Abstract
We hypothesized that human genes differ by their sensitivity to UV exposure. We used somatic mutations detected by genome-wide screens in melanoma and reported in the Catalog Of Somatic Mutations In Cancer (COSMIC).
As a measure of UV sensitivity, we used the number of silent mutations generated by C>T transitions in pyrimidine dimers of a given transcript divided by the number of potential sites for this type of mutations in the transcript. We found that human genes varied by UV sensitivity by two orders of magnitude. We noted that the melanoma associated tumor suppressor gene CDKN2A was among the top five most UV-sensitive genes in the human genome. Melanoma driver genes have a higher UV-sensitivity compared to other genes in the human genome. The difference was more prominent for tumor suppressors compared to oncogene. The results of this study suggest that differential sensitivity of human transcripts to UV light may explain melanoma specificity of some driver genes. Practical significance of the study relates to the fact that differences in UV sensitivity among human genes need to be taken into consideration while predicting melanoma-associated genes by the number of somatic mutations detected in a given gene.
Keywords: UV exposure, mutation rate, melanoma, COSMIC, CDKN2A
1 |. INTRODUCTION
Cutaneous melanoma shows the largest number of somatic mutations per tumor compared to other cancer types (Martincorena & Campbell, 2015). The mutational spectrum of melanoma mutations is characterized by a UV-mutational signature (Alexandrov, Nik-Zainal, Wedge, Campbell, & Stratton, 2013; Petljak et al., 2019). The single most prevalent type of mutations associated with UV signature is cytosine (C) → thymine (T) at dipyrimidine sites through deamination of cytosine-containing cyclobutane pyrimidine dimers (Ikehata & Ono, 2011).
Silent mutations detected in tumor samples, including melanoma, are the second (after missense) most common type of somatic variants. Silent mutations do not change the amino acid sequence. Even though there are anecdotal examples of functionality of silent mutations (Bali & Bebok, 2015; Pagani, Raponi, & Baralle, 2005), the absolute majority of them are expected to be functionally neutral which makes them highly suitable for assessing gene sensitivity to environmental mutagens. Even though mutations in noncoding regions are also mostly functionally neutral they cannot be used for comparative analysis of human transcripts.
The Catalog Of Somatic Mutations In Cancer (COSMIC) (Forbes et al., 2010; Tate et al., 2019) is the largest well annotated collection of somatic variants detected in tumor samples. A considerable fraction of COSMIC data is generated by whole genome screens, making them suitable for comparative analysis of mutability of human genes.
A number of local chromosomal features including nucleosomes, transcription factor binding sites, chromatin accessibility, replication timing and DNA strand have been shown to influence mutation rate (Gonzalez-Perez, Sabarinathan, & Lopez-Bigas, 2019; Morganella et al., 2016; Tomkova, Tomek, Kriaucionis, & Schuster-Bockler, 2018). These studies suggest, therefore, that human genes may possess intrinsically different UV sensitivity.
The goal of this study was to assess the variation in UV sensitivity among human genes and to compare UV sensitivity of melanoma drivers with UV sensitivity of other genes in the human genome.
2 |. MATERIALS AND METHODS
2.1 |. Data
We used somatic mutations reported in the Catalog Of Somatic Mutations In Cancer (COSMIC) (Forbes et al., 2010; Tate et al., 2019). The database was accessed December 26, 2019. To account for possible bias related to unequal targeting of different genes, we used only confirmed somatic mutations from whole genome mutational screens. Mutational data from 15 melanoma studies were used in the analysis (Abaan et al., 2013; Aydin et al., 2014; Berger et al., 2012; Dahlman et al., 2012; Furney et al., 2012; Hodis et al., 2012; Krauthammer et al., 2015; Krauthammer et al., 2012; Kuckein, 1980; Nikolaev et al., 2011; Pleasance et al., 2010; Sanborn et al., 2015; Shain et al., 2015; Wagle et al., 2014; Wei et al., 2011). In total, those studies produced 334,625 unique mutations detected in 733 samples.
2.2 |. Estimations of the number of potential mutational sites for UV-induced mutations
We used transcript IDs provided by COSMIC to retrieve corresponding nucleotide sequences from the Consensus CDS (CCDS) database (Pujar et al., 2018). In total 38,721 unique transcripts linked to 17,924 human genes were identified and used in the analysis. Since UV-mutational signature is context dependent (Lindberg, Bostrom, Elliott, & Larsson, 2019), and the majority of UV-induced substitutions occur in di-pyrimidines (Alexandrov et al., 2013; Petljak et al., 2019), we have identified the number of C>Ts for each of the 16 possible trinucleotides with “C” in the middle. We have also estimated numbers of potential sites where C>T transition in pyrimidine dimers if it happened would produce a silent mutation. These estimates provided us with the numbers of potential sites for UV-induced silent mutations for each individual transcript. To estimate the observed numbers of UV-induced (C>T transitions in pyrimidine dimers) silent mutations we have used COSMIC silent mutations detected by whole genome screens. The data on the number of potential sites and observed number of silent mutations in trinucleotides can be found in supplementary materials (Supplementary Table S1).
2.3 |. Estimation of UV sensitivity
Each human transcript was scanned across all possible trinucleotides with one nucleotide step, which allowed us to identify all trinucleotides in the transcript with “C” in the middle. Among trinucleotides with “C” in the middle we identified trinucleotides where substitution of middle/core “C” to “T” leads to a silent mutation. That gives us the number of potential sites for silent mutation-producing C>T transitions for each of 16 possible trinucelotides in each transcript. As a quantitative measure of UV-sensitivity of a transcript we have used the ratio of the observed number of silent mutation producing C>T substitutions in trinucleotides with pyrimidine dimers: ACC, ACT, CCA, CCC, CCG, CCT, GCC, GCT, TCA, TCC, TCG, and TCT to the number of potential sites in individual transcripts for those type of mutations. Thus UV sensitivity was estimated by using the following formula: UVs(t)=Nobs(C>T silent)/Nsites(C>T silent): where UVs(t) is a UV sensitivity of a given transcript, Nobs(C>T silent) – the observed number of silent mutations generated by C>T substitutions in pyrimidine dimers in a given transcript, Nsites(C>T silent) – the number potential sites in a given transcript where C>T substitutions in pyrimidine dimers would produce silent mutations. C>T substitutions in trinucleotides without pyrimidine dimers: ACA, ACG, GCA, and GCG were considered as not UV-induced and were analyzed separately.
