Abstract
Plants host numerous endophytic microbes which promote plant performance, in particular under stress. A new endophytic fungus was isolated from the leaves of a deciduous wood tree Leucas aspera. Morphological inspection and multilocus phylogeny identified the fungus as a new Trichoderma strain. If applied to Arabidopsis thaliana and Nicotiana attenuata, it mainly colonizes their roots and strongly promotes initial growth of the plants on soil. The fungus grows on high NaCl or mannitol concentrations, and shows predatory capability on the pathogenic fungus Alternaria brassicicola. Colonized Arabidopsis plants tolerate higher salt stress and show lower A. brassicicola spread in roots and shoots, while arbuscular mycorrhiza formation in N. attenuata is not affected by the Trichoderma strain. These beneficial features of the novel Trichoderma strain are important prerequisites for agricultural applications.
Keywords: Trichoderma, plant beneficial endophyte, growth promotion, pathogen protection, hormone induction
Introduction
Trichoderma species are versatile filamentous ascomycetes which are found in nearly all environments. They live in soil, grow on wood as saprophytes, or feed on fungi, plants, animals and insects as parasites (Carsolio et al., 1994; Gautheret et al., 1995; Furukawa et al., 1998; El-Katatny et al., 2000; Rocha-Ramirez et al., 2002; Druzhinina et al., 2011; Li et al., 2013; Mukherjee et al., 2014; Li Destri Nicosia et al., 2015; Berini et al., 2016; Druzhinina and Kubicek, 2016; Rosmana et al., 2016; Karlsson et al., 2017). Various Trichoderma species were shown to protect plants against pathogenic fungi, such as Rhizoctonia solani (Grosch et al., 2007; Zhang and Zhuang, 2020). Therefore, they are commonly used as bio-control agents in agriculture, with more than 250 commercial Trichoderma-based bio-fungicides registered world-wide (Woo et al., 2014). Apart from being used as bio-fungicide, Trichoderma species also stimulate plant growth (Lee et al., 2016) and nutrient uptake under nutrient deficient conditions (Li et al., 2015), often in combination with better stress tolerance of crop plants (Studholme et al., 2013). Other species, such as T. pleuroti (CBS124387) and T. pleuroticola (CBS124383) cause green mold disease in oyster mushroom (Pleurotus ostreatus) farms (Park et al., 2006).
Most of the investigated Trichoderma species colonize either the root surface, or live as endophytes inside root tissues (Samolski et al., 2012; Ruano-Rosa et al., 2016). However, some species were also isolated from the aerial parts of the plants (Bailey and Melnick, 2013). In response, plants often activate defense mechanisms including the biosynthesis of the defense-related phytohormones salicylic acid (SA), jasmonic acid (JA), ethylene (ET) or abscisic acid (ABA) (Contreras-Cornejo et al., 2009; Hermosa et al., 2012; Sivakumaran et al., 2016; Checker et al., 2018). The phytohormones regulate two types of induced resistance in plants, namely, SA-dependent systemic acquired resistance (SAR) and JA/ET-dependent induced systemic resistance (ISR). The signaling events induced by Trichoderma species often result in elevated SA and JA levels in different parts of the plant (Martínez-Medina et al., 2013; Leonetti et al., 2017).
In this study, we wanted to find out if the novel endophytic Trichoderma strain isolated from the leaves of Leucas aspera also interacts with other plant species (Arabidopsis, Nicotiana attenuata) and has beneficial effects in terms of plant growth and alleviation of abiotic and biotic stress. We could show that, although the strain is phylogenetically related to mushroom-infecting T. pleuroti and T. pleuroticola, it efficiently colonizes the roots of the two plant species, strongly promotes their growth on soil during early development and protects them against systemic A. brassicicola spread, while mycorrhiza formation in N. attenuata appears not to be affected. We also evaluated if phytohormones might be involved in the plant-fungus interaction.
Materials and Methods
Growth Medium and Conditions for Seedlings
Seeds of wild-type A. thaliana (ecotype Columbia-0) were surface-sterilized for 8 min in sterilizing solution containing lauryl sarcosine (1%) and Clorix cleaner (23%). Surface-sterilized seeds were washed with sterilized water eight times and placed on Petri dishes with MS medium supplemented with 0.3% gelrite (Murashige and Skoog, 1962). After cold treatment at 4°C for 48–72 h, plates were incubated at 22°C under long day conditions (16 h light/8 h dark; 80 μmol m–2 s–1).
Nicotiana attenuata Torr. ex S. Watson seeds of the 31st generation of an inbred accession originally collected from southwestern Utah were used for all experiments mentioned for this species. Seeds were germinated after surface sterilization and treatment with liquid smoke (1:50 dilution; House of Herbs, Passaic, NY, United States) and 1 mM of gibberellic acid (GA3; Duchefa-Biochemie, The Netherlands) on agar plates containing Gamborg’s B5 medium as previously described in Krügel et al. (2002). Seeds were kept in a growth chamber under a day/night cycle of 16 h (26–28°C)/8 h (24–26°C).
Growth of Fungi and Spore Collection
Based on our previous screen for plant growth-promoting fungi in a field station in India, the new Trichoderma strain was selected for detailed characterization. It was isolated from the leaves of Leucas aspera (Wild.) Link (family: Lamiaceae), a widely distributed medicinal plant reported for its antifungal, antioxidant, antimicrobial and cytotoxic activities (Prajapati et al., 2010; Rajani et al., 2020). The Trichoderma strain was grown on Petri dishes with Kaefer medium (KM) or Potato-Dextrose-Agar (PDA) medium, pH 6.5, at 23°C in the dark (Bains and Tewari, 1987; Hill and Käfer, 2001). Alternaria brassicicola was grown on Potato-Dextrose-Agar (PDA) medium, pH 6.5, 23°C in the dark (Bains and Tewari, 1987). We did not observe any difference of the fungal performance on the two media. Two additional pathogens, Fusarium brachygibbosum and Alternaria spp. isolate Utah 10, native to the natural habitat of N. attenuata isolated in a previous study (Luu et al., 2015), were grown on PDA medium at 26°C in the dark.
For spore collection, sterilized 0.01% Tween 20 solution was poured onto plates with fungi which were grown for less than 2 weeks. Spores were scratched from the agar surface and dispersed in 0.01% Tween 20. The resulting spore suspension was filtered through two layers of nylon membrane (75 μm pore size, Sefar AG, Switzerland), pelleted and washed with sterile distilled water. The A. brassicicola spore concentration was determined in a hemocytometer, while the Trichoderma spore concentration was determined by O.D.600 nm measurements using a spectrometer (BioSpectrometer® basic, Eppendorf, Germany).
For co-cultivation experiments with N. attenuata, the Trichoderma strain was cultivated on PDA plates. Spores of 7–14 day-old cultures were dislodged from the surface with sterile distilled water containing 0.01% Triton X-100. The resulting solution was diluted with distilled water to an O.D.600 nm of 0.250–0.350.
Co-cultivation of A. thaliana and N. attenuata With Trichoderma
Co-cultivation of A. thaliana and fungi was performed according to Johnson et al. (2011) with modifications. A plug (5 mm diameter) from a KM plate containing the fungus or a control plug without the fungus was put on a fresh plate with solid plant nutrient medium (PNM), which contained a layer of a nylon membrane (pore size 75 μm) on the agar surface. The plates were incubated at 23°C for 7 days. Unless specified, four 10 day-old A. thaliana seedlings of equal size were transferred to the plates. They were incubated at 22°C under long day conditions (16 h light/8 h dark; 80 μmol m–2 s–1).
For co-cultivation of A. thaliana with Trichoderma on soil, 1 kg of soil was suspended in 5 L of distilled water overnight. The liquid was removed by filtration and the soil was autoclaved twice. 200 g of the soil was transferred to magenta boxes for co-cultivation assays. The soil was inoculated with a 5 mm plug of KM medium with or without Trichoderma 3 cm below the soil surface in the center of the box. 10 day-old Arabidopsis seedlings were transferred to the soil, and the boxes were kept at 22°C under long day conditions (16 h light/8 h dark; 80 μmol m–2 s–1) for 4 weeks.
Co-cultivation of N. attenuata with Trichoderma was performed in Petri dishes and on soil. For experiments in Petri dishes, sterilized seeds treated with liquid smoke (1:50 dilution; House of Herbs, Passaic, NY, United States) and GA (1 mM GA3; Duchefa-Biochemie, The Netherlands) were incubated for 1 h with a highly diluted spore and hyphae solution before transfer to GB5 medium (see Santhanam et al., 2019 for experimental details). In a second set-up, liquid smoke-and GA-treated seeds were germinated on GB5 medium for 7 days before they were transferred in a circle with 10 seedlings to a new plate. Immediately after transfer roots either received 10 μL sterile distilled water or the same amount of spore solution (O.D.600 nm = 0.2653). One day later an Agar plug of Alternaria spp. Utah 10 was placed in the middle of the plate. Inoculated seedlings were kept at 26°C and 14 h light and 10 h dark cycle for 16 days.
For pot experiments on soil, Trichoderma treated seedlings and controls were transferred to pots and cultivated in a Snijders Chamber with a 16 h light/8 h dark cycle at 65% relative humidity.
To study the effect of Trichoderma strain on arbuscular mycorrhizal fungi (AMF), N. attenuata seedlings were transferred to Teku pots with sand 10 days after germination, and transferred to 10% of the commercial inoculum (BiomycVital, which contains AMF spores and tiny pieces of roots/hyphae in expanded clay)1 after another 12 days. Upon transfer, half of the plants received a Trichoderma spore solution, while control plants received the same amount of distilled water. Plants were watered with hydroponics solution containing 0.05 mM P. Roots were collected for further analysis 8 weeks after transfer.
Nucleic Acid Isolation, Primers, and PCR and Sequencing
Arabidopsis root and fungal tissue were ground in liquid nitrogen, and DNA extraction was performed according to Doyle (1990). RNA from AMF-colonized N. attenuata roots was extracted with the LiCl method according to Kistner and Matamoros (2005). RNA samples were treated with DNAse removal kit (Ambion, Thermo Fisher Scientific, Germany) according to the manufacturer’s instructions and reverse transcribed with Superscript II (Invitrogen, Thermo Fisher Scientific, Germany) and Oligo-dT.
