Abstract
Nucleic acid-based molecular diagnosis has gained special importance for the detection and early diagnosis of genetic diseases as well as for the control of infectious disease outbreaks. The development of systems that allow for the detection and analysis of nucleic acids in a low-cost and easy-to-use way is of great importance. In this context, we present a combination of a nanotechnology-based approach with the already validated dynamic chemical labeling (DCL) technology, capable of reading nucleic acids with single-base resolution. This system allows for the detection of biotinylated molecular products followed by simple detection using a standard flow cytometer, a widely used platform in clinical and molecular laboratories, and therefore, is easy to implement. This proof-of-concept assay has been developed to detect mutations in KRAS codon 12, as these mutations are highly important in cancer development and cancer treatments.
Keywords: Nucleic acid testing (NAT), Fluorescence-activated cell sorting (FACS), Nanotechnology, Dynamic chemical labeling (DCL), Peptide nucleic acid (PNA), KRAS point Mutation
Graphical abstract
1. Introduction
Early, rapid, and effective nucleic acid analysis is currently in demand for the diagnosis of genetically-based and infectious diseases [1,2]. It is a key tool for early diagnosis, accurate prognosis, and control of disease outbreaks. It is noteworthy that it can also be used for treatment management in personalised medicine. The gold standard for nucleic acid analysis is quantitative polymerase chain reaction (qPCR) and reverse-transcription quantitative PCR (RT-qPCR), when RNA is analysed, because of its high specificity, sensitivity, and reproducibility. However, qPCR is not exempt from issues. For instance, during the COVID-19 pandemic, the use, almost exclusively, of PCR-based assays for identifying SARS-CoV-2 created huge problems in terms of material accessibility and the need to work with isolated RNA molecules [3,4]. Thus, developing new methods to rapidly detect and genotype nucleic acids with high specificity is important in the molecular testing field. In this context, technologies that can offer advantages over PCR assays, such as working directly with complex matrix samples while maintaining the high specificity of PCR, are of great interest.
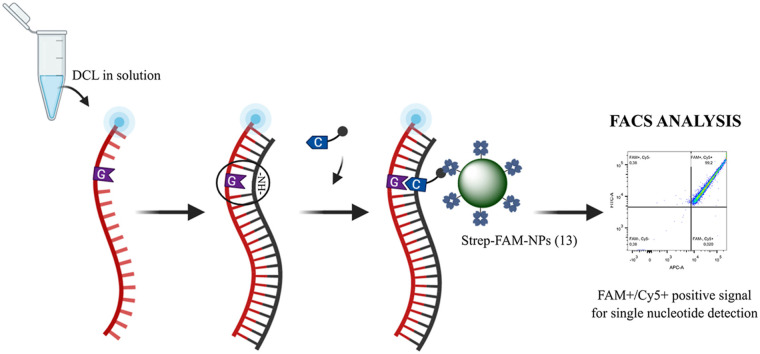
Herein, we have developed a novel strategy for nucleic acid reading with single-base resolution based on the combination of bifunctional polystyrene nanoparticles (PS-NPs) for the capture and analysis of biotinylated products generated by dynamic chemical labelling (DCL) (Fig. 1 A). DCL is an already validated and versatile chemical-based approach that harnesses the Watson and Crick base-pairing rules for templating the dynamic incorporation of an aldehyde-modified nucleobase (SMART-Nucleobase) into the abasic position of an abasic peptide nucleic acid (PNA) probe, also known as a DGL probe (see chemical structure in Supporting Information 1) [5,6]. These two key reagents, the DGL probe and the SMART-Nucleobase, can be modified and tagged, enabling the detection of DCL products by fluorescence, mass spectroscopy, colourimetry, and chemiluminescence. The DCL reaction comprises several steps: in the first step, the DGL probe hybridises with a complementary target nucleic acid forming a DGL:DNA heteroduplex (Fig. 1B, step I). This hybridisation occurs so that the nucleotide under interrogation at the target nucleic acid strand lays opposite to the abasic position of the DGL probe, creating a so-called chemical pocket. In this chemical pocket, a reversible reaction between the aldehyde group of the SMART-Nucleobase and the secondary amine found in the abasic position results in different and unstable iminium intermediate species (Fig. 1B, step II). The intermediate formed following the Watson and Crick base pairing rules is the most stable, allowing the next step, the irreversible reductive amination, which covalently links the SMART-Nucleobase into that position (Fig. 1B, step III). Herein, the DCL product formed will be captured by the PS-NPs for analysis by Fluorescence-Activated Cell Sorting (FACS).
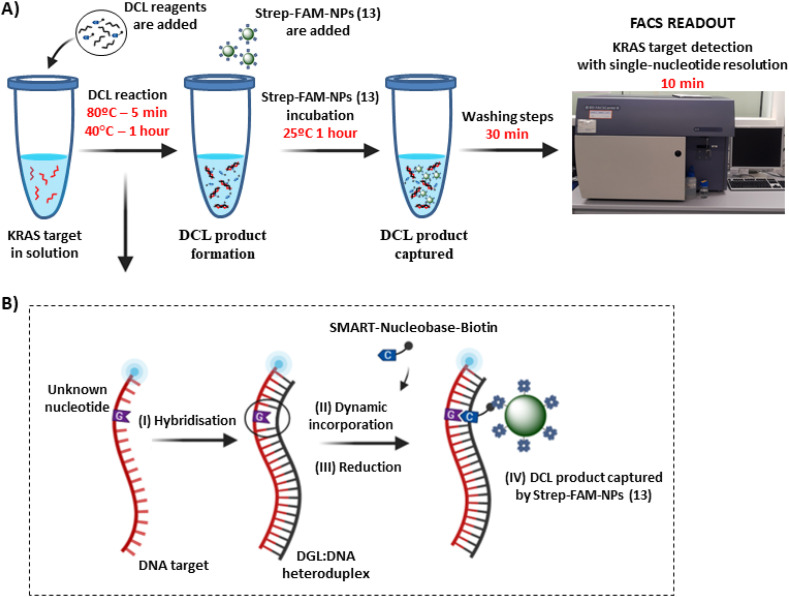
Fig. 1.
Single-nucleotide detection of KRAS using DCL in solution and bifunctional Strep-FAM-NPs (13). A) Work-flow scheme: 1) DCL reagents are added and DCL product is formed; 2) Strep-FAM-NPs (13) are added. These Strep-FAM-NPs (13) capture the DCL product. 3) Washing steps are performed; 4) Readout by FACS analysis. B) Schematic representation of the hybridisation step (I), followed by the dynamic incorporation of the biotin-labelled SMART-Nucleobase (II) and the reductive amination using NaBH3CN (III). Finally, the DCL product is captured by the Strep-FAM-NPs (13) (IV).
