Abstract
The RNA interference (RNAi) drug ARC-520 was shown to be effective in reducing serum hepatitis B virus (HBV) DNA, hepatitis B e antigen (HBeAg) and hepatitis B surface antigen (HBsAg) in HBeAg-positive patients treated with a single dose of ARC-520 and daily nucleosidic analogue (entecavir). To provide insights into HBV dynamics under ARC-520 treatment and its efficacy in blocking HBV DNA, HBsAg, and HBeAg production we developed a multi-compartmental pharmacokinetic–pharamacodynamic model and calibrated it with frequent measured HBV kinetic data. We showed that the time-dependent single dose ARC-520 efficacies in blocking HBsAg and HBeAg are more than 96% effective around day 1, and slowly wane to 50% in 1–4 months. The combined single dose ARC-520 and entecavir effect on HBV DNA was constant over time, with efficacy of more than 99.8%. The observed continuous HBV DNA decline is entecavir mediated, the strong but transient HBsAg and HBeAg decays are ARC-520 mediated. The modeling framework may help assess ongoing RNAi drug development for hepatitis B virus infection.
Subject terms: Computational biology and bioinformatics, Immunology, Diseases, Mathematics and computing
Introduction
Treatment options for chronic hepatitis B virus (HBV) infections are limited to two main drug groups: pegylated interferon- (IFN) and nucleos(t)ide analogues (NAs)1–3. Treatment with IFN induces antiviral activity, immunomodulatory effects, and robust off-treatment responses. These responses, however, vary among patients and induce functional cure, defined as hepatitis B surface antigen (HBsAg) loss, in only Caucasian patients and less than Asian patients. Moreover, IFN treatment is poorly tolerated4–6. By contrast, treatment with NAs is well tolerated and can be life-long but has limited effect in reducing serum HBsAg and hepatitis B e-antigen (HBeAg) production and, in limiting hepatitis B covalently closed circular DNA (cccDNA) persistence and HBV DNA integration1,7,8, all of which play important roles in chronic infections. HBeAg is thought to induce T cell tolerance to both e- and core antigens and to be an important reason for viral persistence9. HBsAgs, besides being used for virion envelopes, form empty non-infectious subviral particles (i.e. without viral genome) whose numbers are at least 1,000-folds higher than those of virions10, and may serve as decoy for antibody responses11. Moreover, they are also assumed to be involved in T cell exhaustion12,13. Functional cure has been proposed as a desirable outcome of treatment. None of the currently licensed therapies can produce this result for a large fraction of chronically infected patients. There is therefore a need for new therapies that target HBsAg production and/or its clearance from circulation14,15.
RNA interference (RNAi) technology has the ability of silencing specific genes and can, therefore, be used for treatment against a large array of infectious agents (see16 for a review on RNAi-based therapies). For hepatitis B infection, small interfering RNAs were designed to hybridize with HBV mRNA inside an infected hepatocyte and, as a result, induce its degradation17–19. ARC-520, the first such small interfering RNA to be tested in clinical trials, was designed with the aim of knocking down the expression of all HBV mRNA, including HBsAg proteins. Experiments in mice and chimpanzees, and a phase II clinical study in patients (Heparc-2001) showed potential for ARC-520 induced HBeAg, HBsAg and HBV DNA titers reduction17,20,21. The Heparc-2001 study showed differential HBsAg reduction among patients based on their HBeAg status and prior exposure to traditional therapy such as NAs20. While ARC-520 has been terminated due to delivery-associated toxicity20, overall results indicate that RNAi-based therapy has the potential of reducing HBsAg and inducing functional cure16,21,22 regardless of the patient’s HBeAg status23,24.
To better understand the effect of RNAi therapies, additional information regarding the host-virus-drug dynamics and therapy outcomes are needed. In this study, we developed mathematical models that best reproduce observed HBV DNA, HBsAg and HBeAg kinetics following a single dose of ARC-520 in five HBeAg-positive patients from the Heparc-2001 study. Mathematical models of hepatitis B infection have been used to study the dynamics of acute, chronic, and occult HBV infections25–29, anti-HBV therapy14,30–35, cell-to-cell transmission36, intracellular interactions36–38, cellular immune responses26,30,39–41, antibody-mediated immune responses11,38,42, HBeAg38,43,44, and HBeAb38 dynamics. We build on previous modeling work, consider the interaction between HBV DNA, HBsAg and HBeAg titers in the presence of a single dose RNAi-based therapy, and use the model to run in silico experiments to predict individual contributions of different drug effects on the dynamics for HBsAg titers.
Methods
Patient data
We use published data from five HBeAg-positive, treatment-naive chronic hepatitis B patients (cohort 7 in20), which are the ones that best responded to ARC-520 therapy. Data consists of serum HBV DNA titers (in IU/ml), HBsAg, and HBeAg concentration (in IU/ml) measured at = days, where and is the day when both daily NA entecavir (ETV) and a single intravenous ARC-520 injection (inoculum of 4 mg/kg) are administrated.
Pharmacokinetics–pharamcodynamics model
We are interested in determining the mechanisms underlying the observed HBV DNA, HBsAg and HBeAg kinetics under combined ETV and ARC-520 therapy. We develop a mathematical model that considers the interactions between infected hepatocytes, I (in cells per ml); total intracellular HBV DNA, D (in copies per ml); serum HBV DNA, V (in IU per ml); serum HBsAg, S (in IU per ml); and serum HBeAg, E (in IU per ml). We assume that infected cells decay at per capita rate , and we exclude cell proliferation (we will relax this assumption later on). We assume intracellular HBV DNA is synthesized at rate and is lost at constant per capita rate . The replication rate summarizes various steps that are not modeled explicitly, such as the transcription of pregenomic RNA (pgRNA) from cccDNA, and the generation of single stranded DNA by reverse transcription. Intracellular HBV DNA is assembled and released into blood as free virions at rate p which are cleared at rate c. To account for the different units of intracellular and serum virus, we use the conversion factor IU/copies45. Lastly, we assume HBsAg and HBeAg are transcribed from cccDNA inside infected hepatocytes and then released into blood at rates and , respectively, and are cleared at per capita rates and , respectively. The model is given by the following model:
| 1 |
Patients were administered daily nucleoside analogous treatment with entecavir starting at day . ETV is known to block reverse transcription of HBV DNA, and therefore inhibit HBV DNA synthesis. We model this (see model (5)) as a constant reduction of the HBV DNA synthesis rate to , where is the ETV efficacy. Experimental studies in humanized mice have shown that serum HBV DNA declines in biphasic manner while HBV-infected cell are not lost in the first months following NA treatment initiation46,47. To account for the biphasic HBV DNA decay in the absence of infected cell killing, we assume that ETV has additional time-dependent inhibitory effects on intracellular HBV DNA synthesis and model it by decreasing further to , where is a constant and t is the time in days post ETV initiation. Moreover, a single ARC-520 dose was administrated at time . Unlike ETV, which was given daily, we model the build-up and clearance of ARC-520 pharmacokinetics over time by considering a two-compartment pharmacokinetic model consisting of drug quantity in the plasma and liver, and , respectively48. The inoculum decays exponentially at rate , where is the plasma drug degradation rate and is the absorption into the liver rate. The drug in the liver decays at rate , identical with the absorption rate49. Following these assumptions, the pharmacokinetic model has the form:
| 2 |
with initial conditions and . This is a linear model which can be solved to give solutions:
| 3 |
Lastly, we assume the relationship between the drug quantity in the liver (t) and drug efficacy to be given by:
| 4 |
where is the maximum drug efficacy, are drug quantities that yield half-maximal effects, and are the infectious events that are affected by ARC-520 therapy, i.e., the transcription of HBV DNA, the transcription of HBsAg, and the transcription of HBeAg, respectively. The effects of ARC-520 on intracellular HBV DNA, HBsAg and HBeAg are modeled as the reduction of intracellular HBV DNA synthesis to , HBsAg production from to , and of HBeAg production from to , respectively. Considered together, models (1) and (4) give the following pharmacokinetics–pharamcodynamics (PK/PD) model:
| 5 |
Data fitting
We used published kinetic HBV DNA, HBsAg, HBeAg data in serum measured from five HBeAg-positive, treatment-naive chronic hepatitis B patients as described in the ‘Patient data’ section.
