibility is that the intact tick protein interacts with an additional receptor to transduce a secondary signal to the MCPs. This hypothesis is supported by studies in other bacterial species that have demonstrated MCPs can respond to external chemical stimuli by binding internal signals (Ni, Huang et al. 2015, Machuca, Liu et al. 2016).
Conclusion:
Overall, our study suggests that chemotaxis contributes to acquisition in the infectious cycle of B. burgdorferi s.s. We hypothesize that Salp12 is deposited into the skin as part of the tick saliva, where it is recognized by resident B. burgdorferi s.s. This recognition contributes to migration of the spirochete into the tick feeding site, and the spirochete enters the tick along with the blood meal. However, our data also suggest that multiple signals may be recognized by the spirochete for acquisition, such as additional tick molecules or cues from wounding. It is possible that identification of additional chemoattractants and targeting these molecules in combination with Salp12 may effectively block pathogen acquisition. This strategy could be used for environmental control of the infectious cycle of vector-borne diseases.
Supplementary Material
Acknowledgements:
The authors would like to thank Dr. Ryuta Uraki for purified mosquito protein AAEL010228, and Dr. M.A. Motaleb for the B. burgdorferi s.s. strains. The authors thank Dr. Yongguo Cao for collaborative input. The authors also thank the NIH and Yale School of Medicine for funding the Orbitrap Fusion (1S10ODOD018034-01) LC MS/MS system located within the Yale MS & Proteomics Resource of the WM Keck Foundation Biotechnology Resource Laboratory; and Edward Voss, Jean Kanyo, and Dr. TuKiet Lam with MS sample preparation, data collection, and helpful discussion, respectively.
Funding: K.E.M. was funded by a James Hudson Brown-Alexander Brown Coxe Fellowship from Yale University and a NIH Immunohematology/Transfusion Medicine Research Training Grant (T32HL007974). This work was supported in part by NIH grants AI1260003 and AI138949 and gifts from the John Monsky and Jennifer Weis Monsky Lyme Disease Research Fund and from the Cohen Foundation. E.F. is an investigator supported by the Howard Hughes Medical Institute.
Abbreviations:
- SGE
Salivary gland extract
- Salp12
Ixodes scapularis salivary gland protein of 12kDa
References:
- Adrion ER, Aucott J, Lemke KW and Weiner JP, 2015. Health care costs, utilization and patterns of care following Lyme disease. PLoS One. 10, e0116767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguita J, Ramamoorthi N, Hovius JW, Das S, Thomas V, Persinski R, Conze D, Askenase PW, Rincon M, Kantor FS and Fikrig E, 2002. Salp15, an ixodes scapularis salivary protein, inhibits CD4(+) T cell activation. Immunity. 16, 849–859. [DOI] [PubMed] [Google Scholar]
- Bakker RG, Li C, Miller MR, Cunningham C and Charon NW, 2007. Identification of specific chemoattractants and genetic complementation of a Borrelia burgdorferi chemotaxis mutant: flow cytometry-based capillary tube chemotaxis assay. Appl Environ Microbiol. 73, 1180–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthold SW, Persing DH, Armstrong AL and Peeples RA, 1991. Kinetics of Borrelia burgdorferi dissemination and evolution of disease after intradermal inoculation of mice. Am J Pathol. 139, 263–273. [PMC free article] [PubMed] [Google Scholar]
- Bockenstedt LK, Gonzalez D, Mao J, Li M, Belperron AA and Haberman A, 2014. What ticks do under your skin: two-photon intravital imaging of Ixodes scapularis feeding in the presence of the lyme disease spirochete. Yale J Biol Med. 87, 3–13. [PMC free article] [PubMed] [Google Scholar]
- Chmelař J, Kotál J, Karim S, Kopacek P, Francischetti IM, Pedra JH and Kotsyfakis M, 2016. Sialomes and mialomes: a systems-biology view of tick tissues and tick-host interactions. Trends Parasitol. 32, 242–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway MJ, Londono-Renteria B, Troupin A, Watson AM, Klimstra WB, Fikrig E and Colpitts TM, 2016. Aedes aegypti D7 saliva protein inhibits dengue virus infection. PLoS Negl Trop Dis. 10, e0004941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coumou J, Narasimhan S, Trentelman JJ, Wagemakers A, Koetsveld J, Ersoz JI, Oei A, Fikrig E and Hovius JW, 2016. Ixodes scapularis dystroglycan-like protein promotes Borrelia burgdorferi migration from the gut. J Mol Med (Berl). 94, 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Wang P, Adusumilli S, Booth CJ, Narasimhan S, Anguita J and Fikrig E, 2009. Antibodies against a tick protein, Salp15, protect mice from the Lyme disease agent. Cell Host Microbe. 6, 482–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Banerjee G, DePonte K, Marcantonio N, Kantor FS and Fikrig E, 2001. Salp25D, an Ixodes scapularis antioxidant, is 1 of 14 immunodominant antigens in engorged tick salivary glands. J Infect Dis. 184, 1056–1064. [DOI] [PubMed] [Google Scholar]
