Abstract
Malaria continues to impose a significant health burden in the continent of Africa with 213 million cases in 2018 alone, representing 93% of cases worldwide. Because of high transmission of malaria within the continent, the selection pressures to develop drug resistance in African parasites are distinct compared to the rest of the world. In light of the spread of resistance to artemisinin conferred by the C580Y mutation in the PfKelch13 propeller domain in Southeast Asia, and its independent emergence in South America, it is important to study genetic determinants of resistance in the African context using African parasites. Through in vitro evolution of Senegalese parasites, we had previously generated the artemisinin-resistant parasites Pikine_R and Thiès_R and established pfcoronin mutations to be sufficient to confer artemisinin resistance in the standard ring-stage survival assay (RSA). In the current study, we used genetic analysis of revertants to demonstrate pfcoronin to be the major driver of elevated RSA in the artemisinin-resistant parasites Pikine_R and Thiès_R evolved in vitro. We interrogated the role of a second gene PF3D7_1433800, which also had mutations in both the Pikine_R and Thiès_R selected lines, but found no evidence of a contribution to reduced susceptibility in the RSA survival assay. Nevertheless, our genetic analysis demonstrates that parasite genetic background is important in the level of pfcoronin mediated RSA survival, and therefore we cannot rule out a role for PF3D7_1433800 in other genetic backgrounds. Finally, we tested the potential synergy between the mutations of pfcoronin and pfkelch13 through the generation of single and double mutants in the Pikine genetic background and found that the contribution of pfcoronin to reduced susceptibility is masked by the presence of pfkelch13. This phenomenon was also observed in the 3D7 background, suggesting that pfcoronin may mediate its effects via the same pathway as pfkelch13. Investigating the biology of proteins containing the beta-propeller domain could further elucidate the different pathways that the parasite could use to attain resistance.
Author summary
Artemisinin-based combination therapies remain a crucial standard treatment in combatting malaria around the world. Given the independent emergence of artemisinin resistance associated with pfkelch13 mutations in Southeast Asia and South America, it is important to study genetic determinants in the African context. Previously, we had generated artemisinin-resistant parasites, referred to as Pikine_R and Thiès_R, by subjecting clinical isolates of parasites from Senegal to artemisinin drug pressure in the laboratory. We found that mutations in the pfcoronin gene are sufficient to result in resistance in a standard laboratory assay. Here we report that pfcoronin mutations are also necessary for the level of artemisinin resistance observed in both Pikine_R and Thiès_R. In diverse parasite strains the level of resistance is also sensitive to genetic background. Genetic analysis revealed that the resistance phenotype of pfkelch13 alone masks any additional contribution from pfcoronin. Future studies should focus on investigating pfcoronin biology.
Introduction
There were 228 million cases of malaria and over 400,000 malaria deaths worldwide in 2018 [1]. Although significant reductions in disease burden have been achieved since the adoption of artemisinin combination therapies (ACTs) in the early 2000s, the greatest burden of malaria, primarily caused by Plasmodium falciparum, continues to occur on the African continent [1]. However, artemisinin drug resistance has emerged in Southeast Asia [2] that is attributed to mutations in the propeller domain of the Kelch13 protein of P. falciparum (PfKelch13). The C580Y mutation in particular, which is approaching fixation in Southeast Asia [3], has recently been reported to have independently emerged in South America [4] and Papua New Guinea [5], threatening the success achieved by ACTs. ACTs largely remain effective in the African continent, despite recent reports of pfkelch13 mutations in Rwanda [6] and Tanzania [7]. Several reports of resistance occurring independently of pfkelch13, suggest more than one genetic pathway for resistance [8–10]. Given that the PfKelch13 C580Y mutation has now been linked to a slowdown of the endocytic machinery for hemoglobin uptake [11], it has yet to be determined if all the different genetic pathways of resistance converge in reducing the uptake of hemoglobin.
The prevalence of malaria, level of immunity, and complexity of infection are very different in the low-transmission setting of Southeast Asia as compared with the African continent. Since Africa has the highest disease burden of P. falciparum malaria, the selection pressures on African parasites are markedly different from those in Southeast Asia [12,13]. Mutations in AP2 mu, encoding the AP-2 complex subunit mu [14], and ubp1, encoding the ubiquitin hydrolase, have been linked to artemisinin resistance only in parasites with African genetic backgrounds [9,15]. This observation implies that mechanistic studies of artemisinin resistance should include parasites from different parts of the African continent.
Our interest is therefore in understanding artemisinin resistance in African parasites. Using in vitro evolution of resistance, coupled with next generation whole genome sequencing, we previously identified two genes (PF3D7_1251200 pfcoronin and PF3D7_1433800 conserved plasmodium protein of unknown function [16]) that had mutations in both the independently selected resistant parasites Pikine_R and Thiès_R [17]. No mutations in pfkelch13 were detected. Mutations in pfcoronin G50E (in Thiès_R) and R100K & E107V (in Pikine_R) were of special interest because of the structural similarity of the WD-40 beta-propeller domain of the actin-bundling protein PfCoronin to the beta-propeller domain of PfKelch13, the known marker of artemisinin resistance in Southeast Asia [2]. Through CRISPR-Cas9 mediated introduction of pfcoronin mutations into their respective parental wildtype genetic backgrounds, we established that the PfCoronin mutations are sufficient to confer reduced artemisinin susceptibility as measured by the gold-standard ring-stage survival assay (RSA) [17].
In the work presented here, we further analyzed the role of pfcoronin in conferring reduced artemisinin sensitivity in the in vitro evolved parasites Thiès_R and Pikine_R. We also studied the role of genetic background in resistance using the standard laboratory strain 3D7. In addition, we investigated the contribution of mutations in PF3D7_1433800, which encodes a conserved Plasmodium protein of unknown function, and we examined potential phenotypic synergy between pfcoronin and pfkelch13 mutations.
Results
PfCoronin is the major driver of reduced susceptibility to artemisinin in the in vitro evolved Senegalese parasites
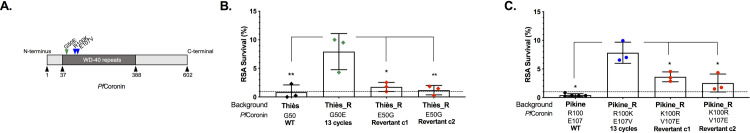
Pikine_R and Thies_R parasites were selected via in vitro evolution from parental Senegalese parasites. Each are resistant to artemisinin and carry mutations in the pfcoronin gene [17]. Using CRISPR-Cas9 gene editing, we successfully reverted the mutations in pfcoronin (Fig 1A) back to the wildtype in both Thiès_R (E50G, S1 Fig) and Pikine_R (K100R & V107E, S2 Fig). When two clones of pfcoronin revertants of Thiès_R and the Thiès wildtype (control) were evaluated using RSA and compared to the RSA phenotype of Thiès_R published previously [17] (7.62 ± 2.52%), we found a significant reduction in RSA survival for both clone 1 (1.36 ± 0.61%, p = 0.013) and clone 2 (1.19 ± 0.98%, p = 0.0098). The Thiès wildtype control had an RSA survival of 0.84 ± 1.22%, p = 0.0097 (Fig 1B). Similarly, in the Pikine genetic background, there was a significant reduction in RSA survival for two clones of pfcoronin revertants of Pikine_R [clone 1 (3.61 ± 1.33%, p = 0.04), clone 2 (2.53 ± 1.66%, p = 0.020)]. The Pikine wildtype control had an RSA survival of 0.43 ± 0.27% (p = 0.019) compared to the RSA phenotype of Pikine_R published previously [17] (7.83 ± 1.70%) (Fig 1C). All RSA survival values, their corresponding standard deviations, and their sequences confirmed post-RSA, are summarized in S1 and S2 Tables. All the revertant parasites had similar dose-response curves to derivatives of artemisinin as well as control compounds in a 72-hour drug assay (S3 Fig and S3 Table). These results confirmed that the PfCoronin mutations R100K & E107V and G50E are the major drivers of reduced artemisinin sensitivity in our in vitro evolved parasites Thiès_R and Pikine_R.
