Abstract
The progression of COVID-19 worldwide can be tracked by identifying mutations within the genomic sequence of SARS-CoV-2 that occur as a function of time. Such efforts currently rely on sequencing the genome of SARS-CoV-2 in patient specimens (direct sequencing) or of virus isolated from patient specimens in cell cultures. A pilot SARS-CoV-2 air sampling study conducted at a clinic within a university student health care center detected the virus vRNA, with an estimated concentration of 0.87 virus genomes L−1 air. To determine whether the virus detected was viable (‘live’), attempts were made to isolate the virus in cell cultures. Virus-induced cytopathic effects (CPE) were observed within two days post-inoculation of Vero E6 cells with collection media from air samples; however, rtRT-PCR tests for SARS-CoV-2 vRNA from cell culture were negative. Instead, three other fast-growing human respiratory viruses were isolated and subsequently identified, illustrating the challenge in isolating SARS-CoV-2 when multiple viruses are present in a test sample. The complete SAR-CoV-2 genomic sequence was nevertheless determined by Sanger sequencing and most closely resembles SARS-CoV-2 genomes previously described in Georgia, USA. Results of this study illustrate the feasibility of tracking progression of the COVID-19 pandemic using environmental aerosol samples instead of human specimens. Collection of a positive sample from a distance more than 2 m away from the nearest patient traffic implies the virus was in an aerosol.
Keywords: aerosol, air sampling, COVID-19
INTRODUCTION
The worldwide spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes coronavirus disease 2019 (COVID-19), has intensified attention to precautions and implementation of infection control processes to reduce its spread in hospitals, clinics and the public. Among the various measures taken to stem the spread of the virus, tracking the progression of the COVID-19 pandemic by analyzing the genomic sequence of SARS-CoV-2 as it spreads internationally (Forster et al., 2020) is a critical one. Currently, such efforts rely on sequencing the genome of SARS-CoV-2 in patient specimens (direct sequencing) or of virus isolated from patient specimens in cell cultures. Direct sequencing of the complete virus genome in patient specimens by common next-generation sequencing (NGS) technology is not always possible for many reasons, e.g., the amount of virus present in any given specimen may be too low to perform whole-genome analyses. Moreover, reliance on human specimens paints an incomplete picture since pre-symptomatic and asymptomatic individuals can also shed infectious SARS-CoV-2 virus (Anderson et al., 2020; Rothe et al., 2020), but many of them do not report to clinics, and thus specimens are not available for analyses. In the absence of testing of all citizens, environmental sampling, which is a non-invasive procedure, may bridge the gap by collecting SARS-CoV-2 shed into air by pre- or asymptomatic people.
Conventionally, infectious respiratory viruses are said to be transmitted mainly through contact with respiratory droplets (> 5 μm). However, evidence is mounting that the virus can be transmitted as fine particles (< 5 μm) (Chia et al., 2020; Liu et al., 2020; Santarpia et al., 2020) present in aerosols formed when a SARS-CoV-2 infected person coughs, sneezes, breathes, or talks (Hsiao et al., 2020). If so, environmental aerosol sampling may help us better understand the transmission process as it allows collection of airborne particles containing SARS-CoV-2 as mentioned above. Nevertheless, the collection of airborne virus for inhalation risk and genome analyses using air samplers is challenging for several reasons (Pan et al., 2019): (A) Certain air samplers such as impingers are inefficient in collecting the smallest size fractions, yet these are where airborne virus particles are often found (Hogan et al., 2005), (B) Filter-based sampling typically desiccates the collected material as air passes through (or by) the filters (Pan et al., 2019) and that drying can result in inactivation of many types of virus, rendering risk analyses impossible (inactivated viruses are non-infectious and cannot cause disease), and (C) Airborne-virus concentrations can be low, either naturally or at the site of collection, making collection and thus detection thereof difficult. The latter was the case for SARS-CoV-2 in one study, wherein its concentration in air inside a patient’s room was low, e.g., mean concentration of 2.86 genome equivalents L−1 of air (Santarpia et al., 2020). To collect enough airborne virus for detection by current molecular techniques, longer sampling times may be needed. However, collection media that desiccate during usage may not be suitable, e.g. gelatin filter or impingers wherein water evaporates during sampling (Nevalainen et al., 1993), as the virus can become inactivated through desiccation.
