Abstract
Simple Summary
Cancer is the second leading cause of death worldwide, right after cardiovascular diseases, and the invasion and metastatization correspond to the foremost cause of cancer-related deaths. Here, we reviewed the state of the art regarding the importance of HOX transcription factors in these last steps of cancer progression and described five of their complex mechanisms of regulation, including the miRs and lncRNAs interference. This information highlights the importance of HOX in the suppression and induction of disease advancement and point out the potential of HOX products as therapeutic targets for diverse cancer types.
Abstract
Invasion and metastasis correspond to the foremost cause of cancer-related death, and the molecular networks behind these two processes are extremely complex and dependent on the intra- and extracellular conditions along with the prime of the premetastatic niche. Currently, several studies suggest an association between the levels of HOX genes expression and cancer cell invasion and metastasis, which favour the formation of novel tumour masses. The deregulation of HOX genes by HMGA2/TET1 signalling and the regulatory effect of noncoding RNAs generated by the HOX loci can also promote invasion and metastasis, interfering with the expression of HOX genes or other genes relevant to these processes. In this review, we present five molecular mechanisms of HOX deregulation by which the HOX clusters products may affect invasion and metastatic processes in solid tumours.
Keywords: HOX, invasion and metastasis, HMGA2/TET1/HOXA signalling pathway, TGFβ signalling, epithelial-to-mesenchymal transition, microRNAs, lncRNAs
1. Introduction
The vast majority of cancer-related mortality in solid tumours is associated with the capacity of the cancer cells to invade and colonize nearby or distant vital organs forming metastasis [1], hallmarks that characterize the last steps of cancer progression [2]. A line of investigation suggests that tumour recurrence and metastization is led by a population of residual cells that survive treatment and are capable to leave their primary location. These cells disperse into the bloodstream, endure pressure in blood vessels, escape immune response, and acclimate to new cellular surroundings in a secondary site [3]. However, the development of new therapies that can eliminate residual tumour cells or prevent their action is conditioned by the incomplete understanding of the mechanisms underlying the long-term survival of these cells following treatment [4].
The metastatic process is complex and involves a multistep journey. The activation of invasion and metastasis is triggered by environmental stimuli, such as aging and circadian disruptions; adhesive signals from extracellular matrix (ECM) components, such as collagen and fibrin; ECM mechanical pressures, including tension and compression; cell–cell interactions; soluble signals, such as growth factors and cytokines; and intratumoural microbiota [5,6]. The invading tumour cells, on the way to the target site, interact with other proteins and cells to overcome the stromal challenges and complete the metastasis key steps of invasion, intravasation, circulation, extravasation and colonization as will be described throughout the text. To add more complexity to this process, secondary sites do not receive invading cancer cells passively. In fact, the host microenvironment, also designated as a premetastatic niche (PMN) or organotropic metastasis, is selectively primed by the primary tumour even before the initiation of the metastatic process. This signalling involves secretory factors and extracellular vesicles that induce vascular leakage, ECM remodelling, and immunosuppression, making the secondary microenvironment selective for the circulating tumour cells (CTCs) [7,8,9]. As common examples of organotropism, we can cite the preferred metastization of breast and prostate tumour cells to lungs, bone and liver; colon and stomach cancer to liver, lung and peritoneum; and lung carcinoma to adrenal glands and liver [10]. Hundreds of genes have been reported to lead this invasive potential, suggesting that primary tumour cells should develop a metastatic genetic signature [11]. Thus, genetic and also epigenetic modifications, accumulated during the primary tumour development and by the adaptations to the microenvironment components contribute to the metastatic process [6]. The genetic modifications involve changes in the primary DNA sequence, such as mutations, while the epigenetic mechanisms relate to chemical modifications of DNA bases and changes in the chromosomal superstructure in which DNA is packaged, such as gene promoter methylations associated with gene silencing [12,13]. In this context, alterations in HOX gene expression have been identified in primary tumours, metastasis and CTCs [14,15].
1.1. HOX Genes Genomic Organization and Transcription
HOX genes are a family of genes that codes for transcription factors characterized by the presence of a conserved DNA sequence designated homeobox [16]. The human genome contains 39 of those genes arranged into four clusters (HOXA, HOXB, HOXC and HOXD) located in distinct chromosomes (7p15, 17q21.2, 12q13, 2q31, respectively). Each cluster presents 9–11 genes that align in 13 paralogous groups, based on sequence homology of their homeoboxes and their position within the cluster [17]. These genes are co-ordinately transcribed following temporal and spatial collinearity with an unidirectional chromatin opening, where the chromosomic order of genes is the same as the order of its expression, forming nested and overlapping expression domains [18], that define the organogenesis along the anterior–posterior axis [19,20]. HOX proteins activity occurs with the formation of multiprotein complexes, the “hoxasomes”, composed by HOX proteins, HOX cofactors and other transcription factors [15]. These cofactors are members of the three amino acids loop extension (TALE) family, which includes PBC members (PBX1–4), HMP members (MEIS1–3 and PREP1–2), and POU family (POU1–6) [21]. The transcription of these genes happens during embryonic development but also in adult cells at lower levels [22] partaking in cellular physiology and tissue homeostasis [19,23].
1.2. HOX Genes and Cancer
Alterations in HOX gene expression have been shown in different cancer types and associated with cancer initiation and progression [15,24,25]. They can interfere with cell differentiation, survival, proliferation, angiogenesis, autophagy, inflammation and apoptosis. They also influence cell movement, migration, invasion, metalloproteinase function and regulate important stemness-related genes [26,27]. More complexly, different HOX genes can activate or inhibit these processes and even exhibit antagonistic behaviours in different tissues.
Thus, HOX genes interfere with the majority of the hallmarks of cancer, whose events must occur in parallel and/or sequentially to promote cancer progression [2]. The uncontrolled cell division that represents cancer initiation, involves not only deregulated control of cell proliferation and resistance to apoptosis, but also changes in energy metabolism in order to fuel cell growth and division. For instance, normal cells under aerobic conditions process glucose, first to pyruvate via glycolysis in the cytosol and thereafter to carbon dioxide in the mitochondria; under anaerobic conditions, glycolysis is favoured. Tumour cells, even in the presence of oxygen limit their energy metabolism largely to a state of aerobic glycolysis, also known as the Warburg effect [2]. At the same time, to ensure nutrient and oxygen delivery, as well as to remove metabolic wastes and carbon dioxide from the constantly growing mass, it is fundamental to form new blood vessels through the angiogenesis process [2].
Another hallmark of cancer influenced by HOX genes, with a dual function in tumorigenesis, is inflammation [2,26]. While in normal cells, the inflammation process is carried out by innate immune cells to fight infections and heal wounds, in cancer cells it can instead inadvertently work to support tumour growth [2]. Cell movement, which drives the migration and invasion processes, is dependent on the supply of matrix-degrading enzymes such as metalloproteinases (group of enzymes that can break down proteins, such as collagen, that are normally found in the spaces between cells). Finally, the presence of cancer stem cells, with extensive growth and differentiation abilities, give phenotypic heterogeneity for tumour cells in order to enable the survival of specific cell subpopulations that are resistant to therapy and capable of regenerating the tumour once therapy has been halted [2].
An example of HOX gene alteration in cancer, relates with the increased expression of HOXA9 in the most aggressive acute leukaemia and its potential as predictive of poor prognosis [28]. Moreover, it was demonstrated that HOXA13 protein physically links to the translation initiation factor eIF4E, frequently overexpressed in cancer and described as a strong promoter of tumour growth and angiogenesis [29]. EIF4E acts on the exportation of specific oncogenes mRNAs from the nucleus to cytoplasm, such as c-Myc, FGF-2, ODC and CCND1. Thus, the deregulation of this HOX gene could facilitate the mRNA nuclear export of eIF4E-dependent oncogene transcripts, as observed in hepatocellular carcinomas (HCCs) [30]. HOX genes are also actively expressed in adipose tissue, which has emerged as an important supportive tissue for cancer proliferation and progression [26]. Half of the KRAS-mutant NSCLC (non-small-cell lung cancers) express the homeobox protein HOXC10, which triggered tumour regression in xenografts and PDX (patient-derived xenografts) models in vivo [31]. Upregulation of HOXA10 expression plays a key role in colorectal cancer development and could be considered as a new biomarker that indicates poor prognosis [32]. HOXC8 upregulation is inversely related to pancreatic ductal adenocarcinoma progression and metastasis formation and therefore could be explored as a marker for pancreatic cancer progression [33]. Moreover, some of the pathways involved in metastasis, from invasion to colonization, are affected by the HOX proteins through mechanisms that modulate their expression, such as HMGA2/TET1/HOXA and TGFβ, or by the interference of microRNAs and lncRNAs regulators [26]. However, further studies are required to consolidate their real potential as therapeutic targets [34].
In this context, several studies are now being performed to evaluate HOX proteins strength as therapeutic targets in cancer therapy. The synthetic peptide HXR9 (recently replaced by HTL001), that blocks the binding of HOX proteins to PBX cofactors and, consequently prevents the binding of these dimers to the DNA, is one of the approaches that are under evaluation [34,35]. Cancer cells’ sensitivity to this peptide is highly correlated to their HOX expression profile, albeit the subset of HOX genes which act as oncosuppressors is not affected [34]. This peptide has shown to be effective in oesophageal and oral squamous cell carcinomas [36,37], melanoma [35,38], ovarian cancer (OC) [39], breast cancer [40], meningioma [41], prostate cancer [42], and leukaemia [43]. The use of RNA interference mechanisms [44,45] and the control of HOX methylation status [46] are additional tools that can control HOX expression with therapeutic function. Therefore, the manipulation of HOX genes expression emerges as a promising strategy to prevent invasion and metastasis.
2. Invasion and Metastasis
Tumour formation and progression are multistep processes that depend on the accumulation of genetic and epigenetic alterations generating cells with great genetic instability and culminating with its uncontrolled proliferation escaping from apoptosis and immune system response [2,47]. Subsequently, the progressive increment of the tumour mass size associated with the activation of metalloproteinases (MMPs), which generates gaps in the barriers between tissues, allow cancer cells to invade and disseminate throughout adjacent normal tissues (invasion process), forming secondary tumours [2,48,49]. The development of metastasis is far more complex because, besides invading adjacent tissues, cancer cells travel through blood and lymphatic vessels, detect the premetastatic niche (PMN), and initiate the formation of new tumour masses in distant sites [6,47,50,51].
Therefore, the metastization process depends on the coordinated support of specific cellular programs such as angiogenesis, alterations of cell adhesion, tumour cell motility and secretion of proteolytic enzymes [52]. The dislocation of cancer cells throughout the blood vessels is achieved through the loss of their cell-to-cell adhesion molecules in a process known as epithelial-mesenchymal transition (EMT) [52]. Cancer cells change from epithelial to mesenchymal phenotype enabling cell motility due cytoskeleton modifications that allow cell movement from primary tumour mass to the blood and lymph vessels (intravasation) [53]. When in circulation, metastatic tumour cells adhere to the basement membrane of a new tissue site (extravasation) by mechanisms not completely understood. This process involves the action of glycoproteins such as fibronectin, type IV collagen and laminin. Subsequently, metastatic tumour cells secrete degradative enzymes, or induce host tissues to produce them, to degrade the ECM of the new tissue site [54]. Finally, cancer cells are driven across the basement membrane and stroma within the region where proteolysis occur, starting its division and establishing tumour masses in this new location with or without the reversion of the mesenchymal phenotype through mesenchymal to epithelial transition (MET) process [1] (Figure 1).
