Bacillus subtilis has a circadian clock, extending the field of chronobiology to nonphotosynthetic bacteria.
Abstract
Circadian clocks create a 24-hour temporal structure, which allows organisms to occupy a niche formed by time rather than space. They are pervasive throughout nature, yet they remain unexpectedly unexplored and uncharacterized in nonphotosynthetic bacteria. Here, we identify in Bacillus subtilis circadian rhythms sharing the canonical properties of circadian clocks: free-running period, entrainment, and temperature compensation. We show that gene expression in B. subtilis can be synchronized in 24-hour light or temperature cycles and exhibit phase-specific characteristics of entrainment. Upon release to constant dark and temperature conditions, bacterial biofilm populations have temperature-compensated free-running oscillations with a period close to 24 hours. Our work opens the field of circadian clocks in the free-living, nonphotosynthetic prokaryotes, bringing considerable potential for impact upon biomedicine, ecology, and industrial processes.
INTRODUCTION
Circadian clocks contribute to the fitness of an organism (1, 2). The absence of characterized circadian rhythms in the first group of cellular organisms to populate Earth is thus notable and unexpected. To challenge this, we selected the nonphotosynthetic, gram-positive bacterium Bacillus subtilis as an experimental model. We chose this organism because of circumstantial observations of rhythms approaching 24 hours, although these were not made under the controlled conditions normally used to study circadian clocks. For example, in single cells of B. subtilis in a microfluidic device, there is pulsed activation of expression of a matrix gene approximately every 24 hours, indicating that, under homogeneous conditions, the stochastic triggering of biofilm formation may follow an internal daily cycle (3). There are cyclic (every ~20 hours) changes in the activity of the promoters of rapA and spo0F, which are important for cell fate decisions between sporulation and growth of B. subtilis (4, 5). These results suggest that there might be daily changes in the expression of genes involved in sensing environmental changes in B. subtilis. Furthermore, B. subtilis is light sensitive, harboring blue- and red-light photoreceptors (6) that could potentially entrain a circadian clock to the 24-hour day. Together, these reports led us to hypothesize that this Eubacterium might entrain to its environment using light and/or temperature signals like other circadian systems.
We developed luciferase reporter strains to conduct high-throughput and noninvasive measurement of gene promoter activity. This approach is used widely to study circadian rhythms in other kingdoms of life (7). The B. subtilis genome lacks homologs of the core clock proteins (KaiA, KaiB, and KaiC) present in cyanobacteria. However, many bacteria including B. subtilis harbor genes encoding Per-Arnt-Sim (PAS) domains, which are structural motifs present in all defined circadian clocks of eukaryotes (8). We reasoned that such genes might encode circadian clock–associated proteins. Of 16 predicted PAS domain–encoding genes in B. subtilis, we first selected the promoter of ytvA to create a bioluminescent reporter strain. ytvA encodes a blue light photoreceptor (9) with a PAS domain accompanied by a PAC domain, which is a common pairing in circadian and sensory/signaling proteins (10). Blue light photoreception is an integral part of circadian systems in all experimental models examined to date (11).
RESULTS
Synchronization to 24-hour light or temperature cycles and free-running rhythms
We identified free-running rhythms of ytvA promoter activity in B. subtilis. Cultures were exposed to 24-hour zeitgeber cycles. Zeitgebers are predictable, recurring environmental signals that biological rhythms use for entrainment or synchronization. In these culture conditions, zeitgeber-sensitive rhythmic processes could be either initiated or synchronized between bacterial cells and, therefore, detected. We first determined whether light can act as a zeitgeber for B. subtilis. Twenty-four-hour light/dark (LD) cycles (12-hour L/12-hour D) were applied to cultures. PytvA::lux gene expression increased during the dark phase and decreased during the light phase (Fig. 1A and fig. S1A). The pattern appears to combine two common features observed in the process of entrainment by a circadian clock. First, the abrupt reversal of expression at the zeitgeber transitions resembles masking (Fig. 1A and fig. S1A) (12). Second, the interaction of two oscillators (the circadian clock and the zeitgeber cycle) each with their own momentum and robustness is suggested by irregular expression of PytvA::lux luciferase from day to day, with a stable pattern appearing only after several days suggesting that the biological oscillator has reached a stable relationship with the 24-hour cycle (Fig. 1A and fig. S1A). A free-running rhythm in ytvA promoter activity occurred when the cultures were released to constant darkness (Fig. 1, B and C, and fig. S1A). This occurred only in strains cultured in glucose-containing media (Fig. 1, B and C, and fig. S2B). The period calculated over a 48-hour window following release to constant conditions was 28.66 ± 1.77 hours. Over 5 days, the period increased in length and damped thereafter. In glucose-free media, damping occurred rapidly on release to constant darkness, precluding the determination of period length in this condition (fig. S2B).
Fig. 1. Entrainment by light and a free-running rhythm in B. subtilis.