Whereas there have been reports regarding the detection of SARS-CoV-2 in air samples (Chia et al., 2020; Liu et al., 2020; Santarpia et al., 2020), it is still unclear whether aerosol transmission is important for the spread of SARS-CoV-2 (Hadei et al., 2020). Here we present the results of a pilot SARS-CoV-2 air sampling study conducted at the Student Health Care Center (SHCC) at the University of Florida (Gainesville, FL, USA), wherein we too detect the virus in an air sample. Further, we present phylogenetic inference of the complete SARS-CoV-2 genomic sequence that was obtained in the study.
METHODOLOGY
Details of the sampling site, air sampling, detection of SARS-CoV-2 genomic RNA, quantification of SARS CoV-2 genomes, Sanger sequencing, and phylogenetic inference are provided as Supplementary Material.
RESULTS
rtRT-PCR analysis detected vRNA in an air sample (Table 1). The amount of virus present in 390 L of sampled air was low (approximately 340 virus genome equivalents). Accordingly, the estimated SARS-CoV-2 concentration was 0.87 virus genomes L−1 air.
Table 1.
Results of RT-PCR test of Air Samples on March 20, 2020
| Sample | Volume (µl) of collection media | RT-PCR test | Ct value | Virus genome equivalents/L of air |
|---|---|---|---|---|
| 1 | 1,489 | + | 39.13 | 0.87 |
| 2 | 1,497 | - | N/A | N/A |
Virus-induced cytopathic effects (CPE) were observed within two days post-inoculation of Vero E6 cells with collection media from air samples 1 and 2 (Fig. 1 B – D), leading to nearly complete destruction of the cell monolayer three days later. rtRT-PCR tests for SARS-CoV-2 vRNA from cell culture were negative, indicating that other virus(es) that outgrew SARS-CoV-2 had been isolated, or that the SARS-CoV-2 that had been detected was non-infectious. Indeed, three respiratory viruses were identified using the Biofire RVP: Influenza A H1N1, Influenza A H3N2, and Human coronavirus OC43 (Fig. 2).
Figure 1.
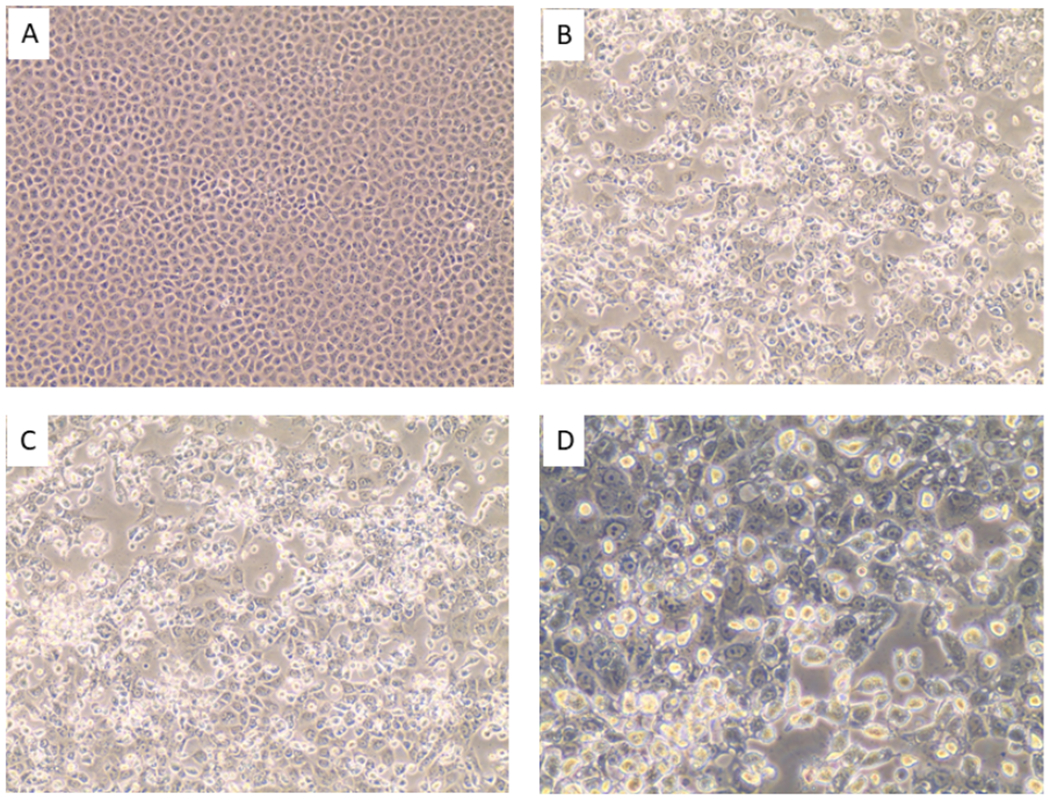
[A] Mock-infected Vero E6 cells. [B] Vero E6 cells inoculated with material collected from air sample 1. [C]. Vero E6 cells inoculated with material from air sample 2. Images A- C photographed at an original magnification of 200X. [D]. Close-up view (400x) of Vero E6 cells inoculated with material collected from air sample 1.
