Abstract
Muscle atrophy is a significant moderator for disease prognosis; as such, interventions to mitigate disuse-induced muscle loss are imperative to improve clinical interventions. Mitochondrial deteriorations may underlie disuse-induced myopathies; therefore, improving mitochondrial quality may be an enticing therapeutic intervention. However, different mitochondria-based treatments may have divergent impacts on the prognosis of disuse atrophy. Therefore, the purpose of this study was to investigate different mitochondria-centered interventions during disuse atrophy in hindlimb unloaded male and female mice. Male and female mice overexpressing peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) or mitochondrially targeted catalase (MCAT) and their respective wild-type (WT) littermate controls were hindlimb unloaded for 7 days to induce disuse atrophy or allowed normal ambulatory activity (cage control; CON). After designated interventions, animals were euthanized, and tissues were collected for measures of mitochondrial quality control and protein turnover. Although PGC-1α overexpression mitigated ubiquitin-proteasome activation (MuRF1 and Atrogin mRNA content), this did not correspond to phenotypic protections from disuse-induced atrophy. Rather, PGC-1α mice appeared to have a greater reliance on autophagic protein breakdown compared with WT mice. In MCAT mice, females exhibited a mitigated response to disuse atrophy; however, this effect was not noted in males. Despite these phenotypic differences, there were no clear cellular signaling differences between MCAT hindlimb unloaded females and MCAT fully loaded females. PGC-1α overexpression does not protect against phenotypic alterations during disuse atrophy but appears to shift catabolic pathways moderating atrophy. However, increased mitochondrially targeted catalase activity appears to blunt disuse atrophy within highly oxidative muscles specifically in female mice.
NEW & NOTEWORTHY We present data suggesting that mitochondria-based interventions may mitigate disuse atrophy. However, the efficacy of mitochondria-based interventions may vary depending on the specific target of the intervention and the sex of the organism. Females appear to be more responsive to increased mitochondrial catalase as a potential therapeutic for mitigating disuse atrophy.
Keywords: catalase, females, mitochondria, muscle atrophy, PGC1α
INTRODUCTION
Skeletal muscle atrophy is a significant comorbidity associated with a variety of pathologies such as cancer, HIV/AIDs, chronic heart failure, renal failure, and intensive care unit (ICU) hospital stays, which also appears to coincide with elevated mortality (18, 32, 68a, 55). Although it is well understood that muscle wasting across these pathologies is detrimental to patient outcomes, effective treatments to mitigate muscle loss remain elusive. This may be due in part to divergent mechanisms contributing to muscle loss, with some muscle pathologies appearing to be more inflammation dependent (e.g., cancer cachexia) (4, 44, 63, 70), whereas others appear to be inflammation independent (such as disuse atrophy) (19). However, one commonality across the various muscle atrophies is the development of mitochondrial aberrations such as increased reactive oxygen species production, decreased ATP production, and altered mitochondrial quality control (7, 10, 51, 69). Specifically, many of these mitochondrial alterations occur with disuse and appear to correspond with muscle atrophy (60, 28, 65a, 50a, 44a). This relationship between mitochondrial stress and muscle size suggests that improving mitochondrial quality may be a therapeutic target to mitigate disuse-induced muscle loss.
Mitochondrial quality comprises multiple features such as ATP production, respiratory control ratio, mitochondrial biogenesis, mitochondrial dynamics, mitochondrial autophagy (i.e., mitophagy), and translation of mitochondrially encoded proteins (54, 71). Deteriorations in any one of these aspects may influence mitochondrial quality and subsequent muscle health. Recent studies have investigated the sufficiency of increasing aspects of mitochondrial quality as treatments for muscle wasting, specifically disuse muscle atrophy (12, 13, 67). Prior studies have found increasing mitochondrial quality control and subsequent content using a transgenic model (peroxisome proliferator-activated receptor gamma coactivator 1-alpha, PGC1α) sufficient to maintain the cross-sectional area of muscle fibers during disuse (12, 13, 67). This may suggest that increasing mitochondrial content and presumably mitochondrial quality control may be sufficient to protect muscle against disuse atrophy. However, this has yet to result in meaningful interventions, perhaps suggesting that there is more to be understood with regard to PGC1α and muscle size. In addition to alterations in mitochondrial content and ATP production, other studies have suggested excessive reactive oxygen species (ROS) production may be the initiating factor in the development of muscle wasting (10), and some data suggest that mitigation of ROS may at least partially protect against disuse-induced muscle loss in animal models (39, 42, 49, 59). However, a direct comparison of different mitochondria-related treatments for disuse muscle atrophy has yet to be described in the scientific literature.
Many studies investigating potential treatments for disuse atrophy have predominantly used male organisms (12, 13, 27, 39, 65, 67). This discrepancy in representation between sexes in preclinical research may partially account for a lack of effective therapeutics in clinical studies. Specific to disuse atrophy, females appear to exhibit differing clinical outcomes compared with males, with females appearing to be more prone to the development of ICU-associated muscle weakness (16, 34). These data may suggest differences in the development and/or molecular underpinnings of disuse muscle atrophy and imply that potential therapeutic options should at least be investigated in both male and female models.
Therefore, the purpose of this study was to investigate the sufficiency of targeting multiple quality control mechanisms in a mouse model of disuse atrophy in both males and females to attenuate atrophy. We hypothesized that mitochondria-based interventions would assuage muscle loss in both male and female mice. However, we did not pre hoc hypothesize if one intervention would be superior to another, nor did we statistically compare differences between males and females. To test our primary hypothesis, we investigated if genetic overexpression of PGC-1α (increasing various components of mitochondrial quality control as well as mitochondrial content) or mitochondrially targeted catalase (MCAT; increasing ROS handling capacities specifically at the mitochondria) would provide protections against disuse atrophy in male and female mice.
METHODS
Animal Interventions
Breeding/genotyping.
All animal protocols were approved by the University of Arkansas Institutional Animal Care and Use Committee (Protocol Nos. 17022 and 17089). Breeder mice for both PGC (muscle specific overexpression of PGC1-α through the creatine kinase promotor) and MCAT (global over expression of mitochondrially targeted catalase) colonies were purchased from Jackson Laboratories (Bar Harbor, ME, Stock Nos. 008231 and 016197, respectively). Three weeks after birth, mice were genotyped for either PGC or MCAT overexpression through PCR as we have previously described (20) and using recommended Jackson Laboratories-suggested genotyping protocols (21a). After genotyping, animals were allocated to either CON or HU conditions (described in Experimental design). Animals were allotted to groups to distribute animals across all groups as much as possible to avoid potential litter-cohort differences. Groups had n = 3–10 depending on colony and group. Sample sizes for each group are listed in figure captions.
Experimental design.
At 8 wk of age, animals underwent transfection with pMitoTimer in the flexor digitorum brevis (FDB) muscle, as we have previously described (detailed in pMitoTimer Electroporation and Analysis) (10). After 2 wk of recovery, disuse atrophy was induced at 10 wk of age using the hindlimb unloading model as previously described (23, 31, 68). Animals’ tails were first sterilized using ethanol wipes and iodine swab sticks. After which, tails were coated with benzoin solution to increase the adhesiveness of the animals’ tail. Athletic tape was then gently wrapped around the animals’ tails, and a specialized hook was connected to the athletic tape. A fishing string was then wrapped around the hook and looped through a custom-designed pulley system to allow for unloading of the hindlimb legs and animal mobility around a specially designed cage. Animals had full use of forelimbs and were able to move around the cage, whereas hindlimbs remained slightly elevated above the cage and were not able to touch the ground. Daily monitoring ensured unloading of hindlimbs as well as no signs of tail necrosis or distress. Animals were hindlimb unloaded for 7 days to induce disuse atrophy. CON animals were allowed to freely move about their cages. This length of disuse has been shown to induce disuse atrophy in both male and female models (5, 12, 23, 24, 26, 29, 33, 36, 41, 56, 61, 64, 73). After disuse or control interventions, animals were anesthetized with 2% isoflurane gas, and tissues were collected for further analysis. Euthanasia was completed by cervical dislocation while animals were still anesthetized with isoflurane. All harvests were completed late morning or early afternoon. Animals were not fasted before tissue harvest. Upon tissue harvest, we noted a mean lower tibial length across all HU groups, with some reaching or approaching statistical significance (male PGCs: 17.16 ± 0.06 mm vs. 17.02 ± 0.02 mm; female PGCs: 16.54 ± 0.09 mm vs. 16.44 ± 0.08 mm; male MCATs: 16.90 ± 0.07 mm vs. 16.72 ± 0.08 mm; female MCATs: 16.77 ± 0.07 mm vs. 16.40 ± 0.06 mm). As such, tibial length was not an appropriate normalizing factor for tissue weights. More so, because the global body mass is expected to change as a result of hindlimb unloading, we did not normalize tissue weights to body mass and have instead opted to present raw tissue weights for this study. Between genotypes, there did not appear to be any inherent differences in muscle mass/tibia lengths.
pMitoTimer Electroporation and Analysis
Briefly, animals were anesthetized with 2% isoflurane mixed with oxygen. Upon anesthesia, 10 µL of 0.36 mg/mL of hyaluronidase dissolved in sterile saline was injected into the right FDB of each mouse. One hour after the hyaluronidase injection, animals were reanesthetized and the right FDBs were injected with 20 µg of pMitoTimer (Addgene, Watertown, MA, Cat. No. 52659) dissolved in 10 µL of sterile saline. After 15 min of recovery, the FDB underwent electroporation with 10 pulses at 75 V/cm, 1 Hz, and 20 ms/pulse. pMitoTimer analysis was completed as previously described (10). At tissue harvest, FDBs were removed and fixed in 4% paraformaldehyde (PFA):phosphate-buffered saline (PBS) solution for 20 min and then rinsed in PBS for 5 min. After washing in PBS, FDBs were carefully spread flat on gelatin-coated microscope slides using forceps. FDBs were mounted with 10% glycerol:PBS solution and a standard microscope cover plate. FDBs were imaged with FITC and TRITC filters using carefully controlled exposure times as previously described (10). FITC and TRITC images were analyzed using Cell Profiler and MATLAB (generous gift of Dr. Z. Yan) as previously described. Images were quantified for red/green ratio and red puncta, measuring mitochondrial oxidative stress (greater red fluorescent indicative of greater mitochondrial stress) and degenerated mitochondria (red fluorescent signal without any green, suggestive of completely degenerated mitochondria tagged for mitophagy). Approximately three images were measured per animal with outcome variables averaged within each animal.
Histological Analysis
During tissue harvest, plantaris muscles were frozen in optimal cutting temperature (OCT) medium. Later, 10-µm cross sections of plantaris muscles were cut with a Leica CM1860 (Leica Biosystems, Buffalo Grove, IL) cryostat microtome and stained for hematoxylin and eosin (H&E) staining as previously described (9). The muscle fiber cross-sectional area (CSA) was measured with Nikon Imaging software. A trained researcher hand-circled ∼100 fibers per animal, fiber areas were then averaged within each animal, and inferential statistics were performed on average CSAs. All circled fibers were free of freezer damage and were clear cross-sectional images of the fiber. All researchers analyzing CSAs were blinded to groups.
mRNA Analysis
mRNA from the gastrocnemius muscle (PGC and MCAT colonies) and soleus muscle (MCAT) were isolated and reverse-transcribed to cDNA using commercial kits as previously described (20, 53). The gastrocnemius muscle was chosen as a mixed fiber muscle (3) that would allow for investigation of disuse atrophy in a muscle that experiences disuse atrophy, yet it is not the most susceptible muscle to disuse atrophy. The soleus was used based on phenotypic evidence, suggesting protections to soleus muscle mass in female MCAT mice (more thoroughly described in the RESULTS section). Taqman probes or primers were used as appropriate to amplify cDNA for genes related to protein turnover and mitochondrial quality control as previously described (20, 53). cDNA was quantified using the −ΔΔCt method and a QuantStudio3 PCR instrument (Thermo Fisher Scientific, Waltham, MA) similar to previously described (20, 53). SYBR primers included Pparα, Mfn1, Mfn2, Opa1, Drp1, Fis1, Mff, and Bnip3 [primer pairs have previously been reported (20)]. Taqman probes included 18s (Clone No. Mm03928990_g1), Pgc1α (Clone No. Mm01208835_m1), Lc3 (Clone No. Mm00458725_g1), Redd1 (Clone No. Mm00512504_g1), Deptor (Clone No. Mm01195339_m1), Catalase (Clone No. Mm00437992_m1), Atrogin (Clone No. Mm00499523_m1), Murf1 (Clone No. Mm01185221_m1), Pax7 (Clone No. Mm01354484_m1), and p62 (Clone No. Mm00448091_m1). All samples were normalized to 18 s, which did not differ between groups.
