Abstract
The diagnosis of primary ciliary dyskinesia (PCD) relies on clinical features and sophisticated studies. The detection of bi-allelic disease-causing variants confirms the diagnosis. However, a standardised genetic panel is not widely available and new disease-causing genes are continuously identified.
To assess the accuracy of untargeted whole-exome sequencing (WES) as a diagnostic tool for PCD, patients with symptoms highly suggestive of PCD were consecutively included. Patients underwent measurement of nasal nitric oxide (nNO) levels, ciliary transmission electron microscopy analysis (TEM) and WES. A confirmed PCD diagnosis in symptomatic patients was defined as a recognised ciliary ultrastructural defect on TEM and/or two pathogenic variants in a known PCD-causing gene.
Forty-eight patients (46% male) were enrolled, with a median age of 10.0 years (range 1.0–37 years). In 36 patients (75%) a diagnosis of PCD was confirmed, of which 14 (39%) patients had normal TEM. A standalone untargeted WES had a diagnostic yield of 94%, identifying bi-allelic variants in 11 known PCD-causing genes in 34 subjects. A nNO<77 nL·min was nonspecific when including patients younger than 5 years (area under the receiver operating characteristic curve (AUC) 0.75, 95% CI 0.60–0.90). Consecutive WES considerably improved the diagnostic accuracy of nNO in young children (AUC 0.97, 95% CI 0.93–1). Finally, WES established an alternative diagnosis in four patients.
In patients with clinically suspected PCD and low nNO levels, WES is a simple, beneficial and accurate next step to confirm the diagnosis of PCD or suggest an alternative diagnosis, especially in preschool-aged children in whom nNO is less specific.
Short abstract
Untargeted whole-exome sequencing in subjects with clinical symptoms highly suggestive of PCD has an excellent diagnostic accuracy and, as prices drop, may be the genetic test of choice for confirming PCD or establishing an alternative diagnosis https://bit.ly/3j2jMbu
Introduction
Primary ciliary dyskinesia (PCD) is a rare, predominantly autosomal recessive genetic disorder, caused by defects in ciliary structure and function. Mutations in PCD-associated genes can lead to impaired ciliary motility, or paucity of cilia, resulting in impaired mucociliary clearance and progressive lung disease [1, 2]. Historically, the gold standard for the diagnosis of PCD had been evidence of an ultrastructural ciliary defect as seen on transmission electron microscopy (TEM) [3]. However, due to high false-negative rates, alternative diagnostic tests were evaluated in the last decade [4–9]. Guidelines for the diagnosis of PCD have recently been published by the American Thoracic Society (ATS) [10] and the European Respiratory Society (ERS) [11]. These include the use of diagnostic modalities such as scrape or brush biopsies for ciliary TEM analysis, high-speed video microscopy with ciliary beat frequency, and the use of nasal nitric oxide (nNO) measurements. These modalities require expertise and each has its limitations [11–14]. Recent advances and availability of genetic diagnostic tools, including the availability of commercial gene panels, can help confirm the diagnosis of PCD, which allows early diagnosis, family planning, genotype–phenotype correlation and possibly individualised treatment in the future. Indeed, the ATS recommends the use of an extended genetic panel for PCD diagnosis in patients fulfilling clinical criteria, with low nNO, superseding the use of TEM [15]. This recommendation is based on expert opinion, as the different panels have not been evaluated in prospective studies. Untargeted whole-exome sequencing (WES) is an efficient and increasingly economical genetic analysis method, which is not limited to a panel of targeted pathogenic variants or known disease-related genes. It is increasingly available to most medical centres and can provide extensive genetic information, which may help in confirming PCD diagnosis or provide alternate diagnoses [16]. The aim of this current study is to assess the diagnostic yield of WES, incorporated into the evaluation of patients with high clinical suspicion of PCD, in a real-life clinical setting.
Methods
The study was approved by the Institutional Review Board, and consent was obtained from patients or their legal guardians. A more detailed explanation of the methods is provided in the online supplement.
Patients
Individuals with a chronic sinopulmonary disease evaluated between 2012 and 2019 in whom cystic fibrosis and immune deficiency were ruled out, were prospectively recruited for this study. Patients were included if they consented to perform WES and had a very high clinical suspicion for PCD: Term born with chronic sinopulmonary symptoms since early childhood and one or more of the following criteria: 1) unexplained bronchiectasis; 2) a condition associated with PCD (situs inversus totalis or any heterotaxic syndrome); and 3) a history of otherwise unexplained neonatal respiratory distress. Using these clinical criteria, we expected a pre-evaluation sensitivity of >70% for a PCD diagnosis in our cohort [5, 17].
Data and sample collection
All subjects underwent detailed clinical assessment, nNO measurements and TEM from nasal brush biopsy, if not previously performed.