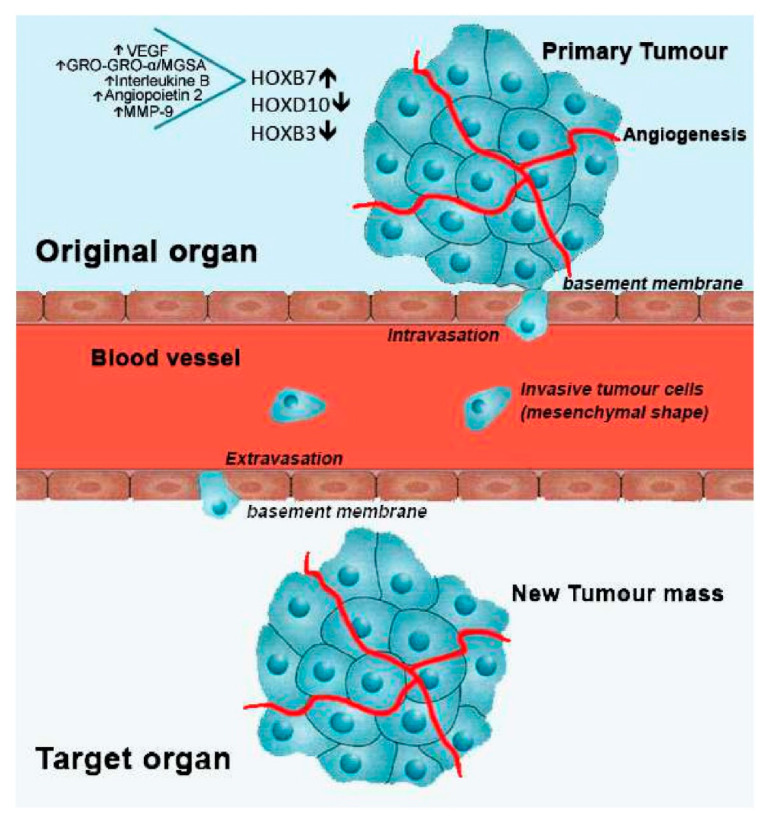
Figure 1.
Metastatic process and involved HOX genes, regarding breast cancer. After cell transformation, a mass is rapidly formed in a specific organ of the body, designated as primary tumour. With the formation of new blood vessels surrounding the primary tumour (angiogenesis), growth is stimulated by the flux of nutrients and oxygen. Then, cells from the primary tumour may acquire motility going through the basement membrane into the blood vessels (intravasation). Once in circulation, metastatic tumour cells stop its progress adhering to the basement membrane of a new tissue site (extravasation). Finally, cancer cells start to proliferate in the new location, forming a new tumour mass. In breast cancer, overexpression of HOXB7, along with the upregulation of the indicated molecules, and downregulation of HOXD10 and HOXB3 have been linked to the metastatic process.
3. Five Mechanisms of HOX Deregulation Affecting Invasion and Metastasis of Cancer Cells
3.1. HMGA2/TET1/HOXA Signalling Pathway
The ten–eleven translocation family (TET) of methylcytosine dioxygenases can induce DNA demethylation and is associated with tumorigenesis in many cancer types, given that its action alters the regulation of transcription and affects the expression of many genes [55,56,57]. These enzymes catalyse the hydroxylation of 5-methylcytosine (5mC) found in methylated DNA sequence, which results in a 5-hydroxymethylcytosine (5hmC). Then, TET can further catalyse oxidation of 5hmC to 5-formylcytosine (5fC) that is also oxidized forming 5-carboxycytosine (5caC). Finally, by the action of a terminal deoxynucleotidyl transferase (TdT), the 5caC is converted to an unmodified cytosine [58] (Figure 2).
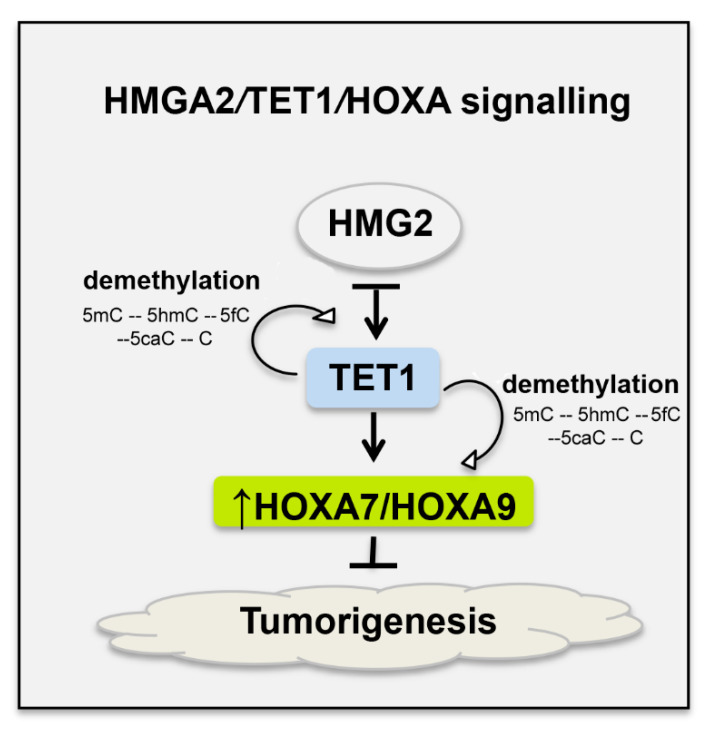
Figure 2.
HMGA2/TET1/HOXA signalling in breast cancer cells. Depletion of the architectural transcription factor high mobility group AT-hook 2 (HMGA2) induces TET1 (ten–eleven translocation methylcytosine dioxygenases). This molecule initiates demethylation of DNA and is associated with tumorigenesis in many cancers. TET1 binds and demethylates its own promoter and the promoters of HOXA7 and HOXA9, increasing their expression, which may result in the inhibition of breast cancer growth and metastasis, already demonstrated for HOXA9 using mouse xenografts [35]. The demethylation promoted by TET1 allows the conversion of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) and this to 5-formylcytosine (5fC), which in turn is converted in to 5-carboxycytosine (5caC) and at the end in an unmodified cytosine (C).
An association was also found between TET enzymes and HOX gene expression. For example, in breast cancer, TET1 can demethylate its own promoter and the promoter of HOXA genes, enhancing its own expression and stimulating HOXA7 and HOXA9 expression (Figure 2) [56]. HOXA9 overexpression has been correlated to a less invasive behaviour of breast cancer cells [56,59]. The TETs are also associated with the nonhistone architectural transcription factor “high mobility group AT-hook 2” (HMGA2) and its inhibition can induce TET1 expression [56]. HMGA2 can modulate the transcription of several genes binding to AT-rich sequences and altering the chromatin structure and consequently affecting a variety of processes, including the cellular cycle, DNA damage repair, apoptosis, senescence, EMT and telomere restoration [58]. Therefore, the effect of HMGA2 in TET1 expression and the subsequent effect of TET1 in HOXA9 expression is known as the HMGA2/TET1/HOXA9 signalling pathway, which has a role in the invasion and metastasis of cancer cells (Figure 2). In breast cancer, when HMGA2 is depleted, an induction of the TET1 and HOXA9 genes occurs inhibiting invasion and metastization [56]. The opposite effect could also occur, given that HMGA2 overexpression reduces TET1 and HOXA9 expression, supporting invasion and metastization [60]. These data highlight tissue-specificity of HOX gene functions, not only in embryonic development but also in cancer [15,17].
3.2. TGFβ Signalling Pathway
HOX proteins interfere with invasion and metastasis of cancer cells due to their ability to affect the expression of TGFβ genes, which are multifunctional cytokines secreted by different cells, with important roles in embryonic development controlling cell behaviour, namely cell proliferation, differentiation, morphogenesis, tissue homeostasis and regeneration [61]. The direct binding of HOX proteins to TGFβ genes promoters, activating their expression, was identified for HOXB7 and HOXC8 [62,63]. This results in the coding of TGFβ ligands, proteins that will be secreted to the ECM [64] (Figure 3). Thus, the dysfunction of TGFβ seems to be also associated with cancer progression, as a consequence of HOX genes expression deregulation [65]. HOXB7 overexpression, for example, induces invasive and metastatic breast cancer by activating the TGFβ signalling pathway [62]. In HCCs, downregulation of HOXB5 inhibits TGFβ, which induces migration and invasion [66]. In NSCLC, HOXC8 promotes proliferation and migration through transcriptional upregulation of TGFβ1 [63]. When in the ECM, TGFβ ligands bind to TGFβ receptor type II in the cell surface and a cascade of processes is initiated (Figure 3), causing modifications in the expression of genes related with cell proliferation, apoptosis, metastasis and angiogenesis [67].
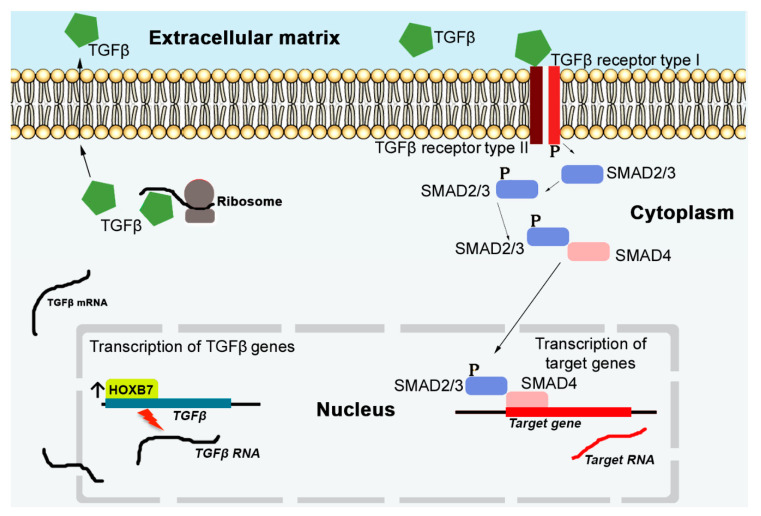
Figure 3.
TGFβ signalling pathway in breast cancer cells. The reception of the TGFβ signals by the TGFβ receptor type II allows the phosphorylation of the TGFβ receptor type I. Subsequently this favour the phosphorylation of SMAD2/3 proteins that can in turn interact with the protein SMAD4. This complex of proteins controls the expression of TGFβ target genes, affecting cellular processes, such as proliferation, apoptosis, metastasis, and angiogenesis. The expression of the TGFβ’s genes is regulated by HOX transcriptional factors, as HOXB7 in breast cancer. After its transcription (nucleus) and translation (cytoplasm), TGFβ ligands are exported to the ECM, where they recognize and are recognized by TGFβ receptors type II.
3.3. HOX Proteins Effect in the Epithelial to Mesenchymal Transition (EMT)
EMT is a fundamental cellular process for non-disease conditions, such as embryogenesis and wound healing; however, it can also favour malignant processes, such as cancer progression [68]. Intercellular structures and cell–cell adhesion molecules are key factors in maintaining a well cohesive normal tissue or even a coherent primary tumour mass. Abnormalities in these structures can lead to cell detachment from the primary tumour and enhance the potential for dissemination and metastatic spread of cancer cells to secondary locations [69].
During EMT, epithelial cells are transcriptionally reprogrammed, which results in a decrease of cell–cell, cell–basement membrane adhesion and an increase of cell migration or invasion [70]. The epithelial cells usually present apical–basal polarity and are maintained together laterally by tight and adherens junctions, formed by epithelial cadherin, CDH1, and desmosomes. Cell junctions with basement membranes are maintained by hemidesmosomes [53] (Figure 4). To maintain these junctions and cell polarity, cells express epithelial adhesion molecules, such as occludin, claudins, α6β4 integrin and cytokeratins. Therefore, for EMT to occur, two parallel processes must take place: (a) repression in the expression of epithelial molecules by EMT-inducing transcription factors expression, namely ZEB, SNAIL and TWIST, and (b) activation of mesenchymal molecules expression, as N-cadherin, vimentin, fibronectin, β1 and β3 integrins and MMPs [53,71]. As a consequence of the EMT process, cells acquire motility and invasive capacities [72] (Figure 4). Thus, EMT has significant importance in epithelial cancers, since it confers a higher tumour-progression potential, as invasion and metastatic capabilities, to cancer cells [53]. This new mesenchymal cell status can be reversible, and cells can recover their epithelial characteristics in a process called mesenchymal-to-epithelial transition (MET) [72].
