Abstract.
Desirable outcomes including rejection- and infection-free kidney transplantation are not guaranteed despite current strategies for immunosuppression and using prophylactic antimicrobial medications. Graft survival depends on factors beyond human leukocyte antigen matching such as the level of immunosuppression, infections, and management of other comorbidities. Risk stratification of transplant patients based on predisposing genetic modifiers and applying precision pharmacotherapy may help improving the transplant outcomes. Unlike certain fields such as oncology in which consistent attempts are being carried out to move away from the “error and trial approach,” transplant medicine is lagging behind in implementing personalized immunosuppressive therapy. The need for maintaining a precarious balance between underimmunosuppression and overimmunosuppression coupled with adverse effects of medications calls for a gene-based guidance for precision pharmacotherapy in transplantation. Technologic advances in molecular genetics have led to increased accessibility of genetic tests at a reduced cost and have set the stage for widespread use of gene-based therapies in clinical care. Evidence-based guidelines available for precision pharmacotherapy have been proposed, including guidelines from Clinical Pharmacogenetics Implementation Consortium, the Pharmacogenomics Knowledge Base National Institute of General Medical Sciences of the National Institutes of Health, and the US Food and Drug Administration. In this review, we discuss the implications of pharmacogenetics and potential role for genetic variants-based risk stratification in kidney transplantation. A single score that provides overall genetic risk, a polygenic risk score, can be achieved by combining of allograft rejection/loss-associated variants carried by an individual and integrated into practice after clinical validation.
Kidney transplantation is the treatment of choice for patients with end-stage renal disease (ESRD).1 Approximately, 100 000 patients are on the kidney transplant waiting list in the United States, but only 21 000 kidney transplantations were performed in 2018.2 Mortality of ESRD patients, who receive kidney transplantation is lower than patients on maintenance dialysis.3 However, compared to general population, mortality of kidney transplant recipients is about 14 times higher in the first year posttransplant and 4 times higher thereafter.4 Furthermore, deceased donor kidney transplant recipients have a 10-year death-censored graft failure of 26% and it is 18% for living donor kidney transplants.5 Several factors influence long-term transplant outcome, including donor age and comorbidity, allograft ischemic time, degree of HLA mismatch, and recipient factors such as response to immunosuppression and the development of donor-specific antibodies.6,7 In general, immunosuppressants have a narrow therapeutic index and exhibit a large intraindividual and interindividual variability of their pharmacokinetics, necessitating a personalized immunosuppressive regimen.8 Other factors also contribute to the suboptimal outcomes in transplant recipients, including cardiovascular disease and infections.9 Complications related to infection could be attenuated by personalizing immunosuppression and antimicrobial treatment.10-13 Furthermore, cardiovascular medications with actionable genetic information are frequently used in kidney transplant recipients.14 Precision prescribing of these medications could improve efficacy, mitigate risk of drug-drug interactions, and improve outcomes. In this review, we discuss the importance of precision medicine in kidney transplantation and the available tools to implement it. We also highlight genetics-based risk stratification and the role of pharmacogenetics in precision prescribing in transplant medicine.
Precision Medicine
The advances in molecular medicine have prompted the call for a new taxonomy of human disease based on molecular biology, which is expected to provide a strong foundation for the future of precision medicine. The term “precision medicine” was advanced by the National Research Council Working Group, which called for establishing a “new taxonomy of human disease based on molecular biology” to replace the classical descriptive diagnostic terms.15 Precision medicine seeks to identify safe and effective treatments based on genetics and environment that are unique to an individual.16,17 Recently, The National Institute of Diabetes and Digestive and Kidney Diseases launched the Kidney Precision Medicine Project with the purpose of understanding and finding new ways to treat chronic kidney disease and acute kidney injury18 (https://kpmp.org/). In transplantation, the advent of genomic and other molecular profiling techniques provides an unprecedented opportunity to apply precision medicine strategies to improve patient outcome. Although precision medicine is a realistic approach, it is not without pitfalls. Any stratified approach to medicine would potentially restrict the number of patients treated with a therapeutic intervention or discriminate against the people who are otherwise healthy.19 Therefore, careful assessment of potential implications of precision medicine is warranted.
Genetics and Immune Response
Role of genetics in immune response is well recognized.20,21 The interplay of innate and adaptive immune response may implicate the outcomes of transplantation including rejection and tolerance.22 Interindividual variations in immune response could be due to heritable genetics and epigenetic factors.23,24 Epigenetics refers to a heritable change in the pattern of gene expression that is mediated by a mechanism specifically not due to alterations in the primary nucleotide sequence.25 Emerging evidence indicates that epigenetic modifications are fundamental to the differentiation and function of immune cells.26 MicroRNAs are noncoding RNAs that mediate posttranscriptional gene regulation. Specific microRNAs have been shown to be associated with kidney allograft rejection, possibly through modifying the expression of certain genes in regulatory T cells.27 Therefore, it appears that the crosstalk between the genes and environment through epigenetics leading to alterations in immune response and transplant outcome.
Donor and Recipient Genetics—Beyond HLA
Introduction of HLA in kidney transplantation resulted in improved clinical outcomes.28 HLA genes are highly polymorphic, and demonstrate the influence of genetic variation in determining long-term transplant outcomes.1 However, even full house matching of HLA loci does not preclude the need for immunosuppression, suggesting the existence of other genetic variations in that need to be considered. Numerous studies have examined the association between genetic variations in immune response genes and transplant outcome with inconsistent findings.29 Similarly, a large-scale genome-wide association study (GWAS) was unable to detect convincing association signals outside of the HLA region.30 Approximately 20% of individuals waitlisted for kidney transplant in the United States are those with failed allograft.31 Furthermore, the incidence of donor-specific HLA antibodies is relatively low (15%–25%) among transplant recipients.7,32 Thus, factors beyond HLA may be responsible for graft failure.
Donor Genetics
Survival of kidney allograft from deceased black donors is shorter, when compared with allografts from white donors.33 Two common variants (G1 and G2) in the last exon of Apolipoprotein L1 (ApoL1) are common in populations of West Sub-Saharan African origin.34 It is believed that these 2 variants account for much of the disparity in rates of ESRD between black patients and white patients.35 Kidney transplant recipients from black deceased donors with 2 high-risk ApoL1 variants experience an earlier allograft failure compared with those with 1 or no ApoL1 high-risk variants.36-38 Although Kidney Donor Profile Index (KDPI) considers all kidneys from deceased black donors as high-risk,37 only a minority of them possess the 2 high-risk ApoL1 variants.39,40 Less than 1% of kidney donors develop ESRD, however it is more common among black versus white donors.41 A faster rate of decline in kidney function after donation has been reported in black living kidney donors with ApoL1 high-risk genotype.42 Furthermore, kidney function after donation in white donors has been reported to be similar to those black donors with low risk ApoL1 genotype, suggesting that the poor kidney outcomes observed in black donors may be attributable to ApoL1 high-risk genotype.41 Kidney allograft donated by a healthy individual with 2 ApoL1 high-risk variants is associated with focal segmental glomerulosclerosis (FSGS) and early allograft failure in recipients.42 Therefore, determining ApoL1 variants may lead to proper risk assessment and improve the current organ allocation system and potentially transplant outcomes. The National Institutes of Health-sponsored APOL1 Long-term Kidney Transplantation Outcomes Network study attempts to improve outcomes after kidney transplantation and to improve the safety of living kidney donation based upon variation in ApoL1 (https://theapollonetwork.org/).43
Other genetic variants that may be considered for precision organ allocation include MHC class I-related chain A (MICA), ATP binding cassette subfamily B member 1 (ABCB1), caveolin-1 (CAV1), and Ficolin-2. MICA is a highly polymorphic gene and implicated in innate immunity.44 Anti-MICA antibodies are associated with acute and chronic rejection in renal transplant recipients.45,46 Donor MICA A5.1 mutation is associated with anti-MICA sensitization and increased proteinuria in kidney transplant recipients.47 Furthermore, the donor MICA rs2596538 G allele carrier status is a predictor of development of cytomegalovirus (CMV) infection during the first post–kidney transplantation year.48 Kidney donor CC genotype at C3435T (rs1045642) of ABCB1 is associated with an increased risk of long-term allograft failure among white recipients.49 Another study found an association of the donor ABCB1 c.1199 G>A (exon 11, rs2229109) allele (GA/AA versus GG: HR = 3.22 [1.14–9.09], P = 0.029) with an increased risk of allograft loss.50 CAV1 is an oncogenic membrane protein associated with cell proliferation, inflammation, and transforming growth factor-beta signaling.51 Common variation in CAV1 was evaluated in 785 white kidney donors and their recipients and replicated in an independent cohort of transplant recipients.51 Donor AA genotype for the CAV1 rs4730751 was associated with 97% increased risk for allograft failure. Graft failure rate for donor genotype AA was 38.6%, genotype CC was 22.3%, and genotype AC was 22.2%.51 Ficolin-2 is involved in maintenance of tissue homeostasis through engaging apoptotic and necrotic cells.52 Ala258Ser variant of Ficolin-2 in donors is associated with lower incidence of severe allograft rejection and graft loss.52 The strength of evidence to support the role of many of the discussed genetic modifiers varies significantly in reported studies with more consistent evidence available for ApoL1 risk variants. We propose that a combination of these variants in addition to ApoL1 may enhance the prognostic prediction.
Recipient Genetics
Recipient immune response genes could also impact outcomes after transplantation.53 Copy number variation in C4, an immune response gene, affects long-term allograft survival.54 A GWAS found an association of acute kidney allograft rejection with protein tyrosine phosphatase receptor type O, a lymphocyte receptor-type tyrosine kinase gene and coiled-coil domain containing 67, a ciliary gene.55 LIM Zinc Finger Domain Containing 1 gene, which encodes a protein involved in cell adhesion and integrin signaling, predicts transplant outcome.56 The risk allele is frequent in individuals from European and African ancestry, but not present in those with East Asian ancestry.56 LIM Zinc Finger Domain Containing 1 locus rs893403 was shown to be associated with kidney allograft rejection in 4 large cohorts involving 2709 transplants.56 Through a genomic collision scenario, outcomes of renal transplant recipients who were homozygous for a deletion polymorphism at chromosome 2q12.3 and had received allografts from donors with at least 1 normal allele were evaluated.56 Genomic collision at chromosome 2q12.3 was associated with 60% higher risk for rejection compared with those without the genomic collision.56 The prevalence of genomic collision at chromosome 2q12.3 is estimated to be 12%–15% in unrelated renal transplantation among individuals with European and African ancestry however not common in individuals with East Asian ancestry.