The primer pairs for amplifying the TEF1 (translation elongation factor 1-alpha) and RPB2 (RNA polymerase II subunit 2) genes from Trichoderma are: TEF1-F: 5′-CATCGAGAAGTTCGAGAAGG-3′; TEF1-R: 5′-AACTTGCAG GCAATGTGG-3′; RPB2-F: 5′-TGGGGWGAYCARAARAAGG-3′; RPB2-R: 5′-CATRATGACSGAATCTTCCTGGT-3′. Each 20 μL PCR reaction contains 2 μL of 10× DreamTaq Buffer (Thermo Fisher Scientific, Germany), 0.2 mM dNTP, 1.0 μM forward/reverse primer, 100 ng genomic DNA template and 1U of DreamTaq DNA Polymerase (Thermo Fisher Scientific, Germany). The reaction was performed in a thermal cycler (Applied Biosystems SimpliAmp Thermal Cycler, Thermo Fischer Scientific, Germany). The initial denaturation step was set at 95°C for 3 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 55°C (TEF1) or 62°C (RPB2) for 30 s, and extension at 72°C for 30 s. The final extension step was set at 72°C for 10 min. The PCR products from at least two independent PCR runs were purified by NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel, Germany). Purified PCR products were sent to Eurofins Genomics for Sanger sequencing. Consensus sequence of TEF1 and RPB2 was deposited to Genbank with accession numbers MT591352 and MT602550, respectively.
To quantify the colonization of Trichoderma under various salt concentrations, Trichoderma TEF1 and A. thaliana RPS (AT1G34030) were detected by qPCR with the following primers: TEF-qF: 5′-TCAAGTCCGTTGAGATGCAC-3′; TEF-qR: 5′-CGTTCTTGACGTTGAAACCA-3′; RPS-qF: 5′- GT CTCCAATGCCCTTGACAT-3′; RPS-qR: 5′- TCTTTCCTCTGCGACCAGTT-3′.
For qPCR analysis of AMF colonization of N. attenuata roots, qPCR reactions were performed on Mx3005P qPCR system (Stratagene, Santa Clara, CA, United States) with Takyon Sybr Green No ROX kit (Eurogentec, Belgium). Primers for NaRAM1, NaPT4, and Rhizophagus irregularis tubulin are from Wang et al. (2018a). Primers for NaEF1 are from Wang et al. (2018b). Primers for RPB2 of Trichoderma are: RPB2-qF: 5′-AGACGTCCATGATCTGCATGAC-3′; RPB2-qR: 5′-TGTCTTGGTCTTGAGTCGCTTG-3′
The genes for A. thaliana Ubiquitin 5, N. attenuata Elongation Factor 1 alpha (NaEF1, Wang et al., 2018b) and A. brassicicola Cutinase 1 were used to monitor A. brassicicola infection in root tissue. The primer pairs for qPCR analysis are: AtUBQ5-qF: 5′-GACGCTTCATCTCGTCC-3′; AtUBQ5-qR: 5′-GTAAACGTAGGTGAGTCCA-3′; AbCUT1-qF: 5′-GACCGAGGAAGCTCAGATGC-3′; AbCUT1-qR: 5′-GCCTGGGATCTTGGAATGC-3′.
Multilocus Phylogenetic Analysis
The nucleotide sequences of TEF1 and RPB2 from 55 Trichoderma species and an outgroup species, Nectria eustromatica, were retrieved from the NCBI Nucleotide public database. The TEF1 and RPB2 sequences of the new Trichoderma strain were obtained from the PCR products. TEF1 and RPB2 genes from the same species were concatenated for combined analysis. In total, 56 concatenated sequences were subjected to alignment using MAFFT v7 online at https://mafft.cbrc.jp/alignment/server (Katoh et al., 2019), with G-INS-i parameters and a scoring matrix of “1PAM/κ = 2” for nucleotide sequences. The resulting alignment was inspected and selected for conserved blocks using Gblocks version 0.91b (Castresana, 2000).
Maximum likelihood analysis was conducted using RaxML-NG v.0.9.0 through web service at https://raxml-ng.vital-it.ch (Kozlov et al., 2019). Using the GTR+FO+G4m model, 2000 distinct ML tree were searched and bootstrapped with 100 replicates. For maximum parsimony analysis, PAUP 4.0a166 was utilized (Swofford, 2002). Heuristic search of 100 replicates was performed with random addition of sequence, and tree bisection-reconnection (TBR) as the branch-swapping algorithm (steepest decent and MulTrees option not in effect). All characters were weighted equally, and gaps were treated as missing character. Bootstrap of 1,000 replicates was undertaken with Maxtrees set as 5,000.
The Bayesian analysis was conducted using MrBayes v3.2.7a (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003). The evolutionary model was set to the general time-reversible model (GTR; Tavare, 1986), and the nucleotide variation rate set to inverse gamma distribution (Yang, 1993). Two simultaneous and independent Markov chain Monte Carlo (mcmc) analyses was run to generate 1 million generations each, while they were sampled for every 10 generations to determine the posterior probability (Geyer, 1991). From the resulting 100,000 sampled trees, the first 25% of them were discarded, and the remaining 75,000 trees were summarized to produce the consensus tree.
The Maximum likelihood boostrap proportions (MLBP), Maximum parsimony bootstrap proportions (MPBP), as well as the Bayesian inference posterior probability (BIPP) from each analysis were combined to the phylogenetic tree from the RAxML analysis using TreeGraph2.15.0–887 beta (Stöver and Müller, 2010). The final tree was created with FigTree v1.4.4 (Rambaut, 2018). The accession numbers of the individual genes are provided in Supplementary Table 1.
Histological Staining and Microscopy
Roots of A. thaliana co-cultivated with the Trichoderma strain for 2 or 7 days were collected and immersed in Wheat Germ Agglutinin, Alexa FluorTM 488 Conjugate (Thermo Fisher Scientific, Germany) for 10 min in dark. Immersed samples were taken out from the staining solution and placed on a glass slide. Water was applied to the slide to wash away excess staining solution and the slide was covered with a cover slip for microscopic inspection with Axio Imager.M2 (Zeiss Microscopy GmbH, Germany). The bright field and fluorescent images were recorded with a monochromatic camera Axiocam 503 mono (Zeiss Microscopy GmbH, Germany). Digital images were processed with the ZEN software (Zeiss Microscopy GmbH, Germany).
For confocal imaging of root colonization, A. thaliana roots co-cultivated with Trichoderma for 2 days were stained with Wheat Germ Agglutinin, Alexa FluorTM488 Conjugate and RH414 [N-(3-Triethylammoniumpropyl)-4-(4-(4-(Diethylamino)phenyl)Butadienyl)Pyridinium Dibromide; Thermo Fischer Scientific, Germany] with the method described above. Samples were imaged using an LSM 880 microscope (Zeiss Microscopy GmbH, Germany) with the 488 nm laser line of an argon multiline laser (11.5 mW). Images were taken with a 40× objective (Plan-Apochromat 40×/0.8). Lambda stacks were created using the 32 channel GaAsP detector followed by Linear Unmixing with the ZEN software. Z-stacks were taken from specific areas of the sample and Maximum Intensity Projections were produced with the ZEN software.
Phytohormone Analyses by LC-MS/MS
Sixteen seedlings from control and co-cultured plates were harvested and separated into root and shoot samples. Mycelium of the Trichoderma strain grown on KM plates was harvested for phytohormone analysis.
Fifty to one hundred thirty milligrams of fresh tissue were extracted and homogenized in 1.5 mL methanol containing 60 ng D4-SA (Santa Cruz Biotechnology, United States), 60 ng D6-JA (HPC Standards GmbH, Germany), 60 ng D6-ABA (Santa Cruz Biotechnology, United States), 12 ng D6-JA-Ile (HPC Standards GmbH), and D5-indole-acetic acid (D5-IAA, OlChemIm s.r.o., Olomouc, Czech Republic) as internal standards. Samples were agitated on a horizontal shaker at room temperature for 10 min. The homogenate was mixed for 30 min and centrifuged at 13,000 rpm for 20 min at 4°C and the supernatant was collected. The homogenate was re-extracted with 500 μL methanol, mixed and centrifuged and the supernatants were pooled. The combined extracts were evaporated under reduced pressure at 30°C and dissolved in 500 μL methanol.
Phytohormone analysis was performed by LC-MS/MS as in Heyer et al. (2018) on an Agilent 1260 series HPLC system (Agilent Technologies) with the modification that a tandem mass spectrometer QTRAP 6500 (SCIEX, Darmstadt, Germany) was used. Since we observed that both the D6-labeled JA and D6-labeled JA-Ile standards (HPC Standards GmbH, Cunnersdorf, Germany) contained 40% of the corresponding D5-labeled compounds, the sum of the peak areas of the D5- and D6- compounds was used for quantification. Details of the instrument parameters and response factors for quantification can be found in Supplementary Table 2.
Indole-acetic acid was quantified using the same LC-MS/MS system with the same chromatographic conditions but with positive mode ionization with an ion spray voltage at 5,500 eV. Multiple reaction monitoring (MRM) was used to monitor analyte parent ion → product ion fragmentations as follows: m/z 176 →130 [collision energy (CE) 19 V; declustering potential (DP) 31 V] for indole-acetic acid (IAA); m/z 181 →133 + m/z 181 →134 + m/z 181 →135 (CE 19 V; DP 31 V) for D5-indole-acetic acid.
Quantification of Mycelial Growth, AMF Colonization, and 11-Carboxyblumenol Levels
Plates with mycelia were scanned with an Epson scanner (Perfection V600 Photo, Epson, Germany), and the files imported into ImageJ (Schindelin et al., 2012). Mycelial coverage on each plate was delineated using a free-hand selection tool and measured with the built-in “Measure” function.
AMF colonization was determined by the “magnified intersections method” described in detail by McGonigle et al. (1990). In brief, roots were cut in about 1 cm pieces and we counted the fungal structures of 150 intersections per sample after staining with Trypan Blue.
For determination of AMF colonization marker 11-carboxyblumenol, three leaf disks per AMF inoculated of the first and second stem leaf were harvested 6 and 8 weeks after AMF inoculation. 11-Carboxyblumenol levels were determined as markers of arbuscule colonization and quantified following the protocol of Wang et al. (2018a).
Statistical Tests
Statistical tests were performed using R studio version 1.1.463 with R version 3.4.4. Figures were plotted using Python 3.7.4 and arranged with LibreOffice Draw 5.1.6.2.