One of the advantages of DCL, compared to standard molecular assays, is its ability to detect nucleic acids directly from body fluids without the need to isolate them. Our group has developed several DCL-based approaches that have been successfully applied to different nucleic acid targets using different biological matrices. For instance, the method was used to genotype single nucleotide polymorphisms related to cystic fibrosis [7], to quantify the circulating miRNAs found in the serum of patients [[8], [9], [10], [11], [12], [13]], and to identify parasitic species by identifying single nucleotide differences within highly homologous regions [14,15]. Most of the latest applications of DCL use DGL probes conjugated to solid supports such as nylon membranes and magnetic beads, so that standard reading platforms that are available in molecular biology laboratories could be used [16].
In this work, we aimed to use nanoparticles but as solid supports to capture and detect the formed heteroduplexes rather than as solid supports to conjugate DGL probes. Therefore, the hybridisation processes can take place in solution, which means that they are thermodynamically favourable when compared to solid-phase processes [[17], [18], [19]]. To achieve this, we used PS-NPs due to their robustness and stability in the biological environment, and their compatibility with standard multistep chemistries, allowing different orthogonal conjugation strategies [[20], [21], [22], [23], [24], [25], [26]]. The novel PS-NPs used in this work are bifunctionalised containing both a streptavidin as the high-affinity partner of biotin, and a fluorophore (5 (6)-carboxyfluorescein, -FAM-) as tracker. These bifunctional PS-NPs developed for capturing the DCL products in solution are henceforth referred to as Strep-FAM-NPs (13)).
We performed this proof-of-concept using KRAS codon 12 p. G12C (c.34G > T) mutation, the most frequent mutation in lung adenocarcinoma [27], as the model. The appearance of point mutations in the KRAS gene triggers cancer development, and represents around 20–30% of all mutations found in human tumours, predominantly adenocarcinomas (pancreatic, colorectal, lung, ovarian, cervical, and biliary tract adenocarcinomas) [2,28,29]. About 99% of mutations in KRAS are point mutations in codons 12, 13, and 61 of exon 2. Additionally, the proof-of-concept has been validated to detect miR-122 sequence, a liver-specific miRNA used as a biomarker of liver damage [12,30], and is also associated with hepatocellular carcinoma development [8,12,13] (see Supporting Information 4 for the validation of this system using miR-122 as a target).
2. Materials and methods
2.1. General
All chemicals and solvents were obtained from Sigma Aldrich. SCD buffer was prepared from 2 × saline sodium citrate (SSC) and 0.1% sodium dodecyl sulphate (SDS) with the pH adjusted to 6.0 using HCl. Fmoc-PEG3–COOH and Fmoc-Lys (Dde)-OH were synthesised as previously described [29]. Fmoc-Glycine-OH and Polaxamer 407 were purchased from Sigma Aldrich. Streptavidin was purchased from VWR. SPPS procedures, DGL probes, and DCL reactions were performed in a TS-100 Thermo-Shaker (Biosan). Flow cytometry studies were conducted using a BD FACSCanto II™ and analysed with the FlowJo software (version 10). DCL reactions analyses in BD FACSCanto II™ were performed using the FITC channel, with a 488 nm excitation laser line and emission collected with a 530/30 nm bandpass filter for FAM detection (shown on the Y-axis of dot-plots), and APC channel for Cy5 detection, (shown in the X-axis of dot-plots), with a 633 nm excitation laser line and emission collected with a 670/14 nm bandpass filter. FACS data were analysed using median fluorescence intensity (MFI) (n = 3) in the APC channel, and data were analysed using one-way ANOVA.
2.2. Preparation of Strep-FAM-NPs (13) for capturing DCL products
PS-NPs (1) (360 nm in size) were synthesised following a dispersion polymerisation process as previously reported [31]. Fmoc-Dde-NPs (8) were prepared according to previously described protocols [23]. Briefly, coupling reactions were carried out in dimethylformamide (DMF) using the Fmoc–NH–protected monomer (50 equiv) with oxyma (50 equiv) and N,N′-diisopropylcarbodiimide (DIC) (50 equiv) as coupling agents (see Supporting Information 2 for synthesis route details).
Dual NP functionalization was achieved via the orthogonally protected amine groups on the PS-NPs. Fluorescein-functionalized NPs (FAM-NPs (10)) were obtained by removing the Dde group from Lys followed by the conjugation of FAM through amide bond formation. Then, the Fmoc protecting group was removed, and the free primary amine PS-NPs were treated with a glutaraldehyde solution (25% (w/v) in phosphate buffered saline (PBS) before streptavidin conjugation (60 mM) in PBS (pH 7.4) and treatment with a sodium cyanoborohydride solution (20 mM) in PBS/EtOH (3:1) for 2 h to obtain bifunctionalised Strep-FAM-NPs (13). Finally, Strep-FAM-NPs (13) were washed with PBS and treated with a quenching solution (40 mM ethanolamine with 1% (w/v) BSA in PBS) and 1% (w/v) polaxamer 407 (details are provided in Supporting Information 2).
2.3. Target selection, probe design, and synthesis
Synthetic single-stranded DNA (ssDNA) oligomers mimicking codon 12 of KRAS wild-type sequence and KRAS containing G12C mutation sequence (ssDNA-KRAS-wt-Cy5, ssDNA-KRAS-wt-biotin, and ssDNA-KRAS-G12C-Cy5) were purchased from Sigma Aldrich (see Table 1 for sequences). All sequences were obtained using BLAST analysis [32].
Table 1.
Sequences of ssDNA, PNA, and DGL probes. KRAS codon 12 is shown in bold. The nucleotide under interrogation (nucleotide located in from of the PNA abasic position) is underlined in ssDNA sequences. The abasic position in the DGL probe is shown as *_*. Wt = wild-type sequence. G12C = presence of a mismatch (G > T) corresponding to the KRAS G12C point mutation. X = amino-PEG-linker; HAc = acetyl group; glu = nucleobase containing a propanoic acid side chain at the gamma position. Chemical structures are available in Supporting Information 1.