Immunoblotting
Immunoblotting of proteins related to muscle protein turnover and mitochondrial quality control was measured as we have previously described (20, 53). Immunoblots were imaged with the Li-Cor Odyssey Fc System (Lincoln, NE) and quantified with Image Studio Software (Li-Cor, Lincoln, NE). Antibodies included PGC-1α (Novus Biologicals, NBP1–04676SS), p62 (Cell Signaling, 5114s), and LC3 (Cell Signaling, 4108).
Statistics
Each transgenic colony and sex was analyzed by paired 2 × 2 ANOVA. Independent factors within each sex included genotype (WT vs. PGC or MCAT) and hindlimb unloading intervention (CON vs. HU). Sex was not a statistical factor due to the excessive comparisons that would result in a 2 × 2 × 2 ANOVA that would not provide meaningful information. More so, some variables (such as protein and mRNA analysis) inherently require one group to be set as the control to compare all other groups. If sex were included in a 2 × 2 × 2 factorial ANOVA, one group would need to be set as this global control and doing so would likely muddle the data and make interpretation of how interventions impacted each sex difficult. Instead, the inclusion of males and females was to investigate how interventions impacted males and females separately.
All F ratios for main effects (factor 1 of genotype with levels of wild-type and transgenic and factor 2 of intervention with levels of control and hindlimb unloading) and interaction (interaction between genotype and intervention) were Tukey-adjusted using the PROC MIXED function within SAS. When a significant interaction F ratio was found within a sex, pairwise comparisons between groups were completed using a Tukey post hoc adjustment. Additionally, based on visual inspection of the data, some outcome variables appeared to provide more meaningful information when analyzed as percent differences from within the genotype and sex control (e.g., comparing percent body weight loss in WT females compared with MCAT females). This type of analysis appeared to be particularly informative when visually inspecting that MCAT mice overall were slightly smaller compared with WT mice (more thoroughly described in the RESULTS section). Because of this, data for MCAT mice were analyzed as percent difference between hindlimb unloaded WT and MCAT mice and statistically analyzed by t tests. This type of analysis was not completed in PGC mice, as there was no phenotypic evidence to suggest different-sized animals that may influence the interpretation of muscle protections (or lack thereof). The significance for all statistical tests was denoted at P < 0.05.
RESULTS
Confirmation of Transgenic Models
PGC transgenic mice had ∼13-fold greater Pgc1α mRNA content than WT mice (P < 0.001, Supplemental Fig. S1, A and B; see https://doi.org/10.6084/m9.figshare.12777098.v2), as well as twofold greater PGC1α protein content than WT mice (P = 0.022, Supplemental Fig. S1C). MCAT transgenic mice had ∼50% greater catalase mRNA content, which was confirmed in both the gastrocnemius and soleus muscles (P = 0.005 and P = 0.024, Supplemental Fig. S1, D–F).
PGC1α Overexpression Did Not Protect against Disuse-Induced Atrophy
Overall, there were no significant interactions between PGC1α overexpression and HU. Hindlimb unloading resulted in ∼9% (∼2.5 g) lower body weights in male mice than in controls (P = 0.001, Table 1). Female HU mice also had a ∼5% (∼1.0 g) lower body weight than CON mice (P = 0.021, Table 1). Male HU mice had ∼30% lower soleus weight (3.6 mg, P < 0.001), ∼23% lower plantaris weight (5.0 mg, P < 0.001), ∼18% lower gastrocnemius weight (26.0 mg, P < 0.001), and 15% lower tibialis anterior weight (8.7 mg, P < 0.001) than CON mice (Table 1). Similarly, female HU mice had ∼28% smaller soleus weight (2.5 mg, P < 0.001), ∼16% smaller plantaris weight (2.3 mg, P < 0.001), ∼15% smaller gastrocnemius weight (14.6 mg, P < 0.001), and ∼10% smaller tibialis anterior weight (4.2 mg, P < 0.001) than CON mice (Table 1). Additionally, in female mice, there was a main effect of genotype, with PGC transgenic mice having ∼9% greater tibialis anterior weights (3.4 mg, P = 0.008) than WT mice (Table 1). There was no effect of HU on EDL mass in male or female mice (P = 0.10 and P = 0.16, respectively, Table 1). Additionally, HU mice had ∼38% and ∼50% less gonadal fat (139.8 mg and 104.4 mg, P < 0.001) in male and female mice, respectively (data not shown).
Table 1.
Body weights and muscle masses from PGC1α colony
| Sex | Group | Body Mass, g | Soleus, mg | Gastrocnemius, mg | Plantaris, mg | Tibialis Anterior (TA), mg | Extensor Digitorum Longus (EDL), mg |
|---|---|---|---|---|---|---|---|
| Males | WT-CON | 26.6 ± 0.8 | 11.2 ± 0.6 | 144.1 ± 3.9 | 21.8 ± 0.5 | 54.7 ± 1.4 | 11.4 ± 0.6 |
| WT-HU | 24.8 ± 0.5* | 8.3 ± 0.5* | 120.3 ± 2.9* | 16.9 ± 0.7* | 47.8 ± 1.3* | 10.9 ± 0.4 | |
| PGC-CON | 27.1 ± 0.8 | 13.0 ± 0.9 | 143.8 ± 3.9 | 22.1 ± 0.7 | 58.3 ± 1.9 | 12.1 ± 0.4 | |
| PGC-HU | 23.8 ± 0.8* | 8.7 ± 0.7* | 115.3 ± 4.5* | 17.0 ± 0.9* | 47.7 ± 2.4* | 10.9 ± 0.4 | |
| Females | WT-CON | 19.9 ± 0.3 | 9.4 ± 0.6 | 99.4 ± 2.6 | 14.5 ± 0.3 | 40.1 ± 1.2 | 8.8 ± 0.3 |
| WT-HU | 18.8 ± 0.5* | 6.5 ± 0.4* | 82.6 ± 1.3* | 11.9 ± 0.4* | 34.5 ± 1.0* | 8.1 ± 0.5 | |
| PGC-CON | 20.2 ± 0.3 | 8.8 ± 0.3 | 95.5 ± 1.6 | 14.7 ± 0.4 | 41.9 ± 0.9# | 9.3 ± 0.5 | |
| PGC-HU | 19.1 ± 0.5* | 6.6 ± 0.3* | 82.9 ± 1.9* | 12.9 ± 0.5* | 39.0 ± 1.2*,# | 8.7 ± 0.4 |
Data are presented as means ± SE. Body weights and muscle masses from the PGC1α portion of the study. Males had the following sample sizes: WT-CON = 9, WT-HU = 10, PGC-CON = 9, and PGC-HU = 9. Females had the following samples sizes: WT-CON = 8, WT-HU = 9, PGC-CON = 9, and PGC-HU = 9. CON, cage control; HU, humanized; MCAT-CON, mitochondrially targeted catalase-cage control; MCAT-HU, mitochondrially targeted catalase-humanized; PGC, peroxisome proliferator-activated receptor gamma coactivator 1-alpha; PGC-CON, peroxisome proliferator-activated receptor gamma coactivator 1-alpha-cage control; PGC-HU, peroxisome proliferator-activated receptor gamma coactivator 1-alpha-humanized; PGC1α, peroxisome proliferator-activated receptor gamma coactivator 1-alpha; WT, wild-type; WT-CON, wild-type-cage control; WT-HU, wild-type-humanized.
Statistical main effect between CON and HU.
Main effect between WT and PGC.
Within the plantaris muscle, HU males had ∼21% lower mean CSA than CON mice, with no effect of genotype (1106 µm2 vs. 850 µm2, P = 0.001, Fig. 1, A and B). In female mice, HU also had a ∼13% lower cross-sectional area compared with that in control mice (846 µm2 vs. 722 µm2, P = 0.029, Fig. 1, C and D). However, in females, a main effect of genotype on cross-sectional area was noted, with PGC transgenic mice having ∼22% greater cross-sectional area than WT mice (890 µm2 vs. 722 µm2, P < 0.001, Fig. 1, C and D). Histograms for CSA can be found in Supplemental Fig. S2 (https://doi.org/10.6084/m9.figshare.12689918.v2).
Fig. 1.
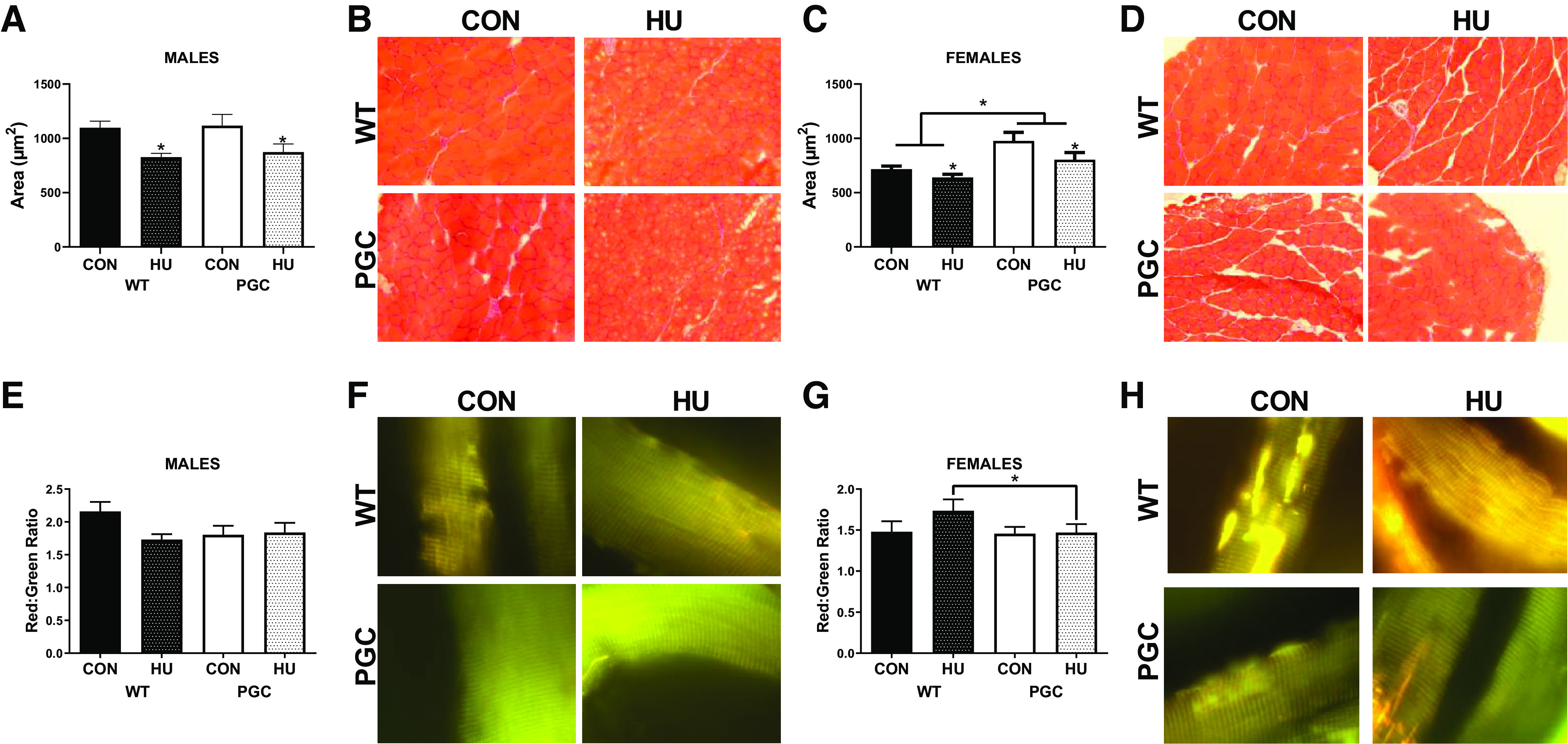
Muscle phenotypic data for males and females from PGC colony portion of this study. A: plantaris cross-sectional area (CSA) quantification for males. B: representative images for CSA for males. C: plantaris cross-sectional area (CSA) quantification for females. D: representative images for CSA for females. E: pMitoTimer red/green ratio in males. F: representative images of pMitoTimer in males. G: pMitoTimer red/green ratio in females. H: representative images of pMitoTimer in females. *Tukey-adjusted P < 0.05. All CSA images were taken at ×20 magnification. pMitoTimer images were taken at ×100 magnification. Males had the following sample sizes: WT-CON = 9, WT-HU = 10, PGC-CON = 9, and PGC-HU = 9. Females had the following samples sizes: WT-CON = 8, WT-HU = 9, PGC-CON = 9, and PGC-HU = 9. PGC, peroxisome proliferator-activated receptor gamma coactivator 1-alpha; PGC-CON, peroxisome proliferator-activated receptor gamma coactivator 1-alpha-cage control; PGC-HU, peroxisome proliferator-activated receptor gamma coactivator 1-alpha-humanized; WT-CON, wild-type-cage control; WT-HU, wild-type-humanized.