CMV infection in graft donors is associated with decreased graft survival.57 A variant of programmed cell death 1 gene, which is involved in viral-induced T-cell exhaustion, is associated with graft survival in patients who had received transplant from CMV-positive donors, whereas no association was found in CMV negative donors.58 Future studies should be designed to examine the benefit of CMV prevention strategies based on genotype to identify who will benefit from prolonged antiviral prophylaxis.58 In a cohort of Hispanic kidney transplant recipients the interferon (IFN)-γ +874 AA genotype was associated with a 3.4-fold increased risk for the CMV infection.59 This may be related to the lower production of IFN-γ in individuals with IFN-γ +874 AA genotype.59 NOD-like receptor family, pyrin domain containing 3 (NLRP3) is involved in inflammatory response. In a retrospective study of 1271 matched donors and recipients, NLRP3 gain of function SNP (rs35829419) in donors was found to be associated with 91% increased risk of biopsy-proven acute rejection. On contrary, loss of function SNP of NLRP3 (rs6672995) in the recipients was associated with a decreased risk for rejection in the first year after renal transplantation.60 Interestingly, tubular epithelial cells express NLRP3 and other inflammatory cytokines including IL-1β and IL-18. A gain of function of NLRP3 may lead to increased expression of these cytokines resulting in kidney injury.60 Polymorphism in genes involved in immune regulation such as regulatory T cells (Treg) function may impact allograft outcomes. In a cohort of 482 black transplant recipients, rs2910164, which can alter the expression of the microRNA (MiR)146A, was associated with acute allograft rejection.27 MiR146A suppresses inflammation through its effect on target genes such as IL1 receptor-associated kinase gene and tumor necrosis factor (TNF) receptor-associated factor gene.27 Thus, rs2910164 variant, which reduces the microRNA expression, may lead to enhanced inflammatory response resulting in increased risk for allograft rejection.27 Other genetic variants involved in immune response such as chemokine receptor (CCR)2 and CCR5,61,62 Cytotoxic T-Lymphocyte Antigen (CTLA)-4,63 Toll-Like Receptor (TLR)3,52 TLR4,64 IL2 Receptor Beta (IL2RB),65 IL6 in donors,66 IL10,67-69 transforming growth factor-beta,70 TNF-α,67-69 CD28,71 and mannose-binding lectin 272 may also influence allograft outcomes. However, these reported association studies are plagued by low sample size studies and confounded by variations in race and ethnicity of the cohorts studied. For instance, polymorphisms of mannose-binding lectin 2 and other complement players including C3 and C4 did not show a consistent association with graft outcomes in different cohorts.73 Lack of adequately powered and validation studies remains as a major barrier for clinical adoption of these genetic variants.
In addition to genes involved in immune system, prothrombotic genetic variants including Factor II, Factor V Leiden, and C677T variant of methylenetetrahydrofolate reductase gene are also associated with acute rejections and notably vascular rejections.74 Given the limited number of such studies, the clinical utility of these genetic variants need further investigation before any recommendation for widespread use can be made. It is also possible that these genetic modifiers may or may not have a pathogenic mechanism. A polygenic risk score (PRS) of allograft rejection/loss-associated variants in an individual can be computed to prognosticate transplant outcomes. At present, the PRSs have low discriminative ability in the general population for the conditions tested.75 A paradigm shift may be needed to change the focus from conventional case-control studies to PRS for a single individual.
A panel of genetic predictors for transplant outcomes is shown in Table 1. Any proposed panel should be dynamically updated based on scientific discoveries.
Table 1.
A panel of genetic predictors for transplant outcomes
| Reference | Gene | Physiologic function | SNP identifier | Associations with clinical outcomes |
|---|---|---|---|---|
| Reeves-Daniel et al36Freedman et al37,38 | ApoL1 | Trypanosome killing function | rs71785313rs60910145rs73885319 | Reduced kidney allograft survival |
| Tonnerre et al47Rohn et al48 | MICA | Stress-induced protein regulated at the cell surface | rs2596538rs67841474 | Anti-MICA sensitization and increased proteinuria in kidney transplant recipients and is a predictor of susceptibility to CMV infection |
| Eikmans et al52 | TLR3 | Cell-bound receptor involved in innate immune system | rs3775296 | Increased acute kidney allograft rejection |
| Hwang et al64 | TLR4 | Binds to endogenous ligands released from damaged tissues and exogenous ligands such as lipopolysaccharide | rs10759932 | Increased rejection-free survival rate |
| Eikmans et al52 | FCN2 | Soluble recognition molecule that can engage apoptotic and necrotic cells | rs7851696 | Reduced incidence of severe kidney allograft rejection and graft loss |
| Steers et al56 | LIMS1 | A minor histocompatibility antigen | rs893403 | Increased kidney allograft rejection |
| Oetting et al27 | MIR146A | Modulated Treg and suppression of inflammatory responses | rs2910164 | Increased kidney allograft rejection |
| Moore et al51 | CAV1 | Involved in cholesterol transport and transmembrane signaling | rs4730751 | Increased kidney allograft failure |
| Forconi et al58 | PD-1 | Involved in the dysfunction of HIV-specific T cell response and CMV-specific CD8 T cells | rs11568821 | Improved kidney allograft survival in recipients from CMV-positive donors |
| Vu et al59 | IFN-γ | Involved in immune response to viral and bacterial infections | rs2430561 | Increased risk for the CMV infection |
| Moore et al49Woillard et al50 | ABCB1 | An efflux pump for intestinal transport of medications including tacrolimus | rs1045642rs2229109 | Increased risk of renal allograft loss |
| Dessing et al60 | NLRP3 | NOD-like receptor family, pyrin domain containing 3 is a member of inflammasome family with a causal role in several inflammatory disorders | rs35829419rs6672995 | Increased acute kidney allograft rejection with rs35829419 andReduced acute kidney allograft rejection with rs6672995 |
| Abdi et al61Cha et al62 | CCR5 | Chemokine receptor specific for the proinflammatory chemokines | rs1799987 | Increased acute kidney allograft rejection |
| Abdi et al61Cha et al62 | CCR2 | Involved in immune response including monocyte recruitment and T cell proliferation | rs1799864 | Increased acute kidney allograft rejection |
| Park et al65 | IL2RB | Stimulating T-cell proliferation through complex of IL2RA-IL2RB-IL2 | rs228942rs228953 | Increased acute kidney allograft rejection episodes |
| Marshall et al66 | IL6 | A pleiotropic cytokine with proinflammatory and anti-inflammatory properties | rs1800795 | Increased acute kidney allograft rejection |
| Sankaran et al68Grinyó et al69 | IL10 | An immunomodulatory cytokine with anti-inflammatory effects | rs1800896 | Increased acute kidney allograft rejection |
| Alakulppi et al67Sankaran et al68Grinyó et al69Sánchez-Fructuoso et al91 | TNF-α | Proinflammatory cytokine | rs1800629 | (rs1800629 in Donor and Recipient)Increased acute kidney allograft rejection episodes(rs1800629 in Recipient)Modulates the effect of ATG treatment |
| Tinckam et al70Hueso et al165 | TGF-β | Anti-inflammatory but profibrotic cytokine | rs1982073rs1800471 | Reduced risk of late acute kidney allograft rejections with rs1800471 and increased kidney allograft subclinical rejection with rs1982073 |
| Pawlik et al71 | CD 28 | A costimulatory molecule involved in T cell-mediated immune response | rs3116496 | Increased acute kidney allograft rejection |
| Golshayan et al72 | MBL2 | Complement-activating MBL, a soluble pattern recognition receptor | rs7096206 rs5030737 rs1800450 rs1800451 | Increased acute kidney allograft rejection |
| Canossi et al63 | CTLA4 | CTLA4 transduces signals that inhibit lymphocyte activation | rs231775 rs3087243 | Reduced acute kidney allograft rejection with rs231775 and increased acute kidney allograft rejection with rs3087243 |
| Heidenreich et al74 | Factor II | Prothrombotic factor | rs1799963 | Increased acute kidney allograft rejection, especially vascular rejections, and early allograft failure |
| Heidenreich et al74 | Factor V Leiden | Prothrombotic factor | rs6025 | Increased acute kidney allograft rejection especially vascular rejections |
| Heidenreich et al74 | MTHFR | Prothrombotic factor | rs1801133 | Increased acute kidney allograft rejection, especially vascular rejections |
| Cartron et al96 | FCGR3A | Encodes the IgG Fc receptor | rs396991 | Increased risk of infection following Rituximab in recipients of liver transplant |
The panel is not exhaustive of all published literature.
ABCB1, ATP binding cassette subfamily B member 1; ApoL1, Apolipoprotein 1; ATG, antithymocyte globulin; CAV1, caveolin-1; CCR, chemokine receptor; CMV, cytomegalovirus; CTLA, cytotoxic T-lymphocyte antigen; IFN-γ, interferon-gamma; IL2RB, IL2 Receptor Beta; MBL, mannose-binding lectin; MICA, MHC class I-related chain A; MiR, microRNA; MTHFR, methylenetetrahydrofolate reductase; NLRP3, NOD-like receptor family, pyrin domain containing 3; SNP, single nucleotide polymorphism; TGF, transforming growth factor; TLR, Toll-Like Receptor; TNF-α, tumor necrosis factor-alpha; Treg, regulatory T cells.
Precision Pharmacology
Genetic factors can explain 20%–95% of interindividual variability in drug response.11 Studies comparing the drug response in monozygotic twins with dizygotic twins indicated the role of genetic variants several decades ago. Half-life of many drugs is different in dizygotic twins, whereas monozygotic twins have similar half-life, suggesting genetic underpinning.76-78 Pharmacogenomics (PGx) is the study of how genes affect a person’s response to drugs, that combines pharmacology and genomics to develop effective, safe medications and doses that will be tailored to a person’s genetic makeup.17,79,80 For instance, variations in genes involved in drug metabolism and transport can affect drug pharmacokinetics, whereas variants in genes encoding for drug-target proteins can impact drug pharmacodynamics.11,17,79,80 During the last decade, the field of pharmacogenetics has evolved into PGx, which involves a shift from a focus on individual candidate gene variants to GWAS.16 Association studies do not address the underlying mechanism necessitating proteome analysis, indicating a role for pharmacoproteomics approach in precision medicine.81 Propelled by advances in molecular genetics, the field of pharmacogenetics is rapidly becoming a reality in clinical practice. Over the past 20 years >20 000 new PGx citations are noted in PubMed.82,83 Furthermore, approximately, 200 Food and Drug Administration approved medications have PGx information available on their labeling.82,83 Inherited variations in about 20 genes have been found to influence clinical response to at least 80 medications.79
Precision Prescribing in Transplant Recipients
Solid organ transplant recipients typically receive induction immunosuppressive therapy at the time of surgery with gradual introduction of maintenance agents. The objective is to mitigate an acute allogeneic response and usually consists of glucocorticoids, T-cell depletion, and B-cell or plasma-cell depletion depending the perceived risk of rejection.84 Over the last several decades there has been significant evolution in the form of induction agents available, however, no head-to-head randomized controlled trial has been conducted to define the most efficacious and safe regimens. Prescribing patterns among the transplant community have thus been led by practice guidelines such as that from the 2009 Kidney Disease Improving Global Outcomes that are deemed “moderate” in strength of evidence.85 In terms of maintenance therapy, the calcineurin inhibitor tacrolimus stands as the “backbone” agent, after having shown superiority over other agents in a prospective and randomized fashion.86 However, side-effect profiles of all maintenance immunosuppressants have effectively preserved a role for each drug in the highly heterogeneous transplant population. There is therefore due need to define and leverage the pharmacodynamics of these agents towards more desirable clinical outcomes. To this end, we herein summarize the pharmacogenetics of various induction and maintenance agents used in the peritransplant and posttransplant settings.
Induction Therapy
Thymoglobulin
Antithymocyte globulin is a polyclonal IgG fraction targeted against human thymocytes derived from rabbits or horses.87 There is however evidence that ATG may work via additional mechanisms such as through expansion of Treg and enhanced IL10 production causing inhibition of TNF-α production by macrophages.88,89 Indeed, TNF-α has been demonstrated in the alloimmune process and there is meta-analytic data that TNF-α polymorphism-308, G/A may influence risk of rejection.90,91 In a retrospective analysis, transplant recipients carrying the risk allele who were not treated with thymoglobulin had a higher risk of rejection compared with those that did.91 It was thus fathomed that ATG may be beneficial in transplant recipients who generate higher levels of TNF-α via this polymorphism.