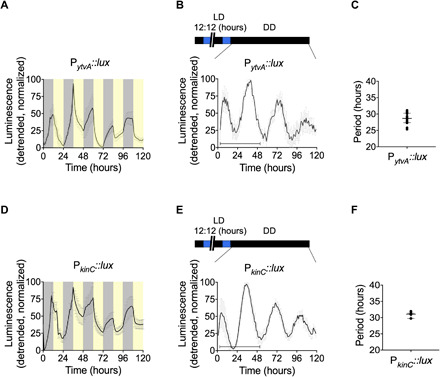
Bioluminescence of PytvA::lux (A) and PkinC::lux (D) under 5 days of entrainment with cycles of darkness and blue light (12-hour D/12-hour L) and after release to constant darkness conditions [DD; (B) and (E)] for 5 days. The temperature was kept constant at 25.5°C. The detrended data are presented as means ± SD. The shading in (A) and (D) shows the timing of the LD cycle (yellow, light phase; gray, dark phase) relative to the bioluminescence. The horizontal bar in (B) and (E) (lower left) shows the time window of 48 hours selected for the analysis of period length. The calculated period length is plotted in (C) and (F); individual data points [(C), N = 16; (F), N = 7] are shown along with the median and interquartile range. See also table S1.
We validated our findings with an additional luciferase reporter strain that uses a promoter from another PAS domain protein coding gene, kinC. KinC is a histidine kinase involved in the regulation of differentiation processes such as biofilm development and sporulation (13). Sporulation is a clock-regulated output in the fungal species Neurospora crassa (14, 15) and Aspergillus (16). Following entrainment in 24-hour LD cycles (Fig. 1D and fig. S1B), PkinC::lux expression has circadian rhythms (Fig. 1, E and F, and fig. S1B). As for the PytvA::lux strain, daily rhythms were detected in cultures containing glucose, but not in those deficient in glucose (fig. S2D). The temporal expression pattern (phase and amplitude) of this reporter was similar to that of PytvA::lux (fig. S3). This is not unexpected based on published observations: The preponderance of gene expression in cyanobacteria at the end of the subjective night relative to other times of day (17) and, furthermore, the congruent expression of these two genes in media containing different carbon sources (18) could suggest shared regulatory pathways. It is possible that, as more clock-regulated genes are identified in B. subtilis, more phases of expression will be identified.
Self-sustained, free-running rhythms in promoter activity of ytvA also occurred following entrainment to temperature cycles (12 hours at 25.5°C/12 hours at 28.5°C). Daily temperature fluctuations are reliable indicators of the time of day in nature, often serving as zeitgebers (recurring cues from the environment that are used by biological rhythms for their synchronization or entrainment) for a circadian clock. Temperature entrained cultures had daily oscillations in ytvA promoter activity (fig. S4). In contrast to cultures entrained using cycles of blue light and darkness, promoter activity was generally higher during the warm phase (correspondent to daytime), which is the converse of its behavior during entrainment to light (compare Fig. 1 and fig. S4). Further, persistent temperature-entrained free-running rhythms were detected only in media lacking glucose (with a period of 23.94 ± 1.64; Fig. 2, A and B). In contrast to the LD-entrained cultures, rhythms were suppressed by media containing glucose as a carbon source (Fig. 2D). Rhythms were also suppressed in the presence of glycerol (Fig. 2C) and in a variety of other media often used to culture B. subtilis (fig. S5). Together with the longer free-running period following light compared to temperature entrainment, this sensitivity to nutritional composition suggests that the circadian clock of B. subtilis responds to the myriad of environmental conditions under which bacteria subsist. Carbon source and availability also affect the free-running rhythm in plants and fungi (19, 20). Further, nutritional composition and environmental conditions such as light and temperature determine how populations grow and differentiate (13, 21). One hundred percent of the cultures exhibiting free-running, circadian rhythms formed a floating biofilm, whose presence was assessed qualitatively by visual observation of a pellicle forming at the air-liquid interface in the well.
Fig. 2. Free-running rhythms in B. subtilis following entrainment in temperature cycles.
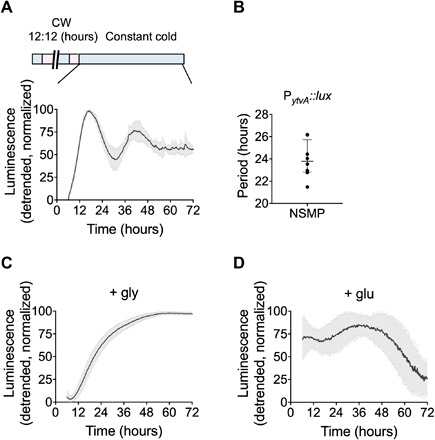
Bioluminescence of PytvA::lux in constant darkness at 25.5°C following 5 days of entrainment in temperature cycles (CW indicates the cold/warm cycle of 12 hours at 25.5°C/12 hours at 28.5°C) is shown. A free-running rhythm is observed in nutrient sporulation medium (NSMP) lacking glucose (A). The detrended data are plotted as means ± SD. The calculated period length of PytvA::lux expression shown in (A) is plotted in (B), where individual data points (N = 8) are shown along with the median and interquartile range. No free-running rhythm is observed in NSMP medium containing glycerol (N = 40) (C) or glucose (N = 15) (D) as a carbon source. See also table S1.