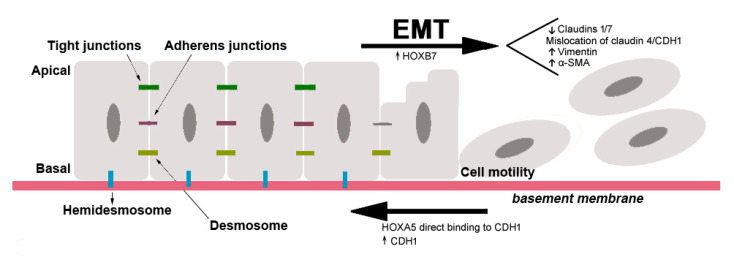
Figure 4.
Examples of HOX genes involved in the activation or inhibition of the epithelial to mesenchymal transition (EMT) in breast cancer. Epithelial cells present apical–basal polarity and are maintained together by tight junctions, adherens junctions and desmosomes. Hemidesmosomes maintain epithelial cells together with basement membrane. At the end of this process, epithelial cells present mesenchymal characteristics as the absence of apical–basal polarity and presence of motility capacity. HOXA5 is known to inhibit the EMT via upregulation of CDH1 in the adherens junctions, while HOXB7 induce the EMT downregulating the epithelial proteins: Claudin-1, Claudin-7, Claudin-4 and CDH1, and upregulating vimentin and α-SMA.
Currently, there are some reports proposing an involvement of HOX proteins in the EMT process, with an impact on the capacity of invasion and metastasis of cancer cells [73,74,75,76]. It has been demonstrated that CDH1 expression is transcriptionally upregulated by the direct binding of HOXA5 to their promoter sequence and that the knockdown of HOXA5 in mammary cells leads to a loss of epithelial traits, increased stemness and cell plasticity, and acquisition of more aggressive phenotypes [77]. HOXC6 was shown to promote invasion of HCCs by driving EMT through the negative regulation of CDH1 expression and being positively associated with vimentin and MMP-9 expression [78,79]. Still in HCC, HOXD9 was shown to interact with the promoter region of ZEB1/ZEB2 increasing its expression. ZEB1 knockdown, in turn, inhibited HOXD9-induced migration and invasion, as well as EMT [78].
HOXA10, which is reported as an EMT inhibitor, when downregulated in endometrial carcinomas favoured their invasive behaviour by reducing the expression of CDH1 [80]. In gastric carcinomas (GC), the downregulation of HOXB9 has been associated with malignancy and metastasis while its re-expression was suggested to cause inhibition of cell proliferation, migration, invasion and induction of MET [73]. In cervical cancers, HOXA10 and HOXB13 were shown to influence the EMT. HOXA10 expression was positively associated with CDH1 and negatively associated with vimentin expression, inhibiting EMT. For HOXB13 the opposite trend was observed with a negative effect on CDH1 and a positive effect on vimentin expression [75]. Additionally, it has been demonstrated that HOXB13 promotes lung adenocarcinoma cell growth, local invasion, and metastasis to the liver in nude mice regulating ABCG1, EZH2 and Slug [78,81]. In breast cancer, HOXB7 overexpression can promote tumour migration and invasion through the induction of EMT in epithelial cells, by reducing the expression of epithelial proteins, as Claudin-1 and Claudin-7, together with mislocalisation of Claudin-4 and CDH1 and increased expression of mesenchymal proteins: vimentin and alpha-smooth muscle actin (α-SMA) [82] (Figure 4). A detailed list of the proposed roles of the HOX proteins in EMT is presented in Table 1.
Table 1.
Proposed role of the HOX proteins in the EMT and MET in distinct cancer types.
| HOX Protein | Type of Cancer | Effect | Reference |
|---|---|---|---|
| HOXA1 | Prostate cancer | Induction of EMT via downregulation of CDH1 and upregulation of MMP-3 and Snail. | [83] |
| HOXA5 | Breast cancer | Inhibition of EMT via upregulation of CDH1. | [77] |
| HOXA9 | Breast Cancer | Less invasive phenotype inclusive in claudin-low cells. | [56,59] |
| HOXA10 | Endometrial carcinoma | Inhibition of EMT via upregulation of CDH1. | [80] |
| Cervical cancer | Inhibition of EMT via upregulation of CDH1 and downregulation of vimentin. | [75] | |
| Oral squamous cell carcinoma | Inhibition of EMT via upregulation of CDH1 and downregulation of N-cadherin. | [84] | |
| Ovarian cancer | Inhibition of EMT via downregulation of vimentin, and MMP-9. | [85] | |
| Glioma | Induction of EMT via activation of Wnt/beta-catenin/TGF-β pathways. | [86] | |
| HOXA13 | Oesophageal squamous cell carcinoma | Induction of EMT via downregulation of CDH1 and upregulation of Snail. | [87] |
| Gastric cancer | Induction of EMT via downregulation of CDH1 and upregulation of N-cadherin and vimentin. | [88] | |
| HOXB5 | Breast cancer | Induction of EMT via downregulation of CDH1 and upregulation of vimentin and Snail2. | [89] |
| Non-small-cell lung cancer | Induction of EMT via activation of Wnt/beta-catenin pathway. | [90] | |
| HOXB7 | Breast cancer | Induction of EMT via downregulation of Claudin-1 and Claudin-7, mislocalisation of Claudin-4 and CDH1, and upregulation of vimentin and α-SMA. | [82] |
| HOXB8 | Colorectal carcinoma | Induction of EMT via downregulation of CDH1 and upregulation of vimentin, N-cadherin, Twist, Zeb1 and Zeb2. | [91] |
| Ovarian cancer | Inhibition of EMT via downregulation of vimentin, and MMP-9. | [85] | |
| Gastric carcinoma | Induction of MET. | [73] | |
| HOXB9 | Lung adenocarcinoma | Induction of EMT via downregulation of CDH1 and upregulation of N-cadherin and vimentin. | [92] |
| Breast cancer | Induction of EMT via activation of the TGF-β pathway. | [93] | |
| HOXB13 | Cervical cancer | Induction of EMT via down regulation of CDH1 and upregulation of vimentin. | [75] |
| Lung adenocarcinoma | Induction of EMT via ABCG1, EZH2 and Slug regulation. | [78,81] | |
| HOXC6 | Hepatocellular carcinoma | Induction of EMT via positive regulation of CDH1 and negative regulation of vimentin and MMP-9. | [78,79] |
| HOXC10 | Ovarian cancer | Induction of EMT via Slug regulation. | [94] |
| Gastric cancer | Induction of EMT via activation of the MAPK pathway. | [95] | |
| HOXD3 | Hepatocellular carcinoma | HOXD3 can directly target the promoter region of VEGFR and increase its expression. | [96] |
| HOXD9 | Hepatocellular carcinoma | Induction of EMT via upregulation of ZEB1 and ZEB2 interaction | [74,78] |
| Colorectal carcinoma | Induction of EMT via upregulation of TWIST and SNAIL. | [76] | |
| HOXD10 | Oesophageal squamous cell carcinoma | Inhibition of EMT via upregulation of CDH1 and downregulation of N-cadherin and vimentin. | [97] |
3.4. MicroRNA’s Interference
MicroRNAs (miRs) are an abundant class of noncoding RNAs (with 21 to 25 nucleotides), which have a crucial role in post-transcriptional gene regulation, including HOX genes expression (Table 2) [98]. This regulation of the HOX genes by miRs is favoured by the location of miRs coding regions in the HOX gene clusters [99]. Thus, the role of the HOX genes in invasion and metastasis of cancer cells might be also dependent on their regulatory miRs. In pancreatic cancer, for example, it has been described that the downregulation of HOXA1, HOXB1 and HOXB3 by miR-10a is associated with the formation of metastasis [100,101]. Furthermore, miR-100 can directly target the 3′-UTR of HOXA1, which reduces the chemotherapy response of small-cell lung cancer [102]. Another example of miR interference is the downregulation of HOXD10 by miR-10b, which favours the invasive and metastatic behaviour of breast cancer cells [103]. Besides, downregulation of HOXD10 by miR-10b also causes an increase in prometastatic gene products, such as MMP-14 and RHOC, contributing to the acquisition of metastatic phenotypes in epithelial OC cells [104]. Moreover, miR-7 and miR-218 epigenetically control tumour suppressor genes RASSF1A and Claudin-6 by downregulating HOXB3 in breast cancer [105]. In HCC, miR-203a negatively targets HOXD3 resulting in an inhibition of cell invasion, metastasis and angiogenesis [96]. On the other hand, miR-224 directly targets and downregulates HOXD10, boosting tumour progression and invasion in HCC [106]. In addition, HOXB7 downregulation promoted by mR-196b-5p leads to metastization in colorectal cancer [107]. Some studies have also connected miR interference with HOXA5 downregulation, considered as a tumour suppressor gene in several cancer contexts [15]. For instance, miR-181d-5p targets CDX2, a transcription factor binding to HOXA5 promoter, consequently leading to EMT in MCF-7 breast cancer cells [108]. Moreover, in NSCLC, miR-196a antagonizes the inhibitory tumour growth effect of HOXA5, partially contributing to an increasingly invasive phenotype [109]. Another noteworthy miR target is HOXC10, shown to be involved in the EMT in both GC and ovarian OC [94,95]. Recent studies have shown that miR-136 and miR-222-3p act as an inhibitor of HOXC10, which impairs the EMT and consequently leads to less perineum and hepatic metastasis in GC and OC, respectively [94,110].
Table 2.
Noncoding RNAs that regulate HOX genes and/or are transcribed from HOX loci with impact on breast cancer progression.
| Non-Coding RNA | HOX Gene Affected | Effect On Breast Cancer Cells | Reference |
|---|---|---|---|
| miR-10b | ↓ HOXD10 | ↑ Aggressiveness | [103] |
| miR-7/-218 | ↓ HOXB3 | ⊗ Cell cycle/clone formation | [105] |
| miR-181d-5p | ↓ HOXA5 | ↑ Aggressiveness, promotes EMT | [108] |
| HOTAIR | ↓ HOXD10 | ↑ Invasion/ metastasis | [103] |
| HOXA-AS2 * | NA | ↑ Oncogenic behaviour | [144] |
| HOTTIP | ↑ HOXA | ↑ oncogenic behaviour | [161] |
| HOXD-AS1 (HAGRL) * | NA | ↑ Oncogenic behaviour | [169] |
⊗, Inhibition; ↑, Increase; ↓, Decrease; *, Noncoding RNAs transcribed from HOX genes loci; NA, Not Applicable.
The relationship between HOX gene expression and miRs regulation is far more complex than initially described. In fact, increasing evidence suggest that HOX proteins can themselves regulate the expression of miRs, which has an impact on invasion and metastasis abilities of cancer cells. In breast cancer and gliomas, HOXD10 downregulation results in the downregulation of miR-7 and upregulation of p21-activated kinase 1 expression (PAK1), which culminates in higher aggressiveness of these cancer types facilitating invasion and metastasis [111].
3.5. lncRNA’s Interference
Over the last decade, several long noncoding RNAs (lncRNAs) were proposed to play essential roles in gene regulation, both at pre- and post-translational levels, and described to contribute to tumour progression and metastasis [112]. A total of 231 lncRNAs have been identified as products of the HOX clusters, amongst which HOXD-AS1, HOXA-AS2, HOTAIRM1, HOTTIP, and HOTAIR, were reported to be involved in cancer regulation [113].