Figure 2.
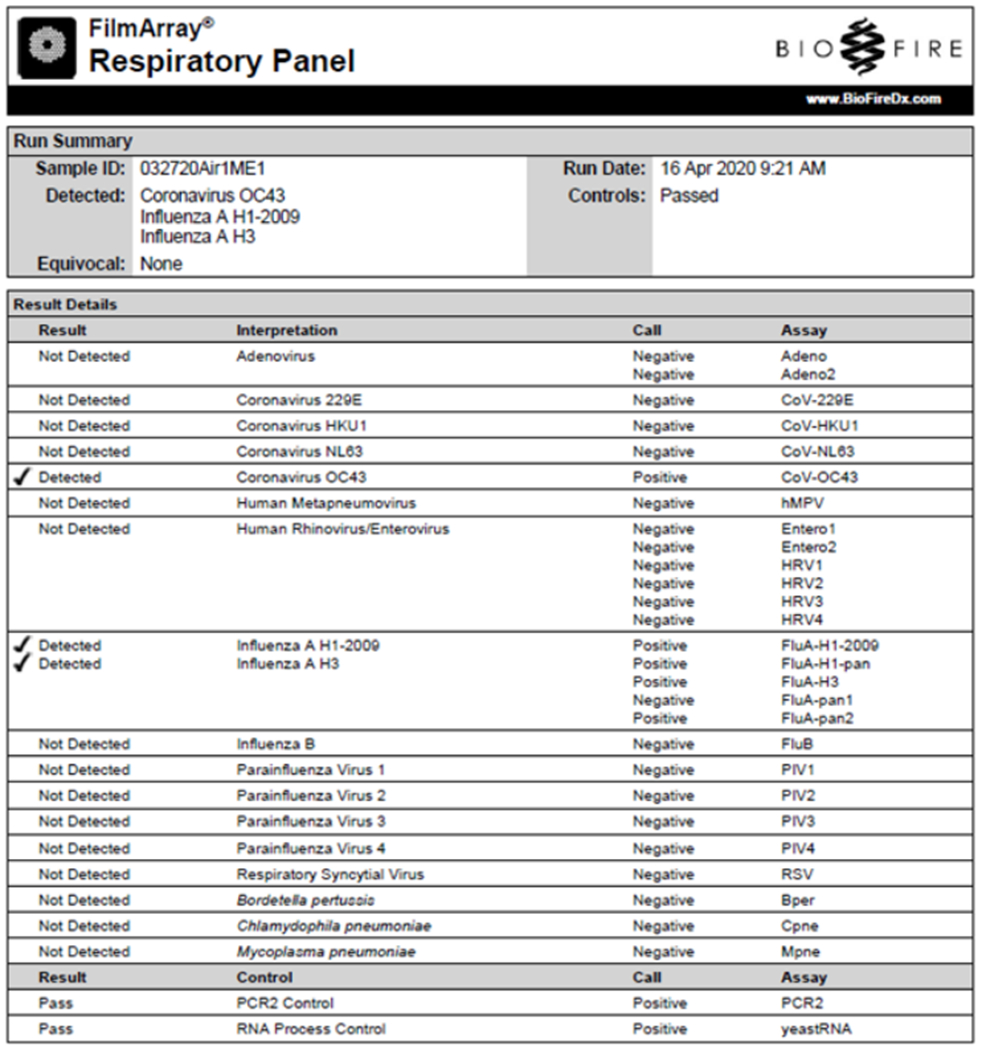
Biofire RVP test results for viruses isolated after inoculation of Vero E6 cells with sample 1. Human coronavirus OC43, and Influenza A H1N1 and H3N2 viruses were identified.