In cooperative patients, nNO sampling was performed with palate closure manoeuvre [18]. In noncooperative patients, nNO was measured during tidal breathing [19]. Both nNO production values of 30 nL·min−1 (90 ppb for uncooperating subjects) and 77 nL·min−1 (233 ppb for uncooperating subjects) have been recommended as cut-off values for evaluation of PCD and were thus both assessed in this study [14, 20].
A nasal brush biopsy was performed under local anaesthesia or during a clinically indicated bronchoscopy and was immediately fixed in glutaraldehyde for TEM analysis. TEM was analysed by a pathologist experienced in the diagnosis of PCD. Ciliary ultrastructure was described as either “hallmark” abnormal, abnormal but suspected secondary ciliary defect [21], inadequate, inconclusive, or normal [10, 11].
Whole-exome analysis
Following DNA extraction from whole blood, exonic sequences were enriched with the SureSelect Human All Exon 50 Mb V5 kit (Agilent Technologies, Santa Clara, CA, USA). Sequences were generated on a HiSeq2500 (Illumina, San Diego, CA, USA) as 125-bp paired-end runs. Read alignment and variant calling were performed with DNAnexus (Palo Alto, CA, USA) with the human genome assembly hg19 (GRCh37) as reference. Data analysis was performed using an in-house bioinformatics pipeline, exomes were covered at above 50× depth.
Definitions
A positive diagnosis [11], was defined as: 1) the presence of a typical clinical phenotype plus a recognised ciliary ultrastructural defect or 2) a typical clinical phenotype and the presence of two pathogenic variants in a known gene causing PCD.
A negative diagnosis was reported in subjects if: 1) an alternative diagnosis was established or 2) all laboratory diagnostic studies were not consistent with PCD.
An unresolved diagnosis was reported in patients with a typical clinical phenotype if nNO levels were low and ciliary ultrastructure was normal or inconclusive and no pathogenic variants in a ciliary gene were found.
Statistical analysis
Proportional differences were assessed using the Chi-squared and Fisher's exact tests for nominal variables and t-test and the Mann–Whitney U-test for continuous variables. The diagnostic accuracy of each laboratory test was determined based on the final diagnosis. Patients with an unresolved diagnosis were not excluded but considered as a negative diagnosis for PCD.
Sensitivity, specificity, positive and negative predictive values and area under the receiver operating characteristic curve were calculated for each diagnostic evaluation as well as a combined evaluation of low nNO levels and WES. All analyses were performed using STATA 15.1 (Stata Corp, TX, USA).
Results
Study population
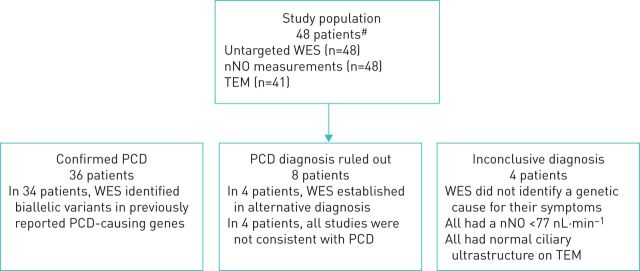
Forty-eight subjects were evaluated in the study (46% male, median age of 10.0 years, range: 1–37 years). Of the 48 subjects included in the study, 36 (75%) were diagnosed with definite PCD, in 4 (14%) patients the diagnosis was inconclusive and in 8 (22%) patients PCD was ruled out (figure 1). Additional patient characteristics are presented in table 1.
FIGURE 1.
Study population and the results of whole-exome sequencing (WES) analysis. nNO: nasal nitric oxide; TEM: transmission electron microscopy; PCD: primary ciliary dyskinesia. #: meeting inclusion criteria (see main text).
TABLE 1.
Characteristics of patients evaluated for primary ciliary dyskinesia
| All patients | Positive | Negative | Inconclusive | |
| Subjects | 48 | 36 | 8 | 4 |
| Age at assessment years median (range) | 10.0 (1–37) | 12.1 (1.0–37) | 9.6 (1–23) | 6.5 (4–10) |
| 0–5 years | 13 (27%) | 7 (21%) | 4 (50%) | 2 (50%) |
| 5–18 years | 27 (56%) | 22 (61%) | 3 (38%) | 2 (50%) |
| >18 years | 8 (17%) | 7 (21%) | 1 (13%) | 0 (0%) |
| Male | 21 (44%) | 14 (39%) | 5 (63%) | 2 (50%) |
| Consanguinity | 34 (71%) | 32 (76%) | 2 (50%) | 4 (100%) |
| Ethnicity | ||||
| Jewish | 13 (27%) | 7 (19%) | 4 (50%) | 2 (50%) |
| Arabic | 35 (73%) | 29 (81%) | 4 (50%) | 2 (50%) |
| Other | ||||
| Bronchiectasis | 46 (96%) | 34 (94%) | 7 (88%) | 4 (100%) |
| Associated conditions# | 11 (23%) | 10 (28%) | 1 (13%) | 0 (0%) |
| Neonatal respiratory distress | 15 (31%) | 13 (36%) | 0 (-%) | 2 (50%) |
| Diagnostic assessment performed | ||||
| nNO measurement | 48 (100%) | 36 (100%) | 8 (100%) | 4 (100%) |
| TEM for ciliary structure | 41 (85%) | 31 (86%) | 6 (75%) | 4 (100%) |
nNO: nasal nitric oxide; TEM: transmission electron microscopy. #: situs abnormalities or retinitis pigmentosa.