Results
Morphological and Phylogenetic Analysis of the New Trichoderma Strain
The Trichoderma strain was isolated from leaves of Leucas aspera (Wild.) Link. We selected this strain for further analysis because we observed in preliminary field experiments that it promotes growth of several crop species. Its morphology shows typical characteristics of Trichoderma species of the harzianum clade. On KM plates, the hyphae cover the entire Petri dish from a single plug (5 mm in diameter) in 3–4 days (Supplementary Figure 1A). During the first 3 days after transfer to a new plate, the hyphae extended and formed conidiophores at the tip of hyphal branches. The conidia grew, replicated, and aggregated at the tip of a conidiophore (Supplementary Figure 1B). After 7 days, mature conidia developed as sphere-like structure composed of numerous individual conidia (Supplementary Figure 1C). The hyphal cell shrank after the conidia were fully developed. This allowed them to detach from the hyphae (Supplementary Figure 1D). The fully matured conidia displayed a green color (Supplementary Figure 1E).
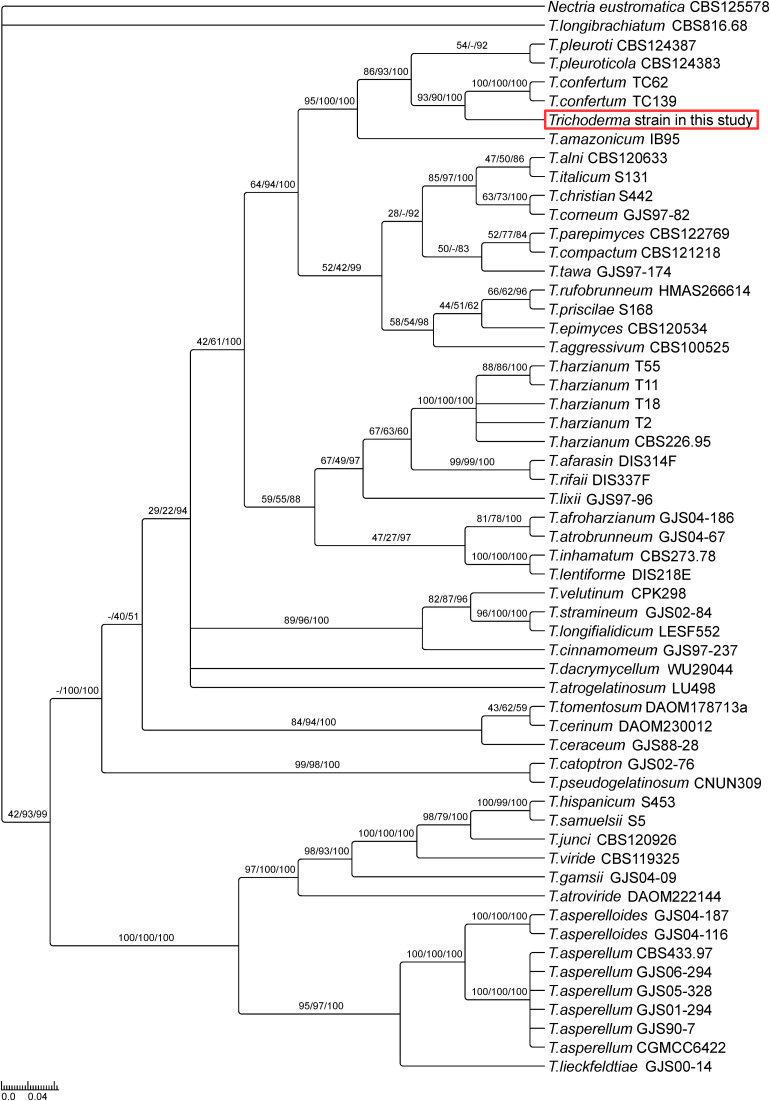
Phylogenetic analysis based on Maximum likelihood, Maximum Parsimony and Bayesian Inference of phylogeny uncovered that the isolated Trichoderma strain belongs to the harzianum clade (Figure 1), closely related to T. confertum TC62 and T. confertum TC139, two strains recently isolated from the soil 2,000 m above sea level in Tibet (Chen and Zhuang, 2017). The multilocus sequence analysis also indicated that the strain is closely related to T. pleuroti and T. pleuroticola, but less compared to T. confertum. In summary, according to the three different analysis methods, the isolated fungus is most probably a new Trichoderma strain closely related to T. confertum.
FIGURE 1.
Phylogeny of selected Trichoderma species suggests a new Trichoderma strain closely related to T. confertum. The phylogenetic tree is based on combined analysis of TEF1 and RPB2 genes using Bayesian inference of phylogeny. Maximum likelihood bootstrap values (MLBP, left), maximum parsimony bootstrap values (MPBP, center) and Bayesian inference posterior probabilities (BIPP, right) are shown at each node. Nectria eustromatica was used as outgroup.
The New Trichoderma Strain Colonizes Arabidopsis and Nicotiana Roots and Promotes Plant Growth
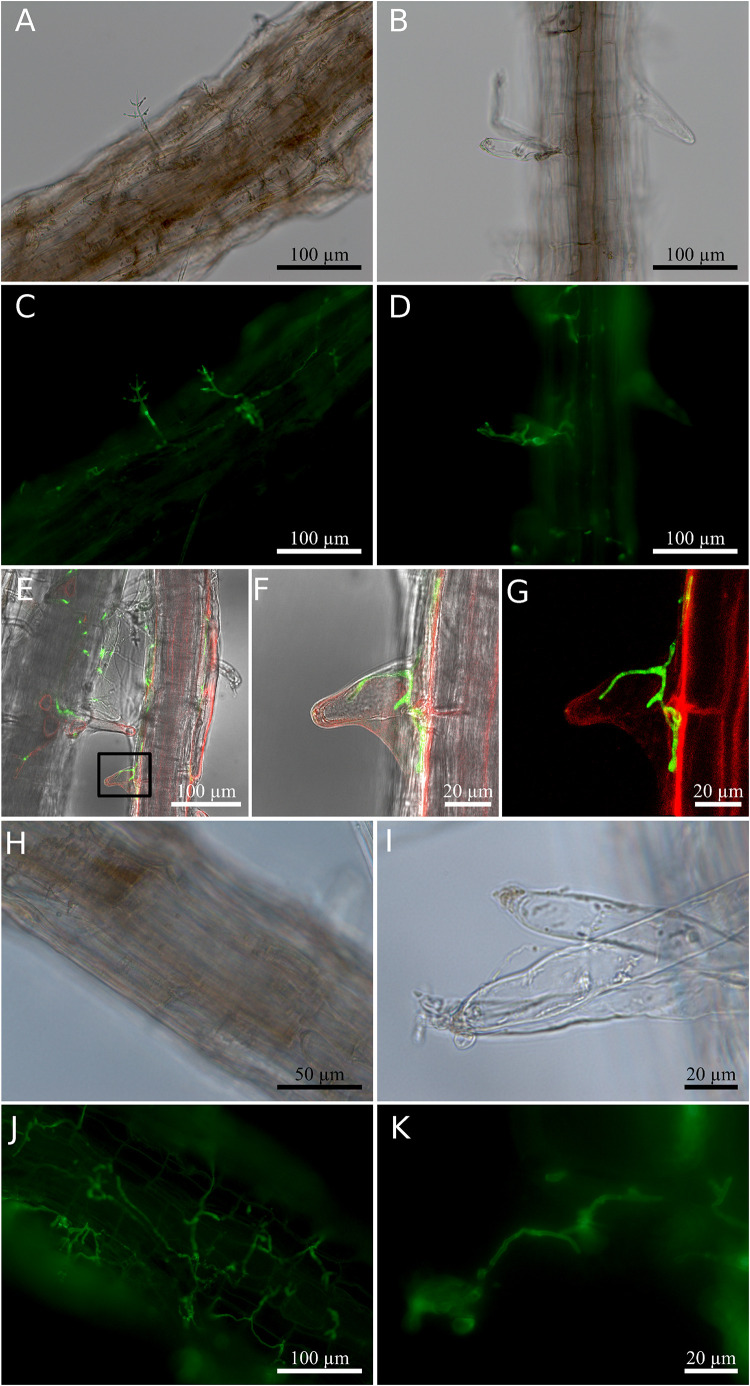
To characterize the endophytic lifestyle of the new strain, and to check which organ of the plant can be colonized by the fungus, it was co-cultivated with the model species, Arabidopsis thaliana. Two days after co-cultivation, hyphae were already detectable on the surface of the roots (Figures 2A,C). Light and confocal microscopy showed that hyphae also invaded into the root hair (Figures 2B,D–G and Supplementary Movie 1). Seven days after co-cultivation, the Arabidopsis roots were highly colonized, and conidiophores were found at the tip of the root hair, although not every root hair contained hyphae or conidiophore (Figures 2H–K). Close inspections revealed that the conidiophores derived from the hyphae in the root hairs. Under these co-cultivation conditions without stress, the aerial parts of the plant were not colonized.
FIGURE 2.
Root colonization of Arabidopsis thaliana by Trichoderma. (A–D) Co-cultivation for 2 days. (A,B) Bright field; (C,D) fluorescence of fungal stain. (E) Confocal images of hyphae inside root hair observed 2 days after co-cultivation. (F) Magnified view of the region enclosed by the small box in (E). (G) Fluorescence signal indicating hyphae (green) and plant cell plasma membrane (red) in (F). (H–K) Co-cultivation for 7 days. (H,I) Bright field; (J,K) fluorescence of fungal stain. Colonized root tissues were stained with WGA Alexa FluorTM 488 conjugate to detect the presence of the fungus, and RH414 was used to visualize the plant plasma membrane. The image shown for the confocal microscopy was chosen from three individual roots of three biological replicates.