The HOX antisense intergenic RNA (HOTAIR) is an lncRNA that correlates with metastasis and poor prognosis in breast, colon and lung cancer [114,115,116]. This lncRNA is transcribed from the antisense strand of the HOXC cluster, repressing the transcription of the HOXD cluster [45]. The induction of HOTAIR expression in breast and lung cancer cell lines promotes growth in soft agar and invasion in Matrigel [45]. For example, in breast cancer, HOTAIR downregulates HOXD10 favouring the invasive and metastatic behaviour (Table 2) [103]. In gliomas, HOTAIR was described to promote invasion through upregulation of MMP-7, MMP-9 and VEGF [117]. In addition, HOTAIR has been identified as a key modulator in chromatin dynamics, partaking simultaneously in histone methylation and deacetylation, but also on the antagonistic process [118,119]. In fact, the knockdown of HOTAIR in GC significantly reverses EMT by increasing the expression of CDH1 and inducing the loss of the PRC2 complex activity (consisting of H3K27 methyltransferase EZH2, SUZ12 and EED) [120]. In cervical cancer, the downregulation of HOTAIR results in an increased expression of CDH1 and a decreased expression of CTNNB1, vimentin, Snail and Twist. In contrast, HOTAIR overexpression promotes VEGF and MMP-9 protein expression [121]. In osteosarcoma, HOTAIR suppression significantly reduces the migration and invasion, decreases the expression of MMP-2 and MMP-9 and increases the expression of CDH1. This occurs through regulation of RAC-α serine/threonine protein kinase target in rapamycin signalling pathway [122], whereas in gliomas, this lncRNA increases MMP-7 and MMP-9 expression by upregulating the Wnt/β-catenin pathway [123]. Moreover, in renal cell carcinomas [124] and HCC [125], upregulation of HOTAIR leads to increased cell invasiveness and metastatic capacity. Therefore, it would be fair to infer the involvement of this lncRNA on the epithelial-mesenchymal transition.
A recent and increasingly number of studies have suggested that HOTAIR can interact with miRNAs to regulate gene expression and cancer progression. In fact, HOTAIR may sponge miRs, by competing for its target sites, thus inhibiting its effects. In breast cancer, HOTAIR was shown to promote invasion and metastasis by sponging miR-601, which consequently downregulated its target, ZEB1 [126]. In OC, HOTAIR actively competes with miR-260 upregulating CCND1 and CCND2 dynamically sprouting OC progression [127]. Moreover, HOTAIR can act as a molecular sponge of miR-204, leading to an increase in HOXC8, promoting proliferation, migration and invasion in oesophageal cancer cells [128], whereas in cholangiocarcinoma, this process occurs through the modulation of miR-204-5p’s interaction with HMGB1 [129]. Notwithstanding, HOTAIR promotes GC cell growth and metastasis by interfering in the miR-217/GPC5 [130], miR-1277-5p/COL5A1 [131], and miR-618/KLF12 axis, being that the last one induces metastasis by promoting the PI3K/ATK signalling pathway [132]. Recent findings have also related HOTAIR interference to Trastuzumab resistance in GC, through the sponging of miR-3030 and consequent upregulation of ERBB4 [133], which is responsible for activating the PI3K/ATK signalling pathway [134]. Several HOTAIR-dependent interferences in miR pathways have also been described in glioma [123], HCC [135,136,137] prostate cancer [138,139] and squamous cell carcinomas [140,141,142,143].
Despite HOTAIR’s extensively described role in carcinogenesis, other lncRNAs transcribed from the HOX clusters should not be overlooked in the cancer context. HOXA cluster antisense RNA 2 (HOXA-AS2), transcribed between the HOXA3 and HOXA4 genes, has been implicated in oncogenic promotion in pancreatic cancer, breast cancer [144,145], colorectal cancer [146], GC [147,148], bladder cancer [149], HCC [150], thyroid cancer [151], gallbladder carcinoma [152], lung cancer [153,154,155], osteosarcoma [156], glioma [157], and leukaemia [158,159]. Another lncRNA transcribed between HOXA1 and HOXA2, the HOXA transcript antisense RNA myeloid-specific 1 (HOTAIRM1), appears upregulated in endometrial and pancreatic cancers, NSCLC, glioma and glioblastoma, but downregulated in HCC, colorectal, OC, GC and papillary thyroid cancers, as well as head and neck tumours [160].
Another lncRNA transcribed from the genomic region of HOXA locus is HOXA distal transcript antisense RNA (HOTTIP) that directly binds the HOXA locus leading to H3K4 methylation and activating the transcription of HOX genes. HOTTIP has been described to exert oncogenic roles in different malignancies such as head and neck tumours, oesophageal carcinomas, colorectal, lung and breast cancers (Table 2) [161]. Furthermore, HOXD cluster antisense RNA 1 (HOXD-AS1) also named HAGRL, transcribed on the antisense direction between HOXD1 and HOXD3, appears to be deregulated in many cancers. In fact, this lncRNA is overexpressed in several malignancies, promoting processes such as cell proliferation, metastasis, and drug resistance [162], acting as a molecular sponge for different miRs [163] or by interfering in chromatin modulation [164]. Amongst these malignancies are cervical cancer [165,166], GC [167], colorectal cancer [168], breast [169], OC [170], and HCC [171,172]. HOXD-AS1 also promotes cell migration and invasion in NSCLC by sponging miR-133b and promoting MMP-9 expression [173], albeit it plays an opposite stand in lung adenocarcinoma by recruiting DNMT1 to the promoter of E2F1, silencing it, and consequently suppressing cell proliferation [164].
4. Conclusions
Invasion and metastasis are the leading causes of cancer-related death, as they reflect the last stages of cancer. Therefore, the development of targeted therapies to prevent and/or lagging invasion and metastasis are crucial to improve the clinical outcomes of cancer patients. HOX genes are in the spotlight of cancer research due to their effects on key molecules related to angiogenesis (VEGFR), invasion (MMPs), cell adhesion (CDH1, vimentins, claudins), and transcription factors involved in the EMT (Twist, Snail, Zeb). These genes were also implicated in the control of mRNAs nuclear export achieved by their impact on eIF4E. In addition, HOX genes were shown to be active in the adipose tissue, an important supportive tissue for tumour progression. However, the identification of HOX targets and their function in different types of cancer is incomplete. Furthermore, microRNAs and lncRNAs transcribed from the HOX loci can also be part of the complex molecular processes by which HOX genes affect invasion and metastasis in cancer. Over the last years, the use of the HXR9 peptide has yielded promising results in different types of tumours and could be an innovative tool in different stages of cancer progression.
5. Future Perspectives
Different aspects of the HOX genes function in cancer have been studied and diverse approaches have already been tested to allow the use of the HOX genes as therapeutic targets for cancer treatment. Here, we propose three take-home messages: (1) HOX have crucial roles in two of the most aggressive processes in cancer development, invasion and metastization, responsible for the majority of cancer deaths; (2) HOX genes can affect invasion and metastization by diverse mechanisms, and possibly more are yet to be discovered; (3) the control of HOX genes expression and/or its targets may represent a promising strategy to prevent invasion and metastasis and decrease the number of cancer-related deaths.
Acknowledgments
We also acknowledge Raul Guizzo for editing assistance and proofreading.
Author Contributions
This manuscript was conceived by A.P., written by all authors (A.P., S.A.d.B.G., J.L.C., A.R.C.-P., R.F.) with support and scientific guidance from R.F. All authors have read and agreed to the published version of the manuscript.
Funding
The work was financed by FEDER (Fundo Europeu de Desenvolvimento Regional) funds through the COMPETE 2020 Operational Programme for Competitiveness and Internationalisation (POCI), Portugal 2020, and by Portuguese funds through FCT (Fundação para a Ciência e a Tecnologia/Ministério da Ciência, Tecnologia e Ensino Superior) in the framework of the project POCI-01-0145-FEDER-030562 (PTDC/BTM-TEC/30562/2017). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Conflicts of Interest
Authors declare there is no conflict of interests.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dillekås H., Rogers M.S., Straume O. Are 90% of deaths from cancer caused by metastases? Cancer Med. 2019;8:5574–5576. doi: 10.1002/cam4.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D., Weinberg R.A. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Massagué J., Obenauf A.C. Metastatic colonization by circulating tumour cells. Nature. 2016;529:298–306. doi: 10.1038/nature17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Damrauer J.S., Phelps S.N., Amuchastegui K., Lupo R., Mabe N.W., Walens A., Kroger B.R., Alvarez J.V. Foxo-dependent Par-4 Upregulation Prevents Long-term Survival of Residual Cells Following PI3K-Akt Inhibition. Mol. Cancer Res. Mcr. 2018;16:599–609. doi: 10.1158/1541-7786.MCR-17-0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tabassum D.P., Polyak K. Tumorigenesis: It takes a village. Nat. Rev. Cancer. 2015;15:473–483. doi: 10.1038/nrc3971. [DOI] [PubMed] [Google Scholar]
- 6.Fares J., Fares M.Y., Khachfe H.H., Salhab H.A., Fares Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transduct. Target. Ther. 2020;5:28. doi: 10.1038/s41392-020-0134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peinado H., Zhang H., Matei I.R., Costa-Silva B., Hoshino A., Rodrigues G., Psaila B., Kaplan R.N., Bromberg J.F., Kang Y., et al. Pre-metastatic niches: Organ-specific homes for metastases. Nat. Rev. Cancer. 2017;17:302–317. doi: 10.1038/nrc.2017.6. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y., Cao X. Characteristics and Significance of the Pre-metastatic Niche. Cancer Cell. 2016;30:668–681. doi: 10.1016/j.ccell.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y., Cao X. Organotropic metastasis: Role of tumor exosomes. Cell Res. 2016;26:149–150. doi: 10.1038/cr.2015.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meleth S., Whitehead N., Evans T.S., Lux L. Technology Assessment on Genetic Testing or Molecular Pathology Testing of Cancers with Unknown Primary Site to Determine Origin. Agency for Healthcare Research and Quality (US); Rockville, MD, USA: 2013. AHRQ Technology Assessments. [PubMed] [Google Scholar]