Fig 1. PfCoronin is the major driver of artemisinin resistance in the in vitro evolved artemisinin resistant parasites Thiès_R and Pikine_R.
A. Map of the PfCoronin protein indicating the positions of mutations in the WD-40 beta propeller domain associated with artemisinin resistance (G50E identified in Thiès_R and R100K and E107V identified in Pikine_R). Ring-stage survival assay (RSA) survival percentage for B. two clones of the reverted pfcoronin G50E mutation in the Thiès_R background (Revertant clone 1 and 2, red), and C. two clones of reverted pfcoronin R100K and E107V mutations in the Pikine_R background (Revertant clone 1 and 2, red) compared to their respective wildtype (WT) parasites (black). Previously published survivals for Thiès_R (green) and Pikine_R (blue) are included [17]. Each data point represents an independent biological replicate, and the dotted line represents the 1% survival threshold, above which survival is considered resistant. Significance level is indicated by the asterisks, *p<0.05, **p<0.01, determined using an unpaired two-tailed student t-test with Welch’s correction for data in B and Mann-Whitney U test for data in C.
Parasite background affects RSA survival of pfcoronin mutants
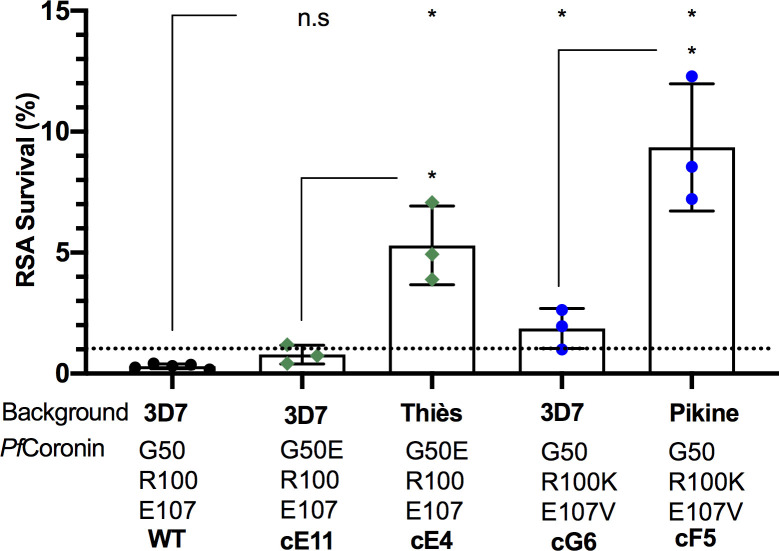
To test if RSA survival estimates for pfcoronin mutations are affected by the genetic background of the parasite, we generated pfcoronin mutants in a 3D7 laboratory parasite and performed RSA (S4 Fig). While the R100K and E107V mutations in PfCoronin were sufficient to pass the 1% RSA survival threshold (1.86 ± 0.86%), which was significantly higher than the 3D7 wildtype (0.30 ± 0.10%, p = 0.036), the PfCoronin G50E mutant did not have increased survival (0.79 ± 0.39%, p = 0.071) (Fig 2 and S1 and S2 Tables). These RSA survival values are significantly lower than those estimated for pfcoronin mutants in Pikine (cF5 survival: 9.35 ± 1.89%, p = 0.030) and Thiès (cE4 survival: 5.30 ± 1.25%, p = 0.035) genetic backgrounds generated previously using CRISPR/Cas9 gene editing [17] (Fig 2). These findings imply that parasite genetic background is indeed important in the observed level of RSA survival after DHA treatment, even with regard to pfcoronin mutations. These results have parallels in the observations for pfkelch13 when the C580Y mutation was introduced into different parasite backgrounds [18].
Fig 2. The level of resistance conferred by mutant pfcoronin is background dependent.
Ring-stage survival assay (RSA) survival percentage of pfcoronin mutants in 3D7 background with either G50E (clone E11, green) or R100K and E107V (clone G6, blue) compared to wildtype (WT) 3D7 (black), and previously published [17] survival values for one of the clones of pfcoronin CRISPR mutants in the Thiès (clone E4, green) and the Pikine (clone F5, blue) backgrounds. Each data point represents an independent biological replicate and the dotted line represents the 1% survival threshold, above which survival is considered resistant. Significance level is indicated by the asterisks, *p<0.05, determined using an unpaired two-tailed student t-test with Welch’s correction.
Mutations in PF3D7_1433800 make a minimal contribution to in vitro artemisinin resistance
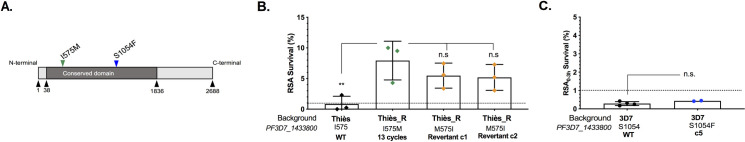
We also evaluated the effect on resistance of mutations in the conserved Plasmodium protein of unknown function encoded in gene PF3D7_1433800, which was the only other gene with mutations in both the independently evolved Thies_R and Pikine_R parasites (I575M in Thiès_R and S1054F in Pikine_R, Fig 3A). This gene has a conserved domain [16] (Fig 3A) and several naturally occurring polymorphisms in both the Thiès and Pikine control parasites (S4 Table), were reported in whole genome sequencing data published previously [17]. Polymorphisms in this gene are found in parasites from all over the world as reported in MalariaGEN [19] (S5 Table). Using a similar approach as that used to test the loss of function as described for pfcoronin, we successfully reverted the PF3D7_1433800 mutation in Thiès_R (I575M) back to the wildtype (M575I) (S5 Fig). The RSA survivals of two clones of PF3D7_1433800 in Thiès_R were not statistically different (Fig 3) compared with the Thiès_R value published previously [17] (clone 1 survival 5.47 ± 2.97%, p = 0.23, clone 2 survival 5.17 ± 3.46%, p = 0.19). We also performed growth assays on all the revertants compared to Thiès_R (S6 Fig). There was no significant short-term growth difference for any of the revertant parasites compared to their parent Thiès_R hence differences in RSA survival were not obscured by a difference in growth rate (S6 Fig).
