Abstract
Aedes aegypti develop in aquatic habitats in which mosquito larvae are exposed to physicochemical elements and microorganisms that may influence their life cycle and their ability to transmit arboviruses. Little is known about the natural bacterial communities associated with A. aegypti or their relation to the biotic and abiotic characteristics of their aquatic habitats. We characterized the physicochemical properties and bacterial microbiota of A. aegypti breeding sites and larvae on Guadeloupe and in French Guiana. In addition, we explored whether geographic location, the type of breeding site and physicochemical parameters influenced the microbiota associated with this mosquito species. We used large-scale 16S rRNA gene sequencing of 160 breeding sites and 147 pools of A. aegypti larvae and recorded 12 physicochemical parameters at the sampled breeding sites. Ordination plots and multiple linear regression were used to assess the influence of environmental factors on the bacterial microbiota of water and larvae. We found territory-specific differences in physicochemical properties (dissolved oxygen, conductivity) and the composition of bacterial communities in A. aegypti breeding sites that influenced the relative abundance of several bacteria genera (e.g., Methylobacterium, Roseoccocus) on the corresponding larvae. A significant fraction of the bacterial communities identified on larvae, dominated by Herbiconiux and Microvirga genera, were consistently enriched in mosquitoes regardless the location. In conclusion, territory-specific differences observed in the biotic and abiotic properties of A. aegypti breeding sites raise concern about the impact of these changes on pathogen transmission by different A. aegypti populations.
Electronic supplementary material
The online version of this article (10.1007/s00248-020-01544-3) contains supplementary material, which is available to authorized users.
Keywords: Aedes aegypti, Breeding site, Physicochemical parameter, Bacterial community
Background
Aedes aegypti (Diptera: Culicidae) is a mosquito vector of arboviruses such as dengue, chikungunya and Zika that constitute major global health problems and threaten the French overseas territories of Guadeloupe and French Guiana [1, 2]. Besides the high susceptibility for these pathogens, this mosquito is a main vector of arboviruses because it is highly anthropophilic and thrives close to humans in urban and peri-urban areas [3, 4]. The species breeds mainly in human-made containers with volumes ranging from a few millilitres to hectolitres, such as drums, plastic buckets, cisterns, flower vases and rubber tyres [4, 5].
Water quality is critical for the choice of oviposition site for gravid females to ensure egg hatching and the development of their progeny from larvae to adults [6–8]. Females select breeding sites according to biotic and abiotic elements in the water, such as organic matter [8], bacteria [9, 10], phosphate, ammonia and potassium [11–13], which are known to be closely related to the abundance of larvae and adults in the field [13–15]. Larvae in their aquatic habitats rely on bacteria communities and organic matter whose composition is highly variable depending on environmental fluctuations [16, 17]. It has been well documented that environmental conditions such as nutritional deficiency, competition and high temperatures (> 30 °C) experienced during larval development can result in lifespan decrease, reduced adult size and increased susceptibility to virus transmission (i.e. Sindbis virus) [18]. In addition, exposure to bacteria in breeding sites during larval development shapes the microbiota of larvae and adult mosquitoes and also affects phenotype traits related to vectorial capacity, such as egg development, lifespan and vector competence [19–21]. For example, larval exposure to an Enterobacteriaceae isolate has been shown to reduce dengue virus dissemination titers in adult mosquitoes [16]. Similarly, exposure to a Bacillus isolate during larval development resulted in significantly increased rates of dengue virus infection and dissemination in the corresponding adults [22]. Thus, the quality of water in which A. aegypti mosquitoes breed might also have a role in determining their susceptibility to human pathogens. These findings highlight the importance of understanding natural variations in the habitat and microbiota of local A. aegypti populations and their potential contributions to adult phenotypic traits of epidemiological interest. However, the complexity and plasticity of A. aegypti–bacteria interactions in breeding sites are still poorly understood, and it is unclear how the bacterial communities in A. aegypti breeding sites are structured in natural settings at different geographical scales.
A handful of studies have reported marked effects of water physicochemical parameters (i.e. pH, dissolved oxygen) on egg hatch and larval physiology [23, 24], but information about their impact on vector competence is still lacking. Field and laboratory studies have shown that dissolved oxygen is positively correlated with larval abundance, while extreme salinity, temperature and pH reduce the abundance and development of larvae [25–30]. Interestingly, a few studies have shown that heavy metals such as iron (Fe), zinc (Zn) and copper (Cu) may be present at different concentrations in A. aegypti breeding sites [31, 32], but their association with the presence and abundance of A. aegypti larvae remains unclear [32]. Abiotic factors can also influence the structure of microbial communities in diverse ecosystems as soil and water. Pollution by heavy metals and changes in salinity [33], temperature [34] and pH [35] can change the metabolic activity and the relative abundance of bacteria. Changes in the microbial composition of the aquatic habitat and gut of mosquitoes have often been attributed to variations in geography [36, 37] or seasonal climatic patterns [38, 39], but no study has addressed whether changes in microbial composition are directly attributable to the physicochemical properties of breeding sites. Despite the omnipresence of A. aegypti mosquitoes in tropical and subtropical regions and their importance in public health, basic understanding of the abiotic factors and the natural microbial communities associated with local populations of the species and the corresponding aquatic habitats is still lacking.
We comprehensively characterized the physicochemical properties and natural bacterial communities associated with A. aegypti breeding sites and larvae on Guadeloupe and in French Guiana. We explored whether features of larval habitats, including geographical location, type of breeding site and the physicochemical parameters of the water (i.e. pH, temperature and turbidity) influence the variation in the bacterial microbiota associated with this mosquito species.
Methods
Collection of Samples
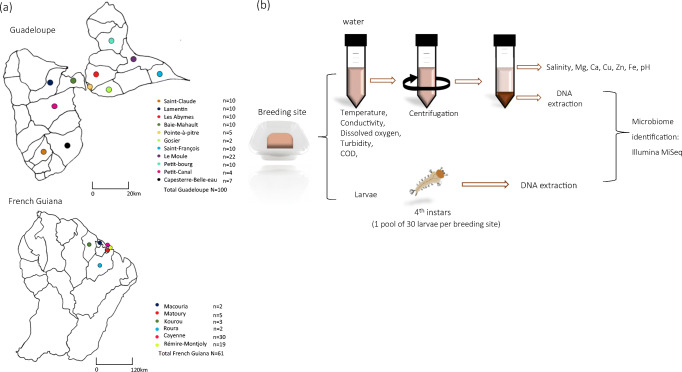
In 2017, A. aegypti breeding sites were selected in urban areas of Guadeloupe and French Guiana (Fig. 1a). At each breeding site (100 on Guadeloupe and 61 in French Guiana), sampling was conducted during the dry season: May and June on Guadeloupe and October and November in French Guiana. Artificial breeding sites (tyres, flower vases, drums, freezers) containing at least 100 A. aegypti larvae and a water volume of at least 700 mL were sampled. A. aegypti larvae were firstly identified in the field based on their specific movements and siphon morphology. The identification was then confirmed on the laboratory using a binocular loupe at a magnification of × 56 (Leica M80, Leica, Nanterre, France) and morphological descriptions [40].
Fig. 1.
Study design. a Location of Aedes aegypti breeding sites sampled on Guadeloupe and in French Guiana. b Workflow for determination of breeding sites physicochemical parameters and for microbiota identification. N number of samples collected in municipalities
The water and larvae were collected separately and taken to the corresponding facilities of Instituts Pasteur of Guadeloupe and French Guiana. The breeding sites were assigned to one of 10 classes according to their typology, in the categories used by local vector control authorities on Guadeloupe and in French Guiana [41]: (i) plant containers (pots, dishes, vases, basins), (ii) buckets (including water cans), (iii) drums, (iv) tyres, (v) cisterns, (vi) small waste (tins), (vii) large waste (freezers, toilet bowls), (viii) boats, (ix) water troughs and (x) gutters.
Physicochemical Analysis of Water at Breeding Sites
Physicochemical analysis was performed on the water collected from breeding sites, as shown in Fig. 1b. For each breeding site, the water temperature (°C) (T), pH and electrical conductivity (μS/cm) were measured directly in the field with a multi-parameter probe (Odeon, PONSEL, France, on Guadeloupe and HANNA Instruments, France, in French Guiana) and an electrical conductivity meter (Cond 3210 + probe TetraCon 325, WTW, Germany). Dissolved oxygen (DO) (mg/L), turbidity (formazin nephelometric unit, FNU) and chemical oxygen demand (COD) (mg/L) were measured in 200 mL of water with no previous sample treatment. The remaining volume of water (~ 500 mL) was centrifuged at 8000 rpm for 10 min at 4 °C. The pellets obtained were preserved at − 20 °C for DNA extraction, and the supernatants were used to determine salinity (g/L), and Fe, Mg, Ca, Cu, Zn all measured in mg/L. All nine physicochemical parameters determined in the laboratory were analysed by accredited standard methods (www.cofrac.fr) at the Laboratory of Environmental Hygiene at the Instituts Pasteur of Guadeloupe and French Guiana.
Bacterial DNA Extraction from Water and Larval Samples
For each breeding site, total DNA extraction was performed on 30 randomly selected fourth-instar larvae of A. aegypti, which were pooled and stored at − 20 °C, and on the pellets obtained from water samples. Total genomic DNA was extracted from the pellets with the NucleoSpin® Soil kit (Macherey-Nagel, USA) according to the manufacturer’s instructions (Fig. 1b). Pooled larvae were surface-sterilized as described previously [42] before DNA extraction. Briefly, each larval pool was first rinsed three times in 2 mL of sterile water, then exposed to 70% ethanol for 10 min. Lastly, the sample was rinsed six times: five in 2 mL of sterile water and once in 2 mL of sterile 0.8% NaCl. DNA extraction and PCR were carried out on each last water rinse to ensure the absence of exogenous bacterial DNA. Subsequently, each larval sample was crushed in 80 μL of sterile phosphate-buffered saline with a bead beater (MM 40, RETSCH, France) at 30 Hz for 30 s. Then, 20 μL of proteinase K (50 μg/mL) and 700 μL of SL1 (buffer lysis) from a NucleoSpin® Soil kit (Macherey-Nagel, USA) were added to homogenates, which were then incubated overnight at 56 °C. Samples were centrifuged for 2 min at 11,000 rpm, and the next steps of the protocol were carried out according to the manufacturer’s instructions. Sterile DNA-free water was used at each extraction as a negative control to check for contamination.