In males, there was no effect of genotype (P = 0.238) or hindlimb unloading (P = 0.987) on red/green ratio of pMitoTimer (Fig. 1, E and F). However, in females, there was an interaction between genotype and hindlimb unloading (P = 0.003). Specifically, WT-HU animals had 46% greater red/green ratios than PGC-HU animals (Fig. 1, G and H). Although not significant (P = 0.15), there was a mean increase in the red/green ratio between WT-CON and WT-HU, suggestive of greater mitochondrial stress. With regard to red puncta, no statistical differences were noted in males or females (data not shown).
Catabolic Signaling Was Differentially Affected in WT Compared with PGC Mice by HU
In males, an interaction was noted in Atrogin mRNA content in the gastrocnemius muscle, with WT-HU mice having ∼3.5-fold greater Atrogin mRNA content than all other groups (P = 0.02, Fig. 2A). Similarly, in female gastrocnemius muscle, an interaction in Atrogin mRNA content was also noted, with WT-HU mice having ∼3.5 fold greater Atrogin mRNA content than all other groups (P = 0.02, Fig. 2B). In males, an interaction was noted in MuRF1 mRNA content, with WT-HU mice having ∼2.5-fold greater MuRF1 mRNA content than all other groups (P = 0.002, Fig. 2C). Females had patterns similar to MuRF1 mRNA content, with WT-HU mice having threefold greater MuRF1 mRNA content than all other groups; however, the interaction did not reach statistical significance (P = 0.08, Fig. 2D). Females also had an effect of genotype on MuRF1 mRNA content, with PGC animals having ∼70% less MuRF1 mRNA content compared with WT mice (P = 0.02, Fig. 2D). In males, PGC mice had approximately twofold greater LC3II protein content than WT that approached statistical significance (P = 0.08, Fig. 2, E and I). In females, PGC mice had approximately threefold greater LC3II content than WT mice (P = 0.008, Fig. 2, F and I). In males, there was a main effect for total LC3 (the summation of LC3II and LC3I bands) protein content (P = 0.040, Fig. 2, G and I), whereby PGC males had ∼75% greater LC3 content than WT. In females, an interaction was noted (Fig. 2, H and I, P = 0.025), whereby PGC-HU animals had approximately threefold greater LC3 protein content than all other groups. In males, main effects of genotype and intervention were found in p62 mRNA content, with HU mice having ∼75% greater p62 mRNA content than CON (P = 0.03, Fig. 2J) and PGC animals having ∼75% less p62 mRNA content compared with WT animals (P = 0.02, Fig. 2J). In females, an effect of treatment was found, with HU mice having ∼65% greater p62 mRNA content than CON (P = 0.01, Fig. 2K). The effect of genotype in females did not reach statistical significance (P = 0.071); however, a mean difference of 50% lower p62 mRNA content was observed in female PGC animals compared with WT animals (Fig. 2K). With regard to the p62 protein content, in males, a main effect of intervention was noted (P = 0.035, Fig. 2, L and I), with HU males having ∼50% less p62 protein content compared with CON males. More so, in females, main effects of genotype (P = 0.021) and intervention (P = 0.041) were detected in p62 protein content (Fig. 2, M and I). In females, HU mice had ∼75% greater p62 protein content than CON mice, and PGC females had ∼75% less p62 protein content compared with WT animals (Fig. 2, M and I).
Fig. 2.
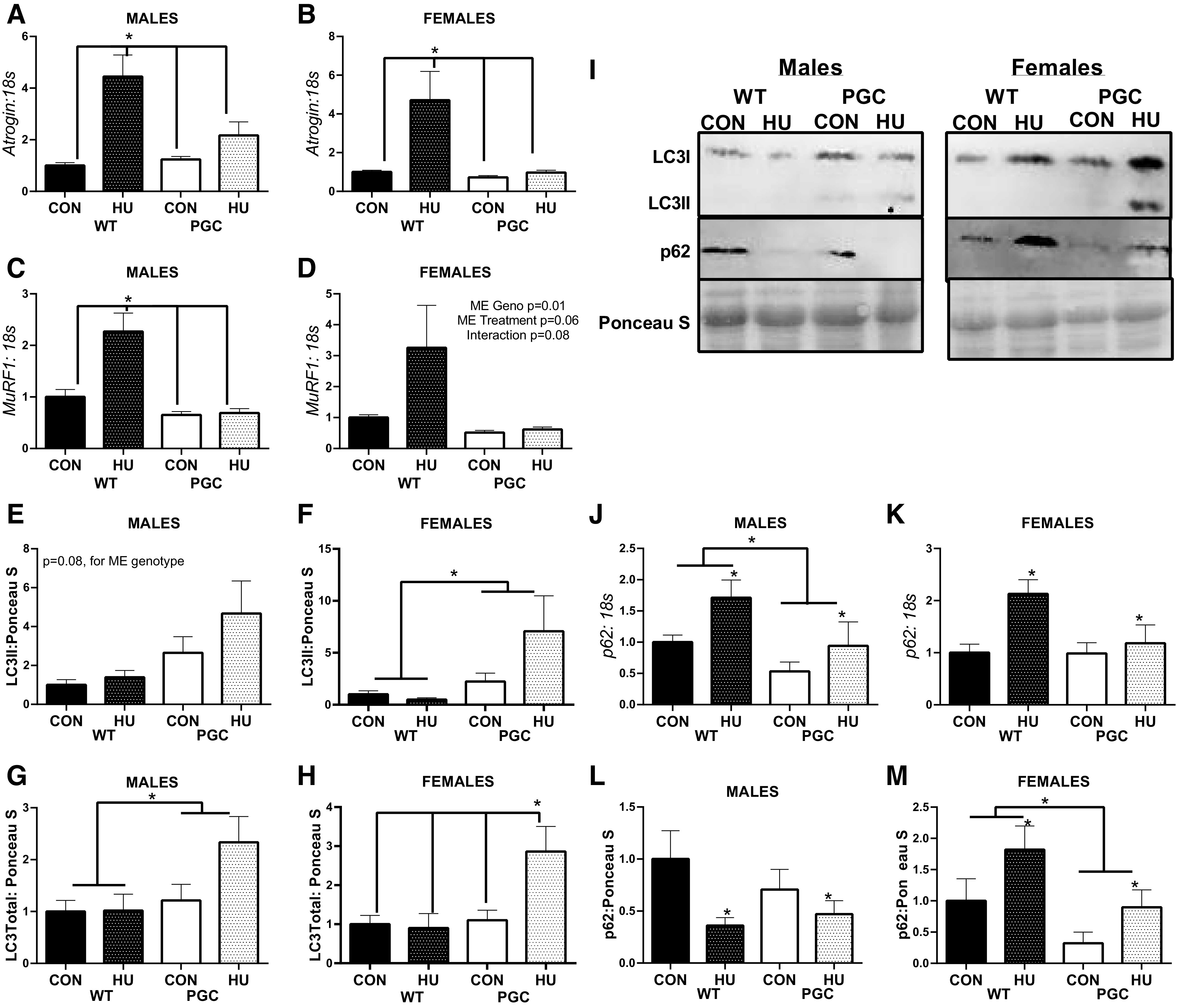
mRNA and protein content of moderators of ubiquitin-proteasome and autophagy-mediated catabolism from the PGC colony portion of this study. A: Atrogin mRNA content in males. B: Atrogin mRNA content in females. C: MurF1 mRNA content in males. D: MurF1 mRNA content in females. E: LC3II protein content in males. F: LCII protein content in females. G: total LC3 protein content in males. H: total LC3 protein content in females. I: representative image of immunoblotting targets. J: p62 mRNA content in males. K: p62 mRNA content in females. L: p62 protein content in males. M: p62 protein content in females. All values are normalized within sex to WT-CON. *Tukey-adjusted P < 0.05. ME denotes main effect. Males had the following sample sizes: WT-CON = 9, WT-HU = 10, PGC-CON = 9, and PGC-HU = 9. Females had the following samples sizes: WT-CON = 8, WT-HU = 9, PGC-CON = 9, and PGC-HU = 9. PGC, peroxisome proliferator-activated receptor gamma coactivator 1-alpha; PGC-CON, peroxisome proliferator-activated receptor gamma coactivator 1-alpha-cage control; PGC-HU, peroxisome proliferator-activated receptor gamma coactivator 1-alpha-humanized; WT-CON, wild-type-cage control; WT-HU, wild-type-humanized.
Other moderators of anabolism Igf1 mRNA, satellite cell marker Pax7 mRNA, and an inhibitor of protein anabolism Redd1 mRNA did not have any interactions between groups (Supplemental Fig. S3; see https://doi.org/0.6084/m9.figshare.12690059.v2). However, there was ∼50% less Igf1, ∼50% greater Pax7, and ∼50% greater Redd1 mRNA in HU animals than CON animals, regardless of genotype (Supplemental Fig. S3). Additionally, markers of mitochondrial dynamics or mitophagy did not demonstrate interactions that would moderate the muscle phenotype (Supplemental Figs. S4 and S5; see https://doi.org/10.6084/m9.figshare.12777137.v1 and https://doi.org/10.6084/m9.figshare.12777152.v1, respectively). However, similar to our prior reports (20), we find genetic overexpression of PGC1α sufficient to result in greater mRNA content of moderators of mitochondrial quality control.
