Abstract
Objective:
We aim to identify prognostic groups within a de novo metastatic cohort, incorporating both anatomic and biologic factors.
Background:
Staging for breast cancer now includes anatomic and biologic factors, although the guidelines for stage IV disease do not account for how these factors may influence outcomes.
Methods:
Adults with de novo metastatic breast cancer were selected from the National Cancer DataBase (2010–2013). Recursive partitioning analysis was used to group patients with similar overall survival (OS) based on clinical T/N stage, tumor grade, ER, PR, HER2, number of metastatic sites, and presence of bone-only metastases. Categories were created by amalgamating homogeneous groups based on 3-year OS rates (stage IVA: >50%, stage IVB: 30%–50%, stage IVC: <30%).
Results:
16,187 patients were identified; median follow-up was 32 months. 65.2% had 1 site of distant metastasis, and 42.9% had bone-only metastases. Recursive partitioning analysis identified the number of metastatic sites (1 vs >1) as the first stratification point, and ER status as the second stratification point for both resulting groups. Additional divisions were made based on HER2 status, PR status, cT stage, tumor grade, and presence of bone-only metastases. After bootstrapping, significant differences in 3-year OS were noted between the 3 groups [stage IVB vs IVA: HR 1.58 (95% confidence interval 1.50–1.67), stage IVC vs IVA: HR 3.54 (95% confidence interval 3.33–3.77)].
Conclusions:
Both anatomic and biologic factors yielded reliable and reproducible prognostic estimates among patients with metastatic disease. These findings support formal stratification of de novo stage IV breast cancer into 3 distinct prognosis groups.
Keywords: metastatic breast cancer, prognosis, stage IV breast cancer, staging
With improved treatment options and survival,1 the number of women living with metastatic breast cancer in the United States (USA) is increasing.2,3 In a review of 21,372 women with stage IV breast cancer in the USA diagnosed in 1988–2011, the median survival increased from 20 months to 26 months.4 Furthermore, the prognosis among patients with metastatic disease can vary significantly with survival known to be influenced by patient demographics, tumor grade, tumor biology, and pattern and extent of metastatic spread.4–6 Although de novo metastatic breast cancer in the USA comprises only 6% of all breast cancer diagnoses each year,7 it presents a unique opportunity to refine prognostic estimates for stage IV disease.
Currently, the American Joint Committee on Cancer (AJCC) staging guidelines for several primary malignancies, including colon, lung, ovarian, and thyroid cancer, include subdivided groups among patients with distant metastatic disease to provide more accurate prognostic estimates.8 For example, patients with thyroid cancer in the USA are stratified using a combination of age and distant metastatic disease to determine the final overall stage: for patients with distant metastatic disease, those aged <55 years old are classified as having stage II disease, compared to those ≥55 years old classified as having stage IV disease.8
With the implementation of the AJCC Cancer Staging Manual eighth edition, prognostic estimates for early-stage breast cancer have been refined by incorporating anatomic and biologic factors into a prognostic stage.9 However, while these same factors may affect prognosis in metastatic disease,5,6 no standard guidelines currently exist to stratify patients with de novo metastatic breast cancer. As such, we aim to evaluate the association of anatomic staging, tumor biology, and pattern of metastatic disease with overall survival (OS) in patients with de novo metastatic breast cancer and to propose a model for prognostic stratification that divides patients into 3 unique subgroups (stages IVA, IVB, and IVC).
METHODS
Adult patients (ages ≥18 years old) diagnosed with clinically or pathologic metastatic breast cancer (cM1 or pM1) between 2004 and 2014 were selected from the National Cancer Data Base (NCDB). Per NCDB guidelines, survival data for patients diagnosed in the most recent database year is masked; therefore, all patients diagnosed in 2014 were excluded. Patients with missing clinical T or N stage, tumor grade, ER (estrogen receptor) status, PR (progesterone receptor) status, HER2 (human-epidermal-growth-factor-receptor-2) status, or site of metastatic disease were excluded. HER2 status has only been reliably collected since 2010, hence all patients diagnosed before 2010 were excluded. By limiting our analysis to patients from 2010 to 2013, we were able to determine 3- and 5-year OS rates. Patients who were not classified based on the World Health Organization histology classification and those who did not undergo any treatment were excluded.
Patient and disease characteristics were summarized with N (%) for categorical variables and median (interquartile range) for continuous variables. The 3- and 5-year OS rates were estimated using the Kaplan-Meier method. OS was defined as the time from diagnosis to death or last follow-up.
A recursive partitioning analysis (RPA)10 was used to group patients with similar OS based on cT stage, cN stage, tumor grade, ER, PR, HER2, number of distant metastatic sites (as determined by the number of tissues/organs involved, not necessarily the number of metastatic tumors), and bone only metastases (yes/no). The splitting criteria was set to a log-rank P-value of 0.10. Recursive partitioning is a classification approach that partitions data based on covariates included in a regression model. It is recursive, meaning that it first partitions the data by the most influential covariate split, and then continues to partition the newly created subgroups based on the influence of covariates. Each subgroup is examined separately, and different covariates may be used to partition different subgroups at the same level of the decision tree. This algorithm stops when a pre-specified criterion is met – in this case, when the log-rank test P-value for an attempted partition is >0.10 for all possible covariates for all subgroups. The final set of subgroups are called terminal nodes. After recursive partitioning, 3-year OS rates were estimated for each terminal node. Patients were grouped based on the association of disease characteristics with 3-year OS. Final groupings of patients with homogeneous survival were created based on amalgamation of terminal nodes and on the 3-year OS rates:
Group A was defined by combining clinical characteristic groups with a 3-year OS rate >50% (all terminal nodes with a 3-year OS rate of >50% were collapsed);
Group B was defined by combining clinical characteristic groups with a 3-year OS rate of 30%–50% (all terminal nodes with a 3-year OS rate of 30%–50% were collapsed);
Group C was defined by combining clinical characteristic groups with a 3-year OS rate <30% (all terminal nodes with a 3-year OS rate of <30% were collapsed).