To confirm the presence of bacterial DNA in each water and larva sample, 25-μL PCR reactions were performed to amplify the 16S rRNA gene with the universal primers 27F (5′ GAGTTTGATCNTGGCGGCTCATCAG 3′) and 1492R (5′ GTNTTACNGCGGCKGCTG 3′), as previously described [43], using DreamTaq DNA Polymerase (Thermo Scientific, USA) according to the manufacturer’s instructions. The presence of PCR amplification fragments was confirmed on 1.5% agarose gel electrophoresis stained with Gel Red (Biotium, USA) and visualized under ultraviolet light.
Illumina MiSeq Paired-End Sequencing and Sequence Processing
The V6–V8 region of the 16S rRNA gene was sequenced with the primers B969F-CS1 5′- ACGCGHNRAACCTTACC-3′ and BA1406R-CS2 5′-ACGGGCRGTGWGTRCAA-3′ [44]. The sequencing technology used was Illumina MiSeq 250-bp paired-ends, conducted at the Quebec Genome Innovation Centre (McGill University, Montreal, Canada), which generated 36,933,412 raw sequences from water (n = 161) and larval samples (n = 145). Sequences were processed with the software USEARCH version 10.0.240 following the UPARSE pipeline [45]. The two paired-end reads with fewer than five mismatches were merged. The maximum allowed ratio between the number of mismatched base pairs and the overlap length was set to 0.3. Reads with low-quality scores were removed, with a maximum expected error value of 1.0. The remaining 12,296,888 high-quality reads were de-replicated and sorted by abundance, and all singletons and chimera were removed. Unique reads were then clustered into operational taxonomic units (OTUs) with the UPARSE OTU clustering method and a 97% identity threshold, with a minimum of two sequences considered to be an OTU. Taxonomic assignment was realized with the Ribosomal Database Project classifier version 16 to remove OTUs identified as chloroplasts. OTUs represented by < 0.005% of the total number of reads were removed. The OTU table was normalized to 20,000 sequences/sample. Sequencing led to adequate coverage of the bacterial communities (Additional file 1).
Data Analysis
Rarefaction curves were generated with the software SHAMAN (SHiny Application for Metagenomic ANalysis, Paris, France, shaman.pasteur.fr) to assess the sufficiency of sequencing [46]. The diversity of OTUs within and between samples was compared with alpha and beta diversity indices, respectively. Alpha-diversity metrics (species richness, equitability, Chao1, Shannon, Simpson diversity indices) and beta-diversity metrics (Bray-Curtis distances matrix) were generated with USEARCH. To summarize and compare the composition of bacterial communities at the different A. aegypti breeding sites and on larvae, bar charts and pie charts were generated showing the distribution of bacterial genera with SHAMAN. The physicochemical parameters and alpha diversity among different breeding sites and localities were compared in Kruskal-Wallis and Mann-Whitney U tests. P values were adjusted with a Benjamini-Hochberg correction.
Principal component analysis was used to explain variation in physicochemical parameters according to container type. A permutational multivariate analysis of variance analysis was conducted with Bray-Curtis distance matrices and 999 random permutations to determine the relations between categorical variables associated with each breeding site and the microbial communities identified from the corresponding water and larvae samples. The categorical variables tested were sampling locality, container type and physicochemical variable. Weighted and unweighted UniFrac distances were used to evaluate diversity among groups with non-metric multidimensional scaling plots in the R program Phyloseq [47]. Volcano plots were generated in SHAMAN to identify significant differences in abundant bacterial taxa in water and larval samples, and significant differences were determined by the Wald test. Canonical correlation analysis was used to identify among the 12 physicochemical parameters, those having a greater influence on the 100 most abundant bacterial genera in water and larval samples. Multivariate regression analysis was used to detect significant correlations between the physicochemical parameters selected from the canonical correspondence analysis and abundant bacteria genera. The results were displayed on a heat map. All tests were conducted with the software XLSTAT-Ecology (XLSTAT 2019.1.2), and the level of statistical significance in all analyses was P ≤ 0.05, except for the results of the multivariate regression analysis, for which the cut-off for significance was P < 0.15 in order to eliminate variables that were less strongly associated with microbial community members.
Results
Differences in Physicochemical Parameters of A. aegypti Breeding Sites on Guadeloupe and in French Guiana
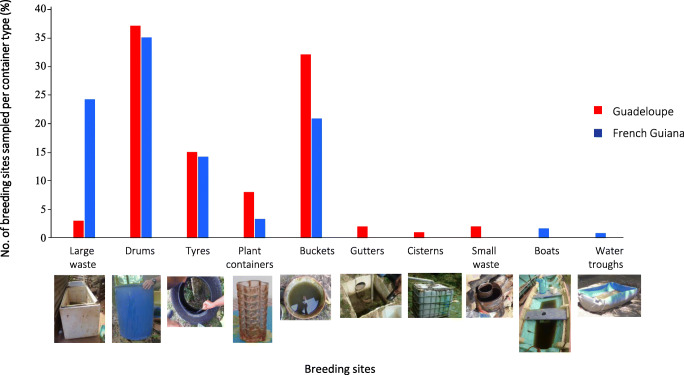
A total of 161 breeding sites in eight container classes on Guadeloupe and seven in French Guiana were investigated (Fig. 2). The most common breeding sites sampled were drums (> 30%), followed by buckets (> 25%) and large waste (~ 25% in French Guiana). Other breeding sites (tyres, plant containers, gutters, small waste, cisterns, boats and water troughs) represented < 15% in the two territories.
Fig. 2.
Breeding sites sampled per container type on Guadeloupe and in French Guiana
No significant differences were detected in pH or temperature according to container class or location (Table 1); however, an effect of container type was observed for electrical conductivity, turbidity, COD and mineral content, which were significantly higher in tyres, buckets and large waste than in drums. Differences were also observed between the two territories in levels of conductivity, dissolved oxygen, Ca and Mg in breeding sites, which were higher on Guadeloupe, and in turbidity, COD, Cu, Fe and Zn content, which were higher in French Guiana.
Table 1.
Physicochemical parameters (mean ± SE) per container class on Guadeloupe (G) and in French Guiana (FG). Significant differences between G and FG for a given parameter and container type are shown in italic. Different letters indicate significant differences for a given parameter among container types in a same locality (G or FG) (P < 0.05). COD, chemical oxygen demand; Cu, copper; Fe, iron; Zn, zinc; Ca, calcium
| Parameter | Locality | Large waste | Small waste | Buckets | Tyres | Drums | Water troughs | Plant containers | Boats | Gutters | Cisterns |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Temperature (°C) |
G | 28 ± 1 | 29 ± 0.9 | 29 ± 1.6 | 30 ± 2 | 29 ± 1.8 | _ | 29 ± 0.9 | _ | 29 ± 0.5 | 26 |
| FG | 28 ± 2 | 27 | 29 ± 2.5 | 28 ± 1.7 | 29 ± 2.1 | 24 | _ | 34 ± 4.9 | _ | _ | |
| pH | G | 8 ± 1 | 8 ± 1 | 8 ± 0.7 | 8 ± 0.5 | 8 ± 0.8 | _ | 8 ± 0.5 | _ | 7 ± 0.4 | 6 |
| FG | 7 ± 0.5 | 7 | 7 ± 0.5 | 7 ± 0.7 | 7 ± 0.3 | 7 | _ | 8 ± 0.9 | _ | _ | |
|
Salinity (g/L) |
G | 0.2 ± 0.2 | 0 | 0 ± 0.1a | 0.1 ± 0.1 | 0 | _ | 0.1 ± 0.1 | _ | 0 | 0 |
| FG | 0.1 ± 0a | 0.2abc | 0abc | 0.1 ± 0.1ab | 0c | 0.1abc | _ | 0.1abc | _ | _ | |
|
Conductivity (μS/cm) |
G | 464 ± 554abc | 299 ± 205abc | 230 ± 215a | 304 ± 148ab | 96 ± 79c | _ | 455 ± 230abc | _ | 202 ± 57abc | 45abc |
| FG | 166 ± 179 | 356 ± 49 | 105 ± 120 | 213 ± 225 | 35 ± 26 | 236 | _ | 199 ± 32.5 | _ | _ | |
| Dissolved oxygen (mg/L) | G | 10 ± 1 | 9 ± 1 | 9 ± 2.3 | 9 ± 1.4 | 9 ± 2.1 | _ | 9 ± 2.5 | _ | 8.3 ± 1.8 | 9 |
| FG | 3 ± 1.7 | 0 | 4 ± 1.54 | 3.4 ± 1.8 | 4 ± 1.3 | 9 | _ | 6 ± 1.3 | _ | ||
|
Turbidity (FNU) |
G | 8 ± 5.7ab | 47 ± 29.8ab | 27 ± 28.2ab | 55 ± 29.9a | 16 ± 20b | _ | 46 ± 31ab | _ | 51 ± 30.7ab | 7ab |
| FG | 49 ± 84a | 133abc | 18 ± 22abc | 120 ± 49b | 7 ± 8c | 10abc | 10 ± 6.7abc | _ | _ | ||
|
Ca (mg/L) |
G | 17 ± 9.2abc | 14 ± 7.8abc | 26 ± 36.2abc | 35 ± 14.8a | 11 ± 11.9b | _ | 35 ± 19 ac | _ | 20 ± 19.1abc | 2abc |
| FG | 11 ± 0.6a | 18ab | 12 ± 17.5ab | 11 ± 7.3ab | 2 ± 1.9b | _ | _ | 20 ± 8.9ab | _ | _ | |
|
Mg (mg/L) |