MCAT Body Weight and Cross-Sectional Area
Across both males and females, there were significant differences detected in muscle mass, with HU groups having ∼7%–35% lower gastrocnemius, plantaris, soleus, and tibialis anterior weights (Table 2). However, upon visual inspection of the data, MCAT females had lower body and muscle weights than WT animals in CON groups that were not robust enough to correspond to a main effect of genotype. An independent t test demonstrated that soleus weights were significantly different between WT-CON and MCAT-CON females (P = 0.022). These differences were not present in males. Therefore, we further analyzed muscle and body weight data in HU animals as percent differences from within genotype CON and compared percent differences between WT-HU and MCAT-HU through an unpaired t test. This analysis denoted that MCAT females tended to have attenuated differences in muscle size compared with WT animals. Specifically, MCAT females had smaller differences between HU and CON with regard to body weight (∼10% lower mass in WT vs. ∼3% lower mass in MCAT, Fig. 3A), soleus mass (∼40% lower mass in WT vs. ∼20% lower mass in MCAT, Fig. 3A), and tibialis anterior mass (∼20% lower mass in WT vs. ∼10% lower mass in MCAT, Fig. 3A) than WT animals (P = 0.012—P = 0.003). However, in males, there were no differences between MCAT-HU and WT-HU mice in percent differences from CON (Fig. 3C). Additionally, in PGC mice, there were no differences between CON and PGC mice using this analysis (Supplemental Fig. S6; see https://doi.org/10.6084/m9.figshare.12777197.v1). In plantaris CSA, there were no differences in males on percent CSA difference (P = 0.881, Fig. 3D). Although not significant, a similar pattern of MCAT females maintaining plantaris cross-sectional area also appeared (∼16% lower CSA in WT females vs. ∼2.5% greater CSA in MCAT females, P = 0.223, Fig. 3E). Based on these findings, we opted to use the soleus muscle as the primary tissue for cell signaling analysis, as this tissue appeared to have the greatest protections in the MCAT females than WT females. Due to the limited tissue availability of the soleus (∼10–14 mg of total tissue for both solei combined), real-time PCR analysis of genes related to musculoskeletal health was analyzed as opposed to immunoblot because more targets could be analyzed.
Table 2.
Body weights and muscle masses from MCAT colony
| Sex | Group | Body Mass, g | Soleus, mg | Gastrocnemius, mg | Plantaris, mg | Tibialis Anterior (TA), mg | Extensor Digitorum Longus (EDL), mg |
|---|---|---|---|---|---|---|---|
| Males | WT-CON | 23.9 ± 0.6 | 10.9 ± 0.3 | 135.0 ± 3.3 | 19.0 ± 0.4 | 50.0 ± 1.4 | 10.2 ± 0.5 |
| WT-HU | 22.5 ± 0.8 | 7.4 ± 0.6* | 108.6 ± 3.0* | 15.8 ± 0.5* | 42.6 ± 1.7* | 9.0 ± 0.4* | |
| MCAT-CON | 22.5 ± 0.5 | 10.6 ± 0.7 | 125.1 ± 2.2 | 17.5 ± 0.4 | 50.2 ± 2.6 | 9.2 ± 0.2 | |
| MCAT-HU | 21.6 ± 0.9 | 6.7 ± 0.7* | 105.4 ± 4.2* | 15.0 ± 1.0* | 42.1 ± 2.6* | 7.9 ± 0.8* | |
| Females | WT-CON | 20.5 ± 0.5 | 9.6 ± 0.5 | 102.3 ± 2.8 | 15.4 ± 0.3 | 40.2 ± 1.5 | 8.2 ± 0.3 |
| WT-HU | 18.4 ± 0.3* | 5.8 ± 0.4* | 81.2 ± 1.7* | 11.6 ± 0.4* | 32.7 ± 0.6* | 7.6 ± 0.3* | |
| MCAT-CON | 20.1 ± 0.2 | 8.0 ± 0.5 | 99.4 ± 2.1 | 13.8 ± 0.5 | 39.7 ± 1.6 | 9.2 ± 0.4 | |
| MCAT-HU | 19.3 ± 0.3* | 6.4 ± 0.4* | 82.4 ± 2.00* | 11.9 ± 0.7* | 37.0 ± 1.1* | 7.6 ± 0.3* |
Data are presented as means ± SE. Body weights and muscle masses from the MCAT portion of the study. Males had the following sample sizes: WT-CON = 8, WT-HU = 7, MCAT-CON = 4, and MCAT-HU = 3. Females had the following samples sizes: WT-CON = 5, WT-HU = 6, MCAT-CON = 6, and MCAT-HU = 6. CON, cage control; HU, humanized; MCAT, mitochondrially targeted catalase; MCAT-CON, mitochondrially targeted catalase-cage control; MCAT-HU, mitochondrially targeted catalase-humanized; PGC-CON, peroxisome proliferator-activated receptor gamma coactivator 1-alpha-cage control; PGC-HU, peroxisome proliferator-activated receptor gamma coactivator 1-alpha-humanized; WT, wild-type; WT-CON, wild-type-cage control; WT-HU, wild-type-humanized.
Statistical main effect between CON and HU.
Main effect between WT and MCAT.
Fig. 3.
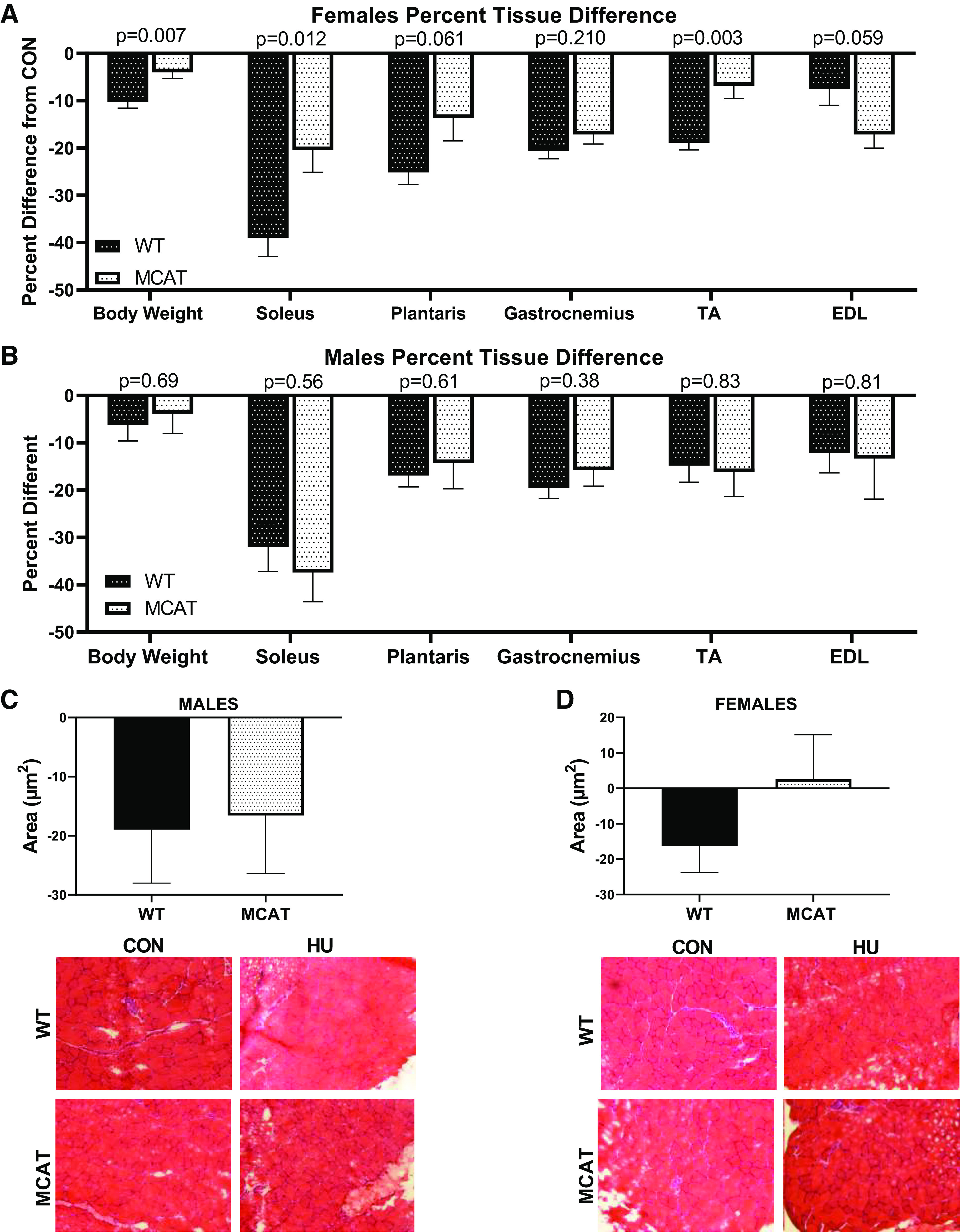
Muscle size data in MCAT male and female mice. A: percent difference tissue loss in female hindlimb-unloaded mice. B: percent difference tissue loss in male hindlimb-unloaded mice. C: percent difference in plantaris fiber cross-sectional area (CSA) in MCAT male mice. D: percent difference in plantaris fiber cross-sectional area (CSA) in MCAT female mice. *Tukey-adjusted P < 0.05. Males had the following sample sizes: WT-CON = 8, WT-HU = 7, MCAT-CON = 4, and MCAT-HU = 3. Females had the following samples sizes: WT-CON = 5, WT-HU = 6, MCAT-CON = 6, and MCAT-HU = 6. MCAT, mitochondrially targeted catalase; MCAT-CON, mitochondrially targeted catalase-cage control; MCAT-HU, mitochondrially targeted catalase-humanized; WT-CON, wild-type-cage control; WT-HU, wild-type-humanized.