Bootstrapping was applied, and this algorithm was repeated 1000 times. Bootstrapping is conducted because it allows for a prediction that is more representative of the true population. For each iteration, a sample was drawn with replacement from the original dataset, and the final patient groups were created based on that sample. Results were summarized and final group assignments were determined based on the groups each disease characteristic combination fell into most often.
The 3- and 5-year OS rates and 95% confidence intervals (CIs) of the defined groups (A/B/C) were estimated using the Kaplan-Meier method. The effect of patient groupings determined by recursive partitioning with bootstrapping on OS was estimated using the Cox Proportional Hazards model, after adjustment for known covariates. To account for the correlation of patients treated at the same hospital, a robust sandwich covariance estimator was included in the adjusted survival model. The RPA tree and KM curve of terminal nodes presented here are for the original cohort. All other analyses are based off the bootstrapped results.
A P-value < 0.05 was considered significant, and no adjustments were made for multiple comparisons. Only patients with complete data were included in each analysis, and effective sample sizes are included for all tables/figures. All statistical analyses were conducted in SAS, version 9.4 (SAS Institute, Cary NC) or R, version 3.3.1 (R Foundation for Statistical Computing, Vienna).
RESULTS
After applying the defined inclusion/exclusion criteria, the final cohort sample size was 16,187 (Supplemental Table 1, http://links.lww.com/SLA/C333). The median follow-up was 32.3 months (95% CI 31.5–33.1), and there were 8610 deaths (53.2% of the cohort). The median patient age was 60 years (interquartile range 51–70 years). Most tumors were ER-positive (74.9%), PR-positive (62.2%), HER2-negative (73.7%), and grade 2–3 (92.5%). The median primary tumor size was 4 cm, and the majority had node-positive disease (72.1%). At initial presentation, most patients had only 1 site of distant metastatic disease (65.2%), and 42.9% had bone-only metastases. Regarding treatments (Supplemental Table 1, http://links.lww.com/SLA/C333), 58% received chemotherapy, 35.7% received radiation therapy, 73.6% of ER-positive or PR-positive patients received endocrine therapy, and 62.7% did not undergo surgery for the primary breast tumor.
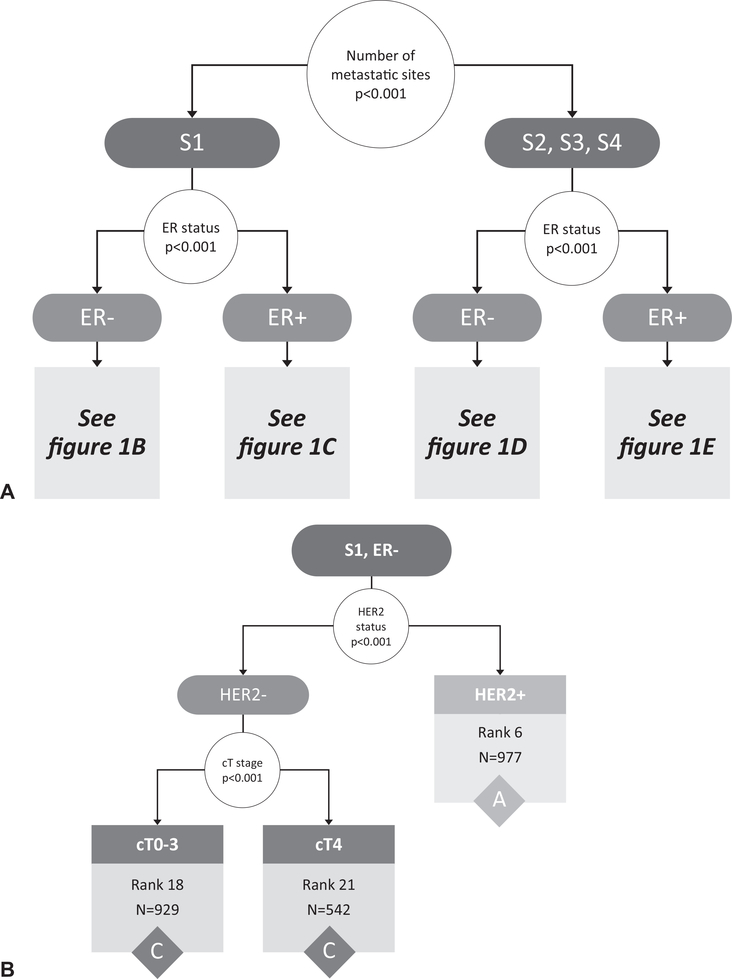
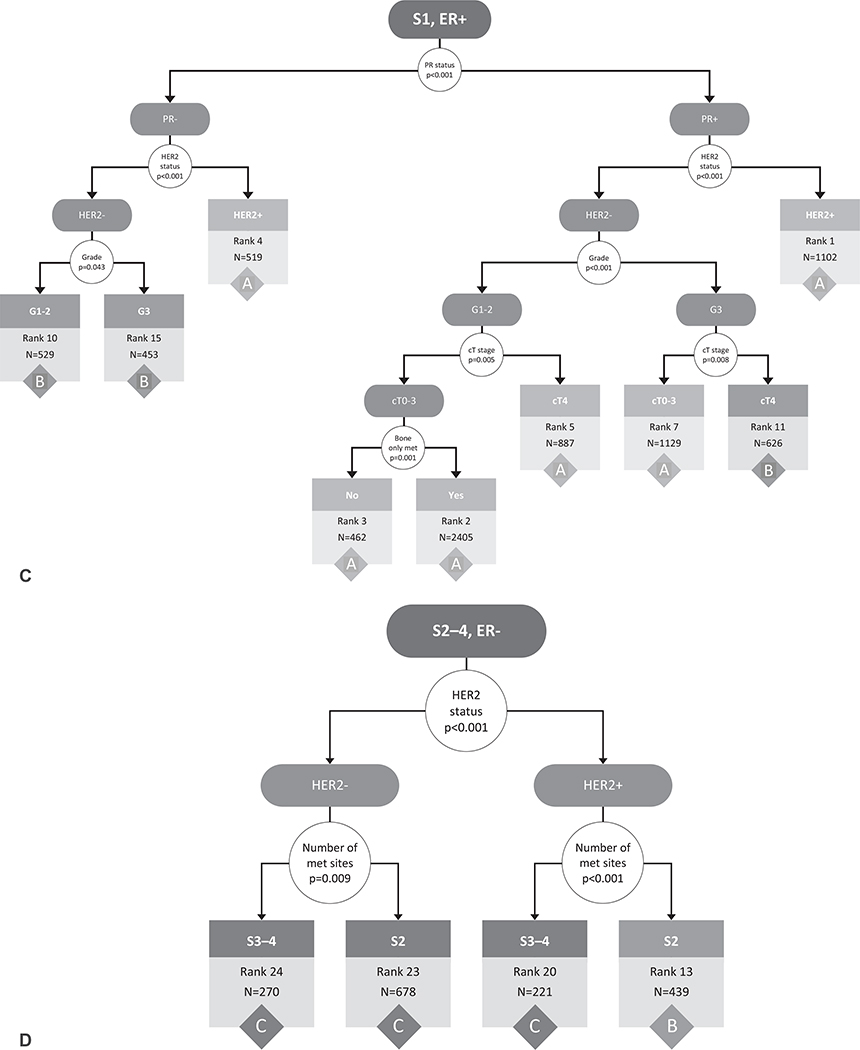
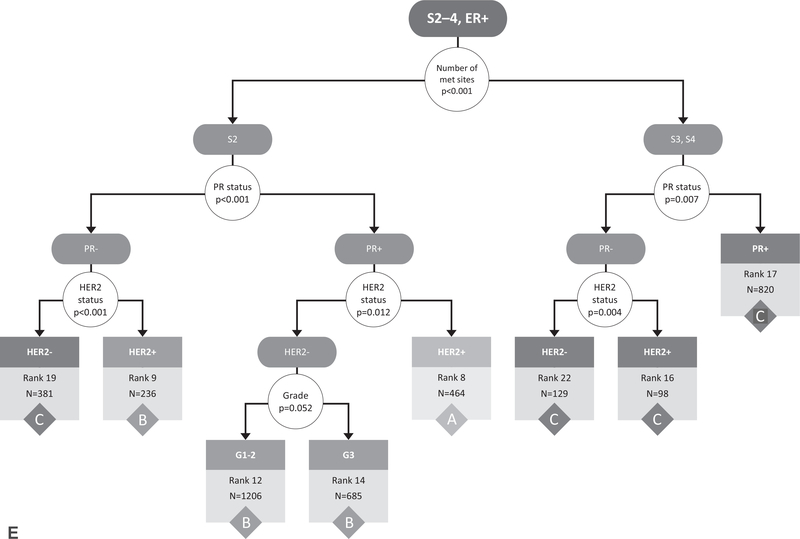
Starting with the entire cohort of 16,187 patients, the RPA on the original cohort before bootstrapping identified the number of metastatic sites (1 vs >1) as the first stratification point (Fig. 1A). The RPA then identified ER status as the second stratification point for both those with 1 site of metastatic disease and those with >1 site of metastatic disease (Fig. 1A). Additional divisions were made based on HER2 status, PR status, cT stage, tumor grade, and presence of bone-only metastases. Although there were 1,123 possible combinations of the included variables (cT, cN, grade, ER, PR, HER2, # metastatic sites, and bone only metastases), the RPA identified 24 subgroups with homogeneous OS (Fig. 1B–E). The OS curves of these 24 subgroups were visualized using the Kaplan-Meier method (Supplemental Fig. 1, http://links.lww.com/SLA/C333).