A third hallmark of circadian clocks is temperature compensation. The period of the free-running circadian rhythm typically has a Q10 close to 1, meaning that it remains relatively stable across a physiologically relevant 10°C temperature range. We entrained B. subtilis cultures to three different temperature cycles for 5 days, after which the cultures were released into constant conditions at the lower temperature of each cycle (Fig. 3, A to C). The free-running periods measured in constant temperatures spanning a 6°C range were not significantly different (P > 0.05; Fig. 3D). Q10 was calculated as 1.03, a moderate undercompensation. At temperatures outside this 6°C range, free-running rhythms were not detected. The amplitude of the oscillation in gene expression was significantly greater at the intermediate temperature relative to both lower and higher temperatures (22.5°C versus 25.5°C, P = 0.0013; 28.5°C versus 25.5°C, P = 0.0311; Fig. 3E). Together with the damping of rhythms outside of this temperature range, we conclude that there is a narrow, optimal temperature range in which the Bacillus cultures can maintain free-running rhythms under our laboratory conditions.
Fig. 3. Circadian rhythms in B. subtilis are temperature compensated.
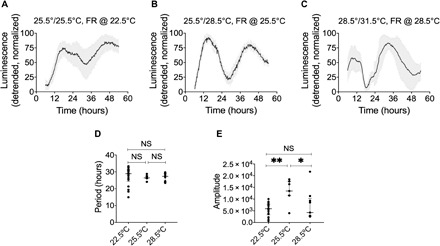
Bioluminescence of PytvA::lux under constant conditions [22.5°C, N = 25 (A); 25.5°C, N = 7 (B); 28.5°C, N = 9 (C)] following 5 days of entrainment with various temperature cycles [(A) 12 hours at 22.5°C/12 hours at 25.5°C; (B) 12 hours at 25.5°C/12 hours at 28.5°C; (C) 12 hours at 28.5°C/12 hours at 31.5°C]. The detrended data are presented as means ± SD. Period (D) and amplitude (E) of the bioluminescent signal of the data from (A) through (C) are shown as single data points, median, and interquartile range. Data were analyzed using ordinary one-way analysis of variance (ANOVA). NS, not significant (P > 0.05); **P = 0.0013, *P = 0.0311. See also table S1.
Phase relationship between the circadian rhythm and the temperature cycle
Our data identify free-running rhythms and their temperature compensation. Perhaps the most important hallmark of circadian rhythms, entrainment, is implied by the synchronization of cultures to 24-hour zeitgeber cycles as shown in Fig. 1 and figs. S1 to S4. We further observed that the free-running rhythm started 180° out of phase in cultures that were entrained in antiphase (fig. S6). We sought to test for explicit features of entrainment, namely, the establishment of a distinct phase of entrainment—meaning when the biological rhythm reliably occurs within each day—according to the zeitgeber cycle. As it is the 24-hour temporal structures of zeitgebers that drove evolution of circadian clocks, systematic entrainment should be a built-in feature of the system. Adaptive entrainment is essential to accommodate circadian entrainment in a constantly changing photoperiodic environment.
To understand entrainment in B. subtilis, we tested the effect of varying the zeitgeber strength—the magnitude of the entrainment signal—on the phase of entrainment, since the two are related in circadian systems in other kingdoms of life (22–25). Most humans, for instance, will entrain earlier in a zeitgeber cycle of higher amplitude. In our experiments with B. subtilis, we used temperature as a zeitgeber. Physical temperature perceived by living organisms is contextual; for example, a 3°C amplitude temperature cycle is perceived as a different amplitude depending on the absolute or ambient temperature. Therefore, our cultures assayed for temperature compensation experienced different zeitgeber strengths at each set of entrainment temperatures. The phase of the first measured oscillation of bioluminescence varied, according to whether the temperature cycled around a lower or higher mean temperature (Fig. 3, A to C). The phase of this first cycle during free run was later at lower temperatures and earlier at higher temperatures. This suggests that circadian phase of B. subtilis depends upon the zeitgeber strength and is not simply driven by the environmental transitions.
We next used “T cycles” (entraining cycles of different length) to distinguish noncircadian, environment-driven synchronization (masking) from circadian entrainment. “T” is defined as the duration of the entire entraining cycle, e.g., on Earth, T is about 24 hours. A general feature of circadian rhythms is a relationship between the period of the rhythm and that of the zeitgeber cycle. A conspecific with a longer free-running period generally entrains later in a 24-hour cycle than will an individual with a shorter period. It follows that a given individual will entrain to a later phase in a shorter cycle and an earlier one in a longer cycle (so-called T cycles) (23, 26–28). In contrast, a “masking” signal synchronizes to the same phase irrespective of the structure of the zeitgeber cycle (e.g., if it is longer or shorter). Entrainment often contains elements of adaptive phase angles that change according to the zeitgeber as well as evidence of masking. We found that B. subtilis entrained systematically later as T cycles became shorter (Fig. 4) despite evidence of masking (fig. S7). The phase angles of the oscillation shifted significantly from a peak of PytvA::lux before the cold to warm transition (T = 24 hours; Fig. 4) to occur at the midpoint of the warm phase of the temperature cycle as the T cycle became shorter (T = 20 hours; Fig. 4). This provides evidence for a robust circadian system in B. subtilis that interprets the zeitgeber cycle as an oscillator rather than simply responding to changes in the environment as a switch.