G | 8.1 ± 10.5abcd | 3.8 ± 2.2abcd | 4.7 ± 8.7a | 3.5 ± 2.2ab | 1.0 ± 01.3c | _ | 7.0 ± 4.7abd | _ | 1.7 ± 0.3abcd | 0abcd |
| FG | 1.4 ± 0.6 | 4.5 | 1.5 ± 2.2 | 1.2 ± 1 | 0.2 ± 0.4 | 2.3 | _ | 2.3 ± 0.5 | _ | _ | |
|
Cu (mg/L) |
G | 0.01 ± 0ab | 0ab | 0ab | 0.02 ± 0a | 0b | _ | 0.01 ± 0ab | _ | 0ab | 0ab |
| FG | 0.03 ± 0abc | 0abc | 3 ± 9.9a | 0.04 ± 0b | 0 ac | _ | 0abc | _ | _ | ||
|
Fe (mg/L) |
G | 0ab | 2.7 ± 2.6ab | 0.5 ± 1ab | 0.8 ± 0.9a | 0.3 ± 0.8b | _ | 0.3 ± 0.3ab | _ | 0.5 ± 0.3ab | 0.1ab |
| FG | 11 ± 26.6abc | 22.9abc | 0.5 ± 0.4a | 10.9 ± 5.4b | 1.3 ± 2.4 ac | 2.7abc | _ | 2.7 ± 0.9abc | _ | _ | |
|
Zn (mg/L) |
G | 0ab | 0ab | 0.3 ± 0.6ab | 1.0 ± 1.3a | 0.5 ± 0.6ab | _ | 0b | _ | 1.3 ± 1.7ab | 3.1ab |
| FG | 4 ± 6.7a | 0.2ab | 0.5 ± 0.6b | 2.7 ± 1.8ab | 1.3 ± 0.9ab | _ | _ | 1.3 ± 1ab | _ | _ | |
|
COD (mg/L) |
G | 21 ± 10.5abc | 56 ± 46abc | 46 ± 45a | 195 ± 168b | 24 ± 30ac | _ | 49 ± 25.6abc | _ | 20 ± 5.1abc | 20abc |
| FG | 229 ± 426a | 529abc | 54 ± 43abc | 216 ± 153ab | 17 ± 35c | 242abc | _ | 242 ± 149.2abc | _ | _ |
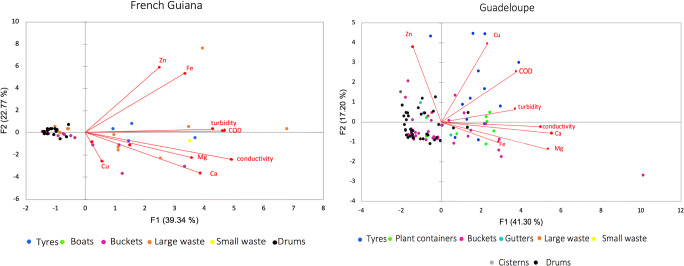
The principal component multivariate analysis (PCA) revealed strong associations between physicochemical variables and breeding sites classes (Fig. 3), with 59% and 62% of the total variance explained by the first two axes on Guadeloupe and in French Guiana, respectively. Only eight physicochemical parameters out of the 12 measured displayed significant variation between sampling sites and/or container-types (Table 1), and were therefore selected for the PCA analysis. Whatever the sampling territory, the two main PCA components (F1 and F2) clearly separated breeding sites classes in two groups: one containing the tyres and the majority of the flowerpots (right area) and one containing the majority of buckets and drums (left area). Such structuration of samples suggests that physicochemical profiles in drums and buckets are similar, but differ from those of plant containers and tyres (PERMANOVA R2 = 0.22 for Guadeloupe, R2 = 0.23 for French Guiana, P < 0.001). Plant containers and tyres had indeed higher COD and conductivity when compared to other breeding sites classes, as reflected by the length and direction of the vectors for these two physicochemical parameters. Finally, the examination of physicochemical parameters associations revealed that across all types of containers, conductivity was positively correlated with Mg (Spearman r = 0.49, P < 0.01) and Ca (Spearman r = 0.62, P < 0.01), while turbidity was associated with COD (Spearman r = 0.59, P < 0.01). Zn content was positively associated with Fe in French Guiana (Spearman r = 0.64, P < 0.01) but negatively on Guadeloupe (Spearman r = − 0.23, P = 0.04).
Fig. 3.
Principal component analysis of eight physicochemical parameters associated with Aedes aegypti breeding sites in French Guiana and on Guadeloupe. Dots correspond to the breeding sites sampled arrayed according to the measured physicochemical parameters. Colour-code indicates different container types. Vectors represent physicochemical parameters and point in the direction of steepest increase of values for the corresponding physicochemical parameter. The water samples from tyres and plant containers are closely related suggesting similar physicochemical profiles and located in the same direction as the vectors, indicating high COD, conductivity and turbidity. Conversely, the water samples from drums and buckets form a cluster on the opposite side of the vectors indicating lower physicochemical values. Ca, Calcium; Mg, Magnesium; Cu, Copper; Fe, Iron; Zn, Zinc; COD, Chemical oxygen demand. Percentages in parenthesis represent the variability explained by each principal component
Bacterial Communities Associated with A. aegypti Breeding Sites
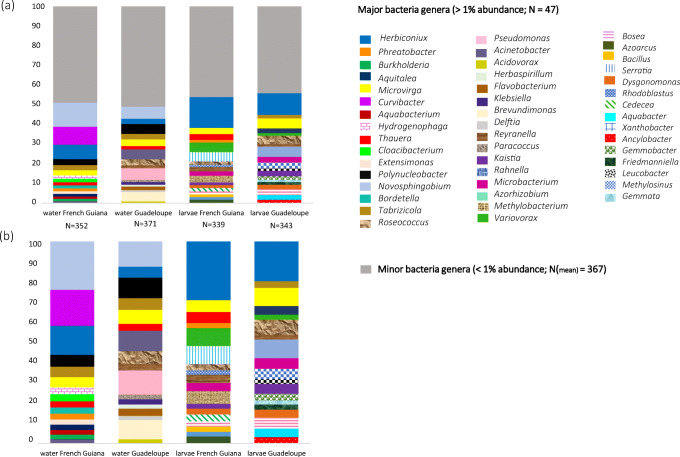
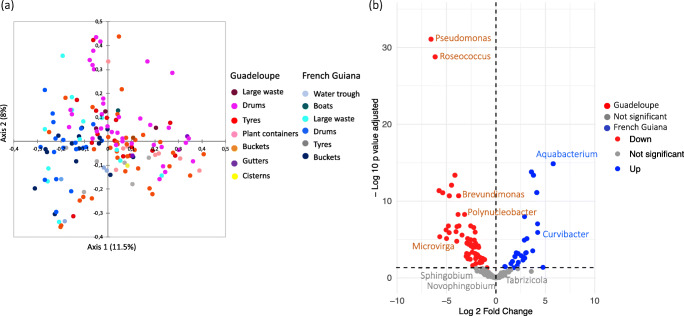
A total of 967 OTUs (117–540 OTUs per sample) belonging to 15 phyla and 376 genera were identified in water collected from A. aegypti breeding sites. The diversity and richness of the bacterial communities across breeding sites and localities were comparable (mean Shannon index 2.5 ± 0.9, mean richness 252.5 ± 0.16, Additional file 2), except in plant containers, which were significantly more diverse than the other classes of breeding site (ANOVA, Tukey post hoc, P < 0.05). The predominant phyla were Proteobacteria (54%), Bacteroidetes (22%), Actinobacteria (10%) and Firmicutes (6%) (Additional file 3). At higher taxonomic resolution, > 80% of the bacterial genera displayed low abundance in water (< 1%). The distribution of the 26 most abundant genera varied considerably among samples, with some genera (e.g., Novosphingobium, Tabrizicola, Acinetobacter) abundant in all containers and locations and others specific to the territory or habitat (Fig. 4, Additional file 4). Non-metric multidimensional scaling unweighted UniFrac analysis and PCoA plots based on Bray-Curtis dissimilarity revealed clear groupings of water samples by territory, showing significant differences in microbial composition between Guadeloupe and French Guiana (PERMANOVA, R2 = 0.2, P < 0.001, Fig. 5a, Additional file 5b). The differences were due mainly to taxa such as Roseoccocus, Pseudomonas and Polynucleobacter, which were significantly more abundant on Guadeloupe (log2 fold-change = 6.5, 5.5 and 5.21, respectively, P < 0.05), while Curvibacter, Aquabacterium and Burkholderia were more abundant in French Guiana (log2 fold-change = 3.3, 2.8 and 2.7 respectively, P < 0.05) (Fig. 5b, Additional file 6). On a finer scale, the bacterial communities in breeding sites were variable, and only slight structuring according to container type was detected (PERMANOVA, R2 = 0.14, P < 0.001, Fig. 5a).
Fig. 4.
Mean relative abundance of bacterial genera found in Aedes aegypti breeding-sites water and larvae in Guadeloupe and French Guiana (a) and relative proportion of major bacteria genera (> 1%) (b)
Fig. 5.
Spatial structuration and variability of microbiota associated with Aedes aegypti breeding site water. a Principal coordinate analysis plots based on Bray Curtis dissimilarity distances. Dots correspond to the breeding sites sampled arrayed according to the similarity of their microbiota. b Volcano plot for differential bacteria genera abundance in water from breeding sites on Guadeloupe and in French Guiana. Each dot represents a bacterial genera found in the samples and the most abundant bacterial genera, with a significant fold-change between the two territories (P < 0.05) are labelled
Bacterial Communities Associated with A. aegypti Larvae
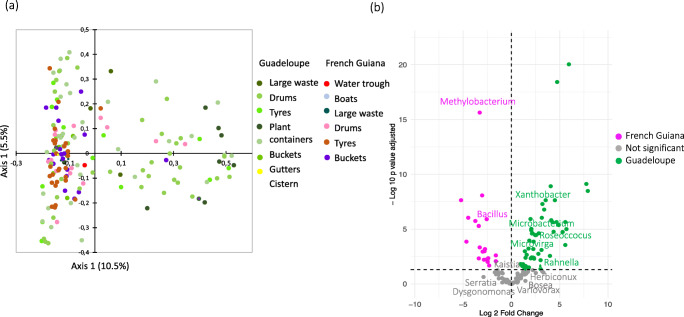
Analysis of the microbiota of A. aegypti larvae revealed 931 OTUs belonging to 15 bacterial phyla and 346 genera. The sampled larvae harboured a wide range of bacterial taxa (up to 170 different species per sample), of which about 16% had a relative abundance > 1%. All bacteria taxa identified in A. aegypti larvae were found in the corresponding breeding sites. As for the water samples, the most abundant phylum in larvae was Proteobacteria (> 50% of all bacteria), followed by Actinobacteria (30%) and Firmicutes (10%) (Additional file 3). The bacterial communities of A. aegypti larvae were less rich (Kruskal-Wallis: P < 0.05) and more homogeneous than those in water from breeding sites, regardless the container type (Additional file 2). No major differences in larval microbiota were detected with respect to species composition, diversity (mean Shannon index 2.5 ± 0.9) or richness (mean 195.2 ± 0.35) in either territory (PERMANOVA, R2 = 0.03, P < 0.001, Additional file 2, Fig. 6a). The most widely spread, abundant bacterial genera were enriched in all the larval samples, regardless of locality, including Bosea (log2 fold-change over water = 3.3, P < 0.05), Herbiconiux (log2 fold-change over water = 3.2, P < 0.05), Bacillus (log2 fold-change over water = 3.5, P < 0.05) and Kaistia (log2 fold-change over water = 2.7, P < 0.05) (Table 2).