| Target nucleic acid sequences. |
Sequence (5′–3′) |
| ssDNA-KRAS-wt-Cy5 | Cy5—TTGGAGCTGGTGGCGTAG |
| ssDNA-KRAS-G12C-Cy5 | Cy5—TTGGAGCTTGTGGCGTAG |
| ssDNA-KRAS-wt-Biotin |
Biotin—TTGGAGCTGGTGGCGTAG |
|
PNA probes. |
Sequence (N–C) |
| PNA-KRAS-wt-Biotin | Biotin—xxCTACGCCACCAGCTCCAA |
| PNA-KRAS-wt-Cy5 |
Cy5—xxCTACGCCACCAGCTCCAA |
|
DGL probes. |
Sequence (N–C) |
| DGL-K12RC | HAc –Cys-xxCTACGCCAgluC*_*AGgluCTCCAA |
SMART-Cytosine-PEG12-Biotin (henceforth referred to as SMART-C-Biotin), SMART-Adenine-Deaza-PEG12-Biotin (henceforth referred to as SMART-A-Biotin), PNAs, and DGLs probes were provided by DESTINA Genomica S.L. PNA and DGL probes were synthesised on a Intavis Bioanalytical Instruments MultiPrep CF synthesiser (Intavis AG GmH, Germany). For this, standard SPPS protocols were followed by successive rounds of coupling of previously activated amino-protected PNA monomers, followed by deprotection of the amino terminal protecting group and washes after each step. Once synthesised, the probes were separated from the resin by treatment with trifluoroacetic acid and precipitation with cold ether. Subsequently, they were purified by HPLC and characterised by HPLC and MALDI-TOF. Two PNAs complementary to KRAS labelled with either Cy5 (PNA-KRAS-wt-Cy5) or biotin (PNA-KRAS-wt-Biotin), and one DGL complementary to KRAS (DGL-K12RC) were designed and synthesised (Table 1). Synthetic single-stranded DNA and probes used for the validation of this system using miR-122 sequence as a target are detailed in Supporting Information 4.
2.4. PNA:DNA and DGL:DNA heteroduplex formation and DCL reaction in human plasma
-
a)
PNA:DNA hybridisation. Hybridisation assays were performed by incubating ssDNA (1 μM) and PNA probes (1 μM) in SCD buffer (pH 6) in a 1.5 mL tube placed in a Thermo-Shaker at 80 °C for 5 min, with shaking at 1400 rpm. Then, the reaction was cooled to 41 °C at 3 °C/min followed by incubation for 1 h.
-
b)
DCL reactions. DCL reactions were performed by incubating ssDNA (1 μM) and DGL probe (1 μM) together with SMART-C-biotin (5 μM) under the same conditions described above for PNA:DNA heteroduplex formation. Once the temperature reached 41 °C, sodium cyanoborohydride was added to a final concentration of 1 mM, and the reaction was incubated at 41 °C for 1 h while shaking at 1400 rpm.
To assess the DCL reaction occurs in a complex biological matrix, a calibration curve was performed in diluted human plasma. Five quantities of target (ssDNA-KRAS-wt-Cy5) were spiked in (2.5 pmol, 12.5 pmol, 25 pmol, 37.5 pmol and 50 pmol), followed by the addition of SCD buffer (pH 6; 1:4) containing 1 μM of DGL probe in a total volume of 50 μL. Then, the reaction was mixed and incubated at 80 °C for 5 min, shaken at 1400 rpm, cooled to 41 °C, and incubated for 1 h. The human plasma samples from healthy donors were provided by the Biobank of the Andalusian Public Health System (agreement number S1900297) approved by the Committee of Ethics of Biomedical Research of Andalusia (study code: 32,160,041).
In all cases, after incubation at 41 °C, 5 × 108 Strep-FAM-NPs (13) were added and incubated at 25 °C for 1 h with shaking at 1400 rpm. For more details about the determination of the number of NPs, see Supporting Information 2.7. Finally, the NPs were washed with PBS 0.1% tween (3 × 200 μL; 5 min at 12,000 rpm)), resuspended in 200 μL of PBS, and analysed by flow cytometry.
3. Results and discussion
3.1. Preparation and characterisation of bifunctional Strep-FAM-NPs (13)
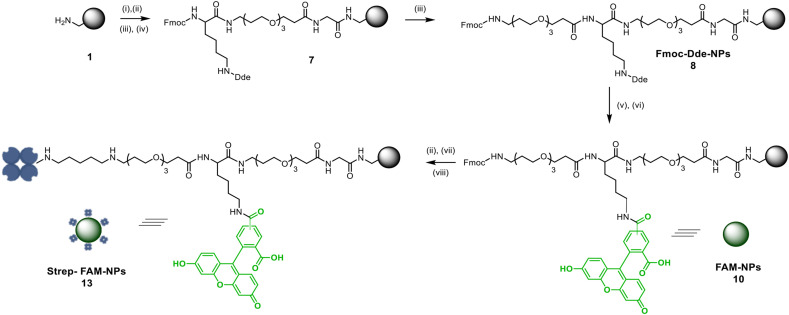
Bifunctional Strep-FAM-NPs (13), designed to capture biotinylated molecular products, were prepared by conjugating the streptavidin moiety and a fluorophore, FAM. These nanoparticles were successfully prepared using standard protocols based on Fmoc solid-phase chemistry with oxyma and N,N-diisopropylcarbodiimide (DIC) as coupling reagents (see synthetic details in Supporting Information 2). The loading of aminomethyl-PS-NPs (1) (0.022 mmol/g) was determined by coupling the Fmoc-Gly OH amino acid. After coupling reactions, their efficiencies were monitored using the Kaiser colorimetric test [33]. To achieve efficient covalent functionalization of both cargoes to PS-NPs in a controlled manner, an orthogonal strategy, based on the full orthogonality between Dde and Fmoc protecting groups was carried out to achieve Fmoc-Dde-NPs (8) (Fig. 2 ). Then, the fluorophore was conjugated by amide bond formation following Dde deprotection (FAM-NPs (10)), and streptavidin was conjugated using glutaraldehyde as a cross-linker, following Fmoc deprotection, to achieve Strep-FAM-NPs (13).
Fig. 2.
Synthetic scheme of aminomethyl PS-NPs bifunctionalisation with FAM and streptavidin (Strep-FAM-NPs (13)). Aminomethyl PS-NPs (1) were functionalized following SPPs standard protocols to obtain bifunctionalised Strep-FAM-NPs (13). Reagents and conditions: (i) Fmoc-Gly-OH, Oxyma, DIC (1400 rpm, 2 h, 60 °C); (ii) 20% piperidine in DMF (1400 rpm, 3 × 20 min, 25 °C); (iii) Fmoc-PEG-OH, Oxyma, DIC (1400 rpm, 2 h, 60 °C); (iv) Fmoc-Lys-Dde (OH), Oxyma, DIC 1400 rpm, 2 h, 60 °C); (v) 3.5 Hydroxylamine, HCl, Imidazole, NMP (1400 rpm, 1 h, 25 °C); (vi) FAM, Oxyma, DIC (1400 rpm, 2 h, 60 °C); (vii) 25% glutaraldehyde in PBS (1400 rpm, 16 h 25 °C); (viii) 0.3 μg/mL streptavidin in PBS (1400 rpm, 25 °C, 16 h).