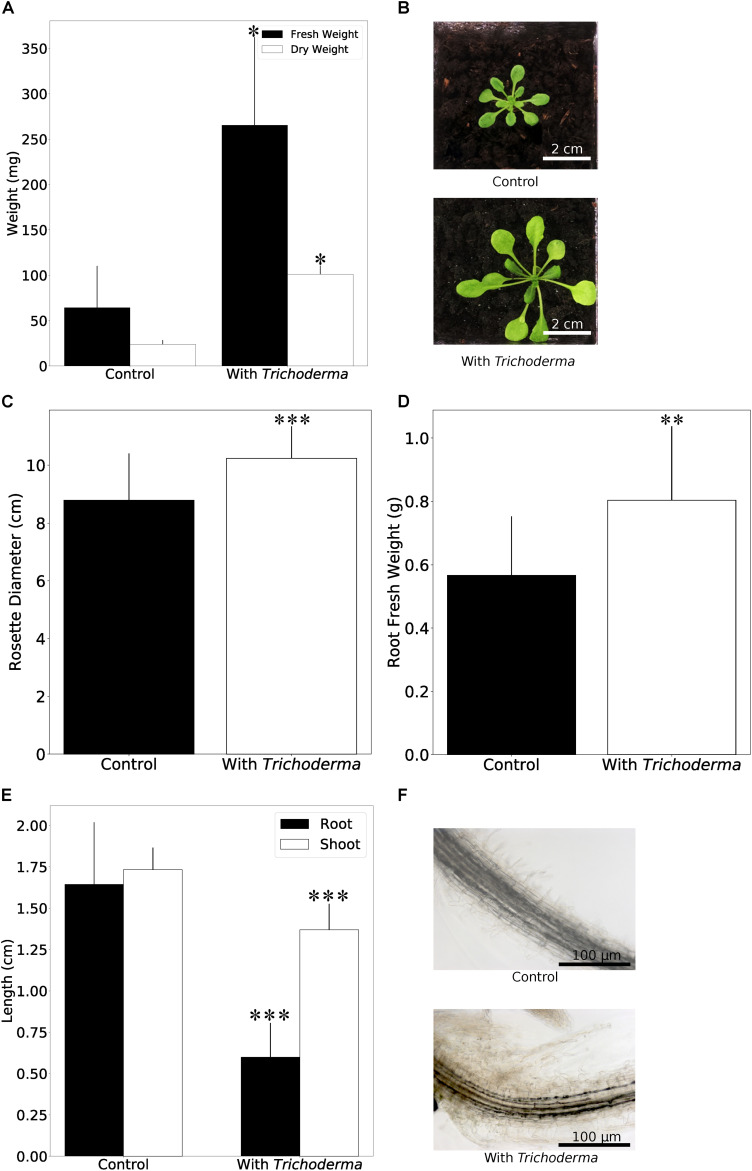
When Arabidopsis plants were co-cultivated on soil in the greenhouse, the germination rate and the performance of the young seedlings were not affected by the fungus. However, we observed a strong initial growth-promoting effect of the fungus on 4-week old Arabidopsis plants, since colonized plants were almost twice as large as the uncolonized controls (Figures 3A,B). Root colonization by the Trichoderma strain was confirmed by microscopy (Supplementary Figure 2). During later stages, the growth difference between colonized and uncolonized plants became less and during flowering time, the growth-stimulating effect of the fungus was barely visible. The number and size of seeds was not significantly different for plants grown with or without the fungus (data not shown). This indicates that the fungus promotes plant growth during early stages of development.
FIGURE 3.
Plant growth performance is influenced by Trichoderma colonization. (A) Fresh and dry weights of A. thaliana grown on soil with or without Trichoderma after 4 weeks. Error bars represent SEs from three independent biological replicates, each with four seedlings. Statistical significance was determined by Welch Two Sample t-test (∗P < 0.05). (B) Growth promotion on A. thaliana on soil after 4 weeks. (C,D) Rosette diameter (C) and root fresh weight (D) of N. attenuata inoculated with or without Trichoderma on soil after 4 weeks. Error bars represent SDs from 39 independent biological replicates for shoots and 18 replicates for roots. Statistical significance was determined by Welch Two Sample t-test (∗∗P < 0.01; ∗∗∗P < 0.001). (E) Shoot and root lengths of N. attenuata 12 days after co-cultivation with Trichoderma (spore solution O.D.600 nm = 0.0135) or without Trichoderma on Petri dishes. Error bars represent SDs from 10 biological replicates. Statistical significance was determined by Welch Two Sample t-test (∗∗∗P < 0.001). (F) Microscopy of N. attenuata roots 12 days on Petri dishes with or without Trichoderma.
Growth promotion was also observed for the model plant N. attenuata (cf. also below). We observed the same colonization efficiencies as described above for Arabidopsis seedlings. Also, the germination rates were similar for inoculated and non-inoculated seeds, and all seedlings were healthy. Similar to Arabidopsis, we observed a stimulatory effect of the fungus on N. attenuata growth after 4 weeks on soil, when both the rosette diameter and root biomass were larger (Figures 3C,D). Comparable to Arabidopsis, the growth-stimulating effect disappeared during later stages of development. However, we observed a clear difference in the response of the two hosts on agar plates during early seedling’s development, where root and shoot development can be monitored in more details. With fungal inoculation, the shoots and roots of 12-day-old Arabidopsis seedlings were bigger in the presence of the fungus (see Figure 5), while 12-day-old colonized N. attenuata had significantly shorter shoots and roots than the uncolonized controls (Figure 3E). We also observed more root hairs beneath the root-shoot junction, where roots are in contact with the fungus (Figure 3F). These effects were not observed for the roots of Arabidopsis seedlings. In conclusion, the fungus has different effects on the early development of the seedlings on agar medium.
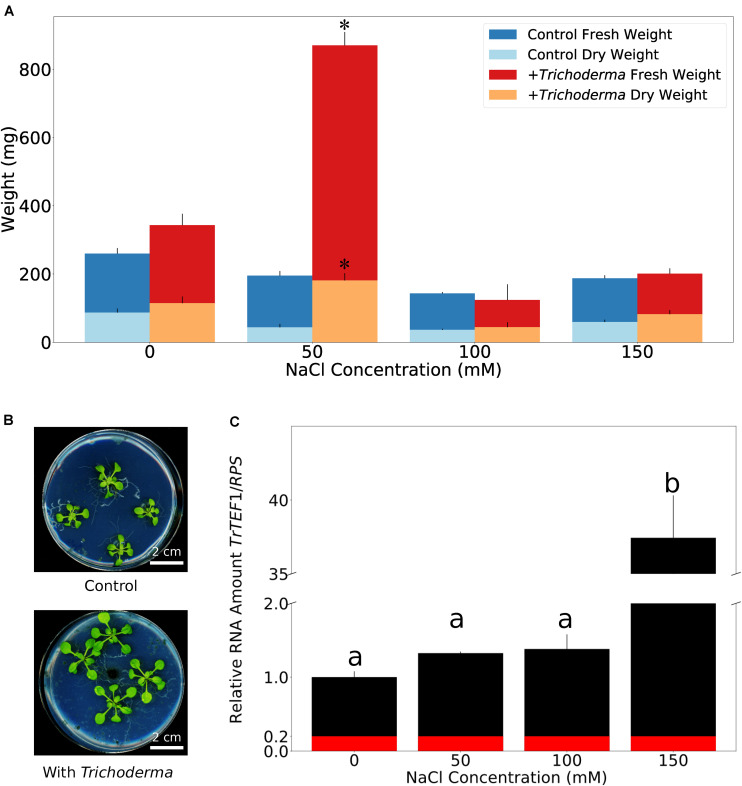
FIGURE 5.
Mild salt condition optimizes fungal colonization and plant growth performance on synthetic medium. (A) Fresh and dry weights of A. thaliana co-cultivated with or without Trichoderma on PNM medium with 0–150 mM NaCl for 5 days. Error bars represent SEs from three independent biological replicates, each with four seedlings. Statistical significance was determined by 2-Way ANOVA. Post-hoc analysis between all groups was further carried out by Tukey HSD test with P < 0.05, and is shown in Supplementary Table 3. Asterisks indicate significant difference in fresh/dry weight of seedlings grown on 50 mM NaCl with or without Trichoderma. (B) Growth phenotype of A. thaliana on PNM medium with 50 mM NaCl 5 days after co-cultivation with or without Trichoderma. (C) Quantification of Trichoderma root colonization on A. thaliana on PNM medium with 0–150 mM NaCl by qPCR. TrTEF1: Trichoderma TEF1; RPS: A. thaliana ribosomal protein S13/S18 family. Error bars represent SEs from three independent biological replicates, each with four seedlings. Statistical significance was determined by Tukey’s HSD test with P < 0.05, and is indicated by different lower-case letters. Red color represents average background value referred from samples without Trichoderma.
The New Trichoderma Strain Is Tolerant Against 100 mM Salt and Mild Salt Conditions Promote the Interaction With the Host on Synthetic Medium
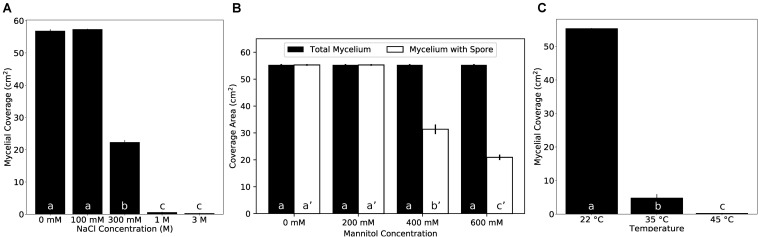
Plant growth promoting fungi and bacteria often also improve the stress tolerance of plants (Qin et al., 2016). Therefore, we first tested if the fungus itself is tolerant against salt and mannitol (osmotic) stress. Fungal growth was not altered on 100 mM NaCl compared to control plates without salt. At 300 mM NaCl, the mycelial growth was reduced by about 50%. At 1 M NaCl, only slowly growing mycelia could be detected after 10 days, and no growth was detectable on 3 M NaCl (Figure 4A and Supplementary Figures 3A–G). Increasing mannitol concentrations did not inhibit mycelial growth, although the production of conidia was reduced on media with >400 mM mannitol (Figure 4B and Supplementary Figures 3H–K). Also high temperature strongly impaired growth of the fungus (Figure 4C and Supplementary Figures 3L–N).
FIGURE 4.
Growth of the Trichoderma strain under various conditions on KM plates. (A) Fungal growth on different NaCl concentrations. (B) Fungal growth on different mannitol concentrations. (C) Fungal growth at different temperatures. Error bars represent SEs from three independent biological replicates. Statistical significance was determined by Tukey’s HSD test with P < 0.05, and is indicated by different lower-case letters.
The ability of the fungus to survive 1 M NaCl intrigued us to find out if salt influences the growth stimulating effect of its host. Unlike on soil, when seedlings were grown on solid PNM medium without addition of NaCl for 5 days, we observed only a small increase in growth and biomass production of the Arabidopsis seedlings in the presence of the fungus. However, application of 50 mM NaCl to the medium strongly promoted growth and biomass production (Figures 5A,B; supported by post-hoc analysis shown in Supplementary Table 3). This was accompanied by a stronger root colonization (Figure 5C). In particular, the lateral roots of the host were massively colonized and the fungus produced large amounts of conidiospores, compared to those on medium without NaCl. On 100 mM and higher NaCl concentrations, the growth of the uncolonized plants was gradually reduced, and growth of Trichoderma-colonized roots was not stimulated any more (Figure 5A and Supplementary Figure 4). Closer inspections uncovered that roots were even more colonized, and the hyphae also appeared on the surface of the areal parts. They were not only detectable at and around the hypocotyl (Supplementary Figure 4) but also on the leaf surface (data not shown). In summary, apparently, the fungus colonizes preferentially the roots. Low NaCl concentrations promoted root colonization and stimulated plant growth, while higher salt concentrations forced the fungus to invade the aerial parts which was associated with a loss of the benefits to the host.
