Abstract
Japanese chestnut (Castanea crenata Sieb. et Zucc.), the only fruit tree species domesticated in Japan, has been cultivated alongside natural stands since prehistorical times. Understanding the genetic diversity of this species and the relationships between cultivated and wild chestnut is important for clarifying its breeding history and determining conservation strategies. We assessed 3 chestnut cultivar populations and 29 wild chestnut populations (618 accessions). Genetic distance analysis revealed that wild populations in the Kyushu region are genetically distant from other populations, whereas other wild and cultivar populations are comparatively similar. Assignment tests suggested that cultivars were relatively similar to populations from central to western Honshu. Bayesian structure analyses showed that wild individuals were roughly classified according to geographical distribution along the Japanese archipelago, except that some wild individuals carried the genetic cluster prevalent in cultivars. Parentage analyses between cultivars and wild individuals identified 26 wild individuals presumed to have a parent–offspring relationship with a cultivar. These results suggested that the genetic structure of some wild individuals in natural stands was influenced by gene flow from cultivars. To conserve wild individuals carrying true “wild” genetic clusters, these individuals should be collected and preserved by ex situ conservation programs.
Subject terms: Plant evolution, Plant domestication, Plant breeding
Introduction
In plant species, there are large phenotypic differences between cultivated and wild forms. Cultivated plants have been selected by humans for food and other useful materials. Cultivated plants generally have good morphotypes for cultivating and harvesting1,2 and bear large fruits or nuts3–5. In contrast, wild individuals distributed in various areas have become adapted to different ecological situations, with the result that wild collections derived from divergent areas have greater genetic diversity than do cultivars in some species6–8. Thus, wild individuals are interesting for plant breeders as a potential source of genes that have been lost during domestication. The domestication process has been studied in many crops, and several key genes selected during domestication have been identified3–5,9,10. Conversely, gene flow from cultivars to wild individuals has influenced the genetic diversity of wild populations of several crops11–15. Thus, it is quite important to co-analyze cultivated and wild populations to clarify the genetic history of both population types.
Chestnut utilization in Japan dates back to the Jomon period (14,000–300 BCE). The oldest known nut remains were found in the Omiya-no-moriura ruins (10,900–10,700 BCE), in which carbonized and peeled nuts were discovered16. When domestication of Japanese chestnut (Castanea crenata Sieb. et Zucc.) began is a difficult but interesting question. Evidence for pre-domestication (called “hansaibai”) of chestnut was found at the location of the Sannai-Maruyama ruins (3500–2000 BCE)17,18. The high frequency of chestnut pollen, wood, and nut remains at this site suggested that people preserved chestnut trees around the village. Interestingly, nut size increased during the early to late Jomon period, and the nut size of several remains from the late Jomon period was as large as those of modern cultivars19,20, suggesting that the effort to improve genetic performance of Japanese chestnut started prehistorically. The nut size is quite different between cultivars and wild individuals: today, the individual nuts of some local cultivars are more than 30 g, but those of wild individuals are generally less than 5 g (Supplementary Fig. 1). The practice of naming cultivars has existed since the Edo period (1603–1867). The oldest chestnut production area from this period was the Tanba region, which encompasses portions of Kyoto, Osaka, and Hyogo Prefectures in western Japan. Many cultivars with large nuts were thought to have been cultivated in the Tanba region and spread throughout Japan by clonal propagation and seed transportation21,22. However, those cultivars and excavated nuts from the Sannai-Maruyama ruins were presumed to be genetically different because there is no genetic information that connects recent cultivars and excavated nuts23.
The chestnut gall wasp, Dryocosmus kuriphilus Yasumatsu, was accidentally introduced from China into Japan around 194024 and has influenced the distribution of both cultivated and wild chestnut. During the 1960s and 1970s, producers cut down local cultivars or naturalized stands and introduced cultivars resistant to chestnut gall wasp. The area of chestnut orchards dramatically increased during that time and was maximized at about 45,000 ha in the late 1970s. On the other hand, wild populations were devastated by chestnut gall wasp until the establishment of Torymus sinensis Kamijo, a natural enemy of Dryocosmus kuriphilus, during the 1980s. For chestnuts in a natural forest setting, insect pollinators, including bumblebees and flies, disperse pollen across long distances25. Even in orchard settings, considerable amounts of pollen are carried in from wild chestnut trees by insect pollinators26. Since the 1970s, the number of actively managed chestnut orchards in mountainous areas has gradually decreased due to the inconvenience of country life, so it is possible that seeds from cultivars are carried by animals and may escape from abandoned orchards. Thus, it is important to clarify how much gene flow has occurred from cultivars to wild populations.
Population genetic studies of wild individuals of chestnut have been done using various chestnut species, including European chestnut (Castanea sativa Mill.), American chestnut (Castanea dentata [Marsh.] Borkh), and Chinese chestnut (Castanea mollissima Bl.)27–32. In any chestnut species, there are exceptional populations that are genetically different from neighboring populations, suggesting that human-mediated seed dispersal has influenced wild natural stands28,29,31,32. So far, however, population genetic studies of Japanese chestnut have been limited to populations from the Tohoku area33 of northeast Japan and classifications of cultivars26,34, and to our knowledge there are no reports describing genetic characterization of wild chestnut populations throughout Japan.
Chestnut is the only domesticated nut species in family Fagaceae. Several of the deciduous species in family Fagaceae are distributed throughout the Japanese archipelago. The genetic population structures of Fagus japonica Maxim., Quercus aliena Blume, Q. serrata Murray, and Q. mongolica var. crispula in Japan roughly distinguish northeast and southwest groups35–37. For F. japonica and Q. serrata, the boundary coincides with the north–south trending Itoigawa–Shizuoka Tectonic Line on the western border of Fossa Magna, a great rift valley. On the other hand, data for Fagus crenata Blume, a dominant species in cool-temperate deciduous forests in Japan, provide clear evidence of genetic divergence between populations along the Japan Sea (Japan Sea lineage) and Pacific (Pacific lineage) sides of Japan38. The nut remains of these species were also discovered from several excavations during the Jomon period, but with the exception of certain Fagus species, nuts of these species contain considerable amounts of tannins39, which need to be removed before eating. Consequently, the deciduous species in the Fagaceae in Japan other than chestnut have not been domesticated for use as food. Therefore, it is interesting to compare the genetic structures of chestnut and these non-domesticated species.
The objectives of this study were to examine the genetic structure of wild chestnut distributed across the main islands of Japan, to examine the genetic relationships between cultivated and wild populations, and to clarify the breeding history of Japanese chestnut cultivars. Because chestnut orchards are found throughout Japan, gene flow from cultivars to wild individuals should be clarified to support future conservation and breeding strategies.