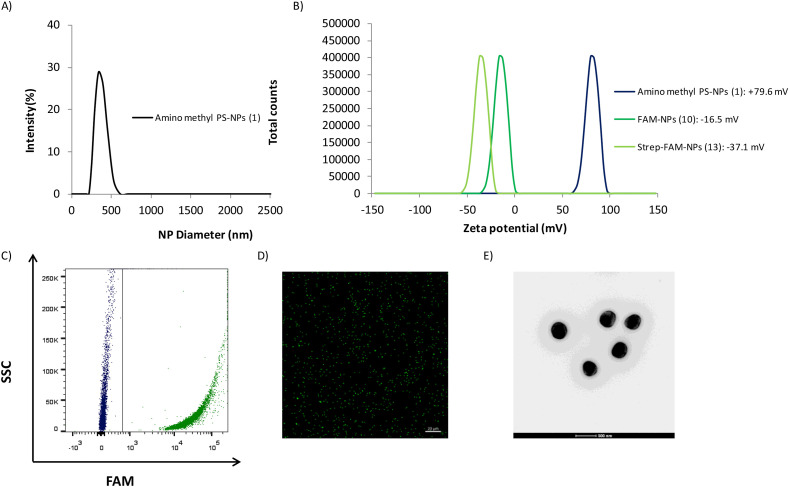
Size distribution was determined by dynamic light scattering (DLS) with a monodisperse nanoparticle population (hydrodynamic diameter was 357.2 ± 0.9 nm with a polydispersity index (PDI) of 0.066) (Fig. 3 A). The zeta potential value of Strep-FAM-NPs (13) in water was slightly negative (−37.1 mV ± 0.8), as compared to FAM-NPs (10) and amino-methyl-NPs (1), which were −16.5 mV ± 0.8 and +79.6 mV ± 0.9, respectively (Fig. 3B). The efficiency of fluorophore conjugation to the nanoparticles was monitored using fluorescence-based techniques. Fig. 3C shows a representative plot of the flow cytometry analysis showing the increase in fluorescence of the Strep-FAM-NPs (13) conjugates concerning unloaded nanoparticles (aminomethyl-PS-NPs (1)). This result was corroborated by confocal microscopy (Fig. 3D), and the size was corroborated by transmission electron microscopy (TEM) analysis (Fig. 3E). The concentration of Strep-FAM-NPs (13) (7,8 × 106 NPs/μL) was calculated according to the spectrophotometric method developed by our group [34] (see more details in Supporting Information 2). The amount of conjugated streptavidin (0.06236 mmol/mL) in the Strep-FAM-NPs (13) was determined by measuring the initial and final concentrations in the reaction vessel using a bicinchoninic acid assay (BCA assay) [35]. To determine the stability of streptavidin conjugation, Strep-FAM-NPs (13) were treated with a 0.4% SDS solution in PBS for 15 min at 70 °C, and the supernatants were analysed by the BCA test, showing the absence of streptavidin and therefore, showing the stability and strength of streptavidin binding to the NPs (see Supporting Information 2.6 for more details).
Fig. 3.
Characterisation of Strep-FAM-NPs (13). (A) Size distribution determined by DLS; (B) Values of the zeta potential of amino-methyl PS-NPs (1) (blue), FAM-NPs (10) (dark green) and Strep-FAM-NPs (13) (green); (C) Scatter plot analysis representative of amino-methyl NPs (1) (blue) and Strep-FAM-NPs (13) (green); (D) Confocal laser microscopy analysis (63 × magnification, 20 μm scale bar, FITC channel) and (E) Evaluation of morphology by transmission electron microscopy (TEM). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Validation of Strep-FAM-NPs (13) to capture biotinylated PNA:DNA heteroduplex
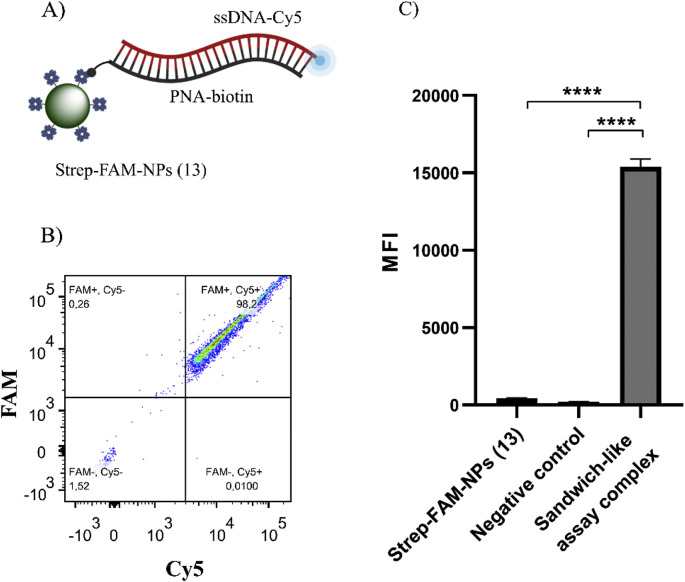
The capacity of Strep-FAM-NPs (13) to capture biotinylated PNA:DNA heteroduplex was evaluated using a sandwich-like assay, and the efficiency was evaluated by flow cytometry analysis. Since Strep-FAM-NPs (13) were FAM-labelled, and the PNA:DNA heteroduplex was fluorescently labelled with Cy5, the efficient binding of the Strep-FAM-NPs (13) to the biotinylated-Cy5-labelled heteroduplex was confirmed by obtaining a double-positive population FAM+/Cy5+. Fig. 4 A shows a schematic of this PNA-Biotin:DNA-Cy5:Strep-FAM-NPs sandwich-like construct. Flow cytometry analysis showed that Strep-FAM-NPs (13) provide a single positive FAM+/Cy5- population and, as a negative control, when the sandwich complex is not complete due to the absence of the biotinylated-labelled probe, the sample run results in an FAM+/Cy5- population, confirming the absence of nonspecific absorption of the target (see Supporting Information 3 for more details). If the biotinylated- and Cy5-labelled heteroduplex is formed, a 98 ± 1% FAM+/Cy5+ population is observed, meaning that this percentage of Strep-FAM-NPs (13) population has captured the heteroduplex (Fig. 4B). FACS results were analysed using MFI (n = 3) in the APC channel, showing a strong MFI increase when the sandwich construct was complete (Fig. 4C). Additionally, this validation has been performed using miR-122 sequence as a target (see Supporting Information 4 for more details).
