Abstract
Objective
Diabetic kidney disease (DKD) is one of the most frequent complications in diabetes associated with substantial morbidity and mortality. To accelerate DKD risk factor discovery, we present an ensemble feature selection approach to identify a robust set of discriminant factors using electronic medical records (EMRs).
Material and Methods
We identified a retrospective cohort of 15 645 adult patients with type 2 diabetes, excluding those with pre-existing kidney disease, and utilized all available clinical data types in modeling. We compared 3 machine-learning-based embedded feature selection methods in conjunction with 6 feature ensemble techniques for selecting top-ranked features in terms of robustness to data perturbations and predictability for DKD onset.
Results
The gradient boosting machine (GBM) with weighted mean rank feature ensemble technique achieved the best performance with an AUC of 0.82 [95%-CI, 0.81–0.83] on internal validation and 0.71 [95%-CI, 0.68–0.73] on external temporal validation. The ensemble model identified a set of 440 features from 84 872 unique clinical features that are both predicative of DKD onset and robust against data perturbations, including 191 labs, 51 visit details (mainly vital signs), 39 medications, 34 orders, 30 diagnoses, and 95 other clinical features.
Discussion
Many of the top-ranked features have not been included in the state-of-art DKD prediction models, but their relationships with kidney function have been suggested in existing literature.
Conclusion
Our ensemble feature selection framework provides an option for identifying a robust and parsimonious feature set unbiasedly from EMR data, which effectively aids in knowledge discovery for DKD risk factors.
Keywords: diabetic kidney disease, risk factor discovery, embedded feature selection, feature stability, ensemble feature selection
BACKGROUND AND SIGNIFICANCE
Diabetic kidney disease (DKD) is one of the most frequent and dangerous microvascular complications in diabetes mellitus (DM), affecting about 20% to 40% of patients with type 1 or type 2 DM.1 It is the leading cause of end-stage renal disease (ESRD), accounting for approximately 50% of the cases in the developed world with major public health and economic implications.2 Systematic screening and monitoring for complications have become a major part of diabetes care management today. Identifying risk factors related to DKD may better stratify patients for tailored monitoring and evidence-based treatment to prevent or slow down disease progression.
Current knowledge on DKD risk factors is derived mainly from hypothesis-driven prospective observational studies with sample size ranging from a few hundred to a few thousand subjects.3–6 To accelerate hypothesis generation for DKD risk factor discovery, more robust and effective observational data-driven approaches are crucial. The electronic medical record (EMR) provides a promising data source for researchers to pragmatically study disease in situ, in particular to discover novel risk factors.7 As a diverse and rich resource and free of study-specific bias, EMR provides an abundance of valuable information delineating patients’ healthcare experience. However, EMR data, similar to other types of biomedical data, contain a considerable amount of irrelevant and redundant features8 and have their own sampling biases.9 To eliminate irrelevant features in modeling, feature selection has been extensively studied in the machine-learning community for many years10–14 and has found successful biomedical applications such as gene and Single Nucleotide Polymorphism selection for biomarker discovery.13 Feature selection has become an increasingly valuable approach7,8,15–17 to reduce overfitting and generalize learning models.18,19 Simplifying a model with fewer features can improve interpretation and shed light on mechanisms underlying a target disease.20 From a computational perspective, reduced feature sets lead to more compact and faster predictive models.
There are broadly 3 classes of feature selection methods: filter, wrapper, and embedded.21 While filter methods work independently of the classifier and use a proxy measure to rank the features, wrapper and embedded methods perform feature selection by optimizing performance of a particular classifier.22 The wrapper method, commonly stepwise regression, has been used for the prediction of DKD or chronic kidney disease (CKD) as most studies used hypothesis-driven approaches23–25 with pre-selected features in which computational cost was not an issue. However, when it comes to mining high-dimensional data, filter or embedded methods are usually preferred due to computational efficiency.12,26–28 To our best knowledge, we have not found published data-driven solutions to DKD risk factor identification that leverage unrestricted EMR data (eg, note concepts, labs, nursing observations) integrated with clinical registries and billing sources. Such integrated data repositories have only recently been linked by clinical data research networks29 and nationally verified for data quality through the PCORnet initiative.30 Another barrier to data-driven DKD risk factor identification could be the strong and multiway correlations within EMR data as well as its heterogeneous nature, which exacerbates the stability or robustness issue in feature selection, resulting in non-reproducible models and misleading rules.31 To address the limitations, a panel of ensemble feature selection methods has been developed.32,33
In this study, we presented an ensemble feature selection framework to identify a robust and parsimonious set of important features that are predictive of DKD by mining an integrated clinical data repository. We combined 3 types of embedded feature selection algorithms with 6 feature ensemble techniques and evaluated both prediction accuracy and stability for these 18 (ie, 3x6) ensemble–feature selection models. We chose the 3 types of embedded feature selection algorithms, namely, elastic net (ElastNet) regressions,34 gradient boosting machine (GBM) trees,35 and deep neural network (DNN),36 because they favor different underlying structures. For evaluation, we rigorously assessed the tradeoff between stability and predictability for each model. The Kuncheva index37 and weighted and relative weighted consistency indices38 were used for measuring stability, and area under receiver operating curve (AUC) was used for evaluating prediction performance. After demonstrating that GBM with ensemble-weighted-mean rank generated the “best” feature set, we evaluated the marginal effects35 of the top features on DKD risk.
METHODS
Diabetes definition
We adopted the surveillance, prevention, and management of diabetes mellitus (SUPREME-DM) definition of diabetes in this study. Diabetes was defined based on: a) use of glucose-lowering medications (insulin or oral hypoglycemic medications); or b) level of hemoglobin A1C of 6.5% or greater, random glucose of 200 mg/dL or greater, or fasting glucose of 126 mg/dL on at least 2 different dates within 2 years; or c) any 2 type 1 and type 2 DM diagnoses given on 2 different days within 2 years; or d) any 2 distinct types of events among a), b), or c); e) excluding any gestational diabetes (temporary glucose rise during pregnancy).39
DKD definition
DKD was defined as diabetes with the presence of microalbuminuria or proteinuria, impaired glomerular filtration rate (GFR), or both.40,41 More specifically, microalbuminuria was defined as albumin-to-creatinine ratio (ACR) being 30 mg/g or greater (similarly, proteinuria was defined as urine protein-to-creatinine ratio being 30 mg/g or greater).40,41 Impaired GFR was defined as the estimated GFR (eGFR)—an age, gender, and race adjusted serum creatinine concentration based on the modification of diet in renal disease (MDRD) equation41—being less than 60 mL/min/1.73m2.