Parameter values
We assume that, prior to therapy initiation, model (5) describes a persistent chronic infection and is at the quasi-equilibrium, given by the initial values , , , and . Initial values for HBV DNA, ; HBsAg, ; and HBeAg, , are set to the patient data prior to the start of therapy, , (day eight prior to the ARC-520 injection). The percentage of HBV-infected hepatocytes is reported to vary between in chronic HBsAg carriers50,51 and in acute infections26,52. Without loss of generality, we arbitrary assume that of hepatocytes are infected at the beginning of treatment. Liver contains approximately hepatocytes, which, when distributed throughout 15 liters of extracellular fluid, gives a total hepatocyte concentration cells/ml53. We set the initial infected hepatocyte population to . Lastly, the pre-treatment level of intracellular HBV DNA in HBeAg positive patients is set to copies/ infected cell, as in54.
Since we assume that model (5) is in chronic equilibrium (for the additional assumption ) before the therapy initiation, parameters , p, , are fixed according to the following formulas:
| 6 |
We start by ignoring the dynamics of infected cells, such as infection of susceptible cells and/or infected cell proliferation (we will relax this assumption in later sections), and assume that infected cells decay due to natural death and immune mediated killing at per capita rate per day, corresponding to a life-span of 250 days (we will later investigate the effect of increasing the killing rate, to include increased immune mediated killing or RNAi induced toxicity and death). The estimated half-life of intracellular HBV DNA is 24 h55, which corresponds to the intracellular HBV DNA decay rate per day. ARC-520’s half-life has been reported to range between 3 and 5 h56, corresponding to decay rates per day; we fix per day. Lastly, we set the initial ARC-520 quantity to the trial dose of mg/kg.
The unknown parameters are . Here, accounts for the total drug effect on HBV DNA production. Since preliminary simulations (not shown) indicate that is time independent, we cannot separate the ETV effects from the ARC-520 effects . We lump them together, and assume a total drug effect, which ranges between . The other parameter ranges are found as follows. The time-dependent inhibitory effects of treatment on intracellular HBV DNA production, g, was estimated from HBV infected humanized mice treated with NA to range between 0.059 and 0.42 per day. We expand this range by searching over the parameter space . There is a wide range of estimates for the free virus clearance rate in serum: as low as 0.69 per day25,33,57; and as high as 21.7 per day58; we search the entire parameter space. The decay rate of HBsAg is bounded between per day, containing previous estimates ranging between 0.057 to 0.58 per day59,60. In previous modeling work44,61 HBeAg decay rate was set to 0.3 per day. We allow for a larger range per day, corresponding to half-lives greater than 5 minutes. We assume that the drug absorption rate ranges between per day. Since ARC-520 was reported to have long lasting effects56, we assume a large range for the half-maximal quantity ; between mg/kg. These ranges are summarized in Table 1.
Table 1.
Variables and parameters in model (5).
| Variables | Description | Units | Initial values | |
|---|---|---|---|---|
| I | Infected hepatocytes | cells/ml | ||
| D | Intracellular HBV DNA | copies/cell | 45054 | |
| V | Free virions | IU/ml | Data at time | |
| S | Serum HBsAg | IU/ml | Data at time | |
| E | Serum HBeAg | IU/ml | Data at time | |
| Parameters | Descriptions | Units | Default values / range | Reference |
| Infected cells decay rate | 1/day | |||
| Inhibitory effects on intracellular HBV production during treatment | 1/day | [0, 1] | ||
| Intracellular HBV DNA synthesis rate | Copies/(cellday) | |||
| Intracellular HBV DNA decay rate | 1/day | 0.69 | 55 | |
| Conversion factor | IU/copies | 1/5.3 | 45 | |
| p | Intracellular HBV DNA release rate | 1/day | ||
| Free virion clearance rate | 1/day | [0, 100] | ||
| HBsAg production rate | IU/(cellday) | |||
| HBeAg production rate | IU/(cellday) | |||
| HBsAg decay rate | 1/day | [0, 200] | ||
| HBeAg decay rate | 1/day | [0, 200] | ||
| Combined ETV and ARC-520 efficacy | unitless | [0.9,1] | ||
| Initial plasma drug quantity | mg/kg | 4 | 20 | |
| d | ARC-520 decay rate | 1/day | 4 | 56 |
| ARC-520 Quantity where is half maximal | mg/kg | |||
| ARC-520 Quantity where is half maximal | mg/kg | |||
| Drug absorption rate | 1/day | [0,1] | ||
Parameters indicated by a * are fitted within the given range.
Optimization algorithm
We estimate the unknown parameters given in Table 1 by minimizing the least squares functional:
| 7 |
for each patient. Functional SSQ describes the distance between HBV DNA, HBsAg, and HBeAg titers , , at times () and populations , and as given by model (5) at times (). As described previously (see Eq. (6)), the before treatment titers at days are used to determine parameters , p, , such that the model’s equilibrium matches the titers exactly. Since we assume that the model stays in equilibrium until treatment initiation, we ignore the titers at time days. Lastly, it should be noted that we assign the same weight to errors in HBV DNA, HBsAg, and HBeAg. Within the parameter space defined in Table 1, we determine optimal parameter fits for each patient by following four steps (code available upon publication):
We create 100 parameter sets using the Latin hypercube samples (LHS) Matlab routine lhsdesign, with random number generator seed two and uniform probability density distribution on each parameter interval. Since the parameter space spans several orders of magnitude in and directions, we replace them with and . Thus, instead of sampling and in , we sample and in . Our preliminary work showed that often yields the best results.Therefore, we replace and sample in the parameter space .
HBV DNA dynamics do not influence HBsAg and HBeAg dynamics. Therefore, we minimize and separately over their corresponding parameter sets and , respectively. We split the LHS into and containing the respective initial parameter guesses and, using Matlab’s fmincon routine to minimize and within the parameter space in Table 1, obtain 100 optimal and parameter sets.
Of the optimal parameter sets found in part two, we choose the ones yielding minimal , as the overall optimal parameter set for the given patient.
- To obtain confidence intervals for the optimal parameter estimates for each patient, we employ a bootstrapping technique. We assume that the best fit parameters yield the true dynamics, and that any discrepancy from the data is due to measurement errors. First, we calculate the residuals
between simulated functions and measured data at times (). Next, we create 1000 data sets for the HBV DNA, HBsAg, and HBeAg data at times , where data at times and are as before and data at the remaining times are obtained by adding a randomly drawn residual (with repetition) to the true value at each time, i.e.8
where , , and is drawn at random from . Lastly, for each data set, we find a new set of optimal parameters by using Matlab’s fmincon with initial parameter guess to minimize and , as described in (2.). This yields 1000 sets of parameters (one for each data sets), and the confidence intervals on the optimal parameters are obtained as the ranges from the 2.5th percentiles to the 97.5th percentiles of the 1000 parameter values.