Fig. 4.
BD FACS results for validation of Strep-FAM-NPs (13) to capture biotinylated PNA:DNA heteroduplex. A) Scheme representing the Strep-FAM-NPs (13) recognizing the biotinylated and Cy5-labelled PNA:DNA heteroduplex. B) FACS results show that the complete sandwich-like assay structure provides an FAM+/Cy5+ population. C) MFI in the APC channel (n = 3) representation and one-way ANOVA analysis. A strong MFI increase is observed when the sandwich-like assay complex is complete.
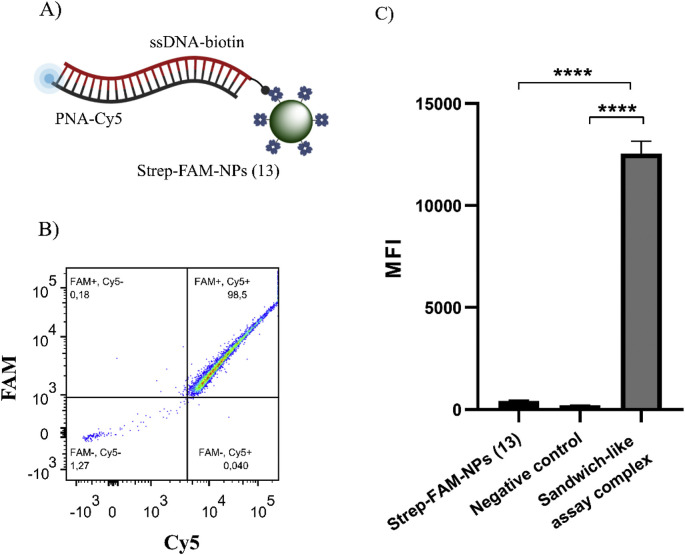
To validate the capacity of the Strep-FAM-NPs (13) to capture any biotinylated target, a second proof-of-concept assay was carried out. For this assay, a biotinylated KRAS nucleic acid sequence (ssDNA-KRAS-wt-biotin) and its complementary PNA labelled with the Cy5 fluorophore (PNA-KRAS-wt-Cy5) were used (see sequences in Table 1). After the formation of the PNA:DNA heteroduplex, the product was captured by Strep-FAM-NPs (13) (Fig. 5 A). Flow cytometry analysis showed that the Strep-FAM-NPs (13) provide an FAM+/Cy5- population similar to the population obtained in the absence of the biotinylated-labelled probe (as a negative control sample) (see Supporting Information 3 for more details). Strep-FAM-NPs (13) can bind to the heteroduplex formed in the presence of the Cy5-labelled PNA probe and its complementary biotinylated ssDNA, resulting in a 97.83 ± 0.70% FAM+/Cy5+ double-positive population (Fig. 5B). The samples were analysed using MFI (n = 3) in the APC channel, showing a strong MFI increase when the sandwich construct was complete (Fig. 5C).
Fig. 5.
BD FACS results for validation of Strep-FAM-NPs (13) to capture biotinylated PNA:DNA heteroduplex, in which DNA is biotinylated. A) Scheme representing the Strep-FAM-NPs (13) recognizing the biotinylated heteroduplex. B) FACS results show that the complete sandwich structure provides an FAM+/Cy5+ population, confirming the DNA detection despite the biotin location. C) MFI in the APC channel (n = 3) representation and one-way ANOVA analysis. A strong MFI increase is observed when the sandwich-like assay complex is complete.
Based on these flow cytometry results obtained using biotinylated molecular products for KRAS, we can confirm that these Strep-FAM-NPs (13) can efficiently capture a PNA:DNA heteroduplex regardless of the biotinylated component position.
3.3. Detection of DCL products by Strep-FAM-NPs (13): KRAS calibration curve in human plasma and KRAS G12C point mutation discrimination
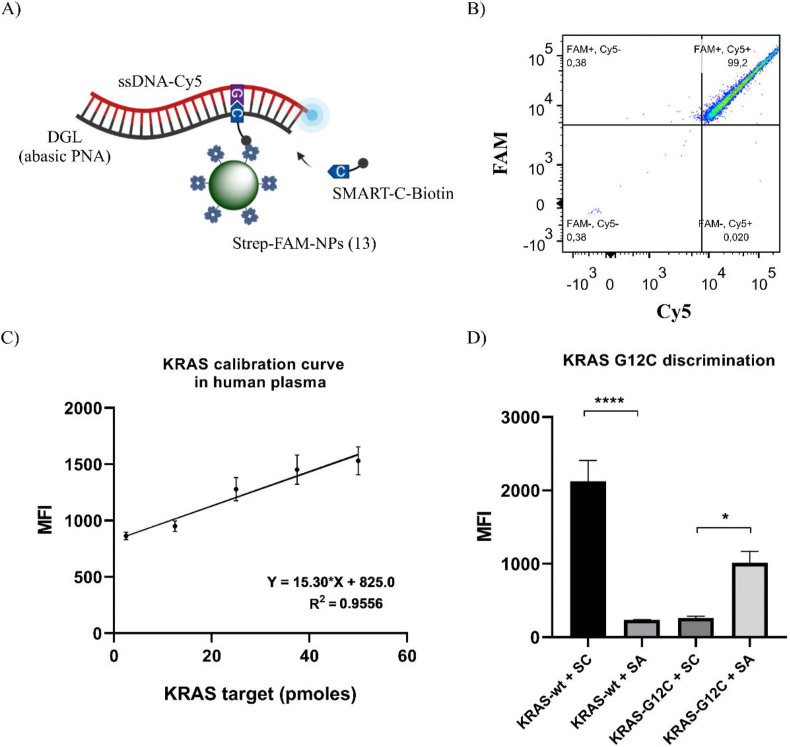
The capacity of the system to detect the KRAS sequence as well as mutations present in codon 12 was evaluated. For this purpose, a DGL probe (DGL-K12RC) was designed to carry an abasic site in front of the first nucleotide of KRAS codon 12. The position of the abasic site means that after hybridisation with a KRAS wild-type nucleic acid sequence, there would be a guanine in front of the abasic position, hence templating the dynamic incorporation of the complementary SMART-C-Biotin. However, when the DGL-K12RC hybridises with a KRAS G12C mutated nucleic acid sequence, there would be a thymine in front of the abasic position, hence templating the dynamic incorporation of the complementary SMART-A-Biotin.