- de Silva AM and Fikrig E, 1995. Growth and migration of Borrelia burgdorferi in Ixodes ticks during blood feeding. Am J Trop Med Hyg. 53, 397–404. [DOI] [PubMed] [Google Scholar]
- de Silva AM and Fikrig E, 1997. Arthropod- and host-specific gene expression by Borrelia burgdorferi. J Clin Invest. 99, 377–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwinson A, Widmer G and McEvoy J, 2016. Glycoproteins and Gal-GalNAc cause Cryptosporidium to switch from an invasive sporozoite to a replicative trophozoite. Int J Parasitol. 46, 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang HY, Dosztanyi Z, El-Gebali S, Fraser M, Gough J, Haft D, Holliday GL, Huang H, Huang X, Letunic I, Lopez R, Lu S, Marchler-Bauer A, Mi H, Mistry J, Natale DA, Necci M, Nuka G, Orengo CA, Park Y, Pesseat S, Piovesan D, Potter SC, Rawlings ND, Redaschi N, Richardson L, Rivoire C, Sangrador-Vegas A, Sigrist C, Sillitoe I, Smithers B, Squizzato S, Sutton G, Thanki N, Thomas PD, Tosatto SC, Wu CH, Xenarios I, Yeh LS, Young SY and Mitchell AL, 2017. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 45, D190–D199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE and Mello CC, 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fuji C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO and Venter JC, 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 390, 580–586. [DOI] [PubMed] [Google Scholar]
- Gern L and Rais O, 1996. Efficient transmission of Borrelia burgdorferi between cofeeding Ixodes ricinus ticks (Acari: Ixodidae). J Med Entomol. 33, 189–192. [DOI] [PubMed] [Google Scholar]
- Hinckley AF, Connally NP, Meek JI, Johnson BJ, Kemperman MM, Feldman KA, White JL and Mead PS, 2014. Lyme disease testing by large commercial laboratories in the United States. Clin Infect Dis. 59, 676–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodzic E, Borjesson DL, Feng S and Barthold SW, 2001. Acquisition dynamics of Borrelia burgdorferi and the agent of human granulocytic ehrlichiosis at the host-vector interface. Vector Borne Zoonotic Dis. 1, 149–158. [DOI] [PubMed] [Google Scholar]
- Kazimírová M and Štibrániová I, 2013. Tick salivary compounds: their role in modulation of host defences and pathogen transmission. Front Cell Infect Microbiol. 3, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Tirloni L, Pinto AF, Moresco J, Yates JR 3rd, da Silva Vaz I Jr. and Mulenga A, 2016. Ixodes scapularis tick saliva proteins sequentially secreted every 24 h during blood feeding. PLoS Negl Trop Dis. 10, e0004323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropec A, Maira-Litran T, Jefferson KK, Grout M, Cramton SE, Gotz F, Goldmann DA and Pier GB, 2005. Poly-N-acetylglucosamine production in Staphylococcus aureus is essential for virulence in murine models of systemic infection. Infect Immun. 73, 6868–6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn BM, 2013. CDC estimates 300,000 US cases of Lyme disease annually. JAMA. 310, 1110. [DOI] [PubMed] [Google Scholar]
- Lin T, Gao L, Zhang C, Odeh E, Jacobs MB, Coutte L, Chaconas G, Philipp MT and Norris SJ, 2012. Analysis of an ordered, comprehensive STM mutant library in infectious Borrelia burgdorferi: insights into the genes required for mouse infectivity. PLoS One. 7, e47532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe ME, 1997. Structure and function of pancreatic lipase and colipase. Annu Rev Nutr. 17, 141–158. [DOI] [PubMed] [Google Scholar]
- Machuca MA, Liu YC, Beckham SA, Gunzburg MJ and Roujeinikova A, 2016. The crystal structure of the tandem-PAS sensing domain of Campylobacter jejuni chemoreceptor Tlp1 suggests indirect mechanism of ligand recognition. J Struct Biol. 194, 205–213. [DOI] [PubMed] [Google Scholar]
- Masuko T, Minami A, Iwasaki N, Majima T, Nishimura S and Lee YC, 2005. Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal Biochem. 339, 69–72. [DOI] [PubMed] [Google Scholar]
- Motaleb MA, Miller MR, Bakker RG, Li C and Charon NW, 2007. Isolation and characterization of chemotaxis mutants of the Lyme disease spirochete Borrelia burgdorferi using allelic exchange mutagenesis, flow cytometry, and cell tracking. Methods Enzymol. 422, 421–437. [DOI] [PubMed] [Google Scholar]