Results
Parameter estimates
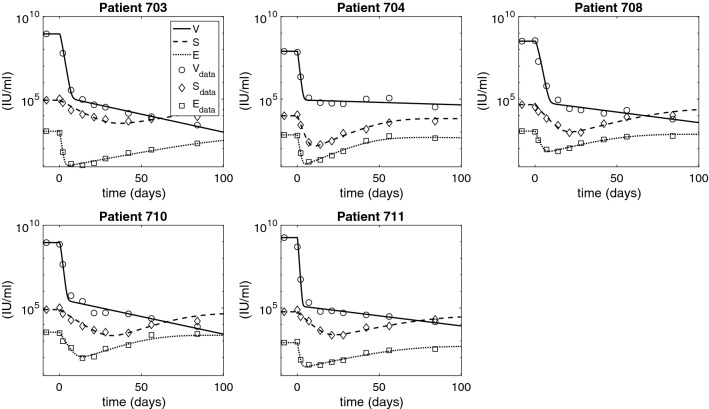
The best parameter estimates, the respective errors (SSQ) and the the 95% confidence intervals obtained by bootstrapping, are given in Table 2. Numerical solutions for each population versus data are shown in Fig. 1 (see also Figs. 2, 3, and 4 for zoomed in results). Table 3 gives the parameters obtained from equilibrium conditions (6).
Table 2.
Estimated parameters, fit errors, and confidence intervals.
| g | c | SSQ | |||||||
|---|---|---|---|---|---|---|---|---|---|
| mg/kg | mg/kg | ||||||||
| 703 | 4.6 | 1.33 | 0.12 | 1.52 | 3.8 | 3.39 | 3.49 | 4.69 | 1.03 |
| 703 | (3.7,5.8) | (1.3,1.6) | (0.11,0.16) | (1.3,1.7) | (4,3.6) | (3.6,3.2) | (3.6,3.4) | (4,5.4) | |
| 704 | 0.3 | 9.27 | 0.6 | 1.35 | 2.94 | 3.25 | 2.96 | 9.81 | 1 |
| 704 | (0,1.4) | (6.8,11) | (0.5,0.7) | (1.1,1.5) | (3,2.7) | (3.4,3.2) | (3,2.8) | (8.5,11.2) | |
| 708 | 2.54 | 1.24 | 0.25 | 0.6 | 3.67 | 3.39 | 2.61 | 6.43 | 1.42 |
| 708 | (0.6,5.1) | (1.1,2.5) | (0.2,0.3) | (0.5,0.8) | (4.2,3.3) | (3.6,3.3) | (2.7,2.5) | (5.2,7.7) | |
| 710 | 4.48 | 1.87 | 0.15 | 0.37 | 3.43 | 3.73 | 3.02 | 8.3 | 1.48 |
| 710 | (2.9,6) | (1.3,2.4) | (0.1,0.18) | (0.2,0.5) | (3.7,3.1) | (4.2,3.4) | (3.2,2.8) | (5.9,11) | |
| 711 | 2.41 | 3.12 | 0.21 | 1.4 | 4.13 | 3.14 | 2.8 | 5.55 | 0.78 |
| 711 | (1.4,3.2) | (2.8,3.3) | (0.2,0.24) | (1.1,1.7) | (4.3,3.9) | (3.2,3) | (2.9,2.7) | (4.7,6.4) | |
| MEAN | 2.87 | 3.37 | 0.27 | 1.05 | 3.59 | 3.38 | 2.98 | 6.96 | 1.14 |
| MEDIAN | 2.54 | 1.87 | 0.21 | 1.35 | 3.67 | 3.39 | 2.96 | 6.43 | 1.03 |
| SD | 1.77 | 3.38 | 0.19 | 0.52 | 0.44 | 0.22 | 0.33 | 2.08 | 0.3 |
Figure 1.
HBV DNA, HBsAg, and HBeAg dynamics over time as given by model (5) (solid curves) versus data (circles). The parameters are given in Tables 1 and 2.
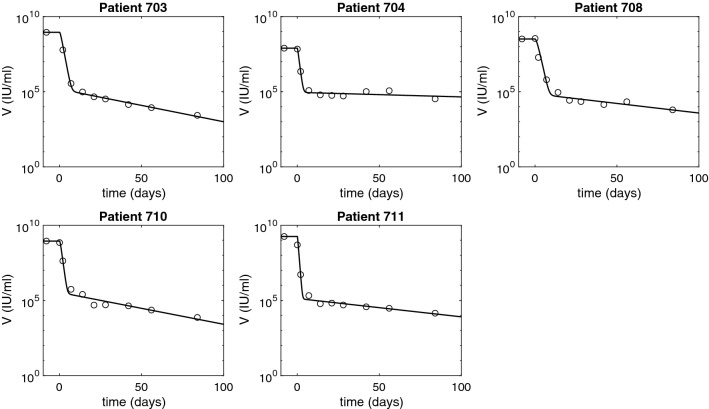
Figure 2.
HBV DNA dynamics over time as given by model (5) (solid curves) versus data (diamonds). The parameters are given in Tables 1 and 2.
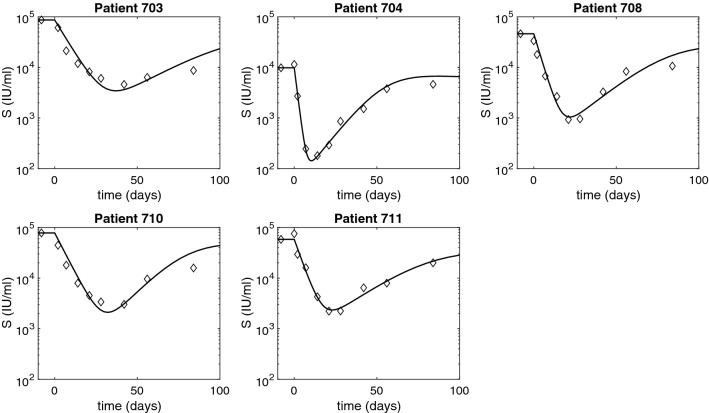
Figure 3.
HBsAg dynamics over time as given by model (5) (solid curves) versus data (diamonds). The parameters are given in Tables 1 and 2.
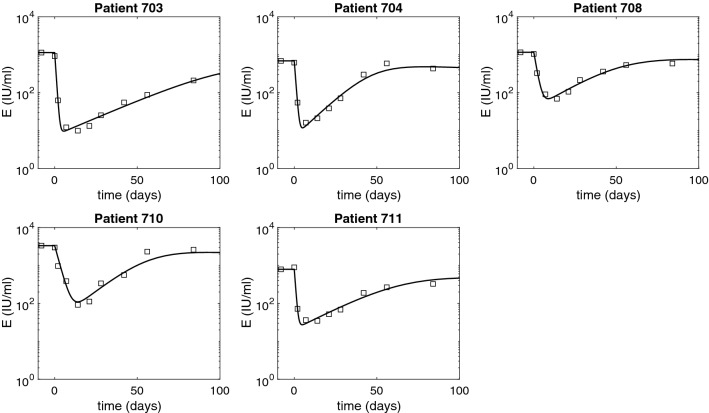
Figure 4.