Rituximab
Rituximab is a humanized chimeric anti-CD20 monoclonal antibody, which is the Food and Drug Administration approved, for the treatment of certain B-cell malignancies. It is believed to work through CD20+ B-cell depletion to influence complement-mediated and antibody-dependent cell-mediated cytotoxicity.92 Its potential use in solid organ transplantation was recognized in 2003 in a series of 4 successful ABO-incompatible living donor transplants where it replaced the traditional practice of pretransplant splenectomy.93 It has subsequently been used in the treatment of posttransplant lymphoproliferative disorder and in rejection.94 Defining the clinical and biologic predictors of efficacy and safety is paramount given the cost and side effects of B-cell depletion. It has been shown in ABO-incompatible living donor liver transplantation that SNPs of the Fc fragment of IgG receptor (FCGR) gene may influence the risk of infection following Rituximab in this setting.95 Indeed certain genotypes in this region have also been shown to correlate with clinical and molecular responses to Rituximab in non-Hodgkins lymphoma.96 The significance of this phenomena on B-cell depletion in renal transplant induction has yet to be established.
Belatacept
Costimulation of the T-cell via the interaction between CD80/CD86 on antigen-presenting cells and CD28 on the T-lymphocyte is a critical activating event in the alloimmune response.97 Belatacept is a CTLA4-Ig fusion protein that exploits the attenuating effect of CTLA4, which blocks the CD28-CD80/CD86 interaction, hence preventing T-cell activation.98,99 The Belatacept and Long-Term Outcomes in Kidney Transplantation trial demonstrated superior patient and graft survival of belatacept over cyclosporine.100 Enthusiasm for this agent, however, was tempered by episodes of histologically severe acute cellular rejection occurring in this trial, which was subsequently found to occur disproportionately in individuals with CD28+ Memory CD8 T cells.101 The hypothesized mechanism of “belatacept resistance” via CD28 is that executes other signaling pathways to enable costimulation independent rejection. Polymorphism in the CD28 gene was shown to be associated with acute kidney allograft rejection.71 Integration of genomics data with pharmacoproteomics analysis may be complimentary in predicting drug response and clinical outcomes.
Maintenance Immunosuppression
Tacrolimus
Tacrolimus is the most common maintenance immunosuppression used in the setting of solid organ transplantation. Currently, we use a standard dose based on body weight, which is titrated to achieve the desired plasma level. Despite close monitoring of the drug plasma level, underimmunosuppression with increased risk of graft rejection and drug toxicity are common.102 The narrow therapeutic index and wide interindividual variability of tacrolimus pharmacokinetics103,104 warrant precision pharmacotherapy, which could prevent graft rejection82 and toxicity.102
Tacrolimus is metabolized by Cytochrome P450 (CYP) 3A and transported in the gut by P-glycoprotein, an efflux pump, encoded by ABCB1 gene. CYP3A4 and CYP3A5 in part explain the interindividual differences of response to calcineurin inhibitors.105 Several studies showed no significant impact for ABCB1 on pharmacokinetics of tacrolimus.105-108 Tacrolimus dose-adjusted trough levels were found to be higher in kidney transplant recipients with genotype of CYP3A5*3/*3 compared with recipients with genotype of *1/*3 plus *1/*1.109 Another study reported that patients with genotype of CYP3A5*1/*1 had dose-adjusted trough concentrations 5.8-fold lower than patients with genotype of CYP3A5*3/*3.110 The authors concluded that up to 45% of the variability of tacrolimus dose requirement is explained by the CYP3A5*1/*3 polymorphisms.110 Higher dose of tacrolimus is needed to achieve target plasma level in black population.111 A recent prospective multicenter study of 2595 kidney transplant recipients showed Native Americans and whites required the lowest median tacrolimus dose, whereas the black recipients required the highest median dose to achieve the therapeutic target.112 The CYP3A5*3 variant was most common in whites with allele frequency of 0.93. It was 0.84 for Native Americans and 0.72 for Asian Americans and 0.3 for black recipients. The CYP3A5*6 and *7 variants are found only in black recipients. The CYP3A5*3 variant was associated with higher dose-normalized tacrolimus trough levels in all 4 populations compared with other gene variants.112 Transplant recipients carrying 1 or 2 CYP3A5*1 alleles (CYP3A5 expressers) need a higher tacrolimus dose compared with CYP3A5 nonexpressers.113 More than 50 studies have shown that individuals with the CYP3A5*1/*1 or CYP3A5*1/*3 genotype have lower dose-adjusted trough level of tacrolimus in comparison with those individuals with the CYP3A5*3/*3 genotype, with *1 carriers requiring 1.5–2 times the standard dose to achieve similar blood levels.114
To examine the clinical implication of testing for the CYP gene variants, a randomized trial of 280 renal transplant recipients who received tacrolimus according to CYP3A5 genotype versus standard practice was conducted.115 The proportion of patients at target level C (0) was higher at day 3 after initiation of tacrolimus.115 However, a randomized controlled trial involving 240 transplant recipients with low immunologic risk showed no change in clinical outcomes when tacrolimus starting dose based on CYP3A5 genotype was adapted.116 Further inclusive studies, providing more generalizable results are warranted. Before conceiving such studies, we should consider the ethics of randomizing an individual to standard dose despite the knowledge that they will achieve subtherapeutic levels of tacrolimus.
According to Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines, patients with CYP3A5 extensive metabolizer or intermediate metabolizer (CYP3A5 expressers) would need higher tacrolimus starting dose, whereas patients with the CYP3A5 nonexpresser, which are poor metabolizers, would need standard tacrolimus starting dose (https://cpicpgx.org). A starting dose 1.5–2 times standard dose, not exceeding 0.3 mg/kg/d in CYP3A5 extensive metabolizer or intermediate metabolizer is recommended to achieve therapeutic target levels. Drug monitoring to guide dose adjustments should be performed.114 Additionally, in whites incorporating CYP3A4*22 genotype into the CPIC recommendation may improve the performance of CYP3A5 genotype adjusted tacrolimus dosing. Tacrolimus dose may be decreased for CYP3A4*22 carriers-CYP3A5 defectives to 0.14 mg/kg/d, whereas it can be allowed to be increasing up to 0.4 mg/kg/d in those with CYP3A4*22 noncarriers-CYP3A5 expresser starting at 0.35 mg/kg/d.117 CYP3A4 and CYP3A5 variants may also predict tacrolimus-related nephrotoxicity. In a study of 95 genotyped recipients, CYP3A4*1/CYP3A5*1 and CYP3A4*1B/CYP3A5*1 variants were found to be more frequently associated with the development of biopsy-proven tacrolimus-related nephrotoxicity than the CYP3A4*1/CYP3A5*3 genotype.118 Additionally, other genetic variants may influence CYP3A4 and CYP3A5 activities. The POR*28 allele (rs1057868) has been shown to be associated with increased in vivo CYP3A5 activity for tacrolimus in those who are CYP3A5 expressers, which indicates an increased CYP3A5 activity for POR*28 carriers. POR*28 homozygosity was found to be associated with a significant higher CYP3A4 activity in those who are CYP3A5 nonexpressers for tacrolimus and cyclosporine.119
Cyclosporine
The effect of variable CYP3A5 expression on cyclosporine dosing, blood pressure, and long-term graft survival in renal transplant patients was evaluated in 399 white patients with stable graft function for >10 weeks posttransplantation.120 The recipient CYP3A5*1 allele was found to have no effect on cyclosporine dose and blood concentrations at trough with and without dose adjustment. Also blood pressure, number of antihypertensive compounds used for treatment, and graft survival were not influenced by CYP3A5*1 allele.120 The impact of variations in the ABCB1, ATP binding cassette subfamily C member 2, solute carrier organic anion transporter family member 1B1, CYP3A4, CYP3A5, or Nuclear Receptor Subfamily 1 Group I Member 2 (NR1I2) genes on the pharmacokinetics of cyclosporine was assessed in 104 pediatric renal transplant candidates. Among children older than 8 years, carriers of the ABCB1 c.1236C>T or c.2677G>T variant allele were found to have approximately 1.3–1.6 times higher oral bioavailability and lower prehepatic extraction ratio of cyclosporine than noncarriers.121 About 30%–37% of the variability in oral bioavailability and prehepatic extraction was explained by the genetic variants. In addition, a corresponding tendency in the dose requirement was found. Overall, the variability in the pharmacokinetics of cyclosporine remained largely unexplained by those investigated genetic variants.121
Mycophenolic Acid (Myfortic)
Myfortic is an inosine monophosphate dehydrogenase inhibitor that should be avoided in individuals with deficiency of hypoxanthine-guanine phosphoribosyl-transferase. Individuals including those with partial deficiency of the enzyme can develop elevated uric acid level resulting in gout, kidney failure, and kidney stones.122 There are limited data about pharmacogenetic testing for myfortic in kidney transplant recipients. However, CPIC recommends pharmacogenetic testing of hypoxanthine phosphoribosyl-transferase 1 gene as it may provide actionable information122,123 such as consideration of using alternative agent in those with hypoxanthine-guanine phosphoribosyl-transferase deficiency.