Fig 3. PF3D7_1433800 makes a minimal contribution to artemisinin resistance.
A. Map of the conserved Plasmodium protein of unknown function (PF3D7_1433800) with the positions for mutations identified in Thiès_R (I575M) and Pikine_R (S1054F) in its conserved domain [16]. B. Ring-stage survival assay (RSA) survival percentage for two clones of revertants (M575I, c1 and c2, orange) of PF3D7_1433800 mutation I575M in Thiès_R compared to our previously published [17] survival for Thiès_R (green) and wildtype (WT) Thiès control (black). C. RSA survival percentage is shown for one clone of PF3D7_1433800 S1054F mutant in the 3D7 background (c5, blue), compared to the WT 3D7 control (black). Each data point represents an independent biological replicate and dotted line represents the 1% survival threshold, above which survival is considered resistant. Significance level is indicated by the asterisks, **p<0.01, determined using an unpaired two-tailed student t-test with Welch’s correction.
Several attempts to revert the PF3D7_1433800 mutation S1054F in Pikine_R and introduce S1054F in the Pikine wildtype failed, which made it difficult to evaluate the effect of this gene in the Pikine background. However, we were successful in introducing this mutation in the 3D7 laboratory strain (S7 Fig) to test the gain of function. The PF3D7_1433800 S1054F mutation in the 3D7 background had survival similar to 3D7 wildtype (Fig 3C), suggesting no evidence of a contribution to reduced sensitivity. Overall, the results from both Thiès_R revertants and 3D7 parasites suggest at most a limited contribution of PF3D7_1433800 mutations to artemisinin resistance, although we cannot rule out an effect in a different genetic background.
The phenotypic contributions of mutations in pfcoronin and pfkelch13 are non-additive
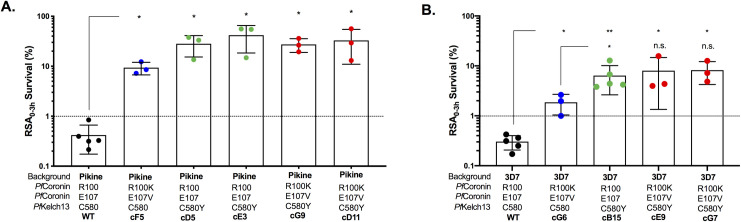
To test the potential synergy between mutations of pfcoronin and pfkelch13, we introduced both of these mutations into parasites of either the Pikine or 3D7 genetic background. As a control, we generated PfKelch13 C580Y parasites (S8B Fig) in the 3D7 background using constructs published previously [20]. Although the C580Y mutation in PfKelch13 has been investigated in isogenic lines of several different genetic backgrounds [18], it has yet to be studied in West African parasites. We obtained two clones of single mutants of pfkelch13 and double mutants of pfkelch13 and pfcoronin in the Pikine background parasites (S8 Fig). Two clones of PfKelch13 C580Y single mutants in Pikine had significantly higher RSA survival values (Fig 4A) compared to the wildtype (wildtype survival: 0.43 ± 0.27%; clone D5 survival: 28.15 ± 12.85%, p = 0.036; and clone E3 survival: 41.75 ± 23.39%, p = 0.036). The RSA survival for C580Y mutants in the Pikine background was higher than what was reported previously for the single mutants of pfcoronin (cF5 survival: 9.35 ± 1.89%%) [17]. The level of RSA survival observed in the double mutants was similar to that of the pfkelch13 single mutants (Fig 4A) (clone G9 survival: 27.27 ± 8.36%, p = 0.036; and clone D11 survival: 32.81 ± 21.83%, p = 0.036), which indicates that the phenotypic contribution of pfcoronin mutation is masked by the presence of the pfkelch13 mutation (Fig 4A).
Fig 4. The contributions of pfcoronin and pfkelch13 to resistance are non-additive.
A. Ring-stage survival assay (RSA) survival percentage for one clone of pfcoronin single mutant (cF5 published previously [17], blue), two clones of pfkelch13 single mutants (cD5 and cE3, green) and two clones of pfcoronin and pfkelch13 double mutants (cG9 and cD11, red) in the Pikine background compared to the wildtype (WT, black). B. Ring-stage survival assay (RSA) survival percentage for one clone of pfcoronin single mutant (cG6, blue), one clone of pfkelch13 single mutant (cB15, green) and two clones of pfcoronin and pfkelch13 double mutants (cE9 and cG7, red) in the 3D7 genetic background compared to the wildtype (WT, black). Each point represents an independent biological replicate and dotted line represents the 1% survival threshold, above which survival is considered resistant. Significance level is indicated by the asterisks, *p<005, **p<0.01, determined using two-tailed Mann-Whitney U test.
We observed similar results in 3D7 when comparing the RSA phenotype of single and double mutants of pfkelch13 and pfcoronin (Fig 4B). Consistent with previously published work [11,20,21], the C580Y mutant in 3D7 had significantly higher RSA survival (6.38 ± 3.74%) compared with the 3D7 wildtype (0.30 ± 0.10%, p = 0.0079) or the 3D7 PfCoronin R100K & E107V single mutant (1.86 ± 0.86%, p = 0.036). Double mutants of pfcoronin and pfkelch13 in 3D7 did not show an increased RSA survival when compared to the single C580Y mutation. The RSA survival of the double mutant was significantly higher than the wildtype (clone E9 survival: 8.01 ± 6.67%, p = 0.036 and clone G7 survival: 8.20 ± 3.97%, p = 0.036). These results are consistent with a non-additive interaction of the two mutations. There were no notable changes in dose-response curves of mutant parasites in the either Pikine or 3D7 genetic backgrounds in a standard 72-hour assay (S3 and S10 Figs and S3 Table).
We tested if RSA phenotype of the single and double mutants in the Pikine genetic background could be explained by a difference in short-term parasite growth of field isolates that grow slower than laboratory strains. We followed the transgenic pfkelch13 and pfcoronin single and double mutants for two reinvasion cycles and compared them to the Pikine wildtype and the PfCoronin R100K & E107V cF5 mutant [17] (S9 Fig). The C580Y single mutants in the Pikine genetic background showed no obvious growth defect. Although the pfcoronin single mutant and the double mutants with pfkelch and pfcoronin had slower apparent growth compared with the wildtype and the pfkelch13 single mutant, none of the parasite short-term growth curves were significantly different (day 4 parasitemia: 1.40 ± 0.30% for Pikine wildtype, 1.89 ± 0.52% for Pikine C580Y cD5, 2.20 ± 0.55% for Pikine C580Y cE3, 1.76 ± 0.27% for Pikine Coronin R100K and E107V cF5, 1.26 ± 0.17% for double mutant cG9 and 1.56 ± 0.48% for double mutant cD11).