HBeAg dynamics over time as given by model (5) (solid curves) versus data (diamonds). The parameters are given in Tables 1 and 2.
Table 3.
Parameters obtained from fitted parameters in Table 2, under equilibrium conditions defined by Eq. (6).
| p | ||||
|---|---|---|---|---|
| 703 | 1.99 | 1206.9 | 1.54 | 2.48 |
| 704 | 1.22 | 861.66 | 0.85 | 1.33 |
| 708 | 0.67 | 613.66 | 1.65 | 0.99 |
| 710 | 2.8 | 1569.85 | 1.63 | 1.77 |
| 711 | 9.37 | 4526.23 | 1.77 | 1.58 |
| MEAN | 3.21 | 1755.66 | 1.49 | 1.63 |
| MEDIAN | 1.99 | 1206.9 | 1.63 | 1.58 |
| SD | 3.54 | 1590.21 | 0.37 | 0.56 |
Previously reported virus clearance rates range from 0.69 per day25,33,57 to 21.7 per day58. We estimate average virus clearance rates among the five patients per day, corresponding to average life-spans of 7.1 h. The fastest free virus clearance rate, per day (life-span of 2.6 h), occurs in patient 704, who has the lowest pre-treatment virus titer. Assuming 50 of hepatocytes are HBV-infected, we estimate an average intracellular HBV DNA release rate per day. Patient 711, who has the highest pre-treatment virus titer, has per day, 2.9 times higher than the average. Under these estimates, the pre-treatment serum virus production rates, , range between 301.5 and 1260 copies/(infected cellday) for patients 703–710, similar to the 200–1000 copies/(infected cellday) reported for acute HBV infection62. Patient 711, however, has a pre-treatment serum virus production rate, copies/(infected cellday), four times larger than in62. Intracellular HBV DNA synthesis rates are copies/(cell day). As with the serum release rate, patient 711 has 2.6-times higher intracellular HBV DNA synthesis than the average, copies/(ml day).
The reported half-life of circulating HBsAg in chronically infected patients is 6.7 days (with a standard deviation of 5.5 days)59, which corresponds to HBsAg decay rates per day. We estimate average HBsAg decay rates per day, corresponding to HBsAg life-span of 5.6 days for patients 703 and 708-711, and per day, corresponding to HBsAg life-span of 1.7 days, for patient 704. The average clearance rates of circulating HBeAg per day, correspond to HBeAg life-spans ranging between 15.8 h and 2.7 days, about one order of magnitude lower than those reported by Loomba et al. for HBsAg59. The decreased HBeAg life-span predicted by our model may be correlated with the emergence of immune events and/or mutation in the core/precore regions44 during ARC-520 treatment. Since we have no data on these events, we did not account for them in our model. Production rates of HBsAg and HBeAg are estimated to be IU/(cell day) and IU/(cell day), respectively.
We estimate high efficacy rates, , for the combined entecavir and ARC-520 effects in blocking HBV DNA synthesis. The additional time-dependent inhibitory effect on intracellular HBV DNA synthesis is on average per day.
The estimated per day, predicts slow transport of ARC-520 from plasma to liver. The half-maximal quantities are small, with average and for the ARC-520 effects on HBsAg and HBeAg, respectively. This implies that the effects of ARC-520 are long-lived, as suggested by Schluep et al.56 who found that RNA inhibitors persist and induce antiviral effects for longer than the drug’s life-span.
Pharmacokinetic–pharmacodynamic model dynamics
The predicted HBV DNA populations as given by model (5) for the estimated parameters follow a biphasic decay with short and sharp first phase corresponding to the removal of HBV DNA followed by long and slow second phase decay due to time dependent treatment induced inhibition of intracellular HBV DNA synthesis and infected cell loss. HBsAg and HBeAg decay at steep rates during the first and days, respectively. After reaching minimum values, on average and orders of magnitude smaller than their initial levels, HBsAg and HBeAg rebound (see Figs. 3 and 4). Once the effects of ARC-520 have completely waned, HBsAg and HBeAg decay at rate .
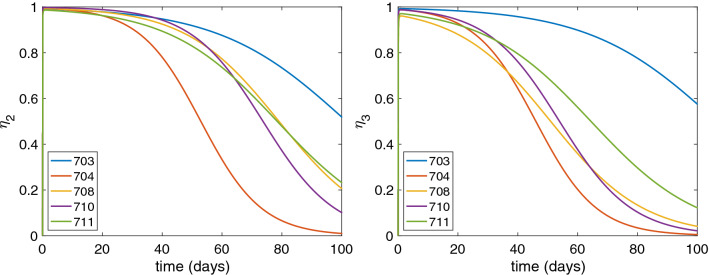
For the estimated parameters, ARC-520 effects and given by model (4) increase from 0 to their maximum values during the first days. The effect of ARC-520 on HBsAg is similar for all patients, with maximal effect at day 1 (ranging between and , which wanes to in 1.8 to 3.4 months (see Fig. 5, left panel). The maximal effect of ARC-520 on HBeAg at day 1 ranges between (patient 708) and (patient 703) and wanes to within 1.5 to 3.5 months (see Fig. 5, right panel). For both HBsAg and HBeAg, the effect of ARC-520 lasts longest in patient 703.
Figure 5.
Efficacy of ARC-520 treatment over time as given by model (4) on (a) HBsAg production, and (b) HBeAg production. The parameters are given in Tables 1 and 2.
In-silico knockout experiments
We are interested in understanding the individual and combined effects of ETV and one-dose of ARC-520 on the dynamics of HBV DNA, HBsAg and HBeAg as given by model (5). We consider the following about the combined ETV and ARC-520 effects on reducing intracellular synthesis, : we either attribute it to ETV alone, ; or split it between the two effects, . Using the parameters obtained from fitting the combination therapy model (5) to the Heparc-2001 clinical trial data20, we conduct in silico experiments to determine how the dynamics change under: in silico monotherapy with entecavir, described by for , , and ; and combined entecavir and ARC-520 treatment, described by for , , and (, , and ) obtained through data fitting.