Azathioprine
Azathioprine is an antimetabolite that has been used for posttransplant immunosuppression.85 As a prodrug, azathioprine should be converted to mercaptopurine. Polymorphic thiopurine methyltransferase (TPMT) inactivates mercaptopurine through methylation. Activity of TPMT can be influenced by genetic variants.124 At least 1 slow metabolizer variant can be found in approximately 10% of whites, which leads to accumulation of toxic metabolites resulting in severe myelosuppression.125 One in 300 whites is homozygous for the allele causes complete deficiency of TPMT activity.125 Genotyping of TPMT may be informative as there are 3 TPMT SNPs accounting for >90% of inactivating alleles.126,127 CPIC guideline recommends that patients with TPMT heterozygous with 1 of alleles *2, *3A, *3B, *3C, and *4 should receive lower initial dose of thiopurine medications. Risk of life-threatening severe myelosuppression exists for patients with the homozygous variant genotype with 2 of the alleles (*2, *3A, *3B, *3C, and *4) during therapy with thiopurine medication. Therefore, significant dose reduction or use of an alternative agent is recommended.125 Nucleoside diphosphate linked moiety X (Nudix)-type motif 15 (NUDT15) is involved in catalyzing the conversion of cytotoxic thioguanine triphosphate metabolites to a less toxic substance, thioguanine monophosphate. R139C variant of NUDT15 is also linked with thiopurine toxicity with consequent severe myelosuppression.128 In individuals who are NUDT15 intermediate metabolizer, a reduction in starting dose should be considered to decrease toxicity. For those who are NUDT15 poor metabolizer, a significant dose reduction or using an alternative agent should be considered.128
Everolimus
Everolimus is a macrolide immunosuppressive agent used in solid organ transplant recipients. It is structurally related to tacrolimus and binds to FK-binding protein and blocks the transduction signal from the IL2 receptor, thus inhibiting T- and B-cell proliferation. In a study of 53 renal transplant patients who had been switched from a regimen consisting of cyclosporin, mycophenolate, mofetil and prednisolone to a calcineurin inhibitor-free regimen consisting of everolimus and prednisolone, polymorphisms in genes coding for ABCB1, CYP3A5, CYP2C8, and Pregnane X Receptor found to have no clinically relevant effect on everolimus pharmacokinetics.129
Precision Prescribing of Nonimmunosuppressive Drugs
Among the drugs that are commonly used in transplant population, there are evidence-based guidelines available for voriconazole, clopidogrel, warfarin, narcotics, simvastatin, and allopurinol. Trough voriconazole concentrations are lower in patients with CYP2C19 ultra-rapid metabolizers compared with poor metabolizers resulting in delay in achieving therapeutic level, which may be critical in a life-threatening infections such as invasive aspergillosis in transplant recipients.130 CPIC guideline recommends that patients with CYP2C19 ultra-rapid or rapid metabolizer status (*17/*17 or *1/*17, respectively) to receive an alternative agent other than voriconazole as therapeutic level may not be achievable. Patients with CYP2C19 poor metabolizer status (2 alleles of either *2 or *3) should use an alternative agent because of high-risk for developing adverse effects.131 Clopidogrel is a prodrug that needs to be activated by CYP2C19.132 According to American College of Cardiology Foundation/American Heart Association Acute Coronary Syndrome guidelines, genetic testing for CYP2C19 loss-of-function alleles may be considered on a case-by-case basis, especially in those with recurrent Acute Coronary Syndrome despite treatment with clopidogrel.133 “Error and trial approach” in case of a life-threatening condition such as acute coronary event especially in transplant recipients may not be advisable. PGx-guided antiplatelet therapy in the highly vulnerable and heavily invested population such as transplant recipients should be considered. The CPIC guideline recommends using an alternative agent in patients with at least 1 decreased function allele because of risk for decreased response. Patients with genotype of increased metabolism should be monitored for increased bleeding risk.134 Warfarin, a vitamin K antagonist, is a commonly used anticoagulation medication with significant interindividual variability and narrow therapeutic index leading to frequent complications due to overdosing and underdosing. Genetic variants in CYP2C9, CYP4F2, and vitamin K epoxide reductase complex subunit 1 can predict the dose needed to meet the therapeutic level.135 Codeine may not be effective in patients who are CYP2D6 poor metabolizers, whereas there is a higher risk for toxicity in those patients who are CYP2D6 ultra-rapid metabolizers.136 Life-threatening side effects have been reported in CYP2D6 ultra-rapid metabolizers including those patients who had received even standard doses of codeine. CYP2D6 is also involved in metabolism in other opioids such as tramadol, hydrocodone and oxycodone, hydromorphone, and oxymorphone.136 Concomitant use of statins such as simvastatin with certain drugs such as cyclosporine may lead to increased blood concentration of simvastatin resulting in myotoxicity.137 An alternative agent or a reduced dose of simvastatin should be prescribed to patients with at least 1 reduced function allele in solute carrier organic anion transporter family member 1B1 (*5, *15, or *17).138 Variants of the HLA-B gene are associated with allopurinol related cutaneous conditions. Patients with at least 1 HLA-B* 58:01 allele are at higher risk for developing allopurinol related cutaneous conditions.139 The CPIC guideline recommends avoiding use of allopurinol in patients with at least 1 HLA-B*58:01 allele.140 A list of commonly used medications with actionable genetic information in transplant population is shown in Table 2.
Table 2.
Gene-drug pairs with sufficient evidence for at least 1 prescribing action to be recommended
| Author | Gene | Medication | Pharmacogenetics implications |
|---|---|---|---|
| Birdwell et al114CPIC166 | CYP3A5 | Tacrolimus | Higher starting dose at 1.5–2 times standard dose, not exceeding 0.3 mg/kg/d in CYP3A5 extensive metabolizer or intermediate metabolizer. |
| Birdwell et al114CPIC166 | CYP3A4 | Tacrolimus | Higher starting dose as above |
| Elens and Haufroid117 | POR | Tacrolimus | POR*28 homozygosity is associated with a significant higher CYP3A4 activity in those who are CYP3A5 nonexpressers |
| Relling et al128CPIC166 | TPMT | Azathioprine | Reduce initial dose in TPMT heterozygous with 1 of alleles *2, *3A, *3B, *3C, and *4 |
| Relling et al128CPIC166 | NUDT15 | Azathioprine | Reduce initial dose for NUDT15 intermediate metabolizer. Consider an alternative agent for NUDT15 poor metabolizer |
| CPIC166 | HPRT1 | Mycophenolic acid | Consider using alternative agent in HGPRT deficiency |
| Crews et al136CPIC166 | CYP2D6 | Codeine Oxycodone | Use alternative analgesics in CYP2D6 poor metabolizers or ultra-rapid metabolizers |
| Moriyama et al131Scott et al134CPIC166 | CYP2C19 | Voriconazole Clopidogrel | Use an alternative agent other than voriconazolein CYP2C19 ultra-rapid or rapid or poor metabolizersUse an alternative agent other than Clopidogrel in patients with at least 1 decreased function allele |
| Johnson et al135CPIC166 | VKORC1 | Warfarin | Consider an alternative oral anticoagulant/calculate warfarin dosing according to CPIC guideline pharmacogenetic algorithma |
| Johnson et al135CPIC166 | CYP2C19 | Warfarin | Consider an alternative oral anticoagulant/calculate warfarin dosing according to CPIC guideline pharmacogenetic algorithm |
| Johnson et al135CPIC166 | CYP4F2 | Warfarin | Consider an alternative oral anticoagulant/calculate warfarin dosing according to CPIC guideline pharmacogenetic algorithm |
| SEARCH Collaborative Group138CPIC166 | SLCO1B1 | Simvastatin | Use an alternative agent or a reduced dose of simvastatin in patients with at least 1 reduced function allele |
| Hershfield et al140CPIC166 | HLA-B*58:01 | Allopurinol | Avoid allopurinol in patients with at least 1 HLA-B*58:01 allele |
aCPIC guideline pharmacogenetic algorithm https://cpicpgx.org/content/guideline/publication/warfarin/2017/28198005.pdf.
CPIC, Clinical Pharmacogenetics Implementation Consortium; CYP, Cytochrome P450; HGPRT, hypoxanthine-guanine phosphoribosyl-transferase; NUDT15, nucleoside diphosphate linked moiety X-type motif 15; SLCO1B1, solute carrier organic anion transporter family member 1B1; TPMT, thiopurine methyltransferase; VKORC1, vitamin K epoxide reductase complex subunit 1.
Pharmacogenetics in Transplantation
Cost of kidney care in the United States is $114 billion per year.141 Cost of allograft failure and return to dialysis is estimated $70 000–$106 000 per year compared with $16 000 per year for those ESRD patients with functioning graft.142 Additionally, >2 million adverse drug reactions with approximately 100 000 associated death occur annually in the United States.143 The cost for adverse drug reactions has been estimated up to $136 billion per year.144 Drugs interactions are very common among kidney transplant recipients in part due to narrow therapeutic index of commonly used medications in transplant population.145,146 In certain fields in medicine such as oncology, due to high side-effect profile and astronomic costs of new biologic and chemotherapy medications, precision medicine is rapidly being implemented in clinical practice.147-149 Despite the enormous cost of caring for transplant patients and vulnerability of these patients, transplant medicine is lagging behind in implementing precision prescribing. Therefore, in addition to potential optimization of transplant outcomes, precision medicine in kidney transplantation may be cost-effective from payer’s standpoint.
GWAS AND GENETIC PANEL TESTING
GWAS is a powerful tool to identify causal genetic variants, by simultaneously analyzing millions of single nucleotide polymorphisms (SNPs) distributed across the genome.30,55 A GWAS conducted by the United Kingdom and Ireland Renal Transplant Consortium and the Wellcome Trust Case Control Consortium-3 failed to identify strong donor or recipient genetic effects outside the HLA region contributing to long- or short-term allograft survival.30 Several reasons could explain the lack of discovery including small sample size and heterogeneous cause for graft loss. Results from the International Genetics and Translational Research in Transplantation Network, a multisite consortium (n = 28 015) with adequate power to capture both rare and common genetic contributions to ESRD and posttransplant outcomes is expected soon.150
It is evident that further studies are required before recommending the utility of genetic variants in clinical setting. Pending new discoveries, a panel of genetic variants could be tested in the research setting for kidney transplant recipients and potential donors consist of genetic variants with pharmacogenetic implications and genetic variants with prognostic value for clinical outcomes. A risk estimate could be derived integrating the genetic, demographic, and clinical data, which if combined to pharmacogenetic of immunosuppressive medications could be a useful tool in clinical setting (Figure 1). This can be achieved by combining a panel of allograft loss-associated variants carried by an individual into a single score that provides overall genetic risk, a PRS.151 The combination of PRSs with clinical risk factors could improve the risk stratification further.152 Efforts are underway to integrate findings from GWAS with expression quantitative trait loci from scRNAseq as well as known regulatory region maps could identify novel genes associated with graft loss.153 In addition to rare renal genetic diseases, there are currently available resources such as Natera (https://www.natera.com/organ-health/renasight-genetic-testing) and Invitae (https://www.invitae.com/en/chronic-kidney-disease/) offering genetic panel testing for patients with chronic kidney disease.154 A transplant genetic panel implicating the rejection risk could be complementary in care to transplant population and potentially improving outcomes. Clinical validation through prospective trials supporting the clinical decision outlined in Figure 1 is required.
FIGURE 1.
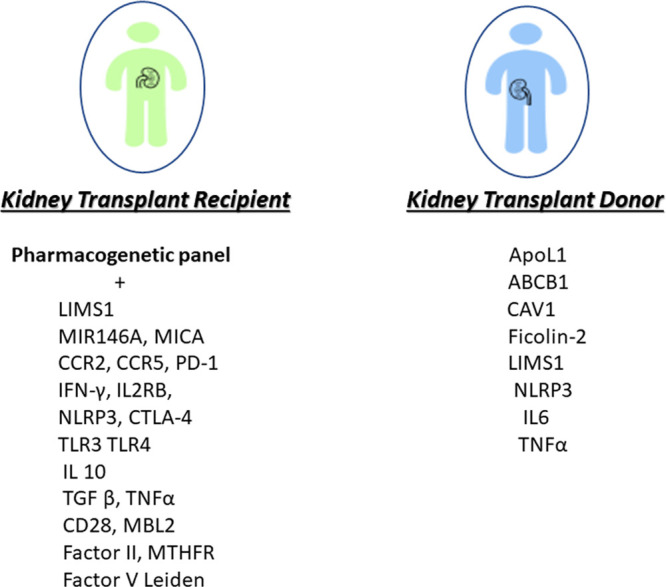
A panel of genetic variants for transplant recipients and donors. This panel functions as an additional tool at disposition of transplant physicians to provide individualized care. Clinical validation through prospective trials supporting the clinical decision outlined is required. ABCB1, ATP binding cassette subfamily B member 1; ApoL1, Apolipoprotein L1; CAV1, caveolin-1; CCR, chemokine receptor; CTLA, Cytotoxic T-Lymphocyte Antigen; IFN-γ, interferon-gamma; IL2RB, IL2, Receptor Beta; MBL, mannose-binding lectin; MICA, MHC class I-related chain A; MiR, microRNA; MTHFR, methylenetetrahydrofolate reductase; NLRP3, NOD-like receptor family, pyrin domain containing 3; TGF-β, transforming growth factor; TLR, Toll-Like Receptor; TNF-α, tumor necrosis factor-alpha.