Results
Basic genetic characteristics of chestnut populations
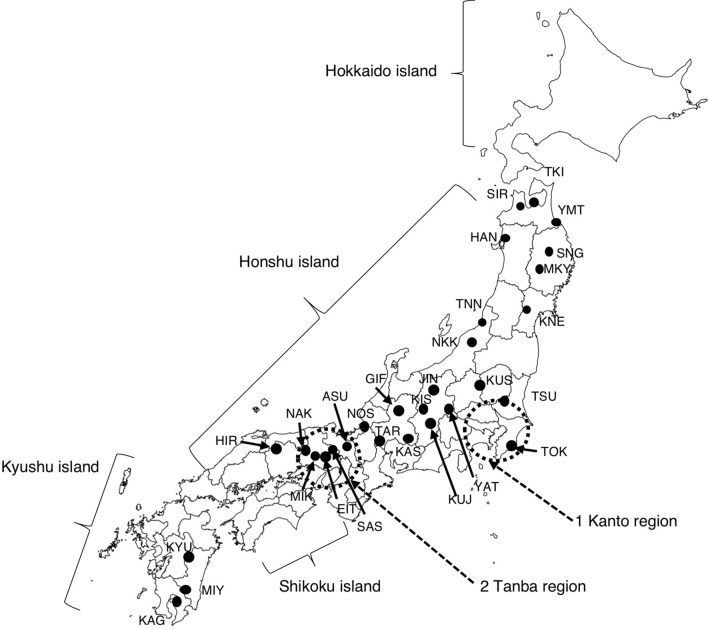
We genotyped chestnuts from 32 populations (618 individuals in total) by using 30 nuclear simple sequence repeat (nSSR) markers (Table 1; Supplementary Tables S1–S3; Fig. 1). Most of the markers had moderate values for genetic diversity (Ho = 0.687 and He = 0.749 for mean values of 30 nSSR markers). The null allele frequencies ranged from 0.013 (PRD84) to 0.097 (EMCs2; Supplementary Table S3), indicating that the markers are reliable enough to use in population genetic studies. Allelic richness (Ar) of the 32 populations ranged from 4.75 to 6.33 (KYU and TAR, respectively; Table 1). The mean Ar value of the cultivar populations (5.46) was similar to that of the wild populations (5.55). Observed and expected heterozygosity of the 32 populations was Ho = 0.605–0.752 and He = 0.596–0.726, respectively (Table 1). Similar to the Ar values, mean heterozygosities of cultivar populations (Ho = 0.708 and He = 0.699) were similar to those of wild populations (Ho = 0.685 and He = 0.681). The inbreeding coefficient (Fis) ranged from − 0.034 to 0.077. Cultivar populations originated in regions other than KAN or TAN (designated “other regions”; OR) and six wild populations (SIR, TSU, TOK, YAT, MIK, and NAK) showed significant Fis values (Table 1).
Table 1.
Grouping and genetic diversity of the 32 chestnut populations investigated in this study.
| Population | Group | No. of trees | Na | Ar | Ho | He | Fis | Chloroplast haplotype | |
|---|---|---|---|---|---|---|---|---|---|
| HAP1 | HAP2 | ||||||||
| KAN | Cultivar | 14 | 5.6 | 5.37 | 0.721 | 0.689 | − 0.011 | 14 | 0 |
| TAN | Cultivar | 21 | 6.0 | 5.28 | 0.724 | 0.685 | − 0.032 | 21 | 0 |
| OR | Cultivar | 26 | 6.9 | 5.66 | 0.687 | 0.716 | 0.060** | 26 | 0 |
| Mean of cultivar populations | 6.3 | 5.46 | 0.708 | 0.699 | |||||
| TKI | Wild | 20 | 5.5 | 4.77 | 0.622 | 0.620 | 0.023 | 20 | 0 |
| SIR | Wild | 20 | 5.6 | 5.05 | 0.625 | 0.635 | 0.042* | 20 | 0 |
| YMT | Wild | 20 | 5.5 | 4.90 | 0.670 | 0.649 | − 0.007 | 20 | 0 |
| HAN | Wild | 20 | 6.2 | 5.42 | 0.677 | 0.668 | 0.013 | 20 | 0 |
| SNG | Wild | 20 | 6.5 | 5.72 | 0.693 | 0.691 | 0.022 | 20 | 0 |
| MKY | Wild | 20 | 6.2 | 5.53 | 0.718 | 0.696 | − 0.007 | 20 | 0 |
| KNE | Wild | 20 | 6.5 | 5.56 | 0.642 | 0.650 | 0.038 | 20 | 0 |
| TNN | Wild | 20 | 5.8 | 5.15 | 0.682 | 0.688 | 0.035 | 20 | 0 |
| NKK | Wild | 20 | 6.3 | 5.59 | 0.718 | 0.681 | − 0.029 | 20 | 0 |
| KUS | Wild | 20 | 6.5 | 5.75 | 0.710 | 0.704 | 0.017 | 20 | 0 |
| TSU | Wild | 17 | 5.8 | 5.33 | 0.651 | 0.669 | 0.057* | 17 | 0 |
| TOK | Wild | 21 | 6.0 | 5.28 | 0.673 | 0.696 | 0.057** | 21 | 0 |
| YAT | Wild | 20 | 6.3 | 5.57 | 0.647 | 0.673 | 0.065** | 20 | 0 |
| JIN | Wild | 19 | 6.4 | 5.78 | 0.714 | 0.695 | 0.000 | 19 | 0 |
| KIS | Wild | 20 | 7.0 | 6.03 | 0.697 | 0.698 | 0.027 | 20 | 0 |
| KUJ | Wild | 18 | 6.2 | 5.60 | 0.680 | 0.684 | 0.035 | 18 | 0 |
| GIF | Wild | 20 | 6.1 | 5.48 | 0.738 | 0.697 | − 0.034 | 20 | 0 |
| KAS | Wild | 21 | 6.8 | 5.98 | 0.708 | 0.711 | 0.029 | 21 | 0 |
| TAR | Wild | 17 | 7.0 | 6.33 | 0.741 | 0.726 | 0.010 | 17 | 0 |
| NOS | Wild | 16 | 6.3 | 5.93 | 0.752 | 0.715 | − 0.020 | 16 | 0 |
| ASU | Wild | 20 | 6.9 | 5.95 | 0.723 | 0.698 | − 0.011 | 20 | 0 |
| SAS | Wild | 18 | 6.6 | 5.97 | 0.720 | 0.705 | 0.006 | 18 | 0 |
| EIT | Wild | 20 | 6.6 | 5.91 | 0.712 | 0.701 | 0.010 | 20 | 0 |
| MIK | Wild | 20 | 6.4 | 5.64 | 0.663 | 0.686 | 0.059** | 20 | 0 |
| NAK | Wild | 20 | 6.6 | 5.87 | 0.682 | 0.719 | 0.077** | 20 | 0 |
| HIR | Wild | 20 | 6.3 | 5.54 | 0.692 | 0.691 | 0.025 | 20 | 0 |
| KYU | Wild | 20 | 5.4 | 4.75 | 0.605 | 0.596 | 0.010 | 0 | 20 |
| MIY | Wild | 18 | 6.2 | 5.59 | 0.691 | 0.682 | 0.016 | 4 | 14 |
| KAG | Wild | 12 | 5.0 | 5.03 | 0.622 | 0.625 | 0.047 | 2 | 10 |
| Mean of wild populations | 6.2 | 5.55 | 0.685 | 0.681 | |||||
Na number of alleles; Ar allelic richness; Ho observed heterozygosity; He expected heterozygosity; Fis fixation index.
*P < 0.05; **P < 0.01.
Figure 1.
Geographic locations of 3 cultivar populations and 29 wild populations used in this study. The cultivar populations are indicated by dotted circles. Wild populations are indicated by dots. The cultivar population from outside Kanto and Tanba (OR; population number 3) is not shown in this figure because these cultivars are distributed all over Japan. The map was created using MapChart (https://mapchart.net).
Genetic relationship among populations
Chloroplast haplotype frequencies of the 618 individuals were determined using 4 chloroplast SSR markers (cpSSRs). Markers Cmcs1 and Cmcs3 each showed a single fragment size in all individuals, whereas Cmcs2 and Cmcs5 each showed two fragment sizes. Only two chloroplast haplotypes (HAP1 and HAP2; Table 2) were identified among the 618 individuals (Table 1). HAP1 was composed of the combination of 143 bp for Cmcs2 and 151 bp for Cmcs5, whereas HAP2 was composed of 142 bp for Cmcs2 and 149 bp for Cmcs5. The combinations of 143 bp for Cmcs2 and 149 bp for Cmcs5, and of 142 bp for Cmcs2 and 151 bp for Cmcs5, were both completely missing, suggesting that these haplotypes differentiated a long time ago and became geographically separated. All of the cultivars and the 26 wild populations on Honshu carried only HAP1. Within the wild populations in the Kyushu region, population KYU carried only HAP2, whereas MIY and KAG carried both HAP1 and HAP2 (Table 1).
