Abstract
Brochoalvelolar lavages (BALs) from patients suffering from hospitalized infections with SARS-CoV-2, other corona viruses (human coronavirus (HCoV)-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1), Influenza virus type A and B, Haemophilus influenzae and Pneumocystis jirovecii were compared cytopathologically.
The aim of the study was to evaluate if the cellular profile detectable in BAL may be specific for the respective pathogens and could lead to diagnosis of COVID-19 even in the absence of PCR results.
Differential cytology and flow cytometry datasets of 62 patients were observed and compared.
We observed a significant association between individual cell pattern changes and the causing pathogen, but no general cell distribution pattern.
The cytology pattern of the BAL fluid in COVID-19 is not specific enough to use it as a sole diagnostic criterion, although it may support clinical decision making
Keywords: bronchoalveolar lavage fluid, COVID-19, cytology profile
1. Introduction
In our hospital, bronchoalveolar lavage fluid (BALF) is frequently obtained to test for the presence of SARS-CoV-2 in patients who require invasive ventilation. Generally, the BALF is also used for cytopathological analysis and has become an important tool in the diagnostics of acute and chronic lung diseases, as distinct cell patterns are indicative for several specific diseases, and this is particularly true for different interstitial lung diseases.[1]
Whilst the cytokine response associated with the single-cell landscape of COVID-19 patients was recently investigated,[2] the cytopathological pattern has not been compared to other infections nor has been used as an additional diagnostic tool in COVID-19 patients despite a timely reminder by K. Alsabi.[3] For this reason we analyzed the cellular components of COVID-19 patients in whom bronchoalveolar lavage (BAL) sampling was performed and compared their cellular profiles to the best possible matching patient groups in which mono-infections with other pathogens as the coronaviruses NL63, HKU1, OC43, and 229E, or Influenza virus type A and B, or Haemophilus influenzae, or Pneumocystis jirovecii have been detected.
2. Methods
A total number of 62 BALFs from patients between 27 and 86 years of age were examined descriptively and retrospectively. BALFs were collected within 2 days after mechanical ventilation was required. For each BALF from a COVID-19 patients the best available matching cases related to age, underlying disease, sex, and confirmed single infections with other pathogens were identified from our database of BALF diagnostics in order to enable the best possible comparison.
Cytopathological diagnostics included the standard histological/cytological stainings Hematoxylin and eosin, Periodic acid–Schiff staining, and May–Grünwald–Giemsa stain. The flow cytometry was performed with fluorescent antibodies from Beckman Coulter (Cyto Stat tetra Chrome CD45-FITC, CD4-RD1, CD8-ECD, CD3-PC5; Cyto Stat tetra Chrome CD45-FITC, CD56-RD1, CD19-ECD, CD3-PC5, IO Test CD 16-PE, IO Test CD 45-FITC, IO Test HLA-DR-PE, IO Test CD3-PC5, 7-AAD Viability Dye; detailed protocols on request, Fig. 1). These antibody sets can be standardized and thus do not display an inter-charge variation. For the cytometry, the BALF volume and color were initially determined. The samples were inspected for blood contaminations, mucus, and ciliated cells before being categorized as BALF. The fluid was then filtered through medical gaze, and the flow through was pelleted by centrifugation at 2.000 g. The cell pellet was resuspended in 1 mL RPMI medium (Lonza, Belgium), a total of 500 μL were used for flow cytometry.
Figure 1.