The low amount of SARS-CoV-2 in the air sample precluded NGS using an Illumina MiSeq platform. But the complete ribonucleotide sequence of the genome of the SARS-CoV-2 strain in the collection medium was determined by Sanger sequencing and was deposited at Genbank (accession no. MT324684.1). To our knowledge, this is the first complete genomic sequence of SARS-CoV-2 determined from an aerosol sample. Maximum likelihood (ML) phylogenetic inference of 6203 complete genome sequences of SARS-CoV-2 as of 11 April 2020 was possible due to presence of phylogenetic signal, as previously reported (Mavian et al., 2020). ML tree revealed that SARS-CoV-2_air_sampler_USA_UF-3_2020-03-25 (air sample) is located at the base of two well-supported clades, together with two other strains from Georgia (EPI_ISL_420786, EPI_ISL_420788) (Fig. 3). One clade contains a majority of Australian strains, one strain from New Zealand, and two from the USA (EPI_ISL_418045 from Idaho, and EPI_ISL_418970 from New York). The second clade includes four strains clustering together with high support: two from the US (EPI_ISL_418897 from Washington State, and EPI_ISL_421426 from New York), one from Australia (EPI_ISL_419904), and one from Taiwan (EPI_ISL_422415). As the Florida and Georgia strains have nearly zero branch distance between each other, it is not possible to infer which seeded the other.
Figure 3.

Maximum likelihood subtree of SARS-CoV-2. Phylogeny of subtree SARS-CoV-2_air_sampler_USA_UF-3_2020-03-25 clusters. Diamonds at nodes indicate ultrafast bootstrap (BB) support >90%.
The result of this pilot environmental sampling provided evidence of a SARS-CoV-2 aerosol at the SHCC, as the sample was collected 3 m away from the nearest patient traffic, i.e., greater than 2 m defined by WHO for aerosol transmission (WHO, 2020). Our findings helped the SHCC administration execute actions that better protect healthcare workers. Following detection of SARS-CoV-2 in air by our March 20 air-sampling study, remediation was accomplished by performing a 2-day decontamination process using a stabilized chlorine dioxide solution (VitalOxide™) delivered through a handheld electrostatic mist sprayer (Clorox Total 360 System®). Air samplings were repeated on March 27, and all rtRT-PCR tests for SARS-CoV-2 vRNA were negative, suggesting that the remediation actions were effective for any SARS-CoV-2 remaining in the air. An additional chlorine dioxide treatment was applied that evening, March 27, and an ultraviolet light unit was added to the air handling system before the clinic space was reopened on March 30. All staff with access to any area of the respiratory clinic even if not directly involved with patient care, were instructed to wear N95 masks. Previously, staff not engaged in patient work wore surgical masks.
DISCUSSION
While the sampling in this study was conducted at a healthcare facility, the same method can be adopted for other types of locations, such as airports, train stations, shopping centers, city centers or churches, i.e., areas where people congregate. This is important, because as pointed out earlier, asymptomatic individuals also shed virus, but they are up to now rarely tested. Furthermore, environmental sampling may result in earlier detection of an (re-)emerging virus than the current practice relying on human specimens availed only after very sick patients start to report to clinics. Note that the concentration of airborne virus in these locations is currently unknown. Thus, it may require a longer sampling time than the 1 hr used in this study.
It should also be pointed out that air samples collected at health clinics that treat patients with respiratory infections typically contain viruses from more than one person (Pan et al., 2017). The amount and variety of viruses collected depends on the duration of sampling, the type of viruses in circulation, and the number of patients in the sampling area, among other factors. Such an aggregated approach may also require modification of currently used phylogenetic analysis based on individual genome sequences if more than one different strain of the same virus is collected and analyzed. Thus, it may allow a holistic risk analysis that considers a suite of viruses, with advanced analyses to determine the variety and quantity of each virus type.
Supplementary Material
ACKNOWLEDGEMENTS
Research reported in this publication was based upon work supported by the National Science Foundation under Grant No. 2030844 and partially supported by National Institute of Environmental Health Sciences of the National Institutes of Health under award number 1R43ES030649. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Science Foundation and the National Institutes of Health. The authors are grateful to Dr. Ronald Berry, Director, and the staff of the UF Student Health Care Center for assistance in sampling at the clinical site.