Table 2.
Chloroplast haplotypes identified in this study.
| Chloroplast haplotype | Cmcs1 (KY951992:82981) | Cmcs2 (KY951992:39981) | Cmcs3 (KY951992:69174) | Cmcs5 KY951992:125578) |
|---|---|---|---|---|
| HAP1 | 99 | 143 | 162 | 151 |
| HAP2 | 99 | 142 | 162 | 149 |
The name of each chloroplast SSR marker is followed by its putative position in the complete Castanea mollissima chloroplast genome (GenBank ID:KY951992) in parentheses.
The table values are allele sizes (bp).
Pairwise Jost’s D values among the 32 populations were calculated using the 30 nSSRs (Supplementary Table S4). The 3 cultivar populations were genetically close to one another (KAN vs TAN [Jost’s D = 0.079], KAN vs OR [0.033], TAN vs OR [0.015]). KAN (a cultivar population) was also close to TOK (a wild population; Jost’s D = 0.025), likely because both originated in the same region (see Fig. 1). The 3 wild populations in Kyushu (KYU, MIY, and KAG) were genetically distant from other populations (0.313–0.651; mean 0.460) but relatively close to each other (KYU vs MIY [0.133], KYU vs KAG [0.197], MIY vs KAG [0.123]). Bottleneck tests of heterozygosity excess were also conducted in the 32 chestnut populations. P values were significant (< 0.05) for TNN, TOK, NOS, and NAK under the two-phase model (TPM) and for NOS under the stepwise mutation model (SMM) (Supplementary Table S5), indicating that some wild populations have undergone a genetic bottleneck.
Population structures
To estimate the optimal number of genetic clusters (K) in STRUCTURE, we calculated ∆K values (Supplementary Table S6). The ∆K values were highest at K = 2 both with and without the LOCPRIOR model. In both models, one cluster corresponded to wild populations in the Kyushu region (blue genetic cluster; Fig. 2) and the other cluster to cultivar populations and the wild populations on Honshu island (orange genetic cluster). In both models, the second-highest ∆K values occurred at K = 3, whereas the values of |L″(K)| and ∆K were lower at K = 4. The values of |L″(K)| and ∆K at K = 5 were higher than those at K = 4. Because the 3 wild populations in Kyushu were genetically distant from the other 29 populations, we also estimated ∆K values using only the 29 populations from outside Kyushu. The ∆K values for this subset were clearly higher at K = 2 and K = 4 (corresponding to K = 3 and K = 5 using all populations) than at other K values, both with and without the LOCPRIOR model. In addition, we applied hierarchal analyses of molecular variance (AMOVA) to estimate the variance among the clusters identified in STRUCTURE (Supplementary Table S7). The results of AMOVA based on two clusters (K = 2) across all 32 populations showed that 23.4% of the total variance was among clusters, 6.1% was among populations within clusters, and 70.5% was within populations. Effects both among clusters and among populations within clusters were significant (P < 0.001). The percentage of variance among clusters based on five clusters (K = 5) in all 32 populations and that based on four clusters (K = 4) in the set of 29 populations excluding populations from Kyushu were small (7.8% and 2.8%, respectively) but significant at P < 0.001.
Figure 2.
Bayesian estimates of population structure based on nSSR data for the 3 cultivar populations and 29 wild populations. Results for K = 2, K = 3, and K = 5 obtained in STRUCTURE with and without LOCPRIOR are shown for comparison. The CLUMPAK online tool (http://clumpak.tau.ac.il/)66,67 was applied to graphically display the results produced by STRUCTURE.
Thus, we constructed additional bar plot diagrams for K = 3 and K = 5 using all 32 populations (Fig. 2) and for K = 2 and K = 4 using the set of 29 populations excluding populations from Kyushu (Supplementary Fig. S2). The results of the clustering at K = 3 and K = 5 using all populations and those at K = 2 and K = 4 using the 29 populations from outside Kyushu were quite similar, so we focused on the clustering results obtained for the full set of populations. The results of the clustering at K = 3 with and without LOCPRIOR were similar. In each case, the accessions were classified into three clusters corresponding to (1) the cultivar populations and the wild populations on central and western Honshu island (“orange”), (2) wild populations on northeast Honshu island (“yellow”), and (3) wild populations in the Kyushu region (“blue”). When K = 5 with and without LOCPRIOR, we further identified a “red” genetic cluster corresponding to the cultivar populations and a “green” one corresponding to wild populations on western Honshu island. Hereafter, the “red”, “yellow”, “orange”, “green”, and “blue” genetic clusters at K = 5 are referred to as the “cultivar”, “northeast Honshu”, “central Honshu”, “western Honshu”, and “Kyushu” clusters, respectively. A neighbor-joining tree was constructed based on the net nucleotide distances between pairs of clusters (Fig. 3). The “cultivar” cluster was genetically similar to the “northeast Honshu”, “central Honshu”, and “western Honshu” clusters, whereas the “Kyushu” cluster was genetically quite distant from the other four clusters. A noticeable contribution of the “cultivar” cluster was observed in some wild individuals in regions ranging from Tohoku to Kyushu. In particular, TSU and TOK (wild populations near Tokyo) carried a high proportion of the “cultivar” cluster (68.3% for TSU and 51.4% for TOK when analyzed without the LOCPRIOR model).
Figure 3.
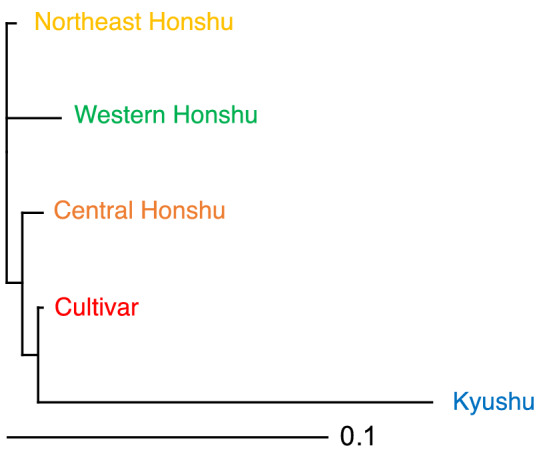
Neighbor-joining tree for the five clusters based on net nucleotide distances for K = 5. The colors of the groups were assigned based on the genetic clusters constructed by Bayesian structure analysis for K = 5 (see Fig. 2).
To validate the genetic structures of wild chestnut populations in Japan, 95 wild chestnut individuals collected by the Forest Tree Breeding Center (FTBC) were combined with the 32 populations in a Bayesian structure analysis without the LOCPRIOR model (Supplementary Table S8). The results were quite similar to those based only on the 32 populations for K = 2, 3, or 5 in that the wild populations were differentiated by region from northeast to southwest, with the exception that the “cultivar” cluster was highly represented in individuals from Akita and Okayama (which are not geographically close) when K = 5 (Supplementary Fig. S3). For K = 5, individuals derived from northeast Honshu (Aomori, Akita, and Miyagi) mainly showed the “northeast Honshu” cluster, whereas those from the western end of Honshu (Tottori, Shimane, Yamaguchi, and Hiroshima), which was not well represented in the analysis of 32 populations, mainly showed the “western Honshu” cluster. The FTBC collection did not include individuals originated in Kyushu, but a noticeable contribution of the “Kyushu” cluster was observed in FTBC individuals derived from Tottori, Shimane, and Yamaguchi, which are geographically close to Kyushu.