Fig. 6.
Spatial structuration and variability of microbiota associated with Aedes aegypti larvae. a Principal coordinate analysis plots based on Bray Curtis dissimilarity distances. Dots correspond to the breeding sites sampled arrayed according to the similarity of their microbiota. b Volcano plot for differential bacteria genera abundance in larvae between Guadeloupe and French Guiana. Each dot represents a bacterial genus found in the samples and the most abundant bacterial genera, with a significant fold-change between the two territories (P < 0.05) are labelled
Table 2.
Bacteria genera enriched in A. aegypti larvae as compared to the corresponding breeding site
| Similar enrichment in both territories (G vs FG, P < 0.05) |
log2 fold-change | More enriched in G (G vs FG, P < 0.05) |
log2 fold-change | More enriched in FG (G vs FG, P < 0.05) |
log2 fold change |
|---|---|---|---|---|---|
| Acidiphilium | 3.024 | Agromyces | 2.143 | Acidisphaera | 5.784 |
| Alsobacter | 3.359 | Ancylobacter* | 2.130 | Bdellovibrio | 5.106 |
| Bacillus | 3.545 | Aquisphaera | 2.062 | Legionella | 3.922 |
| Bosea | 3.333 | Bradyrhizobium | 4.942 | Methylobacterium* | 4.963 |
| Enterococcus | 2.459 | Caldilinea | 3.65 | Methylovirgula | 2.918 |
| Friedmanniella | 2.628 | Defluviicoccus | 3.473 | Nocardia | 3.392 |
| Gemmobacter | 3.275 | Desulfuromusa | 2.15 | Nocardioides | 4.011 |
| Herbiconiux | 3.252 | Gemmata* | 3.304 | Psychroglaciecola | 2.113 |
| Kaistia | 2.754 | Gemmiger | 3.417 | Romboutsia | 2.716 |
| Nitratireductor | 4.233 | Hyphomicrobium | 3.802 | Roseiarcus | 4.034 |
| Nitrolancea | 3.815 | Isoptericola | 2.462 | Roseococcus* | 5.93 |
| Rhodoblastus | 2.33 | Jiangella | 2.95 | Thauera | 3.302 |
| Roseomonas | 5.648 | Kinneretia | 1.929 | ||
| Schlesneria | 2.545 | Leucobacter | 4.972 | ||
| Litorilinea | 3.115 | ||||
| Methylocella | 2.989 | ||||
| Microbacterium* | 2.109 | ||||
| Mycobacterium | 2.062 | ||||
| Pleomorphomonas | 2.109 | ||||
| Rhodobacter | 4.105 | ||||
| Saccharomonospora | 1.328 | ||||
| Sphingobium | 3.207 | ||||
| Xanthobacter* | 3.225 | ||||
| Xanthomonas | 3.14 |
(log2-fold change > 1; P < 0.05)
*Genera significantly more abundant in breeding sites in the corresponding territory G, Guadeloupe; FG, French Guiana
Territory-specific differences in relative abundance were detected for certain bacteria genera. For instance, Methylobacterium and Bacillus were significantly more abundant in larvae in French Guiana (log2 fold-change = 3.2 and 2 respectively, P < 0.05), while Leucobacter, Friedmanniella and Xanthobacter were more strongly associated with larvae on Guadeloupe (log2 fold-change = 5.7, 3.2 and 3.2, respectively, P < 0.05) (Fig. 6b, Table 2, Additional file 6). Interestingly, Xanthobacter, was also significantly more abundant in the water samples from Guadeloupe when compared to those from French Guiana (Additional file 6). Other genera, such as Polynucleobacter and Emticicia, predominated in water from breeding sites, while their relative abundance was significantly lower in the corresponding larvae (log2 fold-change = 9.196, and 7.945 respectively, P < 0.001) (Additional file 6).
Relations Between the Composition of Bacterial Communities and Environmental Variables
We investigated whether differences in microbiota in water (N = 161) and larval (N = 145) samples were associated to by physicochemical variations. A single linear regression analysis identified the physicochemical parameters significantly associated with richness and diversity, and a multiple linear regression analysis of significant factors indicated the chemical parameters that better predicted microbiota structuration. On Guadeloupe, turbidity was significantly correlated with the bacterial diversity (R2 = 0.27, P < 0.05) and richness (R2 = 0.35, P < 0.05) in water samples, as well as with larval bacterial richness (R2 = 0.49, P < 0.05), while COD was slightly correlated with larval bacterial diversity (R2 = 0.10, P < 0.05). In French Guiana, turbidity appeared to be related to the evenness of the breeding site microbiota (R2 = 0.249, P < 0.05), whereas in the larval microbiota neither species richness nor species evenness were associated to physicochemical parameters.
Canonical correlation analysis was used to identify correlations among the 12 environmental variables and the 100 most abundant bacteria genera in water (Additional file 7a) and larvae (Additional file 7b) at breeding sites in each territory. As expected, physicochemical parameters were more correlated with bacteria genera from water than with those from larval samples. The CCA plot indicates strong positive correlation between conductivity, Ca, Mg, oxygen dissolved and several bacteria genera such as Acinetobacter, Pseudomonas, Gemmobacter, Polynucleobacter and Ancylobacter either in water and larvae, while these parameters were weakly or negatively associated to other genera commonly enriched in larvae such as Bacillus and Herbiconiux. Salinity and turbidity were the parameters that correlated the least with bacterial microbiota in water samples, while in larvae, the weakest correlations were obtained with Fe, Zn and salinity. For these reasons, these latter physicochemical parameters were excluded from further analysis.
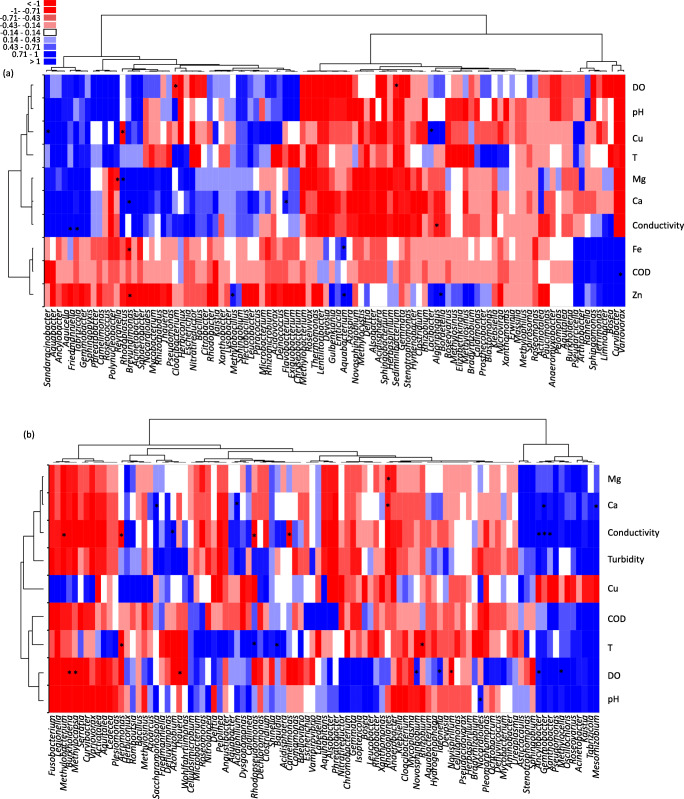
Multiple linear regression was then conducted using the retained potential microbiota determinants and the top 100 most abundant bacteria genera from water and larval samples (Fig. 7). The heatmap shows contrasted associations between DO, pH, Cu, Ca, Mg, conductivity, T and bacterial genera in water samples. Indeed, these parameters are positively correlated to Sandaracinobacter, Aquabacter, Ancylobacter, Aquicella, Friedmanniella, Tabrizicola, Gemmobacter and Sphingopyxis forming a clearly defined clade, while they are negatively correlated to a clade composed of 17 bacteria genera such as Methylobacterium, Emticicia, Delftia and Aquabacterium (Fig. 7). Conversely, Fe, Zn and DCO are negatively correlated with bacteria in water samples, except for a clade where strong positive correlations are observed; these include Curvibacter, Variovorax, Bosea, Limnobacter, Terrimonas, Sphingomonas, Rhanella, Arthrobacter and Pseudarcicella. Regarding larval samples, two clearly defined clades being differentially influenced by physicochemical parameters are observed: (i) a clade composed by genera such as Gemmobacter, Xanthobacter, Pseudomonas, Tabrizicola and Kaistia that is positively influenced by the Mg, Ca, conductivity, turbidity, COD, T, DO, pH and (ii) a clade composed by Fusobacterium, Serratia, Cedecea and Aquitalea that is negatively impacted by these parameters. Interestingly, in these two clades, the associations obtained with Cu were contrasted with respect to those observed with the rest of physicochemical parameters. When considering the most statistically significant correlations (P < 0.05), conductivity and dissolved oxygen were associated to a higher number of bacteria genera in larval samples when compared to the rest of physicochemical parameters (Fig. 7).
Fig. 7.