- 11.Ramaswamy S., Ross K.N., Lander E.S., Golub T.R. A molecular signature of metastasis in primary solid tumors. Nat. Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 12.Kanwal R., Gupta S. Epigenetic modifications in cancer. Clin. Genet. 2012;81:303–311. doi: 10.1111/j.1399-0004.2011.01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al Aboud N.M., Tupper C., Jialal I. StatPearls. StatPearls Publishing LLC.; Treasure Island, FL, USA: 2020. Genetics, Epigenetic Mechanism. [PubMed] [Google Scholar]
- 14.Bhatlekar S., Fields J.Z., Boman B.M. Role of HOX Genes in Stem Cell Differentiation and Cancer. Stem Cells Int. 2018;2018:3569493. doi: 10.1155/2018/3569493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Bessa Garcia S.A., Araújo M., Pereira T., Mouta J., Freitas R. HOX genes function in Breast Cancer development. Biochim. Biophys. Acta Rev. Cancer. 2020;1873:188358. doi: 10.1016/j.bbcan.2020.188358. [DOI] [PubMed] [Google Scholar]
- 16.Scott M.P. Vertebrate homeobox gene nomenclature. Cell. 1992;71:551–553. doi: 10.1016/0092-8674(92)90588-4. [DOI] [PubMed] [Google Scholar]
- 17.Bhatlekar S., Fields J.Z., Boman B.M. HOX genes and their role in the development of human cancers. J. Mol. Med. (Berl. Ger.) 2014;92:811–823. doi: 10.1007/s00109-014-1181-y. [DOI] [PubMed] [Google Scholar]
- 18.Zakany J., Duboule D. The role of Hox genes during vertebrate limb development. Curr. Opin. Genet. Dev. 2007;17:359–366. doi: 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 19.Shah N., Sukumar S. The Hox genes and their roles in oncogenesis. Nat. Rev. Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 20.Luo Z., Rhie S.K., Farnham P.J. The Enigmatic HOX Genes: Can We Crack Their Code? Cancers. 2019;11:323. doi: 10.3390/cancers11030323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Longobardi E., Penkov D., Mateos D., De Florian G., Torres M., Blasi F. Biochemistry of the tale transcription factors PREP, MEIS, and PBX in vertebrates. Dev. Dyn. 2014;243:59–75. doi: 10.1002/dvdy.24016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rux D.R., Wellik D.M. Hox genes in the adult skeleton: Novel functions beyond embryonic development. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2017;246:310–317. doi: 10.1002/dvdy.24482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paço A., de Bessa Garcia S.A., Freitas R. Methylation in HOX Clusters and Its Applications in Cancer Therapy. Cells. 2020;9:1613. doi: 10.3390/cells9071613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagata H., Kozaki K.I., Muramatsu T., Hiramoto H., Tanimoto K., Fujiwara N., Imoto S., Ichikawa D., Otsuji E., Miyano S., et al. Genome-wide screening of DNA methylation associated with lymph node metastasis in esophageal squamous cell carcinoma. Oncotarget. 2017;8:37740–37750. doi: 10.18632/oncotarget.17147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sui B.Q., Zhang C.D., Liu J.C., Wang L., Dai D.Q. HOXB13 expression and promoter methylation as a candidate biomarker in gastric cancer. Oncol. Lett. 2018;15:8833–8840. doi: 10.3892/ol.2018.8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jonkers J., Pai P., Sukumar S. Multiple roles of HOX proteins in Metastasis: Let me count the ways. Cancer Metastasis Rev. 2020;39:661–679. doi: 10.1007/s10555-020-09908-4. [DOI] [PubMed] [Google Scholar]
- 27.Li B., Huang Q., Wei G.H. The Role of HOX Transcription Factors in Cancer Predisposition and Progression. Cancers. 2019;11:528. doi: 10.3390/cancers11040528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Y., Zhou B., Mao F., Xu J., Miao H., Zou Z., Phuc Khoa L.T., Jang Y., Cai S., Witkin M., et al. HOXA9 Reprograms the Enhancer Landscape to Promote Leukemogenesis. Cancer Cell. 2018;34:643–658.e5. doi: 10.1016/j.ccell.2018.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Benedetti A., Graff J.R. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189–3199. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- 30.Cillo C., Schiavo G., Cantile M., Bihl M.P., Sorrentino P., Carafa V., D’Armiento M., Roncalli M., Sansano S., Vecchione R., et al. The HOX gene network in hepatocellular carcinoma. Int. J. Cancer. 2011;129:2577–2587. doi: 10.1002/ijc.25941. [DOI] [PubMed] [Google Scholar]
- 31.Guerra S.L., Maertens O., Kuzmickas R., De Raedt T., Adeyemi R.O., Guild C.J., Guillemette S., Redig A.J., Chambers E.S., Xu M., et al. A Deregulated HOX Gene Axis Confers an Epigenetic Vulnerability in KRAS-Mutant Lung Cancers. Cancer Cell. 2020;37:705–719.e706. doi: 10.1016/j.ccell.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan Y., Sun S., Jiao N., Shu Y., Zhang Y. Upregulation of HOXA10 Protein Expression Predicts Poor Prognosis for Colorectal Cancer. Genet. Test. Mol. Biomark. 2018;22:390–397. doi: 10.1089/gtmb.2017.0240. [DOI] [PubMed] [Google Scholar]
- 33.Adwan H., Zhivkova-Galunska M., Georges R., Eyol E., Kleeff J., Giese N.A., Friess H., Bergmann F., Berger M.R. Expression of HOXC8 is inversely related to the progression and metastasis of pancreatic ductal adenocarcinoma. Br. J. Cancer. 2011;105:288–295. doi: 10.1038/bjc.2011.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morgan R., El-Tanani M., Hunter K.D., Harrington K.J., Pandha H.S. Targeting HOX/PBX dimers in cancer. Oncotarget. 2017;8:32322–32331. doi: 10.18632/oncotarget.15971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morgan R., Pirard P.M., Shears L., Sohal J., Pettengell R., Pandha H.S. Antagonism of HOX/PBX dimer formation blocks the in vivo proliferation of melanoma. Cancer Res. 2007;67:5806–5813. doi: 10.1158/0008-5472.CAN-06-4231. [DOI] [PubMed] [Google Scholar]
- 36.Shen L.Y., Zhou T., Du Y.B., Shi Q., Chen K.N. Targeting HOX/PBX dimer formation as a potential therapeutic option in esophageal squamous cell carcinoma. Cancer Sci. 2019;110:1735–1745. doi: 10.1111/cas.13993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou T., Fu H., Dong B., Dai L., Yang Y., Yan W., Shen L. HOXB7 mediates cisplatin resistance in esophageal squamous cell carcinoma through involvement of DNA damage repair. Thorac. Cancer. 2020;11:3071–3085. doi: 10.1111/1759-7714.13142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Errico M.C., Felicetti F., Bottero L., Mattia G., Boe A., Felli N., Petrini M., Bellenghi M., Pandha H.S., Calvaruso M., et al. The abrogation of the HOXB7/PBX2 complex induces apoptosis in melanoma through the miR-221&222-c-FOS pathway. Int. J. Cancer. 2013;133:879–892. doi: 10.1002/ijc.28097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgan R., Plowright L., Harrington K.J., Michael A., Pandha H.S. Targeting HOX and PBX transcription factors in ovarian cancer. BMC Cancer. 2010;10 doi: 10.1186/1471-2407-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morgan R., Boxall A., Harrington K.J., Simpson G.R., Gillett C., Michael A., Pandha H.S. Targeting the HOX/PBX dimer in breast cancer. Breast Cancer Res. Treat. 2012;136:389–398. doi: 10.1007/s10549-012-2259-2. [DOI] [PubMed] [Google Scholar]
- 41.Ando H., Natsume A., Senga T., Watanabe R., Ito I., Ohno M., Iwami K., Ohka F., Motomura K., Kinjo S., et al. Peptide-based inhibition of the HOXA9/PBX interaction retards the growth of human meningioma. Cancer Chemother. Pharm. 2014;73:53–60. doi: 10.1007/s00280-013-2316-5. [DOI] [PubMed] [Google Scholar]
- 42.Morgan R., Boxall A., Harrington K.J., Simpson G.R., Michael A., Pandha H.S. Targeting HOX transcription factors in prostate cancer. BMC Urol. 2014;14 doi: 10.1186/1471-2490-14-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Z., Zhang Z., Li Y., Arnovitz S., Chen P., Huang H., Jiang X., Hong G.M., Kunjamma R.B., Ren H., et al. PBX3 is an important cofactor of HOXA9 in leukemogenesis. Blood. 2013;121:1422–1431. doi: 10.1182/blood-2012-07-442004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chopra V.S., Mishra R.K. "Mir"acles in hox gene regulation. Bioessays News Rev. Mol. Cell. Dev. Biol. 2006;28:445–448. doi: 10.1002/bies.20401. [DOI] [PubMed] [Google Scholar]
- 45.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L., et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agger K., Cloos P.A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 47.Weinberg R.A. The Biology of Cancer. 2nd ed. Garland Science; New York, NY, USA: 2014. [Google Scholar]
- 48.Merkle C.J. Cancer Nursing: Principles and Practice. 7th ed. Jones and Bartlett Publishers; Sudbury, ON, Canada: 2010. Biology of Cancer. [Google Scholar]
- 49.Kang W., Ferruzzi J., Spatarelu C.-P., Han Y.L., Sharma Y., Koehler S.A., Butler J.P., Roblyer D., Zaman M.H., Guo M., et al. Tumor invasion as non-equilibrium phase separation. bioRxiv. 2020 doi: 10.1101/2020.04.28.066845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stuelten C.H., Parent C.A., Montell D.J. Cell motility in cancer invasion and metastasis: Insights from simple model organisms. Nat. Rev. Cancer. 2018;18:296–312. doi: 10.1038/nrc.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tahtamouni L., Ahram M., Koblinski J., Rolfo C. Molecular Regulation of Cancer Cell Migration, Invasion, and Metastasis. Anal. Cell. Pathol. (Amst.) 2019;2019:1356508. doi: 10.1155/2019/1356508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yilmaz M., Christofori G., Lehembre F. Distinct mechanisms of tumor invasion and metastasis. Trends Mol. Med. 2007;13:535–541. doi: 10.1016/j.molmed.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 53.Dongre A., Weinberg R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 54.Quintero-Fabián S., Arreola R., Becerril-Villanueva E., Torres-Romero J.C., Arana-Argáez V., Lara-Riegos J., Ramírez-Camacho M.A., Alvarez-Sánchez M.E. Role of Matrix Metalloproteinases in Angiogenesis and Cancer. Front. Oncol. 2019;9:1370. doi: 10.3389/fonc.2019.01370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsu C.H., Peng K.L., Kang M.L., Chen Y.R., Yang Y.C., Tsai C.H., Chu C.S., Jeng Y.M., Chen Y.T., Lin F.M., et al. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep. 2012;2:568–579. doi: 10.1016/j.celrep.2012.08.030. [DOI] [PubMed] [Google Scholar]
- 56.Sun M., Song C.X., Huang H., Frankenberger C.A., Sankarasharma D., Gomes S., Chen P., Chen J., Chada K.K., He C., et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc. Natl. Acad. Sci. USA. 2013;110:9920–9925. doi: 10.1073/pnas.1305172110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu J., Li H., Shi M., Zhu Y., Ma Y., Zhong Y., Xiong C., Chen H., Peng C. TET1-mediated DNA hydroxymethylation activates inhibitors of the Wnt/β-catenin signaling pathway to suppress EMT in pancreatic tumor cells. J. Exp. Clin. Cancer Res. Cr. 2019;38:348. doi: 10.1186/s13046-019-1334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang S., Mo Q., Wang X. Oncological role of HMGA2 (Review) Int. J. Oncol. 2019;55:775–788. doi: 10.3892/ijo.2019.4856. [DOI] [PubMed] [Google Scholar]