Mediators of Protein Catabolism in MCAT Mice Did Not Suggest Protections against HU
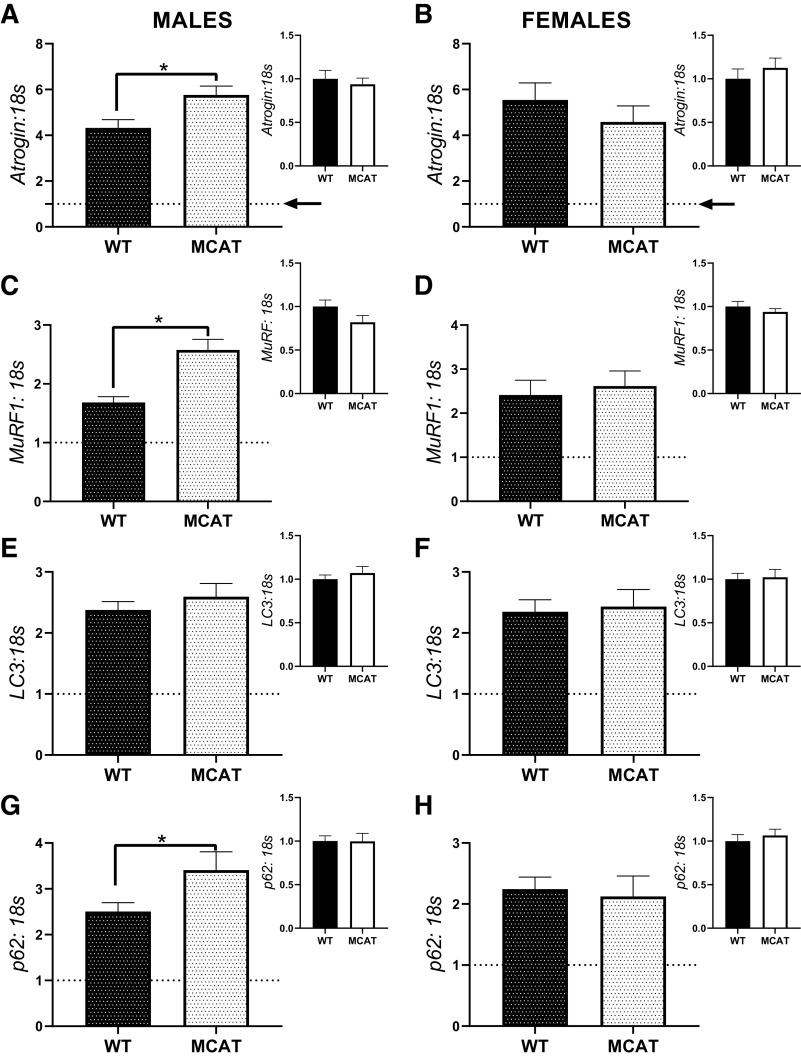
Due to the observation that there appeared to be phenotypic differences in the response to HU between males and females, for mRNA analysis, we compared mRNA responses of WT-HU and MCAT-HU soleus muscle. Additionally, as a supplemental analysis, we also compared contents of mRNAs in WT-CON and MCAT-HU animals. All analyses were performed with t tests. In males, MCAT-HU mice had 33% greater Atrogin mRNA than WT-HU mice (P = 0.046, Fig. 4A), with no differences noted between WT-CON and MCAT-CON animals (P = 0.68). In females, there was no difference in Atrogin mRNA content between WT-HU and MCAT-HU mice (P = 0.37, Fig. 4B). Additionally, there was no difference between WT-CON and MCAT-CON animals (P = 0.40). In males, MCAT-HU mice had ∼50% greater MuRF1 mRNA content than WT-HU mice (P = 0.001, Fig. 4C), though there was no difference between WT-CON and HU-CON animals (P = 0.17, Fig. 4C). However, in females, there was no difference noted in MuRF1 mRNA content (P = 0.68). There were no differences in Lc3 mRNA content between WT-HU and MCAT-HU animals in males or females (P = 0.41 and P = 0.81, respectively); more so there were no differences in Lc3 mRNA content in either males or females between WT-CON and MCAT-CON animals (P = 0.42 and P = 0.85, respectively Fig. 4E). In males, MCAT-HU mice had ∼35% greater p62 mRNA content than WT-HU mice (P = 0.048, Fig. 4G), with no differences noted between WT-CON and MCAT-CON mice (P = 0.97, Fig. 4G). However, in females, there were no differences noted in p62 mRNA content between either WT-HU and MCAT-HU (P = 0.76) or WT-CON and MCAT-CON (P = 0.55, Fig. 4H).
Fig. 4.
mRNA content of moderators of ubiquitin-proteasome and autophagy-mediated catabolism from the soleus muscle of MCAT animals. A: atrogin mRNA response and basal content in males. B: atrogin mRNA response and basal content in females. C: MuRF1 mRNA response and basal content in males. D: MuRF1 mRNA response and basal content in females. E: Lc3 mRNA response and basal content in males. F: Lc3 mRNA response and basal content in females. G: p62 mRNA response and basal content in males. H: p62 mRNA response and basal content in females. Larger graphs show the responses of WT-HU and MCAT-HU, with the inset containing comparisons between WT-CON and MCAT-CON. To demonstrate responses compared with control, 1 (indicating the within genotype basal levels) is graphed with a dotted line. *P < 0.05 from a Student’s t test between genotypes. Males had the following sample sizes: WT-CON = 8, WT-HU = 7, MCAT-CON = 4, and MCAT-HU = 3. Females had the following samples sizes: WT-CON = 5, WT-HU = 6, MCAT-CON = 6, and MCAT-HU = 6. MCAT-CON, mitochondrially targeted catalase-cage control; MCAT-HU, mitochondrially targeted catalase-humanized; WT-CON, wild-type-cage control; WT-HU, wild-type-humanized.
Mediators of Protein Anabolism Did Not Appear to Differ between WT and MCAT Mice
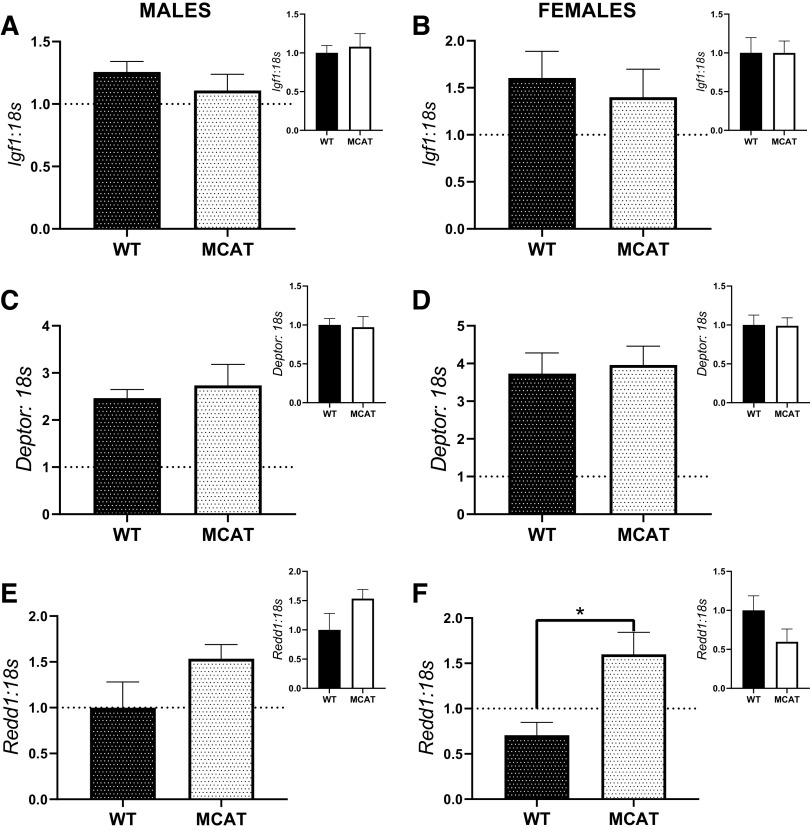
In males, there were no differences in Igf1 mRNA content between either WT-HU and MCAT-HU (P = 0.67) or WT-CON and MCAT-CON mice (P = 0.32, Fig. 5A). Correspondingly, in females, there were no differences in Igf1 mRNA content between either WT-HU and MCAT-HU (P = 0.63) or WT-CON and MCAT-CON mice (P = 0.32, Fig. 5B). Additionally, in males, there were no differences in Deptor mRNA content between either WT-HU and MCAT-HU (P = 0.84) or WT-CON and MCAT-CON mice (P = 0.51, Fig. 5C). Similarly, in females, there were no differences in Deptor mRNA content between either WT-HU and MCAT-HU (P = 0.63) or WT-CON and MCAT-CON mice (P = 0.32, Fig. 5D). In males, there was no difference in Redd1 mRNA content between either WT-HU and MCAT-HU (P = 0.22) or WT-CON and MCAT-CON (P = 0.27, Fig. 5E). In females, MCAT-HU mice had approximately twofold greater Redd1 mRNA content than WT-HU mice (P = 0.013, Fig. 5F). However, there were no statistical differences between female WT-CON and MCAT-CON (P = 0.015, Fig. 5F).
Fig. 5.
mRNA content of moderators of protein anabolism the soleus of MCAT mice. A: Igf1 mRNA content in males. B: Igf1 mRNA content in females. C: deptor mRNA content in males. D: deptor mRNA content in females. E: Redd1 mRNA content in males. F: Redd1 mRNA content in females. Larger graphs show the responses of WT-HU and MCAT-HU, with the inset containing comparisons between WT-CON and MCAT-CON. To demonstrate responses compared with control, 1 (indicating the within genotype basal levels) is graphed with a dotted line. *P < 0.05 from a Student’s t test between genotypes. Males had the following sample sizes: WT-CON = 8, WT-HU = 7, MCAT-CON = 4, and MCAT-HU = 3. Females had the following samples sizes: WT-CON = 5, WT-HU = 6, MCAT-CON = 6, and MCAT-HU = 6. MCAT, mitochondrially targeted catalase; MCAT-CON, mitochondrially targeted catalase-cage control; MCAT-HU, mitochondrially targeted catalase-humanized; WT-CON, wild-type-cage control; WT-HU, wild-type-humanized.
In MCAT Mice, Mitochondrial Stress Had Divergent Responses between Males and Females
In males, there were no statistical differences in red/green ratio response between WT and MCAT animals, P = 0.621, Fig. 6, A and E). However, in females, MCAT-HU females had a divergent response compared with WT-HU females, with MCAT females having ∼42% lower red/green ratio than MCAT-CON; conversely, WT-HU females had ∼30% greater red/green ratio than WT-CON (P = 0.004, Fig. 6, B and F). No differences in response were noted in number of red puncta (a measure of degenerated mitochondria) in males (P = 0.650, Fig. 6, C and E). Although not significant (P = 0.062), MCAT-HU females had fewer red puncta than MCAT-WT animals (Fig. 6, D and F). Neither mitochondrial dynamics nor mitophagy demonstrated differences that would suggest protections against disuse atrophy in female MCAT mice (Supplemental Figs. S7 and S8; see https://doi.org/10.6084/m9.figshare.12777236.v1 and https://doi.org/10.6084/m9.figshare.12777278.v2, respectively).
Fig. 6.
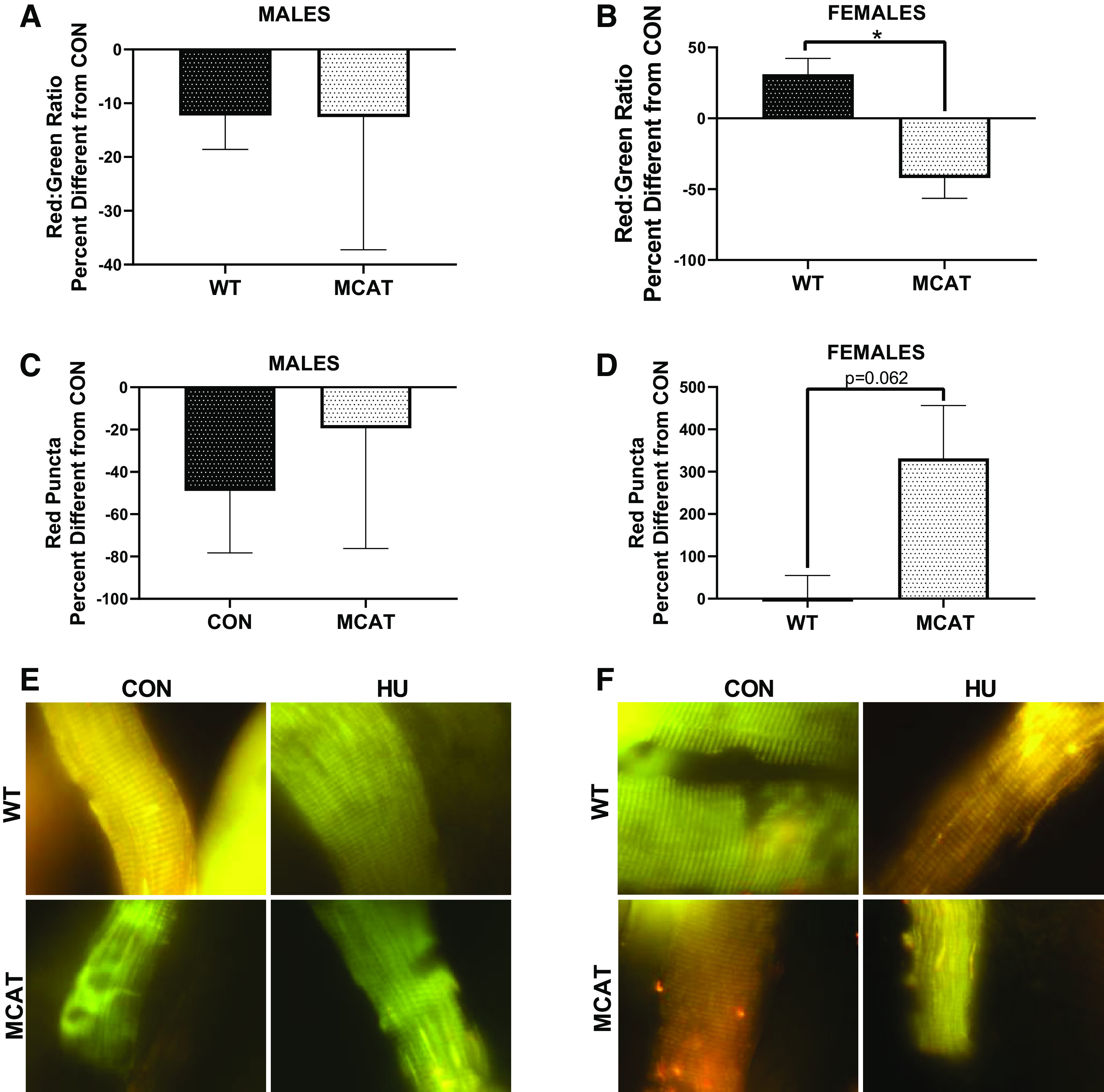
pMitoTimer from males and females from the MCAT portion of this study. A: percent difference between CON and HU animal with each genotype in red/green ratio in males. B: percent difference between CON and HU animals with each genotype in red/green ratio in females. C: percent difference between CON and HU animals with each genotype in red puncta in males. D: percent difference between CON and HU animals with each genotype in red puncta in females. E: representative images of pMitoTimer in males. F: representative images of pMitoTimer in females. To demonstrate responses compared with control, 1 (indicating the within genotype basal levels) is graphed with a dotted line. *Tukey-adjusted P < 0.05. pMitoTimer images were taken at ×100 magnification. Males had the following sample sizes: WT-CON = 8, WT-HU = 7, MCAT-CON = 4, and MCAT-HU = 3. Females had the following samples sizes: WT-CON = 5, WT-HU = 6, MCAT-CON = 6, and MCAT-HU = 6. MCAT, mitochondrially targeted catalase; MCAT-CON, mitochondrially targeted catalase-cage control; MCAT-HU, mitochondrially targeted catalase-humanized; WT-CON, wild-type-cage control; WT-HU, wild-type-humanized.