Study cohort
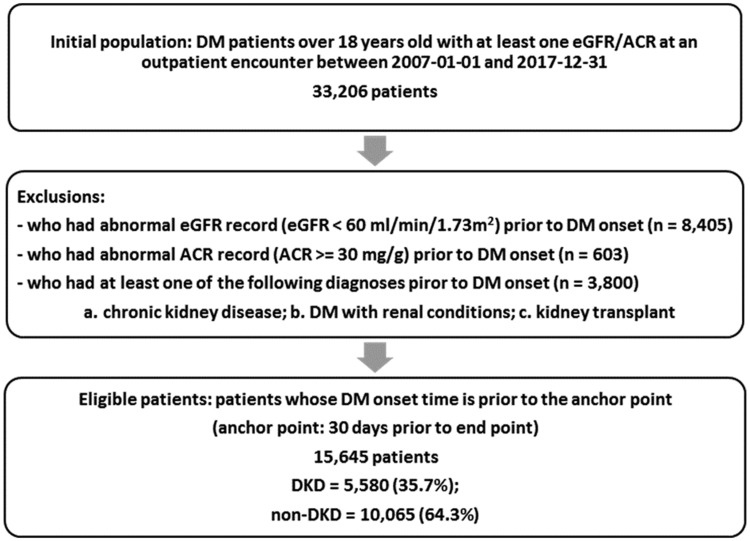
A retrospective cohort was built using de-identified data from November 2007 to December 2017 in the University of Kansas Medical Center’s integrated clinical data repository, Healthcare Enterprise Repository for Ontological Narration (HERON).42 The study did not require IRB approval because data used meet the de-identification criteria specified in the HIPAA Privacy Rule. Data request was approved by the HERON Data Request Oversight Committee. As shown in Figure 1, a total of 33 206 adult DM patients (age ≥ 18) who had at least 1 valid eGFR or ACR record at an outpatient encounter were eligible for this study, so that they were identifiable as DKD present or not. We excluded patients with any kidney disease manifestation (eg, CKD diagnosis, low eGFR, or microalbuminuria) prior to their DM onset. The case group included all DKD patients with their onset time, or endpoint, defined as the first time of their abnormal eGFR or ACR. The control group was defined as DM patients whose eGFR values were always above or equal to 60 mL/min/1.73m2 and have never had microalbuminuria, with their endpoint defined as the last time of their normal eGFR or ACR. Finally, 15 645 patients were included in the final cohort with 5580 (35.7%) DKD patients.
Figure 1.
Study cohort inclusion and exclusion.
Clinical variables
We included 15 types of clinical observations from HERON (Table 1).46 Each category is a mix of categorical and numerical data elements. For laboratory tests and vital signs, numeric values were used while we created binary indicator variables for categorical features. In addition, we decomposed clinical features into more meaningful pieces according to: a) different sources of a diagnosis (ie, primary diagnoses or secondary diagnoses), b) different aspect of a medication fact (ie, drug refill or drug amount), c) different types of encounters where a procedure was ordered or performed (ie, inpatient or outpatient), d) different states of an alert (ie, fired or overridden). These data elements can be extracted from EMR, and they have been explicitly incorporated in our system as an additional attribute called “modifier.”47 Based on our previous investigation, a reasonable choice of level of granularity for medications is at the semantic clinical drug form (SCDF) or semantic clinical brand form (SCBF) level according to the HERON ontology, while diagnoses observations were represented at the ICD 9 or 10 code level.48
Table 1.
Integrated data repository data domain categories
| Domain | Descriptions | Data Type | # of Eligible Featuresa |
|---|---|---|---|
| ALERTS | Includes drug interaction, dose warnings, drug interactions, medication administration warnings, and best practice alerts | Binary | 1230 |
| ALLERGY | Includes documented allergies and reactions | Binary | 60 |
| DEMOGRAPHICS | Basic demographics such as age, gender, race, etc., as well as their reachability, and some geographical information | Binary/ Numeric | 10 |
| DIAGNOSES | Organized using ICD9 and ICD10 hierarchies. Intelligent Medical Objects interface terms are grouped to ICD9 and ICD10 levels. Diagnosis resources are further separated by source of the assignment (eg, EMR, professional billing, technical billing, registry). | Binary | 4769 |
| HISTORY | Contains family, social (ie, smoking), and surgical history from the EMR | Binary/ Numeric | 155 |
| LABORATORY TESTS | Results of a variety of laboratory tests, including cardiology and microbiology findings. Note that the actual lab values are used in modeling, if available (excluding serum creatinine, eGFR, ACR). | Binary/ Numeric | 1745 |
| MEDICATIONS | Includes dispensing, administration, prescriptions, as well as home medication reconciliation at KUH, grouped at semantic clinical drug form (SCDF) or semantic clinical brand form (SCBF) level. Medication resources are further separated by types of medication activity. | Binary | 1240 |
| NAACCR (ABRIDGED) | Includes information from the North American Association Central Cancer Registry translated to layman terms43 | Binary | 151 |
| NCDR | Includes information from the Cath PCI National Cardiovascular Data Registry44 | Binary | 253 |
| NTDS | Includes information entered into the National Trauma Registry45 | Binary | 175 |
| PROCEDURES | Includes CPT professional services and inpatient ICD9 billing procedure codes | Binary | 571 |
| ORDERS | Includes physician orders for non-medications such as culture and imaging orders from the EMR | Binary | 1091 |
| REPORTS | Includes observations from physician notes authored in the EMR using templates that collect structured data elements | Binary | 1081 |
| VIZIENT | (formerly UHC) Includes both billing classifications such as Diagnostic Related Groups (DRG), comorbidities, discharge placement, LOS, and national quality metrics | Binary | 696 |
| VISIT DETAILS | Includes visit types, vital signs collected at the visit, discharge disposition, and clinical services providing care from both EMR and billing | Binary/ Numeric | 480 |
These are not all distinct concepts from the entire HERON system, but only the total number of distinct features that had ever been recorded for at least 1 patient in the study cohort.
For each patient, we extracted the most recent values for all available features from the 15 categories at least 30 days prior to the endpoint. Initially, a total of 84 874 distinct features were available for our study cohort. Among them, 43 487 (51%) were recorded by only <1% of the patients so that we first excluded these rare features to reduce data sparsity. 27 928 (33%) of the features were recorded only for non-DKD patients, which were also dropped to control selection bias. Serum creatinine and albumin were removed from the candidate feature list, as they were part of the eGFR and ACR calculations that determine the target outcome. The final feature space contained 13 557 variables.
Embedded feature selection methods
Embedded feature selection methods leverage the structure of a specified learning model and select features contribute best to the model accuracy. We focused on 3 types of embedded feature selection methods: 1) regularized regression, which assumes an additive structure and benefits linear relationships; 2) tree-based gradient boosting machine, which assumes hierarchical structure and benefits nonlinear relationships; and 3) neural network, which incorporates a structure handling more complex correlations. All 3 methods have demonstrated success in a variety of biomedical studies.49–52
Regularized regression
Regularized logistic regression is a sparsity-inducing feature selection method that implicitly identifies the stronger one within each pair of correlated features.34 Desired sparsity of the final model is controlled by penalizing either absolute or quadratic values of the coefficients, with least absolute shrinkage and selection operator (LASSO) and ElastNet being 2 popular choices.