Patients diagnosed with PCD
Of the 36 individuals diagnosed with PCD, WES identified bi-allelic variants in 34 individuals across 11 previously reported motile cilia genes (tables 2 and E1) (94% diagnostic yield). The specific variants identified in these motile cilia genes are presented in table E1. In one adult patient with a classical PCD phenotype (bronchiectasis, low nNO levels and missing dynein arms on TEM) WES identified two likely pathogenic variants in an axonemal cilia gene (DNAH10), predicted to cause PCD [22] but not previously reported to cause disease. As per our definitions for the purpose of assessment of the diagnostic accuracy of WES (and due to lack of functional studies), this was considered a false negative. WES did not identify any pathogenic variants in one patient with a classic PCD phenotype (chronic purulent otitis media, bronchiectasis, low nNO and positive TEM with absent dynein arms).
Genotype–phenotype
In terms of genotype–phenotype association, patients with variants in LLRC6 and LLRC50 genes had a high prevalence of situs abnormalities, history of neonatal respiratory distress and were associated with both outer and inner dynein arm defects on TEM. Patients with variants in HYDIN, RSPH4A and RSPH9 genes had normal TEM and no situs abnormalities. Other patient characteristics and associated genetic variants are presented in table 2.
TABLE 2.
Primary ciliary dyskinesia causing genes in our cohort and the related prevalence of situs abnormalities and neonatal distress, and associated findings by transmission electron microscopy (TEM) and nasal nitric oxide (nNO) measurements
| Gene (n=34) | Locus | Situs abnormalities | Neonatal respiratory distress | TEM | nNO ppb mean (range) |
| DNAAF3 (n=2) | 19q13 | 0 | 1 | ODA+IDA | 46 (32–60) |
| DNAH11 (n=9) | 7p15–21 | 1 | 3 | Normal | 41.8 (10–79) |
| DNAI1 (n=4) | 9p21–p13 | 1 | 1 | Normal/short ODA | 34.5 (20–70) |
| DNAI2 (n=3) | 17q25.1 | 1 | 2 | Normal/orientation/ODA+ IDA | 41.3 (30–47) |
| DYX1C1 (DNAAF4) (n=1) | 15q21 | 1 | 0 | Short ODA+IDA | 55 |
| HYDIN (n=3) | 16q22 | 0 | 1 | Normal | 59.6 (19–90) |
| LRRC50 (DNAAF1) (n=2) | 16q24 | 2 | 2 | ODA+IDA | 65 |
| LRRC6 (n=6) | 8q24 | 4 | 2 | ODA+IDA | 81.9 (12–328) |
| MCIDAS (n=2) | 5q11 | 0 | 0 | Absence of ciliary cells | 57 (54–60) |
| RSPH4A (n=1) | 6q22 | 0 | 0 | Normal | 38 |
| RSPH9 (n=1) | 6p21 | 0 | 1 | Normal | 48 |
ODA: outer dynein arm; IDA: inner dynein arm.
Patients with an alternative diagnosis
In four subjects, WES established an alternative diagnosis. One subject was diagnosed with immune deficiency due to an autosomal dominant STAT1 gain-of-function variant (OMIM, 614162). TEM, in this patient, initially showed dynein arm and central pair defects, but a repeated nasal brush biopsy demonstrated normal ciliary structure (possibly reflecting a secondary defect observed by the initial TEM). Two additional individuals, from one large consanguineous family, were diagnosed with plasminogen deficiency (OMIM, 217090, PLG gene) confirmed by repeated low plasma plasminogen levels. These individuals had low nNO levels (<30 nL·min−1) on multiple occasions and recurrent inadequate samples for TEM evaluation. In a fourth individual (suffering from situs inversus and mild respiratory symptoms), WES revealed two pathogenic variants in WDR16, previously described to cause laterality disorders without evidence of ciliary dysfunction [23].
Patients with an inconclusive diagnosis
In four subjects (8.3%, mean age 6.5 years, range: 4–10 years), a final diagnosis could not be reached. These four subjects all had a nNO level <77 nL·min−1 and normal ciliary ultrastructure on TEM. WES did not identify a genetic cause for their symptoms. Thus, for the purpose of the diagnostic evaluation of WES, results in these subjects were considered as true negative.