REFERENCES
- Anderson RM, Heesterbeek H, Klinkenberg D and Hollingsworth TD (2020). How Will Country-Based Mitigation Measures Influence the Course of the COVID-19 Epidemic? Lancet 395: 931–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia PY, Coleman KK, Tan YK, Ong SWX, Gum M, Lau SK, Sutjipto S, Lee PH, Son TT, Young BE, Milton DK, Gray GC, Schuster S, Barkham T, De PP, Vasoo S, Chan M, Ang BSP, Tan BH, Leo YS, Ng O-T, Wong MSY and Marimuthu K (2020). Detection of Air and Surface Contamination by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in Hospital Rooms of Infected Patients. medRxiv: 2020.2003.2029.20046557. [Google Scholar]
- Forster P, Forster L, Renfrew C and Forster M (2020). Phylogenetic Network Analysis of SARS-CoV-2 Genomes. Proc. Natl. Acad. Sci. U.S.A 117: 9241–9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadei M, Hopke PK, Jonidi A and Shahsavani A (2020). A Letter About the Airborne Transmission of SARS-CoV-2 Based on the Current Evidence. Aerosol Air Qual. Res 20: 911–914. [Google Scholar]
- Hogan CJJ, Kettleson EM, Lee MH, Ramaswami B, Angenent LT and Biswas P (2005). Sampling Methodologies and Dosage Assessment Techniques for Submicrometer and Ultrafine Virus Aerosol Particles. J. Appl. Microb 99: 1422–1434. [DOI] [PubMed] [Google Scholar]
- Hsiao T-C, Chuang H-C, Griffith SM, Chen S-J and Young L-H (2020). COVID-19: An Aerosol’s Point of View from Expiration to Transmission to Viral-Mechanism. Aerosol Air Qual. Res: 905–910. [Google Scholar]
- Liu Y, Ning Z, Chen Y, Guo M, Liu Y, Gali NK, Sun L, Duan Y, Cai J, Westerdahl D, Liu X, Xu K, Ho K. f., Kan H, Fu Q and Lan K (2020). Aerodynamic Analysis of SARS-CoV-2 in Two Wuhan Hospitals. Nature s 41586-020-2271-3. [DOI] [PubMed] [Google Scholar]
- Mavian C, Marini S, Prosperi M and Salemi M (2020). A Snapshot of SARS-CoV-2 Genome Availability up to 30th March, 2020 and Its Implications. bioRxiv: 2020.2004.2001.020594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevalainen A, Willeke K, Liebhaber F, Pastuszka J, Burge H and Henningson E (1993). Bioaerosol Sampling, In Aerosol Measurement: Principles, Techniques and Applications, Willeke K and Baron PA (Eds.), Van Nostrand Reinhold, New York, NY, pp. 471–492. [Google Scholar]
- Pan M, Bonny TS, Loeb J, Jiang X, Lednicky JA, Eiguren-Fernandez A, Hering S, Fan ZH and Wu CY (2017). Collection of Viable Aerosolized Influenza Virus and Other Respiratory Viruses in a Student Health Care Center through Water-Based Condensation Growth. mSphere 2: e00251–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M, Lednicky JA and Wu CY (2019). Collection, Particle Sizing and Detection of Airborne Viruses. J. Appl. Microb 127: 1596–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, Zimmer T, Thiel V, Janke C, Guggemos W, Seilmaier M, Drosten C, Vollmar P, Zwirglmaier K, Zange S, Wölfel R and Hoelscher M (2020). Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N. Engl. J. Med 382: 970–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarpia JL, Rivera DN, Herrera V, Morwitzer MJ, Creager H, Santarpia GW, Crown KK, Brett-Major D, Schnaubelt E, Broadhurst MJ, Lawler JV, Reid SP and Lowe JJ (2020). Transmission Potential of SARS-CoV-2 in Viral Shedding Observed at the University of Nebraska Medical Center. medRxiv: 2020.2003.2023.20039446. [Google Scholar]
- WHO (2020). Modes of Transmission of Virus Causing COVID-19: Implications for Ipc Precaution Recommendations, In Scientific Brief, WHO, Geneva, Switzerland. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