Fig. 4. Phase angle of entrainment in T cycles.
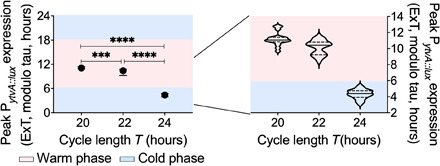
PytvA::lux was cultured under symmetrical temperature cycles [alternations between 25.5°C (50% of cycle) and 28.5°C (50% of cycle)] using different cycle lengths (T) [T20 (a 20-hour zeitgeber cycle): N = 17; T22 (a 22-hour zeitgeber cycle): N = 16; T24 (a 24-hour zeitgeber cycle): N = 26]. The phase [peak of luciferase expression; expressed as external time (ExT), where midnight is 0] shifted to a later phase with shorter temperature cycles. The blue shaded areas indicate the cold phase, and the pink shaded areas indicate the warm phase. The graph on the left shows median period with the interquartile range. The graph on the right is a violin plot of the same data. Phases observed in the different T cycles were compared using ordinary one-way ANOVA. All comparisons were significantly different from each other (****P < 0.0001, ***P = 0.0005).
DISCUSSION
Circadian clocks remain largely unknown in the nonphotosynthetic bacteria, despite bacteria representing about 15% of the living matter on Earth (29). We have identified circadian rhythms in a nonphotosynthetic bacterium. Our experiments using promoters from two PAS domain–containing genes revealed free-running circadian rhythms, systematic entrainment to zeitgeber cycles, and temperature compensation of the circadian period. We conclude that the free-living bacteria B. subtilis has a circadian clock. Why have circadian clocks remained elusive in the bacteria? Data from the purple photosynthetic bacterium Rhodospirillum rubrum suggest that rhythmic processes occur [e.g., enzymatic activity (30)], but these rhythms have not yet been shown to function as a circadian clock. The purple bacteria Rhodobacter sphaeroides, which has KaiB and KaiC orthologs, can also show rhythmic behavior depending on environmental conditions. However, neither of these systems have been tested systematically for the hallmarks of circadian regulation. Recently, Klebsiella aerogenes has been shown to have temperature-entrainable gene expression that can show circa 24-hour rhythms on release to constant conditions (31). In this isolate from the gut microbiome of a patient, rhythms generally occur only in the presence of melatonin, suggesting that these bacteria might not generate free-running rhythms independently of host cues. Twenty-four-hour light cycles modified mediators of pattern formation in Pseudomonas aeruginosa, but no circadian rhythms were observed (32).
Our experiments indicate that robust or detectable circadian rhythms depend upon environmental characteristics such as nutrient supply and ambient temperature. Furthermore, in our conditions, only cultures that form biofilms will show circadian rhythms. This is an interesting observation because biofilms represent a distinct developmental state relative to planktonic cultures. Many microbes will produce biofilms under certain conditions, and they have been associated with pathology. Effectively, biofilms arise when a microbial community shifts programs and produces a sticky matrix, thus creating a mechanism to form a differentiated population. This conditionality of the rhythms might be important for adaptive functions of the clock in bacteria and perhaps the life history of B. subtilis. Conditionality of circadian regulation is common. For example, constant light conditions suppress the circadian clock (33) in almost every case other than photosynthetic organisms, while in plants, many rhythms cease or change their period in constant darkness (34, 35). In Drosophila, a proportion of insects are arrhythmic, with this number depending on the strain (36). Furthermore, in the model fungus N. crassa, rhythms in nonmutant wild-type strains are highly dependent on media composition (37). Together, this indicates that the conditionality of circadian rhythms due to genotype, metabolism, and environmental conditions is common across life. The wealth of information concerning the environmental regulation of metabolism in the Eubacteria makes these organisms an excellent system in which to understand the functions of conditional rhythmicity.
While the rhythms that we report might be regulated by a transcription-translation feedback system, there remain other possibilities. For instance, we cannot exclude the possibility that the rhythms are linked to metabolic cycles because this has been shown in a variety of organisms [e.g., (20)]. It has been speculated that the ultradian rhythms in yeast that are tied to metabolic state (and also broadly integrated with transcriptional regulation) might be related to circadian clocks (38). It is also possible that the presence of rhythms only in population harboring biofilms could indicate some role for the biofilm matrix in maintaining the robustness of the rhythm. It will be informative to investigate whether temperature and light are inputs to one master pacemaker, or whether B. subtilis might have multiple oscillators, as described for a variety of unicellular and multicellular organisms (39, 40). It is also possible that B. subtilis might have either a master oscillator or one or more downstream oscillators that are coupled to and entrained by a main pacemaker (41).
We suggest that the incorporation of temporal structures into industrial, biomedical, and agricultural applications for bacteria might provide important translational opportunities. Our discovery of circadian rhythms in the Eubacteria should motivate future insights into the mechanisms and evolution of circadian rhythms across life.