The New Trichoderma Strain Inhibits Growth of Alternaria and Protects Arabidopsis and Nicotiana Against Alternaria Infection
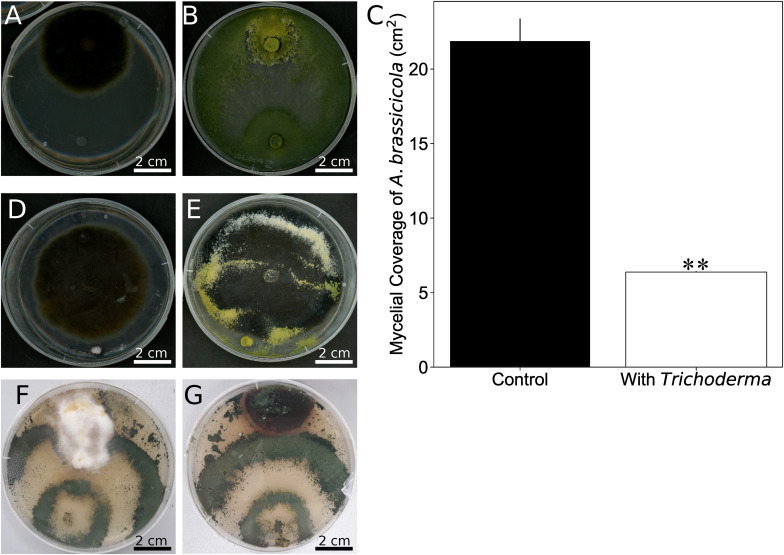
One of the prominent traits of Trichoderma species in the harzianum clade is their potential to act as bio-control agent. After 8 days of co-cultivation of Alternaria brassicicola with the Trichoderma strain on PDA plates, the mycelial coverage of A. brassicicola was reduced by 73% and Trichoderma hyphae grew on top of the A. brassicicola mycelial lawn (Figures 6A–C). To rule out that faster growth of Trichoderma restricts A. brassicicola growth, Trichoderma was added to an agar plate with a 7-day old A. brassicicola culture (Figure 6D). After additional 7 days of co-cultivation, Trichoderma hyphae and spores were again observed on top of the A. brassicicola mycelial lawn (Figure 6E). This supports active predation of A. brassicicola by the new Trichoderma strain (Druzhinina et al., 2018).
FIGURE 6.
Trichoderma inhibits growth of Alternaria brassicicola. (A,B) A plug of A. brassicicola was applied to the upper side of the PDA plate. A plug of KM agar (A) or the Trichoderma strain (B) was put to the lower side of the plate before co-cultivation for 8 days. (C) Quantification of A. brassicicola mycelial coverage from (A,B). Error bars represent SDs from three independent biological replicates. Statistical significance was determined by Welch Two Sample t-test (∗∗P < 0.01). (D) An agar plug with Trichoderma was placed on a PDA plate which contained 7-day old A. brassicicola culture. (E) Seven days after the plug with Trichoderma was placed in (D). (F,G) Co-cultivation of Trichoderma with the native N. attenuata pathogens Fusarium brachygibbosum (F) or Alternaria spp. strain Utah 10 (G).
We further tested Fusarium brachygibbosum and Alternaria spp. Utah isolate 10, two fungal species previously characterized as a native pathogen for N. attenuta (Luu et al., 2015), and co-cultivated them with Trichoderma. Growth of Trichoderma was much faster than that of the two other species, but F. brachygibbosum clearly stopped further growth of Trichoderma when hyphae of the two fungi met, while Alternaria spp. was overgrown by Trichoderma after 3½ weeks of co-cultivation (Figures 6F,G).
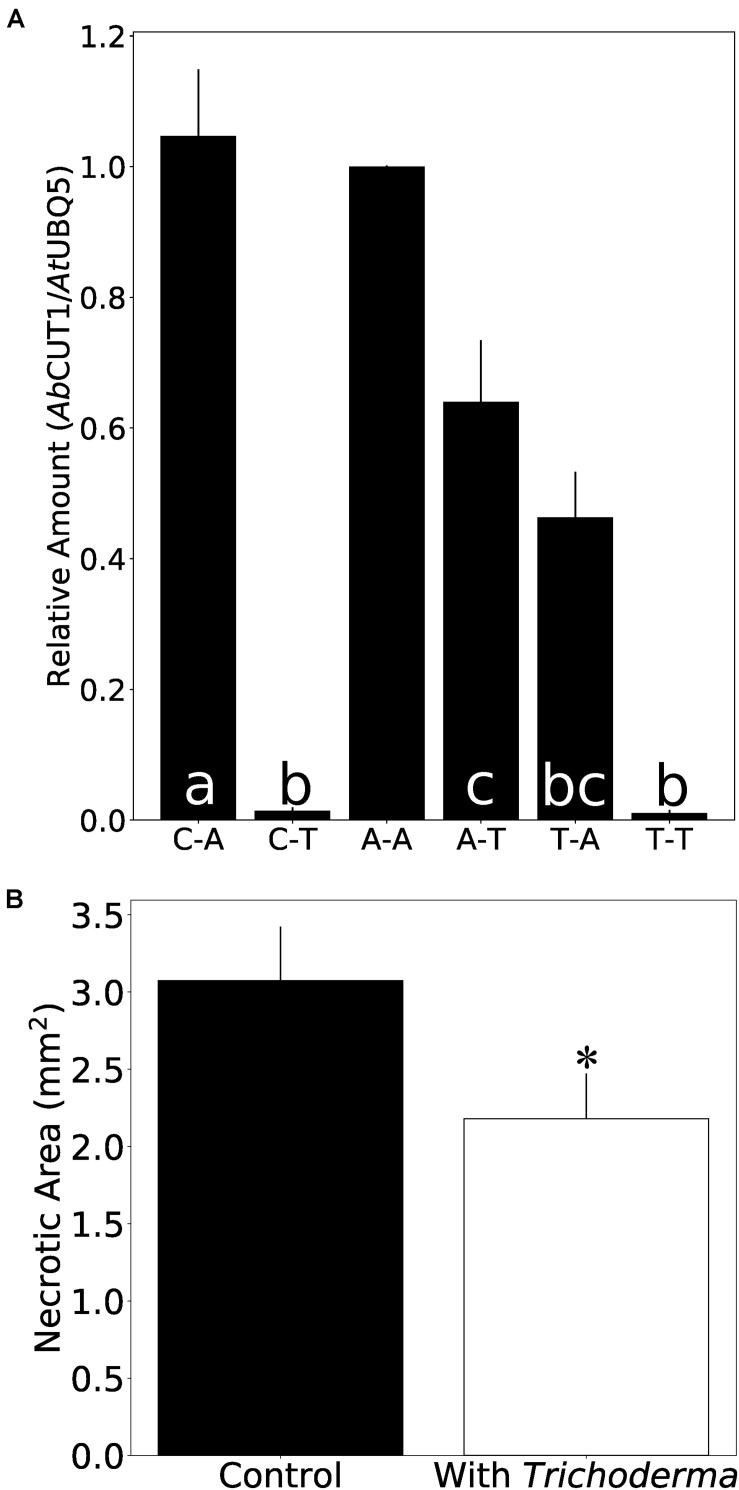
To test if the Trichoderma strain also protects plants from Alternaria infection, Arabidopsis seedlings were first exposed to A. brassicicola (A) or Trichoderma (T) or were mock-treated (C) and then transferred to plates with either A. brassicicola or Trichoderma for additional 7 days. As expected, the highest amount of A. brassicicola DNA was detected in seedlings which were exposed to A. brassicicola only (Figure 7A). Roots which were exposed to Trichoderma either before or after A. brassicicola treatment (A-T) contained less DNA of the fungal pathogen. Furthermore, the seedlings were better protected against A. brassicicola when they were already colonized by Trichoderma before pathogen infection (Figure 7A and Supplementary Figure 5A). Similar results were observed for N. attenuata and Alternaria (Supplementary Figure 5B). This demonstrates that the Trichoderma strain restricts growth of the pathogen in roots of its host plant.
FIGURE 7.
Trichoderma protects Arabidopsis seedlings against A. brassicicola infection. (A) Amount of A. brassicicola CUT1 DNA relative to A. thaliana UBQ5 DNA in root tissue. C-A: co-cultivation with control plug for 7 days, then co-cultivation with A. brassicicola for another 7 days. C-T: co-cultivation with control plug for 7 days, then co-cultivation with Trichoderma for another 7 days. A-A: co-cultivation with A. brassicicola for 7 days, then co-cultivation with A. brassicicola for another 7 days. A-T: co-cultivation with A. brassicicola for 7 days, then co-cultivation with Trichoderma for another 7 days. T-A: co-cultivation with Trichoderma for 7 days, then co-cultivation with A. brassicicola for another 7 days. T-T: co-cultivation with Trichoderma for 7 days, then co-cultivation with Trichoderma for another 7 days. Values from qPCR experiment were normalized to A-A. Error bars represent SEs from three independent biological replicates, each with 6–9 seedlings. Statistical significance was determined by Tukey’s HSD test with P < 0.05, and is indicated by different lower-case letters. (B) Necrosis area on leaflets infected by A. brassicicola. Error bars represent SEs from 35 independent biological replicates. Statistical significance was determined by Welch Two Sample t-test (∗P < 0.05).
To investigate whether Trichoderma also protects the leaves against A. brassicicola infection, 500 colony forming units (CFU) of an A. brassicicola spore suspension were applied to the leaves of Arabidopsis seedlings which were either co-cultivated with the symbiont or mock-treated for 7 days. Four days later, the necrotic zone on the leaves of co-cultivated plants was significantly smaller compared to the non-colonized controls (Figure 7B). Taken together, Trichoderma restricts spread of Alternaria in both roots and shoots.