FIGURE 1.
Recursive partitioning tree created based on the following covariates in the original data cohort: clinical T stage (0/1/2/3/4), clinical N stage (0/1/2/3), tumor grade (1/2/3), ER (+/−), PR (+/−), HER2 (+/−), number of metastatic sites (1/2/3/4), and bone-only metastases (yes/no). The initial partitioning variables (number of metastatic sites and ER status) are shown in (A), whereas the continued partitioning for those with 1 metastatic site (B, C) and those with >1 metastatic site (D, E) are shown separately. Results based on adults with de novo metastatic breast cancer from the National Cancer Data Base (2010–2013). S indicates number of metastatic sites, G indicates grade, B indicates bone only metastases yes/no. White circles represent decision points, with arrows directing to potential outcomes for the decision. Terminal nodes with associated rankings and count (N) within that group, are represented by boxes color-coded according to final stage grouping (A/B/C). Rank assigned based on 3-year survival rates (listed in Table 1) from highest/best to lowest/worst (ie, rank 1 has the highest survival rates and rank 24 has the lowest). ER indicates estrogen receptor; HER2, human epidermal growth factor receptor 2; PR, progesterone receptor.
Review of the 3-year OS rates for the 24 subgroups from the RPA revealed that the patients could be amalgamated into 3 distinct groups: A, B, and C (Table 1). The 3-year OS rate was selected as the final stratification variable for staging, because the median OS of the entire cohort was 32.3 months. Using the 24 subgroups identified by the RPA, 3-year OS rates were noted to range from 5.1% to 70.8% (Table 1). Bootstrapping around the RPA and group amalgamation was used to create the final groupings: group A (proposed stage IVA; N = 7941, 49.1% of patients) was defined as those with clinical characteristics most often associated with a 3-year OS rate >50%; group B (proposed stage IVB; N = 4291, 25.8% of patients) was defined as those with clinical characteristics most often associated with a 3-year OS rate of 30%–50%; and group C (proposed stage IVC; N = 3955, 25.1% of patients) was defined as those with clinical characteristics most often associated with a 3-year OS rate <30%. The division points in 3-year OS rates (>50% vs 30%–50% vs <30%) were selected based on clinical relevance. In addition to the variables used in the RPA, other factors that were significantly different (on univariate analysis) between the 3 groups included patient race/ethnicity, tumor histology, chemotherapy receipt, endocrine therapy receipt, and surgery receipt/type (Supplemental Table 2, http://links.lww.com/SLA/C333).