3 |. RESULTS
3.1 |. Human genes differ by UV-sensitivity
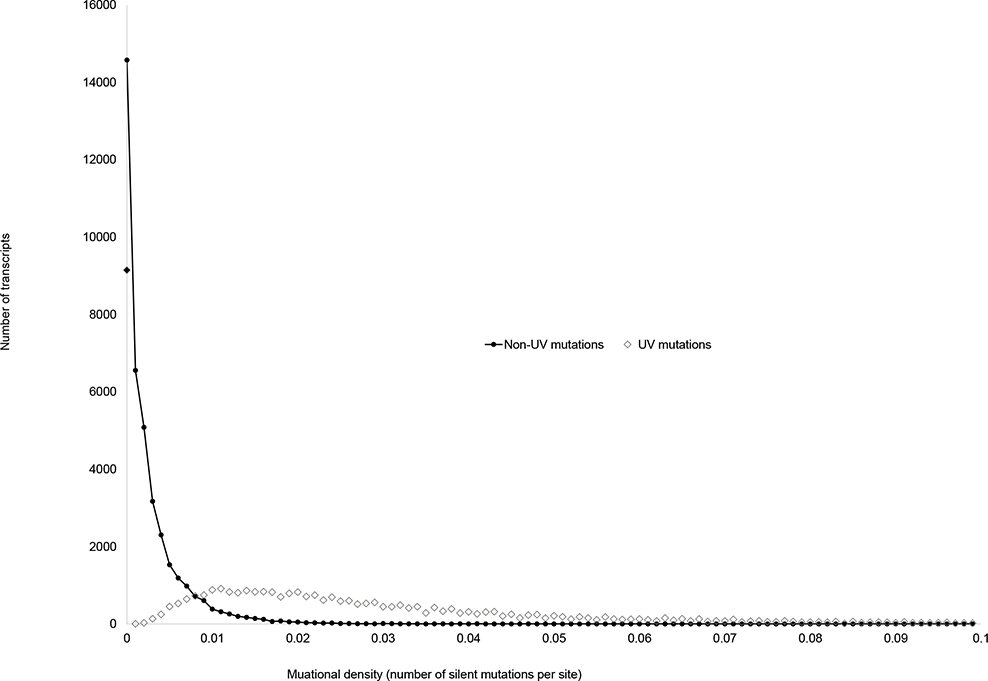
Figure 1 shows the distribution of human transcripts by densities of UV-induced and non UV-induced C>T substitutions. For UV-induced mutations average transcript mutability was 0.02428±0.0001 and for non-UV-induced mutations it was 0.0025±0.00002 (t-test =150.5, N=77,442, p=2.2×10−86). Human transcripts were categorized by mutational densities using 0.001 increment, so the first group included genes with estimated density between 0 and 0.001, the second group included genes with estimated density 0.001–0.002 and so on. We observed drastic differences between the two distributions the density of non UV-induced C>T mutations clustering near zero and the density of UV-induced C>T transitions clustering near 0.01. A relatively large number of observations for UV-induced mutations in the density group between 0 and 0.001 (solid diamond) can be a result of insufficient statistical power for estimation of mutational densities for small human genes. We noted that transcripts without reported silent mutations were two times smaller compared to the genes with at least one reported silent mutation: 951±6 versus 1,924±12. It is likely, therefore, that currently available sample size may be too small to estimate of the densities of silent mutations in small genes reliably.
Figure 1.
The distribution of human transcripts by densities of UV-induced (open diamonds) and not UV-induced (solid circles) silent mutations. Two distributions are drastically different with the average density of UV-induced silent mutations being about ten times higher compared to the average density of non UV-induced silent mutations. In total, 97,435 mutations detected in 733 samples from 15 studies were used in the analysis.
The top 10 most UV-sensitive genes listed from higher to lower UV-sensitivity were CDKN2A, PCP4, POM121L12, STATH, OR13C4, S100Z, OR4C3, HIGD1A, OR13C8, and OR4K2. Supplementary Table S1 shows the assessed UV-sensitivities for individual transcripts.
3.2 |. UV-induced mutations in melanoma driver genes
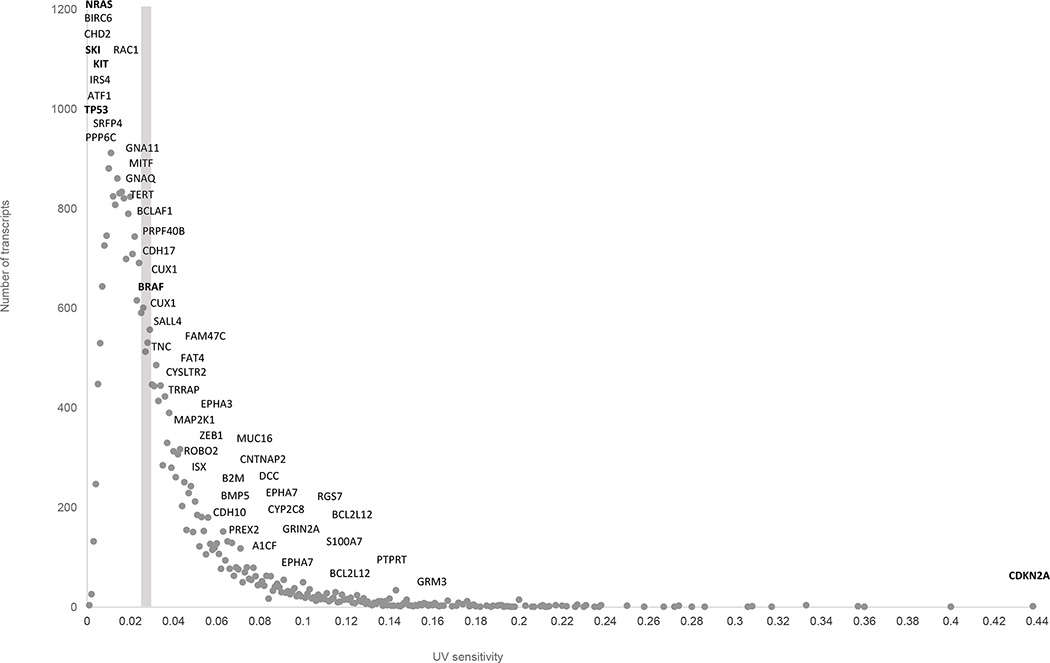
Figure 2 shows the distribution of genes by UV sensitivity. We used the same categorization of transcripts by UV sensitivity (0.001 increment) as for Figure 1. The majority of melanoma drivers have a higher than average UV sensitivity. CDKN2A is an extreme example: this gene shows twenty times higher UV sensitivity compared to overall average UV sensitivity.