Mycorrhiza Formation Is Not Affected by the New Trichoderma Strain in N. attenuata
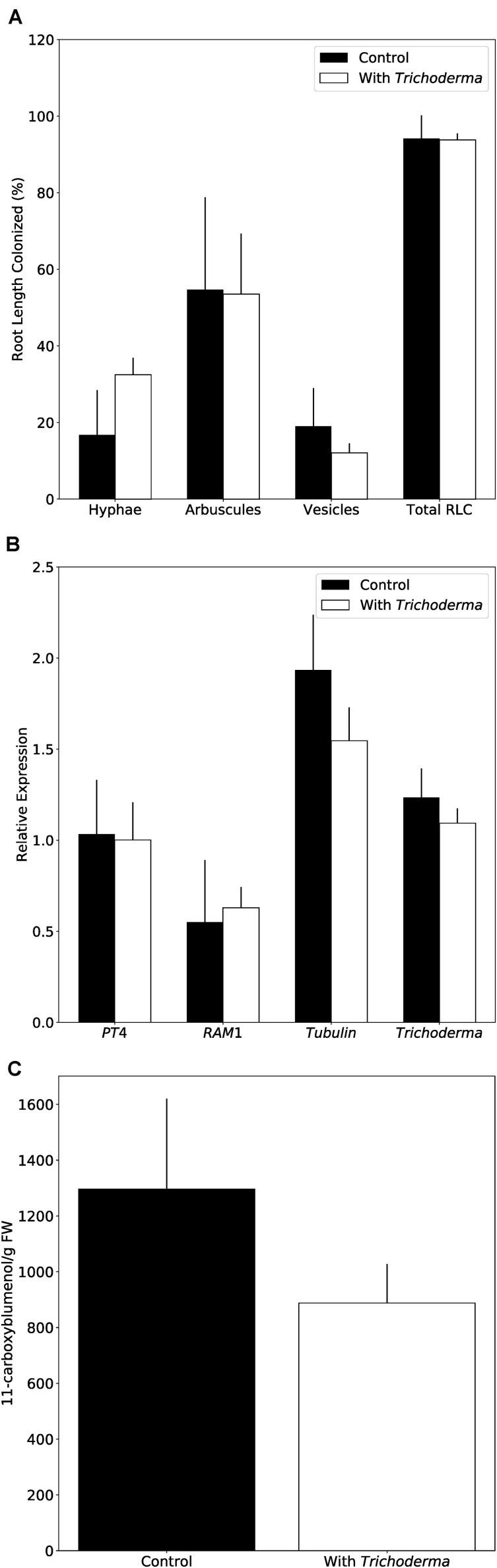
Restriction of Alternaria growth by the new Trichoderma strain indicated a putative use for bioprotection. However, agricultural application requires that other beneficial fungi, such as arbuscular mycorrhizal fungi (AMF) are not affected by the Trichoderma strain. As Arabidopsis is a non-mycorrhizal species, we used the well-established N. attenuata system (Groten et al., 2015). N. attenuata plants grown on soil in the greenhouse were simultaneously inoculated with AMF and Trichoderma. Microscopic observations of the roots and qPCR analyses with fungus-specific markers clearly indicate that AMF and Trichoderma colonize the roots and propagate, without inhibiting each other (Figures 8A,B). In addition, the amounts of 11-carboxyblumenol, a marker for AMF root colonization (Wang et al., 2018a), did not differ between Trichoderma-inoculated and non-inoculated samples (Figure 8C). 11-Carboxyblumenol levels were also similar when plants were pre-inoculated with AMF and after 6 weeks co-cultured with Trichoderma (data not shown). These results suggest that AMF colonization is not affected by the new Trichoderma strain in N. attenuata.
FIGURE 8.
Trichoderma does not interfere with mycorrhiza formation on N. attenuata. (A) AMF colonization 8 weeks after inoculation with or without Trichoderma and AMF simultaneously. Error bars represent SEs from three independent biological replicates. Statistical significance was determined by Welch Two Sample t-test between control and co-culture treatments and was not significantly different. RLC, Root length colonization. (B) AMF marker gene expression determined by qPCR. PT4, Phosphate transporter 4; RAM1,
The Trichoderma Strain Alters Phytohormone Levels in Arabidopsis Roots and Shoots
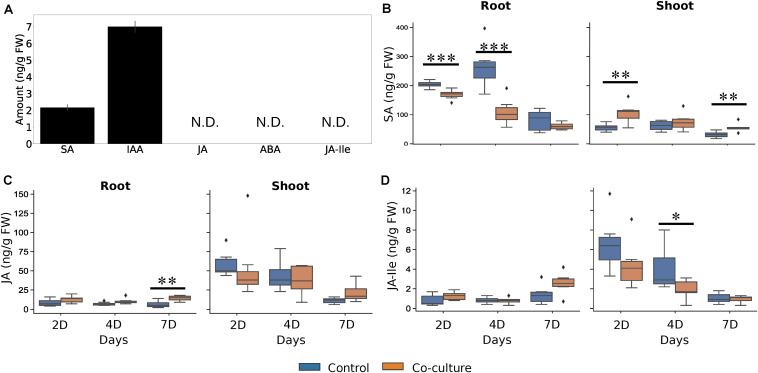
Beneficial plant-microbe interactions often result in altered phytohormone levels, which may lead to better fitness of the host upon pathogen attack but can also influence root colonization due to an altered plant immune system (Jacobs et al., 2011). In mycelial cultures, we detected only trace amounts of auxin (indole-acetic acid, IAA) and SA (Figure 9A). However, SA in Trichoderma-colonized seedlings were significantly reduced in roots and increased in shoots compared to controls (Figure 9B). Metabolites related to the biosynthesis and degradation of JA as well as ABA and IAA also showed some minor changes after Trichoderma colonization, but compared to SA, these changes were rather weak (Figures 9C,D and Supplementary Figure 6). Overall, it appears that the fungus does not produce high hormone levels itself influencing plant performance, but the fungus may activate SA-dependent resistance responses in the plant.
FIGURE 9.
Trichoderma facilitates the allocation of phytohormones in A. thaliana. (A) Phytohormone levels in Trichoderma mycelium. Error bars represent the SEs from six independent biological replicates. N.D., not detected. (B–D) SA (B), JA (C), and JA-Ile (D) levels from control and co-cultured seedlings in roots and shoots 2, 4, and 7 days after co-cultivation. Statistical significance was determined by Welch Two Sample t-test between control and co-culture treatments (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). At least 6 biological replicates were used for measurement, each with 16 seedlings. ABA, abscisic acid; IAA, indole-3-acetic acid; SA, salicylic acid; JA, jasmonic acid; JA-Ile, jasmonoyl-isoleucine conjugate. The diamond shape in the figure represent outliers, which the data points exceed 1.5 times of the inter-quartile range from the 75th percentiles, or lower than 1.5 times of the inter-quartile range from the 25th percentiles. The inter-quartile range is the range between 25th and 75th percentiles.
Discussion
In this study, a new endophytic Trichoderma strain is described. It belongs to the harzianum clade, closely related to T. confertum, T. pleuroti and T. pleuroticola. It survives under salt and osmotic stress, and possesses a strong capability to reduce A. brassicicola growth. The hyphae colonize the root surface and are found in root hairs of A. thaliana. Infection assays showed reduced A. brassicicola spread in roots and shoots of Trichoderma-colonized Arabidopsis plants, while mycorrhiza formation is not affected in N. attenuata. These observations are important for potential application of the endophyte as bio-control agent, and for the development of more effective and versatile bio-control agents.
Numerous Trichoderma species have been reported to stimulate plant growth (Contreras-Cornejo et al., 2009; González-Pérez et al., 2018), and T. atroviride and T. virens have been shown to promote root hair development (Contreras-Cornejo et al., 2015; González-Pérez et al., 2018). Our results highlight the importance of the growth conditions for the investigations of the symbiotic interactions with the new Trichoderma strain. Most importantly, as long as the symbionts grow in soil, we observe growth promotion during early phases of the development in the two tested host species. However, the growth stimulating effect of the fungus was barely or not detectable at all on agar plates, as long as no NaCl is added. A possible scenario could be that the fungus requires low concentrations of NaCl for growth and thus root colonization. If the salt concentration in the medium is too high, the fungus helps the plant by stimulating osmolyte production and Na+ elimination through root exudates (Contreras-Cornejo et al., 2014). We demonstrate that the fungus also tries to escape from the stress by growing on the plant material, since the roots become more colonized with increased salt concentrations. Ultimately, hyphae can also be detected in the aerial parts of the plant, which occurs only when the stress around the roots is high. We assume that the extensive fungal propagation triggers the plant defense machinery to restrict fungal growth and consequently may reduce the host´s investment into growth. While our experiment focusses on the role of NaCl for the symbiosis, there are apparently other growth-stimulating factors in soil. A comparative analysis of the different growth conditions established in this study may help to elucidate critical parameters with agricultural relevance.
Root Colonization Alters Root Architecture
The new Trichoderma strain not only colonizes the root surface, but also penetrates into the root epidermis and resides in the root hairs (Figures 2E–G and Supplementary Movie 1). To the best of our knowledge, this is a new colonization strategy for Trichoderma species and demonstrates that the fungus can also live as endophyte. This finding is further supported by the fact that the fungus was originally isolated from the leaf cells of a tree. Although Trichoderma species have been often reported to colonize plant roots (López-Bucio et al., 2015; Ruano-Rosa et al., 2016), the invasion of hyphae into root cells might indicate a closer symbiosis compared to other Trichoderma strains and species. Reprogramming of root development, inhibition of root growth and stimulating root branching is a typical feature of AMF (Bonfante and Genre, 2010), but also observed for Trichoderma-colonized Arabidopsis roots (e.g., Contreras-Cornejo et al., 2015). Similar to AMF associations, the endophyte might contribute to nutrient and water uptake and allow the plants to reduce their root sizes. Further studies are needed to support this hypothesis. Additionally, an increase in the number of root hairs may lead to a larger surface area for fungal attachment. Its close phylogenetic relationship to Trichoderma species which grow preferentially on mushrooms also demonstrates that minor changes in the Trichoderma genomes allow major changes, enlargements or alterations in their host range or preference.
The New Trichoderma Strain Has Potential as New Bio-Control Agent
The infection assays with A. brassicicola show effective protection of Arabidopsis roots and shoots by Trichoderma. Interestingly, the beneficial fungus also restricted growth of A. brassicicola in the roots, when the roots were already infected by the pathogen (Figure 7A, A-T vs. C-T). This is consistent with the plate experiments in which Trichoderma actively predated A. brassicicola. Propagation of the pathogen in the leaves is also restricted when the roots are colonized by Trichoderma. Different local and systemic plant immune responses against various pathogens in Alternaria-colonized hosts have been reported, however, a general strategy for Trichoderma species is not apparent (Busby et al., 2016; Rai and Agarkar, 2016). Apparently, systemic signals travel from the roots to the leaves, and this is reflected by elevated SA levels in the leaves of Trichoderma-colonized seedlings even before they are exposed to the pathogen (Figure 9B). The higher SA levels in the leaves might indicate that the new Trichoderma strain has the ability to induce SAR. The low or undetectable levels of the defense-related hormones in the mycelium suggest that they are not of fungal origin.