Parentage analysis
To clarify whether cultivars had influenced the genetic structure of wild individuals, we conducted parentage analysis of 557 wild individuals (Table 1) by using 92 major cultivars (Supplementary Table S1) as a putative parent database. We identified 26 wild individuals presumed to have a direct offspring–parent relationship with a cultivar in the database (Table 3). The delta score for putative parents ranged from 1.7 to 19.5. Traditional cultivar ‘Ginyose’ was presumed to be a parent of wild individuals TSU11, NAK13, and NAK17. Recent major cultivar ‘Tsukuba’ was also found to be a putative parent of HAN01, NOS14, and NOS15. Another major cultivar, ‘Ishizuchi’, was found to be a putative parent of KNE02 and TOK3. We also conducted parentage analysis of wild individuals collected by FTBC (Supplementary Table S8) using the database of 92 major cultivars and identified 8 putative parent–offspring pairs between cultivars and wild individuals (Supplementary Table S9). The local cultivars ‘Saimyouji 1′ and/or ‘Saimyouji 2′ were presumed to be a parent of 3 wild individuals from Akita. In addition, 3 wild individuals from Okayama were presumed to be offspring of the traditional cultivar ‘Ginyose’ or of ‘Hiratsuka 4′, a selection from ‘Ginyose’.
Table 3.
Putative parent cultivars of wild individuals identified using CERVUS software.
| Location | Population | Genotype | Putative parent cultivar | Delta |
|---|---|---|---|---|
| Akita | HAN | HAN01 | Tsukuba | 7.3 |
| Akita | HAN | HAN03 | Moriwase | 15.1 |
| Iwate | MKY | MKY08 | Buzen | 2.9 |
| Miyagi | KNE | KNE02 | Ishizuchi | 3.4 |
| Nigata | TNN | TNN06 | Moriwase | 15.9 |
| Nigata | NKK | NKK20 | Kinshuu | 11.5 |
| Ibaraki | TSU | TSU6 | Nakatetanba | 5.2 |
| Ibaraki | TSU | TSU11 | Ginyose | 2.5 |
| Chiba | TOK | TOK3 | Ishizuchi | 10.6 |
| Chiba | TOK | TOK4 | Tajiriginyose | 7.0 |
| Gifu | TAR | TAR15 | Tamatsukuri | 7.9 |
| Fukui | NOS | NOS05 | Moriwase | 14.3 |
| Fukui | NOS | NOS06 | Moriwase | 19.5 |
| Fukui | NOS | NOS14 | Tsukuba | 14.9 |
| Fukui | NOS | NOS15 | Tsukuba | 4.7 |
| Fukui | NOS | NOS16 | Obuse 2 | 3.9 |
| Hyogo | EIT | EIT11 | Kinseki | 13.3 |
| Hyogo | EIT | EIT20 | Imakita | 9.2 |
| Hyogo | MIK | MIK7 | Arima | 15.0 |
| Hyogo | MIK | MIK8 | Dengorou | 8.5 |
| Hyogo | NAK | NAK13 | Ginyose | 3.4 |
| Hyogo | NAK | NAK16 | Arima | 15.1 |
| Hyogo | NAK | NAK17 | Ginyose | 2.5 |
| Hyogo | NAK | NAK18 | Kanotsume | 9.5 |
| Miyazaki | MIY | MIY5 | Waseginzen | 9.7 |
| Miyazaki | MIY | MIY10 | Obiwase | 1.7 |
Assignment of cultivars to wild populations
To infer the origins of the cultivars in this study, 61 cultivars from KAN, TAN, and OR were assigned to the 29 wild populations using GeneClass 2.0. The probabilities of membership are reported in Fig. 4 (KAN and TAN) and Supplementary Table S10 (KAN, TAN, and OR). Overall, cultivars showed higher probabilities of origin in central-to-western populations than in northern and Kyushu populations. In particular, cultivars from the Tanba region were likely to be assigned to central and western populations. For most cultivars, the probabilities of originating from wild populations at the northern end of Honshu and from wild populations in the Kyushu region were zero or nearly zero. The populations TOK, KAS, TAR, NOS, SAS, EIT, and NAK showed higher probability scores than the other populations.
Figure 4.
Heatmap showing the probabilities of cultivars from KAN and TAN having originated in each wild population, estimated by an exclusion test.
Analysis of isolation by distance
To clarify the correlation between genetic differentiation and geographical distance in wild populations, the 26 wild populations on Honshu were used for isolation-by-distance analysis. A significant correlation between genetic distance and geographic distance was detected by the Mantel test when all possible pairs of the 26 wild populations were included (r2 = 0.356, P < 0.001; Fig. 5). When the analysis was limited to pairs that included either TSU or TOK, both of which are closely related to cultivars, there was no significant correlation between genetic differentiation and geographical distance (r2 = 0.002, P < 0.736).
Figure 5.
Analysis of isolation by distance among the 26 populations on Honshu island based on a Mantel test with 9999 random permutations. White dots indicate pairs without TSU or TOK, whereas black dots indicate pairs including either TSU or TOK The Mantel test based on all pairs showed significant correlation between genetic and geographic distances (r2 = 0.356, p = 0.001).
Discussion
In this study, we examined the genetic diversity and genetic relationships among 29 wild populations and 3 cultivated populations. Genetic diversity parameters such as heterozygosity and allelic richness were similar in cultivated and wild populations, and no significant evidence of genetic bottleneck was identified in cultivated populations. In perennial fruit crops, some species maintained high levels of genetic diversity during domestication, such as apple and grape40,41, whereas Chinese cherry showed moderate loss of genetic diversity in cultivated accessions8. This kind of comparison depends on the geographic diversity of the cultivars and individuals collected. We identified a total of 242 alleles from 61 cultivars and 385 alleles from 557 wild individuals. Although the number of cultivars actually being propagated in orchards in Japan is less than about 50, the number of wild individuals available is enormous because they are naturally distributed all over Japan. Thus, it should be possible to broaden the genetic diversity of cultivars by introducing favorable alleles from wild populations.
The wild populations in Kyushu are genetically distant from those in other regions and from the cultivar populations (Table 1, Fig. 3). In Bayesian structure analysis using K = 3 (Fig. 2), accessions were classified into (1) wild populations in northeast Honshu (“yellow”), (2) cultivars (regardless of location) and wild populations in central and western Honshu (“orange”), and (3) wild populations in the Kyushu region (“blue”). Interestingly, in some individuals from Kyushu populations MIY and KAG, we identified introgressions of genetic clusters from Honshu (“yellow” and “orange”) into the genetic cluster from Kyushu (“blue”), suggesting that seeds and scions have been imported into Kyushu from Honshu island. Human-mediated seed and scion dispersal have been common in chestnut26,28,29,31,32. The altitude of MIY is 109–212 m above sea level (asl) and MIY is located close to a Satoyama landscape (secondary woodlands and grasslands adjacent to human settlement)42, where human activities are presumed to have had an impact on the distribution of trees. A similar result was obtained from chloroplast haplotype analysis, as MIY and KAG carried not only HAP2 (presumed to have originated in Kyushu) but also HAP1 (Table 1).
The genetic diversity of KYU was lowest among the populations used in this study: this population had only the “Kyushu” genetic cluster and carried only HAP2. The “Kyushu” genetic cluster was rarely observed in cultivars except for ‘Obiwase’ in Miyazaki Prefecture and ‘Nanatate’ in Kochi Prefecture (both from population OR). As the altitude of KYU is high (1037–1074 m asl), it might have had less chance to receive chestnut seed or pollen from other regions and cultivated populations. The “Kyushu” genetic cluster was also detected at low levels in some wild chestnut individuals collected by FTBC in Yamaguchi, Shimane, Hiroshima, and Tottori Prefectures, which are located on the western end of Honshu close to Kyushu island (Supplementary Fig. S2). In a previous study, HAP2 was also identified in Korean cultivar ‘Pochun B-1′43. As the Korean peninsula is located not far north of Kyushu island, it may be possible that wild chestnuts in Korea have a genetic structure similar to those on Kyushu island.