When we investigate in silico ETV monotherapy targeting HBV DNA intracellular synthesis, , we can analytically derive the solutions of model (5) by considering . , and . The infected cell population becomes , the intracellular HBV DNA:
| 9 |
and extracellular HBV DNA:
| 10 |
The equations for HBeAg is given by:
| 11 |
and for HBeAg is given by:
| 12 |
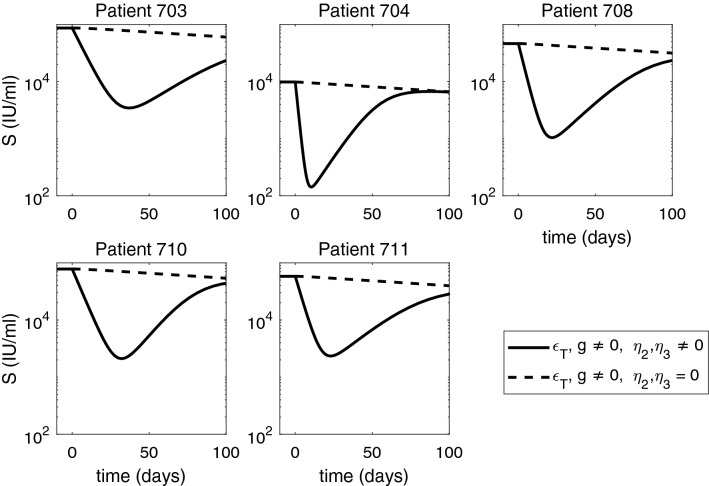
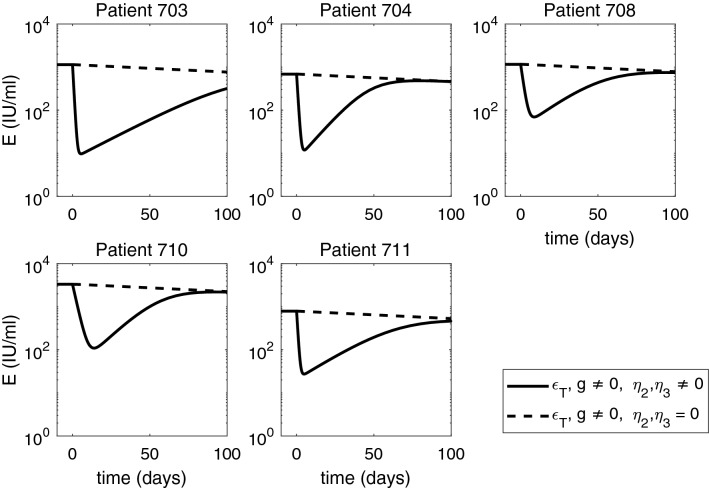
Note that both S(t) and E(t) are independent of . HBV DNA follows a biphasic decay with short and sharp first phase corresponding to the removal of free virus followed by a slow second phase decay due to time dependent treatment induced inhibition of intracellular HBV DNA synthesis and removal of infected cells (see Fig. 6, dashed curves). Serum antigen levels remain elevated for all three populations (see Figs. 7 and 8 , dashed curves).
Figure 6.
Short-term HBV DNA dynamics under ETV monotherapy (dashed curves), and combined ETV and ARC-520 therapy (solid curves), as given by model (5). Parameters are given in Tables 1 and 2. Additionally, and for ETV monotherapy. Note that both axes are plotted on log scale and that the two graphs overlap.
Figure 7.
Short-term HBsAg dynamics under ETV monotherapy (dashed curves), and combined ETV and ARC-520 therapy (solid curves), as given by model (5). Parameters are given in Tables 1 and 2. Additionally, and for ETV monotherapy. Note that both axes are plotted on log scale.
Figure 8.
Short-term HBeAg dynamics under ETV monotherapy (dashed curves), and combined ETV and ARC-520 therapy (solid curves), as given by model (5). Parameters are given in Tables 1 and 2. Additionally, and for ETV monotherapy. Note that both axes are plotted on log scale.
When we consider that the treatment that blocks intracellular HBV DNA synthesis, , comes from both ETV and ARC-520, we recover the solutions of model (5) for combination therapy given by , , and . Both HBsAg and HBeAg decay at a steep rate during the first and days, respectively. After reaching minimum values, on average and orders of magnitude smaller than their initial levels, HBsAg and HBeAg rebound to their respective ETV monotherapy levels (see Figs. 7 and 8, solid curves).
Sensitivity of model predictions with respect to changes in the infected cell population’s initial condition
Previous estimates for the percentage of HBV-infected hepatocytes vary between in chronic HBsAg carriers50,51 and in acute infections26,52. We have derived our results by assuming that during chronic HBeAg-positive cases half of the liver is infected. Here, we investigate how changes in the size of the initial infected cell population alter our predictions. Analytical investigations show that the dynamics of the viral proteins HBsAg and HBeAg are not influenced by the initial size of the infected cell population, . After treatment initiation , and and (based on the equilibrium assumption (6)). Therefore, the equations for S and E:
| 13 |
and
| 14 |
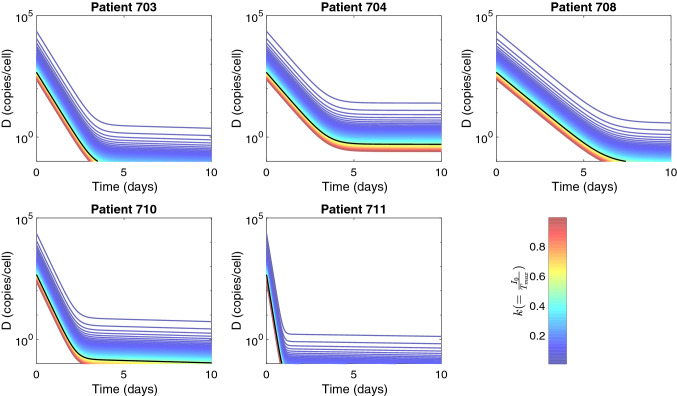
are independent of . Moreover, for and we find that intracellular HBV DNA D depends on (see Fig. 9) but HBV DNA in serum does not.
Figure 9.
Intracellular HBV DNA dynamics of model (5) for and . Solid black lines show the dynamics for , which was used in data fitting. Other parameters used are given in Tables 1, 2, and .
Long-term predictions and the need for uninfected hepatocyte dynamics
We assumed above that infected hepatocytes have a fixed life-span of 250 days. In this section, we are relaxing this assumption and investigate long-term HBV DNA and HBsAg dynamics when increased hepatocyte loss (due to either drug toxicity, or immune-mediated killing) is being considered. When we model it by increasing the infected cell death rate in (5) we obtain the following: long-term dynamics of S and E under ETV monotherapy predict that HBsAg decreases below 1 IU/ml months for per day, years for per year, and years for per day, following the initiation of therapy. Since ETV and other nucleoside analogues do not trigger cccDNA removal (and consequently HBsAg and HBeAg removal), the fast loss of HBsAg predicted by model (5) for higher killing rates is not realistic. In this section, we include the dynamics of uninfected and infected cell populations and investigate changes in predictions for increased killing rate We incorporate uninfected hepatocytes T which get infected by free virus at rate , as modeled previously26,39,63. Note that we ignore the age of the infection and assume that once a cell becomes infected, it is producing virus (for a PDE model extension in a hepatitis C virus infection, see64,65). Both uninfected and infected hepatocytes proliferate according to a logistic term with maximal growth rate and and carrying capacity . In chronic HBV infections, cccDNA persist under long-term nucleoside analogues treatment66. Since the average cccDNA number of untreated HBeAg positive patients is 2.58 copies per infected cell54, infected hepatocytes may have two infected off springs. On the other hand, it has been suggested that cccDNA is destabilized by cell division or even lost during mitosis66. We account for this by assuming that a fraction of proliferating infected hepatocytes have one infected and one uninfected offspring, and the remaining infected hepatocytes have two infected offsprings. The new model is given by:
| 15 |
Liver regenerates rapidly after injury. To account for fast proliferation during chronic disease, we assume that hepatocytes’ maximum proliferation rate is per day, and per day, corresponding to doubling time of (up to) 16 h26,67. The infectivity rate is at the lower end of previously fitted values11, IU/(ml day); we include a death rate for the uninfected hepatocyte population, per day68, identical to that in model (5); and set the fraction of infected hepatocytes that have one uninfected and one infected offspring to . Initial conditions of uninfected and infected hepatocytes are set such that the model is in equilibrium prior to treatment with , and , , and as in Table 1. This leads to almost all hepatocytes being infected.