However, according to current ATS criteria [10] these subjects might be diagnosed as patients with PCD. Thus, an additional evaluation was performed considering WES results in these patients as false negative (see below).
Accuracy of the diagnostic evaluation
As a standalone test, WES had the highest accuracy with an AUC of 0.97 (95% CI 0.93–1), followed by nNO<30 nL·min−1 (90 ppb) with AUC of 0.84 (95% CI 0.71–0.98). The sensitivity and specificity of WES for the diagnosis of PCD were 94.4% (95% CI 81.3–99.3) and 100% (95% CI 73.5–100), respectively (table 3).
TABLE 3.
Diagnostic accuracy of nasal nitric oxide (nNO), transmission electron microscopy (TEM) and genetic evaluation for diagnosis of primary ciliary dyskinesia
| nNO <77 nL·min−1 | nNO <30 nL·min−1 | TEM | WES | |
| All subjects, including unresolved diagnosis (n=57) | 48 | 48 | 41 | 48 |
| Sensitivity (95% CI) | 100 (89.1–100) | 93.8 (79.2–99.2) | 62.2 (44.8–77.5) | 94.4 (81.3–99.3) |
| Specificity (95% CI) | 50.0 (21.1–78.9) | 75.0 (42.8–94.5) | 85.7 (57.2–98.2) | 100 (73.5–100) |
| ROC area (95% CI) | 0.75 (0.60–0.90) | 0.84 (0.71–0.98) | 0.74 (0.62–0.86) | 0.97 (0.93–1.0) |
WES: untargeted whole-exome sequencing; ROC: receiver operating characteristic.
In view of current ATS criteria, suggesting a diagnosis of PCD in symptomatic patients and low nNO levels, with no need of auxiliary testing, the diagnostic accuracy of WES according to current ATS criteria was also evaluated. This showed a reduced diagnostic accuracy with an AUC of 0.91 (95% CI 0.85–0.98) due to a reduced sensitivity of 83.3% (95% CI 67.2–93.6).
WES was also assessed as an auxiliary test following nNO. The added value of WES was particularly evident in young subjects (≤5 year), in whom nNO<77 nL·min−1 (233 ppb) alone had an AUC of 0.67 (95% CI 0.46–0.87), while the combination of nNO<77 nL·min−1 and consecutive WES yielded an AUC of 0.97 (95% CI 0.93–1.00) (table 4).
TABLE 4.
Diagnostic accuracy of nasal nitric oxide (nNO)
| nNO <77 nL·min−1 | nNO <77 nL·min−1 and WES | |||
| ≤5 years | >5 years | All | ||
| All subjects, including unresolved diagnosis (n=48) | ||||
| Sensitivity (95% CI) | 100 (47.8–100) | 100 (87.2–100) | 100 (89.1–100) | 93.8 (79.2–99.2) |
| Specificity (95% CI) | 33.3 (4.3–77.7) | 66.7 (22.3–95.7) | 50.0 (21.1–78.9) | 100 (73.5–100) |
| ROC area (95% CI) | 0.67 (0.46–0.87) | 0.83 (0.63–1.00) | 0.75 (0.60–0.90) | 0.97 (0.93–1.0) |
| Subjects with unresolved diagnosis excluded (n=40) | ||||
| Sensitivity (95% CI) | 100 (47.8–100) | 100 (87.2–100) | 100 (89.1–100) | 93.8 (79.2–99.2) |
| Specificity (95% CI) | 50.0 (6.8–93.2) | 100 (39.8–100) | 75 (34.9–96.8) | 100 (63.1–100) |
| ROC area (95% CI) | 0.75 (0.47–1.0) | 1.0 | 0.88 (0.72–1.0) | 0.97 (0.93–1.0) |
WES: untargeted whole-exome sequencing; ROC: receive operating characteristic.
Excluding patients with unresolved diagnosis from the analysis (as they may be regarded as true negative or false negative [10]) considerably improved the diagnostic abilities of nNO, especially in individuals >5 years of age (table 4).
Cost comparison WES versus commercially available genetic panel for PCD
The current cost of WES is estimated at USD 1000, with clinical WES prices ranging between USD 500–5000 [24], depending on insurance participation.
The cost of a commercial genetic panel for PCD is estimated at USD 300 (personal communication). The use of WES in our study group yielded a genetic diagnosis in 39 subjects (PCD and non-PCD). Theoretically, a commonly used specific commercial genetic PCD panel [25] for these 48 subjects would have cost 3.3-times less but would have resulted in genetic diagnosis in 31 subjects (1.25-times less). Naturally, custom-made panels are also available, which may increase the diagnostic yield as well as the cost.