With the exception of those in Kyushu, the wild populations were genetically similar and carried only HAP1 (Fig. 3; Table 1). On Honshu island, isolation-by-distance analysis showed continuous patterns of genetic differentiation from one end to the other. Genetic diversity tends to decrease from south to north in the Northern Hemisphere, as is common in deciduous tree species44. On the other hand, Bayesian structure analysis with and without LOCPRIOR showed a clear relationship between genetic structure and geographical distribution (Fig. 2). For K = 3, wild chestnut populations on Honshu island were divided at the middle of central Honshu, roughly corresponding to the boundary of Fossa Magna, a major tectonic depression. Genetic diversification related to Fossa Magna was also reported in F. japonica35, Q. mongolica var. crispula37, and Q. serrata36,37. The genetic boundary of Q. aliena showed a rather different pattern and was located between Chubu and Kinki Districts, about 200 km west of Fossa Magna36. Following yet another pattern, F. crenata showed genetic divergence between the Japan Sea lineage and the Pacific lineage38. Quercus variabilis and Q. phillyraeoides, which are mainly distributed south of Fossa Magna, were presumed to be unaffected by this boundary. Also, Q. acutissima Carruth is not thought to have been present in the main archipelago of Japan during the last glacial maximum45 and shows monomorphism of cpDNA in Japan46, suggesting that it was heavily influenced by human activity rather than by natural migration. To summarize these results, Fossa Magna commonly affected the genetic diversification of deciduous species in family Fagaceae that were naturally and widely distributed across this boundary, except for Q. aliena and F. crenata. The barrier caused by high mountain ranges and large deposits of volcanic rocks might have prevented the migration of seed37.
For K = 5 in the Bayesian structure analysis, the cultivar populations shared the “red” (“cultivar”) genetic cluster with some wild chestnut populations, especially in the Kanto region (TOK and TSU; Fig. 2). We suspect that the “red” cluster in these wild populations is related to gene flow from cultivars to natural stands. So far, there have been no reports that cultivars were selected from wild individuals in the Kanto region (TOK and TSU), whereas several reports suggest that cultivars were selected in the Tanba region of western Honshu21,22. Although genetic differentiation and geological distance among wild populations on Honshu are significantly correlated (r2 = 0.356; Fig. 5), analysis of pairs that included TOK or TSU showed no significant correlation (r2 = 0.002), suggesting that these populations were affected by non-natural (i.e., human-mediated) transportation of genetic clusters from other regions. Furthermore, in Ibaraki Prefecture (the location of TSU), chestnut production and the presence of orchards dramatically increased from the 1940s to the 1960s, as wild individuals were replaced by cultivars21. Trees in these orchards might have hybridized with wild individuals in TSU. The direct parent–offspring relationships between cultivars and wild individuals identified in several regions also suggest gene flow from cultivars to wild populations in these regions (Table 3, Supplementary Table S10). In fact, the “cultivar” (“red”) genetic cluster was observed in some wild individuals in regions ranging from Tohoku to Kyushu (TSU, TOK, MKY, KNE, TNN, KUJ, TAR, NOS, SAS, EIT, MIK, NAK, MIY, and others; Fig. 2) when analyzed both with and without the LOCPRIOR model, suggesting that the present wild chestnut populations throughout Japan have been influenced by gene flow from cultivars. Interestingly, all of the wild populations showing significant values in bottleneck analysis under TPM (TNN, TOK, NOS, and NAK) carried considerable portions of the “cultivar” genetic cluster. These sites might have been somewhat influenced by human activities. Large amounts of timber from wild chestnut were used for railway construction47 in the early twentieth century. Chestnut timber is thought to have been in short supply by the mid-1910s48, and it was reported that trees distributed in forests near railway lines and other convenient places had been cut down and exhausted for railway construction49.
Gene flow from cultivars to wild populations has been identified in some fruit tree species. In the Japanese archipelago, pear shows extensive introgression of prehistorically naturalized P. pyrifolia into the threatened native species P. ussuriensis, which was identified in northern Tohoku12. With rare exceptions, wild pear trees have only been found in human-disturbed secondary forests or pasturelands, so native individuals have likely sustained a genetic influence from cultivated or escaped trees. Therefore, true native P. ussuriensis is distributed in limited areas and is endangered. In apple, some studies have investigated crop-to-wild gene flow from modern cultivars (Malus domestica Borkh.) imported from western countries into native species (Malus sieversii and Malus orientalis)15. In grape, a wild population in Georgia had a high proportion of hermaphrodite vines, a character specific to cultivar genotypes, indicating gene flow from cultivated to wild types50.
Genetic studies of current chestnut populations in combination with information from preserved nut remnants can reveal clues as to how and where chestnut might have been domesticated. It is obvious that one component of the domestication syndrome in Japanese chestnut is large nut (seed) size, as in other species51. Although previous reports clarified that nut size increased during the early to late Jomon (4000–1000 BCE)17,18, Motoki suggested that the nuts discovered mainly in ruins in northeast Japan were not directly influenced by native cultivars that originated in the Tanba region, which is geographically far from northeast Japan23. In Bayesian structure analysis with K = 3, cultivars were closer to wild populations in central to western Honshu, the latter including the Tanba region, than to those in northeast Honshu, which includes Sannai-Maruyama ruin (Aomori Prefecture), Aota ruin, and Yachi ruin (Niigata Prefecture), where large nuts were excavated. Thus, cultivars from the Tanba region would not have been selected directly from the nuts from these ruins. On the other hand, the results of Bayesian analysis and an assignment test suggested that the candidate origin of the cultivars was central to western Honshu, though the Tanba region (western Honshu)is more likely, based on the literature21,22. In any case, it is quite difficult to clarify the detailed origin of cultivars by comparisons to wild populations that were influenced by gene flow from cultivars, because the direction of gene flow is opposite to that typically seen during breeding and domestication processes (i.e., from wild populations into cultivars).
In cereals, it took several 1000 years to consistently obtain the non-shattering spikelet phenotype, a defining characteristic of domesticated crops45. Among fruit tree species, apple is a good example of a species whose domestication appears to have taken more than a few 1000 years. The cultivated species M. domestica originated in the Tian Shan mountains from M. sieversii. During the dispersal of apple from Asia to Europe along the Silk Route, hybridization with and introgression from Caucasian and European Malus species contributed to form present-day apple cultivars52. Slow domestication of a species suggests that there were periods of pre-domestication cultivation53, or “hansaibai”, which is suggested by Kitagawa and Yasuda17 to have occurred in Japanese chestnut at the location of the Sannai-Maruyama ruin. It is possible that the effort to improve genotypes via pre-domestication cultivation and domestication of chestnut was initiated more than once, and in several regions, because large nut remains were discovered in several ruins from different eras.
In conclusion, this study of both cultivars and wild populations of chestnut indicates the presence of gene flow from cultivars to wild forms and reveals a possible history of cultivar breeding. The genetic relationships between cultivars and wild individuals clarified in this study will be useful for both conservation and breeding. From the perspective of chestnut breeding, wild individuals distributed in Kyushu could be valuable genetic resources because they are genetically quite distant from current cultivars. Climate change is bringing new problems of black spot nut and nut rot to chestnut, and the materials from Kyushu might be helpful for chestnut production in hot areas. For the purposes of conservation, it is important to preserve wild individuals from Kyushu that are divergent from cultivars because gene flow from cultivars to natural stands was observed in this study. To achieve this goal, wild individuals carrying true “wild” genetic clusters should be collected and preserved by ex situ conservation programs such as the gene banks of public institutions.