Fig. 6A shows the structure of the DCL product captured by Strep-FAM-NPs (13). The capacity of these bifunctional nanoparticles for capturing the DCL product was evaluated by flow cytometry. Strep-FAM-NPs (13) and the incomplete construct provide an FAM+/Cy5- population (see Supporting Information 3 for more details). When the fluorescent heteroduplex DNA-KRAS-wt-Cy5:DGL-K12RC is formed, SMART-C-Biotin is incorporated into the abasic site and the Strep-FAM-NPs (13) are able to capture the complex, resulting in an 98.23 ± 1.06% FAM+/Cy5+ population (Fig. 6B), indicating the detection of the KRAS-wild-type sequence. Additionally, the detection of DCL products by Strep-FAM-NPs (13) has been validated using miR-122 sequence as a target (see Supporting Information 4 for more details). These results confirm that DCL process has been successfully performed in solution and DCL products were captured by Strep-FAM-NPs (13). Some constraints that affect the efficiency of DNA hybridisation on surfaces such as nonspecific adsorption of the attached oligonucleotides to the solid surface [[17], [18], [19]] have been avoided by using this approach.
Fig. 6.
KRAS detection and G12C point mutation discrimination using DCL in solution and bifunctional Strep-FAM-NPs (13). A) Scheme of the biotinylated DCL product formed. B) FACS analysis: When the SMART-C-Biotin is incorporated into the abasic site, the population is FAM+/Cy5+, indicative of the detection of the KRAS wild-type sequence. C) Calibration curve by synthetic KRAS-wt-Cy5 sequence in human plasma. The dots represent the mean and SEM (n = 3) of MFI in the APC channel. D) Specificity assay for the discrimination of the KRAS G12C point mutation. SC = SMART-C-Biotin. SA = SMART-A-Biotin. MFI values in the APC channel from each condition were compared using one-way ANOVA.
To ensure the analytical capability of Strep-FAM-NPs (13) for capturing and detecting KRAS target in a complex biological medium, a calibration curve was generated by the spike in different quantities of ssDNA-KRAS-wt-Cy5 sequence (2.5 pmol, 12.5 pmol, 25 pmol, 37.5 pmol and 50 pmol) in human plasma, followed by a DCL reaction in a total volume of 50 μL. Each condition was analysed using MFI in the APC channel. The plot of MFI versus the target quantities showed a linear regression, confirming the suitability of the Strep-FAM-NPs (13) for capturing and detecting the wild-type KRAS target sequence (Fig. 6C). The experiments were repeated three times for each condition (n = 3), showing a coefficient of variation ranging from 5% to 15%. The limit of detection was 2.5 ± 0.09 pmoles (12.5 nM) (showing an average MFI signal to background ratio of 2.20 ± 0.08), corresponding to 1.5055E + 12 molecules of target.
To assess the specificity of the system to discriminate the presence of the KRAS G12C point mutation, two DCL reactions were performed using the ssDNA-KRAS-wt-Cy5 sequence and the ssDNA-KRAS-G12C-Cy5 sequence together with both SMART-C-Biotin and SMART-A-Biotin. The results showed a strong MFI increase in the APC channel when the incorporation of the SMART-Nucleobase was specific (SMART-C-Biotin in the KRAS wild-type sequence, and SMART-A-Biotin in the KRAS G12C mutated sequence) (Fig. 6D).
These results have showed that the Strep-FAM-NPs (13) are suitable for capturing nucleic acids in a complex biological matrix and, by using FACS analysis, the system is capable to detect the presence of the KRAS G12C point mutation. The methodology developed in this work can be performed within 3 h by using widely available reading platforms such as the flow cytometer with a limit of detection of 2.5 pmoles (12.5 nM). Although further optimisation has still to be done, for instance to increase the sensibility of this approach, this proof-of-concept could be adaptable and comparable to other existing methods currently used to detect point mutations [[36], [37], [38], [39]].
4. Conclusions
Herein, we have presented the development of bifunctional nanoparticles for capturing DCL products, in which hybridisation processes has been performed in solution-phase, enabling nucleic acid detection with single-base resolution using flow cytometry. This is the first time that DCL processes have been performed in solution (meaning that hybridisation processes are thermodynamically favourable) and DCL products have been read using bead-based platforms. To do so, novel bifunctional nanoparticles, Strep-FAM-NPs (13), containing streptavidin and fluorescein, were developed and characterised. These initial proof-of-concept experiments have demonstrated that these novel bifunctional nanoparticles are able to efficiently capture biotinylated and fluorescently labelled PNA:DNA heteroduplex. The system can detect DCL products in a complex biological matrix such as human plasma, and it is specific to discriminate single-base variations, in this case, discriminating a KRAS wild-type sequence and a KRAS G12C mutated sequence.
The methodology developed in this work can be performed within 3 h by using flow cytometry, widely used in molecular laboratories, with a limit of detection of 2.5 pmoles (12.5 nM). For these reasons, we consider this methodology can be adaptable and comparable to other existing methodologies used to detect point mutations. Furthermore, the wide possibility to conjugate cargoes of different natures such as different fluorophores as well as molecules to the PS-NPs, together with the DCL technology, will lead to the development of novel strategies for nucleic acid analysis with single-base resolution.
Moreover, this study, which can be adapted to other chemistries used in molecular testing, as it enables the capture of biotinylated molecular products produced in solution, opens up opportunities to develop novel assay formats for nucleic acid genotyping and detection of high homologous sequencing directly from body fluids or amplicons obtained by endpoint PCR.