Discussion
In this study, we evaluated, in a real-life setting, the yield of untargeted WES as a diagnostic tool for PCD. Our findings demonstrate that untargeted WES in subjects with clinical symptoms highly suggestive of PCD has excellent sensitivity and specificity for diagnosis. Furthermore, untargeted WES allows identification of new PCD-causing genes, novel variants in known PCD genes, and diagnosis of alternative genetic disorders. We propose that WES should be the genetic diagnostic tool of choice in subjects with high clinical suspicion of PCD and low nNO levels and or abnormal/equivocal results using other available diagnostic modalities.
PCD is a rare condition with a prevalence of 1:10 000–20 000, which has nonspecific signs and symptoms shared with many other conditions, and thus requires evaluation in centres with expertise in its diagnosis and treatment. The diagnosis is complicated by the fact that the tests employed in the diagnostic process of PCD are cumbersome, expensive, require expertise not universally available, and there is no single test that can be considered a gold standard for diagnosis. Bearing these hurdles in mind, our study validates current expert opinion-based guidelines [20, 26], incorporating a combination of clinical history, nNO measurement and WES as a highly accurate pathway in establishing a diagnosis of PCD.
Though the genetic approach to PCD diagnosis has developed in recent years, the evidence for its use, as a diagnostic tool is still scarce. Therefore, most guidelines, both American and European recommend it as an auxiliary tool to confirm a PCD diagnosis [10, 27, 28]. Over 40 PCD-causing genes have been identified to date, but in only 65–75% of the patients, a genetic diagnosis is reached using traditional tools [17, 29]. The genotype–phenotype association has been described [30, 31], providing a clinical rationale to achieve a genetic diagnosis, especially with current progression towards precision medicine.
The use of WES as part of the routine diagnostic pathway in patients with suspected PCD has not been reported previously. Several studies evaluated the ability of targeted and untargeted WES to detect pathogenic variants in patients previously diagnosed with PCD, with an overall yield of 50–76% [29, 32–34]. Our prospective evaluation, using an untargeted WES approach in patients with clinical symptoms highly suggestive of PCD, provided a genetic diagnosis in 34 out of 36 individuals with a confirmed PCD diagnosis (94% yield).
In a heterogeneous disease, such as PCD with the constant discovery of new causative genes, the use of WES is practical. Furthermore, WES is becoming increasingly affordable, it does not require expertise present only in PCD centres and the data generated may be repeatedly accessed as genetic databases expand, further improving its future diagnostic yield. Indeed, in three individuals in the current study, only a repeated assessment of undiagnostic WES (as genetic databases improved in power and precision) allowed us to establish a genetic diagnosis.
At least eight (14%) subjects in this study benefited from the unbiased WES approach, as opposed to a PCD-targeted genetic panel testing: in four individuals WES provided an alternative diagnosis, and in four other subjects, pathogenic variants in PCD-causing genes not included in a commercially available panel were identified [25]. Still, when evaluating the cost of WES compared to the current PCD-targeted genetic panel, WES was three times more expensive, while genetically diagnosing an additional 25% patients. Therefore, our data suggest that until prices of WES are reduced, it would be cost-effective to perform WES only after receiving nondiagnostic PCD-targeted genetic panel results. Importantly, the financial burden of misdiagnosing patients should also be considered, as this often leads to unnecessary diagnostic tests and treatments, as was recently reported [35].
The nNO is consistently low in cooperative older patients with PCD (>5 years old) [6], with a sensitivity and specify >95% for the diagnosis of PCD, outperforming TEM or TEM with focused genetic testing [14]. Even in infants <1 year of age, tidal breathing nNO has been shown to have a diagnostic value [36] but is not routinely recommended for use <5 years of age due to low specificity. In line with the aforementioned studies, we have found excellent predictive values using 77 or 30 nL·min−1 cut-offs for PCD diagnosis. The nNO in our cohort was both feasible and useful in supporting the clinical diagnosis of PCD. Nonetheless, caution should be exercised using nNO as the final standalone test to diagnose PCD, due to possible false-positive results as exemplified in recent reports [37, 38] as well as in the current study. This is particularly true when evaluating children <5 years in whom low nNO values are nonspecific. In the young patients in our cohort, the addition of WES increased the diagnostic yield significantly, as compared to standalone nNO measurements. An early definitive genetic diagnosis allows disease-directed treatment at a specialised clinic, which may potentially improve long-term outcomes [39]. Furthermore, it provides the family with a genetic cause for their child's symptoms, the option of family planning, and in the future may direct personalised treatment. Finally, it should be noted that our cohort did not include patients with genotypes associated with higher levels of nNO [40], potentially lowering the false-negative predictive value of high nNO in other patient populations, and thus additionally adding to the usefulness of WES.