Figure 2.
The distribution of human transcripts by UV-sensitivity. Dark gray circles indicate the number of transcripts in a given category of UV sensitivity. Names of known melanoma genes are shown with the positions corresponding to their UV-sensitivity. The gray vertical bar indicates the average UV sensitivity for all human transcripts.
3.3 |. Nucleotide context analysis
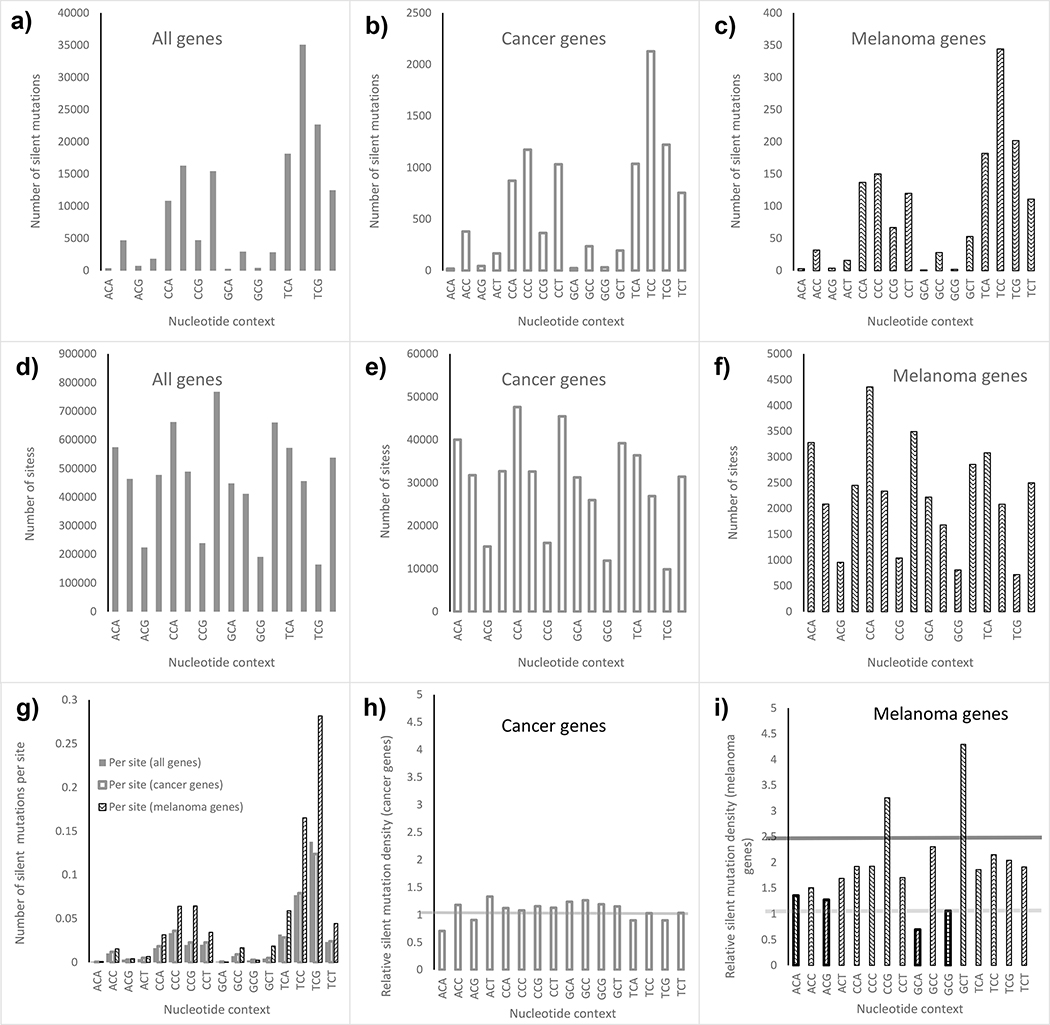
It is known that mutability is generally context dependent (Rogozin, Malyarchuk, Pavlov, & Milanesi, 2005). For that reason, for the transcript level analysis we estimated UV sensitivity of individual transcripts by taking into account nucleotide context. Figure 3 summarizes the results of the analysis. Melanoma drivers were analyzed separately from other cancer genes. Melanoma drivers were then compared to all human genes. We found that distributions of the observed numbers of silent mutations generated by the core C>T substitutions across 16 genotypes are similar across three groups of genes (Figure 3, upper panel). The distributions of the number of potential sites for silent mutations resulting from C>T transitions in core “Cs” were also similar between 3 groups (middle panel, figures 3d–f). However the relative densities of silent mutations among melanoma drivers was higher compared to the density of silent mutations for other cancer genes, as well as for all genes in the human genome (Figure 3g). We also computed relative silent mutation densities using all human transcripts as a reference group. If the relative density is close to one the mutational density in the group of the genes is similar to a typical gene in the human genome. For all known cancer related genes mutational densities were similar to the average human gene (Figure 3h). However melanoma drivers show a much higher density of silent mutation (Figure 3i). This is true only for C>T transitions in pyrimidine dimers (UV-induced substitutions) while non UV-induced substitutions (those occurring in non-pyrimidine triplets: ACA, ACG, GCA and GCG) do not differ from the whole genome average (yellow horizontal line, Figure 3i).