Credit Author Statement
ARR & MALG: Designed, performed the experiments and wrote the paper. RAGC: Performed the experiments. VCC: Designed and characterised the nanoparticles. FJLD: Synthesis of SMART-Nucleobase. JJGM: Probe synthesis and purification. MAF: Designed synthesis pathways for SMART Nucleobases and optimisation of protocols for abasic PNA synthesis. RMSM: Designed the experiments, wrote and edited the paper. JJDM: Designed the experiments, wrote and edited the paper.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was supported by the Spanish Ministry of Economy and Competitiveness (grant number BIO2016-80519). The authors are members of the NANOCARE network (RED2018-102469-T) funded by the STATE INVESTIGATION AGENCY. ARR thanks the Spanish Ministry of Education for PhD funding (scholarship FPU15/06418). FJ López-Delgado thanks the Spanish Ministry of Economy and Competitiveness for the Torres Quevedo fellowship (PTQ-16- 08597). These studies were approved and supported by DestiNA Genomics Ltd. Schemes in Fig. 1, Fig. 4, Fig. 5, Fig. 6 have been partially created using BioRender.com.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.talanta.2021.122092.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Patrinos G.P., Danielson P.B., Ansorge W.J. Elsevier Ltd; 2017. Molecular Diagnostics: Past, Present, and Future. [DOI] [Google Scholar]
- 2.Tan C., Du X. KRAS mutation testing in metastatic colorectal cancer. World J. Gastroenterol. 2012;18:5171–5180. doi: 10.3748/wjg.v18.i37.5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akst J. RNA extraction kits for COVID-19 tests are in short supply in US. The scientist. https://www.the-scientist.com/news-opinion/rna-extraction-kits-for-covid-19-tests-are-inshort-supply-in-us-67250 Updated 11 March 2020. [Accessed 25 Sept 2020]. Available from:
- 4.Fomsgaard Anna S., Rosenstierne Maiken W. An alternative workflow for molecular detection of SARS-CoV-2 – escape from the NA extraction kit-shortage, Copenhagen, Denmark. Euro Surveill. 2020;25(14) doi: 10.2807/1560-7917.ES.2020.25.14.2000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowler F.R., Diaz-Mochon J.J., Swift M.D., Bradley M. DNA analysis by dynamic chemistry. Angew. Chem. Int. Ed. 2010;49:1809–1812. doi: 10.1002/anie.200905699. [DOI] [PubMed] [Google Scholar]
- 6.Bradley M., Díaz-Mochón J.J. 2009. Nucleobase Characterisation. WO2009037473 A2. [Google Scholar]
- 7.Bowler F.R., Reid P.A., Boyd C., Diaz-Mochon J.J., Bradley M. Dynamic chemistry for enzyme-free allele discrimination in genotyping by MALDI-TOF mass spectrometry. Anal. Methods. 2011;3:1656–1663. doi: 10.1039/c1ay05176h. [DOI] [Google Scholar]
- 8.Rissin D.M., López-Longarela B., Pernagallo S., Ilyine H., Vliegenthart A.D.B., Dear J.W., Díaz-Mochón J.J., Duffy D.C. Polymerase-free measurement of microRNA-122 with single base specificity using single molecule arrays: detection of drug-induced liver injury. PloS One. 2017;12:1–15. doi: 10.1371/journal.pone.0179669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marín-Romero A., Robles-Remacho A., Tabraue-ChAvez M., López-Longarela Ba, Sánchez-Martín R.M., Guardia-Monteagudo J.J., Fara M.A., López-Delgado F.J., Pernagallo S., Díaz-Mochón J.J. A PCR-free technology to detect and quantify microRNAs directly from human plasma. Analyst. 2018;143:5676–5682. doi: 10.1039/c8an01397g. [DOI] [PubMed] [Google Scholar]
- 10.Delgado-Gonzalez A., Robles-Remacho A., Marin-Romero A., Detassis S., Lopez-Longarela B., Lopez-Delgado F.J., de Miguel-Perez D., Guardia-Monteagudo J.J., Fara M.A., Tabraue-Chavez M., Pernagallo S., Sanchez-Martin R.M., Diaz-Mochon J.J. PCR-free and chemistry-based technology for miR-21 rapid detection directly from tumour cells. Talanta. 2019;200:51–56. doi: 10.1016/j.talanta.2019.03.039. [DOI] [PubMed] [Google Scholar]
- 11.Detassis S., Grasso M., Tabraue-Chávez M., Marín-Romero A., López-Longarela B., Ilyine H., Ress C., Ceriani S., Erspan M., Maglione A., Díaz-Mochón J.J., Pernagallo S., Denti M.A. New platform for the direct profiling of microRNAs in biofluids. Anal. Chem. 2019;91:5874–5880. doi: 10.1021/acs.analchem.9b00213. [DOI] [PubMed] [Google Scholar]
- 12.López-Longarela B., Morrison E.E., Tranter J.D., Chahman-Vos L., Léonard J.F., Gautier J.C., Laurent S., Lartigau A., Boitier E., Sautier L., Carmona-Saez P., Martorell-Marugan J., Mellanby R.J., Pernagallo S., Ilyine H., Rissin D.M., Duffy D.C., Dear J.W., Díaz-Mochón J.J. Direct detection of miR-122 in hepatotoxicity using dynamic chemical labeling overcomes stability and isomiR challenges. Anal. Chem. 2020;92:3388–3395. doi: 10.1021/acs.analchem.9b05449. [DOI] [PubMed] [Google Scholar]
- 13.Marín-Romero A., Tabraue-Chávez M., Dear J.W., Sánchez-Martín R.M., Ilyine H., Guardia-Monteagudo J.J., Fara M.A., López-Delgado F.J., Díaz-Mochón J.J., Pernagallo S. Amplification-free profiling of microRNA-122 biomarker in DILI patient serums, using the luminex MAGPIX system. Talanta. 2020;219:121265. doi: 10.1016/j.talanta.2020.121265. [DOI] [PubMed] [Google Scholar]
- 14.Angélica Luque-González M., Tabraue-Chávez M., López-Longarela B., María Sánchez-Martín R., Ortiz-González M., Soriano-Rodríguez M., Antonio García-Salcedo J., Pernagallo S., José Díaz-Mochón J. Identification of Trypanosomatids by detecting Single Nucleotide Fingerprints using DNA analysis by dynamic chemistry with MALDI-ToF. Talanta. 2018;176:299–307. doi: 10.1016/j.talanta.2017.07.059. [DOI] [PubMed] [Google Scholar]
- 15.Tabraue-Chávez M., Luque-González M.A., Marín-Romero A., Sánchez-Martín R.M., Escobedo-Araque P., Pernagallo S., Díaz-Mochón J.J. A colorimetric strategy based on dynamic chemistry for direct detection of Trypanosomatid species. Sci. Rep. 2019;9:1–13. doi: 10.1038/s41598-019-39946-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venkateswaran S., Luque-González M.A., Tabraue-Chávez M., Fara M.A., López-Longarela B., Cano-Cortes V., López-Delgado F.J., Sánchez-Martín R.M., Ilyine H., Bradley M., Pernagallo S., Díaz-Mochón J.J. Novel bead-based platform for direct detection of unlabelled nucleic acids through Single Nucleobase Labelling. Talanta. 2016;161:489–496. doi: 10.1016/j.talanta.2016.08.072. [DOI] [PubMed] [Google Scholar]