Heat map of correlations found by multivariate regression analysis between the 100 most abundant bacteria genera associated to Aedes aegypti breeding sites water (a) and larvae (b) (columns), and the physicochemical parameters retained after canonical correlation analysis (rows). Blue squares indicate a positive correlation in relative abundance, white squares indicate absence of correlation and red squares indicate negative correlations. The intensity of the colour corresponds to the magnitude of the (log)-fold change. Asterisks indicate significant correlations between a given bacteria genera and a physicochemical parameter (P < 0.05). COD chemical oxygen demand, DO dissolved oxygen, Cu copper, Fe iron, Zn zinc, Ca calcium
Discussion
The variability of A. aegypti-associated bacteria may trigger a differential influence on adult mosquito phenotypic traits such as survival and vector competence [16]. Hence, the characterization of habitat features influencing microbiota structuration is of major interest for risk assessments and vector control. In this study, we found that (i) the physicochemical properties and bacterial communities of A. aegypti breeding sites on Guadeloupe differ substantially from those in French Guiana, (ii) the container type strongly influences the physicochemical parameters but not the microbiota at breeding sites, (iii) the microbiota associated with A. aegypti larvae, even if influenced by that of the breeding sites, was highly conserved in both territories, and (iv) dissolved oxygen and conductivity are strongly associated with the composition of A. aegypti microbiota. The characterization was based on comprehensive sampling of 161 breeding sites conducted during the dry season in both territories to ensure that containers were not rinsed by rainwater and to better observe permanent breeding sites. We found that drums were among the most common A. aegypti breeding sites on Guadeloupe and in French Guiana, as was observed in the neighbouring regions of Martinique and Suriname [48–50]. Drums are used extensively to store water because of the irregular water supply, which provides an excellent habitat for A. aegypti.
An appropriate aquatic habitat for A. aegypti is regulated by biotic and abiotic factors and their interactions. As expected, the temperature and pH were generally around 29 °C and 7.5, respectively, regardless of the type of container and the territory, as they have been shown to be favourable for larval development and survival [51–53]. Dissolved oxygen, electrical conductivity, salinity, COD, turbidity and the concentrations of Ca, Mg, Fe, Zn and Cu depended on the territory and/or the container type, the highest values being associated with tyres and plant containers. The high content of mineral and organic compounds may be due either to the small volume of containers (i.e. tyres), the presence of plants or the container material (i.e. metal), which could result in a higher ion content, turbidity, COD and less dissolved oxygen [54–56]. Our results confirm that A. aegypti does not breed only in clear water, as it can breed in containers with high turbidity (> 133 FNU), high DCO (> 242 mg/L) and low dissolved oxygen, nearly reaching anoxia, like Culex quinquefasciatus mosquitoes [57]. These findings are consistent with reports of successful A. aegypti development in septic tanks and drums, indicating tolerance of high organic pollution [58], which, in the absence of a preferred habitat, allows A. aegypti to extend its niche and breed in marginal habitats to maintain its population, especially during unfavourable seasons. The differences found in the physicochemical parameters of breeding sites raise questions about their possible impact on the structure and composition of A. aegypti communities. Such interactions remain largely unexplored but should be considered, as habitat disturbance can alter the microbiota and thus affect adult vector phenotypic traits [59].
In agreement with other studies, we found that the predominant microbiota at A. aegypti breeding sites was Proteobacteria [17, 42, 60–67]. Significant differences in the relative abundance of bacteria such as Pseudomonas were recorded between Guadeloupe and French Guiana. Our regression analysis revealed positive correlations with Mg, Ca and conductivity, which is consistent with the higher abundance of these proteobacteria on Guadeloupe as they are known to be more abundant in environments with high electrical conductivity [68]. Of the proteobacteria, Roseococcus, Pseudomonas and Brevundimonas in particular are strict aerobic bacteria [68–70], which could explain the correlations obtained in our study between their abundance and higher dissolved oxygen, as seen on Guadeloupe. Curvibacter was particularly abundant in French Guiana breeding sites. These bacteria are frequently detected in iron-rich environments dominated by chemolithoautotrophic species or contaminated with toxic metals [71, 72]. In our study, Curvibacter were positively correlated with Fe and Zn, and negatively with dissolved oxygen, conditions that were characteristic of French Guiana, where large waste like abandoned, rusty freezers composed 24% of breeding sites sampled (only 3% of Guadeloupe breeding sites were large waste). Metal containers may have increased metal concentrations in the stagnant water contained therein as seen elsewhere [73]. Conversely, when considering the entire bacterial communities, the container type did not significantly influence the structure of the microbiota in breeding sites water or larvae.
By contrast to water samples, no major differences in species composition were observed in the microbiota associated with A. aegypti larvae between Guadeloupe and French Guiana (Fig. 5a). This is presumably due to host selection for microbial communities that can colonize the larval gut environment, which is consistent with the lower alpha diversity recorded in larvae when compared to water samples. The shared microbiota of A. aegypti larvae consisted of abundant genera such as Herbiconiux, Bosea, Bacillus and Kaitsia, which were more prevalent in the larvae than in the water from the corresponding breeding sites. Herbiconiux can degrade the cellulose and xylan found in the gut of some insects, which may explain the abundance of this ubiquitous genus in larval samples [74, 75]. The three other genera (Bosea, Bacillus and Kaistia) may be part of the core microbiota of mosquitoes, as they have also been identified in the gut of both larvae and adults specimens from other Culex and Aedes species [17, 62, 63, 76–78]. The role of these commonly found bacteria in gut vector biology may have led to their evolutionary conservation. While the functional roles of these genera are still largely unknown, Bacillus are suspected of affecting the fertility of the mosquitoes [79], and their abundance and distribution were found to be associated to male mosquitoes and their particular feeding dynamics [80]. Interestingly, Bacillus thuringiensis subsp. israelensis (Bti) produce proteins with insecticide properties, which make it a widely used biological control agent [81].
Differences in the microbiota associated with A. aegypti larvae and those in breeding sites are also due to the relative abundance of bacteria which development is constrained by the larval gut environment. In our study, the aerobic bacteria Polynucleobacter and Emticicia bacteria were significantly more abundant in water samples from Guadeloupe than from French Guiana, but the relative abundance of these genera in the larvae were significantly lower than in the water they came from for both territories. As suggested by Coon and colleagues [82], the gut hypoxia, which is probably a cue for growth and moulting of larval mosquitoes, may constrain the growth of such aerobic bacteria. Nevertheless, how insects “select” and acquire from breeding sites the microorganisms that become part of their semi-stable microbiota is still not well understood.
Finally, we observed territory-specific differences in the relative abundance of certain bacteria genera in A. aegypti larvae (i.e. Methylobacterium, Xanthobacter, Roseoccocus, Microbacterium, Microvirga, and Pseudomonas) that were consistent with the differences between breeding sites on Guadeloupe and in French Guiana. These findings confirm that the larval microbiota are significantly affected by the microbial communities in water at breeding sites [16, 19]. As some bacteria from larvae can be transstadially transmitted to adult mosquitoes [83], the influence of breeding sites on mosquito microbiota raises questions about the transmission of pathogens. We found that Pseudomonas was more abundant at breeding sites and larvae on Guadeloupe than in French Guiana, whereas Serratia was more abundant in water and larvae from French Guiana. Pseudomonas and Serratia are genera commonly found both larvae and adult mosquitoes [42, 61, 64, 84–87], suggesting that the territory-specific differences observed on relative abundance of these genera at the breeding sites may be reflected in the microbiota of the corresponding adults and shape their transmission potential for pathogens.
Interestingly, contrasted abundances of Serratia and Pseudomonas genera were also found in adult A. aegypti from the Caribbean island of Grenada [88] but further studies using a standardized methodology would be required to assess their natural variability in mosquito populations across the Caribbean. Bacteria such as Serratia marcescens have been found to increase the susceptibility of A. aegypti females for dengue virus, while Pseudomonas rhodesiae can inhibit La Crosse virus replication in Aedes albopictus cells [89, 90]. Bacillus is another bacteria genus whose relative abundance was higher in French Guiana larvae than in those from Guadeloupe. This bacteria is also commonly found in adult mosquitoes [83] and has been found to decrease their susceptibility to P. falciparum infection [91]. It is noteworthy to mention that the vector competence experiments cited above used mosquitoes that were reared in laboratory conditions, and it is unknown whether the influence of Serratia spp., Pseudomonas spp. and Bacillus spp. on pathogen transmission by mosquitoes is maintained in natural conditions. Laboratory studies have also shown that A. aegypti from Guadeloupe and French Guiana have similar vector competence for arboviruses such as Zika and chikungunya [92, 93]; however, as larval exposure to bacteria can alter the vectorial capacity of this species [16], the differences we observed between Guadeloupe and French Guiana in terms of microbiota from breeding sites and larvae, may result in differences in the vectorial capacity of the corresponding A. aegypti populations.
Conclusions
The physicochemical parameters at A. aegypti breeding sites depend on the type of container and the territory sampled; however, the only major differences on breeding site physicochemical profiles at a broader scale (between the two territories) were associated with differences in the relative abundance of genera in A. aegypti microbiota. How and to what extent geographical variation in microbiota at breeding sites and larvae affects the vectorial capacity of A. aegypti for human pathogens across the globe remain open questions.
Electronic supplementary material
Rarefaction curves of OTU diversity for each sample. Each sample contains nearly 20, 000 sequences to ensure equal sampling depth. (PDF 1961 kb) (PDF 1960 kb)
Mean values for bacterial diversity and richness indices associated with water at breeding sites and A. aegypti larvae. (PDF 50 kb) (PDF 49 kb)
Relative abundance of the most abundant phyla. Bar shows mean relative abundance of the bacterial taxa sequenced from A. aegypti larvae and water samples from French Guiana and Guadeloupe. (PDF 1189 kb) (PDF 1188 kb)
Relative abundance of the most abundant genera. Bar shows mean relative abundance of the bacterial taxa sequenced from A. aegypti larvae and water samples from French Guiana and Guadeloupe. (PDF 1176 kbp) (PDF 1175 kb)
Spatial structuration and variability of microbiota associated with water at breeding sites and A. aegypti larvae. Non-metric multidimensional scaling plots based on UniFrac distances for water (A,B) and larvae (C,D) according to type of breeding site. (PDF 241 kb) (PDF 240 kb)
Significant differences on bacteria genera abundance according to sample type (water vs. larval samples) and/or the territory (XLS 34 kb). (PPTX 56 kb)
Canonical correlation analysis of 12 physicochemical variables and the 100 most abundant bacteria genera in Ae. aegypti breeding sites water (a) and larvae (b). COD, chemical oxygen demand; DO, dissolved oxygen; Cu: Copper; Fe: Iron; Zn: Zinc; Ca: Calcium. (PDF 33 kb) (XLSX 33 kb)
Acknowledgements
The authors thank Cédric Ramdini at ARS Guadeloupe for supporting collection of mosquitoes. We thank the inhabitants of Guadeloupe and French Guiana for allowing us to sample around their homes. We are grateful to the hygiene and environment laboratories of Guadeloupe and French Guiana for their assistance in measuring water physicochemical parameters. We thank Judith Moudouong for helping to establish the metagenomic scripts.