Figure 3.
a-c) The number of reported silent mutations in 16 possible trinucleotides with “C” in the middle for different categories of genes. d-f) The number of sites of silent mutations in 16 possible trinucleotides with “C” in the middle for different categories of genes. g) The numbers of silent mutations per site across trinucleotides across different gene categories. h-i) Relative mutation densities in all COSMIC-defined cancer genes (h) and melanoma genes (i). For melanoma genes we separately analyzed non UV-induced (light bars) and UV-induced (dark bars) C>T substitutions producing silent mutations. Horizontal lines shows averages for non UV-induced (light gray) and UV-induced (dark gray) silent mutations.
3.4 |. UV-sensitivity of tumor suppressors and oncogenes
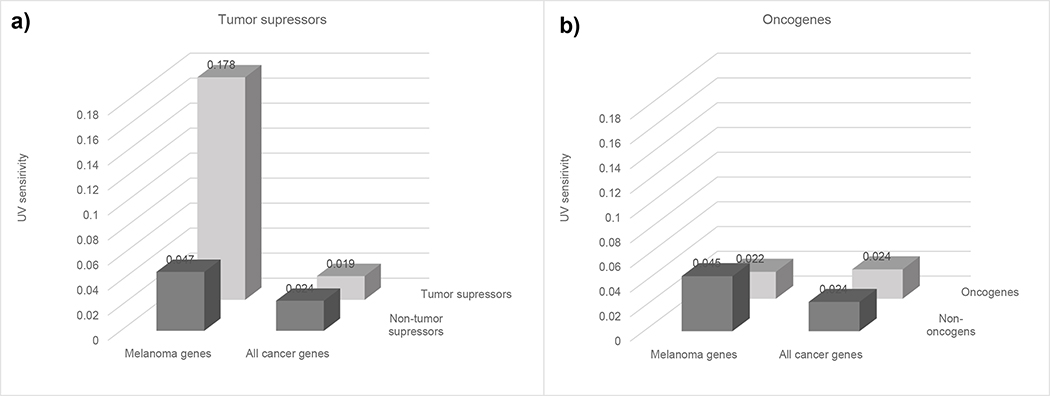
We further investigated UV sensitivity of melanoma drivers by subdividing them into tumor suppressors (TSs) and oncogenes. The majority of functional mutations in tumor suppressors are expected to be loss-of-function mutations while the majority of functional mutations in oncogenes are expected to be gain-of-function mutations. Since it is much easier to destroy a function by random mutations than to generate a novel function, loss-of-function mutations that are predominant in TS tend to occur at multiple sites. Gain-of-function mutations, on the other hand, are more specific and tend to occur at a single position. Therefore, one can expect that TSs will show a stronger association with UV-sensitivity compared to oncogenes. We estimated UV-sensitivities of melanoma-associated TSs and melanoma oncogenes and compared them with UV sensitivities of all known TSs and oncogenes. We used UniprotKB database annotations (UniProt, 2019) to identify human tumor suppressors and oncogenes. Figure 4 shows the results of the analysis. The average UV sensitivity of known melanoma TSs: CDKN2A, PTPRT, GRIN2A, DCC, and GRM3 was 0.178±0.042, which was significantly higher than the overall average - 0.024±0.001, as well as the average UV sensitivity of all TSs 0.024±0.04: corresponding t-tests were 3.6 and 3.8 and corresponding p-values 0.0006 and 0.0002. Melanoma specific and all-cancer oncogenes showed similar UV sensitivities (Figure 4b).
Figure 4.
UV sensitivity of tumor suppressors and oncogenes grouped by cancer specificity (melanoma versus all cancers).
3.5 |. Partitioning of melanoma mutations by mutational signatures
We used Mutational Signatures in Cancer (MuSiCa) (Diaz-Gay et al., 2018) software to classify melanoma mutations according to contributions of known mutational signatures. All mutations detected by genome wide screens were used in the analysis. We found that the absolute majority of mutations (76.4%) are assigned to the signature 7 - “UV light” (Table 1). Aside from UV light, several other signatures were found including age, POLE and alkylating agents. At the same time, the proportion of C>T transitions in pyrimidine dimers (the type of mutations we considered to be UV light-induced) among all mutations was 80.1%. The vast majority of mutations appear UV-light related.
Table 1.
Partitioning of genome wide screen detected somatic mutations in cutaneous melanoma by known mutational signatures.