Four individuals with symptoms suggestive of PCD and low nNO levels underwent WES without a definitive genetic diagnosis. This may be the result of mutations in intronic regions, not covered by WES or due to novel unidentified PCD-causing genes. Moreover, mutations may have been missed due to the calling algorithms used for analysis. Future re-analysis using improved algorithms and use of exome trios may allow the identification of additional disease-causing mutations [41].
Some limitations of this study should be noted. First, in view of our strict inclusion criteria, the population in which the WES was assessed had a very high pre-evaluation likelihood for a PCD diagnosis [17]. Thus, our results are relevant for a specific population of patients with high clinical suspicion for PCD or patients with suggestive symptoms and low nNO levels and may not reflect patients with mild symptoms undergoing routine evaluation for chronic respiratory symptoms. Secondly, as there is no single gold-standard test for diagnosis of PCD, in the current study (similar to other studies on this rare disorder) WES results were used as part of the diagnostic pathway. However, only genetic mutations that would have been reported by appropriate genetic panels, were regarded as true positive. This difficulty was previously discussed by the 2016 ERS diagnostic guidelines concluding that since no single test rules out PCD, a diagnostic modality can be evaluated even though it is used in the diagnostic pathway by comparing the accuracy of the test with the patient's final outcome [11]. Thirdly, the specific population in which the diagnostic modality was assessed. The Hadassah Medical Center, is a regional referral centre for chronic pulmonary conditions specialising in PCD diagnosis. The population basis is 1.5 million residents of the larger Jerusalem area and its surroundings. The Arab Muslim residents, who have a high degree of consanguinity, comprise ∼25% of this population but represent 73% of our cohort. Thus, the predictive value of our approach should be tested in other populations.
In conclusion, we have shown that in patients with a highly suggestive clinical history, WES has a very high accuracy and is beneficial for PCD diagnosis, especially in young children. Therefore, as prices of WES drop and expertise increases, consecutive testing of WES following low nNO levels or in patients with highly suggestive symptoms, provides an objective, practical and definitive tool for diagnosing patients with PCD.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00213-2020.SUPPLEMENT (144.3KB, pdf)
Footnotes
This article has supplementary material available from openres.ersjournals.com.
Conflict of interest: A. Gileles-Hillel has nothing to disclose.
Conflict of interest: H. Mor-Shaked has nothing to disclose.
Conflict of interest: D. Shoseyov has nothing to disclose.
Conflict of interest: J. Reiter has nothing to disclose.
Conflict of interest: R. Tsabari has nothing to disclose.
Conflict of interest: A. Hevroni has nothing to disclose.
Conflict of interest: M. Cohen-Cymberknoh has nothing to disclose.
Conflict of interest: I. Amirav has nothing to disclose.
Conflict of interest: S. Brammli-Greenberg has nothing to disclose.
Conflict of interest: A. Horani has nothing to disclose.
Conflict of interest: E. Kerem has nothing to disclose.
Conflict of interest: O. Breuer has nothing to disclose.
Support statement: The study was funded by the Israeli Chief Scientist grant no. 3–9907. Funding support for research was provided to A. Gileles-Hillel by the Israeli Science Foundation grant no. 2779/19. Funding support for research was provided to A. Horani by US NIH-NHLBI grant K08HL150223. Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Noone PG, Leigh MW, Sannuti A, et al. . Primary ciliary dyskinesia: diagnostic and phenotypic features. Am J Respir Crit Care Med 2004; 169: 459–467. doi: 10.1164/rccm.200303-365OC [DOI] [PubMed] [Google Scholar]
- 2.Mullowney T, Manson D, Kim R, et al. . Primary ciliary dyskinesia and neonatal respiratory distress. Pediatrics 2014; 134: 1160–1166. doi: 10.1542/peds.2014-0808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Afzelius BA. A human syndrome caused by immotile cilia. Science 1976; 193: 317–319. doi: 10.1126/science.1084576 [DOI] [PubMed] [Google Scholar]
- 4.Jackson CL, Behan L, Collins SA, et al. . Accuracy of diagnostic testing in primary ciliary dyskinesia. Eur Respir J 2016; 47: 837–848. doi: 10.1183/13993003.00749-2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behan L, Dimitrov BD, Kuehni CE, et al. . PICADAR: a diagnostic predictive tool for primary ciliary dyskinesia. Eur Respir J 2016; 47: 1103–1112. doi: 10.1183/13993003.01551-2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leigh MW, Hazucha MJ, Chawla KK, et al. . Standardizing nasal nitric oxide measurement as a test for primary ciliary dyskinesia. Ann Am Thorac Soc 2013; 10: 574–581. doi: 10.1513/AnnalsATS.201305-110OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stannard WA, Chilvers MA, Rutman AR, et al. . Diagnostic testing of patients suspected of primary ciliary dyskinesia. Am J Respir Crit Care Med 2010; 181: 307–314. doi: 10.1164/rccm.200903-0459OC [DOI] [PubMed] [Google Scholar]