Noninvasive Transplant Immune Monitoring
Solid-organ transplantation is effectively genomic transplantation—a concept depicted by Lo and colleagues who demonstrated that donor-derived cell-free DNA (dd-cfDNA) is present in the plasma of kidney and liver transplant recipients.155 They envisioned that dd-cfDNA might be used as a diagnostic tool for detecting transplant rejection. Indeed, distinctive graft and recipient genotype SNPs have been exploited to barcode donor DNA circulating in recipient serum for this purpose. This approach was first demonstrated as proof of concept in a retrospective analysis of heart transplant recipients in 2011156. Genome transplant dynamic methodology was subsequently clinically validated in solid organ transplantation.157 A multicenter study of renal allograft recipients evaluated the role of circulating dd-cfDNA in blood for diagnosis of acute rejection.158 The assay uses targeted amplification and sequencing of SNPs to quantify donor and recipient DNA contributions. The study showed that plasma levels of dd-cfDNA can discriminate active rejection status of the renal allograft. Extending this concept further to incorporate epigenetic analyses may unravel distinct “signatures” of allograft states such as rejection, infection, or fibrosis. Furthermore, the sheer granularity of epigenetic methods may decipher new and more accurate categories of allograft diseases than the nebulous clinical definitions currently in use.
Pharmacomicrobiomics
Human gut harbors a complex community of >100 trillion microbial cells, which constitute the gut microbiota.159 The gut microbiome encodes about 3.3 million genes, which is 150 times more genes than our own genome.160 The symbiotic gut microbiota provides complementary biologic and metabolic functions that cannot be performed by humans.161,162 There is a growing evidence that gut bacteria can affect the response to drugs by modulating either efficacy or toxicity.163 Pharmacomicrobiomics is an emerging field that investigates the interplay of microbiome variation and drugs response.164 Future investigations should consider gut microbiome in delivering precision therapies in kidney transplantation.
FUTURE DIRECTION AND CONCLUSION
Precision pharmacotherapy in conjuncture with genotype-based risk stratification of transplant recipients and donors may help with donor selection, identification of high-risk recipients, and individualization of pharmacotherapy. Efficient drug monitoring may not function as an alternative for gene-based guidance in pharmacotherapy of transplant recipients. Incorporation of genetic predictors into routine clinical practice may be challenging for physicians in part due to perceived difficulty with interpretation of genetic information. Integration of clinical decision support tools with electronic health records (EHRs) can facilitate the use of available actionable genetic information. Nephrologists have been traditionally advocating precision prescribing based on the level of kidney function. Adjustment of dose of a drug according to glomerular filtration rate through an alert system in EHR is an example of precision prescribing. Similarly, relevant genetic information can be incorporated to EHR and provide guidance to clinicians for precision prescribing (Figure 2). The concept of personalized medicine based on individual patient characteristics, including genetics, molecular markers, and environmental factors, rather than on population averages is attractive (Figures 3 and 4). Precision medicine through incorporation of available genetic information into clinical practice to individualize care for kidney transplant recipients is a realistic hope and on the horizon in the light of ever-decreasing cost of genetic testing and advances in molecular diagnostics. Lack of high-quality data derived from traditional case-control studies remains a barrier for routine use of PRSs in the clinical practice. However, it is noteworthy that precision medicine may a blind spot for conventional randomized trials considering the current low discriminative ability of PRSs in the general population. Increasing access to large datasets has fostered data-driven sciences that are poised to transform personalized medicine.
FIGURE 2.
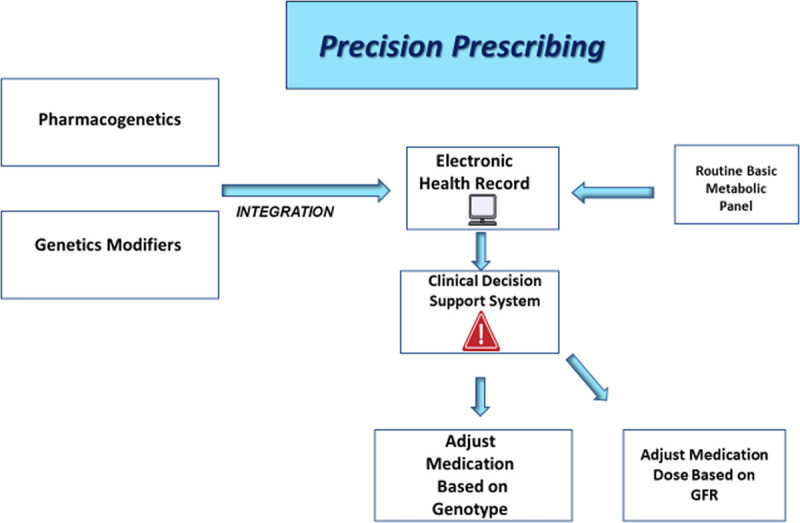
Integration of clinical decision support (CDS) tools with electronic health records (EHRs) can facilitate the use of available actionable genetic information. Adjustment of dose of a drug according to glomerular filtration rate (GFR) through an alert system in EHR is an example of precision prescribing. Similarly, relevant genetic information can be incorporated to EHR and provide guidance to clinicians for precision prescribing. Further studies are required to validate the proposed model.
FIGURE 3.
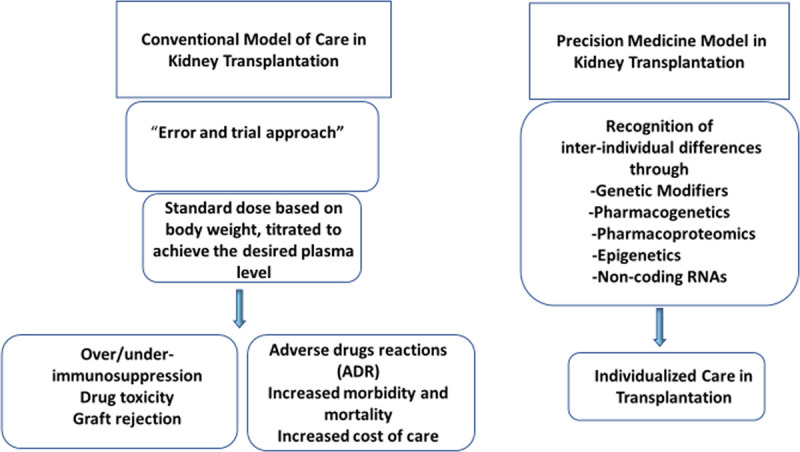
Recognition of interindividual differences is becoming possible through integration of pharmacogenetics, pharmacoproteomics, epigenetics, and noncoding RNAs data into clinical practice. Further studies are required to validate the proposed model. ADR, adverse drug reaction.
FIGURE 4.
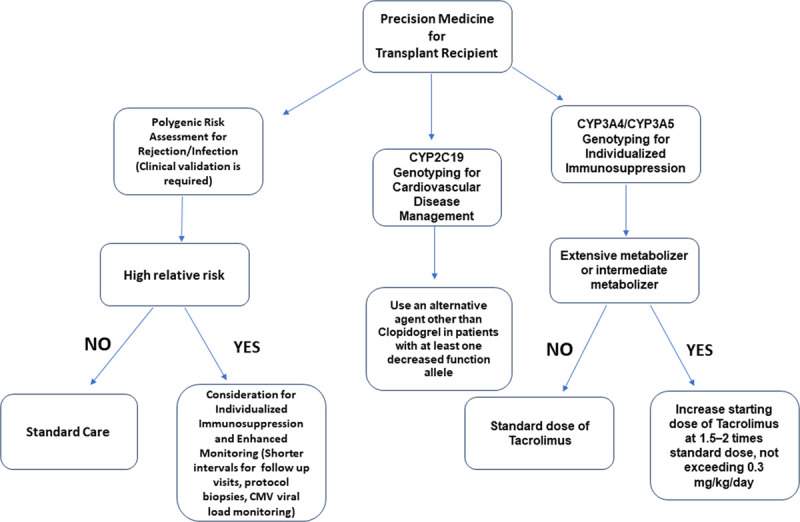
A single score that provides overall genetic risk, a polygenic risk score (PRS) can be achieved by combining of allograft rejection/loss associated-variants carried by an individual and in conjuncture with pharmacogenetics may be integrated into practice after clinical validation through prospective clinical trial supporting the clinical decision outlined. CMV, cytomegalovirus; CYP, Cytochrome P450.
Footnotes
Published online 7 January, 2021.
Ehsan Nobakht and Muralidharan Jagadeesan contributed equally.
The authors declare no funding.
S.D. is the clinical laboratory director of PMCDx, Inc. The other authors declare no conflicts of interest.
E.N. participated in writing of the article and tables/figures preparation. M.J. participated in the writing of the article as co-first author. R.P. and J.B. participated in the writing of the article. S.D. participated in conceiving of the presented idea and writing of the article.
REFERENCES
- 1.Suthanthiran M, Strom TB. Renal transplantation. N Engl J Med. 1994; 331:365–376 [DOI] [PubMed] [Google Scholar]
- 2.Annual Report of the U.S. Organ Procurement and Transplantation Network and the Scientific Registry of Transplant Recipients. Department of Health and Human Services, Health Resources and Services Administration, Healthcare Systems Bureau, Division of Transplantation; United Network for Organ Sharing; University Renal Research and Education Association. Available at https://unos.org/data/transplant-trends/. Accessed August 9, 2020
- 3.Port FK, Wolfe RA, Mauger EA, et al. Comparison of survival probabilities for dialysis patients vs cadaveric renal transplant recipients. JAMA. 1993; 270:1339–1343 [PubMed] [Google Scholar]
- 4.Arend SM, Mallat MJ, Westendorp RJ, et al. Patient survival after renal transplantation; more than 25 years follow-up. Nephrol Dial Transplant. 1997; 12:1672–1679 [DOI] [PubMed] [Google Scholar]
- 5.Hart A, Smith JM, Skeans MA, et al. OPTN/SRTR 2016 annual data report: kidney. Am J Transplant. 2018; 18:18–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hariharan S, Johnson CP, Bresnahan BA, et al. Improved graft survival after renal transplantation in the United States, 1988 to 1996. N Engl J Med. 2000; 342:605–612 [DOI] [PubMed] [Google Scholar]
- 7.Loupy A, Jordan SC. Transplantation: donor-specific HLA antibodies and renal allograft failure. Nat Rev Nephrol. 2013; 9:130–131 [DOI] [PubMed] [Google Scholar]
- 8.McCune JS, Bemer MJ. Pharmacokinetics, pharmacodynamics and pharmacogenomics of immunosuppressants in allogeneic haematopoietic cell transplantation: part I Clin Pharmacokinet. 2016; 55:525–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ojo AO, Hanson JA, Wolfe RA, et al. Long-term survival in renal transplant recipients with graft function. Kidney Int. 2000; 57:307–313 [DOI] [PubMed] [Google Scholar]
- 10.Shamila K, Emily AB. Common infections in kidney transplant recipients. Clin J Am Soc Nephrol. 2012; 7:2058–2070 [DOI] [PubMed] [Google Scholar]
- 11.Evans WE, McLeod HL. Pharmacogenomics–drug disposition, drug targets, and side effects. N Engl J Med. 2003; 348:538–549 [DOI] [PubMed] [Google Scholar]
- 12.Vince N, Poschmann J, Josien R, et al. 23rd Nantes Actualités Transplantation: “genomics and immunogenetics of kidney and inflammatory diseases-lessons for transplantation.” Transplantation. 2019; 103:857–861 [DOI] [PubMed] [Google Scholar]
- 13.van Gelder T, van Schaik RH, Hesselink DA. Pharmacogenetics and immunosuppressive drugs in solid organ transplantation. Nat Rev Nephrol. 2014; 10:725–731 [DOI] [PubMed] [Google Scholar]
- 14.Cavallari LH, Mason DL. Cardiovascular pharmacogenomics – implications for patients with chronic kidney disease. 2016; 23:82–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyatt CM, Schlondorff D. Precision medicine comes of age in nephrology: identification of novel biomarkers and therapeutic targets for chronic kidney disease. Kidney Int. 2016; 89:734–737 [DOI] [PubMed] [Google Scholar]
- 16.Nelson MR, Bacanu SA, Mosteller M, et al. Genome-wide approaches to identify pharmacogenetic contributions to adverse drug reactions. Pharmacogenomics J. 2009; 9:23–33 [DOI] [PubMed] [Google Scholar]
- 17.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999; 286:487–491 [DOI] [PubMed] [Google Scholar]
- 18.Kidney Precision Medicine Project. Available at https://www.kpmp.org/. Accessed August 9, 2020.