- 59.Li M., Li X., Zhuang Y., Wang Y., Burow M.E., Collins-Burow B., Xue M., Song C., Shan B. Induction of HOXA9 expression in three-dimensional organotypic culture of the Claudin-low breast cancer cells. Oncotarget. 2016;7:51503–51514. doi: 10.18632/oncotarget.10491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Novak P., Jensen T., Oshiro M.M., Wozniak R.J., Nouzova M., Watts G.S., Klimecki W.T., Kim C., Futscher B.W. Epigenetic inactivation of the HOXA gene cluster in breast cancer. Cancer Res. 2006;66:10664–10670. doi: 10.1158/0008-5472.CAN-06-2761. [DOI] [PubMed] [Google Scholar]
- 61.Massagué J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu S., Jin K., Hui Y., Fu J., Jie C., Feng S., Reisman D., Wang Q., Fan D., Sukumar S., et al. HOXB7 promotes malignant progression by activating the TGFβ signaling pathway. Cancer Res. 2015;75:709–719. doi: 10.1158/0008-5472.CAN-14-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu H., Zhang M., Xu S., Zhang J., Zou J., Yang C., Zhang Y., Gong C., Kai Y., Li Y. HOXC8 promotes proliferation and migration through transcriptional up-regulation of TGFβ1 in non-small cell lung cancer. Oncogenesis. 2018;7:1. doi: 10.1038/s41389-017-0016-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nüchel J., Ghatak S., Zuk A.V., Illerhaus A., Mörgelin M., Schönborn K., Blumbach K., Wickström S.A., Krieg T., Sengle G., et al. TGFB1 is secreted through an unconventional pathway dependent on the autophagic machinery and cytoskeletal regulators. Autophagy. 2018;14:465–486. doi: 10.1080/15548627.2017.1422850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Akhurst R.J., Hata A. Targeting the TGFβ signalling pathway in disease. Nat. Rev. Drug Discov. 2012;11:790–811. doi: 10.1038/nrd3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun J.-P., Ge Q.-X., Ren Z., Sun X.-F., Xie S.-P. Down-regulation of HOXB5 inhibits TGF-β-induced migration and invasion in hepatocellular carcinoma cells via inactivation of the PI3K/Akt pathway. RSC Adv. 2018;8:41415–41421. doi: 10.1039/C8RA06860G. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Costanza B., Umelo I.A., Bellier J., Castronovo V., Turtoi A. Stromal Modulators of TGF-β in Cancer. J. Clin. Med. 2017;6:7. doi: 10.3390/jcm6010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Škovierová H., Okajčeková T., Strnádel J., Vidomanová E., Halašová E. Molecular regulation of epithelial-to-mesenchymal transition in tumorigenesis (Review) Int. J. Mol. Med. 2018;41:1187–1200. doi: 10.3892/ijmm.2017.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jiang W.G., Sanders A.J., Katoh M., Ungefroren H., Gieseler F., Prince M., Thompson S.K., Zollo M., Spano D., Dhawan P., et al. Tissue invasion and metastasis: Molecular, biological and clinical perspectives. Semin. Cancer Biol. 2015;35:S244–S275. doi: 10.1016/j.semcancer.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 70.Gheldof A., Berx G. Cadherins and epithelial-to-mesenchymal transition. Prog. Mol. Biol. Transl. Sci. 2013;116:317–336. doi: 10.1016/b978-0-12-394311-8.00014-5. [DOI] [PubMed] [Google Scholar]
- 71.Lin X., Shang X., Manorek G., Howell S.B. Regulation of the Epithelial-Mesenchymal Transition by Claudin-3 and Claudin-4. PLoS ONE. 2013;8:e67496. doi: 10.1371/journal.pone.0067496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mittal V. Epithelial Mesenchymal Transition in Tumor Metastasis. Annu. Rev. Pathol. 2018;13:395–412. doi: 10.1146/annurev-pathol-020117-043854. [DOI] [PubMed] [Google Scholar]
- 73.Chang Q., Zhang L., He C., Zhang B., Zhang J., Liu B., Zeng N., Zhu Z. HOXB9 induction of mesenchymal-to-epithelial transition in gastric carcinoma is negatively regulated by its hexapeptide motif. Oncotarget. 2015;6:42838–42853. doi: 10.18632/oncotarget.5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lv X., Li L., Lv L., Qu X., Jin S., Li K., Deng X., Cheng L., He H., Dong L. HOXD9 promotes epithelial-mesenchymal transition and cancer metastasis by ZEB1 regulation in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. Cr. 2015;34:133. doi: 10.1186/s13046-015-0245-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75.Saha S.S., Chowdhury R.R., Mondal N.R., Roy S., Sengupta S. Expression signatures of HOX cluster genes in cervical cancer pathogenesis: Impact of human papillomavirus type 16 oncoprotein E7. Oncotarget. 2017;8:36591–36602. doi: 10.18632/oncotarget.16619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu M., Xiao Y., Tang W., Li J., Hong L., Dai W., Zhang W., Peng Y., Wu X., Wang J., et al. HOXD9 promote epithelial-mesenchymal transition and metastasis in colorectal carcinoma. Cancer Med. 2020;9:3932–3943. doi: 10.1002/cam4.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Teo W.W., Merino V.F., Cho S., Korangath P., Liang X., Wu R.C., Neumann N.M., Ewald A.J., Sukumar S. HOXA5 determines cell fate transition and impedes tumor initiation and progression in breast cancer through regulation of E-cadherin and CD24. Oncogene. 2016;35:5539–5551. doi: 10.1038/onc.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu M., Zhan J., Zhang H. HOX family transcription factors: Related signaling pathways and post-translational modifications in cancer. Cell. Signal. 2020;66:109469. doi: 10.1016/j.cellsig.2019.109469. [DOI] [PubMed] [Google Scholar]
- 79.Li P.-D., Chen P., Peng X., Ma C., Zhang W.-J., Dai X.-F. HOXC6 predicts invasion and poor survival in hepatocellular carcinoma by driving epithelial-mesenchymal transition. Aging. 2018;10:115–130. doi: 10.18632/aging.101363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yoshida H., Broaddus R., Cheng W., Xie S., Naora H. Deregulation of the HOXA10 homeobox gene in endometrial carcinoma: Role in epithelial-mesenchymal transition. Cancer Res. 2006;66:889–897. doi: 10.1158/0008-5472.CAN-05-2828. [DOI] [PubMed] [Google Scholar]
- 81.Zhan J., Wang P., Li S., Song J., He H., Wang Y., Liu Z., Wang F., Bai H., Fang W., et al. HOXB13 networking with ABCG1/EZH2/Slug mediates metastasis and confers resistance to cisplatin in lung adenocarcinoma patients. Theranostics. 2019;9:2084–2099. doi: 10.7150/thno.29463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu X., Chen H., Parker B., Rubin E., Zhu T., Lee J.S., Argani P., Sukumar S. HOXB7, a homeodomain protein, is overexpressed in breast cancer and confers epithelial-mesenchymal transition. Cancer Res. 2006;66:9527–9534. doi: 10.1158/0008-5472.CAN-05-4470. [DOI] [PubMed] [Google Scholar]
- 83.Wang H., Liu G., Shen D., Ye H., Huang J., Jiao L., Sun Y. HOXA1 enhances the cell proliferation, invasion and metastasis of prostate cancer cells. Oncology Reports. Oncol. Rep. 2015;34:1203–1210. doi: 10.3892/or.2015.4085. [DOI] [PubMed] [Google Scholar]
- 84.Carrera M., Bitu C.C., de Oliveira C.E., Cervigne N.K., Graner E., Manninen A., Salo T., Coletta R.D. HOXA10 controls proliferation, migration and invasion in oral squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2015;8:3613–3623. [PMC free article] [PubMed] [Google Scholar]
- 85.Kim M., Suh D.H., Choi J.Y., Lee S., Bae J.R., Kim K., No J.H., Kim Y.B. Mutually exclusive antiproliferative effect of cell line-specific HOX inhibition in epithelial ovarian cancer cell lines: SKOV-3 vs RMUG-S. J. Cell. Mol. Med. 2020;24:3246–3251. doi: 10.1111/jcmm.14993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Duan R., Han L., Wang Q., Wei J., Chen L., Zhang J., Kang C., Wang L. HOXA13 is a potential GBM diagnostic marker and promotes glioma invasion by activating the Wnt and TGF-β pathways. Oncotarget. 2015;6:27778–27793. doi: 10.18632/oncotarget.4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shi Q., Shen L., Dong B., Fu H., Kang X., Dai L., Yang Y., Yan W., Chen K.N. Downregulation of HOXA13 sensitizes human esophageal squamous cell carcinoma to chemotherapy. Thorac. Cancer. 2018;9:836–846. doi: 10.1111/1759-7714.12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qin Z., Chen Z., Weng J., Li S., Rong Z., Zhou C. Elevated HOXA13 expression promotes the proliferation and metastasis of gastric cancer partly via activating Erk1/2. Oncotargets Ther. 2019;12:1803–1813. doi: 10.2147/OTT.S196986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee J.Y., Hur H., Yun H.J., Kim Y., Yang S., Kim S.I., Kim M.H. HOXB5 Promotes the Proliferation and Invasion of Breast Cancer Cells. IJBS. 2015;11:701–711. doi: 10.7150/ijbs.11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang B., Li N., Zhang H. Knockdown of homeobox B5 (HOXB5) inhibits cell proliferation, migration, and invasion in non-small cell lung cancer cells through inactivation of the Wnt/beta-catenin pathway. Oncol. Res. 2018;26:37–44. doi: 10.3727/096504017X14900530835262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang T., Lin F., Sun X., Jiang L., Mao R., Zhou S., Shang W., Bi R., Lu F., Li S. HOXB8 enhances the proliferation and metastasis of colorectal cancer cells by promoting EMT via STAT3 activation. Cancer Cell Int. 2019;19:3. doi: 10.1186/s12935-018-0717-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhan J., Wang P., Niu M., Wang Y., Zhu X., Guo Y., Zhang H. High expression of transcriptional factor HoxB9 predicts poor prognosis in patients with lung adenocarcinoma. Histopathology. 2015;66:955–965. doi: 10.1111/his.12585. [DOI] [PubMed] [Google Scholar]
- 93.Chiba N., Comaills V., Shiotani B., Takahashi F., Shimada T., Tajima K., Winokur D., Hayashida T., Willers H., Brachtel E., et al. Homeobox B9 induces epithelial-to-mesenchymal transition-associated radioresistance by accelerating DNA damage responses. Proc. Natl. Acad. Sci. USA. 2012;109:2760–2765. doi: 10.1073/pnas.1018867108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Peng Y., Li Y., Li Y., Wu A., Fan L., Huang W., Fu C., Deng Z., Wang K., Zhang Y., et al. HOXC10 promotes tumour metastasis by regulating the EMT-related gene Slug in ovarian cancer. Aging (Albany NY) 2020;12:19375–19398. doi: 10.18632/aging.103824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guo C., Hou J., Ao S., Deng X., Lyu G. HOXC10 upregulation promotes gastric cancer cell proliferation and metastasis through MAPK pathway. Chin. J. Cancer Res. 2017;29:572–580. doi: 10.21147/j.issn.1000-9604.2017.06.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang L., Tong D., Guo Q., Wang X., Wu F., Li Q., Yang J., Zhao L., Qin Y., Liu Y., et al. HOXD3 targeted by miR-203a suppresses cell metastasis and angiogenesis through VEGFR in human hepatocellular carcinoma cells. Sci. Rep. 2018;8:2431. doi: 10.1038/s41598-018-20859-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang J., Liu S., Zhang D., Ma Z., Sun L. Homeobox D10, a tumor suppressor, inhibits the proliferation and migration of esophageal squamous cell carcinoma. J. Cell Biochem. 2019;120:13717–13725. doi: 10.1002/jcb.28644. [DOI] [PubMed] [Google Scholar]