| Signature | Proposed Etiology | % of mutations |
|---|---|---|
| 7 | UV light | 0.764 |
| 23 | Unknown (liver cancer) | 0.069 |
| 1 | Age | 0.062 |
| 10 | POLE (ultra-hypermutation) | 0.034 |
| 11 | Alkylating agents | 0.022 |
| 12 | Unknown (liver cancer) | 0.01 |
| 17 | Unknown (different cancers) | 0.009 |
| 2 | APOBEC | 0.008 |
| 21 | Unknown (stomach cancer / MSI) | 0.007 |
| 4 | Smoking | 0.006 |
| 3 | BRCA1 / BRCA2 (failure of DNA DSBR / large INDELs) | 0.005 |
| 22 | Aristolochic acid | 0.004 |
| 5 | Unknown (all cancer types) | 0 |
| 6 | Defective DNA MMR / MSI (small INDELs) | 0 |
| 8 | Unknown (breast cancer and medulloblastoma) | 0 |
| 9 | POLH (CLL, BCL) | 0 |
| 13 | APOBEC | 0 |
| 14 | Unknown (uterine cancer and glioma / hypermutation) | 0 |
| 15 | Defective DNA MMR (small INDELs) | 0 |
| 16 | Unknown (liver cancer) | 0 |
| 18 | Unknown (different cancers) | 0 |
| 19 | Unknown (pilocytic astrocytoma) | 0 |
| 20 | Defective DNA MMR (small INDELs) | 0 |
| 24 | Aflatoxin | 0 |
| 25 | Unknown (Hodgkin lymphoma) | 0 |
| 26 | Defective DNA MMR (small INDELs) | 0 |
| 27 | Unknown (kidney clear cell carcinomas / small INDELs) | 0 |
| 28 | Unknown (stomach cancer) | 0 |
| 29 | Tobacco chewing | 0 |
| 30 | Unknown (breast cancer) | 0 |
4 |. DISCUSSION
We found a large variation in UV sensitivity among human transcripts (Figures 1 and 2). Cyclin Dependent Kinase Inhibitor 2A, CDKN2A (transcripts ENST00000530628 and ENST00000579755) was identified as a having a highest UV sensitivity. CDKN2A is a key gene controlling cell cycle progression. CDKN2A regulates expression of TP53 (Foulkes, Flanders, Pollock, & Hayward, 1997; Zhao, Choi, Lee, Bode, & Dong, 2016). TP53 plays a central role in cell survival (Deben, Deschoolmeester, Lardon, Rolfo, & Pauwels, 2016; Yamamoto & Iwakuma, 2018). Based on the role of CDKN2A in regulation of cell cycle/apoptosis and its unusually high UV-sensitivity one can hypothesize that inactivation of CDKN2A by UV-induced mutations is often the first driving step in melanoma development. Inactivation of CDKN2A will lead to a higher cell proliferation and downregulation of apoptosis in the mutant clone. Increased cell proliferation and decreased apoptosis will accelerate accumulation of mutations in other driver genes further driving tumorigenesis.
We and others demonstrated that several germline polymorphisms in CDKN2A are associated with increased risk of melanoma (Begg et al., 2005; Berwick et al., 2006; Orlow et al., 2007). Those findings support the idea that loss-of-function somatic mutations in CDKN2A may initiate melanoma. Since CDKN2A has extremely high mutational sensitivity to UV-light one can expect that loss-of-function somatic mutations can be the first initiation event in melanoma development. On the other hand, the study by Martincorena et al. (Martincorena et al., 2015) found no positive selection for CDKN2A mutation in normal skin suggesting that CDKN2A inactivation may also provide selective advantage at later stages of clonal evolution.
Cancer associated genes can be roughly divided into the genes with cancer specific effect and common (pan-cancer) genes. TP53 is a classic example of a common driver which is frequently mutated in all cancer types. However, other drivers are found to be mutated only in specific cancer types (e.g. CANT1 mutated in prostate cancers only) (Gerhardt et al., 2011). Since all cancer types have common cancer-related features including increase proliferation rate, suppressed apoptosis, angiogenesis, and avoiding immune host response (Hanahan & Weinberg, 2011; Pavlova & Thompson, 2016), the existence of cancer type specific drivers is rather puzzling. Several explanations for existence of cancer specific driver genes have been proposed. It is generally accepted that cancer specificity of driver genes reflects biological differences between normal human tissues: selective advantages somatic mutations depend on tissue environment (Tiong & Yeang, 2018). Schaefer and Serrano demonstrated that cancer/tissue specificity of drivers can be influenced by environmental factors like viral infection which are frequently tissue specific (Schaefer & Serrano, 2016). Tissue specificity of drivers may also be related to histone modifications and chromatin architecture (Lim, Mun, Kim, & Kim, 2017). The results of our analysis suggest that cancer specificity can be a result of differential sensitivity of human genes to the cancer-inducing mutagens. Differential sensitivity to cancer-inducing mutagen may act as a selective factor for the recruitment of driver genes from the pool of the genes that potentially could act as drivers. It is known that human genome contains a large number of genes influencing cancer-related functions. According to the gene ontology (GO) database (accessed February, 10, 2020) there are 778 genes modulating cell proliferation and 1,201 genes modulating cell survival. Those and other genes modulating cancer-related functions can be considered as a pool of genes from which cancer-specific driver can be recruited. In case of melanoma, one can expect that genes with a highest UV sensitivity will have a better chance to be recruited as cancer drivers because they are more likely to get mutated. The outlined reasoning is especially relevant to tumor suppressors for which multiple sites for loss-of-function mutations are available and UV exposure can produce them. Oncogenes, on the other hand, can be activated by a mutation at a single site only and a nucleotide substitution required for the gain-of-function mutation is not necessarily a C>T transition in pyrimidine dimer which is a principal target for UV exposure. A higher UV-sensitivity of melanoma TS compared to melanoma oncogenes is consistent with the idea that melanoma drivers are recruited by a tumor cell based on their UV sensitivity. It does not mean, of course, that all melanoma drivers will have high UV sensitivity. In fact UV sensitivity of many known melanoma drivers including NRAS, TP53, KIT and BRAF is lower than average. Interestingly, all those drivers except TP53 are oncogenes. TP53 has a dual nature and can act as tumor suppressor or oncogene depending on the type of mutation it acquires (Hainaut & Pfeifer, 2016). The higher UV-sensitivity of melanoma TSs can be related to the differences between TSs and other genes in the expression levels. It is known that gene mutability negatively correlates with the gene expression level. (Hodis et al., 2012; Lawrence et al., 2013) Since expression of TSs tends to be suppressed in tumors (Wang, Wu, Rajasekaran, & Shin, 2018), this can contribute to a higher mutability of TSs compared to other genes. Some studies also indicate that expression directed repair may be important. (Pleasance et al., 2010)
Based on the studied mechanism of UV-induced mutations (Bose, 2016; Cadet & Douki, 2018; Dusenbery, McCormick, & Smith, 1983; Kaplan, 1978), UV light is a principal source of C>T transitions in pyrimidine dimers. Nevertheless, we cannot completely rule out a contribution of other sources of mutations including spontaneous mutagenesis (Ohno, 2019) or the effect of non-UV mutagens, e.g. temozolomide treatment (Daniel et al., 2019). In an observational study like this it is impossible to adequately estimate effects of different sources of somatic mutations in melanoma. However, the contribution of sources of mutations other than UV light in melanoma was shown to be small. (Alexandrov et al., 2020; Birkeland et al., 2018; Phillips, 2018)
We used silent mutations to account for effects of clonal selection when estimating UV sensitivity of individual transcripts. In contrast to silent mutations, missense mutations are influenced by both mutability and selection. Consequently, one needs to account for differences in UV sensitivity when predicting how many missense mutations one can expect in the transcript. Therefore, the practical significance of this study relates to the prediction of the expected number of somatic mutations in genes. The prediction of the expected number of somatic mutations is essential in identification of cancer related genes based on the number of somatic mutations detected in tumor samples (Gorlov et al., 2018; Lawrence et al., 2013). In cited and other similar studies (Martincorena et al., 2017; Vineis, 2003) the excess of nonsynonymous mutations in a gene is used as an indicator of its cancer relevance. The excess of somatic mutations, however, can also reflect a gene’s high inherent mutability, e.g. abnormally high UV-sensitivity in case of melanoma. Thus, differences in UV sensitivity need to be taken into account when evaluating the expected number of nonsynonymous substitutions in a gene in the analysis of melanoma samples.