TABLE 1.
Characteristics and 3-yr Survival Rates for all Terminal Nodes in Figure 1
| Rank | 3-yr Survival Rate | Sample Size | Characteristics | Group |
|---|---|---|---|---|
| 1 | 0.708 | 1102 | S1/ER+/PR+/HER2+ | A |
| 2 | 0.666 | 2405 | S1/ER+/PR+/HER2−/G1−2/cT0–3/B1 | A |
| 3 | 0.606 | 462 | S1/ER+/PR+/HER2−/G1–2/cT0–3/B0 | A |
| 4 | 0.600 | 519 | S1/ER+/PR−/HER2+ | A |
| 5 | 0.588 | 887 | S1/ER+/PR+/HER2−/G1–2/cT4 | A |
| 6 | 0.551 | 977 | S1/ER−/HER2+ | A |
| 7 | 0.551 | 1129 | S1/ER+/PR+/HER2−/G3/cT0–3 | A |
| 8 | 0.503 | 464 | S2/ER+/PR+/HER2+ | A |
| 9 | 0.489 | 236 | S2/ER+/PR−/HER2+ | B |
| 10 | 0.481 | 529 | S1/ER+/PR−/HER2−/G1–2 | B |
| 11 | 0.453 | 626 | S1/ER+/PR+/HER2−/G3/cT4 | B |
| 12 | 0.440 | 1206 | S2/ER+/PR+/HER2−/G1–2 | B |
| 13 | 0.427 | 439 | S2/ER−/HER2+ | B |
| 14 | 0.372 | 685 | S2/ER+/PR+/HER2−/G3 | B |
| 15 | 0.363 | 453 | S1/ER+/PR−/HER2−/G3 | B |
| 16 | 0.291 | 98 | S3−4/ER+/PR−/HER2+ | C |
| 17 | 0.281 | 820 | S3−4/ER+/PR+ | C |
| 18 | 0.280 | 929 | S1/ER−/HER2−/cT0–3 | C |
| 19 | 0.202 | 381 | S2/ER+/PR−/HER2− | C |
| 20 | 0.195 | 221 | S3−4/ER−/HER2+ | C |
| 21 | 0.170 | 542 | S1/ER−/HER2−/cT4 | C |
| 22 | 0.137 | 129 | S3−4/ER+/PR−/HER2− | C |
| 23 | 0.081 | 678 | S2/ER−/HER2− | C |
| 24 | 0.051 | 270 | S3−4/ER−/HER2− | C |
Analysis based on adults with de novo metastatic breast cancer from the national cancer data base (2010–2013).
Characteristics are listedin orderof appearance withinrecursive partitioning (RPA) Tree. S indicates number ofmetastatic sites (eg,S1 = 1 metastatic site),Gindicatestumorgrade, B indicates bone only metastases (B1 = yes; B0 = no). Only characteristics that were specified in the RPA tree are listed.
ER indicates estrogen receptor; HER2, human epidermal growth factor receptor 2; PR, progesterone receptor.
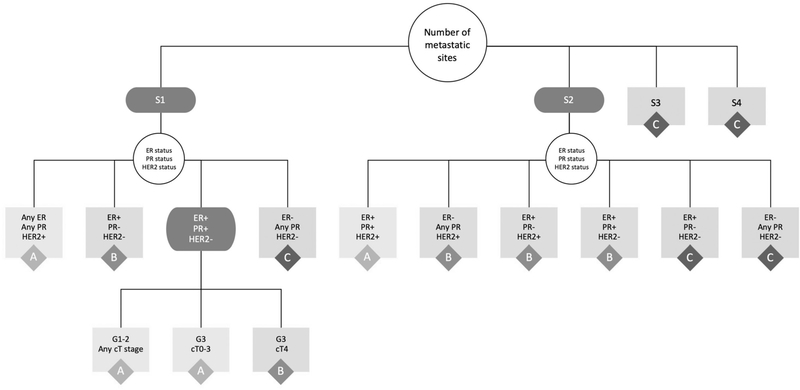
Within group A, the largest proportion of patients (N = 2405 of 7941) were noted to have the following characteristics: ER-positive, PR-positive, HER2-negative, grade 1–2, cT0–3, and bone-only metastases. Within group B, the largest proportion of patients (N = 1206 of 4291) were noted to have the following characteristics: 2 sites of metastatic disease, ER-positive, PR-positive, HER2-negative, and grade 1–2. Within group C, the largest proportion of patients (N = 929 of 3955) were noted to have the following characteristics: 1 site of metastatic disease, ER-negative, HER2-negative, and cT0–3. The second largest proportion of patients within group C (N = 820) were noted to have the following characteristics: 3–4 sites of metastatic disease (meaning 3–4 different, distant organ systems involved), ER-positive, and PR-positive. To improve the potential clinical utility of the proposed stratification scheme, the characteristics of these groups were then used to collapse the population into the fewest number of groups that would maintain the final stage assignment (A/B/C), similar to how the groupings are presented in the most recent version of the AJCC staging manual (Fig. 2 and Table 2).
FIGURE 2.