- 19.Willis JC, Lord GM. Immune biomarkers: the promises and pitfalls of personalized medicine. Nat Rev Immunol. 2015; 15:323–329 [DOI] [PubMed] [Google Scholar]
- 20.Marson A, Housley WJ, Hafler DA. Genetic basis of autoimmunity. J Clin Invest. 2015; 125:2234–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orrù V, Steri M, Sole G, et al. Genetic variants regulating immune cell levels in health and disease. Cell. 2013; 155:242–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim IK, Bedi DS, Denecke C, et al. Impact of innate and adaptive immunity on rejection and tolerance. Transplantation. 2008; 86:889–894 [DOI] [PubMed] [Google Scholar]
- 23.Brodin P, Davis MM. Human immune system variation. Nat Rev Immunol. 2017; 17:21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mas V, Le T, Maluf D. Epigenetics in kidney transplantation: current evidence, predictions, and future research directions Transplantation. 2016; 100:23–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dwivedi RS, Herman JG, McCaffrey TA, et al. Beyond genetics: epigenetic code in chronic kidney disease. Kidney Int. 2011; 79:23–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suárez-Álvarez B, Baragaño Raneros A, Ortega F, et al. Epigenetic modulation of the immune function: a potential target for tolerance. Epigenetics. 2013; 8:694–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oetting W, Schladt D, Dorr C, et al. Analysis of 75 candidate SNPs associated with acute rejection in kidney transplant recipients: validation of rs2910164 in microRNA MIR146A Transplantation Direct. 2019; 103:1591–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheigh JS, Chami J, Stenzel KH, et al. Renal transplantation between HLA identical siblings. Comparison with transplants from HLA semi-identical related donors. N Engl J Med. 1977; 296:1030–1034 [DOI] [PubMed] [Google Scholar]
- 29.Simmonds MJ. Using genetic variation to predict and extend long-term kidney transplant function. Transplantation. 2015; 99:2038–2048 [DOI] [PubMed] [Google Scholar]
- 30.Hernandez-Fuentes MP, Franklin C, Rebollo-Mesa I, et al. ; United Kingdom and Ireland Renal Transplant Consortium (UKIRTC) and the Wellcome Trust Case Control Consortium (WTCCC)-3. Long- and short-term outcomes in renal allografts with deceased donors: a large recipient and donor genome-wide association study. Am J Transplant. 2018; 18:1370–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magee JC, Barr ML, Basadonna GP, et al. Repeat organ transplantation in the United States, 1996–2005. Am J Transplant. 2007; 75 Pt 21424–1433 [DOI] [PubMed] [Google Scholar]
- 32.Everly MJ, Rebellato LM, Haisch CE, et al. Incidence and impact of de novo donor-specific alloantibody in primary renal allografts. Transplantation. 2013; 95:410–417 [DOI] [PubMed] [Google Scholar]
- 33.United States Renal Data System.. 2016 USRDS Annual Data Report: epidemiology of kidney disease in the United States. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases 2019Available at https://www.usrds.org/annual-data-report. Accessed August 9, 2020.
- 34.Genovese G, Friedman DJ, Ross MD, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010; 329:841–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parsa A, Kao WH, Xie D, et al. ; AASK Study Investigators; CRIC Study Investigators; APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med. 2013; 369:2183–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reeves-Daniel AM, DePalma JA, Bleyer AJ, et al. The APOL1 gene and allograft survival after kidney transplantation. Am J Transplant. 2011; 11:1025–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freedman BI, Pastan SO, Israni AK, et al. APOL1 genotype and kidney transplantation outcomes from deceased African American donors. Transplantation. 2016; 100:194–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freedman BI, Julian BA, Pastan SO, et al. Apolipoprotein L1 gene variants in deceased organ donors are associated with renal allograft failure. Am J Transplant. 2015; 15:1615–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giulio G, David JF, Michael DR, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010; 329:841–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tzur S, Rosset S, Shemer R, et al. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet. 2010; 128:345–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lentine KL, Schnitzler MA, Xiao H, et al. Racial variation in medical outcomes among living kidney donors. N Engl J Med. 2010; 363:724–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doshi MD, Ortigosa-Goggins M, Garg AX, et al. APOL1 genotype and renal function of black living donors. J Am Soc Nephrol. 2018; 29:1309–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freedman BI, Moxey-Mims M. The APOL1 long-term kidney transplantation outcomes network-APOLLO. Clin J Am Soc Nephrol. 2018; 13:940–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bahram S, Mizuki N, Inoko H, et al. Nucleotide sequence of the human MHC class I MICA gene. Immunogenetics. 1996; 44:80–81 [DOI] [PubMed] [Google Scholar]
- 45.Mizutani K, Terasaki P, Bignon JD, et al. Association of kidney transplant failure and antibodies against MICA. Hum Immunol. 2006; 67:683–691 [DOI] [PubMed] [Google Scholar]
- 46.Zou Y, Stastny P, Süsal C, et al. Antibodies against MICA antigens and kidney-transplant rejection. N Engl J Med. 2007; 357:1293–1300 [DOI] [PubMed] [Google Scholar]
- 47.Tonnerre P, Gérard N, Chatelais M, et al. MICA variant promotes allosensitization after kidney transplantation. J Am Soc Nephrol. 2013; 24:954–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rohn H, Tomoya Michita R, Schwich E, et al. The donor major histocompatibility complex class I chain-related molecule A allele rs2596538 G predicts cytomegalovirus viremia in kidney transplant recipients. Front Immunol. 2018; 9:917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore J, McKnight AJ, Döhler B, et al. Donor ABCB1 variant associates with increased risk for kidney allograft failure. J Am Soc Nephrol. 2012; 23:1891–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woillard JB, Gatault P, Picard N, et al. A donor and recipient candidate gene association study of allograft loss in renal transplant recipients receiving a tacrolimus-based regimen. Am J Transplant. 2018; 18:2905–2913 [DOI] [PubMed] [Google Scholar]
- 51.Moore J, McKnight AJ, Simmonds MJ, et al. Association of caveolin-1 gene polymorphism with kidney transplant fibrosis and allograft failure. JAMA. 2010; 303:1282–1287 [DOI] [PubMed] [Google Scholar]
- 52.Eikmans M, de Canck I, van der Pol P, et al. The functional polymorphism Ala258Ser in the innate receptor gene ficolin-2 in the donor predicts improved renal transplant outcome. Transplantation. 2012; 94:478–485 [DOI] [PubMed] [Google Scholar]
- 53.Gautreaux MD, Freedman BI. Genotypic variation and outcomes in kidney transplantation: donor and recipient effects. Kidney Int. 2013; 84:431–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bay JT, Schejbel L, Madsen HO, et al. Low C4 gene copy numbers are associated with superior graft survival in patients transplanted with a deceased donor kidney. Kidney Int. 2013; 84:562–569 [DOI] [PubMed] [Google Scholar]
- 55.Ghisdal L, Baron C, Lebranchu Y, et al. Genome-wide association study of acute renal graft rejection. Am J Transplant. 2017; 17:201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steers NJ, Li Y, Drace Z, et al. Genomic mismatch at LIMS1 locus and kidney allograft rejection. N Engl J Med. 2019; 380:1918–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gatault P, Halimi JM, Forconi C, et al. CMV infection in the donor and increased kidney graft loss: impact of full HLA-I mismatch and posttransplantation CD8(+) cell reduction. Am J Transplant. 2013; 13:2119–2129 [DOI] [PubMed] [Google Scholar]
- 58.Forconi C, Gatault P, Miquelestorena-Standley E, et al. Polymorphism in programmed cell death 1 gene is strongly associated with lung and kidney allograft survival in recipients from CMV-positive donors. J Heart Lung Transplant. 2017; 36:315–324 [DOI] [PubMed] [Google Scholar]
- 59.Vu D, Shah T, Ansari J, et al. Interferon-gamma gene polymorphism +874 A/T is associated with an increased risk of cytomegalovirus infection among Hispanic renal transplant recipients. Transpl Infect Dis. 2014; 16:724–732 [DOI] [PubMed] [Google Scholar]
- 60.Dessing MC, Kers J, Damman J, et al. Donor and recipient genetic variants in NLRP3 associate with early acute rejection following kidney transplantation. Sci Rep. 2016; 6:36315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abdi R, Tran TB, Sahagun-Ruiz A, et al. Chemokine receptor polymorphism and risk of acute rejection in human renal transplantation. J Am Soc Nephrol. 2002; 13:754–758 [DOI] [PubMed] [Google Scholar]
- 62.Cha RH, Yang SH, Kim HS, et al. Genetic interactions between the donor and the recipient for susceptibility to acute rejection in kidney transplantation: polymorphisms of CCR5. Nephrol Dial Transplant. 2009; 24:2919–2925 [DOI] [PubMed] [Google Scholar]
- 63.Canossi A, Aureli A, Delreno F, et al. Influence of cytotoxic T-lymphocyte antigen-4 polymorphisms on acute rejection onset of cadaveric renal transplants. Transplant Proc. 2013; 45:2645–2649 [DOI] [PubMed] [Google Scholar]
- 64.Hwang YH, Ro H, Choi I, et al. Impact of polymorphisms of TLR4/CD14 and TLR3 on acute rejection in kidney transplantation. Transplantation. 2009; 88:699–705 [DOI] [PubMed] [Google Scholar]
- 65.Park SJ, Yoon YC, Kang SW, et al. Impact of IL2 and IL2RB genetic polymorphisms in kidney transplantation. Transplant Proc. 2011; 43:2383–2387 [DOI] [PubMed] [Google Scholar]
- 66.Marshall SE, McLaren AJ, McKinney EF, et al. Donor cytokine genotype influences the development of acute rejection after renal transplantation. Transplantation. 2001; 71:469–476 [DOI] [PubMed] [Google Scholar]
- 67.Alakulppi NS, Kyllönen LE, Jäntti VT, et al. Cytokine gene polymorphisms and risks of acute rejection and delayed graft function after kidney transplantation. Transplantation. 2004; 78:1422–1428 [DOI] [PubMed] [Google Scholar]
- 68.Sankaran D, Asderakis A, Ashraf S, et al. Cytokine gene polymorphisms predict acute graft rejection following renal transplantation. Kidney Int. 1999; 56:281–288 [DOI] [PubMed] [Google Scholar]