Materials and methods
Plant materials and DNA extraction
Thirty-two populations (618 accessions), including 3 cultivated populations and 29 wild populations, were used in this study (Table 1, Supplementary Tables S1 and S2, and Fig. 1). The three cultivated populations were composed of local cultivars originated in the Kanto region (KAN), Tanba region (TAN), and other regions (OR). All of the sampling sites for wild populations excluded chestnut orchards. Individual trees were selected at least 20 m apart so as to minimize the sampling of close relatives. The heights of most trees were 5 to 15 m, which is typical for wild trees of Castanea crenata because damage from the white-striped longicorn beetle (Batocera lineolata) shortens the life of C. crenata in Japan.
Ninety-five wild chestnut individuals collected by the FTBC as part of a forest tree gene bank program were used to validate the genetic structure of wild chestnut populations (Supplementary Table S7). The FTBC program aimed to collect excellent trees for wood products, so the collections would likely be genetically biased, but they included the western part of Japan (Tottori, Shimane, Hiroshima, and Yamaguchi), where sampling of wild chestnut populations had not previously been done. We did not include this collection in the main analysis because of the potential for bias and the low number of individuals.
Genomic DNA was extracted from young leaves and buds with a DNeasy Plant Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions.
Genetic markers
The 618 accessions and the wild chestnut individuals collected by FTBC were genotyped for 30 nSSRs54–56 (Supplementary Table S3) and 4 cpSSRs57 (Cmcs1, Cmcs2, Cmcs3, and Cmcs5; Table 2). PCR amplification was performed in 10 µL containing 5 µL of 2 × Green GoTaq G2 Hot Start Master Mix (0.4 mM each dNTP, Taq DNA polymerase, and 4 mM MgCl2, pH 8.5; Promega, Madison, WI, USA), 20 pmol of each forward primer labeled with a fluorescent dye (5-FAM or 5-HEX) and unlabeled reverse primer, and 2.5 ng of genomic DNA. Amplification was performed in 35 cycles of 94 °C for 1 min, 55 °C for 1 min, and 72 °C for 2 min. PCR products were separated and detected with a 3130xl Genetic Analyzer (Life Technologies, Carlsbad, CA, USA). The size of each amplified band was determined by comparison with a set of internal-standard DNA fragments (400HD ROX, Life Technologies) in GeneMapper software v. 5.0 (Life Technologies).
Basic genetic characteristics
The observed heterozygosity (Ho) and expected heterozygosity (He) were calculated in GenAlEx v. 6.5 software58, and inbreeding coefficients (Fis) and allelic richness (Ar, n = 12) were calculated using FSTAT Version 2.9.359. Significant deviations from Hardy–Weinberg equilibrium, as indicated by deviations of Fis from zero, were tested by randomization using FSTAT. The frequencies of null alleles were estimated using CERVUS version 3.0 software60. Pairwise genetic distances based on a Jost’s D matrix among the 32 populations were calculated with GenoDive version 3.0 software61. To detect signatures of bottlenecks in the populations, we used the program INEST 2.262 to assess excess heterozygosity in each population. We ran the model using 100,000 coalescent simulations, and tested significance using the Wilcoxon signed-rank test, calculated based on 1,000,000 permutations under both the stepwise mutation model (SMM) and the two-phase model (TPM).
Analysis of population structure
Bayesian statistical inference of the population structure was performed by use of STRUCTURE 2.3.4 software63 with the correlated model for allele frequency using no prior population information (USEPOPINFO = 0), both with and without the LOCPRIOR model. The LOCPRIOR model allows for sample group information to be used to aid in the clustering process64. This model has been found to detect genetic structure at lower levels of divergence than the regular mode. The analysis was run 10 times for each value of K (number of inferred ancestral populations) from 2 to 8 for 1,000,000 iterations after a burn-in period of 1,000,000 iterations. Evanno et al.’s criterion of ∆K was used to estimate the appropriate K value65. Furthermore, genetic variance among clusters was also used to perform AMOVA66,67 in GenoDive version 3.0 software61. Significance levels were determined using 10,000 permutations. Because the populations from Kyushu were clearly genetically distant from the others, the values of ∆K and variance among clusters were also calculated using the 29 populations from outside of Kyushu. The CLUMPAK online tool68 was applied to calculate the ∆K values and to graphically display the results produced by STRUCTURE. A neighbor-joining tree based on net nucleotide distances was generated using POPULATIONS v. 1.2.3069. The 618 accessions were used for the main analysis. For validation purposes, the 95 wild chestnut individuals collected by FTBC were co-analyzed with the 618 accessions. The results of the validation are shown in Supplementary Figure S2.
Data analyses
To identify possible parent cultivars of wild individuals, putative parent–offspring relationships were calculated with CERVUS60 software, which uses a maximum-likelihood-based approach to infer parentage. The genotype data of the 30 nSSRs for 92 major cultivars (Supplementary Table S1) were used as the cultivar database of candidate parents. The paternity analyses were carried out to infer whether each wild accession had a parent–offspring relationship with each of cultivars. The parameters of the simulated genotypes were the following: “offspring” 100,000; “candidate parents” 100; “prop. sampled” 0.10; “prop. loci typed” 1.00; and “prop. loci mistyped” 0.02. Because the true parents of wild individuals could be outside of our database, we applied strict assignments using a 99% confidence threshold (95% for default parameter) for delta LOD scores to minimize the type-II error69 (when a paternity or maternity is attributed to a sampled individual, in this case a cultivar in the database, whereas the true parent is outside the set of sampled individuals). The average probability of non‐exclusion of an incorrect father using this approach was 1.5 × 10−7.
Assignment tests have been used to infer the origin of cultivars in several crops70–72. Assignment based on Bayesian criteria for computation for the 92 cultivars was carried out in GeneClass 2.073 by using the wild populations as reference sets. We applied an exclusion test using the Monte Carlo resampling method74. The genotypes were generated by Markov Chain Monte Carlo simulations of 10,000 individuals for each of the wild populations. When the probability that the wild populations in a region were sources of a cultivar was less than a given threshold (α = 0.01), the populations in that region were excluded from candidate sources for the tested cultivar.
For the 26 wild populations on Honshu island, we tested isolation by distance. Mantel tests75 were used to estimate the correlation between (FST/(1 − FST)) and the geographic distances (km) between sampled populations based on 10,000 randomizations using GenAlEx v. 6.5 software. Because wild populations TSU and TOK carried substantial portions of the “cultivar” genetic cluster, correlation of genetic distance and FST/(1 − FST) was also calculated for pairs including either TSU or TOK.
Supplementary Information
Acknowledgements
This work was supported by JSPS KAKENHI Grant Number 20K15524. We thank all of the researchers involved in the sampling of wild chestnut individuals.
Author contributions
S.N. and H.I. designed the experiments. S.N. performed the experiments. S.T., N.T., and Y.T. contributed reagents and analysis tools. S.N., N.T., H.I., M.K.K., K.I., and T.S. prepared the plant materials. S.N. and H.I. wrote the paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-80696-1.