MATERIALS AND METHODS
Strains and strain construction
All B. subtilis strains used in this study are derived from stock 168 (Jena), a domesticated but biofilm-proficient isolate (42). The promoter regions of ytvA and kinC genes were amplified using oligos ytvA_SacI_FW (5′- AGATCTGAGCTCCTTCATCATCACCTTCCTAAAG-3′)–ytvA_SalI_REV (5′- CTCGAGGTCGACTTAGGCCGTCAGCTTGCTATG-3′) and kinC_SacI_FW (5′- AGATCTGAGCTCTTTGTTTAATGACTGGAGAAATC-3′)–kinC_SalI_REV (5′- CTCGAGGTCGACTGCCGCTTGTGTTTCTCTAC-3′), respectively. Polymerase chain reaction products were digested with SacI and SalI enzymes (Thermo Fisher Scientific) and cloned into the corresponding sites of pAH321 harboring the promoterless luxABCDE genes (43). The vectors were verified by sequencing the cloned fragment and were subsequently transformed into B. subtilis 168 using natural competence (44). Integration of the reporter cassettes into amyE locus was verified by the lack of amylase activity on 1% (w/v) starch containing Lysogeny broth (LB) plates (45) and the presence of luminescence in the transformed strains.
Growth conditions
B. subtilis that had not been previously exposed to entrainment conditions was inoculated for overnight culture in LB medium [tryptone (10 g liter−1), yeast extract (5 g liter−1), and NaCl (5 g liter−1)]. Strains were subsequently grown as on a variety of media, as described in Results. Nutrient sporulation medium (NSMP) (46) [Nutrient broth (8 g liter−1) (Difco), 1 μM FeCl3, 700 μM CaCI2, 50 μM MnCl2, 1 mM MgCl2, and 100 mM potassium phosphate] was used without carbon source or supplemented either with 2.56% (v/v) glycerol or 0.1% (w/v) glucose (Figs. 1 to 4 and figs. S1 to S3 and S5). The following media were used in fig. S4: modified MSgg medium [5 mM potassium phosphate, 100 mM Mops, 2 mM MgCl2, 700 μM CaCl2, 50 μM MnCl2, 100 μM FeCl3, 1 μM ZnCl2, 2 μM thiamine HCl, 2.56% (v/v) glycerol, 0.5% (w/v) monosodium glutamate, and 50 μM l-tryptophan]; LB supplemented with 1 mM MnCl2; 2× SG medium [Nutrient broth (16 g liter−1) (Difco), KCl (2 g liter−1), MgSO4 7H2O (0.5 g liter−1), 1 mM Ca(NO3)2, 0.1 mM MnCl2·4H2O, and 1 μM FeSO4] either with or without 0.1% (w/v) glucose; 10% (v/v) NSMP supplemented with NaCl (5 g liter−1); chemically defined medium (CDM35) as described in Ponomarova et al., 2017 (47). The NSMP, MSgg, and CDM35 media were made fresh from stock solutions on the day of the experiment, and the stock solution for iron was freshly prepared every 2 weeks.
For all luminometry experiments, white 96-well plates (Nunclon Delta, Thermo Fisher Scientific) were used, with each well inoculated with approximately 5 × 105 cells. Plates were sealed with a transparent, evaporation-free cover (Optical Adhesive Covers, Applied Biosystems, Life Technologies). For experiments with temperature entrainment, cultures were exposed to temperature cycles for 5 days, after which the cultures were released to conditions corresponding to the cooler temperature. We measured bioluminescence (Berthold Centro LB960 XS3) for 1 s every 10 min. All experiments were carried out in temperature-controlled incubators (Panasonic MIR-154). For experiments with blue light entrainment, cultures were grown in NSMP medium without or with 0.1% (w/v) glucose and were exposed to a 12-hour darkness/12-hour blue light cycle for 5 days, followed by release into constant darkness. The temperature was kept constant at 25.5°C during these experiments. Bioluminescence was measured for 1 s each hour. The plate was ejected from the machine between readings, for exposure to blue light (light-emitting diodes with peak emission at 450 nm; Barthelme, Nürnberg, Germany) at a photon flux density of 35 μE m−2 s−1.
Cell growth under entrained and free-running conditions
The presence of a biofilm was assessed qualitatively, by eye, as a pellicle forming at the air-liquid interface in the well. To start to understand the state of our rhythmic, biofilm forming cultures, we determined cell number from day 4 (1 day before the end of the entraining cycle) and into day 7 (the second day in constant conditions). Cultures grown in 96-well plates (as for luminometry experiment; Fig. 2) were exposed to a temperature cycle (12 hours at 25.5°C/12 hours at 28.5°C) for 5 days, after which the cultures were released to constant temperature of 25.5°C. Cells were harvested every 12 hours, starting 30 hours before release to constant conditions until 42 hours after release. Samples were sonicated mildly (Diagenode Bioruptor, USA) at low power (130 W) for 12 s, for 2 cycles, with a 5-s pause between cycles according to a protocol modified from Dragoš et al. (48). Sonicated cells were examined by light microscopy (Leica, Germany) to confirm disruption of biofilm and cell viability. The sonicated cells were plated on LB agar and grown overnight at 37°C. The number of colony-forming units was counted on the following day. Figure S8 shows that the cell growth was stable before release to constant conditions, whereupon it increased approximately threefold. In the 42 hours of constant conditions, the cell number gradually decreased about 50%.