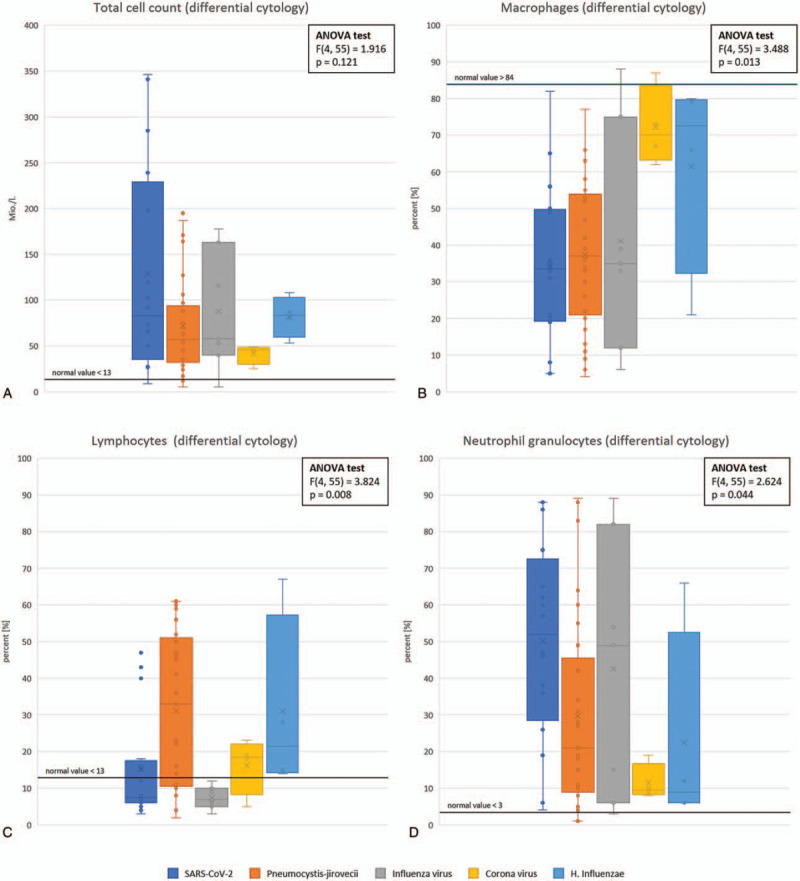
Comparison of cytological parameter measured by staining and flow cytometry of COVID-19 bronchoalveolar lavage fluids and matched groups of bronchoalveolar lavage fluids from patients with other single infections.
Multiplexing PCR was performed to detect respiratory pathogens using the QiaStat Respiratory Pathogens cartridge (Qiagen, Hilden, Germany). SARS-CoV-2 was detected using the Altona-RT-PCR assay as described previously,[4] and P. jirovecii was also analyzed by existing and published protocols.[5,6]
For all data, ANOVA testing was applied to test for statistical significance, while taking into account the bias of small study cohorts.
3. Results
For SARS-CoV-2 differential cytology by classical stainings was available from 16 patients at the age of 43 to 80 years, of which flow cytometry was available in 10 patients. This group was compared to patients infected with other human coronaviruses (HCoVs) (i.e., the matching group contained patients with confirmed HKU1, OC43, NL63, or 229E infection, irrespective of the virus subgrouping) with differential cytology available in 4 patients, of which 3 had also results for flow cytometry. The group with Influenza virus mono-infections consisted of 9 patients at the age of 52 to 77 years, of which in 4 cases a flow cytometry dataset was available. Regarding H. influenzae differential cytology by classical stainings and flow cytometry were available for 4 patients at the age of 34 to 75 years. Finally, the P. jirovecii infected group consisted of 29 patients (age ranging from 27–79 years), of which differential cytology by classical stainings and flow cytometry data were available for 25 patients. All measurements were compared to accepted normal ranges.[7]
Differential cytology by flow cytometry revealed that the total cell count of all examined pathogen positive BALFs was below the normal value of 13 million cells per liter BALF (Fig. 1) and that the macrophages in the BALF of all groups were below the normal value of 84% from the total cell population. According to the ANOVA test there is no significant difference in the total cell count between the pathogen positive BALF (F(4,55) = 1.916, P = .1208), whereas the ANOVA test shows that the results regarding the macrophages are significant for each pathogen (F(4,55) = 3.488, P = .013; Fig. 1).