Naturally occurring polymorphisms of pfcoronin and PF3D7_1433800 occur throughout African countries and Southeast Asia but are distinct from the mutations discovered in Pikine_R or Thiès_R
Using the Pf3K MalariaGEN database of naturally occurring polymorphisms across geographic sites from historic monogenomic samples [19], we examined polymorphisms in pfcoronin, PF3D7_1433800, and pfkelch13. In pfcoronin, 37 naturally occurring SNPs were identified, with most of these occurring at a low allele frequency (< 0.1) and in limited geographic locations. Only two polymorphisms, S183G and V424I, had frequencies > 0.1. In contrast to pfcoronin, PF3D7_1433800 is a highly polymorphic gene with 361 SNPs identified across geographic sites (S5 Table), but again most have allele frequencies < 0.1. A subset of sites, particularly in Southeast Asia, have higher allelic frequencies. Analysis of the pfkelch13 gene identified few polymorphic sites with the exception of the C580Y polymorphism with an allele frequency of 0.7 in Cambodian parasites.
When we compared the naturally occurring polymorphisms to those identified in our in vitro selection experiments, none of the nucleotide substitutions or amino acid changes identified in Pikine_R or Thiès_R were present among the naturally occurring SNPs in pfcoronin or PF3D7_1433800. This observation implies that the mutations occurred de novo in the course of selection.
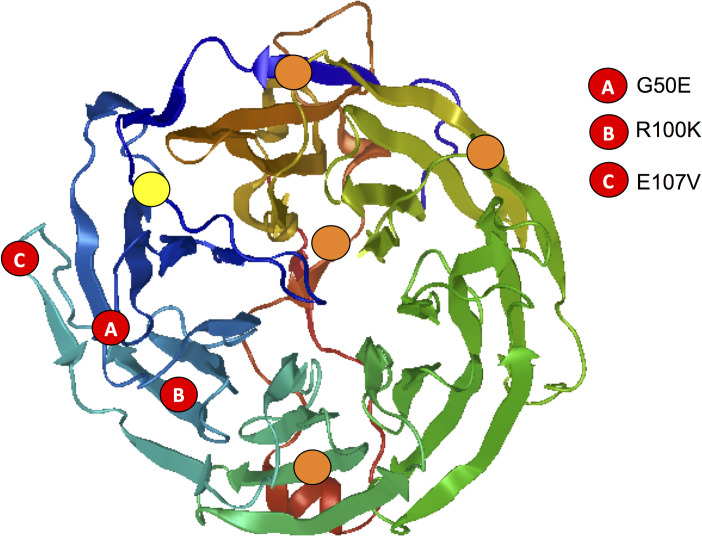
pfcoronin resistance mutations are on the opposite side of actin binding sites in Coronin
We generated the structure of PfCoronin WD-40 domain through homology modeling [22] using the structurally similar [23] crystal structure of Toxoplasma gondii Coronin (4OZU.pdb) [24]. When we mapped the mutations in PfCoronin associated with reduced artemisinin susceptibility and the putative actin-binding sites on the structure of the PfCoronin WD-40 domain, the resistance mutations (G50E, R100K and E107V) aligned on the opposite side of most of the putative actin-binding sites (6L-8K, R23, K126-K127, R197 –E201, K282 –D284, L307 –R310, S352 –I353) [24] (Fig 5). The only conserved actin binding site between mouse coronin 1A, T. gondii Coronin, and PfCoronin is arginine at position 23 (indicated in yellow, Fig 5).
Fig 5. Mutations in PfCoronin conferring resistance to artemisinin are at opposite ends of the putative actin binding sites.
Structure of WD-40 domain of Plasmodium falciparum generated through SWISS-MODEL homology modeling [22] using the crystal structure of the WD-40 domain of Toxoplasma gondii Coronin (4OZU.pdb) [24], which is 43% identical with 99% coverage to the the WD-40 domain of P. falciparum. The predicted actin binding sites are indicated in orange and conserved actin binding site from mouse coronin 1A (R23) [24] indicated in yellow. The mutations that confer artemisinin resistance [17] are indicated in red.
Discussion
In this study, we established that mutations in pfcoronin are the major drivers of reduced artemisinin sensitivity in our in vitro evolved Senegalese parasite lines Thiès_R and Pikine_R. When mutations in pfcoronin were reverted back to the wildtype in both Thiès_R and Pikine_R, we observed significantly reduced parasite survival in the ring-stage survival assay (RSA) in both genetic backgrounds (Fig 1). This supports our previously published observation of a gain of function associated with the introduction of pfcoronin mutations into wildtype parasites [17]. The resistance contribution of pfcoronin mutations was dependent on parasite genetic background, consistent with observations made with pfkelch13 [18]. Although mutations R100K and E107V were sufficient to surpass the 1% threshold of resistance in RSA (Fig 2), the level of resistance for pfcoronin mutants was much higher in Senegalese parasites [17] than in the 3D7 laboratory strain. We found no evidence of resistance contribution for mutations in the only other gene, PF3D7_1433800, commonly found in the in vitro evolved artemisinin resistant parasites (Fig 3). Upon comparison of naturally occurring polymorphisms of this gene and pfcoronin, PF3D7_1433800 was found to be highly polymorphic with 361 non-synonymous mutations found throughout the African region and Southeast Asia (S5 Table). Further work in other African genetic backgrounds will be needed to confirm the lack of a contribution of this gene by itself or in combination with pfcoronin mutations. Taken together, our work suggests that mutations in pfcoronin are both necessary and sufficient for in vitro artemisinin resistance in Pikine_R and Thiès_R, making them the major driver of artemisinin resistance in vitro.
We investigated the potential interaction between the mutations in PfCoronin (R100K and E107V) and PfKelch13 (C580Y) through studies of single and double mutants in both Pikine and 3D7 genetic backgrounds. There was a lack of synergy between the mutations of PfCoronin and PfKelch13 (Fig 4). In fact, the pfcoronin gene was found to be masked by pfkelch13. The level of resistance was much higher in the contemporary African parasites compared to laboratory strain 3D7 also of African ancestry, emphasizing the importance of studying genetic factors present using recent clinical isolates. Any potential interaction between pfcoronin and pfkelch13 needs further examination, especially in the African parasites.
The C580Y mutation has recently been linked to reduced expression of PfKelch13 protein [11,25], resulting in a slowdown of the endocytic machinery for hemoglobin uptake from the host cell without affecting binding interactions of the protein [11]. The artemisinin-resistance phenotype that we observe with PfCoronin could also result from differences in the level of functional protein in the mutants compared to the wildtype. We plan to explore this in the future alongside binding partner interactions of PfCoronin mutants. Considering the role of actin in endocytosis in Plasmodium as well as higher eukaryotes [26–28], PfCoronin mediated actin-bundling could be connected to endocytosis mediated artemisinin resistance. Although the resistance mutations are on the opposite side of the putative actin bundling sites (Fig 5), we cannot rule out the potential involvement of the mutations in impairing or aiding actin bundling, which is the major known function of PfCoronin [23,24,29]. It is noteworthy that T. gondii Coronin has a role in endocytosis and membrane recycling, a process crucial in the endoplasmic reticulum (ER) stress response, independent of its function in actin bundling [24]. This function remains unexplored in P. falciparum and might suggest an involvement of PfCoronin in the endocytic machinery to attain artemisinin resistance similar to PfKelch13 without involving actin bundling [11].