Data analysis
Graphing
Bioluminescence traces were baseline detrended using the open-access web tool BioDare2 (https://biodare2.ed.ac.uk) (49), and values were normalized between 0 and 100%. GraphPad Prism 8.1 (GraphPad Software, La Jolla, CA) was used to plot all graphs.
Calculation of free-running period using nonlinear modeling
For the analysis of the free-running period using continuous luminometry measurements, the period was calculated by analysis in the R programming language (50). To describe and parameterize the data, a nonlinear model was constructed, which performs a decay trend correction and fits a cosine-based function to the signal by using a nonlinear least squares (nls) method (51, 52). The model assumes an exponentially decaying baseline signal and an exponentially decaying oscillating (cosine) signal
with t = time (in hours) from the start of the experiment. Here, a0 is the amplitude (maximum) of the baseline signal, and k0 is the decay rate of the baseline signal (in hour−1). The shape of the baseline is consistent with, e.g., a first-order decay of the B. subtilis population during the experiment or a depletion of an essential nutrient. Furthermore, a1 is the amplitude of the oscillation, k1 is the decay rate (in hour−1) of the oscillation, T is the period of the oscillation (in hours), and θ the phase of the signal (in hours) at the start of the experiment. The advantage of such a physical-biological model is that all model parameters have a correspondent biological reference. Under most experimental conditions, the decay rates are positive and the period is about 24 hours. Some data were detrended (baseline detrending) using BioDare2 (53) before this calculation of free-running period.
The nls method in R requires sufficiently well-chosen starting values of all six model parameters a0, k0, a1, k1, T, and θ. For most experiments, the oscillatory part of the signal is much weaker than the baseline signal, hence a0 ≫ a1. Therefore, the amplitude a0 was set at the maximum value of the raw signal y(t), so that . Assuming that a1 ≪ a0, the raw signal is approximately an exponentially decaying signal, y(t) ≈ a0 · exp(− k0 · t). Hence, ; therefore, the decay rate k0 can be estimated as the negative of the slope of for t [via linear regression without an intercept using the R function lm (52)]. To have a crude estimate of the remaining parameters a1, k1, T, and θ, we calculated a baseline-corrected signal that shows damped oscillations around zero. The oscillation amplitude was estimated as the maximum of the absolute value of ycorr within the first 24 hours, therefore . The phase-shift θ was estimated as the time at which the maximum value of ycorr within the first 24 hours occurs, therefore . The oscillation period T was estimated initially as the difference in time between the maximum value of y within the first 24 hours and the maximum value of y within the second 24 hours such that . Last, since , the decay rate of the oscillating signal was roughly estimated as the negative of the slope of versus t (linear regression without intercept using the R function lm). Applying the nls function on the full nonlinear model using the set of starting values resulted in a set of least squares estimates of the parameters a0, k0, a1, k1, T, and θ, as well as SEs and P values for each parameter.
As a measure of “goodness of fit,” Akaike’s An Information Criterion (AIC) (54) was used, by subtracting the baseline AIC from the final model AIC. Bonferroni multiple testing correction was applied on calculated P values.
Phase angle determination of the T cycle series
Once stable entrainment was observed, three entraining cycles were used for analysis of entrained phase. Each individual signal was trend corrected by subtraction of a second-order polynomial fit of the raw data (fig. S9, A and B). As the masking peak is the most dominant feature in the signal, a fitted curve would mainly be a reflection of the zeitgeber. Therefore, to find the circadian component in the overall signal, we applied a fitting procedure, a combination of a sine [] and cosine [] using the least square error method (Python numpy.linalg.lstsq) excluding data from the warm phase that shows extreme masking (fig. S9C) from the fitting process (fig. S9D). The resulting sine curve is a thus a representation of the underlying circadian component (fig. S9E).