DISCUSSION
To our knowledge, this is one of the first studies to investigate multiple mitochondrially targeted genetic interventions in the prevention or attenuation for disuse-induced atrophy in both males and females. Phenotypically, MCAT expression in females appeared to blunt disuse-induced muscle atrophy, with particular protection in the soleus muscle; yet, these protections did not translate to males. This protection may be attributed to a differential response in mitochondrial oxidative stress of MCAT unloaded female mice when compared with WT mice. Specifically, WT unloaded females had greater mitochondrial oxidative stress (measured by pMitoTimer), whereas MCAT unloaded females had decreased oxidative stress. Conversely, we found no differences in phenotypic musculoskeletal outcomes between WT and PGC mice, though a shift in catabolic pathway utilization was observed. Taken together, these data appear to suggest that mitochondrial interventions to mitigate or prevent disuse atrophy may be more nuanced than previously believed.
Similar to prior reports, we found overexpression of PGC1α sufficient to mitigate induction of the ubiquitin proteasome system (6, 12, 13, 29, 56, 57, 67). However, compared with other studies, we did not find genetic overexpression of PGC1α to protect against disuse atrophy regarding either muscle mass or muscle cross-sectional area. There are a few potential reasons for these differences in findings. Prior studies that have found PGC1α effective for disuse atrophy have used relatively older mice (24 wk of age compared with 10 wk of age in our study) (12, 13) or have used more aggressive interventions such as denervation (57). Therefore, the efficacy of greater PCG1α may be dependent on the age of the animal or the model system. For example, other transgenic models have demonstrated divergent efficacy for mitigating muscle loss depending on the age of the organism (4b). As such, it is plausible that the different results for PGC1α efficacy could be partially attributed to interactions between PGC1α signaling cascades and the age of the organism. Conversely, other research has found that local transfection of PGC1α is sufficient to alter cellular signaling cascades associated with atrophy (similar to our findings); however, these differences in cellular signaling cascades were not sufficient to mitigate aging-induced atrophy (72). Moreover, regarding denervation-induced atrophy, potential differences may be attributed to subtle differences in cellular signaling cascades specific to denervation compared with disuse. For example, recent studies have found denervation and disuse to modify slightly different atrogenes (8). Although both disuse and denervation induce similar cellular signaling cascades, these modest differences between the two models may partially explain differences noted between our study and previous research (57). Additionally, this same study only used Atrogin and MuRF1 content as surrogates for atrophy, without measuring phenotypic outcomes (57). Our data demonstrate that using only cellular signaling without phenotypic outcomes is not sufficient to determine the efficacy of atrophy-attenuation interventions and should be considered for future intervention-based research. Contradicting PGC1α being protective, other studies have found PGC1α-knockout animals to blunt disuse-induced muscle atrophy (66). It seems unlikely that both increased PGC1α and decreased PGC1α can equally protect against disuse atrophy, suggesting that, at minimum, the interactions between PGC1α and musculoskeletal health are more complicated than previously described. Regardless, our data suggest that promotion of greater mitochondrial content, via PGC1α, is insufficient to protect against disuse-induced atrophy.
Although PGC1α overexpression is sufficient to blunt the ubiquitin-proteasome responses to atrophy, this does not seem to be the case with other catabolic pathways, notably autophagy. In fact, our data tend to suggest that autophagy induction, particularly LC3 protein content, may be the driving catabolic pathway in PGC mice. For example, we observed strong main effects or interactions for LC3 protein content in hindlimb unloaded males and females. This finding aligns with prior studies from our group and others demonstrating PGC1α to drive LC3 content (20, 35). Of note, females comparatively exhibited greater markers of autophagy induction than males at both the RNA and the protein level (approximately threefold greater LC3 content in HU females compared with ∼50% greater LC content in males). Females are generally thought to have greater mitochondrial content (15, 40) and presumably PGC1α. As PGC1α is known to partially mediate autophagy (20, 35), these differences in markers of autophagy may be partially mediated by sex-specific differences in mitochondrial density. Also plausible is that these differences in autophagy may also be inherent differences in muscle phenotypes between males and females (52). Sex differences in mitochondrial biogenesis and subsequent autophagy appear to agree with other research finding potential sex differences in catabolic signaling cascades (45, 47) and may suggest the potential need for therapeutics tailored to sex-specific responses to atrophy. Yet, these studies on sex differences during disuse atrophy are still in the relatively early stages and more studies are needed in preclinical models to fully characterize sex differences in disuse atrophy.
Although we did not observe any phenotypic protections against disuse atrophy in PGC mice, we did see a protection in female MCAT mice. It is possible that MCAT protections are specific to females, though we should acknowledge that the limited sample size in males restricts our ability to infer strong conclusions on the potential protective role of additional catalase in males. These protections in females may be in part due to mitochondrial protections in response to disuse in females. Specifically, we saw WT-HU females having greater red/green ratios [indicative of mitochondrial stress (30)] in response to disuse, whereas MCAT-HU females had lower red/green ratios in response to disuse. Contrastingly, in males, there appeared to be no influence of 7 days of unloading on mitochondrial oxidative stress. Taken together, these results imply that mitochondrial alterations during disuse may differ between sexes and, correspondingly, and result in different responses to mitochondria-based interventions. Prior research has provided the preliminary evidence to suggest that females may be more responsive to ROS-specific interventions. For example, females are known to have greater mitochondrial content than males (15, 40), and mitochondria are the primary reactive oxygen species (ROS) generation sites within the cell (50). Therefore, it is plausible that females may have a greater capacity to generate ROS during myopathies than males, particularly in mitochondrially dense muscles such as the soleus. This exacerbated disease severity has been reported in mice with genetically greater mitochondrial content in relation to insulin resistance (14, 38). Correspondingly, because of this potential for greater ROS generation, females may be more responsive to mitochondria-related treatments to mitigate ROS generation. This would align with our data finding robust protections from disuse atrophy in the soleus muscle of female MCAT mice. However, we recognize that this hypothesis requires further validation.
Specific to MCAT itself, prior studies have found MCAT mice to protect against other musculoskeletal pathologies such as insulin resistance, cardiomyopathies, and aging-induced muscle loss (65b, 30a, 31a, 4b, 58). Though there has been controversy within the scientific literature on MCAT’s efficacy for muscle pathologies (31b, 25), these protections are thought to be at least partially due to MCAT’s control over oxidative phosphorylation (4a). As previously mentioned, different transgenic strains appear to interact differently depending on the age of the model organism. This appears to also be true with MCAT mice. For example, MCAT appears protective for cardiomyopathies in older mice, but not younger mice (4b). Therefore, it is plausible that because of the age of our mice, we are not maximizing the potential therapeutic potential of mitochondria-targeted catalase and we may have seen greater protections in older animals. Yet, this hypothesis would require further investigation.
Although we noted MCAT females to have some phenotypic protection from disuse atrophy, we did not find any robust cellular signaling cascade differences between WT and MCAT female mice that would clearly account for phenotypic differences noted between WT and MCAT female mice. If anything, MCAT mice appeared to have augmented atrophic signaling compared with WT mice. This was noted in Atrogin (males), MuRF1 (males), and p62 (males). Due to catalase’s role as a ROS scavenger, it is likely that any protections from MCAT mice were due to functional differences in mitochondrial respiration and subsequent ROS emission and/or neutralization. This hypothesis would align with the data we found in pMitoTimer, suggesting protections in females but not males. Additionally, while these differences in catabolic signaling do not necessarily explain phenotypic protections to muscle size, they do highlight signaling differences between males and females within this particular strain of mice and emphasize the need to investigate muscle pathologies and subsequent interventions in both males and females.
We should acknowledge that this study was not statistically designed to make comparisons between male and female mice. Due to the multifactorial nature of the study, we believed completing paired sets of experiments in male and female mice would allow for us to determine the efficacy of these interventions in male mice as well as female mice. However, given the nature of the analysis (i.e., no three way interaction comparison), we cannot make strong inferences on either differential or equivalent responses to interventions between males and females. Regardless, that one intervention appeared efficacious in females and not in males implies that more research should at minimum be completed in male and female models and, where appropriate, be designed to compare differences between males and females.
In conclusion, we find that mitochondrially targeted catalase appears sufficient to mitigate muscle atrophy, yet this effect appears to be sex-specific. Additionally, we find overexpression of PGC1α insufficient to protect against disuse muscle atrophy. Hindlimb unloading resulted in similar muscle losses in both PGC and WT animals, despite PGC animals having substantially less ubiquitin-proteasome activation. However, PGC hindlimb unloaded animals appear to preferentially rely on autophagy-mediated catabolism compared with the ubiquitin-proteasome system, particularly in females. Conversely, we find that overexpression of mitochondrially targeted catalase was sufficient to mitigate disuse response in female mice. This effect appeared to be more robust in oxidative muscles such as the soleus and appears to be at least partially moderated by reduced oxidative stress within the muscle. Taken together, our results suggest different mechanisms contributing to disuse atrophy in males and females and imply the need for different therapeutic targets in males and females. However, more studies are needed to further understand potential sex and age interactions with catalase during disuse muscle pathologies. Regardless, these findings further support the necessity of investigating prospective muscle therapeutics in both male and female models.