Summary of the results from the recursive partitioning analysis into the fewest groups, while still maintaining the final overall prognostic stage group (A/B/C). Results based on adults with de novo metastatic breast cancer from the National Cancer Data Base (2010–2013). S indicates number of metastatic sites, G indicates grade. Black boxes represent decision points, while white and gray boxes represent potential outcomes for the decision. Gray boxes also represent terminal nodes with the final stage grouping (A/B/C). ER indicates estrogen receptor; HER2, human epidermal growth factor receptor 2; PR, progesterone receptor.
TABLE 2.
Summary of Characteristics of the Recursive Partitioning Analysis (RPA) Groupings After Condensing Into the Fewest Groups Possible
| When the Number of Metastatic Sites is... | And Biomarker Status is... | And the Grade and cT Stage is... | Then the Prognostic Stage Group is... |
|---|---|---|---|
| 1 | Any ER, any PR, HER2+ | Any grade, any cT stage | IVA |
| ER+,PR−, HER2− | Any grade, any cT stage | IVB | |
| ER+,PR+, HER2− | Grade 1–2, any cT stage | IVA | |
| Grade 3, cT0–3 | IVA | ||
| Grade 3, cT4 | IVB | ||
| ER−, any PR, HER2− | Any grade, any cT stage | IVC | |
| 2 | ER+, PR+, HER2+ | Any grade, any cT stage | IVA |
| ER−, any PR, HER2+ | Any grade, any cT stage | IVB | |
| ER+,PR−, HER2+ | Any grade, any cT stage | IVB | |
| ER+,PR+, HER2− | Any grade, any cT stage | IVB | |
| ER+/PR−/HER2− | Any grade, any cT stage | IVC | |
| ER−, any PR, HER2− | Any grade, any cT stage | IVC | |
| ≥3 | Any ER, any PR, any HER2 | Any grade, any cT stage | IVC |
Analysis based on adults with de novo metastatic breast cancer from the national cancer data base (2010–2013).
ER indicates estrogen receptor; HER2, human epidermal growth factor receptor 2; PR, progesterone receptor.
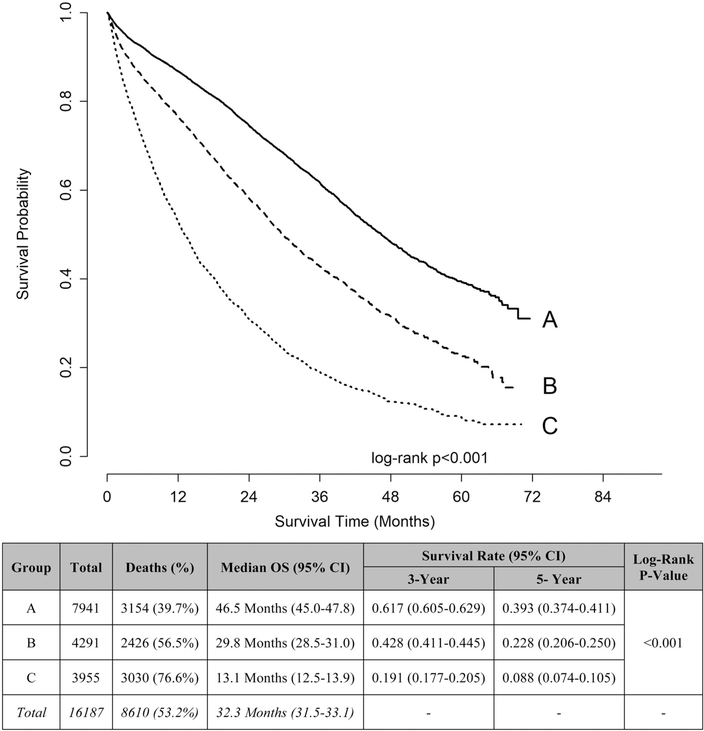
The OS curves of the 3 amalgamated groups were visualized using the Kaplan-Meier method (log-rank P-value < 0.001) based on the original data set (Supplemental Fig. 2, http://links.lww.com/SLA/C333). To provide the most accurate representation of the population, the Kaplan-Meier curves were then recreated based on the bootstrapped RPA and further defined by 3-year OS rates (Fig. 3). The median OS for group A was 46.5 months, group B was 29.8 months, and group C was 13.1 months. The 3-year OS rate for group A was 61.7%, group B was 42.8%, and group C was 19.1%. Similar differences in 5-year OS rates were noted.
FIGURE 3.
Kaplan-Meier curves and unadjusted overall survival rates of amalgamated groups based on bootstrapped recursive partitioning analysis and further defined by 3-yr overall survival (OS) rates. Group A defined by combining clinical characteristic groups with 3-yr OS rate >50%. Group B defined by combining clinical characteristic groups with 3-yr OS rate 30%–50%. Group C defined by combining clinical characteristic groups with 3-yr OS rate <30%. Analysis based on adults with de novo metastatic breast cancer from the National Cancer Data Base (2010–2013).
To determine the effect of these newly defined groups (A/B/C) on OS, a Cox Proportional Hazards model was utilized and allowed for adjustment of clinically relevant covariates (including patient age, race/ethnicity, sex, education level, income level, insurance status, facility type and location, Charlson/Deyo Comorbidity score, chemotherapy receipt, radiation receipt, and surgery receipt/type). After adjustment, the newly defined staging subgroups remained a significant factor associated with OS; group B versus A: HR 1.58 (95% CI 1.50– 1.67), group C versus A: HR 3.54 (95% CI 3.33–3.77) (all P-values < 0.001) (Supplemental Table 3, http://links.lww.com/SLA/C333).