A limitation of our approach is that we may have overestimated the number of UV-induced mutations by assuming that all C>T transitions in pyrimidine dimers are induced by UV light. It is known that some other factors can also cause C>T transitions in pyrimidine dimers. Those factors include defects in DNA mismatch repair (Li, Pearlman, & Hsieh, 2016), error-prone polymerase POLE (Park & Pursell, 2019), and alkylating agents. (Fu, Calvo, & Samson, 2012) Indeed, we observed effects of these factors in the analysis of contribution of different mutational signatures (Table 1). The contribution of these other sources of C>T substitutions to pyrimidine dimers is, however, small.
In conclusion, we found a substantial variation in UV sensitivity among human genes and identified gene characteristics associated with it. Differences in UV sensitivity need to be taken into consideration when predicting how many potentially functional nonsynonymous mutations one can expect to detect in a gene which is essential step in identification of cancer-related genes based on the number of somatic mutations in them.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the National Institutes of Health [U19 CA148127 to C.I.A., P01 CA206980-01A1 to M.B. and N.E.T., CA233524 to N.E.T., P30 CA008748 core grant to MSK]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors gratefully acknowledge helpful discussion from the members of InterMEL consortium.
Funding information
National Institutes of Health, Grant/Award Numbers: ID U19 CA148127, ID CA206980-01A1, ID P30 CA008748.
Footnotes
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
DATA AVAILABILITY STATEMENT
The data on the observed number of silent mutations in individual trinucleotides and the number of corresponding trinucleotides in each transcript are shown in the Supplementary Table. The code used for the estimation of the number of potential sites for individual transcripts is available from the corresponding author.
REFERENCES
- Abaan OD, Polley EC, Davis SR, Zhu YJ, Bilke S, Walker RL, … Meltzer PS (2013). The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology. Cancer Res, 73(14), 4372–4382. doi: 10.1158/0008-5472.CAN-12-3342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, … Consortium P (2020). The repertoire of mutational signatures in human cancer. Nature, 578(7793), 94–101. doi: 10.1038/s41586-020-1943-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, & Stratton MR (2013). Deciphering signatures of mutational processes operative in human cancer. Cell Rep, 3(1), 246–259. doi: 10.1016/j.celrep.2012.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin IT, Melamed RD, Adams SJ, Castillo-Martin M, Demir A, Bryk D, … Celebi JT (2014). FBXW7 mutations in melanoma and a new therapeutic paradigm. J Natl Cancer Inst, 106(6), dju107. doi: 10.1093/jnci/dju107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bali V, & Bebok Z (2015). Decoding mechanisms by which silent codon changes influence protein biogenesis and function. Int J Biochem Cell Biol, 64, 58–74. doi: 10.1016/j.biocel.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg CB, Orlow I, Hummer AJ, Armstrong BK, Kricker A, Marrett LD, … Melanoma Study G (2005). Lifetime risk of melanoma in CDKN2A mutation carriers in a population-based sample. J Natl Cancer Inst, 97(20), 1507–1515. doi: 10.1093/jnci/dji312 [DOI] [PubMed] [Google Scholar]
- Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, … Garraway LA (2012). Melanoma genome sequencing reveals frequent PREX2 mutations. Nature, 485(7399), 502–506. doi: 10.1038/nature11071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berwick M, Orlow I, Hummer AJ, Armstrong BK, Kricker A, Marrett LD, … Group GEMS (2006). The prevalence of CDKN2A germ-line mutations and relative risk for cutaneous malignant melanoma: an international population-based study. Cancer Epidemiol Biomarkers Prev, 15(8), 1520–1525. doi: 10.1158/1055-9965.EPI-06-0270 [DOI] [PubMed] [Google Scholar]
- Birkeland E, Zhang S, Poduval D, Geisler J, Nakken S, Vodak D, … Lonning PE (2018). Patterns of genomic evolution in advanced melanoma. Nat Commun, 9(1), 2665. doi: 10.1038/s41467-018-05063-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose JL (2016). Chemical and UV Mutagenesis. Methods Mol Biol, 1373, 111–115. doi: 10.1007/7651_2014_190 [DOI] [PubMed] [Google Scholar]
- Cadet J, & Douki T (2018). Formation of UV-induced DNA damage contributing to skin cancer development. Photochem Photobiol Sci, 17(12), 1816–1841. doi: 10.1039/c7pp00395a [DOI] [PubMed] [Google Scholar]
- Dahlman KB, Xia J, Hutchinson K, Ng C, Hucks D, Jia P, … Pao W (2012). BRAF(L597) mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer Discov, 2(9), 791–797. doi: 10.1158/2159-8290.CD-12-0097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel P, Sabri S, Chaddad A, Meehan B, Jean-Claude B, Rak J, & Abdulkarim BS (2019). Temozolomide Induced Hypermutation in Glioma: Evolutionary Mechanisms and Therapeutic Opportunities. Front Oncol, 9, 41. doi: 10.3389/fonc.2019.00041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deben C, Deschoolmeester V, Lardon F, Rolfo C, & Pauwels P (2016). TP53 and MDM2 genetic alterations in non-small cell lung cancer: Evaluating their prognostic and predictive value. Crit Rev Oncol Hematol, 99, 63–73. doi: 10.1016/j.critrevonc.2015.11.019 [DOI] [PubMed] [Google Scholar]