For P. jirovecii and H. influenzae groups the lymphocytes percentages are above of the normal value of 13% of total cells for most of the BALFs, whereas the SARS-CoV-2 group and the other corona virus group display just a slight increase of lymphocytes, which is below the one reached in the BALFs of the P. jirovecii and the H. influenzae groups. In contrast, the lymphocytes count of BALFs with Influenza virus detection was mostly in the normal range. These differential cytology results have been confirmed as significant (F(4,55) = 3.824, P = .008), but not for the flow cytometry findings(F(4,41) = 1.712, P = .166).
For all measured samples the mean number of neutrophil granulocytes exceeds the normal value with the exception of “common” Corona viruses, which is significant (F(4, 55) = 2.624, P = .044).
In the groups infected with H. influenzae or corona viruses other than SARS-CoV-2, the number of CD3+ T-cells is almost exclusively in the normal range of 63% to 83% (flow cytometry, Fig. 2 ). In contrast, the percentages in the SARS-CoV-2 and P. jirovecii group are in the range of 45% up to values of 95%. The CD3+ T-cell amount in BALFs from patients with Influenza virus infection ranges from normal to decreased values of 37% (F(4,41) = 3.422, P = .017).
Figure 2.

Comparison of flow cytometry data obtained from bronchoalveolar lavage fluids of COVID-19 patients matched to other groups with single infections.
Figure 2 (Continued).

Comparison of flow cytometry data obtained from bronchoalveolar lavage fluids of COVID-19 patients matched to other groups with single infections.
The proportion of CD4+ T-cells in the BALFs of the groups with SARS-CoV-2 and H. influenzae are mostly in the normal range of 40% to 70% (flow cytometry, Fig. 2 ). In contrast, the amount of the CD4+ T-cells in the Influenza virus and the Corona viruses infected groups are below the normal range, whereas the percentages of CD4+ T-cells are distributed from 1% to 80% in P. jirovecii positive BALFs (F(4,41) = 3.959, P = .008). Also the numbers of CD8+ T-cells in the BALFs of the P. jirovecii group vary widely from 3% to 82%. For the other pathogens, the values are in the normal range of 20% to 40% except few deviations. Regarding the T4/T8 ratio in SARS-CoV-2 and H. influenzae groups there is a normal distribution from 1.1 to 3.5 with some slightly increased values up to 5. In contrast, the T4/T8 ratio of the Influenza virus and the Corona viruses group are lowered with values between 0.3 and 2.1 (flow cytometry, Fig. 2 ). Patients suffering from P. jirovecii infection show more variability with regard to their T4/T8 ratio, but this finding is not significant (F(4,41) = 0.479, P = .751).
With the exception of 1 Influenza virus positive BALF (flow cytometry, Fig. 2 ), all B cell values are in the normal range < 4%. The NK cell values of all pathogen groups are also in the normal range between 2% and 14% (flow cytometry, Fig. 2 ).
In addition, the overall cell viability was investigated (flow cytometry, Fig. 2 ). Thereby, the numbers of non-vital cells in the BALFs with SARS-CoV-2 are mostly above the threshold of 20%, reaching values of up to 60%. In case of H. influenza infections, some samples contain up to 40% of non-vital cells, whereas P. jirovecii, Influenza virus and the Corona viruses groups do not show an increased number of non-vital cells. The ANOVA test shows significant result for the pathogens (non-vital cells: F(4,41) = 6.580, P = .0003; vital cells: F(4,41) = 5.224, P = .002).
Finally, in none of the cases increased amounts of eosinophilic granulocytes, plasma cells, and mast cells were observed, respectively.