Without loss of generality, we investigate the dynamics for patient 703 under combination therapy for a continuum of values. Our hypothesis is that NA monotherapy cannot lead to HBsAg loss. In order to obtain infected cell persistence (under NA monotherapy), we need to decrease (for a fixed ) as increases (a threshold required for infected cells persistence is given in Fig. 10). Therefore, HBsAg persistence under increased infected cell killing (as seen in NA treatment) may be explained by high ratio of infected to uninfected cell proliferation. Other events, such as HBV DNA integration, adaptive immune responses, such as cytolytic and non-cytolytic effects, and/or antibody neutralization11,26 may also explain HBsAg persistence under infected cell (and potentially cccDNA) loss. This is especially true for HBeAg negative patients and NA experienced, HBeAg-positive patients.
Figure 10.
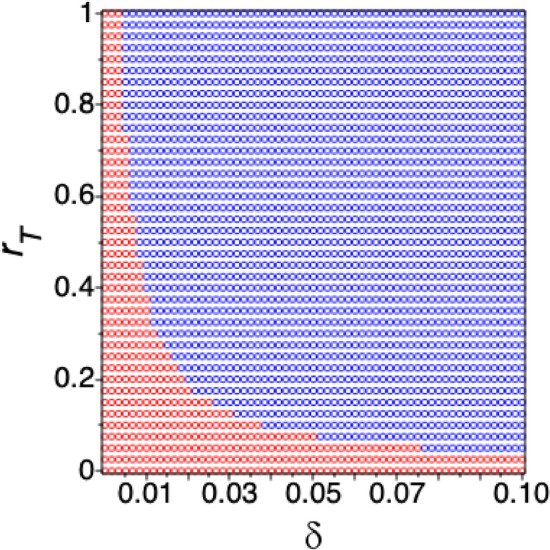
() ranges where infected cells given by model (15) are cleared (blue dots) or persist (red dots) under ETV monotherapy. Here per day, ml/(IU day), per day, initial conditions and are set such that the model is in chronic equilibrium in the absence of treatment. The other parameters are given in Tables 1 and 2 for patient 703.
Discussion
Reaching functional cure with current anti-HBV therapies in patients with chronic hepatitis B infection is hindered difficult by the lack of approved direct anti-HBsAg treatment and the presence of large numbers of HBsAg in the blood of infected patients69,70. Therapies silencing viral translation through RNA interference technology17,20,21,71, inhibiting HBsAg release via nucleic acid polymers72–74, and inducing neutralization of HBsAg via specific antibodies75,76 have shown different levels of success69,70. Understanding the relative effects in reducing HBV DNA, HBsAg and HBeAg titers of these new approaches alone, and in combination with traditional nucles(t)ide analogues, is particularly important in informing the development of new generation anti-HBsAg therapies.
To help in this endeavor, we developed mathematical models describing the HBV DNA, HBsAg and HBeAg in the presence of a silencing RNAi drug called ARC-520. We used the models and clinical trial data from treatment naive, HBeAg-positive patients that receive a one time ARC-520 injection and daily nucleoside analogue treatment with entecavir20, to determine the efficacy of ARC-520 and nucleoside therapies on the short and long-term dynamics of HBV DNA, HBsAg, and HBeAg. To the best of our knowledge, we report for the first time that the time-dependent ARC-520 effects on HBsAg and HBeAg are more than 96 effective around day 1, and slowly wane to 50 in 1.8-3.4 months and 1.5-3.5 months, respectively. The combined ARC-520 and entecavir effect on HBV DNA is constant over time, with efficacy of more than , which is similar to other nucleoside analogues trials.
A simplified version of the model, which ignored the dynamics of hepatocyte proliferation and infection, was sufficient to explain the short-term (about 100 days) dynamics observed in five patients in the current study. In the long-term, however, infected cells may die at faster rates, due to either drug toxic effects or increased immune killing. Lowering infected hepatocyte’s life-span to 100 (10) days, however, resulted in fast HBsAg removal, with decay below 1 IU/ml in 4.2 years ( 5.3 months). This loss, however, was in contradiction with clinical reports of low percentages of patients clearing HBsAg during long-term nucleoside analogues treatment6, suggesting that more complex models are needed for long-term (several years) predictions. To determine under what conditions increased infected cells death does not spill over into unrealistic HBsAg and HBeAg loss under long-term nucleoside analogue therapy, we extended model (5) to include infected and uninfected cell dynamics. We assumed lower infected cells life-span (100 and 10 days), included division of both infected and uninfected populations, and determined that long-term HBsAg and HBeAg persistence under long-term HBV DNA clearance can be explained by high ratios of infected to uninfected division rates. Therefore, high ratio of infected to uninfected division rates, which correspond to the infection of the entire liver and may be indicative of scenarios where HBsAg seroclearance will not happen. Interestingly, we and others have associated high ratios of infected to uninfected division rates to triphasic HBV DNA decay under treatments with nucleoside analogues, a sign of suboptimal drug response33,35. Whether infected hepatocytes indeed proliferate faster than uninfected hepatocytes remains under investigation.
While modeling results suggest that one-dose of ARC-520, in combination of daily entecavir, has limited long-term effects, we did not consider whether a transient reduction of HBsAg and HBeAg leads to the appearance of anti-HBs or anti-HBe antibodies, removal of immune-exhaustion, and eventual functional cure. Recent studies found that large levels of HBsAg might cause dysfunctional programming of HBsAg-specific B cells through persistent stimulation77. It has been suggested that therapeutic vaccines containing one (PreS2) or two (PreS1 or PreS2) envelope proteins together with serum HBsAg reducing drug therapies are needed in order to induce high levels of anti-HB antibodies, which may correlate with functional cure78–80. We ignored the level of immune modulation following RNAi based therapy, such as cytolytic and non-cytolytic T cell functions and antibody responses, which is a model limitation, and therefore, we cannot say whether such effects were induced at higher rates during the transient HBsAg loss.
Our study has limitations. We only used the data on HBeAg-positive patients (cohort 7 in20) since they best responded to ARC-520 therapy. Moreover, we did not model HBV DNA integration, which has been reported as a source of HBsAg production, especially in HBeAg-negative and NA-experienced HBeAg-positive patients with low cccDNA20. As kinetic HBV data from next generation RNAi therapy capable of inducing stronger HBsAg reduction in both HBeAg-negative and HBeAg-positive patients becomes available21,81,82, we aim to adapt our modeling framework to include HBV DNA integration.
In conclusion, we developed a mathematical model and used it together with patient data, to estimate the time-dependent ARC-520 efficacies in blocking HBsAg and HBeAg productions. Additional data and theoretical efforts are needed to determine whether RNAi therapies have a feedback effect on the reversal of immune exhaustion, immunomodulatory immune responses, and potential functional cure.
Acknowledgements
SK and SMC acknowledge funding from National Science Foundation Grant No. 1813011. HD acknowledges funding from NIH Grant Nos. R01AI144112 and R01AI146917. We thank Christine Wooddell and the anonymous reviewers for their valuable comments.