- Diaz-Gay M, Vila-Casadesus M, Franch-Exposito S, Hernandez-Illan E, Lozano JJ, & Castellvi-Bel S (2018). Mutational Signatures in Cancer (MuSiCa): a web application to implement mutational signatures analysis in cancer samples. BMC Bioinformatics, 19(1), 224. doi: 10.1186/s12859-018-2234-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusenbery RL, McCormick SC, & Smith PD (1983). Drosophila mutations at the mei-9 and mus(2)201 loci which block excision of thymine dimers also block induction of unscheduled DNA synthesis by methyl methanesulfonate, ethyl methanesulfonate, N-methyl-N-nitrosourea, UV light and X-rays. Mutat Res, 112(4), 215–230. doi: 10.1016/0167-8817(83)90008-1 [DOI] [PubMed] [Google Scholar]
- Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, … Futreal PA (2010). COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res, 38(Database issue), D652–657. doi: 10.1093/nar/gkp995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes WD, Flanders TY, Pollock PM, & Hayward NK (1997). The CDKN2A (p16) gene and human cancer. Mol Med, 3(1), 5–20. [PMC free article] [PubMed] [Google Scholar]
- Fu D, Calvo JA, & Samson LD (2012). Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer, 12(2), 104–120. doi: 10.1038/nrc3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furney SJ, Turajlic S, Fenwick K, Lambros MB, MacKay A, Ricken G, … Marais R (2012). Genomic characterisation of acral melanoma cell lines. Pigment Cell Melanoma Res, 25(4), 488–492. doi: 10.1111/j.1755-148X.2012.01016.x [DOI] [PubMed] [Google Scholar]
- Gerhardt J, Steinbrech C, Buchi O, Behnke S, Bohnert A, Fritzsche F, … Kristiansen G (2011). The androgen-regulated Calcium-Activated Nucleotidase 1 (CANT1) is commonly overexpressed in prostate cancer and is tumor-biologically relevant in vitro. Am J Pathol, 178(4), 1847–1860. doi: 10.1016/j.ajpath.2010.12.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Perez A, Sabarinathan R, & Lopez-Bigas N (2019). Local Determinants of the Mutational Landscape of the Human Genome. Cell, 177(1), 101–114. doi: 10.1016/j.cell.2019.02.051 [DOI] [PubMed] [Google Scholar]
- Gorlov IP, Pikielny CW, Frost HR, Her SC, Cole MD, Strohbehn SD, … Amos CI (2018). Gene characteristics predicting missense, nonsense and frameshift mutations in tumor samples. BMC Bioinformatics, 19(1), 430. doi: 10.1186/s12859-018-2455-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainaut P, & Pfeifer GP (2016). Somatic TP53 Mutations in the Era of Genome Sequencing. Cold Spring Harb Perspect Med, 6(11). doi: 10.1101/cshperspect.a026179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, & Weinberg RA (2011). Hallmarks of cancer: the next generation. Cell, 144(5), 646–674. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, … Chin L (2012). A landscape of driver mutations in melanoma. Cell, 150(2), 251–263. doi: 10.1016/j.cell.2012.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikehata H, & Ono T (2011). The mechanisms of UV mutagenesis. J Radiat Res, 52(2), 115–125. doi: 10.1269/jrr.10175 [DOI] [PubMed] [Google Scholar]
- Kaplan RW (1978). Electiveness of photorepair, influence of dark-repair on shape of dose-response curves, and high-dose decline, in UV-induced colour mutations of Serratia. Mutat Res, 49(3), 357–370. doi: 10.1016/0027-5107(78)90106-9 [DOI] [PubMed] [Google Scholar]
- Krauthammer M, Kong Y, Bacchiocchi A, Evans P, Pornputtapong N, Wu C, … Halaban R (2015). Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat Genet, 47(9), 996–1002. doi: 10.1038/ng.3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, … Halaban R (2012). Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet, 44(9), 1006–1014. doi: 10.1038/ng.2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuckein D (1980). [Computed tomography in cases of pancreas neoplasms (author’s transl)]. Rontgenblatter, 33(10), 520–524. [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, … Getz G (2013). Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature, 499(7457), 214–218. doi: 10.1038/nature12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Pearlman AH, & Hsieh P (2016). DNA mismatch repair and the DNA damage response. DNA Repair (Amst), 38, 94–101. doi: 10.1016/j.dnarep.2015.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B, Mun J, Kim YS, & Kim SY (2017). Variability in Chromatin Architecture and Associated DNA Repair at Genomic Positions Containing Somatic Mutations. Cancer Res, 77(11), 2822–2833. doi: 10.1158/0008-5472.CAN-16-3033 [DOI] [PubMed] [Google Scholar]
- Lindberg M, Bostrom M, Elliott K, & Larsson E (2019). Intragenomic variability and extended sequence patterns in the mutational signature of ultraviolet light. Proc Natl Acad Sci U S A, 116(41), 20411–20417. doi: 10.1073/pnas.1909021116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, & Campbell PJ (2015). Somatic mutation in cancer and normal cells. Science, 349(6255), 1483–1489. doi: 10.1126/science.aab4082 [DOI] [PubMed] [Google Scholar]
- Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, … Campbell PJ (2017). Universal Patterns of Selection in Cancer and Somatic Tissues. Cell, 171(5), 1029–1041 e1021. doi: 10.1016/j.cell.2017.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, … Campbell PJ (2015). Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science, 348(6237), 880–886. doi: 10.1126/science.aaa6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morganella S, Alexandrov LB, Glodzik D, Zou X, Davies H, Staaf J, … Nik-Zainal S (2016). The topography of mutational processes in breast cancer genomes. Nat Commun, 7, 11383. doi: 10.1038/ncomms11383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev SI, Rimoldi D, Iseli C, Valsesia A, Robyr D, Gehrig C, … Antonarakis SE (2011). Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet, 44(2), 133–139. doi: 10.1038/ng.1026 [DOI] [PubMed] [Google Scholar]