Availability of Data and Materials
The datasets generated and analysed during the current study are available in the NCBI Sequence Read Archive repository under the BioProject with accession code PRJNA600474 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA600474).
Authors’ Contributions
AVR, CG, ID, PC and SB designed the research. Mosquito sampling collections were undertaken by LH, AVR, AG, CD, DG and JG. GL and SR determined water physicochemical parameters. LH, AD and JG performed the metagenomics experiments. LH, YR and AVR performed data analysis, prepared the figures and wrote the manuscript, with contributions from SB, PC, ID and CG.
Funding Information
This work was supported by Action Concertées Inter Pasteuriennes (Grant ACIP 01-2016) and by the Programme Opérationnel FEDER-Guadeloupe-Conseil Régional 2014–2020 (grant 2015-FED-192). LH was funded by a PhD scholarship from La Région Guadeloupe and her missions were supported by the Calmette & Yersin program from the Institut Pasteur Department of International Affairs.
Compliance with Ethical Standards
Competing Interests
The authors declare that they have no conflict of interest.
Ethics Approval
Not applicable.
Consent for Publication
Not applicable.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lyza Hery, Email: lhery@pasteur-guadeloupe.fr.
Amandine Guidez, Email: aguidez@pasteur-cayenne.fr.
Audrey-Anne Durand, Email: audrey-anne.durand@iaf.inrs.ca.
Christelle Delannay, Email: cdollin@pasteur-guadeloupe.fr.
Josiann Normandeau-Guimond, Email: josiann.guimond@iaf.inrs.ca.
Yann Reynaud, Email: yreynaud@pasteur-guadeloupe.fr.
Jean Issaly, Email: jissaly@pasteur-cayenne.fr.
Daniella Goindin, Email: daniella.goindin@gmail.com.
Grégory Legrave, Email: gregory.legrave@hotmail.fr.
Joel Gustave, Email: joel.gustave@ars.sante.fr.
Stéphanie Raffestin, Email: sraffestin@pasteur-cayenne.fr.
Sebastien Breurec, Email: sbreurec@gmail.com.
Philippe Constant, Email: philippe.constant@iaf.inrs.ca.
Isabelle Dusfour, Email: isabelle.dusfour@pasteur.fr.
Claude Guertin, Email: Claude.Guertin@iaf.inrs.ca.
Anubis Vega-Rúa, Email: avega@pasteur.fr.
References
- 1.CIRE Antilles Guyane. Zika virus surveillance in the Antilles Guyane - epidemiological situation. Epidemial Week 7 of 2017. https://www.santepubliquefrance.fr/content/download/41605/848036. Accessed 22 Feb 2020
- 2.Aubry P. Dengue outbreaks in the French West-Indies in a context of arbovirosis emergence and reemergence. Bull Acad Natl Med. 2008;192:781–793. [PubMed] [Google Scholar]
- 3.Souza-Neto JA, Powell JR, Bonizzoni M. Aedes aegypti vector competence studies: a review. Infect Genet Evol. 2019;67:191–209. doi: 10.1016/j.meegid.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization, Special Programme for Research, Training in Tropical Diseases (2009) Dengue: guidelines for diagnosis, treatment, prevention and control. Epidemic, & Pandemic Alert https://www.who.int/tdr/publications/training-guideline-publications/dengue-diagnosis-treatment/en/. Accessed 19 July 2019
- 5.Getachew D, Tekie H, Gebre-Michael T, Balkew M, Mesfin A. Breeding sites of Aedes aegypti: potential dengue vectors in Dire Dawa, East Ethiopia. Interdiscip Perspect Infect Dis. 2015;2015:1–8. doi: 10.1155/2015/706276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentley MD, Day JF. Chemical ecology and behavioral aspects of mosquito oviposition. Annu Rev Entomol. 1989;34(1):401–421. doi: 10.1146/annurev.en.34.010189.002153. [DOI] [PubMed] [Google Scholar]
- 7.Harrington LC, Ponlawat A, Edman JD, Scott TW, Vermeylen F (2008) Influence of container size, location, and time of day on oviposition patterns of the dengue vector Aedes Aegypti in Thailand. Vector-Borne and Zoonotic Dis 8(3):415–424. 10.1089/vbz.2007.0203 [DOI] [PMC free article] [PubMed]
- 8.Barrera R, Amador M, Clark GG. Ecological factors influencing Aedes aegypti (Diptera: Culicidae) productivity in artificial containers in Salinas, Puerto Rico. J Med Entomol. 2006;43(3):484–492. doi: 10.1603/0022-2585(2006)43[484:efiaad]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 9.Ponnusamy L, Schal C, Wesson DM, Arellano C, Apperson CS. Oviposition responses of Aedes mosquitoes to bacterial isolates from attractive bamboo infusions. Parasit Vectors. 2015;8(1):486. doi: 10.1186/s13071-015-1068-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trexler JD, Apperson CS, Zurek L, Gemeno C, Schal C, Kaufman M, Walker E, Watson DW, Wallace L. Role of bacteria in mediating the oviposition responses of Aedes Albopictus (Diptera: Culicidae) J Med Entomol. 2003;40:841–848. doi: 10.1603/0022-2585-40.6.841. [DOI] [PubMed] [Google Scholar]
- 11.Darriet F, Corbel V. Influence des engrais de type NPK sur l’oviposition d’Aedes aegypti. Parasite. 2008;15(1):89–92. doi: 10.1051/parasite/2008151089. [DOI] [PubMed] [Google Scholar]
- 12.Mutero CM, Wekoyela P, Githure J, Konradsen F. Ammonium sulphate fertiliser increases larval populations of Anopheles Arabiensis and culicine mosquitoes in rice fields. Acta Trop. 2004;89(2):187–192. doi: 10.1016/j.actatropica.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Onchuru TO. Ajamma YU, Burugu M, Kaltenpoth M, Masiga D, Villinger J. Chemical parameters and bacterial communities associated with larval habitats of Anopheles, Culex and Aedes mosquitoes (Diptera: Culicidae) in western Kenya. Int J Trop Insect Sci. 2016;36(3):146–160. [Google Scholar]
- 14.Burke R, Barrera R, Lewis M, Kluchinsky T, Claborn D. Septic tanks as larval habitats for the mosquitoes Aedes aegypti and Culex quinquefasciatus in playa-Playita, Puerto Rico. Med Vet Entomol. 2010;24(2):117–123. doi: 10.1111/j.1365-2915.2010.00864.x. [DOI] [PubMed] [Google Scholar]
- 15.Murrell EG, Damal K, Lounibos LP, Juliano SA. Distributions of competing container mosquitoes depend on detritus types, nutrient ratios, and food availability. Ann Entomol Soc Am. 2011;104:688–698. doi: 10.1603/AN10158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickson LB, Jiolle D, Minard G, Moltini-Conclois I, Volant S, Ghozlane A, Lambrechts L. Carryover effects of larval exposure to different environmental bacteria drive adult trait variation in a mosquito vector. Sci Adv. 2017;3(8):e1700585. doi: 10.1126/sciadv.1700585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minard G, Mavingui P, Moro CV. Diversity and function of bacterial microbiota in the mosquito holobiont. Parasit Vectors. 2013;6(1):146. doi: 10.1186/1756-3305-6-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alto BW, Lounibos LP (2013) Vector competence for arboviruses in relation to the larval environment of mosquitoes, pp. 81–101. In Takken W., Koenraadt C. J. M. (eds.), Ecology of parasite-vector interactions, vol. 3 Wageningen academic publishers, Wageningen, NL
- 19.Coon KL, Vogel KJ, Brown MR, Strand MR. Mosquitoes rely on their gut microbiota for development. Mol Ecol. 2014;23(11):2727–2739. doi: 10.1111/mec.12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Souza R, Virginio F, Suesdek L, Baruf JB, Genta FA. Microorganism-based larval diets affect mosquito development, size and nutritional reserves in the yellow fever mosquito Aedes aegypti (Diptera: Culicidae) Front Physiol. 2019;10:152. doi: 10.3389/fphys.2019.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jupatanakul N, Sim S, Dimopoulos G. The insect microbiome modulates vector competence for arboviruses. Viruses. 2014;6(11):4294–4313. doi: 10.3390/v6114294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moltini-Conclois I, Stalinski R, Tetreau G, Després L, Lambrechts L. Larval exposure to the bacterial insecticide Bti enhances dengue virus susceptibility of adult Aedes aegypti mosquitoes. Insects. 2018;9(4):193. doi: 10.3390/insects9040193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Judson CL. The physiology of hatching of aedine mosquito eggs: hatching stimulus. Ann Entomol Soc Am. 1960;53(5):688–691. [Google Scholar]
- 24.Clark TM, Vieira MA, Huegel KL, Flury D, Carper M. Strategies for regulation of hemolymph pH in acidic and alkaline water by the larval mosquito Aedes aegypti (L.)(Diptera; Culicidae) J Exp Biol. 2007;210(24):4359–4367. doi: 10.1242/jeb.010694. [DOI] [PubMed] [Google Scholar]
- 25.Clark TM, Flis BJ, Remold SK. Differences in the effects of salinity on larval growth and developmental programs of a freshwater and a euryhaline mosquito species (Insecta: Diptera, Culicidae) J Exp Biol. 2004;207(13):2289–2295. doi: 10.1242/jeb.01018. [DOI] [PubMed] [Google Scholar]
- 26.Rueda LM, Patel KJ, Axtell RC, Stinner RE. Temperature-dependent development and survival rates of Culex quinquefasciatus and Aedes aegypti (Diptera: Culicidae) J Med Entomol. 1990;27(5):892–898. doi: 10.1093/jmedent/27.5.892. [DOI] [PubMed] [Google Scholar]
- 27.Clark TM, Flis BJ, Remold SK. pH tolerances and regulatory abilities of freshwater and euryhaline Aedine mosquito larvae. J Exp Biol. 2004;207(13):2297–2304. doi: 10.1242/jeb.01021. [DOI] [PubMed] [Google Scholar]
- 28.Devi NP. Physico-chemical assessment of Na larvae in outskirts of Dehradun City, Uttarakhand. J Commun Disord. 2014;46(3):29–39. [Google Scholar]