DISCUSSION
In this study, we propose a novel prognostic stratification for patients with de novo metastatic breast cancer that further divides this patient population into 3 distinct groups: IVA, IVB, and IVC. To accomplish this, we utilized a RPA to amalgamate patients into 3 discrete groups with similar OS based on the prognostic variables used for early stage breast cancer (T stage, N stage, tumor grade, and biomarkers), and select features of the metastatic disease. Although other variables in our analysis were also associated with prognosis, we aimed to keep our modeling in alignment with the current staging guidelines for early-stage breast cancer (namely, TNM, ER, PR, HER2, and grade)8 and added 2 variables related to the metastatic population (number of metastatic sites and bone-only metastases) that have been shown in prior studies to be associated with prognosis.4–6 Similar to the staging guidelines for early-stage breast cancer, we propose that our model be applied to any de novo stage IV breast cancer patient who plans to pursue standard of care treatment. Our proposed stratification model will enable more accurate prognostic estimates that will inform both providers and patients, similar to how staging has been used for early-stage breast cancer discussions, and among stage IV patients in other disease sites which further stratify patients into stage IV risk categories. In addition, our model may potentially inform treatment selection for patients with de novo metastatic breast cancer, by allowing providers to better balance the toxicities of treatment to patient comorbidities and life expectancies.
Cancer staging systems serve to accurately and concisely summarize disease extent and prognosis, thus facilitating communication between providers and to patients to enable individualized treatment decisions.11 Implementation of the AJCC eighth edition in 2018 significantly improved the prognostic estimates provided by breast cancer staging, allowing more individualized counseling for early-stage breast cancer patients.9,12,13 However, the current staging system does not refine staging protocols for patients with metastatic disease, with all M1 patients still combined into a single stage, independent of anatomic and biologic factors.
Although the consolidation of all patients with metastatic breast cancer into a single stage implies a similar prognosis, survival has been shown to vary widely among patients with metastatic disease. Using large, national database analyses of both the NCDB and SEER, earlier studies have shown that the prognosis for patients with de novo metastatic breast cancer depends on intrinsic tumor biology and anatomic factors.4,5,14,15 Specifically, hormone receptor (HR)-positive disease, HER2-positive disease, fewer metastatic sites, and the presence of bone-only metastases have been shown to predict improved survival,15–17 factors we also identified to correlate with OS. In a study of 7575 patients with de novo stage IV breast cancer, the median OS for those with triple negative (HR-negative, HER2-negative) disease was <20 months compared to >40 months for those with HR-positive/HER2-negative disease.6 Based on the prognostic heterogeneity among this patient population, a need for more accurate prognostic estimates for patients with de novo metastatic breast cancer exists that is not met by the current staging guidelines. Although prognostic tools exist for breast cancer patients with brain metastases,18,19 these models have largely not been tested in the entire population of patients with de novo stage IV disease.
In other types of primary malignancies, stratification of metastatic disease into distinct subgroups has been shown to better predict prognosis. The International Association for the Study of Lung Cancer recently proposed subdividing metastatic non-small cell lung cancer patients according to number of metastatic sites.20 Similarly, in colorectal cancer, the AJCC seventh edition’s sub-classification of the M1 staging category into M1a and M1b to reflect single versus multiple metastatic sites has been shown to improve prognostication.21 Other studies have used RPA to sub-classify patients with metastatic disease into prognostic groups in prostate cancer, upper urinary tract carcinomas, and soft tissue sarcomas, yielding more accurate prognostic estimates.22–24 Finally, other studies in nonmetastatic cancer have also demonstrated the validity of using RPA with bootstrapping to propose or refine prognostic groups.25–28
Our stratification model similarly improves the prognostication of metastatic breast cancer staging, with implications both for patient counseling and treatment. The median OS in our study population was 32.3 months, which is similar to other studies.4 However, the median OS among our 3 subgroups ranged widely from 13.1 to 46.5 months, suggesting that use of this system could substantially improve discrimination among prognostic groups. With this improved discrimination, selecting appropriate patients for systemic and/or local-regional treatments, or clinical trials, may also be enhanced. For example, recent evidence suggests that treatment strategies targeted towards patients with ≤5 sites of metastatic disease may improve survival outcomes.29 Other studies have suggested that certain subgroups, including those with bone-only metastases,30,31 smaller primary tumors,4,32 lower metastatic burden,32 and HR-positivity,31,33 may benefit from resection of the primary breast tumor. More specifically, Soran et al. recently demonstrated that patients with ER/PR positive, HER2 negative disease, age <55, and solitary bone metastases had an improved OS.31 Although resection of the primary tumor remains controversial, surgical rates have remained relatively stable.34 However, given that local-regional treatment may selectively benefit those with fewer metastatic sites or with tumor biology amenable to systemic therapies,29 our proposed stratification could also become informative in surgical treatment planning and future research studies.
Similar to breast cancer staging for early-stage disease,35 genomic profiling and molecular markers will likely become critical tools for assessing prognosis for patients with metastatic disease. For example, circulating tumor cells (CTCs) have been proposed as a prognostic tool for staging advanced disease.36,37 Cristofanilli et al compared the survival rates of patients with metastatic breast cancer based on the number of CTCs and found that those with ≥5 CTCs had a worse survival.37 In a study of patients with ER-positive metastatic breast cancer, hybrid capture-based genomic profiling was carried out on ctDNA from peripheral blood, and 89% of mutations detected in tissue were also detected in ctDNA, suggesting this technology may serve as an alternative or complementary approach to tissue-based genomic testing for select patients.38 Moving forward, these tools may eventually be used not only to tailor treatments in a predictive fashion, but also to provide prognostic information.