- 17.Ravan H., Kashanian S., Sanadgol N., Badoei-Dalfard A., Karami Z. Strategies for optimizing DNA hybridization on surfaces. Anal. Biochem. 2014;444:41–46. doi: 10.1016/j.ab.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 18.Irving D., Gong P., Levicky R. DNA surface hybridization: comparison of theory and experiment. J. Phys. Chem. B. 2010;114:7631–7640. doi: 10.1021/jp100860z. [DOI] [PubMed] [Google Scholar]
- 19.Levicky R., Horgan A. Physicochemical perspectives on DNA microarray and biosensor technologies. Trends Biotechnol. 2005;23:143–149. doi: 10.1016/j.tibtech.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Yusop R.M., Unciti-Broceta A., Johansson E.M.V., Sánchez-Martín R.M., Bradley M. Palladium-mediated intracellular chemistry. Nat. Chem. 2011;3:239–243. doi: 10.1038/nchem.981. [DOI] [PubMed] [Google Scholar]
- 21.Tsakiridis A., Alexander L.M., Gennet N., Sanchez-Martin R.M., Livigni A., Li M., Bradley M., Brickman J.M. Microsphere-based tracing and molecular delivery in embryonic stem cells. Biomaterials. 2009;30:5853–5861. doi: 10.1016/j.biomaterials.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez-Martin R.M., Alexander L., Muzerelle M., Cardenas-Maestre J.M., Tsakiridis A., Brickman J.M., Bradley M. Microsphere-mediated protein delivery into cells. Chembiochem. 2009;10:1453–1456. doi: 10.1002/cbic.200900136. [DOI] [PubMed] [Google Scholar]
- 23.Cárdenas-Maestre J.M., Pérez-Lõpez A.M., Bradley M., Sánchez-Martín R.M. Microsphere-based intracellular sensing of caspase-3/7 in apoptotic living cells, Macromol. Biosci. 2014;14:923–928. doi: 10.1002/mabi.201300525. [DOI] [PubMed] [Google Scholar]
- 24.Altea-Manzano P., Unciti-Broceta J.D., Cano-Cortes V., Ruiz-Blas M.P., Valero-Griñan T., Diaz-Mochon J.J., Sanchez-Martin R. Tracking cell proliferation using a nanotechnology-based approach. Nanomedicine. 2017;12:1591–1605. doi: 10.2217/nnm-2017-0118. [DOI] [PubMed] [Google Scholar]
- 25.Borger J.G., Cardenas-Maestre J.M., Zamoyska R., Sanchez-Martin R.M. Novel strategy for microsphere-mediated DNA transfection. Bioconjugate Chem. 2011;22:1904–1908. doi: 10.1021/bc200289n. [DOI] [PubMed] [Google Scholar]
- 26.Cano-Cortes M.V., Navarro-Marchal S.A., Ruiz-Blas M.P., Diaz-Mochon J.J., Marchal J.A., Sanchez-Martin R.M. A versatile theranostic nanodevice based on an orthogonal bioconjugation strategy for efficient targeted treatment and monitoring of triple negative breast cancer. Nanomed. Nanotechnol. Biol. Med. 2020;24:102120. doi: 10.1016/j.nano.2019.102120. [DOI] [PubMed] [Google Scholar]
- 27.Li S., Balmain A., Counter C.M. A model for RAS mutation patterns in cancers: finding the sweet spot. Nat. Rev. Canc. 2018;18:767–777. doi: 10.1038/s41568-018-0076-6. [DOI] [PubMed] [Google Scholar]
- 28.Rajalingam K., Schreck R., Rapp U.R., Albert Š. Ras oncogenes and their downstream targets. Biochim. Biophys. Acta Mol. Cell Res. 2007;1773:1177–1195. doi: 10.1016/j.bbamcr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 29.Schubbert S., Shannon K., Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Canc. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 30.Starkey Lewis P.J., Dear J., Platt V., Simpson K.J., Craig D.G.N., Antoine D.J., French N.S., Dhaun N., Webb D.J., Costello E.M., Neoptolemos J.P., Moggs J., Goldring C.E., Park B.K. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology. 2011;54:1767–1776. doi: 10.1002/hep.24538. [DOI] [PubMed] [Google Scholar]
- 31.Unciti-Broceta A., Johansson E.M.V., Yusop R.M., Sánchez-Martín R.M., Bradley M. Synthesis of polystyrene microspheres and functionalization with pd0 nanoparticles to perform bioorthogonal organometallic chemistry in living cells. Nat. Protoc. 2012;7:1207–1218. doi: 10.1038/nprot.2012.052. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J., Madden T.L. PowerBLAST: a new network BLAST application for interactive or automated sequence analysis and annotation. Genome Res. 1997;7:649–656. doi: 10.1101/gr.7.6.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaiser E., Colescott R.L., Bossinger C.D., Cook P.I. Color test for detection of free terminal amino groups in the solid-phase synthesis of peptides. Anal. Biochem. 1970;34:595–598. doi: 10.1016/0003-2697(70)90146-6. [DOI] [PubMed] [Google Scholar]
- 34.Unciti-Broceta J.D., Cano-Cortés V., Altea-Manzano P., Pernagallo S., Díaz-Mochón J.J., Sánchez-Martín R.M. Number of nanoparticles per cell through a spectrophotometric method - a key parameter to assess nanoparticle-based cellular assays. Sci. Rep. 2015;5:1–10. doi: 10.1038/srep10091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith P.K., Krohn R.I., Hermanson G.T., Mallia A.K., Gartner F.H., Provenzano M.D., Fujimoto E.K., Goeke N.M., Olson B.J., Klenk D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 36.Nordgård O., Oltedal S., Janssen E.A.M., Gilje B., Kørner H., Tjensvoll K., Smaaland R. Comparison of a PNA clamp PCR and an ARMS/Scorpion PCR assay for the detection of K-ras mutations. Diagn. Mol. Pathol. 2012;21:9–13. doi: 10.1097/PDM.0b013e31821e59dc. [DOI] [PubMed] [Google Scholar]
- 37.Van Haele M., Vander Borght S., Ceulemans A., Wieërs M., Metsu S., Sagaert X., Weynand B. Rapid clinical mutational testing of KRAS, BRAF and EGFR: a prospective comparative analysis of the Idylla technique with high-throughput next-generation sequencing. J. Clin. Pathol. 2020;73:35–41. doi: 10.1136/jclinpath-2019-205970. [DOI] [PubMed] [Google Scholar]
- 38.Matsunaga M., Kaneta T., Miwa K., Ichikawa W., Fujita K.I., Nagashima F., Furuse J., Kage M., Akagi Y., Sasaki Y. A comparison of four methods for detecting KRAS mutations in formalin-fixed specimens from metastatic colorectal cancer patients. Oncol. Lett. 2016;12:150–156. doi: 10.3892/ol.2016.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolton L., Reiman A., Lucas K., Timms J., Cree I.A. KRAS mutation analysis by PCR: a comparison of two methods. PloS One. 2015;10:1–13. doi: 10.1371/journal.pone.0115672. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.