Another feature of this new strain is its ability to sustain beneficial microbe interaction with plants. Although pathogen progression in root tissue is hindered by the new Trichoderma strain, the presence of the fungus does not interfere with AMF colonization. Recently, Metwally and Al-Amri (2020) showed an interactive role of Trichoderma viride and AMF on growth and pigment content of onion plants, however, due to the small number of AMF−Trichoderma–host plant combinations that have been investigated so far, general conclusions on those tripartite interactions are not possible (cf. Szczałba et al., 2019). Those studies are important for a successful bio-control agent, as Trichoderma species are also competitors of beneficial microbes (Sood et al., 2020), which could impair plant growth or yield.
Conclusion
In conclusion, the new Trichoderma strain might be a useful tool as bio-control agent, since it stimulates the plant immune system against pathogen infection, but at the same time does not interfere with other beneficial microbial interactions, such as mycorrhizal formation. Its growth promoting ability in soil provides additional benefit in agricultural application. Furthermore, the experimental set-up allows us to address further questions to understand the role of this fungus on plant performance, especially why the fungus is successful in promoting plant growth in soil but not on minimal medium, and how it influences the balance between growth and stress responses under different environmental conditions.
Data Availability Statement
The datasets generated for this study can be found in the online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
Y-HT organized the project, performed the experiments, collected the samples and data, analyzed the results, plotted the figures, and wrote up the study. HR performed the soil and salt experiments on Arabidopsis. KG performed the experiments on Nicotiana. PR isolated the Trichoderma strain. AF assisted in the microscopy. MR measured the phytohormones. IB, KN, RU, and RO edited the manuscript. RO organized the project and wrote up the study. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Veit Grabe from the Max Planck Institute for Chemical Ecology, Jena, for his assistance on confocal microscopy.
Funding. The work was supported by the CRC1127.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.573670/full#supplementary-material
References
- Bailey B. A., Melnick R. L. (2013). “The endophytic Trichoderma,” in Trichoderma: Biology and Applications, eds Mukherjee P. K., Horwitz B. K., Singh U. S., Mukherjee M., Schmoll M. (Wallingford: CAB International; ), 152–172. 10.1079/9781780642475.0000 [DOI] [Google Scholar]
- Bains P. S., Tewari J. P. (1987). Purification, chemical characterization and host-specificity of the toxin produced by Alternaria brassicae. Physiol. Mol. Plant Pathol. 30 259–271. 10.1016/0885-5765(87)90039-7 [DOI] [Google Scholar]
- Berini F., Caccia S., Franzetti E., Congiu T., Marinelli F., Casartelli M., et al. (2016). Effects of Trichoderma viride chitinases on the peritrophic matrix of Lepidoptera. Pest Manag. Sci. 72 980–989. 10.1002/ps.4078 [DOI] [PubMed] [Google Scholar]
- Bonfante P., Genre A. (2010). Mechanisms underlying beneficial plant–fungus interactions in mycorrhizal symbiosis. Nat. Commun. 1 1–11. 10.1038/ncomms1046 [DOI] [PubMed] [Google Scholar]
- Busby P. E., Ridout M., Newcombe G. (2016). Fungal endophytes: modifiers of plant disease. Plant Mol. Biol. 90 645–655. 10.1007/s11103-015-0412-0 [DOI] [PubMed] [Google Scholar]
- Carsolio C., Gutiérrez A., Jiménez B., Van Montagu M., Herrera-Estrella A. (1994). Characterization of ech-42, a Trichoderma harzianum endochitinase gene expressed during mycoparasitism. Proc. Natl. Acad. Sci. U. S. A. 91 10903–10907. 10.1073/pnas.91.23.10903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17 540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Checker V. G., Kushwaha H. R., Kumari P., Yadav S. (2018). “Role of phytohormones in plant defense: signaling and cross talk,” in Molecular Aspects of Plant-Pathogen Interaction, eds Singh A., Singh I. K. (Singapore: Springer; ), 159–184. 10.1007/978-981-10-7371-7_7 [DOI] [Google Scholar]
- Chen K., Zhuang W.-Y. (2017). Seven new species of Trichoderma from soil in China. Mycosystema 36 1441–1462. 10.13346/j.mycosystema.170134 [DOI] [Google Scholar]
- Contreras-Cornejo H. A., López-Bucio J. S., Méndez-Bravo A., Macías-Rodríguez L., Ramos-Vega M., Guevara-García ÁA., et al. (2015). Mitogen-activated protein kinase 6 and ethylene and auxin signaling pathways are involved in Arabidopsis root-system architecture alterations by Trichoderma atroviride. Mol. Plant-Microbe Interact. 28 701–710. 10.1094/MPMI-01-15-0005-R [DOI] [PubMed] [Google Scholar]
- Contreras-Cornejo H. A., Macías-Rodríguez L., Alfaro-Cuevas R., López-Bucio J. (2014). Trichoderma spp. improve growth of Arabidopsis seedlings under salt stress through enhanced root development, osmolite production, and Na+ elimination through root exudates. Mol. Plant-Microbe Interact. MPMI 27 503–514. 10.1094/MPMI-09-13-0265-R [DOI] [PubMed] [Google Scholar]
- Contreras-Cornejo H. A., Macías-Rodríguez L., Cortés-Penagos C., López-Bucio J. (2009). Trichoderma virens, a plant beneficial fungus, enhances biomass production and promotes lateral root growth through an auxin-dependent mechanism in Arabidopsis. Plant Physiol. 149 1579–1592. 10.1104/pp.108.130369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. J. (1990). Isolation of plant DNA from fresh tissue. Focus. 12 13–15. [Google Scholar]
- Druzhinina I. S., Chenthamara K., Zhang J., Atanasova L., Yang D., Miao Y., et al. (2018). Massive lateral transfer of genes encoding plant cell wall-degrading enzymes to the mycoparasitic fungus Trichoderma from its plant-associated hosts. PLoS Genet. 14:e1007322. 10.1371/journal.pgen.1007322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druzhinina I. S., Kubicek C. P. (eds) (2016). Environmental and Microbial Relationships, 3rd Edn Berlin: Springer International Publishing, 10.1007/978-3-319-29532-9 [DOI] [Google Scholar]
- Druzhinina I. S., Seidl-Seiboth V., Herrera-Estrella A., Horwitz B. A., Kenerley C. M., Monte E., et al. (2011). Trichoderma: the genomics of opportunistic success. Nat. Rev. Microbiol. 9 749–759. 10.1038/nrmicro2637 [DOI] [PubMed] [Google Scholar]
- El-Katatny M., Somitsch W., Robra K., El-Katatny M., Gubitz G. (2000). Production of chitinase and β-1,3-glucanase by Trichoderma harzianum for control of the phytopathogenic fungus. Sclerotium rolfsii. Food Technol. Biotechnol. 38 173–180. [Google Scholar]
- Furukawa H., Kusne S., Sutton D. A., Manez R., Carrau R., Nichols L., et al. (1998). Acute invasive sinusitis due to Trichoderma longibrachiatum in a liver and small bowel transplant recipient. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 26 487–489. 10.1086/516317 [DOI] [PubMed] [Google Scholar]
- Gautheret A., Dromer F., Bourhis J. H., Andremont A. (1995). Trichoderma pseudokoningii as a cause of fatal infection in a bone marrow transplant recipient. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 20 1063–1064. 10.1093/clinids/20.4.1063 [DOI] [PubMed] [Google Scholar]
- Geyer C. J. (1991). “Markov chain monte carlo maximum likelihood,” in Computing Science and Statistics: Proceedings of the 23rd Symposium on the Interface, ed. Keramidas E. M. (Fairfax, VA: Interface Foundation; ), 156–163. [Google Scholar]
- González-Pérez E., Ortega-Amaro M. A., Salazar-Badillo F. B., Bautista E., Douterlungne D., Jiménez-Bremont J. F. (2018). The Arabidopsis-Trichoderma interaction reveals that the fungal growth medium is an important factor in plant growth induction. Sci. Rep. 8:16427. 10.1038/s41598-018-34500-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosch R., Lottmann J., Rehn V. N. C., Rehn K. G., Mendonça-Hagler L., Smalla K., et al. (2007). Analysis of antagonistic interactions between Trichoderma isolates from Brazilian weeds and the soil-borne pathogen Rhizoctonia solani. J. Plant Dis. Prot. 114 167–175. 10.1007/BF03356213 [DOI] [Google Scholar]
- Groten K., Nawaz A., Nguyen N. H. T., Santhanam R., Baldwin I. T. (2015). Silencing a key gene of the common symbiosis pathway in Nicotiana attenuata specifically impairs arbuscular mycorrhizal infection without influencing the root-associated microbiome or plant growth. Plant Cell Environ. 38 2398–2416. 10.1111/pce.12561 [DOI] [PubMed] [Google Scholar]
- Hermosa R., Viterbo A., Chet I., Monte E. (2012). Plant-beneficial effects of Trichoderma and of its genes. Microbiol. Read. Engl. 158 17–25. 10.1099/mic.0.052274-0 [DOI] [PubMed] [Google Scholar]
- Heyer M., Reichelt M., Mithöfer A. (2018). A holistic approach to analyze systemic jasmonate accumulation in individual leaves of Arabidopsis rosettes upon wounding. Front. Plant Sci. 9:1569. 10.3389/fpls.2018.01569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T., Käfer E. (2001). Improved protocols for Aspergillus minimal medium: trace element and minimal medium salt stock solutions. Fungal Genet Newsl 48 20–21. 10.4148/1941-4765.1173 [DOI] [Google Scholar]
- Huelsenbeck J. P., Ronquist F. (2001). MRBAYES: bayesian inference of phylogenetic trees. Bioinforma. Oxf. Engl. 17 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Jacobs S., Zechmann B., Molitor A., Trujillo M., Petutschnig E., Lipka V., et al. (2011). Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the Fungus Piriformospora indica. Plant Physiol. 156 726–740. 10.1104/pp.111.176446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. M., Sherameti I., Ludwig A., Nongbri P., Sun C., Lou B., et al. (2011). Protocols for Arabidopsis thaliana and Piriformospora indica co-cultivation – A model system to study plant beneficial traits. Endocytobiosis Cell Res. 21 101–113. [Google Scholar]