WD-40 beta-propeller domains, which are understudied in apicomplexans, are conserved protein structures in eukaryotes that support a wide range of cellular functions [30–33], similar to the beta-propeller domains found in Kelch-like proteins [34]. We still do not know the exact function of PfKelch13 despite its implication in the ring-stage endocytosis machinery [11], nor do we understand the biological mechanism of artemisinin resistance mediated by PfCoronin. Although widespread resistance-conferring mutations in pfkelch13 are yet to be reported on the African continent, our in vitro work shows that C580Y parasites in the Pikine genetic background exhibit substantial survival in the RSA similar to the Cambodian isolates [2,18,35]. This is unlike Tanzanian F32 parasites which had much lower RSA survival with M476I pfkelch13 and FCB with C580Y pfkelch13 [18], raising additional concerns that West African expressing variants of pfcoronin and/or pfkelch13 could be competitive in the field. This emphasizes the need to gather RSA survival estimates for contemporary African parasites. Investigating the biology of beta propeller domain-containing proteins and the possible genetic interactions between different players in artemisinin resistance could further elucidate mechanisms of resistance.
Methods
In vitro parasite culture
All laboratory strains [3D7 (MR4)], culture-adapted field isolates from Thiès (SenTh032.09) and Pikine (SenP019.04), previously published DHA selected lines Thiès_R and Pikine_R [17], and the subsequently generated CRISPR parasite lines were cultured in O+ red blood cells (RBC) (Interstate Blood Bank (IBB), Memphis, TN) with complete RPMI1640 media (Gibco, Waltham, MA) supplemented with 10% O+ serum (IBB). Parasites were placed in a modular incubator and gassed with 1% O2/5% CO2/94% N2 mixture before incubating at 37°C.
RSA (Ring-stage Survival Assay)
RSA was performed as described previously [2,35] using 0–3 hour post reinvasion rings that were highly synchronized and exposed to 700 nM DHA or DMSO for 6 hours. Rings were synchronized with 5% D-sorbitol treatment. At 66 hours after the drug washout, parasitemia was assessed by Giemsa-stained smear microscopy of thin smears. The RSA survival percentage was calculated by dividing the parasitemia in DHA-treated parasites over the DMSO-treated control parasitemia. A minimum of two to three biological replicates of RSA were conducted for each parasite line with two technical replicates per biological replicate. Smears were blinded and at least 10,000 RBC counted per replicate.
Drug sensitivity assays with SYBR Green I
Drug sensitivity assays were performed as previously described [36]. Briefly, ring-stage parasites were grown to 0.8–1% parasitemia in 2% hematocrit in 40μL total volume in 384-well plates. Parasite growth was determined by SYBR Green I staining (Lonza, Visp, Switzerland) of parasite DNA in the trophozoite stage, usually 72 hours after plating. For parasites that grew slowly (double mutants of pfcoronin and pfkelch13 in Pikine background), parasites were stained after 96 hours in culture. The dose-response curves for standard anti-malarial drugs (DHA, ART, MQ; Sigma-Aldrich, St. Louis, MO) were generated from a 12-point dilution series of drugs, carried out in triplicate, centered on expected EC50 reported in the literature; with three biological replicates performed for each drug. A SpectraMax M5 (Molecular Devices, Sunnyvale, CA) plate reader was used to measure fluorescence, and data were analyzed with GraphPad Prism version 6 (GraphPad Software, La Jolla, CA). EC50 was calculated using nonlinear regression with the log(inhibitor) vs. response with a four-parameter variable slope curve-fitting equation.
Gene editing using CRISPR-Cas9
Guide sequences targeting the loci of interest were designed using the online tool available via benchling.com. For introducing pfcoronin mutations in 3D7 background, we used the strategy successfully implemented previously in Pikine and Thiès parasite backgrounds [17]. For reverting pfcoronin G50E, gRNA containing the mutation was used (S1 Fig). We used previously published strategies for reverting pfcoronin R100K and E107V, including the gRNA 1 sequence (S2 Fig). All gRNAs were individually annealed and ligated into the BbsI digested pDC2-Cas9-U6-hDHFR plasmid, generously provided by Marcus Lee (Wellcome Sanger Institute, Hinxton, UK), or the pUF1Cas9-U6-DHODH generated using the Cas9 plasmid generated previously [20], both of which contain the U6 snRNA polymerase III promoter and regions for the expression of Cas9 enzyme and human DHFR or yeast DHODH drug selection cassette as described previously [37,38]. For generating pfkelch13 C580Y mutants, previous constructs [20] were used for the 3D7 background. After several failed transfection attempts with these constructs in the Pikine background, the same gRNA was annealed and ligated into pUF1Cas9-U6-DHODH. Homology regions of about 500bp containing either the SNP of interest or wildtype sequence (Primers used in S6 Table) were cloned into Zeroblunt TOPO vector (Thermo Scientific, Waltham, MA). Site-directed mutagenesis was performed using Quickchange II mutagenesis kit (Agilent, Santa Clara, CA) to scramble guide targeting sequence with shield mutations in the homology region. For C580Y homology region for Pikine background, 500bp homology with SNP and shield mutations was amplified from the construct described previously [20].
Transfection was performed on 5–8% sorbitol synchronized rings from 3D7, SenTh032.09.13.1 (Thiès_R), Sen P019.04 (Pikine wildtype) or SenP019.04.13.1 (Pikine_R) parasites with 50μg of Cas9 plasmid and 50μg of plasmid containing the homology region using the Bio-Rad Gene Pulser at 0.31 kV and 960 μF as described previously [39]. After the transfection, RBCs were plated at 5% hematocrit in complete media. Transfected parasites were allowed to recover for either 8 hours (for 3D7) or overnight (for Senegalese parasite backgrounds) before the addition of 5 nM WR99210 (for pDC2-Cas9-U6-hDHFR plasmid) (Jacobus Pharmaceutical, NJ) or 500nM DSM1 (for pUF1-Cas9-U6-yDHODH plasmid) (Millipore Sigma, Germany). Drug selection was continued for 96 hours for all but the pfkelch13 mutant generation in Pikine background. For C580Y transfections in Pikine background, drug selection was continued for three weeks. For all transfections, parasite recovery was monitored by microscopy twice a week. Once parasitemia was more than 1%, the loci of interests were genotyped by Sanger sequencing performed by either Psomagen USA (Cambridge, MA) or Genewiz (Cambridge, MA) using the bulk transfectant gDNA. After confirming successful transfection, dilutional cloning was performed in 96-well plate to obtain at least two confirmed CRISPR edited parasite clones.
To confirm the genotype of the edited parasites, parasite DNA was extracted from in vitro cultures using QIAamp DNA blood kits (Qiagen, Hilden, Germany), per manufacturer’s instructions. Primers were ordered from Integrated DNA technologies (IDT, Newark, NJ) to amplify relevant candidate gene amplicons of 500–800bp lengths (S6 Table). Primers used for PCR are listed in S6 Table. For the verification of transfectants, primer combinations within and outside of the homology region were used to avoid amplifying the residual plasmid from transfection. PCR amplification was conducted using the Phusion High-Fidelity DNA Polymerase (NEB, Ipswich, MA) following standard procedure. Amplicons were cleaned using ZymoKit DNA purification kit (Irvine, CA) per manufacturer’s instructions and sent for Sanger sequencing to Genewiz.