- Ohno M (2019). Spontaneous de novo germline mutations in humans and mice: rates, spectra, causes and consequences. Genes Genet Syst, 94(1), 13–22. doi: 10.1266/ggs.18-00015 [DOI] [PubMed] [Google Scholar]
- Orlow I, Begg CB, Cotignola J, Roy P, Hummer AJ, Clas BA, … Group GEMS (2007). CDKN2A germline mutations in individuals with cutaneous malignant melanoma. J Invest Dermatol, 127(5), 1234–1243. doi: 10.1038/sj.jid.5700689 [DOI] [PubMed] [Google Scholar]
- Pagani F, Raponi M, & Baralle FE (2005). Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proc Natl Acad Sci U S A, 102(18), 6368–6372. doi: 10.1073/pnas.0502288102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park VS, & Pursell ZF (2019). POLE proofreading defects: Contributions to mutagenesis and cancer. DNA Repair (Amst), 76, 50–59. doi: 10.1016/j.dnarep.2019.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova NN, & Thompson CB (2016). The Emerging Hallmarks of Cancer Metabolism. Cell Metab, 23(1), 27–47. doi: 10.1016/j.cmet.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petljak M, Alexandrov LB, Brammeld JS, Price S, Wedge DC, Grossmann S, … Stratton MR (2019). Characterizing Mutational Signatures in Human Cancer Cell Lines Reveals Episodic APOBEC Mutagenesis. Cell, 176(6), 1282–1294 e1220. doi: 10.1016/j.cell.2019.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips DH (2018). Mutational spectra and mutational signatures: Insights into cancer aetiology and mechanisms of DNA damage and repair. DNA Repair (Amst), 71, 6–11. doi: 10.1016/j.dnarep.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, … Stratton MR (2010). A comprehensive catalogue of somatic mutations from a human cancer genome. Nature, 463(7278), 191–196. doi: 10.1038/nature08658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujar S, O’Leary NA, Farrell CM, Loveland JE, Mudge JM, Wallin C, … Pruitt KD (2018). Consensus coding sequence (CCDS) database: a standardized set of human and mouse protein-coding regions supported by expert curation. Nucleic Acids Res, 46(D1), D221–D228. doi: 10.1093/nar/gkx1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Malyarchuk BA, Pavlov YI, & Milanesi L (2005). From context-dependence of mutations to molecular mechanisms of mutagenesis. Pac Symp Biocomput, 409–420. [PubMed] [Google Scholar]
- Sanborn JZ, Chung J, Purdom E, Wang NJ, Kakavand H, Wilmott JS, … Cho RJ (2015). Phylogenetic analyses of melanoma reveal complex patterns of metastatic dissemination. Proc Natl Acad Sci U S A, 112(35), 10995–11000. doi: 10.1073/pnas.1508074112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer MH, & Serrano L (2016). Cell type-specific properties and environment shape tissue specificity of cancer genes. Sci Rep, 6, 20707. doi: 10.1038/srep20707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shain AH, Garrido M, Botton T, Talevich E, Yeh I, Sanborn JZ, … Bastian BC (2015). Exome sequencing of desmoplastic melanoma identifies recurrent NFKBIE promoter mutations and diverse activating mutations in the MAPK pathway. Nat Genet, 47(10), 1194–1199. doi: 10.1038/ng.3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, … Forbes SA (2019). COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res, 47(D1), D941–D947. doi: 10.1093/nar/gky1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiong KL, & Yeang CH (2018). Explaining cancer type specific mutations with transcriptomic and epigenomic features in normal tissues. Sci Rep, 8(1), 11456. doi: 10.1038/s41598-018-29861-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkova M, Tomek J, Kriaucionis S, & Schuster-Bockler B (2018). Mutational signature distribution varies with DNA replication timing and strand asymmetry. Genome Biol, 19(1), 129. doi: 10.1186/s13059-018-1509-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt C (2019). UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res, 47(D1), D506–D515. doi: 10.1093/nar/gky1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vineis P (2003). Cancer as an evolutionary process at the cell level: an epidemiological perspective. Carcinogenesis, 24(1), 1–6. doi: 10.1093/carcin/24.1.1 [DOI] [PubMed] [Google Scholar]
- Wagle N, Van Allen EM, Treacy DJ, Frederick DT, Cooper ZA, Taylor-Weiner A, … Garraway LA (2014). MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov, 4(1), 61–68. doi: 10.1158/2159-8290.CD-13-0631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LH, Wu CF, Rajasekaran N, & Shin YK (2018). Loss of Tumor Suppressor Gene Function in Human Cancer: An Overview. Cell Physiol Biochem, 51(6), 2647–2693. doi: 10.1159/000495956 [DOI] [PubMed] [Google Scholar]
- Wei X, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J, … Samuels Y (2011). Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet, 43(5), 442–446. doi: 10.1038/ng.810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, & Iwakuma T (2018). Regulators of Oncogenic Mutant TP53 Gain of Function. Cancers (Basel), 11(1). doi: 10.3390/cancers11010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Choi BY, Lee MH, Bode AM, & Dong Z (2016). Implications of Genetic and Epigenetic Alterations of CDKN2A (p16(INK4a)) in Cancer. EBioMedicine, 8, 30–39. doi: 10.1016/j.ebiom.2016.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.