- 69.Grinyó J, Vanrenterghem Y, Nashan B, et al. Association of four DNA polymorphisms with acute rejection after kidney transplantation. Transpl Int. 2008; 21:879–891 [DOI] [PubMed] [Google Scholar]
- 70.Tinckam K, Rush D, Hutchinson I, et al. The relative importance of cytokine gene polymorphisms in the development of early and late acute rejection and six-month renal allograft pathology. Transplantation. 2005; 79:836–841 [DOI] [PubMed] [Google Scholar]
- 71.Pawlik A, Dabrowska-Zamojcin E, Dziedziejko V, et al. Association between IVS3 + 17T/C CD28 gene polymorphism and the acute kidney allograft rejection. Transpl Immunol. 2014; 30:84–87 [DOI] [PubMed] [Google Scholar]
- 72.Golshayan D, Wójtowicz A, Bibert S, et al. ; Swiss Transplant Cohort Study. Polymorphisms in the lectin pathway of complement activation influence the incidence of acute rejection and graft outcome after kidney transplantation. Kidney Int. 2016; 89:927–938 [DOI] [PubMed] [Google Scholar]
- 73.Michielsen LA, van Zuilen AD, Muskens IS, et al. Complement polymorphisms in kidney transplantation: critical in graft rejection? Am J Transplant. 2017; 17:2000–2007 [DOI] [PubMed] [Google Scholar]
- 74.Heidenreich S, Junker R, Wolters H, et al. Outcome of kidney transplantation in patients with inherited thrombophilia: data of a prospective study. J Am Soc Nephrol. 2003; 14:234–239 [DOI] [PubMed] [Google Scholar]
- 75.Lewis CM, Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med. 2020; 12:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vesell ES, Page JG. Genetic control of drug levels in man: antipyrine. Science. 1968; 161:72–73 [DOI] [PubMed] [Google Scholar]
- 77.Vesell ES, Page JG. Genetic control of drug levels in man: phenylbutazone. Science. 1968; 159:1479–1480 [DOI] [PubMed] [Google Scholar]
- 78.Vesell ES, Passananti GT, Greene FE, et al. Genetic control of drug levels and of the induction of drug-metabolizing enzymes in man: individual variability in the extent of allopurinol and nortriptyline inhibition of drug metabolism. Ann N Y Acad Sci. 1971; 179:752–773 [DOI] [PubMed] [Google Scholar]
- 79.Relling MV, Evans WE. Pharmacogenomics in the clinic. Nature. 2015; 526:343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roden DM, McLeod HL, Relling MV, et al. Pharmacogenomics. Lancet. 2019; 394:521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chambliss AB, Chan DW. Precision medicine: from pharmacogenomics to pharmacoproteomics. Clin Proteomics. 2016; 13:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Adams SM, Crisamore KR, Empey PE. Clinical pharmacogenomics: applications in nephrology. Clin J Am Soc Nephrol. 2018; 13:1561–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Garg KB, Ganguli I, Das R, et al. Spectrum of Lactobacillus species present in healthy vagina of Indian women. Indian J Med Res. 2009; 129:652–657 [PubMed] [Google Scholar]
- 84.Wiseman AC. Induction therapy in renal transplantation: why? What agent? What dose? We may never know. Clin J Am Soc Nephrol. 2015; 10:923–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kidney Disease: Improving. Global Outcomes (KDIGO) Transplant Work Group. KDIGO clinical practice guideline for the care of kidney transplant recipients. Am J Transplant. 2009; 9Suppl 3S1–155 [DOI] [PubMed] [Google Scholar]
- 86.Ekberg H, Tedesco-Silva H, Demirbas A, et al. ; ELITE-Symphony Study. Reduced exposure to calcineurin inhibitors in renal transplantation. N Engl J Med. 2007; 357:2562–2575 [DOI] [PubMed] [Google Scholar]
- 87.Mohty M, Gaugler B. Mechanisms of action of antithymocyte globulin: old dogs with new tricks! Leuk Lymphoma. 2008; 49:1664–1667 [DOI] [PubMed] [Google Scholar]
- 88.Lopez M, Clarkson MR, Albin M, et al. A novel mechanism of action for anti-thymocyte globulin: induction of CD4+CD25+Foxp3+ regulatory T cells. J Am Soc Nephrol. 2006; 17:2844–2853 [DOI] [PubMed] [Google Scholar]
- 89.Brennan FM, Green P, Amjadi P, et al. Interleukin-10 regulates TNF-alpha-converting enzyme (TACE/ADAM-17) involving a TIMP-3 dependent and independent mechanism. Eur J Immunol. 2008; 38:1106–1117 [DOI] [PubMed] [Google Scholar]
- 90.Park S, Kim SK, Kang SW, et al. Association between tumor necrosis factor-alpha (TNF-alpha) polymorphism (-308, G/A) and acute rejection of solid organ allograft: a meta-analysis. Int J Clin Exp Med. 2016; 9:17060–17068 [Google Scholar]
- 91.Sánchez-Fructuoso AI, Pérez-Flores I, Valero R, et al. The polymorphism -308G/A of tumor necrosis factor-α gene modulates the effect of immunosuppressive treatment in first kidney transplant subjects who suffer an acute rejection. J Immunol Res. 2016; 2016:2197595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cerny T, Borisch B, Introna M, et al. Mechanism of action of rituximab. Anticancer Drugs. 2002; 13Suppl 2S3–10 [DOI] [PubMed] [Google Scholar]
- 93.Tydén G, Kumlien G, Fehrman I. Successful ABO-incompatible kidney transplantations without splenectomy using antigen-specific immunoadsorption and rituximab. Transplantation. 2003; 76:730–731 [DOI] [PubMed] [Google Scholar]
- 94.Chauhan K, Mehta AA. Rituximab in kidney disease and transplant. Animal Model Exp Med. 2019; 2:76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sakai H, Tanaka Y, Tazawa H, et al. Effect of Fc-γ receptor polymorphism on rituximab-mediated B cell depletion in ABO-incompatible adult living donor liver transplantation. Transplant Direct. 2017; 3:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cartron G, Dacheux L, Salles G, et al. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood. 2002; 99:754–758 [DOI] [PubMed] [Google Scholar]
- 97.Allison JP. CD28-B7 interactions in T-cell activation. Curr Opin Immunol. 1994; 6:414–419 [DOI] [PubMed] [Google Scholar]
- 98.Walunas TL, Lenschow DJ, Bakker CY, et al. CTLA-4 can function as a negative regulator of T cell activation. Immunity. 1994; 1:405–413 [PubMed] [Google Scholar]
- 99.Larsen CP, Pearson TC, Adams AB, et al. Rational development of LEA29Y (belatacept), a high-affinity variant of CTLA4-Ig with potent immunosuppressive properties. Am J Transplant. 2005; 5:443–453 [DOI] [PubMed] [Google Scholar]
- 100.Vincenti F, Rostaing L, Grinyo J; et al. Belatacept and long-term outcomes in kidney transplantation N Eng J Med. 2016; 374:333–343 [DOI] [PubMed] [Google Scholar]
- 101.Cortes-Cerisuelo M, Laurie SJ, Mathews DV, et al. Increased pretransplant frequency of CD28+ CD4+ TEM predicts belatacept-resistant rejection in human renal transplant recipients. Am J Transplant. 2017; 17:2350–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Coto E, Tavira B. Pharmacogenetics of calcineurin inhibitors in renal transplantation. Transplantation. 2009; 883 SupplS62–S67 [DOI] [PubMed] [Google Scholar]
- 103.Phupradit A, Vadcharavivad S, Ingsathit A, et al. Impact of POR and CYP3A5 polymorphisms on trough concentration to dose ratio of tacrolimus in the early post-operative period following kidney transplantation. Ther Drug Monit. 2018; 40:549–557 [DOI] [PubMed] [Google Scholar]
- 104.Hu C, Yin WJ, Li DY, et al. Evaluating tacrolimus pharmacokinetic models in adult renal transplant recipients with different CYP3A5 genotypes. Eur J Clin Pharmacol. 2018; 74:1437–1447 [DOI] [PubMed] [Google Scholar]
- 105.Jacobson PA, Oetting WS, Brearley AM, et al. ; DeKAF Investigators. Novel polymorphisms associated with tacrolimus trough concentrations: results from a multicenter kidney transplant consortium. Transplantation. 2011; 91:300–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Haufroid V, Wallemacq P, VanKerckhove V, et al. CYP3A5 and ABCB1 polymorphisms and tacrolimus pharmacokinetics in renal transplant candidates: guidelines from an experimental study. Am J Transplant. 2006; 6:2706–2713 [DOI] [PubMed] [Google Scholar]
- 107.Tsuchiya N, Satoh S, Tada H, et al. Influence of CYP3A5 and MDR1 (ABCB1) polymorphisms on the pharmacokinetics of tacrolimus in renal transplant recipients. Transplantation. 2004; 78:1182–1187 [DOI] [PubMed] [Google Scholar]
- 108.Op den Buijsch RA, Christiaans MH, Stolk LM, et al. Tacrolimus pharmacokinetics and pharmacogenetics: influence of adenosine triphosphate-binding cassette B1 (ABCB1) and cytochrome (CYP) 3A polymorphisms. Fundam Clin Pharmacol. 2007; 21:427–435 [DOI] [PubMed] [Google Scholar]
- 109.Hesselink DA, Schaik RHN, Heiden IP, et al. Genetic polymorphisms of the CYP3A4, CYP3A5, and MDR-1 genes and pharmacokinetics of the calcineurin inhibitors cyclosporine and tacrolimus. Clin Pharmacol Ther. 2003; 74:245–254 [DOI] [PubMed] [Google Scholar]
- 110.Haufroid V, Mourad M, Van Kerckhove V, et al. The effect of CYP3A5 and MDR1 (ABCB1) polymorphisms on cyclosporine and tacrolimus dose requirements and trough blood levels in stable renal transplant patients. Pharmacogenetics. 2004; 14:147–154 [DOI] [PubMed] [Google Scholar]
- 111.Beermann KJ, Ellis MJ, Sudan DL, et al. Tacrolimus dose requirements in African-American and Caucasian kidney transplant recipients on mycophenolate and prednisone. Clin Transplant. 2014; 28:762–767 [DOI] [PubMed] [Google Scholar]
- 112.Mohamed ME, Schladt DP, Guan W, et al. ; DeKAF Genomics and GEN03 Investigators. Tacrolimus troughs and genetic determinants of metabolism in kidney transplant recipients: a comparison of four ancestry groups. Am J Transplant. 2019; 19:2795–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Terrazzino S, Quaglia M, Stratta P, et al. The effect of CYP3A5 6986A>G and ABCB1 3435C>T on tacrolimus dose-adjusted trough levels and acute rejection rates in renal transplant patients: a systematic review and meta-analysis. Pharmacogenet Genomics. 2012; 22:642–645 [DOI] [PubMed] [Google Scholar]
- 114.Birdwell KA, Decker B, Barbarino JM, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP3A5 genotype and tacrolimus dosing. Clin Pharmacol Ther. 2015; 98:19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Thervet E, Loriot MA, Barbier S, et al. Optimization of initial tacrolimus dose using pharmacogenetic testing. Clin Pharmacol Ther. 2010; 87:721–726 [DOI] [PubMed] [Google Scholar]
- 116.Shuker N, Bouamar R, van Schaik RH, et al. A randomized controlled trial comparing the efficacy of Cyp3a5 genotype-based with body-weight-based tacrolimus dosing after living donor kidney transplantation. Am J Transplant. 2016; 16:2085–2096 [DOI] [PubMed] [Google Scholar]
- 117.Elens L, Haufroid V. Genotype-based tacrolimus dosing guidelines: with or without CYP3A4*22? Pharmacogenomics. 2017; 18:1473–1480 [DOI] [PubMed] [Google Scholar]