Gradient boosting machine
GBM is an ensemble learning technique that effectively combines weak and simple learners into a stronger one.35 As a base learner, we used decision trees, which can handle nonlinear relations and correlations with their hierarchical structure. GBM is sensitive to hyper-parameters, and thus a grid of parameter configurations for the learning rate, number of trees, and depth of trees are considered for tuning within each training stage.
Deep neural network
DNN uses a cascade of multiple layers of nonlinear processing units to transform and extract features.36 Lower layers select simple features and use them to learn more abstract concepts for constructing the upper layers. It is difficult to fine-tune all hyperparameters for DNN. We experimented on using 100, 200, 300, 400, and 500 neurons with 2 and 3 hidden layers. For better computational efficiency, we used “rectifier” as the activation function and set the number of epochs at 100.
All algorithms for embedded feature selection were implemented in R, on open-source language for statistical applications. Particularly, we used h2o library53 implementations for LASSO, ElastNet, and DNN and xgboost library54 for GBM. Missing values were handled internally specific to each library implementation.
Experimental design
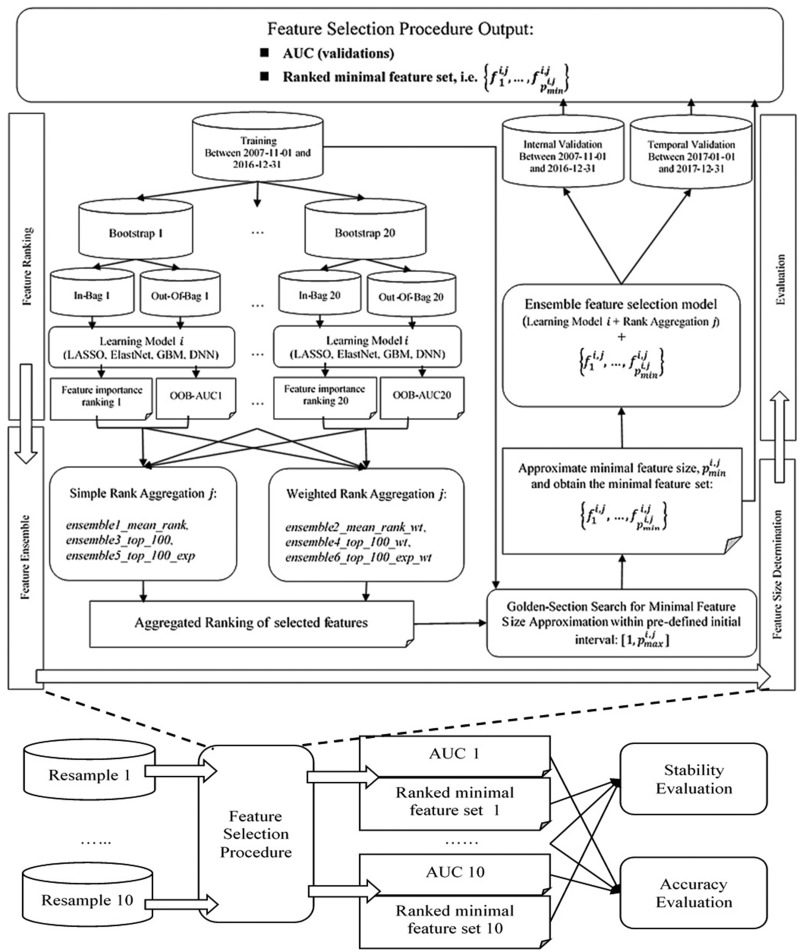
The overall data set was split into training, internal validation, and temporal validation sets, with internal validation being 30% randomly held-out patients with endpoints occurring before January 1, 2017, and temporal validation containing patients with endpoints occurring after January 1, 2017. The flowchart in Figure 2 shows the experimental process. At the training stage, we first applied each embedded feature selection method on 20 bootstrap samples, ranked the features in accordance with their importance (feature ranking), and aggregated the rankings with different ensemble techniques (feature ensemble). Then, for each combination of feature selection method and ensemble technique, or ensemble–feature selection model, we conducted a golden-section search to estimate the minimal feature size sufficient for that particular classifier to achieve a close-to-optimal accuracy based on 5-fold cross validation within training (feature size estimation). Finally, we evaluated prediction accuracy on both validation sets and feature stability across the 10 resamples (evaluation). Note that we required the union of each 20 bootstrap samples to cover the overall training set to ensure that the sample space has been thoroughly searched.
Figure 2.
Flowchart for the experimental design.
Feature ranking
LASSO and ElastNet rank the feature importance based on the magnitude of normalized coefficients. Feature importance for GBM is measured as its contribution to the improvement of prediction accuracy. More specifically, it is the averaged importance taken across all boosted trees, with each tree-specific importance calculated as the cumulative improvement of AUC attributed to splitting by that feature weighted by the number of observations the node is responsible for, which was then normalized to a percentage. In terms of DNN, we adopted Gedeon’s method for assigning ranks to features, which is measured as the weights connecting the input features to the first 2 hidden layers.55
Feature ensemble
To improve stability of selected feature sets against data perturbation, we performed feature ensemble on bootstrap samples: feature selection is run on several random subsamples of the training data, and the different ranks of features are aggregated into a more robust ranking.32 More formally, for each of the feature selection methods described above, we obtained lists of feature rankings, , each provided by a bootstrap sample, and then aggregated them by computing a score, , as
| (1) |
where denotes a bootstrap-dependent weight, and we tested the following rank aggregation functions, which are straightforward to implement.
Simple Aggregation is a group of methods that set all in equation (1) to 1, and the lower the calculated value is, the better the ensemble rank is.
Ensemble-mean13: averaging the rank of features over the bootstrap samples, ie, (ensemble1_mean_rank);
Ensemble-stability (top ) selection11: assigning a “hard” membership of a feature as whether or not it is among the top , ie, if ; otherwise (ensemble3_top_100);
Ensemble-exponential11: assigning a “soft” membership of a feature as how high up the rank is relative to top by applying an exponentially decreasing function to the rank, ie, (ensemble5_top_100_exp).
Weighted Aggregation, on the other hand, assigned weight to a bootstrap in accordance with the out-of-bag (OO) validated AUC, ie, -. In other words, we wanted to give more credit to the bootstrap samples with better prediction performance.