References
- 1.Paran I, van der Knaap E. Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper. J. Exp. Bot. 2007;58:3841–3852. doi: 10.1093/jxb/erm257. [DOI] [PubMed] [Google Scholar]
- 2.Tan L, et al. Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 2008;40:1360–1364. doi: 10.1038/ng.197. [DOI] [PubMed] [Google Scholar]
- 3.Kaga A, Isemura T, Tomooka N, Vaughan DA. The genetics of domestication of the azuki bean (Vigna angularis) Genetics. 2008;178:1013–1036. doi: 10.1534/genetics.107.078451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frary A, et al. fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- 5.Juniper BE, Mabberley DJ. The Story of the Apple. Oregon, USA: Timber Press Portland; 2006. [Google Scholar]
- 6.Lam HM, et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 2010;42:1053–1059. doi: 10.1038/ng.715. [DOI] [PubMed] [Google Scholar]
- 7.Miller AJ, Gross BL. From forest to field: perennial fruit crop domestication. Am. J. Bot. 2011;98:1389–1414. doi: 10.3732/ajb.1000522. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, et al. Genetic diversity and domestication footprints of Chinese cherry [Cerasus pseudocerasus (Lindl.) G. Don] as revealed by nuclear microsatellites. Front. Plant Sci. 2018;9:238. doi: 10.3389/fpls.2018.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Studer A, Zhao Q, Ross-Ibarra J, Doebley J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 2011;43:1160–1163. doi: 10.1038/ng.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simons KJ, et al. Molecular characterization of the major wheat domestication gene Q. Genetics. 2006;172:547–555. doi: 10.1534/genetics.105.044727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arias D, Rieseberg L. Gene flow between cultivated and wild sunflowers. Theor. Appl. Genet. 1994;89:655–660. doi: 10.1007/BF00223700. [DOI] [PubMed] [Google Scholar]
- 12.Iketani H, et al. Introgression between native and prehistorically naturalized (archaeophytic) wild pear (Pyrus spp.) populations in Northern Tohoku, Northeast Japan. Conserv. Genet. 2010;11:115–126. doi: 10.1007/s10592-009-0009-8. [DOI] [Google Scholar]
- 13.O'Connor K, Powell M, Nock C, Shapcott A. Crop to wild gene flow and genetic diversity in a vulnerable Macadamia (Proteaceae) species in New South Wales, Australia. Biol. Conserv. 2015;191:504–511. doi: 10.1016/j.biocon.2015.08.001. [DOI] [Google Scholar]
- 14.De Andrés M, et al. Genetic diversity of wild grapevine populations in Spain and their genetic relationships with cultivated grapevines. Mol. Ecol. 2012;21:800–816. doi: 10.1111/j.1365-294X.2011.05395.x. [DOI] [PubMed] [Google Scholar]
- 15.Cornille A, et al. New insight into the history of domesticated apple: secondary contribution of the European wild apple to the genome of cultivated varieties. PLoS Genet. 2012;8:e1002703. doi: 10.1371/journal.pgen.1002703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou F, et al. Chestnut farming with Chinese, Japanese and Eurojapanese hybrid cultivars. In: Beccaro G, Alma A, Bounous G, Loranjo JC, et al., editors. Chestnut Handbook: Crop & Forest Management. Boca Raton: CRC Press; 2019. pp. 182–201. [Google Scholar]
- 17.Kitagawa J, Yasuda Y. The influence of climatic change on chestnut and horse chestnut preservation around Jomon sites in Northeastern Japan with special reference to the Sannai-Maruyama and Kamegaoka sites. Quat. Int. 2004;123:89–103. doi: 10.1016/j.quaint.2004.02.011. [DOI] [Google Scholar]
- 18.Tsuji K, Tsuji S, Minaki M. Seed and fruit remains and plant utilization during the early to middle Jomon periods at the Sannai-maruyama site, Aomori Prefecture, northern Japan. Jpn. J. Hist. Bot. 2006;2:101–120. [Google Scholar]
- 19.Minaki M. Enlargement of the fruit size of Castanea crenata Sieb. et Zucc since the Jomon Period (since ca. 10000 yBP) in Japan. Jpn. J. Histor. Bot. 1994;2:3–10. [Google Scholar]
- 20.Yoshikawa J. Change in the size of Castanea crenata fruits during the Jomon period. Jpn. J. Hist. Bot. 2011;18:57–63. [Google Scholar]
- 21.Isaki M. Theory and Actual Condition of Chestnut Cultivation. Tokyo, Japan: Hakuyusya; 1978. [Google Scholar]
- 22.Nishio S, et al. Use of population structure and parentage analyses to elucidate the spread of native cultivars of Japanese chestnut. Tree Genet. Genomes. 2014;10:1171–1180. doi: 10.1007/s11295-014-0751-z. [DOI] [Google Scholar]
- 23.Motoki Y. Chestnuts and Civilization in Japan. Ohtsu, Japan: Kaiseisya Press; 2015. [Google Scholar]
- 24.Oho N, Umeya K. Occurrence of the chestnut gall wasp in the people’s republic of China. Shokubutsu Boeki (Plant Protection) 1975;29:463–464. [Google Scholar]
- 25.Hasegawa Y, Suyama Y, Seiwa K. Variation in pollen-donor composition among pollinators in an entomophilous tree species, Castanea crenata, revealed by single-pollen genotyping. PLoS ONE. 2015;10:e0120393. doi: 10.1371/journal.pone.0120393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishio S, et al. Estimation of effective pollen dispersal distance for cross-pollination in chestnut orchards by microsatellite-based paternity analyses. Sci. Hortic. 2019;250:89–93. doi: 10.1016/j.scienta.2019.02.037. [DOI] [Google Scholar]
- 27.Mattioni C, et al. Landscape genetics structure of European sweet chestnut (Castanea sativa Mill): indications for conservation priorities. Tree Genet. Genomes. 2017;13:39. doi: 10.1007/s11295-017-1123-2. [DOI] [Google Scholar]
- 28.Mattioni C, Martin MA, Pollegioni P, Cherubini M, Villani F. Microsatellite markers reveal a strong geographical structure in European populations of Castanea sativa (Fagaceae): evidence for multiple glacial refugia. Am. J. Bot. 2013;100:951–961. doi: 10.3732/ajb.1200194. [DOI] [PubMed] [Google Scholar]
- 29.Gailing O, Nelson CD. Genetic variation patterns of American chestnut populations at EST-SSRs. Botany. 2017;95:799–807. doi: 10.1139/cjb-2016-0323. [DOI] [Google Scholar]
- 30.Liu W, Kang M, Tian H, Huang HW. A range wide geographic pattern of genetic diversity and population structure of Castanea mollissima populations inferred from nuclear and chloroplast microsatellites. Tree Genet. Genomes. 2013;9:975–987. doi: 10.1007/s11295-013-0610-3. [DOI] [Google Scholar]
- 31.Martin MA, et al. Landscape genetic structure of chestnut (Castanea sativa Mill.) in Spain. Tree Genet. Genomes. 2012;8:127–136. doi: 10.1007/s11295-011-0427-x. [DOI] [Google Scholar]
- 32.Poljak I, et al. Genetic diversity of the sweet chestnut (Castanea sativa Mill.) in Central Europe and the western part of the Balkan Peninsula and evidence of marron genotype introgression into wild populations. Tree Genet. Genomes. 2017;13:18. doi: 10.1007/s11295-017-1107-2. [DOI] [Google Scholar]
- 33.Tanaka T, Yamamoto T, Suzuki M. Genetic diversity of Castanea crenata in northern Japan assessed by SSR markers. Breeding Sci. 2005;55:271–277. doi: 10.1270/jsbbs.55.271. [DOI] [Google Scholar]
- 34.Nishio S, et al. Genetic diversity of Japanese chestnut cultivars assessed by SSR markers. Breeding Sci. 2011;61:109–120. doi: 10.1270/jsbbs.61.109. [DOI] [Google Scholar]
- 35.Hiraoka K, Tomaru N. Population genetic structure of Fagus japonica revealed by nuclear microsatellite markers. Int. J. Plant Sci. 2009;170:748–758. doi: 10.1086/599072. [DOI] [Google Scholar]
- 36.San Jose-Maldia L, et al. Geographic patterns of genetic variation in nuclear and chloroplast genomes of two related oaks (Quercus aliena and Q. serrata) in Japan: implications for seed and seedling transfer. Tree Genet. Genomes. 2017;13:121. doi: 10.1007/s11295-017-1202-4. [DOI] [Google Scholar]