- 29.Sultana A, Hasan S, Hossain M, Alim A, Al Mamun M, Bashar K. Larval breeding habitats and ecological factors influence the species composition of mosquito (Diptera: Culicidae) in the parks of Dhaka city, Bangladesh. Bangladesh J Zool. 2017;45(2):111–122. [Google Scholar]
- 30.Navarro DM, De Oliveira PES, Potting RPJ, Brito AC, Fital SJF, Santana AG. The potential attractant or repellent effects of different water types on oviposition in Aedes aegypti L.(Dipt., Culicidae) J Appl Entomol. 2003;127(1):46–50. [Google Scholar]
- 31.Dom NC, Ahmad AH, Ismail R. Habitat characterization of Aedes sp. breeding in urban hotspot area. Procedia Soc Behav Sci. 2013;85:100–109. [Google Scholar]
- 32.Mireji PO, Keating J, Hassanali A, Mbogo CM, Nyambaka H, Kahindi S, Beier JC. Heavy metals in mosquito larval habitats in urban Kisumu and Malindi, Kenya, and their impact. Ecotox Environ Safe. 2008;70(1):147–153. doi: 10.1016/j.ecoenv.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Székely AJ, Berga M, Langenheder S. Mechanisms determining the fate of dispersed bacterial communities in new environments. ISME J. 2013;7(1):61–71. doi: 10.1038/ismej.2012.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G, Cornejo-Castillo FM. Structure and function of the global ocean microbiome. Science. 2015;348(6237):1261359. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- 35.Liu S, Ren H, Shen L, Lou L, Tian G, Zheng P, Hu B. pH levels drive bacterial community structure in sediments of the Qiantang River as determined by 454 pyrosequencing. Front Microbiol. 2015;6:285. doi: 10.3389/fmicb.2015.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coon KL, Brown MR, Strand MR. Mosquitoes host communities of bacteria that are essential for development but vary greatly between local habitats. Mol Ecol. 2016;25(22):5806–5826. doi: 10.1111/mec.13877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boissière A, Tchioffo MT, Bachar D, Abate L, Marie A, Nsango SE, Morlais I (2014) Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with plasmodium falciparum infection. PLoS Pathog 8(5):e1002742. 10.1371/journal.ppat.1002742 [DOI] [PMC free article] [PubMed]
- 38.Akorli J, Gendrin M, Pels NAP, Yeboah-Manu D, Christophides GK, Wilson MD. Seasonality and locality affect the diversity of Anopheles gambiae and Anopheles coluzzii midgut microbiota from Ghana. PLoS One. 2016;11(6):e0157529. doi: 10.1371/journal.pone.0157529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Osei-Poku J, Mbogo CM, Palmer WJ, Jiggins FM. Deep sequencing reveals extensive variation in the gut microbiota of wild mosquitoes from Kenya. Mol Ecol. 2012;21(20):5138–5150. doi: 10.1111/j.1365-294X.2012.05759.x. [DOI] [PubMed] [Google Scholar]
- 40.Clark-Gil S, Darsie F. The mosquitoes of Guatemala, their identification, distribution and bionomics, with keys to adult females and larvae. Mosquito Systematics. 1983;15(3):151–284. [Google Scholar]
- 41.Hanson S, Brin F, Martine D, Coezy M, Boulemar E, Mounichy A & Chingan J (2015) Bilan des actions de lutte anti vectorielle et de prévention développées dans le cadre de l’épidémie de chikungunya en Guadeloupe. sentinelles971.com
- 42.Zouache K, Raharimalala FN, Raquin V, Tran-Van V, Raveloson LHR, Ravelonandro P, Mavingui P. Bacterial diversity of field-caught mosquitoes, Aedes albopictus and Aedes aegypti, from different geographic regions of Madagascar. FEMS Microbiol Ecol. 2011;75(3):377–389. doi: 10.1111/j.1574-6941.2010.01012.x. [DOI] [PubMed] [Google Scholar]
- 43.Feldman RA, Black MB, Cary CS, Lutz RA, Vrijenhoek RC. Molecular phylogenetics of bacterial endosymbionts and their vestimentiferan hosts. Mol Mar Biol Biotechnol. 1997;6(3):268–277. [PubMed] [Google Scholar]
- 44.Comeau AM, Li WK, Tremblay J-É, Carmack EC, Lovejoy C (2011) Arctic Ocean microbial community structure before and after the 2007 record sea ice minimum. PLoS One 6(11):e27492. 10.1371/journal.pone.0027492 [DOI] [PMC free article] [PubMed]
- 45.Edgar R. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 46.Quereda JJ, Dussurget O, Nahori MA, Ghozlane A, Volant S, Dillies MA, Cossart P. Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc Natl Acad Sci. 2016;113(20):5706–5711. doi: 10.1073/pnas.1523899113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agence de santé Guadeloupe, Saint-Martin, Saint-Barthélemy (2019) Les réponses face à la recrudescence des cas de dengue en Guadeloupe. https://www.lemoule.fr/toute-l-actualite/884-les-reponses-face-a-la-recrudescence-des-cas-de-dengue-en-guadeloupe-janvier-2019. Accessed 15 Oct 2019
- 49.Yebakima A, Schucht G, Vernerey M, Mouchet J. Situation d’Aedes aegypti en Martinique et considération sur la stratégie de lutte. Cahiers ORSTOM, série Entom méd Parasitol. 1979;17(4):213–219. [Google Scholar]
- 50.Hiwat H, Doerdjan K, Kerpens M, Samjhawan A, Sardjoe P, Soekhoe T (2013) Importance of domestic water containers as Aedes aegypti breeding sites in Suriname, implications for dengue control. Acad J Sur:4403–4407
- 51.Nayaka BS. Physico-chemical characteristics of larval habitat waters of mosquitoes in and around Bangalore, Karnataka, India. Int J Entomol Res. 2018;3(2):177–179. [Google Scholar]
- 52.Amarasing LD, Dalpadado DR. Vector mosquito diversity and habitat variation in a semi urbanized area of Kelaniya in Sri Lanka. Int J Entomol Res. 2014;2(1):15–21. [Google Scholar]
- 53.Garcia-Sánchez DC, Pinilla GA, Quintero J. Ecological characterization of Aedes aegypti larval habitats (Diptera: Culicidae) in artificial water containers in Girardot, Colombia. J Vector Ecol. 2017;42(2):289–297. doi: 10.1111/jvec.12269. [DOI] [PubMed] [Google Scholar]
- 54.Dom NC, Ahmad P, Mokhtar M, Rajan Assessment of heavy metal concentration on Aedes mosquito breeding sites in urban area, Malaysia. Int J Mosq Res. 2017;4:12–19. [Google Scholar]
- 55.Kumar M, Puri A (2012) A review of permissible limits of drinking water. Indian J Occup Environ Med 16(1):40–44. 10.4103/0019-5278.99696 [DOI] [PMC free article] [PubMed]
- 56.Lokhande RS, Singare PU, Pimple DS. Pollution in water of Kasardi River flowing along Taloja industrial area of Mumbai, India. World Environ. 2011;1(1):6–13. [Google Scholar]
- 57.Kudom AA. Larval ecology of Anopheles coluzzii in Cape Coast, Ghana: water quality, nature of habitat and implication for larval control. Malar J. 2015;14(1):447. doi: 10.1186/s12936-015-0989-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mackay AJ, Amador M, Diaz A, Smith J, Barrera R (2009) Dynamics of Aedes aegypti and Culex quinquefasciatus in septic tanks. J Am Mosquito Contr 25(4):409–417. 10.2987/09-5888.1 [DOI] [PubMed]
- 59.Bennett KL, Almanza A, McMillan WO, Saltonstall K, Vdovenko EL, Vinda JS et al (2019) Habitat disturbance and the organization of bacterial communities in Neotropical hematophagous arthropods. PLoS One 14(9). 10.1371/journal.pone.0222145 [DOI] [PMC free article] [PubMed]
- 60.Dada N, Jumas-Bilak E, Manguin S, Seidu R, Stenström TA, Overgaard HJ. Comparative assessment of the bacterial communities associated with Aedes aegypti larvae and water from domestic water storage containers. Parasit Vectors. 2014;7(1):391. doi: 10.1186/1756-3305-7-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yadav KK, Bora A, Datta S, Chandel K, Gogoi HK, Prasad KS, Veer V. Molecular characterization of midgut microbiota of Aedes albopictus and Aedes aegypti from Arunachal Pradesh, India. Parasit Vectors. 2015;8(1):641. doi: 10.1186/s13071-015-1252-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang X, Liu T, Wu Y, Zhong D, Zhou G, Su X, Chen XG. Bacterial microbiota assemblage in Aedes albopictus mosquitoes and its impacts on larval development. Mol Ecol. 2018;27(14):2972–2985. doi: 10.1111/mec.14732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chandel K, Mendki MJ, Parikh RY, Kulkarni G, Tikar SN, Sukumaran D, Veer V. Midgut microbial community of Culex quinquefasciatus mosquito populations from India. PLoS One. 2013;8(11):e80453. doi: 10.1371/journal.pone.0080453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gimonneau G, Tchioffo MT, Abate L, Boissière A, Awono-Ambéné PH, Nsango SE, Christen R, Morlais I. Composition of Anopheles coluzzii and Anopheles gambiae microbiota from larval to adult stages. Infect Genet Evol. 2014;28:715–724. doi: 10.1016/j.meegid.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 65.Wang Y, Gilbreath TM, III, Kukutla P, Yan G, Xu J. Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS One. 2011;6(9):e24767. doi: 10.1371/journal.pone.0024767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Osei-Poku J, Mbogo CM, Palmer WJ, Jiggins FM. Deep sequencing reveals extensive variation in the gut microbiota of wild mosquitoes from Kenya. Mol Ecol. 2012;21(20):5138–5150. doi: 10.1111/j.1365-294X.2012.05759.x. [DOI] [PubMed] [Google Scholar]
- 67.Zotzmann S, Steinbrink A, Schleich K, Frantzmann F, Xoumpholphakdy C, Spaeth M, Moro CV, Mavingui P, Klimpel S. Bacterial diversity of cosmopolitan Culex pipiens and invasive Aedes japonicus from Germany. Parasitol Res. 2017;116(7):1899–1906. doi: 10.1007/s00436-017-5466-2. [DOI] [PubMed] [Google Scholar]