Ensemble-weighted-mean13: taking weighted average of the ranks over the bootstrap experiment (ensemble2_mean_rank_wt);
Ensemble-weighted-stability (top ) selection: applying the weights and average over the classic ensemble-stability measurement (ensemble4_top_100_wt);
Ensemble-weighted-exponential: applying the weights and average over the classic ensemble-exponential measurement (ensemble5_top_100_exp_wt).
Feature size estimation
Since the performance of a classifier can be influenced by the number of features56 and DNN, GBMs are designed to “combine” rather than “eliminate” the weaker features. It is a natural assumption that these types of feature selection methods would prefer more features than LASSO and ElastNet. To ensure a fair comparison, we proposed an iterative golden-section search procedure57 for approximating a minimal feature size, which can achieve an accuracy that is significantly close to the optimum using the Delong test.58 More details of the search algorithm are available in Supplementary Material – Appendix A.
Evaluation protocol
Stability measures can be feature-focused and subset-focused based on their evaluation scope; or subset-size-biased and subset-size-unbiased according to their ability to adjust for feature set of varying sizes. We randomly resampled times () and applied the Kuncheva index ()37 and weighted consistency index (WCI)6 to compare stability for fixed feature sizes and relative weighted consistency index (WCIrel)38 to compare stability for flexible features sizes adapted to different feature selection algorithms (Supplementary Material – Appendix B).
Accuracy is evaluated by AUC on both internal and temporal validation sets. To control for overfitting, we made sure that both validation sets were never seen by the classifiers or feature selectors.
RESULTS
Summaries of the patient population are presented in Table 2, who were primarily white with a mean age of 59 and evenly distributed between male and female. It appears that even though patients’ basic demographics did not vary significantly between training and validation sets, DKD rate dropped significantly (P-value < .001) from 44.4% (training) and 45.5% (internal validation) to 10.1% (temporal validation). When we broke down the cohort by DM onset year, we observed that patients in Temporal holdout were relatively newer to our health system (52.7% vs. 19.4% DM onset after 2014, P-value < .001). It suggests that the DKD rate discrepancy could be caused by significantly shorter follow-up periods for patients in the Temporal holdout set.
Table 2.
Patient characteristics of training and validation sets
| Demographic Characteristics | Training (2007-2016) | Internal Validation (2007-2016) | Temporal Validation (2017) |
|---|---|---|---|
| N | 8098 | 3461 | 4086 |
| % DKD | 44.4 | 45.5 | 10.1 |
| Mean age in years (SD) | 59.2 (14.1) | 59.2 (14.0) | 58.5 (13.9) |
| % Male | 49.2 | 49.1 | 50.0 |
| Race | |||
| % White | 68.2 | 67.8 | 64.5 |
| % Black | 20.5 | 21.1 | 23.4 |
| % Asian | 1.7 | 1.7 | 2.0 |
| % American Indian/ Pacific Islander | 0.7 | 0.7 | 0.6 |
| % Two races | 0.2 | 0.2 | 0.3 |
| % Other | 8.5 | 8.6 | 8.5 |
| % Unknown | 0.4 | 0.4 | 1.1 |
| DM onset | |||
| % DM onset after 2014 | 19.6 | 19.4 | 52.7 (P < .001) |
| % DM onset after 2015 | 10.0 | 10.1 | 32.8 (P < .001) |
| % DM onset after 2016 | 3.3 | 3.0 | 17.4 (p < .001) |
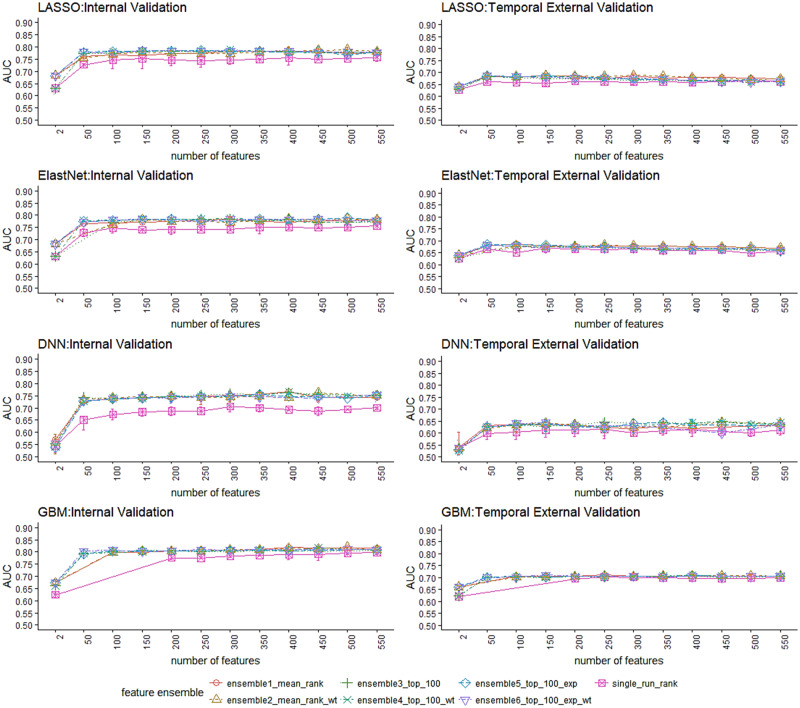
Figure 3 illustrates that the feature ensemble methods (ensemble 1 through 5) performed better than a single-run selection (single_run_rank) consistently over different feature set sizes and classifiers. In most cases, the ensemble methods performed better with fewer features, suggesting their capability of eliminating irrelevant/redundant features. More specifically, the ensemble methods focusing on stabilizing top-ranked features (ensemble3_top_100, ensemble4_top_100_wt) yielded better accuracy with a smaller feature set, while the methods with emphasis on stabilizing overall feature rankings (ensemble1_mean_rank, ensemble2_mean_rank_wt) achieved better accuracy with a larger feature set. On the other hand, the improvement of ensemble methods over single-run has a different manifestation among different classifiers, with LASSO and ElastNet showing smaller discrepancies than GBM and DNN. The weighted-aggregation methods did not yield significant improvement in accuracy over the simple-aggregation methods, even though the former is designed to bias towards rankings with better performance within bootstraps. Although the AUC on temporal validation drops significantly from the internal validation in general, GBM outperformed the other 3 models significantly on both validation sets.
Figure 3.
AUC vs. number of selected features for different feature selection combos.