GRANTS
This study was funded by the National Institutes of Health Award numbers R15AR069913/AR/NIAMS and P20GM125503.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
M.E.R-C., T.A.W., and N.P.G. conceived and designed research; M.E.R-C., S.L., W.S.H., L.T.J., L.C.W., and N.P.G. performed experiments; M.E.R-C., L.T.J., L.C.W., and M.G.A. analyzed data; M.E.R-C., S.L., W.S.H., T.A.W., and N.P.G. interpreted results of experiments; M.E.R-C. prepared figures; M.E.R-C. and N.P.G. drafted manuscript; M.E.R-C., S.L., W.S.H., L.T.J., L.C.W., M.G.A., T.A.W., and N.P.G. edited and revised manuscript; M.E.R-C., S.L., W.S.H., L.T.J., L.C.W., M.G.A., T.A.W., and N.P.G. approved final version of manuscript.
REFERENCES
- 3.Armstrong RB, Phelps RO. Muscle fiber type composition of the rat hindlimb. Am J Anat 171: 259–272, 1984. doi: 10.1002/aja.1001710303. [DOI] [PubMed] [Google Scholar]
- 4.Assi M, Derbré F, Lefeuvre-Orfila L, Rébillard A. Antioxidant supplementation accelerates cachexia development by promoting tumor growth in C26 tumor-bearing mice. Free Radic Biol Med 91: 204–214, 2016. [DOI] [PubMed] [Google Scholar]
- 4a.Barbeau P-A, Miotto PM, Holloway GP. Mitochondrial-derived reactive oxygen species influence ADP sensitivity, but not CPT-I substrate sensitivity. Biochem J 475: 2997–3008, 2018. doi: 10.1042/BCJ20180419. [DOI] [PubMed] [Google Scholar]
- 4b.Basisty N, Dai DF, Gagnidze A, Gitari L, Fredrickson J, Maina Y, Beyer RP, Emond MJ, Hsieh EJ, MacCoss MJ, Martin GM, Rabinovitch PS. Mitochondrial-targeted catalase is good for the old mouse proteome, but not for the young: ‘reverse’ antagonistic pleiotropy? Aging Cell 15: 634–645, 2016. doi: 10.1111/acel.12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bialek P, Morris C, Parkington J, St Andre M, Owens J, Yaworsky P, Seeherman H, Jelinsky SA. Distinct protein degradation profiles are induced by different disuse models of skeletal muscle atrophy. Physiol Genomics 43: 1075–1086, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brault JJ, Jespersen JG, Goldberg AL. Peroxisome proliferator-activated receptor gamma coactivator 1alpha or 1beta overexpression inhibits muscle protein degradation, induction of ubiquitin ligases, and disuse atrophy. J Biol Chem 285: 19460–19471, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brocca L, Pellegrino MA, Desaphy JF, Pierno S, Camerino DC, Bottinelli R. Is oxidative stress a cause or consequence of disuse muscle atrophy in mice? A proteomic approach in hindlimb-unloaded mice. Exp Physiol 95: 331–350, 2010. [DOI] [PubMed] [Google Scholar]
- 8.Brocca L, Toniolo L, Reggiani C, Bottinelli R, Sandri M, Pellegrino MA. FoxO-dependent atrogenes vary among catabolic conditions and play a key role in muscle atrophy induced by hindlimb suspension. J Physiol 595: 1143–1158, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown JL, Lee DE, Rosa-Caldwell ME, Brown LA, Perry RA, Haynie WS, Huseman K, Sataranatarajan K, Van Remmen H, Washington TA, Wiggs MP, Greene NP. Protein imbalance in the development of skeletal muscle wasting in tumour-bearing mice. J Cachexia Sarcopenia Muscle 9: 987–1002, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown JL, Rosa-Caldwell ME, Lee DE, Blackwell TA, Brown LA, Perry RA, Haynie WS, Hardee JP, Carson JA, Wiggs MP, Washington TA, Greene NP. Mitochondrial degeneration precedes the development of muscle atrophy in progression of cancer cachexia in tumour-bearing mice. J Cachexia Sarcopenia Muscle 8: 926–938, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cannavino J, Brocca L, Sandri M, Bottinelli R, Pellegrino MA. PGC1-α over-expression prevents metabolic alterations and soleus muscle atrophy in hindlimb unloaded mice. J Physiol 592: 4575–4589, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cannavino J, Brocca L, Sandri M, Grassi B, Bottinelli R, Pellegrino MA. The role of alterations in mitochondrial dynamics and PGC-1α over-expression in fast muscle atrophy following hindlimb unloading. J Physiol 593: 1981–1995, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi CS, Befroy DE, Codella R, Kim S, Reznick RM, Hwang YJ, Liu ZX, Lee HY, Distefano A, Samuel VT, Zhang D, Cline GW, Handschin C, Lin J, Petersen KF, Spiegelman BM, Shulman GI. Paradoxical effects of increased expression of PGC-1alpha on muscle mitochondrial function and insulin-stimulated muscle glucose metabolism. Proc Natl Acad Sci U S A 105: 19926–19931, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colom B, Alcolea MP, Valle A, Oliver J, Roca P, García-Palmer FJ. Skeletal muscle of female rats exhibit higher mitochondrial mass and oxidative-phosphorylative capacities compared to males. Cell Physiol Biochem 19: 205–212, 2007. [DOI] [PubMed] [Google Scholar]
- 16.De Jonghe B, Sharshar T, Lefaucheur JP, Authier FJ, Durand-Zaleski I, Boussarsar M, Cerf C, Renaud E, Mesrati F, Carlet J, Raphaël JC, Outin H, Bastuji-Garin S; Groupe de Réflexion et d’Etude des Neuromyopathies en Réanimation . Paresis acquired in the intensive care unit: a prospective multicenter study. JAMA 288: 2859–2867, 2002. [DOI] [PubMed] [Google Scholar]
- 18.Evans WJ Skeletal muscle loss: cachexia, sarcopenia, and inactivity. Am J Clin Nutr 91: 1123S–1127S, 2010. [DOI] [PubMed] [Google Scholar]
- 19.Gao Y, Arfat Y, Wang H, Goswami N. Muscle atrophy induced by mechanical unloading: mechanisms and potential countermeasures. Front Physiol 9: 235, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greene NP, Lee DE, Brown JL, Rosa ME, Brown LA, Perry RAJ, Henry JN, Washington TA. Mitochondrial quality control, promoted by PGC-1α, is dysregulated by Western diet-induced obesity and partially restored by moderate physical activity in mice. Physiol Rep 3: e12470, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.The Jackson Laboratory Protocol 26531 - Tg(CAG-OTC/CAT)4033Prab-alternate1 (Online) https://www.jax.org/Protocol?stockNumber=016197&protocolID=26531 [2020 Nov 04].
- 23.Jamart C, Raymackers JM, Li An G, Deldicque L, Francaux M. Prevention of muscle disuse atrophy by MG132 proteasome inhibitor. Muscle Nerve 43: 708–716, 2011. [DOI] [PubMed] [Google Scholar]
- 24.Jang J, Park J, Chang H, Lim K. l-Carnitine supplement reduces skeletal muscle atrophy induced by prolonged hindlimb suspension in rats. Appl Physiol Nutr Metab 41: 1240–1247, 2016. doi: 10.1139/apnm-2016-0094. [DOI] [PubMed] [Google Scholar]
- 25.Johnson JM, Ferrara PJ, Verkerke ARP, Coleman CB, Wentzler EJ, Neufer PD, Kew KA, de Castro Brás LE, Funai K. Targeted overexpression of catalase to mitochondria does not prevent cardioskeletal myopathy in Barth syndrome. J Mol Cell Cardiol 121: 94–102, 2018. doi: 10.1016/j.yjmcc.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones SW, Hill RJ, Krasney PA, O’Conner B, Peirce N, Greenhaff PL. Disuse atrophy and exercise rehabilitation in humans profoundly affects the expression of genes associated with the regulation of skeletal muscle mass. FASEB J 18: 1025–1027, 2004. doi: 10.1096/fj.03-1228fje. [DOI] [PubMed] [Google Scholar]
- 27.Kanazashi M, Tanaka M, Nakanishi R, Maeshige N, Fujino H. Effects of astaxanthin supplementation and electrical stimulation on muscle atrophy and decreased oxidative capacity in soleus muscle during hindlimb unloading in rats. J Physiol Sci 69: 757–767, 2019. doi: 10.1007/s12576-019-00692-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang C, Ji LL. Muscle immobilization and remobilization downregulates PGC-1α signaling and the mitochondrial biogenesis pathway. J Appl Physiol (1985) 115: 1618–1625, 2013. doi: 10.1152/japplphysiol.01354.2012. [DOI] [PubMed] [Google Scholar]
- 29.Kang C, Ji LL. PGC-1α overexpression via local transfection attenuates mitophagy pathway in muscle disuse atrophy. Free Radic Biol Med 93: 32–40, 2016. doi: 10.1016/j.freeradbiomed.2015.12.032. [DOI] [PubMed] [Google Scholar]
- 30.Laker RC, Xu P, Ryall KA, Sujkowski A, Kenwood BM, Chain KH, Zhang M, Royal MA, Hoehn KL, Driscoll M, Adler PN, Wessells RJ, Saucerman JJ, Yan Z. A novel MitoTimer reporter gene for mitochondrial content, structure, stress, and damage in vivo. J Biol Chem 289: 12005–12015, 2014. doi: 10.1074/jbc.M113.530527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Lark DS, Kang L, Lustig ME, Bonner JS, James FD, Neufer PD, Wasserman DH. Enhanced mitochondrial superoxide scavenging does not improve muscle insulin action in the high fat-fed mouse. PLoS One 10: e0126732, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawler JM, Song W, Demaree SR. Hindlimb unloading increases oxidative stress and disrupts antioxidant capacity in skeletal muscle. Free Radic Biol Med 35: 9–16, 2003. doi: 10.1016/S0891-5849(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 31a.Lee H-Y, Choi CS, Birkenfeld AL, Alves TC, Jornayvaz FR, Jurczak MJ, Zhang D, Woo DK, Shadel GS, Ladiges W, Rabinovitch PS, Santos JH, Petersen KF, Samuel VT, Shulman GI. Targeted expression of catalase to mitochondria prevents age-associated reductions in mitochondrial function and insulin resistance. Cell Metab 12: 668–674, 2010. doi: 10.1016/j.cmet.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31b.Lee HY, Lee JS, Alves T, Ladiges W, Rabinovitch PS, Jurczak MJ, Choi CS, Shulman GI, Samuel VT, Ladiges W, Rabinovitch PS, Santos JH, Petersen KF, Samuel VT, Shulman GI. Mitochondrial-targeted catalase protects against high-fat diet-induced muscle insulin resistance by decreasing intramuscular lipid accumulation. Diabetes 66: 2072–2081, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leitner LM, Wilson RJ, Yan Z, Gödecke A. Reactive oxygen species/nitric oxide mediated inter-organ communication in skeletal muscle wasting diseases. Antioxid Redox Signal 26: 700–717, 2017. doi: 10.1089/ars.2016.6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levine S, Biswas C, Dierov J, Barsotti R, Shrager JB, Nguyen T, Sonnad S, Kucharchzuk JC, Kaiser LR, Singhal S, Budak MT. Increased proteolysis, myosin depletion, and atrophic AKT-FOXO signaling in human diaphragm disuse. Am J Respir Crit Care Med 183: 483–490, 2011. doi: 10.1164/rccm.200910-1487OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lipes J, Mardini L, Jayaraman D. Sex and mortality of hospitalized adults after admission to an intensive care unit. Am J Crit Care 22: 314–319, 2013. doi: 10.4037/ajcc2013225. [DOI] [PubMed] [Google Scholar]