Author contributions
All authors conceived the study and performed the analyses. S.K. wrote the code. S.K. and S.M.C. wrote the manuscript. All authors reviewed and revised the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rijckborst V, Janssen HL. The role of interferon in hepatitis B therapy. Curr. Hepat. Rep. 2010;9:231–238. doi: 10.1007/s11901-010-0055-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, et al. Combination therapy based on pegylated interferon alfa improves the therapeutic response of patients with chronic hepatitis B who exhibit high levels of hepatitis B e-antigen at 24 weeks: a retrospective observational study. Medicine. 2019;98:e17022. doi: 10.1097/MD.0000000000017022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Razavi-Shearer D, et al. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol. Hepatol. 2018;3:383–403. doi: 10.1016/S2468-1253(18)30056-6. [DOI] [PubMed] [Google Scholar]
- 4.Chu C-M, Liaw Y-F. Hepatitis B surface antigen seroclearance during chronic HBV infection. Antivir. Ther. 2010;15:133–143. doi: 10.3851/IMP1497. [DOI] [PubMed] [Google Scholar]
- 5.Zhang W, Zhang D, Dou X, et al. Consensus on pegylated interferon alpha in treatment of chronic hepatitis B. J. Clin. Transl. Hepatol. 2018;6:1. doi: 10.14218/JCTH.2017.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agarwal K, et al. EASL 2017: clinical practice guidelines on the management of hepatitis B virus infection. J. Hepatol. 2017;67:370–398. doi: 10.1016/j.jhep.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 7.Fung J, Lai C-L, Seto W-K, et al. Nucleoside/nucleotide analogues in the treatment of chronic hepatitis B. J. Antimicrob. Chemother. 2011;66:2715–2725. doi: 10.1093/jac/dkr388. [DOI] [PubMed] [Google Scholar]
- 8.Papatheodoridis G, Vlachogiannakos I, Cholongitas E, et al. Discontinuation of oral antivirals in chronic hepatitis B: a systematic review. Hepatology. 2016;63:1481–1492. doi: 10.1002/hep.28438. [DOI] [PubMed] [Google Scholar]
- 9.Chen MT, et al. A function of the hepatitis B virus precore protein is to regulate the immune response to the core antigen. Proc. Natl. Acad. Sci. USA. 2004;101:14913–14918. doi: 10.1073/pnas.0406282101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prange R. Host factors involved in hepatitis B virus maturation, assembly, and egress. Med. Microbiol. Immunol. 2012;201:449–461. doi: 10.1007/s00430-012-0267-9. [DOI] [PubMed] [Google Scholar]
- 11.Ciupe SM, Ribeiro RM, Perelson AS. Antibody responses during hepatitis B viral infection. PLoS Comput. Biol. 2014;10:e1003730. doi: 10.1371/journal.pcbi.1003730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wieland SF, Chisari FV. Stealth and cunning: hepatitis B and hepatitis C viruses. J. Virol. 2005;79:9369–9380. doi: 10.1128/JVI.79.15.9369-9380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertoletti A, Ferrari C. Innate and adaptive immune responses in chronic hepatitis B virus infections: towards restoration of immune control of viral infection. Gut. 2012;61:1754–1764. doi: 10.1136/gutjnl-2011-301073. [DOI] [PubMed] [Google Scholar]
- 14.Neumann AU, Phillips S, Levine I, et al. Novel mechanism of antibodies to hepatitis B virus in blocking viral particle release from cells. Hepatology. 2010;52:875–885. doi: 10.1002/hep.23778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaillant A. Rep 2139: antiviral mechanisms and applications in achieving functional control of HBV and HDV infection. ACS Infect. Dis. 2018;5:675–687. doi: 10.1021/acsinfecdis.8b00156. [DOI] [PubMed] [Google Scholar]
- 16.Setten RL, Rossi JJ, Han S-P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019;18:421–446. doi: 10.1038/s41573-019-0017-4. [DOI] [PubMed] [Google Scholar]
- 17.Wooddell CI, et al. Hepatocyte-targeted RNAi therapeutics for the treatment of chronic hepatitis B virus infection. Mol. Ther. 2013;21:973–985. doi: 10.1038/mt.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia Y, Liang TJ. Development of direct-acting antiviral and host-targeting agents for treatment of hepatitis B virus infection. Gastroenterology. 2019;156:311–324. doi: 10.1053/j.gastro.2018.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trubetskoy VS, et al. Phosphorylation-specific status of RNAi triggers in pharmacokinetic and biodistribution analyses. Nucleic Acids Res. 2017;45:1469–1478. doi: 10.1093/nar/gkw828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wooddell CI, et al. RNAi-based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci. Transl. Med. 2017;9:eaan0241. doi: 10.1126/scitranslmed.aan0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuen M-F, et al. RNA interference therapy with ARC-520 results in prolonged hepatitis B surface antigen response in patients with chronic hepatitis B infection. Hepatology. 2020;72:19–31. doi: 10.1002/hep.31008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van den Berg F, et al. Advances with RNAi-based therapy for hepatitis B virus infection. Viruses. 2020;12:851. doi: 10.3390/v12080851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang L, Kottilil S, Wilson E. Strategies to eliminate HBV infection: an update. Future Virol. 2020;15:35–51. doi: 10.2217/fvl-2019-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.French J, Locarnini S, Zoulim F. Direct-acting antivirals and viral rna targeting for hepatitis B cure. Curr. Opin. HIV AIDS. 2020;15:165–172. doi: 10.1097/COH.0000000000000622. [DOI] [PubMed] [Google Scholar]
- 25.Nowak M, Bonhoeffer S, Hill A, et al. Viral dynamics in hepatitis B virus infection. Proc. Natl. Acad. Sci. USA. 1996;93:4398–4402. doi: 10.1073/pnas.93.9.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciupe S, Ribeiro R, Nelson P, et al. The role of cells refractory to productive infection in acute hepatitis B viral dynamics. Proc. Natl. Acad. Sci. USA. 2007;104:5050–5055. doi: 10.1073/pnas.0603626104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goyal A, Ribeiro RM, Perelson AS. The role of infected cell proliferation in the clearance of acute HBV infection in humans. Viruses. 2017;9:350. doi: 10.3390/v9110350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciupe SM. Modeling the dynamics of hepatitis B infection, immunity, and drug therapy. Immunol. Rev. 2018;285:38–54. doi: 10.1111/imr.12686. [DOI] [PubMed] [Google Scholar]
- 29.Ciupe SM, Catllá AJ, Forde J, Schaeffer DG. Dynamics of hepatitis B virus infection: what causes viral clearance? Math. Popul. Stud. 2011;18:87–105. doi: 10.1080/08898480.2011.564563. [DOI] [Google Scholar]
- 30.Ji, Y. et al. A mathematical model for anti-HBV infection treatment with lamivudine and curative effect prediction. In Control and Automation, 2007. ICCA 2007. IEEE International Conference on, 2485–2488 (IEEE, 2007).