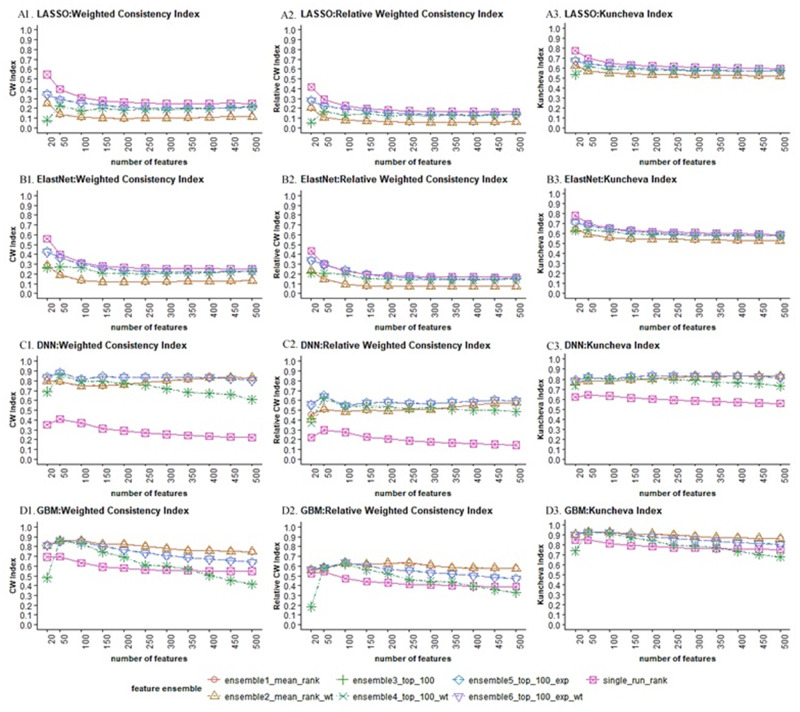
In terms of the stability of feature selection (Figure 4), DNN (C1-C3) and GBM (D1-D3) generated more stable feature sets than LASSO (A1-A3) and ElastNet (B1-B3) with respect to all 3 measures, which is not surprising, as LASSO and ElastNet have been reported to be less stable dealing with highly correlated data.31 Interestingly, while feature ensemble methods improved stability for GBM and DNN, they turned out to aggravate the instability of LASSO and ElastNet. Comparing among the feature ensemble methods, the ensemble-(weighted)-exponential (ensemble5_top_100_exp, ensemble6_top_100_exp_wt) gave a more stable feature set than ensemble-(weighted)-stability (ensemble3_top_100, ensemble4_ top_100_exp) and ensemble-(weighted)-mean (ensemble1_mean, ensemble2_mean_wt) for most of the selection algorithms (A, B, C), while GBM (D) appeared to direct towards a totally opposite direction.
Figure 4.
Stability vs. number of selected features for different feature selection combos.
Except for DNN, all ensemble–feature selection models saw decreasing stability as feature size got larger, indicating that the top-ranked features were more robust against data perturbation. It appears that DNN and GBM generated feature sets steadily stable even with increasing size when we combined DNN with ensemble-(weighted)-exponential and GBM with ensemble-(weighted)-mean methods. It is also worth noting that all 3 stability measures showed consistent results, where (second column) was more conservative than the other 2 measures.
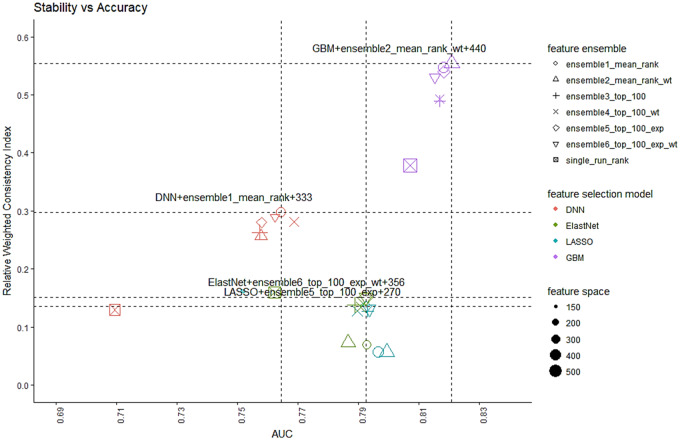
Figure 5 summarizes the tradeoff between accuracy and stability of all 24 ensemble–feature selection models under consideration, trained with their minimal robust feature sets. Interestingly, to arrive at the best balancing point between stability and accuracy, LASSO and ElastNet work the best with ensemble methods that concentrate on stabilizing the top features, while DNN and GBM favor the methods maintaining an overall stability. Conceptually, we would prefer the combination located at the top-right corner, which indicates both high accuracy and stability. To quantify that, we calculated the Euclidean distance from each point to the origin (AUC = 0.5, = 0.0) and picked the one with the largest distance, which is the combination of GBM and ensemble-weighted mean with a minimal feature size of 440.
Figure 5.
Stability vs. AUC tradeoff for different feature selection combos.
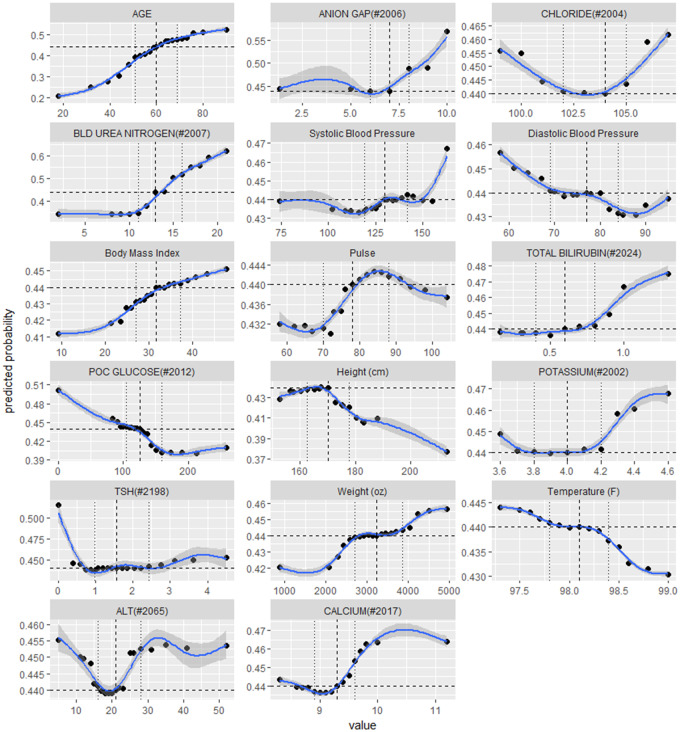
Finally, we took a closer look at these 440 features and their marginal effects on DKD risk. Figure 6 displays the partial dependence plots35 for the 17 numerical features among the top 20, consisting of a patient’s age at prediction point, 9 labs, and 7 vital signs. Additionally, there are 3 binary features among the top 20: Surescript Encounter and 2 fired alerts, which are protective against DKD (not shown in Figure 6 but available in Supplementary Material – Appendix C). In Figure 6, the 3 vertical lines correspond to the 25th, 50th, and 75th percentiles of the feature values, while the horizontal line marks the predicted DKD risk corresponding to value at 50th serving as a baseline. For example, 11, 13, 16 are the 3 quartiles of the blood urea nitrogen (BUN) measured in mg/dL. Compared with the predicted probability at the baseline, an increase of BUN from 13 mg/dL to 16 mg/dL would increase the DKD risk from 0.44 to 0.51, or by 14%.