- Karlsson M., Atanasova L., Jensen D., Zeilinger S. (2017). “Necrotrophic mycoparasites and their genomes,” in The Fungal Kingdom, eds Heitman J., Howlett B., Crous P., Stukenbrock E., James T., Gow N. (Washington, DC: ASM Press; ), 1005–1026. 10.1128/microbiolspec.FUNK-0016-2016 [DOI] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistner C., Matamoros M. (2005). “RNA isolation using phase extraction and LiCl precipitation,” in Lotus Japonicus Handbook, ed. Márquez A. J. (Berlin: Springer-Verlag; ), 123–124. 10.1007/1-4020-3735-X_9 [DOI] [Google Scholar]
- Kozlov A. M., Darriba D., Flouri T., Morel B., Stamatakis A. (2019). RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinforma. Oxf. Engl. 35 4453–4455. 10.1093/bioinformatics/btz305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krügel T., Lim M., Gase K., Halitschke R., Baldwin I. T. (2002). Agrobacterium-mediated transformation of Nicotiana attenuata, a model ecological expression system. Chemoecology 12 177–183. 10.1007/PL00012666 [DOI] [Google Scholar]
- Lee S., Yap M., Behringer G., Hung R., Bennett J. W. (2016). Volatile organic compounds emitted by Trichoderma species mediate plant growth. Fungal Biol. Biotechnol. 3:7. 10.1186/s40694-016-0025-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonetti P., Zonno M. C., Molinari S., Altomare C. (2017). Induction of SA-signaling pathway and ethylene biosynthesis in Trichoderma harzianum-treated tomato plants after infection of the root-knot nematode Meloidogyne incognita. Plant Cell Rep. 36 621–631. 10.1007/s00299-017-2109-0 [DOI] [PubMed] [Google Scholar]
- Li R.-X., Cai F., Pang G., Shen Q.-R., Li R., Chen W. (2015). Solubilisation of phosphate and micronutrients by Trichoderma harzianum and its relationship with the promotion of tomato plant growth. PloS One 10:e0130081. 10.1371/journal.pone.0130081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Fu K., Gao S., Wu Q., Fan L., Li Y., et al. (2013). Impact on bacterial community in midguts of the Asian corn borer larvae by transgenic Trichoderma strain overexpressing a heterologous chit42 gene with chitin-binding domain. PloS One 8:e55555. 10.1371/journal.pone.0055555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Destri Nicosia M. G., Mosca S., Mercurio R., Schena L. (2015). Dieback of Pinus nigra seedlings caused by a strain of Trichoderma viride. Plant Dis. 99 44–49. 10.1094/PDIS-04-14-0433-RE [DOI] [PubMed] [Google Scholar]
- López-Bucio J., Pelagio-Flores R., Herrera-Estrella A. (2015). Trichoderma as biostimulant: exploiting the multilevel properties of a plant beneficial fungus. Sci. Hortic. 196 109–123. 10.1016/j.scienta.2015.08.043 [DOI] [Google Scholar]
- Luu V. T., Schuck S., Kim S.-G., Weinhold A., Baldwin I. T. (2015). Jasmonic acid signalling mediates resistance of the wild tobacco Nicotiana attenuata to its native Fusarium, but not Alternaria, fungal pathogens. Plant Cell Environ. 38 572–584. 10.1111/pce.12416 [DOI] [PubMed] [Google Scholar]
- Martínez-Medina A., Fernández I., Sánchez-Guzmán M. J., Jung S. C., Pascual J. A., Pozo M. J. (2013). Deciphering the hormonal signalling network behind the systemic resistance induced by Trichoderma harzianum in tomato. Front. Plant Sci. 4:206. 10.3389/fpls.2013.00206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle T. P., Miller M. H., Evans D. G., Fairchild G. L., Swan J. A. (1990). A new method which gives an objective measure of colonization of roots by vesicular—arbuscular mycorrhizal fungi. New Phytol. 115 495–501. 10.1111/j.1469-8137.1990.tb00476.x [DOI] [PubMed] [Google Scholar]
- Metwally R. A., Al-Amri S. M. (2020). Individual and interactive role of Trichoderma viride and arbuscular mycorrhizal fungi on growth and pigment content of onion plants. Lett. Appl. Microbiol. 70 79–86. 10.1111/lam.13246 [DOI] [PubMed] [Google Scholar]
- Mukherjee A. K., Sampath Kumar A., Kranthi S., Mukherjee P. K. (2014). Biocontrol potential of three novel Trichoderma strains: isolation, evaluation and formulation. 3 Biotech 4 275–281. 10.1007/s13205-013-0150-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15 473–497. 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- Park M. S., Bae K. S., Yu S. H. (2006). Two new species of Trichoderma associated with green mold of oyster mushroom cultivation in Korea. Mycobiology 34 111–113. 10.4489/MYCO.2006.34.3.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapati M. S., Patel J. B., Modi K., Shah M. B. (2010). Leucas aspera: a review. Pharmacogn. Rev. 4 85–87. 10.4103/0973-7847.65330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y., Druzhinina I. S., Pan X., Yuan Z. (2016). Microbially mediated plant salt tolerance and microbiome-based solutions for saline agriculture. Biotechnol. Adv. 34 1245–1259. 10.1016/j.biotechadv.2016.08.005 [DOI] [PubMed] [Google Scholar]
- Rai M., Agarkar G. (2016). Plant–fungal interactions: what triggers the fungi to switch among lifestyles? Crit. Rev. Microbiol. 42 428–438. 10.3109/1040841X.2014.958052 [DOI] [PubMed] [Google Scholar]
- Rajani P., Rajasekaran C., Vasanthakumari M. M., Olsson S. B., Ravikanth G., Uma Shaanker R., et al. (2020). Inhibition of plant pathogenic fungi by endophytic Trichoderma spp. through mycoparasitism and volatile organic compounds. Microbiol. Res 242:126595. 10.1016/j.micres.2020.126595 [DOI] [PubMed] [Google Scholar]
- Rambaut A. (2018). FigTree. Available at: http://tree.bio.ed.ac.uk/software/figtree/ (accessed June 11, 2020). [Google Scholar]
- Rocha-Ramirez V., Omero C., Chet I., Horwitz B. A., Herrera-Estrella A. (2002). Trichoderma atroviride G-protein alpha-subunit gene tga1 is involved in mycoparasitic coiling and conidiation. Eukaryot. Cell 1 594–605. 10.1128/ec.1.4.594-605.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. (2003). MRBAYES 3: bayesian phylogenetic inference under mixed models. Bioinforma. Oxf. Engl. 19 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Rosmana A., Nasaruddin N., Hendarto H., Hakkar A. A., Agriansyah N. (2016). Endophytic association of Trichoderma asperellum within Theobroma cacao suppresses vascular streak dieback incidence and promotes side graft growth. Mycobiology 44 180–186. 10.5941/MYCO.2016.44.3.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruano-Rosa D., Prieto P., Rincón A., Gómez Rodríguez M. V., Valderrama R., Barroso J., et al. (2016). Fate of Trichoderma harzianum in the olive rhizosphere: time course of the root colonization process and interaction with the fungal pathogen Verticillium dahliae. BioControl 61 269–282. 10.1007/s10526-015-9706-z [DOI] [Google Scholar]
- Samolski I., Rincón A. M., Pinzón L. M., Viterbo A., Monte E. (2012). The qid74 gene from Trichoderma harzianum has a role in root architecture and plant biofertilization. Microbiol. Read. Engl. 158 129–138. 10.1099/mic.0.053140-0 [DOI] [PubMed] [Google Scholar]
- Santhanam R., Menezes R. C., Grabe V., Li D., Baldwin I. T., Groten K. (2019). A suite of complementary biocontrol traits allows a native consortium of root-associated bacteria to protect their host plant from a fungal sudden-wilt disease. Mol. Ecol. 28 1154–1169. 10.1111/mec.15012 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumaran A., Akinyemi A., Mandon J., Cristescu S. M., Hall M. A., Harren F. J. M., et al. (2016). ABA suppresses Botrytis cinerea elicited NO production in tomato to influence H2O2 generation and increase host susceptibility. Front. Plant Sci. 7:709. 10.3389/fpls.2016.00709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood M., Kapoor D., Kumar V., Sheteiwy M. S., Ramakrishnan M., Landi M., et al. (2020). Trichoderma: the “secrets” of a multitalented biocontrol agent. Plants Basel Switz. 9:762. 10.3390/plants9060762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöver B. C., Müller K. F. (2010). TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11:7. 10.1186/1471-2105-11-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studholme D. J., Harris B., Le Cocq K., Winsbury R., Perera V., Ryder L., et al. (2013). Investigating the beneficial traits of Trichoderma hamatum GD12 for sustainable agriculture-insights from genomics. Front. Plant Sci. 4:258. 10.3389/fpls.2013.00258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. (2002). PAUP∗. Phylogenetic Analysis Using Parsimony (∗and Other Methods). Version 4.0b10. 10.1111/j.0014-3820.2002.tb00191.x [DOI] [PubMed] [Google Scholar]
- Szczałba M., Kopta T., Gs̨stoł M., Sêkara A. (2019). Comprehensive insight into arbuscular mycorrhizal fungi, Trichoderma spp. and plant multilevel interactions with emphasis on biostimulation of horticultural crops. J. Appl. Microbiol. 127 630–647. 10.1111/jam.14247 [DOI] [PubMed] [Google Scholar]
- Tavare S. (1986). Some probabilistic and statistical problems on the analysis of DNA sequences. Lect. Math. Life Sci. 17, 57–86. [Google Scholar]
- Wang M., Schäfer M., Li D., Halitschke R., Dong C., McGale E., et al. (2018a). Blumenols as shoot markers of root symbiosis with arbuscular mycorrhizal fungi. eLife 7:e37093. 10.7554/eLife.37093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Wilde J., Baldwin I. T., Groten K. (2018b). Nicotiana attenuata’s capacity to interact with arbuscular mycorrhiza alters its competitive ability and elicits major changes in the leaf transcriptome. J. Integr. Plant Biol. 60 242–261. 10.1111/jipb.12609 [DOI] [PubMed] [Google Scholar]
- Woo S. L., Ruocco M., Vinale F., Nigro M., Marra R., Lombardi N., et al. (2014). Trichoderma-based products and their widespread use in agriculture. Open Mycol. J. 8 71–126. 10.2174/1874437001408010071 [DOI] [Google Scholar]
- Yang Z. (1993). Maximum-likelihood estimation of phylogeny from DNA sequences when substitution rates differ over sites. Mol. Biol. Evol. 10 1396–1401. 10.1093/oxfordjournals.molbev.a040082 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhuang W.-Y. (2020). Trichoderma brevicrassum strain TC967 with capacities of diminishing cucumber disease caused by Rhizoctonia solani and promoting plant growth. Biol. Control 142:104151 10.1016/j.biocontrol.2019.104151 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.