Our study limitations include those associated with retrospective studies based on large databases. When using the NCDB, the definition of cM1 and pM1 disease follows the guidelines outlined in the AJCC staging manual.8 Based on how the data is coded in the NCDB, it is not possible to ascertain if the tumor characteristics listed in NCDB, such as tumor grade and receptor status, are based on biopsy from the primary tumor or distant sites. However, this study’s strengths lie in its use of novel methodology across a large sample of breast cancer patients. Furthermore, this model is reproducible and adaptable for future use, making it amenable to changes and additions incorporated in subsequent staging editions. It is important to note that our proposed staging system is based on a population that largely received treatment for their breast cancer, similar to how the latest staging guidelines for early-stage breast cancer were developed.8,39 In addition to externally validating this stratification model in more contemporary cohorts and other databases (national and institutional), future directions include examining the survival benefit derived from local and systemic therapy among our proposed subgroups. This will be particularly important in the HER2-positive subtypes, given the findings of both the CLEOPATRA and EMILIA trials which demonstrated improved survival for stage IV patients receiving novel therapies.40,41
In conclusion, our prognostic stratification model reliably discriminates outcomes in women with de novo metastatic breast cancer into 3 distinct prognostic groups. Within these groups, factors including anatomic staging, biomarker status, and extent of metastatic disease were associated with 3-year OS. Clinical implementation of this staging system for de novo stage IV breast cancer will enable more tailored patient counseling, individualized multidisciplinary treatment plans, and improved overall care among the growing population of survivors with metastatic disease. Inclusion of this staging system into the national guidelines will improve prognostication for patients with de novo metastatic breast cancer and will allow better patient selection for clinical trials and treatment interventions.
Supplementary Material
ACKNOWLEDGMENT
The authors would like to acknowledge and thank Lauren Halligan for her contributions in preparation of the figures for this manuscript.
Dr. R. Greenup is supported by the National Institutes of Health Office of Women’s Research Building Interdisciplinary Research Careers in Women’s Health K12HD043446 (PI: Andrews); Dr. O. Fayanju is supported by the National Institutes of Health (NIH) under Award Number 1K08CA241390 (PI: Fayanju). This work was in part supported by Duke Cancer Institute through NIH grant P30CA014236 (PI: Kastan) for the Biostatistics Core.
Dr. E.S. Hwang serves on the NCI Breast Cancer Steering Committee and the NCCN Breast Cancer Prevention Committee; Dr. J. Plichta is a recipient of research funding by the Color Foundation (PI: Plichta).
Footnotes
The authors report no conflicts of interest.
The authors report no proprietary or commercial interest in any product mentioned or concept discussed in this article.
The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
The National Cancer Data Base (NCDB) is a joint project of the Commission on Cancer (CoC) of the American College of Surgeons and the American Cancer Society. The CoC’s NCDB and the hospitals participating in the CoC NCDB are the source of the de-identified data used herein; they have not verified and are not responsible for the statistical validity of the data analysis or the conclusions derived by the authors.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.annalsofsurgery.com).
REFERENCES
- 1.Caswell-Jin JL, Plevritis SK, Tian L, et al. Change in survival in metastatic breast cancer with treatment advances: meta-analysis and systematic review. JNCI cancer spectrum. 2018;2:ky062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mariotto AB, Etzioni R, Hurlbert M, et al. Estimation of the number of women living with metastatic breast cancer in the United States. Cancer Epidemiol Biomarkers Prev. 2017;26:809–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sundquist M, Brudin L, Tejler G. Improved survival in metastatic breast cancer 1985–2016. Breast. 2017;31:46–50. [DOI] [PubMed] [Google Scholar]
- 4.Thomas A, Khan SA, Chrischilles EA, et al. Initial surgery and survival in stage IV breast cancer in the United States, 1988–2011. JAMA Surg 2016;151:424–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leone BA, Vallejo CT, Romero AO, et al. Prognostic impact of metastatic pattern in stage IV breast cancer at initial diagnosis. Breast Cancer Res Treat. 2017;161:537–548. [DOI] [PubMed] [Google Scholar]
- 6.Wu SG, Li H, Tang LY, et al. The effect of distant metastases sites on survival in de novo stage-IV breast cancer: a SEER database analysis. Tumour Biol. 2017;39:1010428317705082. [DOI] [PubMed] [Google Scholar]
- 7.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- 8.Hortobagyi GN, Connolly JL, D’Orsi CJ, et al. AJCC Cancer Staging Manual. 8th ed. New York, NY: Springer International Publishing; 2016. [Google Scholar]
- 9.Plichta JK, Campbell BM, Mittendorf EA, et al. Anatomy and breast cancer staging: is it still relevant? Surg Oncol Clin N Am 2018;27:51–67. [DOI] [PubMed] [Google Scholar]
- 10.Strobl C, Malley J, Tutz G. An introduction to recursive partitioning: rationale, application, and characteristics of classification and regression trees, bagging, and random forests. Psychol Methods. 2009;14:323–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gospodarowicz MK, Miller D, Groome PA, et al. The process for continuous improvement of the TNM classification. Cancer. 2004;100:1–5. [DOI] [PubMed] [Google Scholar]
- 12.Hortobagyi GN, Connolly JL, D’Orsi CJ, et al. AJCC Cancer Staging Manual. 8th ed. New York, NY: 2016. [Google Scholar]
- 13.Wang M, Chen H, Wu K, et al. Evaluation of the prognostic stage in the 8th edition of the American Joint Committee on Cancer in locally advanced breast cancer: an analysis based on SEER 18 database. Breast. 2018;37:56–63. [DOI] [PubMed] [Google Scholar]
- 14.Press DJ, Miller ME, Liederbach E, et al. De novo metastasis in breast cancer: occurrence and overall survival stratified by molecular subtype. Clin Exp Metastasis. 2017;34:457–465. [DOI] [PubMed] [Google Scholar]