Figure 6.
Top demographic, lab and vital features and their partial dependence effects on predicting DKD risk.
DISCUSSION
Quality criteria for feature selection should combine both accuracy and stability. Accuracy alone would select a feature set that is predictive of the outcome but could be sensitive to overfitting. Stability alone can be naively increased by considering a fixed set of common features, which would lead to poor predictive models.
Feature ranking aggregation is an ensemble technique that is independent of the feature selection algorithm, which can be conveniently implemented to improve both accuracy and stability of the existing feature selection methods. It is unexpected that neither ElastNet, which was designed to remedy the unstableness of LASSO by allowing the selection of correlated features in pairs, nor ElastNet in combination with feature ensemble techniques seemed to be more stable than LASSO. In contrast, GBM and DNN benefited more from the feature ensemble and outperformed LASSO and ElastNet with respect to both accuracy and stability.
Among the 440 features identified by the “best” ensemble–feature selection model, more than half of them are labs and vitals that contribute the most to the discriminant power. Some features are known predictors of DKD such as age, body mass index (BMI), BUN, and systolic blood pressure (SBP), while their associations with DKD risk can be further interpolated on a continuous scale. For example, even within the normal range of BUN, ie, 7 to 20 mg/dL, the model still suggests an aggressive risk increase when BUN is above 11 mg/dL. We have also identified factors that were not widely incorporated in the state-of-art multivariate predictive models for DKD, but had been sporadically examined for their association with kidney functions in separate univariate analyses. For example, our model identified serum anion gap to be associated with increased DKD risk when its value is above 6 mEg/L, which is consistent with the findings in59, who found the serum anion gap to be a potential early marker for CKD, and the mean anion gaps for patients with eGFR at 15 to 29, 30 to 44, and 45 to 59 mL/min/1.73m2 are 6.07, 6.78, and 6.55 mEg/L, respectively. Since the anion gap is a calculated difference between primary measured citation (sodium and potassium) and primary measured anions (chloride and bicarbonate), it would be straightforward to explain the positive relation between potassium and DKD risk. However, the effect of chloride is trickier, since low chloride may lead to a high anion gap if sodium drops at a slower rate, and indirectly increase DKD risk, but meanwhile hyperchloremia also could occur early in chronic renal failure.60 This complexity is captured and described by our model with a nonlinear relationship. Furthermore, some counterintuitive relationships have also drawn our attention, eg, the inverse relationship with diastolic blood pressure (DBP). With further literature research, we learned that low DBP of < 70 mmHG was identified to increase all-cause mortality of CKD patients.61 Due to page limit, we elaborated on only the top 20 features in this article and left the full feature list and their marginal effects to Supplementary Material – Appendix C.
Moreover, the hypothesis-free approach can lead to identification of novel risk factors, not previously considered in the literature (Supplementary Material – Appendix D). Here we highlighted some “new” features with relatively large marginal effects. Drivers for DKD risk included: alerts that encapsulate drug–drug combinations (antidiabetics and non-cardioselective betablockers), general medicine admission service and liver transplant status, acute renal failure University HealthSystem Consortium (UHC) Diagnosis Related Groups (DRG) status, diabetes diagnoses with noted complication, outpatient prescription fill of potassium chloride extended release, and low CathPCI pre-procedure hemoglobin. More pronounced protective factors included: outpatient refill adherence noted as a Surescript encounter, especially prednisone fills, alerts that the patient should be on a diabetic or general care plan while inpatient, diabetes diagnoses without complication, an emergency medicine visit, lack of allergies or a tetracycline allergy, low stenosis noted in CathPCI findings, a low E/A ratio from echocardiography, health maintenance letters generated, negative endocrine findings in physician notes, and cellulitis or dysphagia diagnoses. Many of the findings are clinically plausible, and next steps will include expanded clinical interpretation with case review and developing approaches to model interpretation with clinicians and ultimately patients.
It is worth noting that some “unconventional” features were selected, such as drug–drug interaction (DDI) alerts, Surescript encounters implying medication adherence, concepts extracted from physician notes, etc.—information that already exists in most EMR systems but that has been under-utilized in observational research. In recent years, DDI alerts and outpatient medication dispensing (Surescript) have received increasing recognition. DDI alerts can be considered as an abstraction representing composite interactions of factors. For example, a DDI alert of antidiabetics and non-cardioselective betablockers implies that the patient has a cardiovascular condition or potentially may be affected by the DDI or both. Outpatient medication, on the other hand, plays a critical role in completing patients’ health information, in which existing common data models (eg, PCORnet and OHDSI) have explicit requirements on collecting outpatient medication data.62,63
Our experiment has demonstrated the capability of a hypothesis-free and machine-learning-based approach for identifying potential risk factors of DKD. This framework will not only significantly reduce the effort in preselecting features manually, but will also enable knowledge discovery from a much larger feature space. As a result, we could efficiently identify a stable feature list with better diversity covering a broader range of patients’ health information, which in turn, would merit a more general characterization of a DKD cohort and create a possible venue for designing better intervention strategies to improve patient care.
There are still several limitations to this study. First, the drop of prediction performance on temporal validation, expected though as the set is strongly biased towards non-DKD patients [45.4% DKD patients in internal validation set; 10.1% DKD patients in temporal validation set (Table 2)], but still indicates a lack of robustness against population shift. Further investigation on finding causal risk factors is in our horizon. Second, we ensembled features only to overcome sampling variations but have not accounted for the differences in the feature selection algorithms, eg, GBM tends to learn better with numerical features, while LASSO picks out more binary features. Finally, as we needed to retune hyperparameters for each bootstrapped sample at feature ranking stage as well as for various feature sets at feature size determination stage, it was only computationally feasible to experiment with a limited number of parameters for GBM and DNN.
CONCLUSION
We developed an ensemble feature selection framework for identifying robust DKD risk factors by balancing predictability and stability. We combined 4 state-of-art feature selection algorithms with 6 feature ensemble techniques as candidate feature selection methods, re-designed the golden-section search for approximating the minimal feature set size for each method, and selected the final set with the best stability and predictability. The final robust set of 440 features achieved an AUC of 0.82 [95% CI, 0.81–0.83] on internal validation and 0.71 [95% CI, 0.68–0.73] on temporal validation. This study explored a more exhaustive list of EMR data integrated with external registries and identified numerous potential risk factors that would merit further investigations.