- 8.Shoemark A, Dixon M, Corrin B, et al. . Twenty-year review of quantitative transmission electron microscopy for the diagnosis of primary ciliary dyskinesia. J Clin Pathol 2012; 65: 267–271. doi: 10.1136/jclinpath-2011-200415 [DOI] [PubMed] [Google Scholar]
- 9.Pifferi M, Montemurro F, Cangiotti AM, et al. . Simplified cell culture method for the diagnosis of atypical primary ciliary dyskinesia. Thorax 2009; 64: 1077–1081. doi: 10.1136/thx.2008.110940 [DOI] [PubMed] [Google Scholar]
- 10.Shapiro AJ, Davis SD, Polineni D, et al. . Diagnosis of primary ciliary dyskinesia. An official American Thoracic Society clinical practice guideline. Am J Respir Crit Care Med 2018; 197: e24–e39. doi: 10.1164/rccm.201805-0819ST [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lucas JS, Barbato A, Collins SA. European Respiratory Society guidelines for the diagnosis of primary ciliary dyskinesia. Eur Respir J 2016; 49: 1601090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shoemark A, Frost E, Dixon M, et al. . Accuracy of immunofluorescence in the diagnosis of primary ciliary dyskinesia. Am J Respir Crit Care Med 2017; 196: 94–101. doi: 10.1164/rccm.201607-1351OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shapiro AJ, Leigh MW. Value of transmission electron microscopy for primary ciliary dyskinesia diagnosis in the era of molecular medicine: genetic defects with normal and non-diagnostic ciliary ultrastructure. Ultrastruct Pathol 2017; 41: 373–385. doi: 10.1080/01913123.2017.1362088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shapiro AJ, Josephson M, Rosenfeld M, et al. . Accuracy of nasal nitric oxide measurement as a diagnostic test for primary ciliary dyskinesia. A systematic review and meta-analysis. Ann Am Thorac Soc 2017; 14: 1184–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shapiro AJ, Zariwala MA, Ferkol T, et al. . Diagnosis, monitoring, and treatment of primary ciliary dyskinesia: PCD foundation consensus recommendations based on state of the art review. Pediatr Pulmonol 2016; 51: 115–132. doi: 10.1002/ppul.23304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petersen BS, Fredrich B, Hoeppner MP, et al. . Opportunities and challenges of whole-genome and -exome sequencing. BMC Genet 2017; 18: 14. doi: 10.1186/s12863-017-0479-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leigh MW, Ferkol TW, Davis SD, et al. . Clinical features and associated likelihood of primary ciliary dyskinesia in children and adolescents. Ann Am Thorac Soc 2016; 13: 1305–1313. doi: 10.1513/AnnalsATS.201511-748OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.American Thoracic Society, European Respiratory Society ATS/ERS recommendations for standardized procedures for the online and offline measurement of exhaled lower respiratory nitric oxide and nasal nitric oxide, 2005. Am J Respir Crit Care Med 2005; 171: 912–930. doi: 10.1164/rccm.200406-710ST [DOI] [PubMed] [Google Scholar]
- 19.Mateos-Corral D, Coombs R, Grasemann H, et al. . Diagnostic value of nasal nitric oxide measured with non-velum closure techniques for children with primary ciliary dyskinesia. J Pediatr 2011; 159: 420–424. doi: 10.1016/j.jpeds.2011.03.007 [DOI] [PubMed] [Google Scholar]
- 20.O'Connor MG, Griffiths A, Iyer NP, et al. . Summary for clinicians: diagnosis of primary ciliary dyskinesia. Ann Am Thorac Soc 2019; 16: 171–174. doi: 10.1513/AnnalsATS.201810-693CME [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dixon M, Shoemark A. Secondary defects detected by transmission electron microscopy in primary ciliary dyskinesia diagnostics. Ultrastruct Pathol 2017; 41: 390–398. doi: 10.1080/01913123.2017.1365990 [DOI] [PubMed] [Google Scholar]
- 22.Pazour GJ, Agrin N, Walker BL, et al. . Identification of predicted human outer dynein arm genes: candidates for primary ciliary dyskinesia genes. J Med Genet 2006; 43: 62–73. doi: 10.1136/jmg.2005.033001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ta-Shma A, Perles Z, Yaacov B, et al. . A human laterality disorder associated with a homozygous WDR16 deletion. Eur J Hum Genet 2015; 23: 1262–1265. doi: 10.1038/ejhg.2014.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarze K, Buchanan J, Taylor JC, et al. . Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet Med 2018; 20: 1122–1130. doi: 10.1038/gim.2017.247 [DOI] [PubMed] [Google Scholar]
- 25.Invitae Primary Ciliary Dyskinesia Panel. www.invitae.com/en/physician/tests/04101/#info-panel-resourcespp Date last accessed: February 21, 2020. Date last updated: August 2020.