Parasite growth assay
Parasite growth assays were conducted over the course of five days. Synchronized rings were plated at 0.5% initial parasitemia and 2% HCT in 6-well plates. Parasitemia was monitored daily using MACSQuant Analyzer Flow Cytometer (Miltenyi Biotec, Germany) through SYBR Green I staining (Lonza, Visp, Switzerland) for 30 minutes at 37°C over the course of five days with two to three biological replicates per parasite line.
Statistical analysis
All statistical analyses were first performed in groups of two using parametric unpaired two-tailed t-test with Welch’s correction. If this resulted in a significantly different F test to compare variances for the pair, making the results of the Welch’s correction unreliable, a non-parametric Mann-Whitney test was conducted using GraphPad Prism version 6.
PfCoronin homology modeling
PfCoronin homology modeling was generated using SWISS-MODEL automated server [22] using Toxoplasma gondii Coronin crystal structure (4ozu.1) [24] (43.26% identity, 99% coverage, 0.77 GMQE, -2.49 QMEAN) was downloaded and visualized using Protean 3D software Version 12.0 (DNASTAR package, Madison, WI).
Supporting information
A. CRISPR gene editing strategy for generating pfcoronin revertants in the SenTh032.09.13.1 (Thiès_R) background. Homology region with primer sequences underlined, pfcoronin mutated site indicated in red, shield mutations in green, protospacer adjacent motif (PAM) sequences highlighted in yellow. B. Sanger sequencing confirmation of CRISPR edited parasite gDNA highlighting the target SNP in red and shield mutations in green compared to the parent and Thiès_R.
(DOCX)
A. CRISPR gene editing strategy for generating pfcoronin revertants in the SenP019.04.13.1 (Pikine_R) background. Homology region with primer sequences underlined, pfcoronin mutated sites indicated in red, shield mutations in green, protospacer adjacent motif (PAM) sequences highlighted in yellow. B. Sanger sequencing confirmation of CRISPR edited parasite gDNA highlighting the target SNPs in red and shield mutations in green compared to the parent and Pikine_R.
(DOCX)
Parasite drug sensitivity was measured by 72-hour in vitro assays with SYBR Green. For parasites that grew slowly (double mutants of pfcoronin and pfkelch13 in the Pikine background), parasites were stained after 96 hours in culture. Representative EC50 dose-response curves from three biological replicates are presented.
(TIFF)
G50E mutation corresponding to clone E11 and B. R100K/E107V mutations corresponding to clone G6 compared to 3D7 wildtype.
(DOCX)
A. CRISPR gene editing strategy for generating PF3D7_1433800 revertants in the SenTh032.09.13.1 (Thiès_R) background. Homology region with primer sequences underlined, PF3D7_1433800 mutated site indicated in red, shield mutations in green, protospacer adjacent motif (PAM) sequences highlighted in yellow. B. Sanger sequencing confirmation of CRISPR edited parasite gDNA highlighting the target SNP in red and shield mutations in green compared to the parent and Thiès_R.
(DOCX)
Normalized growth curve from two biological replicates of Thiès background parasites referred to in main Figs 1B and 3B followed through two reinvasion cycles for growth comparison. Statistical analyses presented no significant difference in growth for any of the parasites.
(TIFF)
A. CRISPR gene editing strategy for generating PF3D7_1433800 S1054F in the 3D7 background after several attempts to knock-in or revert the SNP failed in SenPik19.04 (Pikine background parasite). Homology region with primer sequences underlined, PF3D7_1433800 mutated site indicated in red, shield mutations in green, protospacer adjacent motif (PAM) sequences highlighted in yellow. One of the shield mutations modified the PAM sequence. B. Sanger sequencing confirmation of CRISPR edited clonal parasite gDNA highlighting the target SNP in red and shield mutations in green compared to the parent. The region is highly AT rich resulting in high Sanger sequencing background.
(DOCX)
A. Confirmation of pfcoronin editing to generate R100K & E107V mutants in 3D7 background using constructs described previously for the Pikine background1. B. Confirmation of pfkelch13 editing to generate C580Y mutants in both 3D7 and Pikine backgrounds. Target SNP(s) are highlighted in red and shield mutations in green and are compared to the wildtype (WT) sequence.
(DOCX)
Normalized growth curve from three biological replicates of Pikine background parasites from Fig 4A followed through two reinvasion cycles for growth comparison. Statistical analyses presented no significant difference in growth for any of the parasites.
(TIFF)
Parasite drug sensitivity was measured by 72-hour in vitro assays with SYBR Green (See methods for details). Representative EC50 dose-response curves from three biological replicates are presented.
(TIFF)
(DOCX)
Parasites were sequenced from DMSO treated group after RSA was completed. Representative chromatogram from one of the biological replicates is presented. SNP and shield mutation positions are shown in red and green, respectively, for CRISPR edited parasites as well as their respective WT sequences.
(DOCX)
(DOCX)
(DOCX)
Geographical distribution of naturally occurring non-synonymous polymorphisms of pfcoronin, PF3D7_1433800 and pfkelch13 in African countries (green columns) and Southeast Asia (orange columns) based on PF3K database.
(XLSX)
For mutagenesis (mut) primers, shield mutations are highlighted in green and targeted mutation(s), if present within the range of the primer, are highlighted in red.