FUNDING
YH is supported by the Major Research Plan of the National Natural Science Foundation of China (Key Program, Grant No. 91746204), the Science and Technology Development in Guangdong Province (Major Projects of Advanced and Key Techniques Innovation, Grant No. 2017B030308008), and Guangdong Engineering Technology Research Center for Big Data Precision Healthcare (Grant No. 603141789047). The dataset used for analysis described in this study was obtained from KUMC’s HERON clinical data repository, which was supported by institutional funding and by the KUMC CTSA grant UL1TR002366 from NCRR/NIH.
CONTRIBUTORS
ML, YH, LRW, and XS designed and conceptualized the overall study. XS performed cohort extraction, data cleaning, and model development. ML, LRW, AY, and XS contributed to the evaluation of experimental results. ML and LRW contributed in data extraction. AY and DR advised regarding motivation of the clinical design and domain for the study and DKD definition. All authors reviewed the manuscript critically for scientific content, and all authors gave final approval of the manuscript for publication.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Conflict of interest statement. The authors declared no conflict of interest.
Supplementary Material
REFERENCES
- 1. Frederik P, Peter R. Diagnosis of diabetic kidney disease: state of the art and future perspective. Kidney Int Suppl 2018; 8: 2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Katherine RT, George LB, Rudolf WB. Diabetic kidney disease: a report from and ADA consensus conference. Diabetes Care 2014; 37 (10): 2864–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zoppini G, Targher G, Chonchol M, et al. Predictors of estimated GFR decline in patients with type 2 diabetes and preserved kidney function. Clin J Am Soc Nephrol: CJASN 2012; 7 (3): 401–8. [DOI] [PubMed] [Google Scholar]
- 4. Ueda H, Ishimura E, Shoji T, et al. Factors affecting progression of renal failure in patients with type 2 diabetes. Diabetes Care 2003; 26 (5): 1530–4. [DOI] [PubMed] [Google Scholar]
- 5. Rossing K, Christensen PK, Hovind P, et al. Progression of nephropathy in type 2 diabetic patients. Kidney Int 2004; 66 (4): 1596–605. [DOI] [PubMed] [Google Scholar]
- 6. Yokoyama H, Kanno S, Takahashi S, et al. Determinants of decline in glomerular filtration rate in nonproteinuric subjects with or without diabetes and hypertension. Clin J Am Soc Nephrol 2009; 4 (9): 1432–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huaidong C, Wei C, Chenglin L, et al. Relational network for knowledge discovery through heterogeneous biomedical and clinical features. Sci Rep 2016; 6: 29915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Elizabeth SC, Indra NS. Mining the electronic health record for disease knowledge. Methods Mol Biol 2014; 1159: 269–86. [DOI] [PubMed] [Google Scholar]
- 9. Weber GM. How many patients are “normal”? Only 1.55%. AMIA Jt Summits Transl Sci Proc 2013; 2013: 79.. [PubMed] [Google Scholar]
- 10. Dash M, Liu H. Feature selection for classification. IDA 1997; 1 (3): 131–56. [Google Scholar]
- 11. Guyon I, Elisseeff A. An introduction to variable and feature selection. J Mach Learn Res 2003; 3: 1157–82. [Google Scholar]
- 12. Liu H, Yu L. Toward integrating feature selection algorithms for classification and clustering. IEEE Trans. Knowl Data Eng 2005; 17 (4): 491–502. [Google Scholar]
- 13. Saeys Y, Inza I, Larranaga P. A review of feature selection techniques in bioinformatics. Bioinformatics 2007; 23 (19): 2507–17. [DOI] [PubMed] [Google Scholar]
- 14. Yang S, Yuan L, CLai YC, et al. Feature grouping and selection over an undirected graph In: Proc. 18th ACM SIGKDD Int'l Conf. Knowledge Discovery and Data Mining (KDD '12); 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Drotar P, Gazda J, Smekal Z. An experimental comparison of feature selection methods on two-class biomedical datasets. Comput Biol Med 2015; 66: 1–10. [DOI] [PubMed] [Google Scholar]
- 16. Hui Y, Jonanthan MG. A hybrid model for automatic identification of risk factors of heart disease. J Biomed Inform 2015; 58: 171–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiamei L, Cheng X, Weifeng Y, et al. Multiple similarity effective solutions exist for biomedical feature selection and classification problems. Sci Rep 2017; 7: 12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cosmin AB, Fei X, Lucy V, et al. Pneumonis identification using statistical feature selection. J Am Med Inform Assoc 2012; 5 (1): 817–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bermingham ML, Pongwong R, Spiliopoulou A, et al. Application of high-dimensional feature selection: evaluation for genomics prediction in man. Sci Rep 2015; 5: 10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Anne-Claire H, Pierre G, Jean-Philippe V. The influence of feature selection methods on accuracy, stability and interpretability of molecular signatures. PLoS One 2011; 6 (12): e28210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Isabelle G, Andr E. An introduction to variable and feature selection. JMLR 2003; 3: 1157–82. [Google Scholar]
- 22. Thomas A, Thibault H, Yves Van de P, et al. Robust biomarker identification for cancer diagnosis with ensemble feature selection methods. Bioinformatics 2010; 26 (3): 392–8. [DOI] [PubMed] [Google Scholar]
- 23. Meg JJ, Jun H, Mark W, et al. Prediction of kidney-related outcomes in patients with type 2 diabetes. American Journal of Kidney Disease 2012; 5: 770–8. [DOI] [PubMed] [Google Scholar]
- 24. Mian W, Junxi L, Lei Z, et al. A non-laboratory-based risk score for predicting diabetic kidney disease in Chinese patients with type 2 diabetes. Oncotarget 2017; 8 (60): 102550–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin C-C, Li C-I, Liu C-S, et al. Development and validation of risk prediction model for end-stage renal disease in patients with type 2 diabetes. Sci Rep 2017; 7 (1): 10177.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baumgartnet C, Osl M, Netzer M, et al. Bioinformatic-driven search for metabolic biomarkers in disease. J Clin Bioinformatics 2011; 1 (1): 2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jonathan B, Samah JF, Cynthia AB, et al. Classification of radiology reports for falls in an HIV study cohort. J Am Med Inform Assoc 2016; 23: e113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Salma J, Sukriti G, Asheesh S, et al. Predicting neurological Adverse Drug Reactions based on biological, chemical and phenotypic properties of g machine learning models. Sci Rep 2017; 7: 872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Waitman LR, Aaronson LS, Nadkarni PM, et al. The greater plains collaborative: a PCORnet clinical research data network. J Am Med Inform Assoc 2014; 21 (4): 637–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fleurence RL, Curtis LH, Califf RM, et al. Launching PCORnet, a national patient-centered clinical research network. JAMIA 2014; 21 (4): 578–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shivapratap G, Truyen T, Tu Dinh N, et al. Stabilizing high-dimensional prediction models using feature graphs. IEEE Journal of Biomedical and Health Informatics 2015; 19 (3): 1044–52. [DOI] [PubMed] [Google Scholar]