- 26.Shoemark A, Dell S, Shapiro A, et al. . ERS and ATS diagnostic guidelines for primary ciliary dyskinesia: similarities and differences in approach to diagnosis. Eur Respir J 2019; 54: 1901066. doi: 10.1183/13993003.01066-2019 [DOI] [PubMed] [Google Scholar]
- 27.Kuehni CE, Lucas JS. Diagnosis of primary ciliary dyskinesia: summary of the ERS Task Force report. Breathe (Sheff) 2017; 13: 166–178. doi: 10.1183/20734735.008517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lucas JS, Barbato A, Collins SA, et al. . European Respiratory Society guidelines for the diagnosis of primary ciliary dyskinesia. Eur Respir J 2017; 49: 1601090. doi: 10.1183/13993003.01090-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fassad MR, Patel MP, Shoemark A, et al. . Clinical utility of NGS diagnosis and disease stratification in a multiethnic primary ciliary dyskinesia cohort. J Med Genet 2019; 5: 322. [DOI] [PubMed] [Google Scholar]
- 30.Best S, Shoemark A, Rubbo B, et al. . Risk factors for situs defects and congenital heart disease in primary ciliary dyskinesia. Thorax 2019; 74: 203–205. doi: 10.1136/thoraxjnl-2018-212104 [DOI] [PubMed] [Google Scholar]
- 31.Davis SD, Ferkol TW, Rosenfeld M, et al. . Clinical features of childhood primary ciliary dyskinesia by genotype and ultrastructural phenotype. Am J Respir Crit Care Med 2015; 191: 316–324. doi: 10.1164/rccm.201409-1672OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andjelkovic M, Minic P, Vreca M, et al. . Genomic profiling supports the diagnosis of primary ciliary dyskinesia and reveals novel candidate genes and genetic variants. PLoS ONE 2018; 13: e0205422. doi: 10.1371/journal.pone.0205422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marshall CR, Scherer SW, Zariwala MA, et al. . Whole-exome sequencing and targeted copy number analysis in primary ciliary dyskinesia. G3 (Bethesda) 2015; 5: 1775–1781. doi: 10.1534/g3.115.019851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emiralioglu N, Taskiran EZ, Kosukcu C, et al. . Genotype and phenotype evaluation of patients with primary ciliary dyskinesia: first results from Turkey. Pediatr Pulmonol 2019; 55: 383–393. doi: 10.1002/ppul.24583 [DOI] [PubMed] [Google Scholar]
- 35.Kouis P, Papatheodorou SI, Middleton N, et al. . Cost-effectiveness analysis of three algorithms for diagnosing primary ciliary dyskinesia: a simulation study. Orphanet J Rare Dis 2019; 14: 142. doi: 10.1186/s13023-019-1116-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams PS, Tian X, Zahid M, et al. . Establishing normative nasal nitric oxide values in infants. Respir Med 2015; 109: 1126–1130. doi: 10.1016/j.rmed.2015.07.010 [DOI] [PubMed] [Google Scholar]
- 37.Lavie M, Amirav I. In defense of high-speed video microscopy in evaluating patients with suspected primary ciliary dyskinesia. Am J Respir Crit Care Med 2019; 200: 1181–1183. doi: 10.1164/rccm.201904-0773LE [DOI] [PubMed] [Google Scholar]
- 38.Shapiro AJ, Davis SD, Leigh MW, et al. . Limitations of nasal nitric oxide testing in primary ciliary dyskinesia. Am J Respir Crit Care Med 2020; 202: 476–477. doi: 10.1164/rccm.202003-0835LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen-Cymberknoh M, Weigert N, Gileles-Hillel A, et al. . Clinical impact of Pseudomonas aeruginosa colonization in patients with primary ciliary dyskinesia. Respir Med 2017; 131: 241–246. doi: 10.1016/j.rmed.2017.08.028 [DOI] [PubMed] [Google Scholar]
- 40.Knowles MR, Ostrowski LE, Leigh MW, et al. . Mutations in RSPH1 cause primary ciliary dyskinesia with a unique clinical and ciliary phenotype. Am J Respir Crit Care Med 2014; 189: 707–717. doi: 10.1164/rccm.201311-2047OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bustamante-Marin XM, Horani A, Stoyanova M, et al. . Mutation of CFAP57 causes primary ciliary dyskinesia by disrupting the asymmetric targeting of a subset of ciliary inner dynein arms. PLoS Genet 2020; 8: 1008691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00213-2020.SUPPLEMENT (144.3KB, pdf)