- 37.Okaura T, Quang ND, Ubukata M, Harada K. Phylogeographic structure and late quaternary population history of the Japanese oak Quercus mongolica var. crispula and related species revealed by chloroplast DNA variation. Genes Genet. Syst. 2007;82:465–477. doi: 10.1266/ggs.82.465. [DOI] [PubMed] [Google Scholar]
- 38.Hiraoka K, Tomaru N. Genetic divergence in nuclear genomes between populations of Fagus crenata along the Japan Sea and Pacific sides of Japan. J. Plant Res. 2009;122:269–282. doi: 10.1007/s10265-009-0217-9. [DOI] [PubMed] [Google Scholar]
- 39.Shimada T. Nutrient compositions of acorns and horse chestnuts in relation to seed-hoarding. Ecol. Res. 2001;16:803–808. doi: 10.1046/j.1440-1703.2001.00435.x. [DOI] [Google Scholar]
- 40.Myles S, et al. Genetic structure and domestication history of the grape. Proc. Natl. Acad. Sci. USA. 2011;108:3530–3535. doi: 10.1073/pnas.1009363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gross BL, Henk AD, Richards CM, Fazio G, Volk GM. Genetic diversity in Malus× domestica (Rosaceae) through time in response to domestication. Am. J. Bot. 2014;101:1770–1779. doi: 10.3732/ajb.1400297. [DOI] [PubMed] [Google Scholar]
- 42.Takeuchi K, et al. Satoyama: The Traditional Rural Landscape of Japan. New York, USA: Springer; 2003. [Google Scholar]
- 43.Nishio S, et al. Genetic evidence that Chinese chestnut cultivars in Japan are derived from two divergent genetic structures that originated in China. PLoS ONE. 2020;15:e0235354. doi: 10.1371/journal.pone.0235354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomaru N. Genetic diversity and population genetic structure in widespread deciduous broad-leaved trees in Japan. Earth Environ. 2013;18:119–126. [Google Scholar]
- 45.Zhang X, et al. Phylogeography of the temperate tree species Quercus acutissima in China: Inferences from chloroplast DNA variations. Biochem. Syst. Ecol. 2015;63:190–197. doi: 10.1016/j.bse.2015.10.010. [DOI] [Google Scholar]
- 46.Saito Y, et al. Genetic variation in Quercus acutissima Carruth., in traditional Japanese rural forests and agricultural landscapes, revealed by chloroplast microsatellite markers. Forests. 2017;8:451. doi: 10.3390/f8110451. [DOI] [Google Scholar]
- 47.Kitagawa J, Nakagawa T, Okuda M, Fukuoka M, Yasuda Y. Correcting misperceptions about the history of Castanea stands in satoyama in Japan. Econ. Bot. 2008;62:594–603. doi: 10.1007/s12231-008-9022-y. [DOI] [Google Scholar]
- 48.Yamaguchi A. The government railways and the procurement of railway sleepers in prewar Japan. In: Sugiyama S, editor. Economic History of Energy and Environment. New York: Springer; 2015. pp. 31–68. [Google Scholar]
- 49.Imai K. Use of chestnut timber for railway construction. In: Imai K, editor. Chestnut. Tokyo: Houseidaigaku Shuppankyoku; 2014. pp. 59–71. [Google Scholar]
- 50.Riaz S, et al. Genetic diversity analysis of cultivated and wild grapevine (Vitis vinifera L.) accessions around the Mediterranean basin and Central Asia. BMC Plant Biol. 2018;18:137. doi: 10.1186/s12870-018-1351-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Larson G, et al. Current perspectives and the future of domestication studies. Proc. Natl. Acad. Sci. USA. 2014;111:6139–6146. doi: 10.1073/pnas.1323964111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cornille A, Giraud T, Smulders MJ, Roldán-Ruiz I, Gladieux P. The domestication and evolutionary ecology of apples. Trends Genet. 2014;30:57–65. doi: 10.1016/j.tig.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 53.Harris, D. R. & Hillman, G. An evolutionary continuum of people-plant interaction. Foraging and farming: the evolution of plant exploitation, 11–26 (1989).
- 54.Buck EJ, Hadonou M, James CJ, Blakesley D, Russell K. Isolation and characterization of polymorphic microsatellites in European chestnut (Castanea sativa Mill.) Mol. Ecol. Notes. 2003;3:239–241. doi: 10.1046/j.1471-8286.2003.00410.x. [DOI] [Google Scholar]
- 55.Kubisiak TL, et al. A transcriptome-based genetic map of Chinese chestnut (Castanea mollissima) and identification of regions of segmental homology with peach (Prunus persica) Tree Genet. Genomes. 2013;9:557–571. doi: 10.1007/s11295-012-0579-3. [DOI] [Google Scholar]
- 56.Nishio S, et al. Novel genomic and EST-derived SSR markers in Japanese chestnuts. Sci. Hortic. 2011;130:838–846. doi: 10.1016/j.scienta.2011.09.012. [DOI] [Google Scholar]
- 57.Sebastiani F, Carnevale S, Vendramin G. A new set of mono-and dinucleotide chloroplast microsatellites in Fagaceae. Mol. Ecol. Notes. 2004;4:259–261. doi: 10.1111/j.1471-8286.2004.00635.x. [DOI] [Google Scholar]
- 58.Peakall R, Smouse PE. (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes. 2006;6:288–295. doi: 10.1111/j.1471-8286.2005.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goudet J. FSTAT (version 1.2): a computer program to calculate F-statistics. J. Hered. 1995;86:485–486. doi: 10.1093/oxfordjournals.jhered.a111627. [DOI] [Google Scholar]
- 60.Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- 61.Meirmans PG. GenoDive version 3.0: easy-to-use software for the analysis of genetic data of diploids and polyploids. Mol. Ecol. Res. 2020;20:1126–1131. doi: 10.1111/1755-0998.13145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chybicki IJ, Burczyk J. Simultaneous estimation of null alleles and inbreeding coefficients. J. Hered. 2009;100:106–113. doi: 10.1093/jhered/esn088. [DOI] [PubMed] [Google Scholar]
- 63.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Res. 2009;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 66.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015;15:1179–1191. doi: 10.1111/1755-0998.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langella, O. Populations 1.2.31. Population genetic software CNRS UPR9034. [accessed 2020, September, 8] Available from http://bioinformatics.org/~tryphon/populations/ (2002).
- 69.Gerber S, Mariette S, Streiff R, Bodénès C, Kremer A. Comparison of microsatellites and amplified fragment length polymorphism markers for parentage analysis. Mol. Ecol. 2000;9:1037–1048. doi: 10.1046/j.1365-294x.2000.00961.x. [DOI] [PubMed] [Google Scholar]
- 70.Goto S, Takahashi M, Matsumoto A, Ieiri R, Tsumura Y. Genetic relationships and origin of commercial clones of Nangouhi, a vegetatively propagated cultivar of hinoki cypress (Chamaecyparis obtusa) Breed. Sci. 2008;58:411–418. doi: 10.1270/jsbbs.58.411. [DOI] [Google Scholar]
- 71.Lopes M, Mendonça D, Rodrigues dos Santos M, Eiras-Dias J, da Câmara Machado A. New insights on the genetic basis of Portuguese grapevine and on grapevine domestication. Genome. 2009;52:790–800. doi: 10.1139/G09-048. [DOI] [PubMed] [Google Scholar]
- 72.Ganopoulos I, Aravanopoulos F, Tsaftaris A. Genetic differentiation and gene flow between wild and cultivated Prunus avium: an analysis of molecular genetic evidence at a regional scale. Plant Biosyst. 2013;147:678–685. doi: 10.1080/11263504.2012.751065. [DOI] [Google Scholar]
- 73.Piry S, et al. GENECLASS2: a software for genetic assignment and first-generation migrant detection. J. Hered. 2004;95:536–539. doi: 10.1093/jhered/esh074. [DOI] [PubMed] [Google Scholar]
- 74.Paetkau D, Slade R, Burden M, Estoup A. Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Mol. Ecol. 2004;13:55–65. doi: 10.1046/j.1365-294X.2004.02008.x. [DOI] [PubMed] [Google Scholar]
- 75.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.