- 32. Randall W, Taghi MK, David D, et al. An extensive comparison of feature ranking aggregation techniques in bioinformatics In: IEEE Information Reuse and Integration (IRI), 2012 IEEE 13th International Conference, Las Vegas, NV, USA: 2012: P377–4. [Google Scholar]
- 33. Kolde R, Laur S, Adler P, et al. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics 2012; 28 (4): 573–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jie G, Zhenan S, Shuiwang J, et al. Feature selection based on structured sparsity: a comprehensive study. IEEE Trans Neural Netw Learn Syst 2017; 28 (7): 1490–507. [DOI] [PubMed] [Google Scholar]
- 35. Friedman J. Greedy boosting approximation: a gradient boosting machine. Ann Statist 2001; 29 (5): 1189–232. [35] [Google Scholar]
- 36. Yann L, Yoshua B, Geoggery H. Deep learning. Nature 2015; 521: 436–44. [DOI] [PubMed] [Google Scholar]
- 37. Kuncheva LI. A stability index for feature selection In: Proceedings of the 25th IASTED International Multi-Conference on Artificial Intelligence and Applications, AIAP 2007, Anaheim, CA, USA: ACTA Press; 2007: P390–5. [Google Scholar]
- 38. Somol P, Novovičová J. Evaluating stability and comparing output of feature selectors that optimize feature subset cardinality. IEEE Trans Pattern Anal Mach Intell 2010; 32 (11): 1921–2939. [DOI] [PubMed] [Google Scholar]
- 39. Nichols GA. Construction of a multisite datalink using electronic health records for the identification, surveillance, prevention, and management of diabetes mellitus: The SUPREME-DM project. Prev Chronic Dis 2012; 9: 110311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.KDOQI. KDOQI clinical practice guidelines and clinical practice recommendations for diabetes and chronic kidney disease. Am J Kidney Dis 2007; 49 (2)(suppl 2): S12–154. American Diabetes Association. Standards of medical care in diabetes—2010. Diabetes Care 2010; 33 (suppl 1): S11–61. [DOI] [PubMed]
- 41. Levey AS, Coresh J, Greene T, et al. , Chronic Kidney Disease Epidemiology Collaboration. Using standardized serum creatinine values in the modification of diet in renal disease study equation for estimating glomerular filtration rate. Ann Intern Med 2006; 145 (4): 247–54. [DOI] [PubMed] [Google Scholar]
- 42. Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc 2010; 17 (2): 124–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weir HK, Johnson CJ, Mariotto AB, et al. Evaluation of North American Association of Central Cancer Registries’ (NAACCR) data for us in population-based cancer survival studies. J Natl Cancer Inst Monogr 2014; 2014 (49): 198–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moussa I, Hermann A, Messenger JC, et al. The NCDR CathPCI Registry: a US national persepective on care and outcomes for percutaneous coronary intervention. Heart 2013; 99 (5): 297–303. [DOI] [PubMed] [Google Scholar]
- 45. Benedict CN, Wendi L, Renase K, et al. History and development of trauma registry: lessons from developed to developing countries. World J Emerg. Surg 2006; 1: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Damle R, Alavi K. The university healthsystem consortium clinical database: an emerging resource in colorectal surgery research. Sem Colon Rectal Surg, Big Data Colorectal Surg 2016; 27 (2): 92–5. [Google Scholar]
- 47. Informatics for Integrating Biology and the Bedside (i2b2). i2b2 Software Documentation, i2b2 Cell Messaging, Data Repository (CRC) Cell. I2b2 Software Version: 1.7.08. Document Version: 1.7.08-004.
- 48. Xing S, Lemuel RW, Yong H, et al. Building predictive models for diabetic kidney disease: an exploration of ontology-based EMR data abstraction. In: American Medical Informatics Association 2018 Annual Conference. 2018. Submitted.
- 49. Jieping Y, Jun L. Sparse methods for biomedical data. SIGKDD Explor 2013; 14 (1): 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. He K, Li Y, Zhu J, et al. Component-wise gradient boosting and false discovery control in survival analysis with high-dimensional covariates. Bioinformatics 2016; 32 (1): 50–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li Y, Chen CY, Wasserman WW. Deep feature selection: theory and application to identify enhancers and promoters. J Comput Biol 2016; 23 (5): 322–36. [DOI] [PubMed] [Google Scholar]
- 52. Koyner JL, Carey KA, Edelson DP, Churpek MM. The development of a machine learning in patient acute kidney injury prediction model. CCM 2018. doi: 10.1097/CCM.0000000000003123. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 53. Kraljevic T. h2o: R Interface for ‘H2O’. R package version 3.18.0.11. 2018. https://CRAN.R-project.org/package=h2o. Accessed March, 2018.
- 54. Tianqi C, Tong H, Michael B, et al. xgboost: Extreme Gradient Boosting. R package version 0.6.4.1. 2018. https://CRAN.R-project.org/package=xgboost. Accessed March, 2018.
- 55. Gedeon TD. Data mining of inputs: analyzing magnitude and functional measures. Int J Neural Syst 1997; 8 (2): 209–18. [DOI] [PubMed] [Google Scholar]
- 56. Jianping H, Zixiang X, James L, et al. Optimal number of features as a function of sample size for various classification rules. Bioinformatics 2005; 21 (8): 1509–15. [DOI] [PubMed] [Google Scholar]
- 57. Press WH, Teukolsky SA, Vetterling WT, et al. Section 10.2. Golden Section Search in One Dimension”, Numerical Recipes: The Art of Scientific Computing 2007. 3rd ed New York: Cambridge University Press, ISBN 978-0-521-88068-8. [Google Scholar]
- 58. Elisabeth RD, David MD, Daniel LC. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988; 44: 837–45. [PubMed] [Google Scholar]
- 59. Matthew KA, Thomas HH, Michal LM. The serum anion gap is altered in early kidney disease and associates with mortality. Kidney Int 2012; 82 (6): 701–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Robert GL. Serum chloride and bicarbonate levels in chronic renal failure. JAMA Int. Med 1979; 139 (10): 1091–2. [PubMed] [Google Scholar]
- 61. Kovesdy CP, Bleyer AJ, Molnar MZ, et al. Blood pressure and mortality in U.S. veterans with chronic kidney disease: a cohort study. Ann Intern Med 2013; 159 (4): 233–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fleurence RL, Curtis LH, Califf RM, Platt R, Selby JV, Brown JS. Launching PCORnet, a national patient-centered clinical research network. J Am Med Inform Assoc 2014; 21 (4): 578–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stephanie JR, Patrick BR, Donal JO, et al. Development and evaluation of a common data model enabling active drug safety surveillance using diaparate healthcare databases. J Am Med Inform Assoc 2010; 17 (6): 652–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.