(DOCX)
Acknowledgments
We thank Dr. Jose-Juan Lopez-Rubio for PfKelch13 C580Y CRISPR plasmids. We also thank Cory L. Schlesener for generating plasmids for PF3D7_1433800 CRISPR experiments and members of Neafsey lab Angela Early and Akanksha Khorgade for help with PF3K database SNP analyses. We would like to acknowledge the members of Wirth Lab for critically reading the manuscript and providing invaluable support throughout the project.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
Funding for this project was provided by National Institutes of Health Grant R01AI099105 (to D.F.W. and D.L.H.), Grant OPP1156051 from the Bill and Melinda Gates Foundation (to D.F.W) and Harvard Defeating Malaria Initiative at Harvard University via institutional support networks. Funders played no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO. World Malaria Report 2019. Geneva: World Health Organization, 2019. [Google Scholar]
- 2.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505(7481):50–5. Epub 2013/12/20. 10.1038/nature12876 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaw MT, Emran NA, Lin Z. Updates on k13 mutant alleles for artemisinin resistance in Plasmodium falciparum. J Microbiol Immunol Infect. 2018;51(2):159–65. Epub 2017/07/18. 10.1016/j.jmii.2017.06.009 . [DOI] [PubMed] [Google Scholar]
- 4.Chenet SM, Akinyi Okoth S, Huber CS, Chandrabose J, Lucchi NW, Talundzic E, et al. Independent Emergence of the Plasmodium falciparum Kelch Propeller Domain Mutant Allele C580Y in Guyana. J Infect Dis. 2016;213(9):1472–5. Epub 2015/12/23. 10.1093/infdis/jiv752 . [DOI] [PubMed] [Google Scholar]
- 5.Prosser C, Meyer W, Ellis J, Lee R. Resistance screening and trend analysis of imported falciparum malaria in NSW, Australia (2010 to 2016). PLoS One. 2018;13(5):e0197369 Epub 2018/05/31. 10.1371/journal.pone.0197369 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tacoli C, Gai PP, Bayingana C, Sifft K, Geus D, Ndoli J, et al. Artemisinin Resistance-Associated K13 Polymorphisms of Plasmodium falciparum in Southern Rwanda, 2010–2015. Am J Trop Med Hyg. 2016;95(5):1090–3. Epub 2016/11/04. 10.4269/ajtmh.16-0483 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bwire GM, Ngasala B, Mikomangwa WP, Kilonzi M, Kamuhabwa AAR. Detection of mutations associated with artemisinin resistance at k13-propeller gene and a near complete return of chloroquine susceptible falciparum malaria in Southeast of Tanzania. Sci Rep. 2020;10(1):3500 Epub 2020/02/28. 10.1038/s41598-020-60549-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukherjee A, Bopp S, Magistrado P, Wong W, Daniels R, Demas A, et al. Artemisinin resistance without pfkelch13 mutations in Plasmodium falciparum isolates from Cambodia. Malar J. 2017;16(1):195 Epub 2017/05/13. 10.1186/s12936-017-1845-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henrici RC, van Schalkwyk DA, Sutherland CJ. Modification of pfap2mu and pfubp1 Markedly Reduces Ring-Stage Susceptibility of Plasmodium falciparum to Artemisinin In Vitro. Antimicrob Agents Chemother. 2019;64(1). Epub 2019/10/23. 10.1128/AAC.01542-19 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sutherland CJ, Lansdell P, Sanders M, Muwanguzi J, van Schalkwyk DA, Kaur H, et al. pfk13-Independent Treatment Failure in Four Imported Cases of Plasmodium falciparum Malaria Treated with Artemether-Lumefantrine in the United Kingdom. Antimicrob Agents Chemother. 2017;61(3). Epub 2017/02/01. 10.1128/AAC.02382-16 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birnbaum J, Scharf S, Schmidt S, Jonscher E, Hoeijmakers WAM, Flemming S, et al. A Kelch13-defined endocytosis pathway mediates artemisinin resistance in malaria parasites. Science. 2020;367(6473):51–9. Epub 2020/01/04. 10.1126/science.aax4735 . [DOI] [PubMed] [Google Scholar]
- 12.Bushman M, Antia R, Udhayakumar V, de Roode JC. Within-host competition can delay evolution of drug resistance in malaria. PLoS Biol. 2018;16(8):e2005712 Epub 2018/08/22. 10.1371/journal.pbio.2005712 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miotto O, Amato R, Ashley EA, MacInnis B, Almagro-Garcia J, Amaratunga C, et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet. 2015;47(3):226–34. 10.1038/ng.3189 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomavo S, Slomianny C, Meissner M, Carruthers VB. Protein trafficking through the endosomal system prepares intracellular parasites for a home invasion. PLoS Pathog. 2013;9(10):e1003629 Epub 2013/11/10. 10.1371/journal.ppat.1003629 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams T, Ennuson NAA, Quashie NB, Futagbi G, Matrevi S, Hagan OCK, et al. Prevalence of Plasmodium falciparum delayed clearance associated polymorphisms in adaptor protein complex 2 mu subunit (pfap2mu) and ubiquitin specific protease 1 (pfubp1) genes in Ghanaian isolates. Parasit Vectors. 2018;11(1):175 Epub 2018/03/14. 10.1186/s13071-018-2762-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37(Database issue):D539–43. Epub 2008/10/30. 10.1093/nar/gkn814 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demas AR, Sharma AI, Wong W, Early AM, Redmond S, Bopp S, et al. Mutations in Plasmodium falciparum actin-binding protein coronin confer reduced artemisinin susceptibility. Proc Natl Acad Sci U S A. 2018;115(50):12799–804. Epub 2018/11/14. 10.1073/pnas.1812317115 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Straimer J, Gnadig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, et al. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347(6220):428–31. Epub 2014/12/17. 10.1126/science.1260867 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malaria GENPfCP. Genomic epidemiology of artemisinin resistant malaria. Elife. 2016;5 Epub 2016/03/05. 10.7554/eLife.08714 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio JJ. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol. 2014;32(8):819–21. 10.1038/nbt.2925 . [DOI] [PubMed] [Google Scholar]
- 21.Mbengue A, Bhattacharjee S, Pandharkar T, Liu H, Estiu G, Stahelin RV, et al. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 2015;520(7549):683–7. Epub 2015/04/16. 10.1038/nature14412 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(Web Server issue):W252–8. Epub 2014/05/02. 10.1093/nar/gku340 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olshina MA, Angrisano F, Marapana DS, Riglar DT, Bane K, Wong W, et al. Plasmodium falciparum coronin organizes arrays of parallel actin filaments potentially guiding directional motility in invasive malaria parasites. Malar J. 2015;14:280 Epub 2015/07/19. 10.1186/s12936-015-0801-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salamun J, Kallio JP, Daher W, Soldati-Favre D, Kursula I. Structure of Toxoplasma gondii coronin, an actin-binding protein that relocalizes to the posterior pole of invasive parasites and contributes to invasion and egress. FASEB J. 2014;28(11):4729–47. Epub 2014/08/13. 10.1096/fj.14-252569 . [DOI] [PubMed] [Google Scholar]
- 25.Yang T, Yeoh LM, Tutor MV, Dixon MW, McMillan PJ, Xie SC, et al. Decreased K13 Abundance Reduces Hemoglobin Catabolism and Proteotoxic Stress, Underpinning Artemisinin Resistance. Cell Rep. 2019;29(9):2917–28 e5. Epub 2019/11/28. 10.1016/j.celrep.2019.10.095 . [DOI] [PubMed] [Google Scholar]
- 26.Mooren OL, Galletta BJ, Cooper JA. Roles for actin assembly in endocytosis. Annu Rev Biochem. 2012;81:661–86. Epub 2012/06/06. 10.1146/annurev-biochem-060910-094416 . [DOI] [PubMed] [Google Scholar]