- 35.Lira VA, Okutsu M, Zhang M, Greene NP, Laker RC, Breen DS, Hoehn KL, Yan Z. Autophagy is required for exercise training-induced skeletal muscle adaptation and improvement of physical performance. FASEB J 27: 4184–4193, 2013. doi: 10.1096/fj.13-228486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maki T, Yamamoto D, Nakanishi S, Iida K, Iguchi G, Takahashi Y, Kaji H, Chihara K, Okimura Y. Branched-chain amino acids reduce hindlimb suspension-induced muscle atrophy and protein levels of atrogin-1 and MuRF1 in rats. Nutr Res 32: 676–683, 2012. doi: 10.1016/j.nutres.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 38.Miura S, Kai Y, Ono M, Ezaki O. Overexpression of peroxisome proliferator-activated receptor gamma coactivator-1alpha down-regulates GLUT4 mRNA in skeletal muscles. J Biol Chem 278: 31385–31390, 2003. doi: 10.1074/jbc.M304312200. [DOI] [PubMed] [Google Scholar]
- 39.Momken I, Stevens L, Bergouignan A, Desplanches D, Rudwill F, Chery I, Zahariev A, Zahn S, Stein TP, Sebedio JL, Pujos-Guillot E, Falempin M, Simon C, Coxam V, Andrianjafiniony T, Gauquelin-Koch G, Picquet F, Blanc S. Resveratrol prevents the wasting disorders of mechanical unloading by acting as a physical exercise mimetic in the rat. FASEB J 25: 3646–3660, 2011. doi: 10.1096/fj.10-177295. [DOI] [PubMed] [Google Scholar]
- 40.Montero D, Madsen K, Meinild-Lundby AK, Edin F, Lundby C. Sexual dimorphism of substrate utilization: Differences in skeletal muscle mitochondrial volume density and function. Exp Physiol 103: 851–859, 2018. doi: 10.1113/EP087007. [DOI] [PubMed] [Google Scholar]
- 41.Morales MG, Abrigo J, Acuña MJ, Santos RA, Bader M, Brandan E, Simon F, Olguin H, Cabrera D, Cabello-Verrugio C. Angiotensin-(1-7) attenuates disuse skeletal muscle atrophy in mice via its receptor, Mas. Dis Model Mech 9: 441–449, 2016. doi: 10.1242/dmm.023390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mortreux M, Riveros D, Bouxsein ML, Rutkove SB. A moderate daily dose of resveratrol mitigates muscle deconditioning in a martian gravity analog. Front Physiol 10: 899, 2019. doi: 10.3389/fphys.2019.00899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neves RX, Rosa-Neto JC, Yamashita AS, Matos-Neto EM, Riccardi DM, Lira FS, Batista ML Jr, Seelaender M. White adipose tissue cells and the progression of cachexia: inflammatory pathways. J Cachexia Sarcopenia Muscle 7: 193–203, 2016. doi: 10.1002/jcsm.12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a.Nikawa T, Ishidoh K, Hirasaka K, Ishihara I, Ikemoto M, Kano M, Kominami E, Nonaka I, Ogawa T, Adams GR, Baldwin KM, Yasui N, Kishi K, Takeda S. Skeletal muscle gene expression in space-flown rats. FASEB J 18: 522–524, 2004. doi: 10.1096/fj.03-0419fje. [DOI] [PubMed] [Google Scholar]
- 45.Oliván S, Calvo AC, Manzano R, Zaragoza P, Osta R. Sex differences in constitutive autophagy. BioMed Res Int 2014: 652817, 2014. doi: 10.1155/2014/652817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piekarski A, Khaldi S, Greene E, Lassiter K, Mason JG, Anthony N, Bottje W, Dridi S. Tissue distribution, gender- and genotype-dependent expression of autophagy-related genes in avian species. PLoS One 9: e112449, 2014. doi: 10.1371/journal.pone.0112449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Powers SK Can antioxidants protect against disuse muscle atrophy? Sports Med 44, Suppl 2: S155–S165, 2014. doi: 10.1007/s40279-014-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Powers SK, Smuder AJ, Judge AR. Oxidative stress and disuse muscle atrophy: cause or consequence? Curr Opin Clin Nutr Metab Care 15: 240–245, 2012. doi: 10.1097/MCO.0b013e328352b4c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50a.Powers SK, Wiggs MP, Duarte JA, Zergeroglu AM, Demirel HA. Mitochondrial signaling contributes to disuse muscle atrophy. Am J Physiol Endocrinol Metab 303: E31–E39, 2012. doi: 10.1152/ajpendo.00609.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosa-Caldwell ME, Brown JL, Perry RA Jr, Shimkus KL, Shirazi-Fard Y, Brown LA, Hogan HA, Fluckey JD, Washington TA, Wiggs MP, Greene NP. Regulation of mitochondrial quality following repeated bouts of hindlimb unloading. Appl Physiol Nutr Metab 45: 264–274, 2020. doi: 10.1139/apnm-2019-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosa-Caldwell ME, Greene NP. Muscle metabolism and atrophy: let’s talk about sex. Biol Sex Differ 10: 43, 2019. doi: 10.1186/s13293-019-0257-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosa-Caldwell ME, Lee DE, Brown JL, Brown LA, Perry RA Jr, Greene ES, Carvallo Chaigneau FR, Washington TA, Greene NP. Moderate physical activity promotes basal hepatic autophagy in diet-induced obese mice. Appl Physiol Nutr Metab 42: 148–156, 2017. doi: 10.1139/apnm-2016-0280. [DOI] [PubMed] [Google Scholar]
- 54.Russell AP, Foletta VC, Snow RJ, Wadley GD. Skeletal muscle mitochondria: a major player in exercise, health and disease. Biochim Biophys Acta 1840: 1276–1284, 2014. doi: 10.1016/j.bbagen.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 55.Ryan AM, Power DG, Daly L, Cushen SJ, Ní Bhuachalla Ē, Prado CM. Cancer-associated malnutrition, cachexia and sarcopenia: the skeleton in the hospital closet 40 years later. Proc Nutr Soc 75: 199–211, 2016. doi: 10.1017/S002966511500419X. [DOI] [PubMed] [Google Scholar]
- 56.Sacheck JM, Hyatt JP, Raffaello A, Jagoe RT, Roy RR, Edgerton VR, Lecker SH, Goldberg AL. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J 21: 140–155, 2007. doi: 10.1096/fj.06-6604com. [DOI] [PubMed] [Google Scholar]
- 57.Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, Goldberg AL, Spiegelman BM. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci USA 103: 16260–16265, 2006. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, Coskun PE, Ladiges W, Wolf N, Van Remmen H, Wallace DC, Rabinovitch PS. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science 308: 1909–1911, 2005. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- 59.Servais S, Letexier D, Favier R, Duchamp C, Desplanches D. Prevention of unloading-induced atrophy by vitamin E supplementation: links between oxidative stress and soleus muscle proteolysis? Free Radic Biol Med 42: 627–635, 2007. doi: 10.1016/j.freeradbiomed.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shenkman BS, Nemirovskaya TL, Belozerova IN, Mazin MG, Matveeva OA. Mitochondrial adaptations in skeletal muscle cells in mammals exposed to gravitational unloading. J Gravit Physiol 9: 159–162, 2002. [PubMed] [Google Scholar]
- 61.Stevenson EJ, Giresi PG, Koncarevic A, Kandarian SC. Global analysis of gene expression patterns during disuse atrophy in rat skeletal muscle. J Physiol 551: 33–48, 2003. doi: 10.1113/jphysiol.2003.044701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Talbert EE, Metzger GA, He WA, Guttridge DC. Modeling human cancer cachexia in colon 26 tumor-bearing adult mice. J Cachexia Sarcopenia Muscle 5: 321–328, 2014. doi: 10.1007/s13539-014-0141-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Talbert EE, Smuder AJ, Min K, Kwon OS, Szeto HH, Powers SK. Immobilization-induced activation of key proteolytic systems in skeletal muscles is prevented by a mitochondria-targeted antioxidant. J Appl Physiol (1985) 115: 529–538, 2013. doi: 10.1152/japplphysiol.00471.2013. [DOI] [PubMed] [Google Scholar]
- 65.Theeuwes WF, Gosker HR, Langen RCJ, Pansters NAM, Schols AMWJ, Remels AHV. Inactivation of glycogen synthase kinase 3β (GSK-3β) enhances mitochondrial biogenesis during myogenesis. Biochim Biophys Acta Mol Basis Dis 1864, 9 Pt B: 2913–2926, 2018. doi: 10.1016/j.bbadis.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 65a.Trevino MB, Zhang X, Standley RA, Wang M, Han X, Reis FCG, Periasamy M, Yu G, Kelly DP, Goodpaster BH, Vega RB, Coen PM. Loss of mitochondrial energetics is associated with poor recovery of muscle function but not mass following disuse atrophy. Am J Physiol Endocrinol Metab 317: E899–E910, 2019. doi: 10.1152/ajpendo.00161.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65b.Umanskaya A, Santulli G, Xie W, Andersson DC, Reiken SR, Marks AR. Genetically enhancing mitochondrial antioxidant activity improves muscle function in aging. Proc Natl Acad Sci USA 111: 15250–15255, 2014. doi: 10.1073/pnas.1412754111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vainshtein A, Desjardins EM, Armani A, Sandri M, Hood DA. PGC-1α modulates denervation-induced mitophagy in skeletal muscle. Skelet Muscle 5: 9, 2015. doi: 10.1186/s13395-015-0033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang J, Wang F, Zhang P, Liu H, He J, Zhang C, Fan M, Chen X. PGC-1α over-expression suppresses the skeletal muscle atrophy and myofiber-type composition during hindlimb unloading. Biosci Biotechnol Biochem 81: 500–513, 2017. doi: 10.1080/09168451.2016.1254531. [DOI] [PubMed] [Google Scholar]
- 68.Washington TA, White JP, Davis JM, Wilson LB, Lowe LL, Sato S, Carson JA. Skeletal muscle mass recovery from atrophy in IL-6 knockout mice. Acta Physiol (Oxf) 202: 657–669, 2011. doi: 10.1111/j.1748-1716.2011.02281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68a.Weijs PJ, Looijaard WG, Dekker IM, Stapel SN, Girbes AR, Oudemans-van Straaten HM, Beishuizen A. Low skeletal muscle area is a risk factor for mortality in mechanically ventilated critically ill patients. Crit Care 18: R12, 2014. doi: 10.1186/cc13189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.White JP, Puppa MJ, Sato S, Gao S, Price RL, Baynes JW, Kostek MC, Matesic LE, Carson JA. IL-6 regulation on skeletal muscle mitochondrial remodeling during cancer cachexia in the ApcMin/+ mouse. Skelet Muscle 2: 14, 2012. doi: 10.1186/2044-5040-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamashita AS, das Neves RX, Rosa-Neto JC, Lira FD, Batista ML Jr, Alcantara PS, Otoch JP, Seelaender M. White adipose tissue IFN-γ expression and signalling along the progression of rodent cancer cachexia. Cytokine 89: 122–126, 2017. doi: 10.1016/j.cyto.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 71.Yan Z, Lira VA, Greene NP. Exercise training-induced regulation of mitochondrial quality. Exerc Sport Sci Rev 40: 159–164, 2012. doi: 10.1097/JES.0b013e3182575599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yeo D, Kang C, Gomez-Cabrera MC, Vina J, Ji LL. Intensified mitophagy in skeletal muscle with aging is downregulated by PGC-1alpha overexpression in vivo. Free Radic Biol Med 130: 361–368, 2019. doi: 10.1016/j.freeradbiomed.2018.10.456. [DOI] [PubMed] [Google Scholar]
- 73.Zhang P, Li W, Liu H, Li J, Wang J, Li Y, Chen X, Yang Z, Fan M. Dystrophin involved in the susceptibility of slow muscles to hindlimb unloading via concomitant activation of TGF-β1/Smad3 signaling and ubiquitin-proteasome degradation in mice. Cell Biochem Biophys 70: 1057–1067, 2014. doi: 10.1007/s12013-014-0023-4. [DOI] [PubMed] [Google Scholar]