Supplementary Material
Acknowledgments
We thank B. Aronson for critical discussions on this work and O. P. Kuipers for comments and support during the early stages of the project. Funding: Work in the lab of M.M. is supported by the Volkswagen Foundation (Life? Funding Program: “The Fourth Dimension”) and the Friedrich Bauer Stiftung and the Verein zur Förderung von Wissenschaft und Forschung of the LMU Munich. A.N.D. is grateful to U.K. BBSRC for funding (Institute Strategic Programme GEN BB/P013511/1), and A.T.K. was supported by the Danish National Research Foundation (DNRF137) for the Center for Microbial Secondary Metabolites. Author contributions: M.M. and J.B. initiated the project. All authors (M.M., J.B., A.N.D., Z.E.-C., F.S., and A.T.K.) discussed experimental protocols. Z.E.-C., F.S., J.B., and A.T.K. performed experiments. Z.E.-C., F.S., and J.B. analyzed data. All authors (M.M., J.B., A.N.D., Z.E.-C., F.S., and A.T.K.) interpreted the data and wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Data related to this paper is available on https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/MCANDY Eelderink-Chen, Bosman et al., 2020, raw data. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIAL
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/2/eabe2086/DC1
REFERENCES AND NOTES
- 1.Woelfle M. A., Ouyang Y., Phanvijhitsiri K., Johnson C. H., The adaptive value of circadian clocks: An experimental assessment in cyanobacteria. Curr. Biol. 14, 1481–1486 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Dodd A. N., Salathia N., Hall A., Kévei E., Tóth R., Nagy F., Hibberd J. M., Millar A. J., Webb A. A. R., Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309, 630–633 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Norman T. M., Lord N. D., Paulsson J., Losick R., Memory and modularity in cell-fate decision making. Nature 503, 481–486 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischofs I. B., Hug J. A., Liu A. W., Wolf D. M., Arkin A. P., Complexity in bacterial cell-cell communication: Quorum signal integration and subpopulation signaling in the Bacillus subtilis phosphorelay. Proc. Natl. Acad. Sci. U.S.A. 106, 6459–6464 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuchina A., Espinar L., Garcia-Ojalvo J., Süel G. M., Reversible and noisy progression towards a commitment point enables adaptable and reliable cellular Decision-Making. PLOS Comput. Biol. 7, e1002273 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Der Steen J. B., Hellingwerf K. J., Activation of the general stress response of Bacillus subtilis by visible light. Photochem. Photobiol. 91, 1032–1045 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Millar A. J., Carré I. A., Strayer C. A., Chua N. H., Kay S. A., Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267, 1161–1163 (1995). [DOI] [PubMed] [Google Scholar]
- 8.Ponting C. P., Aravind L., PAS: A multifunctional domain family comes to light. Curr. Biol. 7, R674–R677 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Ávila-Pérez M., Hellingwerf K. J., Kort R., Blue light activates the σB-dependent stress response of Bacillus subtilis via YtvA. J. Bacteriol. 188, 6411–6414 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kay S. A., PAS, present, and future: Clues to the origins of circadian clocks. Science 276, 753–754 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Devlin P. F., Signs of the time: Environmental input to the circadian clock. J. Exp. Bot. 53, 1535–1550 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Mrosovsky N., Masking: History, definitions, and measurement. Chronobiol. Int. 16, 415–429 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Lopez D., Vlamakis H., Kolter R., Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol. Rev. 33, 152–163 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Merrow M. W., Dunlap J. C., Intergeneric complementation of a circadian rhythmicity defect: Phylogenetic conservation of structure and function of the clock gene frequency. EMBO J. 13, 2257–2266 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y., Merrow M., Loros J. J., Dunlap J. C., How temperature changes reset a circadian oscillator. Science 281, 825–829 (1998). [DOI] [PubMed] [Google Scholar]
- 16.Greene A. V., Keller N., Haas H., Bell-Pedersen D., A circadian oscillator in Aspergillus spp. regulates daily development and gene expression. Eukaryot. Cell 2, 231–237 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y., Tsinoremas N. F., Johnson C. H., Lebedeva N. V., Golden S. S., Ishiura M., Kondo T., Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 9, 1469–1478 (1995). [DOI] [PubMed] [Google Scholar]
- 18.Zhu B., Stülke J., SubtiWiki in 2018: From genes and proteins to functional network annotation of the model organism Bacillus subtilis. Nucleic Acids Res. 46, D743–D748 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loros J. J., Feldman J. F., Loros J. J., Loss of temperature compensation of circadian period length in the frq-9 mutant of Neurospora crassa. J. Biol. Rhythms 1, 187–198 (1986). [DOI] [PubMed] [Google Scholar]
- 20.Haydon M. J., Mielczarek O., Robertson F. C., Hubbard K. E., Webb A. A. R., Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 502, 689–692 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sonenshein A. L., Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5, 917–927 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Eelderink-Chen Z., Mazzotta G., Sturre M., Bosman J., Roenneberg T., Merrow M., A circadian clock in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 107, 2043–2047 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roenneberg T., Merrow M., The circadian clock and human health. Curr. Biol. 26, R432–R443 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Pittendrigh C. S., Daan S., A functional analysis of circadian pacemakers in nocturnal rodents – IV. Entrainment: Pacemaker as clock. J. Comp. Physiol. 106, 291–331 (1976). [Google Scholar]