- 27.Galletta BJ, Mooren OL, Cooper JA. Actin dynamics and endocytosis in yeast and mammals. Curr Opin Biotechnol. 2010;21(5):604–10. Epub 2010/07/20. 10.1016/j.copbio.2010.06.006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smythe WA, Joiner KA, Hoppe HC. Actin is required for endocytic trafficking in the malaria parasite Plasmodium falciparum. Cell Microbiol. 2008;10(2):452–64. Epub 2007/10/20. 10.1111/j.1462-5822.2007.01058.x . [DOI] [PubMed] [Google Scholar]
- 29.Tardieux I, Liu X, Poupel O, Parzy D, Dehoux P, Langsley G. A Plasmodium falciparum novel gene encoding a coronin-like protein which associates with actin filaments. FEBS Lett. 1998;441(2):251–6. Epub 1999/01/12. 10.1016/s0014-5793(98)01557-9 . [DOI] [PubMed] [Google Scholar]
- 30.Clemen CS, Eichinger L, Rybakin V. The coronin family of proteins. New York, N.Y. Austin, Tex: Springer; Landes Bioscience; 2008. xvi, 139 p. p. [DOI] [PubMed] [Google Scholar]
- 31.Jain BP, Pandey S. WD40 Repeat Proteins: Signalling Scaffold with Diverse Functions. Protein J. 2018;37(5):391–406. Epub 2018/08/03. 10.1007/s10930-018-9785-7 . [DOI] [PubMed] [Google Scholar]
- 32.Schapira M, Tyers M, Torrent M, Arrowsmith CH. WD40 repeat domain proteins: a novel target class? Nat Rev Drug Discov. 2017;16(11):773–86. Epub 2017/10/14. 10.1038/nrd.2017.179 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brehar FM, Dragomir MP, Petrescu GED, Gorgan RM. Fighting Cancer Stem Cell Fate by Targeting LIS1 a WD40 Repeat Protein. Front Oncol. 2019;9:1142 Epub 2019/11/22. 10.3389/fonc.2019.01142 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hudson AM, Cooley L. Phylogenetic, structural and functional relationships between WD- and Kelch-repeat proteins. Subcell Biochem. 2008;48:6–19. Epub 2008/10/18. 10.1007/978-0-387-09595-0_2 . [DOI] [PubMed] [Google Scholar]
- 35.Witkowski B, Amaratunga C, Khim N, Sreng S, Chim P, Kim S, et al. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect Dis. 2013;13(12):1043–9. Epub 2013/09/17. 10.1016/S1473-3099(13)70252-4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson JD, Dennull RA, Gerena L, Lopez-Sanchez M, Roncal NE, Waters NC. Assessment and continued validation of the malaria SYBR green I-based fluorescence assay for use in malaria drug screening. Antimicrob Agents Chemother. 2007;51(6):1926–33. Epub 2007/03/21. 10.1128/AAC.01607-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng CL, Siciliano G, Lee MC, de Almeida MJ, Corey VC, Bopp SE, et al. CRISPR-Cas9-modified pfmdr1 protects Plasmodium falciparum asexual blood stages and gametocytes against a class of piperazine-containing compounds but potentiates artemisinin-based combination therapy partner drugs. Mol Microbiol. 2016;101(3):381–93. Epub 2016/04/14. 10.1111/mmi.13397 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim MY, LaMonte G, Lee MC, Reimer C, Tan BH, Corey V, et al. UDP-galactose and acetyl-CoA transporters as Plasmodium multidrug resistance genes. Nat Microbiol. 2016:16166 Epub 2016/09/20. 10.1038/nmicrobiol.2016.166 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deitsch K, Driskill C, Wellems T. Transformation of malaria parasites by the spontaneous uptake and expression of DNA from human erythrocytes. Nucleic Acids Res. 2001;29(3):850–3. Epub 2001/02/13. 10.1093/nar/29.3.850 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. CRISPR gene editing strategy for generating pfcoronin revertants in the SenTh032.09.13.1 (Thiès_R) background. Homology region with primer sequences underlined, pfcoronin mutated site indicated in red, shield mutations in green, protospacer adjacent motif (PAM) sequences highlighted in yellow. B. Sanger sequencing confirmation of CRISPR edited parasite gDNA highlighting the target SNP in red and shield mutations in green compared to the parent and Thiès_R.
(DOCX)
A. CRISPR gene editing strategy for generating pfcoronin revertants in the SenP019.04.13.1 (Pikine_R) background. Homology region with primer sequences underlined, pfcoronin mutated sites indicated in red, shield mutations in green, protospacer adjacent motif (PAM) sequences highlighted in yellow. B. Sanger sequencing confirmation of CRISPR edited parasite gDNA highlighting the target SNPs in red and shield mutations in green compared to the parent and Pikine_R.
(DOCX)
Parasite drug sensitivity was measured by 72-hour in vitro assays with SYBR Green. For parasites that grew slowly (double mutants of pfcoronin and pfkelch13 in the Pikine background), parasites were stained after 96 hours in culture. Representative EC50 dose-response curves from three biological replicates are presented.
(TIFF)
G50E mutation corresponding to clone E11 and B. R100K/E107V mutations corresponding to clone G6 compared to 3D7 wildtype.
(DOCX)
A. CRISPR gene editing strategy for generating PF3D7_1433800 revertants in the SenTh032.09.13.1 (Thiès_R) background. Homology region with primer sequences underlined, PF3D7_1433800 mutated site indicated in red, shield mutations in green, protospacer adjacent motif (PAM) sequences highlighted in yellow. B. Sanger sequencing confirmation of CRISPR edited parasite gDNA highlighting the target SNP in red and shield mutations in green compared to the parent and Thiès_R.
(DOCX)
Normalized growth curve from two biological replicates of Thiès background parasites referred to in main Figs 1B and 3B followed through two reinvasion cycles for growth comparison. Statistical analyses presented no significant difference in growth for any of the parasites.
(TIFF)
A. CRISPR gene editing strategy for generating PF3D7_1433800 S1054F in the 3D7 background after several attempts to knock-in or revert the SNP failed in SenPik19.04 (Pikine background parasite). Homology region with primer sequences underlined, PF3D7_1433800 mutated site indicated in red, shield mutations in green, protospacer adjacent motif (PAM) sequences highlighted in yellow. One of the shield mutations modified the PAM sequence. B. Sanger sequencing confirmation of CRISPR edited clonal parasite gDNA highlighting the target SNP in red and shield mutations in green compared to the parent. The region is highly AT rich resulting in high Sanger sequencing background.
(DOCX)
A. Confirmation of pfcoronin editing to generate R100K & E107V mutants in 3D7 background using constructs described previously for the Pikine background1. B. Confirmation of pfkelch13 editing to generate C580Y mutants in both 3D7 and Pikine backgrounds. Target SNP(s) are highlighted in red and shield mutations in green and are compared to the wildtype (WT) sequence.
(DOCX)
Normalized growth curve from three biological replicates of Pikine background parasites from Fig 4A followed through two reinvasion cycles for growth comparison. Statistical analyses presented no significant difference in growth for any of the parasites.
(TIFF)
Parasite drug sensitivity was measured by 72-hour in vitro assays with SYBR Green (See methods for details). Representative EC50 dose-response curves from three biological replicates are presented.
(TIFF)
(DOCX)
Parasites were sequenced from DMSO treated group after RSA was completed. Representative chromatogram from one of the biological replicates is presented. SNP and shield mutation positions are shown in red and green, respectively, for CRISPR edited parasites as well as their respective WT sequences.
(DOCX)
(DOCX)
(DOCX)
Geographical distribution of naturally occurring non-synonymous polymorphisms of pfcoronin, PF3D7_1433800 and pfkelch13 in African countries (green columns) and Southeast Asia (orange columns) based on PF3K database.
(XLSX)
For mutagenesis (mut) primers, shield mutations are highlighted in green and targeted mutation(s), if present within the range of the primer, are highlighted in red.
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.