- 98.Tanzer A., Amemiya C.T., Kim C.B., Stadler P.F. Evolution of microRNAs located within Hox gene clusters. J. Exp. Zool. Part B Mol. Dev. Evol. 2005;304:75–85. doi: 10.1002/jez.b.21021. [DOI] [PubMed] [Google Scholar]
- 99.Yekta S., Tabin C.J., Bartel D.P. MicroRNAs in the Hox network: An apparent link to posterior prevalence. Nat. Rev. Genet. 2008;9:789–796. doi: 10.1038/nrg2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Weiss F.U., Marques I.J., Woltering J.M., Vlecken D.H., Aghdassi A., Partecke L.I., Heidecke C.D., Lerch M.M., Bagowski C.P. Retinoic acid receptor antagonists inhibit miR-10a expression and block metastatic behavior of pancreatic cancer. Gastroenterology. 2009;137:2136–2145.e7. doi: 10.1053/j.gastro.2009.08.065. [DOI] [PubMed] [Google Scholar]
- 101.Ohuchida K., Mizumoto K., Lin C., Yamaguchi H., Ohtsuka T., Sato N., Toma H., Nakamura M., Nagai E., Hashizume M., et al. MicroRNA-10a is overexpressed in human pancreatic cancer and involved in its invasiveness partially via suppression of the HOXA1 gene. Ann. Surg. Oncol. 2012;19:2394–2402. doi: 10.1245/s10434-012-2252-3. [DOI] [PubMed] [Google Scholar]
- 102.Xiao F., Bai Y., Chen Z., Li Y., Luo L., Huang J., Yang J., Liao H., Guo L. Downregulation of HOXA1 gene affects small cell lung cancer cell survival and chemoresistance under the regulation of miR-100. Eur. J. Cancer. 2014;50:1541–1554. doi: 10.1016/j.ejca.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 103.Ma L. Role of miR-10b in breast cancer metastasis. Breast Cancer Res. 2010;12:210. doi: 10.1186/bcr2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nakayama I., Shibazaki M., Yashima-Abo A., Miura F., Sugiyama T., Masuda T., Maesawa C. Loss of HOXD10 expression induced by upregulation of miR-10b accelerates the migration and invasion activities of ovarian cancer cells. Int. J. Oncol. 2013;43:63–71. doi: 10.3892/ijo.2013.1935. [DOI] [PubMed] [Google Scholar]
- 105.Li Q., Zhu F., Chen P. miR-7 and miR-218 epigenetically control tumor suppressor genes RASSF1A and Claudin-6 by targeting HoxB3 in breast cancer. Biochem. Biophys. Res. Commun. 2012;424:28–33. doi: 10.1016/j.bbrc.2012.06.028. [DOI] [PubMed] [Google Scholar]
- 106.Li Q., Ding C., Chen C., Zhang Z., Xiao H., Xie F., Lei L., Chen Y., Mao B., Jiang M., et al. miR-224 promotion of cell migration and invasion by targeting Homeobox D 10 gene in human hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2014;29:835–842. doi: 10.1111/jgh.12429. [DOI] [PubMed] [Google Scholar]
- 107.Stiegelbauer V., Vychytilova-Faltejskova P., Karbiener M., Pehserl A.M., Reicher A., Resel M., Heitzer E., Ivan C., Bullock M., Ling H., et al. miR-196b-5p Regulates Colorectal Cancer Cell Migration and Metastases through Interaction with HOXB7 and GALNT5. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017;23:5255–5266. doi: 10.1158/1078-0432.CCR-17-0023. [DOI] [PubMed] [Google Scholar]
- 108.Wang H., Wei H., Wang J., Li L., Chen A., Li Z. MicroRNA-181d-5p-Containing Exosomes Derived from CAFs Promote EMT by Regulating CDX2/HOXA5 in Breast Cancer. Mol. Nucleic Acids. 2020;6:654–667. doi: 10.1016/j.omtn.2019.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu X., Lu K., Wang K., Sun M., Zhang E., Yang J., Yin D., Liu Z., Zhou J., Liu Z., et al. MicroRNA-196a promotes non-small cell lung cancer cell proliferation and invasion through targeting HOXA5. BMC Cancer. 2012;12:348. doi: 10.1186/1471-2407-12-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zheng J., Ge P., Liu X., Wei J., Wu G., Li X. MiR-136 inhibits gastric cancer-specific peritoneal metastasis by targeting HOXC10. Tumour Biol. 2017;39 doi: 10.1177/1010428317706207. [DOI] [PubMed] [Google Scholar]
- 111.Reddy S.D., Ohshiro K., Rayala S.K., Kumar R. MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res. 2008;68:8195–8200. doi: 10.1158/0008-5472.CAN-08-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jiang M.C., Ni J.J., Cui W.Y., Wang B.Y., Zhuo W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am. J. Cancer Res. 2019;9:1354–1366. [PMC free article] [PubMed] [Google Scholar]
- 113.Botti G., De Chiara A., Di Bonito M., Cerrone M., Malzone M., Collina F., Cantile M. Noncoding RNAs within the HOX gene network in tumor pathogenesis and progression. J. Cell Physiol. 2018;234:395–413. doi: 10.1002/jcp.27036. [DOI] [PubMed] [Google Scholar]
- 114.Kogo R., Shimamura T., Mimori K., Kawahara K., Imoto S., Sudo T., Tanaka F., Shibata K., Suzuki A., Komune S., et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 115.Chisholm K.M., Wan Y., Li R., Montgomery K.D., Chang H.Y., West R.B. Detection of long non-coding RNA in archival tissue: Correlation with polycomb protein expression in primary and metastatic breast carcinoma. PLoS ONE. 2012;7:e47998. doi: 10.1371/journal.pone.0047998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Loewen G., Jayawickramarajah J., Zhuo Y., Shan B. Functions of lncRNA HOTAIR in lung cancer. J. Hematol. Oncol. 2014;7:90. doi: 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhao W.H., Yuan H.Y., Ren X.Y., Huang K., Guo Z.Y. Association between expression of HOTAIR and invasiveness of gliomas, and its predictive value. Adv. Clin. Exp. Med. Off. Organ Wroc. Med Univ. 2019;28:1179–1183. doi: 10.17219/acem/99527. [DOI] [PubMed] [Google Scholar]
- 118.Tan S.K., Pastori C., Penas C., Komotar R.J., Ivan M.E., Wahlestedt C., Ayad N.G. Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol. Cancer. 2018;17:74. doi: 10.1186/s12943-018-0822-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Song Y., Wang R., Li L.W., Liu X., Wang Y.F., Wang Q.X., Zhang Q. Long non-coding RNA HOTAIR mediates the switching of histone H3 lysine 27 acetylation to methylation to promote epithelial-to-mesenchymal transition in gastric cancer. Int. J. Oncol. 2019;54:77–86. doi: 10.3892/ijo.2018.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim H.J., Lee D.W., Yim G.W., Nam E.J., Kim S., Kim S.W., Kim Y.T. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int. J. Oncol. 2015;46:521–530. doi: 10.3892/ijo.2014.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li E., Zhao Z., Ma B., Zhang J. Long noncoding RNA HOTAIR promotes the proliferation and metastasis of osteosarcoma cells through the AKT/mTOR signaling pathway. Exp. Ther. Med. 2017;14:5321–5328. doi: 10.3892/etm.2017.5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Angelopoulou E., Paudel Y.N., Piperi C. Critical role of HOX transcript antisense intergenic RNA (HOTAIR) in gliomas. J. Mol. Med. (Berl.) 2020;98:1525–1546. doi: 10.1007/s00109-020-01984-x. [DOI] [PubMed] [Google Scholar]
- 124.Xia M., Yao L., Zhang Q., Wang F., Mei H., Guo X., Huang W. Long noncoding RNA HOTAIR promotes metastasis of renal cell carcinoma by up-regulating histone H3K27 demethylase JMJD3. Oncotarget. 2017;8:19795–19802. doi: 10.18632/oncotarget.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Abbastabar M., Sarfi M., Golestani A., Khalili E. lncRNA involvement in hepatocellular carcinoma metastasis and prognosis. EXCLI J. 2018;17:900–913. doi: 10.17179/excli2018-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang Y., Gong G., Xu J., Zhang Y., Wu S., Wang S. Long noncoding RNA HOTAIR promotes breast cancer development by targeting ZEB1 via sponging miR-601. Cancer Cell Int. 2020;20:320. doi: 10.1186/s12935-020-01410-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chang L., Guo R., Yuan Z., Shi H., Zhang D. LncRNA HOTAIR Regulates CCND1 and CCND2 Expression by Sponging miR-206 in Ovarian Cancer. Cell Physiol. Biochem. 2018;49:1289–1303. doi: 10.1159/000493408. [DOI] [PubMed] [Google Scholar]
- 128.Wang A.H., Tan P., Zhuang Y., Zhang X.T., Yu Z.B., Li L.N. Down-regulation of long non-coding RNA HOTAIR inhibits invasion and migration of oesophageal cancer cells via up-regulation of microRNA-204. J. Cell. Mol. Med. 2019;23:6595–6610. doi: 10.1111/jcmm.14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lu M., Qin X., Zhou Y., Li G., Liu Z., Yue H., Geng X. LncRNA HOTAIR suppresses cell apoptosis, autophagy and induces cell proliferation in cholangiocarcinoma by modulating the miR-204-5p/HMGB1 axis. Biomed. Pharm. 2020;130:110566. doi: 10.1016/j.biopha.2020.110566. [DOI] [PubMed] [Google Scholar]
- 130.Dong X., He X., Guan A., Huang W., Jia H., Huang Y., Chen S., Zhang Z., Gao J., Wang H. Long non-coding RNA Hotair promotes gastric cancer progression via miR-217-GPC5 axis. Life Sci. 2019;217:271–282. doi: 10.1016/j.lfs.2018.12.024. [DOI] [PubMed] [Google Scholar]
- 131.Wei Z., Chen L., Meng L., Han W., Huang L., Xu A. LncRNA HOTAIR promotes the growth and metastasis of gastric cancer by sponging miR-1277-5p and upregulating COL5A1. Gastric Cancer. 2020;23:1018–1032. doi: 10.1007/s10120-020-01091-3. [DOI] [PubMed] [Google Scholar]
- 132.Xun J., Wang C., Yao J., Gao B., Zhang L. Long Non-Coding RNA HOTAIR Modulates KLF12 to Regulate Gastric Cancer Progression via PI3K/ATK Signaling Pathway by Sponging miR-618. Onco Targets. 2019;12:10323–10334. doi: 10.2147/OTT.S223957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bie L., Luo S., Li D., Wei Y., Mu Y., Chen X., Wang S., Guo P., Lu X. HOTAIR Competitively Binds MiRNA330 as a Molecular Sponge to Increase the Resistance of Gastric Cancer to Trastuzumab. Curr. Cancer Drug Targets. 2020;20:700–709. doi: 10.2174/1568009620666200504114000. [DOI] [PubMed] [Google Scholar]
- 134.Xu J., Gong L., Qian Z., Song G., Liu J. ERBB4 promotes the proliferation of gastric cancer cells via the PI3K/Akt signaling pathway. Oncol. Rep. 2018;39:2892–2898. doi: 10.3892/or.2018.6343. [DOI] [PubMed] [Google Scholar]
- 135.Gong X., Zhu Z. Long Noncoding RNA HOTAIR Contributes to Progression in Hepatocellular Carcinoma by Sponging miR-217-5p. Cancer Biother. Radiopharm. 2020;35:387–396. doi: 10.1089/cbr.2019.3070. [DOI] [PubMed] [Google Scholar]
- 136.Yang T., He X., Chen A., Tan K., Du X. LncRNA HOTAIR contributes to the malignancy of hepatocellular carcinoma by enhancing epithelial-mesenchymal transition via sponging miR-23b-3p from ZEB1. Gene Dev. 2018;670:114–122. doi: 10.1016/j.gene.2018.05.061. [DOI] [PubMed] [Google Scholar]