4. Conclusion
In conclusion, following the analysis of BALF cytological profiles an increased number of non-vital cells in the BALF may direct to the diagnosis of SARS-CoV-2 infection, which is likely more aggressive than the other pathogens examined, especially in direct comparison to other Corona virus types. In addition, BALs with SARS-CoV-2 also show increased CD8+ T-cell values. BALs of the “common” corona viruses (HCoV-229E, HCoV-OC43, NL63, HKU1) show normal cell numbers except a reduced number of CD4+ T-cells. T
Any data presented here are mainly descriptive and intended to trigger further research. In any case, BALF maybe a proper specimen for in depth diagnostics[8] and also cytopathological observations may be confirmative for COVID-19 and could lead to SARS-CoV-2 testing if not previously initiated. In the present situation of the ongoing pandemic and the early phase of a second wave it is however important also to collect and describe as many relevant information on clinical courses and check how these parts of the COVID-19 puzzle fit together. It is meanwhile a given (although not broadly accepted) fact that we have to live with the virus for a long time and thus we should learn whenever we can, not at least as we currently have not always had the chance to walk the preferred way of controlled clinical studies.
Author contributions
Conceptualization: Verena Schildgen, Oliver Schildgen.
Data curation: Verena Schildgen, Michael Brockmann, Oliver Schildgen.
Formal analysis: Vanessa Vedder, Verena Schildgen, Jessica Lüsebrink, Michael Brockmann, Oliver Schildgen.
Investigation: Vanessa Vedder, Birgitta Domscheit, Ramona Liza Tillmann, Jessica Lüsebrink, Wolfram Windisch, Christian Karagiannidis, Michael Brockmann.
Methodology: Verena Schildgen, Jessica Lüsebrink.
Supervision: Verena Schildgen, Wolfram Windisch, Christian Karagiannidis, Oliver Schildgen.
Writing – original draft: Oliver Schildgen.
Writing – review & editing: Oliver Schildgen.
Footnotes
Abbreviations: BAL = bronchoalveolar lavage, BALF = bronchoalveolar lavage fluid, HCoV = human coronavirus.
How to cite this article: Vedder V, Schildgen V, Lüsebrink J, Tillmann RL, Domscheit B, Windisch W, Karagiannidis C, Brockmann M, Schildgen O. Differential cytology profiles in bronchoalveolar lavage (BAL) in COVID-19 patients: a descriptive observation and comparison with other corona viruses, Influenza virus, Haemophilus influenzae, and Pneumocystis jirovecii. Medicine. 2021;100:1(e24256).
This study was performed in agreement with vote from the ethical committee of the Private University of Witten/Herdecke (votes 122/2016 and 58/2019).
The authors have no funding and conflicts of interest to disclose.
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
References
- [1].Meyer KC, Raghu G. Bronchoalveolar lavage for the evaluation of interstitial lung disease: is it clinically useful? Eur Respir J 2011;38:761–9. [DOI] [PubMed] [Google Scholar]
- [2].Liao M, Liu Y, Yuan J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med 2020;26:842–4. [DOI] [PubMed] [Google Scholar]
- [3].Drak Alsibai K. Detection of hemosiderin-laden macrophages in bronchoalveolar lavage fluid of COVID-19 patients: is Perls stain a potential indicator of oxidative alveolar damage? Acta Cytol 2020;64:617–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Malecki M, Lusebrink J, Teves S, et al. Pharynx gargle samples are suitable for SARS-CoV-2 diagnostic use and save personal protective equipment and swabs. Infect Control Hosp Epidemiol 2020;1–2. doi: 10.1017/ice.2020.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dunaiski CM, Janssen L, Erzinger H, et al. Inter-specimen imbalance of mitochondrial gene copy numbers predicts clustering of Pneumocystis jirovecii isolates in distinct subgroups. J Fungi 2018;4:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Prickartz A, Lusebrink J, Khalfaoui S, et al. Low titer Pneumocystis jirovecii infections: more than just colonization? J Fungi 2016;2: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Costabel U. Atlas der bronchoalveolaren: Lavage. 1994;Stuttgart, New York: Thieme Verlag, ISBN 3-13-110501-1. [Google Scholar]
- [8].Gualano G, Musso M, Mosti S, et al. Usefulness of bronchoalveolar lavage in the management of patients presenting with lung infiltrates and suspect COVID-19-associated pneumonia: A case report. Int J Infect Dis 2020;97:174–6. [DOI] [PMC free article] [PubMed] [Google Scholar]