- Motaleb MA, Miller MR, Li C, Bakker RG, Goldstein SF, Silversmith RE, Bourret RB and Charon NW, 2005. CheX is a phosphorylated CheY phosphatase essential for Borrelia burgdorferi chemotaxis. J Bacteriol. 187, 7963–7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan S, Coumou J, Schuijt TJ, Boder E, Hovius JW and Fikrig E, 2014. A tick gut protein with fibronectin III domains aids Borrelia burgdorferi congregation to the gut during transmission. PLoS Pathog. 10, e1004278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni B, Huang Z, Wu YF, Fan Z, Jiang CY and Liu SJ, 2015. A novel chemoreceptor MCP2983 from Comamonas testosteroni specifically binds to cis-aconitate and triggers chemotaxis towards diverse organic compounds. Appl Microbiol Biotechnol. 99, 2773–2781. [DOI] [PubMed] [Google Scholar]
- Novak EA, Sekar P, Xu H, Moon KH, Manne A, Wooten RM and Motaleb MA, 2016. The Borrelia burgdorferi CheY3 response regulator is essential for chemotaxis and completion of its natural infection cycle. Cell Microbiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahl A, Kuhlbrandt U, Brune K, Rollinghoff M and Gessner A, 1999. Quantitative detection of Borrelia burgdorferi by real-time PCR. J Clin Microbiol. 37, 1958–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrican LA, 1997. Acquisition of Lyme disease spirochetes by cofeeding Ixodes scapularis ticks. Am J Trop Med Hyg. 57, 589–593. [DOI] [PubMed] [Google Scholar]
- Pedra JH, Narasimhan S, Deponte K, Marcantonio N, Kantor FS and Fikrig E, 2006. Disruption of the salivary protein 14 in Ixodes scapularis nymphs and impact on pathogen acquisition. Am J Trop Med Hyg. 75, 677–682. [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G and Nielsen H, 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 8, 785–786. [DOI] [PubMed] [Google Scholar]
- Porter SL, Wadhams GH and Armitage JP, 2011. Signal processing in complex chemotaxis pathways. Nat Rev Microbiol. 9, 153–165. [DOI] [PubMed] [Google Scholar]
- Ramamoorthi N, Narasimhan S, Pal U, Bao F, Yang XF, Fish D, Anguita J, Norgard MV, Kantor FS, Anderson JF, Koski RA and Fikrig E, 2005. The Lyme disease agent exploits a tick protein to infect the mammalian host. Nature. 436, 573–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuijt TJ, Coumou J, Narasimhan S, Dai J, Deponte K, Wouters D, Brouwer M, Oei A, Roelofs JJ, van Dam AP, van der Poll T, Van’t Veer C, Hovius JW and Fikrig E, 2011. A tick mannose-binding lectin inhibitor interferes with the vertebrate complement cascade to enhance transmission of the lyme disease agent. Cell Host Microbe. 10, 136–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Yang Z, Geng Y, Wolinsky LE and Lovett MA, 1998. Chemotaxis in Borrelia burgdorferi. J Bacteriol. 180, 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih CM, Chao LL and Yu CP, 2002. Chemotactic migration of the Lyme disease spirochete (Borrelia burgdorferi) to salivary gland extracts of vector ticks. Am J Trop Med Hyg. 66, 616–621. [DOI] [PubMed] [Google Scholar]
- States SL, Huang CI, Davis S, Tufts DM and Diuk-Wasser MA, 2017. Co-feeding transmission facilitates strain coexistence in Borrelia burgdorferi, the Lyme disease agent. Epidemics. 19, 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze CW, Zhang K, Kariu T, Pal U and Li C, 2012. Borrelia burgdorferi needs chemotaxis to establish infection in mammals and to accomplish its enzootic cycle. Infect Immun. 80, 2485–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Raddi G, Liu J, Charon NW and Li C, 2011. Chemoreceptors and flagellar motors are subterminally located in close proximity at the two cell poles in spirochetes. J Bacteriol. 193, 2652–2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Sultan S, Yerke A, Moon KH, Wooten RM and Motaleb MA, 2017. Borrelia burgdorferi CheY2 is dispensable for chemotaxis or motility but crucial for the infectious life cycle of the spirochete. Infect Immun. 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang Y, Adusumilli S, Liu L, Narasimhan S, Dai J, Zhao YO and Fikrig E, 2011. Molecular interactions that enable movement of the Lyme disease agent from the tick gut into the hemolymph. PLoS Pathog. 7, e1002079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomic data are available via the ProteomeXchange Consortium and the PRIDE partner repository (http://www.proteomexchange.org/) under the identifier PXD011273.