- 137.Zhang C., Xu L., Deng G., Ding Y., Bi K., Jin H., Shu J., Yang J., Deng H., Wang Z., et al. Exosomal HOTAIR promotes proliferation, migration and invasion of lung cancer by sponging miR-203. Sci. China Life Sci. 2020;63:1265–1268. doi: 10.1007/s11427-019-1579-x. [DOI] [PubMed] [Google Scholar]
- 138.Zhang J.-J., Zhou X.H., Zhou Y., Wang Y.G., Qian B.Z., He A., Shen Z., Hu H.Y., Yao Y. Bufalin suppresses the migration and invasion of prostate cancer cells through HOTAIR, the sponge of miR-520b. Acta Pharm. Sin. 2019;40:1228–1236. doi: 10.1038/s41401-019-0234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang N., Jiang Y., Lv S., Wen H., Wu D., Wei Q., Dang Q. HOTAIR expands the population of prostatic cancer stem-like cells and causes Docetaxel resistance via activating STAT3 signaling. Aging (Albany NY) 2020;12:12771–12782. doi: 10.18632/aging.103188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yu G.J., Sun Y., Zhang D.W., Zhang P. Long non-coding RNA HOTAIR functions as a competitive endogenous RNA to regulate PRAF2 expression by sponging miR-326 in cutaneous squamous cell carcinoma. Cancer Cell Int. 2019;19:270. doi: 10.1186/s12935-019-0992-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang W., Wu D., He X., Hu X., Hu C., Shen Z., Lin J., Pan Z., He Z., Lin H., et al. CCL18-induced HOTAIR upregulation promotes malignant progression in esophageal squamous cell carcinoma through the miR-130a-5p-ZEB1 axis. Cancer Lett. 2019;460:18–28. doi: 10.1016/j.canlet.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 142.Li T., Qin Y., Zhen Z., Shen H., Cong T., Schiferle E., Xiao S. Long non-coding RNA HOTAIR/microRNA-206 sponge regulates STC2 and further influences cell biological functions in head and neck squamous cell carcinoma. Cell Prolif. 2019;52:e12651. doi: 10.1111/cpr.12651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tao D., Zhang Z., Liu X., Zhang Z., Fu Y., Zhang P., Yuan H., Liu L., Cheng J., Jiang H. LncRNA HOTAIR promotes the invasion and metastasis of oral squamous cell carcinoma through metastasis-associated gene 2. Mol. Carcinog. 2020;59:353–364. doi: 10.1002/mc.23159. [DOI] [PubMed] [Google Scholar]
- 144.Wu J., Li M., Zhang Y. Long noncoding RNA HOXA-AS2 regulates the expression of SCN3A by sponging miR-106a in breast cancer. J. Cell Biochem. 2019;120:14465–14475. doi: 10.1002/jcb.28706. [DOI] [PubMed] [Google Scholar]
- 145.Fang Y., Wang J., Wu F., Song Y., Zhao S., Zhang Q. Long Non-Coding Rna Hoxa-As2 Promot. Prolif. Invasion Breast Cancer By Act. A Mir-520c-3p Sponge. Oncotarget. 2017;8:46090–46103. doi: 10.18632/oncotarget.17552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tong G., Wu X., Cheng B., Li L., Li X., Li Z., Nong Q., Chen X., Liu Y., Wang S. Knockdown of HOXA-AS2 suppresses proliferation and induces apoptosis in colorectal cancer. Am. J. Transl. Res. 2017;9:4545–4552. [PMC free article] [PubMed] [Google Scholar]
- 147.Ding J., Xie M., Lian Y., Zhu Y., Peng P., Wang J., Wang L., Wang K. Long noncoding RNA HOXA-AS2 represses P21 and KLF2 expression transcription by binding with EZH2, LSD1 in colorectal cancer. Oncogenesis. 2017;6:e288. doi: 10.1038/oncsis.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Xie M., Sun M., Zhu Y.N., Xia R., Liu Y.W., Ding J., Ma H.W., He X.Z., Zhang Z.H., Liu Z.J., et al. Long noncoding RNA HOXA-AS2 promotes gastric cancer proliferation by epigenetically silencing P21/PLK3/DDIT3 expression. Oncotarget. 2015;6:33587–33601. doi: 10.18632/oncotarget.5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang F., Wu D., Chen J., Chen S., He F., Fu H., Wu Q., Liu S., Li X., Wang W. Long non-coding RNA HOXA-AS2 promotes the migration, invasion and stemness of bladder cancer via regulating miR-125b/Smad2 axis. Exp. Cell Res. 2019;375:1–10. doi: 10.1016/j.yexcr.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 150.Zhang Y., Xu J., Zhang S., An J., Zhang J., Huang J., Jin Y. HOXA-AS2 Promotes Proliferation and Induces Epithelial-Mesenchymal Transition via the miR-520c-3p/GPC3 Axis in Hepatocellular Carcinoma. Cell Physiol. Biochem. 2018;50:2124–2138. doi: 10.1159/000495056. [DOI] [PubMed] [Google Scholar]
- 151.Xia F., Chen Y., Jiang B., Du X., Peng Y., Wang W., Huang W., Feng T., Li X. Long Noncoding RNA HOXA-AS2 Promotes Papillary Thyroid Cancer Progression by Regulating miR-520c-3p/S100A4 Pathway. Cell Physiol. Biochem. 2018;50:1659–1672. doi: 10.1159/000494786. [DOI] [PubMed] [Google Scholar]
- 152.Zhang P., Cao P., Zhu X., Pan M., Zhong K., He R., Li Y., Jiao X., Gao Y. Upregulation of long non-coding RNA HOXA-AS2 promotes proliferation and induces epithelial-mesenchymal transition in gallbladder carcinoma. Oncotarget. 2017;8:33137–33143. doi: 10.18632/oncotarget.16561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zheng F.X., Wang X.Q., Zheng W.X., Zhao J. Long noncoding RNA HOXA-AS2 promotes cell migration and invasion via upregulating IGF-2 in non-small cell lung cancer as an oncogene. Eur. Rev. Med. Pharm. Sci. 2019;23:4793–4799. doi: 10.26355/eurrev_201906_18064. [DOI] [PubMed] [Google Scholar]
- 154.Liu Y., Lin X., Zhou S., Zhang P., Shao G., Yang Z. Long noncoding RNA HOXA-AS2 promotes non-small cell lung cancer progression by regulating miR-520a-3p. Biosci. Rep. 2019;39:BSR20190283. doi: 10.1042/BSR20190283. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 155.Cui T.J., Lin G.S., Dai Y.M., Zheng J.P., Chen Z., Chen Q., Zheng Y., Lin X. LncRNA HOXA-AS2 regulates microRNA-216a-5p to promote malignant progression of non-small cell lung cancer. Eur. Rev. Med. Pharm. Sci. 2019;23:264–273. doi: 10.26355/eurrev_201908_18656. [DOI] [PubMed] [Google Scholar]
- 156.Wang Y., Zhang R., Cheng G., Xu R., Han X. Long non-coding RNA HOXA-AS2 promotes migration and invasion by acting as a ceRNA of miR-520c-3p in osteosarcoma cells. Cell Cycle. 2018;17:1637–1648. doi: 10.1080/15384101.2018.1489174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gao Y., Yu H., Liu Y., Liu X., Zheng J., Ma J., Gong W., Chen J., Zhao L., Tian Y., et al. Long Non-Coding RNA HOXA-AS2 Regulates Malignant Glioma Behaviors and Vasculogenic Mimicry Formation via the MiR-373/EGFR Axis. Cell Physiol. Biochem. 2018;45:131–147. doi: 10.1159/000486253. [DOI] [PubMed] [Google Scholar]
- 158.Zhao H., Zhang X., Frazão J.B., Condino-Neto A., Newburger P.E. HOX antisense lincRNA HOXA-AS2 is an apoptosis repressor in all trans retinoic acid treated NB4 promyelocytic leukemia cells. J. Cell. Biochem. 2013;114:2375–2383. doi: 10.1002/jcb.24586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhao Q., Zhao S., Li J., Zhang H., Qian C., Wang H., Liu J. TCF7L2 activated HOXA-AS2 decreased the glucocorticoid sensitivity in acute lymphoblastic leukemia through regulating HOXA3/EGFR/Ras/Raf/MEK/ERK pathway. Biomed. Pharm. 2019;109:1640–1649. doi: 10.1016/j.biopha.2018.10.046. [DOI] [PubMed] [Google Scholar]
- 160.Zhao Y., Wang W., Guan C., Hu Z., Liu L., Li W., Jiang X. Long noncoding RNA HOTAIRM1 in human cancers. Clin. Chim. Acta. 2020;511:255–259. doi: 10.1016/j.cca.2020.10.011. [DOI] [PubMed] [Google Scholar]
- 161.Ghafouri-Fard S., Dashti S., Taheri M. The HOTTIP (HOXA transcript at the distal tip) lncRNA: Review of oncogenic roles in human. Biomed. Pharmacother. 2020;127:110158. doi: 10.1016/j.biopha.2020.110158. [DOI] [PubMed] [Google Scholar]
- 162.Zheng L., Chen J., Zhou Z., He Z. Knockdown of long non-coding RNA HOXD-AS1 inhibits gastric cancer cell growth via inactivating the JAK2/STAT3 pathway. Tumor. Biol. 2017;39:1010428317705335. doi: 10.1177/1010428317705335. [DOI] [PubMed] [Google Scholar]
- 163.Zhang Y., Dun Y., Zhou S., Huang X.H. LncRNA HOXD-AS1 promotes epithelial ovarian cancer cells proliferation and invasion by targeting miR-133a-3p and activating Wnt/β-catenin signaling pathway. Biomed. Pharm. 2017;96:1216–1221. doi: 10.1016/j.biopha.2017.11.096. [DOI] [PubMed] [Google Scholar]
- 164.Guo X., Chen Z., Zhao L., Cheng D., Song W., Zhang X. Long non-coding RNA-HAGLR suppressed tumor growth of lung adenocarcinoma through epigenetically silencing E2F1. Exp. Cell Res. 2019;382:111461. doi: 10.1016/j.yexcr.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 165.Chen S., Li K. HOXD-AS1 facilitates cell migration and invasion as an oncogenic lncRNA by competitively binding to miR-877-3p and upregulating FGF2 in human cervical cancer. BMC Cancer. 2020;20:924. doi: 10.1186/s12885-020-07441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chi C., Mao M., Shen Z., Chen Y., Chen J., Hou W. HOXD-AS1 Exerts Oncogenic Functions and Promotes Chemoresistance in Cisplatin-Resistant Cervical Cancer Cells. Hum. Gene. 2018;29:1438–1448. doi: 10.1089/hum.2017.256. [DOI] [PubMed] [Google Scholar]
- 167.Ye Y., Yang S., Han Y., Sun J., Xv L., Wu L., Ming L. HOXD-AS1 confers cisplatin resistance in gastric cancer through epigenetically silencing PDCD4 via recruiting EZH2. Open Biol. 2019;9:190068. doi: 10.1098/rsob.190068. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 168.Yan F., Ma Y., Liu L., Li L., Deng J., Sun J. Long Noncoding RNA HOXD-AS1 Promotes the Proliferation, Migration, and Invasion of Colorectal Cancer via the miR-526b-3p/CCND1 Axis. J. Surg. Res. 2020;255:525–535. doi: 10.1016/j.jss.2020.05.078. [DOI] [PubMed] [Google Scholar]
- 169.Li Y., Han X., Li Q., Wang C., Lou Z., Wang X. Long noncoding RNA HOXD-AS1 induces epithelial-mesenchymal transition in breast cancer by acting as a competing endogenous RNA of miR-421. J. Cell Biochem. 2019;120:10633–10642. doi: 10.1002/jcb.28353. [DOI] [PubMed] [Google Scholar]
- 170.Dong S., Wang R., Wang H., Ding Q., Zhou X., Wang J., Zhang K., Long Y., Lu S., Hong T., et al. HOXD-AS1 promotes the epithelial to mesenchymal transition of ovarian cancer cells by regulating miR-186-5p and PIK3R3. J. Exp. Clin. Cancer Res. 2019;38:110. doi: 10.1186/s13046-019-1103-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ji W., Wang Q., Yang J. LncRNA HOXD-AS1 promotes the metastasis of human hepatocellular carcinoma via modulating miR-326/SLC27A4. Cancer Cell Int. 2020;20:161. doi: 10.1186/s12935-020-01217-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sun J., Guo Y., Bie B., Zhu M., Tian H., Tian J., Li J., Yang Y., Ji F., Kong G., et al. Silencing of long noncoding RNA HOXD-AS1 inhibits proliferation, cell cycle progression, migration and invasion of hepatocellular carcinoma cells through MEK/ERK pathway. J. Cell Biochem. 2020;121:443–457. doi: 10.1002/jcb.29206. [DOI] [PubMed] [Google Scholar]
- 173.Xia H., Jing H., Li Y., Lv X. Long noncoding RNA HOXD-AS1 promotes non-small cell lung cancer migration and invasion through regulating miR-133b/MMP9 axis. Biomed. Pharm. 2018;106:156–162. doi: 10.1016/j.biopha.2018.06.073. [DOI] [PubMed] [Google Scholar]