- 118.Kuypers DRJ, Jonge H, Naesens M, et al. CYP3A5 and CYP3A4 but not MDR1 single-nucleotide polymorphisms determine long-term tacrolimus disposition and drug-related nephrotoxicity in renal recipients. Clin Pharmacol Ther. 2007; 82:711–725 [DOI] [PubMed] [Google Scholar]
- 119.Elens L, Hesselink DA, Bouamar R, et al. Impact of POR*28 on the pharmacokinetics of tacrolimus and cyclosporine A in renal transplant patients. Ther Drug Monit. 2014; 36:71–79 [DOI] [PubMed] [Google Scholar]
- 120.Kreutz R, Zürcher H, Kain S, et al. The effect of variable CYP3A5 expression on cyclosporine dosing, blood pressure and long-term graft survival in renal transplant patients. Pharmacogenetics. 2004; 14:665–671 [DOI] [PubMed] [Google Scholar]
- 121.Fanta S, Niemi M, Jönsson S, et al. Pharmacogenetics of cyclosporine in children suggests an age-dependent influence of ABCB1 polymorphisms. Pharmacogenet Genomics. 2008; 18:77–90 [DOI] [PubMed] [Google Scholar]
- 122.Cajka T, Smilowitz JT, Fiehn O. Validating quantitative untargeted lipidomics across nine liquid chromatography-high-resolution mass spectrometry platforms. Anal Chem. 2017; 89:12360–12368 [DOI] [PubMed] [Google Scholar]
- 123.Godley MD, Godley SH, Dennis ML, et al. The effect of assertive continuing care on continuing care linkage, adherence and abstinence following residential treatment for adolescents with substance use disorders. Addiction. 2007; 102:81–93 [DOI] [PubMed] [Google Scholar]
- 124.Black AJ, McLeod HL, Capell HA, et al. Thiopurine methyltransferase genotype predicts therapy-limiting severe toxicity from azathioprine. Ann Intern Med. 1998; 129:716–718 [DOI] [PubMed] [Google Scholar]
- 125.Wilck N, Matus MG, Kearney SM, et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature. 2017; 551:585–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schaeffeler E, Fischer C, Brockmeier D, et al. Comprehensive analysis of thiopurine S-methyltransferase phenotype-genotype correlation in a large population of German-Caucasians and identification of novel TPMT variants. Pharmacogenetics. 2004; 14:407–417 [DOI] [PubMed] [Google Scholar]
- 127.Yates CR, Krynetski EY, Loennechen T, et al. Molecular diagnosis of thiopurine S-methyltransferase deficiency: genetic basis for azathioprine and mercaptopurine intolerance. Ann Intern Med. 1997; 126:608–614 [DOI] [PubMed] [Google Scholar]
- 128.Relling MV, Schwab M, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium guideline for thiopurine dosing based on TPMT and NUDT15 genotypes: 2018 update. Clin Pharmacol Ther. 2019; 105:1095–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Moes DJ, Press RR, den Hartigh J, et al. Population pharmacokinetics and pharmacogenetics of everolimus in renal transplant patients. Clin Pharmacokinet. 2012; 51:467–480 [DOI] [PubMed] [Google Scholar]
- 130.Guinea J, Escribano P, Marcos-Zambrano LJ, et al. Therapeutic drug monitoring of voriconazole helps to decrease the percentage of patients with off-target trough serum levels. Med Mycol. 2016; 54:353–360 [DOI] [PubMed] [Google Scholar]
- 131.Moriyama B, Owusu Obeng A, Barbarino J, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP2C19 and voriconazole therapy. Clin Pharmacol Ther. 2018; 103:349. [DOI] [PubMed] [Google Scholar]
- 132.Cavallari LH, Lee CR, Beitelshees AL, et al. Multisite investigation of outcomes with implementation of CYP2C19 genotype-guided antiplatelet therapy after percutaneous coronary intervention. JACC Cardiovasc Interv. 2018; 11:181–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jneid H, Anderson JL, Wright RS, et al. 2012 ACCF/AHA focused update of the guideline for the management of patients with unstable angina/Non-ST-elevation myocardial infarction (updating the 2007 guideline and replacing the 2011 focused update): a report of the American College of Cardiology Foundation/American Heart Association Task Force on practice guidelines. Circulation. 2012; 126:875–910 [DOI] [PubMed] [Google Scholar]
- 134.Scott SA, Sangkuhl K, Stein CM, et al. ; Clinical Pharmacogenetics Implementation Consortium. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013; 94:317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Johnson JA, Caudle KE, Gong L, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for pharmacogenetics-guided warfarin dosing: 2017 update. Clin Pharmacol Ther. 2017; 102:397–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Crews KR, Gaedigk A, Dunnenberger HM, et al. ; Clinical Pharmacogenetics Implementation Consortium. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin Pharmacol Ther. 2014; 95:376–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Study of the Effectiveness of Additional Reductions in Cholesterol and Homocysteine (SEARCH) Collaborative Group. Intensive lowering of LDL cholesterol with 80 mg versus 20 mg simvastatin daily in 12 064 survivors of myocardial infarction: a double-blind randomised trial. Lancet. 2010; 376:1658–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Link E, Parish S, Armitage J; SEARCH Collaborative Group. SLCO1B1 variants and statin-induced myopathy—a genomewide study. N Eng J Med. 2008; 359:789–799 [DOI] [PubMed] [Google Scholar]
- 139.Shuen-lu H, Wen-Hung C, Lieh-Bang L, et al. HLA-B5801 allele as a genetic marker for severe cutaneous adverse reactions caused by allopurinol. Proc Natl Acad Sci U S A. 2005; 102:4134–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hershfield MS, Callaghan JT, Tassaneeyakul W, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for human leukocyte antigen-B genotype and allopurinol dosing. Clin Pharmacol Ther. 2013; 93:153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.U.S. Department of Health & Human Services. doi: 10.3109/15360288.2015.1037530. Available at https://www.hhs.gov/about/news/2019/07/10/hhs-launches-president-trump-advancing-american-kidney-health-initiative.html. Accessed August 9, 2020. [DOI] [PubMed]
- 142.James A, Mannon RB. The cost of transplant immunosuppressant therapy: is this sustainable? Curr Transplant Rep. 2015; 2:113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA. 1998; 279:1200–1205 [DOI] [PubMed] [Google Scholar]
- 144.Johnson JA, Bootman JL. Drug-related morbidity and mortality. A cost-of-illness model. Arch Intern Med. 1995; 155:1949–1956 [PubMed] [Google Scholar]
- 145.Taber DJ, Pilch NA, Bratton CF, et al. Medication errors and adverse drug events in kidney transplant recipients: incidence, risk factors, and clinical outcomes. Pharmacotherapy. 2012; 32:1053–1060 [DOI] [PubMed] [Google Scholar]
- 146.Fernando B, Veronica C, Ignacio GC, et al. A systematic approach to assess the burden of drug interactions in adult kidney transplant patients. Current Drug Safety. 2016; 11:156–163 [DOI] [PubMed] [Google Scholar]
- 147.Jameson JL, Longo DL. Precision medicine–personalized, problematic, and promising. N Engl J Med. 2015; 372:2229–2234 [DOI] [PubMed] [Google Scholar]
- 148.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010; 464:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Varnai R, Szabo I, Tarlos G, et al. Pharmacogenomic biomarker information differences between drug labels in the United States and Hungary: implementation from medical practitioner view. Pharmacogenomics J. 2020; 20:380–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fishman CE, Mohebnasab M, van Setten J, et al. Genome-wide study updates in the international genetics and translational research in transplantation network (iGeneTRAiN). Front Genet. 2019; 10:1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chatterjee N, Shi J, García-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat Rev Genet. 2016; 17:392–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018; 19:581–590 [DOI] [PubMed] [Google Scholar]
- 153.Müller-Deile J, Jobst-Schwan T, Schiffer M. Moving beyond GWAS and eQTL analysis to validated hits in chronic kidney disease. Cell Metab. 2019; 29:9–10 [DOI] [PubMed] [Google Scholar]
- 154.Carriazo S, Ortiz A, Perez-Gomez MV. Diagnostic utility of exome sequencing for kidney disease. N Engl J Med. 2019; 380:2078. [DOI] [PubMed] [Google Scholar]
- 155.Lo YM, Tein MS, Pang CC, et al. Presence of donor-specific DNA in plasma of kidney and liver-transplant recipients. Lancet. 1998; 351:1329–1330 [DOI] [PubMed] [Google Scholar]
- 156.Snyder TM, Khush KK, Valantine HA, et al. Universal noninvasive detection of solid organ transplant rejection. Proc Natl Acad Sci U S A. 2011; 108:6229–6234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Grskovic M, Hiller DJ, Eubank LA, et al. Validation of a clinical-grade assay to measure donor-derived cell-free DNA in solid organ transplant recipients. J Mol Diagn. 2016; 18:890–902 [DOI] [PubMed] [Google Scholar]
- 158.Bloom RD, Bromberg JS, Poggio ED, et al. ; Circulating Donor-Derived Cell-Free DNA in Blood for Diagnosing Active Rejection in Kidney Transplant Recipients (DART) Study Investigators. Cell-free DNA and active rejection in kidney allografts. J Am Soc Nephrol. 2017; 28:2221–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ramezani A, Massy ZA, Meijers B, et al. Role of the gut microbiome in uremia: a potential therapeutic target. Am J Kidney Dis. 2016; 67:483–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhu B, Wang X, Li L. Human gut microbiome: the second genome of human body. Protein Cell. 2010; 1:718–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jia W, Li H, Zhao L, et al. Gut microbiota: a potential new territory for drug targeting. Nat Rev Drug Discov. 2008; 7:123–129 [DOI] [PubMed] [Google Scholar]
- 162.Paul R, Raj DS. Metabolic synergy to uremic toxicity: a tale of symbiosis and dysbiosis in CKD. Nephrol Self-Assessment. Program. 2019; 18 [Google Scholar]
- 163.Sharma A, Buschmann MM, Gilbert JA. Pharmacomicrobiomics: the holy grail to variability in drug response? Clin Pharmacol Ther. 2019; 106:317–328 [DOI] [PubMed] [Google Scholar]
- 164.Alexander JL, Wilson ID, Teare J, et al. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol. 2017; 14:356–365 [DOI] [PubMed] [Google Scholar]
- 165.Hueso M, Navarro E, Moreso F, et al. Relationship between subclinical rejection and genotype, renal messenger RNA, and plasma protein transforming growth factor-beta1 levels. Transplantation. 2006; 81:1463–1466 [DOI] [PubMed] [Google Scholar]
- 166.Clinical Pharmacogenetics Implementation Consortium (CPIC). Available at https://cpicpgx.org/genes-drugs/. Accessed August 9, 2020.