- 68.Ribas F, Perramon J, Terradillos A, Frias J, Lucena F. The Pseudomonas group as an indicator of potential regrowth in water distribution systems. J Appl Microbiol. 2000;88(4):704–710. doi: 10.1046/j.1365-2672.2000.01021.x. [DOI] [PubMed] [Google Scholar]
- 69.Yurkov V, Schoepp B, Verméglio A. Photoinduced electron transfer and cytochrome content in obligate aerobic phototrophic bacteria from genera Erythromicrobium, Sandaracinobacter, Erythromonas, Roseococcus and Erythrobacter. Photosynth Res. 1998;57(2):117–128. [Google Scholar]
- 70.Ryan MP, Pembroke JT. Brevundimonas spp: emerging global opportunistic pathogens. Virulence. 2018;9(1):480–493. doi: 10.1080/21505594.2017.1419116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schmidt B, Sánchez LA, Fretschner T, Kreps G, Ferrero MA, Siñeriz F, Szewzyk U. Isolation of Sphaerotilus-Leptothrix strains from iron bacteria communities in Tierra del Fuego wetlands. FEMS Microbiol Ecol. 2014;90(2):454–466. doi: 10.1111/1574-6941.12406. [DOI] [PubMed] [Google Scholar]
- 72.Fortunato CS, Huber JA. Coupled RNA-SIP and metatranscriptomics of active chemolithoautotrophic communities at a deep-sea hydrothermal vent. ISME J. 2016;10:1925–1938. doi: 10.1038/ismej.2015.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dom NC, Ahmad P, Mokhtar MA, Rajan S. Assessment of heavy metal concentration on Aedes mosquito breeding sites in urban area, Malaysia. Int J Mosq Res. 2017;4:12–19. [Google Scholar]
- 74.Kim BC, Park DS, Kim H, Oh HW, Lee KH, Shin KS, Bae KS. Herbiconiux moechotypicola sp. nov., a xylanolytic bacterium isolated from the gut of hairy long-horned toad beetles, Moechotypa diphysis (Pascoe) Int J Syst Evol Microbiol. 2012;62(1):90–95. doi: 10.1099/ijs.0.028357-0. [DOI] [PubMed] [Google Scholar]
- 75.Hu X, Yu J, Wang C, Chen H. Cellulolytic bacteria associated with the gut of Dendroctonus armandi larvae (Coleoptera: Curculionidae: Scolytinae) Forests. 2014;5(3):455–465. [Google Scholar]
- 76.Zotzmann S, Steinbrink A, Schleich K, Frantzmann F, Xoumpholphakdy C, Spaeth M, Klimpel S. Bacterial diversity of cosmopolitan Culex pipiens and invasive Aedes japonicus from Germany. Parasitol Res. 2017;116(7):1899–1906. doi: 10.1007/s00436-017-5466-2. [DOI] [PubMed] [Google Scholar]
- 77.Duguma D, Rugman-Jones P, Kaufman MG, Hall MW, Neufeld JD, Stouthamer R, Walton WE. Bacterial communities associated with Culex mosquito larvae and two emergent aquatic plants of bioremediation importance. PLoS One. 2013;8(8):e72522. doi: 10.1371/journal.pone.0072522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ramirez JL, Souza-Neto J, Cosme RT, Rovira J, Ortiz A, Pascale JM, Dimopoulos G. Reciprocal tripartite interactions between the Aedes aegypti midgut microbiota, innate immune system and dengue virus influences vector competence. PLoS Negl Trop Dis. 2012;6(3):e1561. doi: 10.1371/journal.pntd.0001561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fouda MA, Hassan MI, Al-Daly AG, Hammad KM. Effect of midgut bacteria of Culex pipiens L. on digestion and reproduction. J Egypt Soc Parasitol. 2001;31(3):767–780. [PubMed] [Google Scholar]
- 80.Rani A, Sharma A, Rajagopal R, Adak T, Bhatnagar RK. Bacterial diversity analysis of larvae and adult midgut microflora using culture-dependent and culture-independent methods in lab-reared and field-collected Anopheles stephensi-an Asian malarial vector. BMC Microbiol. 2009;9(1):96. doi: 10.1186/1471-2180-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guégan M, Zouache K, Démichel C, Minard G, Potier P, Mavingui P, Moro CV. The mosquito holobiont: fresh insight into mosquito-microbiota interactions. Microbiome. 2018;6(1):49. doi: 10.1186/s40168-018-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Coon KL, Valzania L, McKinney DA, Vogel KJ, Brown MR, Strand MR. Bacteria-mediated hypoxia functions as a signal for mosquito development. Proc Natl Acad Sci. 2017;114(27):E5362–E5369. doi: 10.1073/pnas.1702983114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Scolari F, Casiraghi M, & Bonizzoni M (2019). Aedes spp. and their microbiota: a review. Frontiers in microbiology, 10, 2036. 10.3389/fmicb.2019.02036 [DOI] [PMC free article] [PubMed]
- 84.Sharma P, Sharma S, Maurya RK, De TD, Thomas T, Lata S, Dixit R. Salivary glands harbor more diverse microbial communities than gut in Anopheles culicifacies. Parasit Vectors. 2014;7(1):235. doi: 10.1186/1756-3305-7-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chavshin AR, Oshaghi MA, Vatandoost H, Chavshin AR, Oshaghi MA, Vatandoost H, Pourmand MR, Raeisi A, Enayati AA, ... & Ghoorchian S (2012) Identification of bacterial microflora in the midgut of the larvae and adult of wild caught Anopheles stephensi: a step toward finding suitable paratransgenesis candidates. Acta Trop.121:129–134. 10.1016/j.actatropica.2011.10.015 [DOI] [PubMed]
- 86.Luxananil P, Atomi H, Panyim S, Imanaka T. Isolation of bacterial strains colonizable in mosquito larval guts as novel host cells for mosquito control. J Biosci Bioeng. 2001;92(4):342–345. doi: 10.1263/jbb.92.342. [DOI] [PubMed] [Google Scholar]
- 87.Tetreau G, Grizard S, Patil CD, Tran FH, Stalinski R, Laporte F, Moro CV. Bacterial microbiota of Aedes aegypti mosquito larvae is altered by intoxication with Bacillus thuringiensis israelensis. Parasit Vectors. 2018;11(1):121. doi: 10.1186/s13071-018-2741-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ramos-Nino ME, Fitzpatrick DM, Eckstrom KM, Tighe S, Hattaway LM, Hsueh AN, Stone DM, Dragon JA, Cheetham S. Metagenomic analysis of Aedes aegypti and Culex quinquefasciatus mosquitoes from Grenada, West Indies. PLoS One. 2020;15(4):e0231047. doi: 10.1371/journal.pone.0231047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Apte-Deshpande A, Paingankar M, Gokhale MD, Deobagkar DN. Serratia odorifera a midgut inhabitant of Aedes aegypti mosquito enhances its susceptibility to dengue-2 virus. PLoS One. 2012;7(7):e40401. doi: 10.1371/journal.pone.0040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dennison NJ, Jupatanakul N, Dimopoulos G. The mosquito microbiota influences vector competence for human pathogens. Curr Opin Insect Sci. 2014;3:6–13. doi: 10.1016/j.cois.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tchioffo MT, Boissière A, Churcher TS, Abate L, Gimonneau G, Nsango SE, Awono-Ambene PH, Christen R, Berry A, Morlais I. Modulation of malaria infection in Anopheles gambiae mosquitoes exposed to natural midgut bacteria. PLoS One. 2013;8:e81663. doi: 10.1371/journal.pone.0081663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chouin-Carneiro T, Vega-Rua A, Vazeille M, Yebakima A, Girod R, Goindin D, Dupont-Rouzeyrol M, Lourenço-de-Oliveira R, Failloux AB. Differential susceptibilities of Aedes aegypti and Aedes albopictus from the Americas to Zika virus. PLoS Negl Trop Dis. 2016;10(3):e0004543. doi: 10.1371/journal.pntd.0004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Girod R, Gaborit P, Marrama L, Etienne M, Ramdini C, Rakotoarivony I, Dollin C, Carinci R, Issaly J, Dusfour I, Gustave J, Yp-Tcha MM, Yébakima A, Failloux AB and Vazeille M (2011) High susceptibility to Chikungunya virus of Aedes aegypti from the French West Indies and French Guiana. Trop Med Int Health16:134–139. 10.1111/j.1365-3156.2010.02613.x [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rarefaction curves of OTU diversity for each sample. Each sample contains nearly 20, 000 sequences to ensure equal sampling depth. (PDF 1961 kb) (PDF 1960 kb)
Mean values for bacterial diversity and richness indices associated with water at breeding sites and A. aegypti larvae. (PDF 50 kb) (PDF 49 kb)
Relative abundance of the most abundant phyla. Bar shows mean relative abundance of the bacterial taxa sequenced from A. aegypti larvae and water samples from French Guiana and Guadeloupe. (PDF 1189 kb) (PDF 1188 kb)
Relative abundance of the most abundant genera. Bar shows mean relative abundance of the bacterial taxa sequenced from A. aegypti larvae and water samples from French Guiana and Guadeloupe. (PDF 1176 kbp) (PDF 1175 kb)
Spatial structuration and variability of microbiota associated with water at breeding sites and A. aegypti larvae. Non-metric multidimensional scaling plots based on UniFrac distances for water (A,B) and larvae (C,D) according to type of breeding site. (PDF 241 kb) (PDF 240 kb)
Significant differences on bacteria genera abundance according to sample type (water vs. larval samples) and/or the territory (XLS 34 kb). (PPTX 56 kb)
Canonical correlation analysis of 12 physicochemical variables and the 100 most abundant bacteria genera in Ae. aegypti breeding sites water (a) and larvae (b). COD, chemical oxygen demand; DO, dissolved oxygen; Cu: Copper; Fe: Iron; Zn: Zinc; Ca: Calcium. (PDF 33 kb) (XLSX 33 kb)
Data Availability Statement
The datasets generated and analysed during the current study are available in the NCBI Sequence Read Archive repository under the BioProject with accession code PRJNA600474 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA600474).