- 25.Stothard E. R., McHill A. W., Depner C. M., Birks B. R., Moehlman T. M., Ritchie H. K., Guzzetti J. R., Chinoy E. D., Le Bourgeois M. K., Axelsson J., Wright K. P. Jr., Circadian entrainment to the natural light-dark cycle across seasons and the weekend. Curr. Biol. 27, 508–513 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruce V., Environmental entrainment of circadian rhythms. Cold Spring Harb. Symp. Quant. Biol. 25, 29–48 (1960). [DOI] [PubMed] [Google Scholar]
- 27.Hoffmann K., Zur beziehung zwischen phasenlage und spontanfrequenz bei der endogenen Tagesperiodik. Zeitschrift fur Naturforsch. B 18, 154–157 (1963). [Google Scholar]
- 28.Merrow M., Brunner M., Roenneberg T., Assignment of circadian function for the Neurospora clock gene frequency. Nature 399, 584–586 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Bar-On Y. M., Phillips R., Milo R., The biomass distribution on Earth. Proc. Natl. Acad. Sci. U.S.A. 115, 6506–6511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Praag E., Agosti R. D., Bachofen R., Rhythmic activity of uptake hydrogenase in the prokaryote Rhodospirillum rubrum. J. Biol. Rhythms 15, 218–224 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Paulose J. K., Cassone C. V., Graniczkowska K. B., Cassone V. M., Entrainment of the circadian clock of the enteric bacterium Klebsiella aerogenes by temperature cycles. iScience 19, 1202–1213 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kahl L. J., Price-Whelan A., Dietrich L. E. P., Light-mediated decreases in cyclic di-GMP levels inhibit structure formation in Pseudomonas aeruginosa biofilms. J. Bacteriol. 202, e00117-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abraham D., Dallmann R., Steinlechner S., Albrecht U., Eichele G., Oster H., Restoration of circadian rhythmicity in circadian clock – deficient mice in constant light. J. Biol. Rhythms 21, 169–176 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Millar A. J., Straume M., Chory J., Chua N. H., Kay S. A., The regulation of circadian period by phototransdutction pathways in Arabidopsis. Science 267, 1163–1166 (1995). [DOI] [PubMed] [Google Scholar]
- 35.Wang Z.-Y., Tobin E. M., Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Beauchamp M., Bertolini E., Deppisch P., Steubing J., Menegazzi P., Helfrich-Förster C., Closely related fruit fly species living at different latitudes diverge in their circadian clock anatomy and rhythmic behavior. J. Biol. Rhythms 33, 602–613 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Kim T.-S., Logsdon B. A., Park S., Mezey J. G., Lee K., Quantitative trait loci for the circadian clock in Neurospora crassa. Genetics 177, 2335–2347 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Causton H. C., Feeney K. A., Ziegler C. A., O’Neill J. S., Metabolic cycles in yeast share features conserved among circadian rhythms. Curr. Biol. 25, 1056–1062 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bell-Pedersen D., Cassone V. M., Earnest D. J., Golden S. S., Hardin P. E., Thomas T. L., Zoran M. J., Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat. Rev. Genet. 6, 544–556 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bell-Pedersen D., Crosthwaite S. K., Lakin-Thomas P. L., Merrow M., Økland M., The Neurospora circadian clock: Simple or complex? Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 1697–1709 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pittendrigh C., Bruce V., Kaus P., On the significance of transients in daily rhythms. Proc. Natl. Acad. Sci. U.S.A. 44, 965–973 (1958). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gallegos-Monterrosa R., Mhatre E., Kovács Á. T., Specific Bacillus subtilis 168 variants form biofilms on nutrient-rich medium. Microbiology 162, 1922–1932 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Schmalisch M., Maiques E., Nikolov L., Camp A. H., Chevreux B., Muffler A., Rodriguez S., Perkins J., Losick R., Small genes under sporulation control in the Bacillus subtilis genome. J. Bacteriol. 192, 5402–5412 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kunst F., Rapoport G., Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177, 2403–2407 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.C. R. Harwood, S. M. Cutting, Molecular Biological Methods for Bacillus (Wiley & Sons Ltd., 1990). [Google Scholar]
- 46.Fortnagel P., Freese E., Analysis of sporulation mutants. II. Mutants blocked in the citric acid cycle. J. Bacteriol. 95, 1431–1438 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ponomarova O., Gabrielli N., Sévin D. C., Mülleder M., Zirngibl K., Bulyha K., Andrejev S., Kafkia E., Typas A., Sauer U., Ralser M., Patil K. R., Yeast creates a niche for symbiotic lactic acid bacteria through nitrogen overflow. Cell Syst. 5, 345–357.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dragoš A., Kiesewalter H., Martin M., Hsu C.-Y., Hartmann R., Wechsler T., Eriksen C., Brix S., Drescher K., Stanley-Wall N., Kümmerli R., Kovács Á. T., Division of labor during biofilm matrix production. Curr. Biol. 28, 1903–1913.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore A., Zielinski T., Millar A. J., Online period estimation and determination of rhythmicity in circadian data, using the BioDare data infrastructure. Methods Mol. Biol. 1158, 13–44 (2014). [DOI] [PubMed] [Google Scholar]
- 50.R. Core Team, R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing Vienna, Austria (2018).
- 51.D. M. Bates, D. G. Watts, Nonlinear Regression Analysis and Its Applications (Wiley, 1988). [Google Scholar]
- 52.D. M. Bates, J. M. Chambers, Nonlinear models, in Statistical Models in S, J. M. Chambers, T. J. Hastie, Eds. (Wadsworth & Brooks/Cole, 1992). [Google Scholar]
- 53.Zielinski T., Moore A. M., Troup E., Halliday K. J., Millar A. J., Strengths and limitations of period estimation methods for circadian data. PLOS ONE 9, e96462 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Y. Sakamoto, M. Ishiguro, G. Kitagawa, Akaike Information Criterion Statistics (D. Reidel Publishing Company, 1986). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/2/eabe2086/DC1