- 15.Lobbezoo DJ, van Kampen RJ, Voogd AC, et al. Prognosis of metastatic breast cancer subtypes: the hormone receptor/HER2-positive subtype is associated with the most favorable outcome. Breast Cancer Res Treat. 2013;141:507–514. [DOI] [PubMed] [Google Scholar]
- 16.Tao L, Chu L, Wang LI, et al. Occurrence and outcome of de novo metastatic breast cancer by subtype in a large, diverse population. Cancer Causes Control 2016;27:1127–1138. [DOI] [PubMed] [Google Scholar]
- 17.Howlader N, Cronin KA, Kurian AW, et al. Differences in breast cancer survival by molecular subtypes in the United States. Cancer Epidemiol Biomarkers Prev. 2018;27:619–626. [DOI] [PubMed] [Google Scholar]
- 18.Subbiah IM, Lei X, Weinberg JS, et al. Validation and development of a modified breast graded prognostic assessment as a tool for survival in patients with breast cancer and brain metastases. J Clin Oncol. 2015;33:2239–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Scodan R, Massard C, Jouanneau L, et al. Brain metastases from breast cancer: proposition of new prognostic score including molecular subtypes and treatment. J Neurooncol. 2012;106:169–176. [DOI] [PubMed] [Google Scholar]
- 20.Eberhardt WE, Mitchell A, Crowley J, et al. The IASLC lung cancer staging project: proposals for the revision of the M descriptors in the forthcoming eighth edition of the TNM classification of lung cancer. J Thorac Oncol. 2015;10:1515–1522. [DOI] [PubMed] [Google Scholar]
- 21.Kennecke H, Yu J, Gill S, et al. Effect of M1a and M1b category in metastatic colorectal cancer. Oncologist. 2014;19:720–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang S, Kim HS, Kim S, et al. Post-metastasis survival in extremity soft tissue sarcoma: a recursive partitioning analysis of prognostic factors. Eur J Cancer. 2014;50:1649–1656. [DOI] [PubMed] [Google Scholar]
- 23.Abdel-Rahman O Assessment of the prognostic value of the 8th AJCC staging system for patients with clinically staged prostate cancer; a time to sub-classify stage IV? PLoS One. 2017;12:e0188450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abdel-Rahman O Revisiting the prognostic heterogeneity of AJCC stage IV carcinomas of the upper urinary tract. Clin Genitourin Cancer. 2018. [DOI] [PubMed] [Google Scholar]
- 25.Adam MA, Thomas S, Roman SA, et al. Rethinking the current American Joint Committee on Cancer TNM staging system for medullary thyroid cancer. JAMA Surg. 2017;152:869–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keane FK, Chen YH, Tishler RB, et al. Population-based validation of the recursive partitioning analysis-based staging system for oropharyngeal cancer. Head Neck. 2016;38:1530–1538. [DOI] [PubMed] [Google Scholar]
- 27.Abdel-Rahman O Dissecting the heterogeneity of stage III non-small-cell lung cancer through incorporation of grade and histology. Future Oncol (London England) 2017;13:2811–2821. [DOI] [PubMed] [Google Scholar]
- 28.Yuan SQ, Chen YT, Huang ZP. Equipping the 8th Edition American Joint Committee on cancer staging for gastric cancer with the 15-node minimum: a population-based study using recursive partitioning analysis. J Gastrointest Surg 2017;21:1591–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palma DA, Olson R, Harrow S, et al. Stereotactic ablative radiotherapy versus standard of care palliative treatment in patients with oligometastatic cancers (SABR-COMET): a randomised, phase 2, open-label trial. Lancet. 2019;393:2051–2058. [DOI] [PubMed] [Google Scholar]
- 30.Rapiti E, Verkooijen HM, Vlastos G, et al. Complete excision of primary breast tumor improves survival of patients with metastatic breast cancer at diagnosis. J Clin Oncol. 2006;24:2743–2749. [DOI] [PubMed] [Google Scholar]
- 31.Soran A, Ozmen V, Ozbas S, et al. Randomized trial comparing resection of primary tumor with no surgery in stage IV breast cancer at presentation: protocol MF07–01. Ann Surg Oncol. 2018;25:3141–3149. [DOI] [PubMed] [Google Scholar]
- 32.Harris E, Barry M, Kell MR. Meta-analysis to determine if surgical resection of the primary tumour in the setting of stage IV breast cancer impacts on survival. Ann Surg Oncol. 2013;20:2828–2834. [DOI] [PubMed] [Google Scholar]
- 33.Meimarakis G, Ruttinger D, Stemmler J, et al. Prolonged overall survival after pulmonary metastasectomy in patients with breast cancer. Ann Thorac Surg. 2013;95:1170–1180. [DOI] [PubMed] [Google Scholar]
- 34.Lane WO, Thomas SM, Blitzblau RC, et al. Surgical resection of the primary tumor in women with de novo stage IV breast cancer: contemporary practice patterns and survival analysis. Ann Surg. 2019;269:537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mittendorf EA, Bartlett JMS, Lichtensztajn DL, et al. Incorporating biology into breast cancer staging: American Joint Committee on Cancer, Eighth Edition, Revisions and Beyond. Am Soc Clin Oncol Educ Book. 2018;38:38–46. [DOI] [PubMed] [Google Scholar]
- 36.Larsson AM, Jansson S, Bendahl PO, et al. Longitudinal enumeration and cluster evaluation of circulating tumor cells improve prognostication for patients with newly diagnosed metastatic breast cancer in a prospective observational trial. Breast Cancer Res. 2018;20:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cristofanilli M, Pierga JY, Reuben J, et al. The clinical use of circulating tumor cells (CTCs) enumeration for staging of metastatic breast cancer (MBC): international expert consensus paper. Crit Rev Oncol Hematol 2019;134:39–45. [DOI] [PubMed] [Google Scholar]
- 38.Chung JH, Pavlick D, Hartmaier R, et al. Hybrid capture-based genomic profiling of circulating tumor DNA from patients with estrogen receptor-positive metastatic breast cancer. Ann Oncol. 2017;28:2866–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giuliano AE, Connolly JL, Edge SB, et al. Breast cancer—major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017. [DOI] [PubMed] [Google Scholar]
- 40.Baselga J, Cortes J, Kim SB, et al. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med. 2012;366:109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verma S, Miles D, Gianni L, et al. Trastuzumab emtansine for HER2-positive advanced breast cancer. N Engl J Med. 2012;367:1783–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.