- 31.Gourley S, Kuang Y, Nagy J. Dynamics of a delay differential model of hepatitis B virus. J. Biol. Dyn. 2008;2:140–53. doi: 10.1080/17513750701769873. [DOI] [PubMed] [Google Scholar]
- 32.Eikenberry S, Hews S, Nagy J, et al. The dynamics of a delay model of HBV infection with logistic hepatocyte growth. Math. Biosci. Eng. 2009;6:283–99. doi: 10.3934/mbe.2009.6.283. [DOI] [PubMed] [Google Scholar]
- 33.Dahari H, Shudo E, Ribeiro RM, et al. Modeling complex decay profiles of hepatitis B virus during antiviral therapy. Hepatology. 2009;49:32–38. doi: 10.1002/hep.22586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewin SR, Ribeiro RM, Walters T, et al. Analysis of hepatitis B viral load decline under potent therapy: complex decay profiles observed. Hepatology. 2001;34:1012–1020. doi: 10.1053/jhep.2001.28509. [DOI] [PubMed] [Google Scholar]
- 35.Carracedo Rodriguez A, Chung M, Ciupe SM. Understanding the complex patterns observed during hepatitis B virus therapy. Viruses. 2017;9:117. doi: 10.3390/v9050117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goyal A, Murray JM. Modelling the impact of cell-to-cell transmission in hepatitis B virus. PLoS ONE. 2016;11:e0161978. doi: 10.1371/journal.pone.0161978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray JM, Goyal A. In silico single cell dynamics of hepatitis B virus infection and clearance. J. Theor. Biol. 2015;366:91–102. doi: 10.1016/j.jtbi.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 38.Goyal A, Chauhan R. The dynamics of integration, viral suppression and cell-cell transmission in the development of occult hepatitis B virus infection. J. Theor. Biol. 2018;455:269–280. doi: 10.1016/j.jtbi.2018.06.020. [DOI] [PubMed] [Google Scholar]
- 39.Ciupe S, Ribeiro R, Nelson P, et al. Modeling the mechanisms of acute hepatitis B virus infection. J. Theor. Biol. 2007;247:23–35. doi: 10.1016/j.jtbi.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long C, Qi H, Huang S-H. Mathematical modeling of cytotoxic lymphocyte-mediated immune response to hepatitis B virus infection. J. Biomed. Biotechnol. 2008;2008:1–9. doi: 10.1155/2008/743690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim H, Kwon H, Jang T, et al. Mathematical modeling of triphasic viral dynamics in patients with HBeAg-positive chronic hepatitis B showing response to 24-week clevudine therapy. PLoS ONE. 2012;7:e50377. doi: 10.1371/journal.pone.0050377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yousfi N, Hattaf K, Tridane A. Modeling the adaptive immune response in HBV infection. J. Math. Biol. 2011;63:933–957. doi: 10.1007/s00285-010-0397-x. [DOI] [PubMed] [Google Scholar]
- 43.Hews S, Eikenberry S, Nagy JD, et al. Rich dynamics of a hepatitis B viral infection model with logistic hepatocyte growth. J. Math. Biol. 2010;60:573–590. doi: 10.1007/s00285-009-0278-3. [DOI] [PubMed] [Google Scholar]
- 44.Kadelka S, Ciupe SM. Mathematical investigation of HBeAg seroclearance. Math. Biosci. Eng. 2019;16:7616–7658. doi: 10.3934/mbe.2019382. [DOI] [PubMed] [Google Scholar]
- 45.National Clinical Guideline Centre, UK. Hepatitis B (chronic): diagnosis and management of chronic hepatitis B in children, young people and adults (2013). [PubMed]
- 46.Uchida T, et al. Persistent loss of hepatitis B virus markers in serum without cellular immunity by combination of peginterferon and entecavir therapy in humanized mice. Antimicrob. Agents Chemother. 2017;61:e00725-17. doi: 10.1128/AAC.00725-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Canini, L. et al. Understanding hepatitis B virus dynamics and the antiviral effect of interferon-alpha treatment in humanized chimeric mice. bioRxiv (2020). [DOI] [PMC free article] [PubMed]
- 48.Gabrielsson J, Weiner D. Pharmacokinetic and Pharmacodynamic Data Analysis: Concepts and Applications. Boca Raton: CRC Press; 2001. [Google Scholar]
- 49.Felmlee, M. A., Morris, M. E. & Mager, D. E. Mechanism-based pharmacodynamic modeling. In Computational Toxicology, 583–600 (Springer, 2012). [DOI] [PMC free article] [PubMed]
- 50.Volz T, et al. Impaired intrahepatic hepatitis B virus productivity contributes to low viremia in most HBeAg-negative patients. Gastroenterology. 2007;133:843–852. doi: 10.1053/j.gastro.2007.06.057. [DOI] [PubMed] [Google Scholar]
- 51.Rodríguez-Iñigo E, Mariscal L, Bartolomé J, et al. Distribution of hepatitis B virus in the liver of chronic hepatitis C patients with occult hepatitis B virus infection. J. Med. Virol. 2003;70:571–580. doi: 10.1002/jmv.10432. [DOI] [PubMed] [Google Scholar]
- 52.Guidotti LG, et al. Viral clearance without destruction of infected cells during acute HBV infection. Science. 1999;284:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]
- 53.Sherlock S, Dooley J, et al. Diseases of the Liver and Biliary System. Hoboken: Wiley; 2002. [Google Scholar]
- 54.Wursthorn K, et al. Peginterferon alpha-2b plus adefovir induce strong cccDNA decline and hbsag reduction in patients with chronic hepatitis B. Hepatology. 2006;44:675–684. doi: 10.1002/hep.21282. [DOI] [PubMed] [Google Scholar]
- 55.Xu C, et al. Interferons accelerate decay of replication-competent nucleocapsids of hepatitis B virus. J. Virol. 2010;84:9332–9340. doi: 10.1128/JVI.00918-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schluep T, et al. Safety, tolerability, and pharmacokinetics of ARC-520 injection, an rna interference-based therapeutic for the treatment of chronic hepatitis B virus infection, in healthy volunteers. Clin. Pharmacol. Drug Dev. 2017;6:350–362. doi: 10.1002/cpdd.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ishida Y, et al. Acute hepatitis B virus infection in humanized chimeric mice has multiphasic viral kinetics. Hepatology. 2018;68:473–484. doi: 10.1002/hep.29891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dandri M, et al. Virion half-life in chronic hepatitis B infection is strongly correlated with levels of viremia. Hepatology. 2008;48:1079–1086. doi: 10.1002/hep.22469. [DOI] [PubMed] [Google Scholar]
- 59.Loomba R, et al. Discovery of half-life of circulating hepatitis B surface antigen in patients with chronic hepatitis B infection using heavy water labeling. Clin. Infect. Dis. 2019;69(3):542–545. doi: 10.1093/cid/ciy1100. [DOI] [PubMed] [Google Scholar]
- 60.Shekhtman L, et al. Modelling hepatitis D virus RNA and HBsAg dynamics during nucleic acid polymer monotherapy suggest rapid turnover of HBsAg. Sci. Rep. 2020;10:1–7. doi: 10.1038/s41598-020-64122-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ciupe SM, Hews S. Mathematical models of e-antigen mediated immune tolerance and activation following prenatal HBV infection. PLoS ONE. 2012;7:e39591. doi: 10.1371/journal.pone.0039591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whalley SA, et al. Kinetics of acute hepatitis B virus infection in humans. J. Exp. Med. 2001;193:847–854. doi: 10.1084/jem.193.7.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nowak MA, et al. Viral dynamics in hepatitis B virus infection. Proc. Natl. Acad. Sci. USA. 1996;93:4398–4402. doi: 10.1073/pnas.93.9.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rong L, et al. Analysis of hepatitis C virus decline during treatment with the protease inhibitor danoprevir using a multiscale model. PLoS Comput. Biol. 2013;9:e1002959. doi: 10.1371/journal.pcbi.1002959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Churkin A, Lewkiewicz S, Reinharz V, Dahari H, Barash D. Efficient methods for parameter estimation of ordinary and partial differential equation models of viral hepatitis kinetics. Mathematics. 2020;8:1483. doi: 10.3390/math8091483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Allweiss L, Dandri M. The role of cccDNA in HBV maintenance. Viruses. 2017;9:156. doi: 10.3390/v9060156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lodish H, et al. Molecular Cell Biology. New York: Macmillan; 2008. [Google Scholar]
- 68.Duncan AW, Dorrell C, Grompe M. Stem cells and liver regeneration. Gastroenterology. 2009;137:466–481. doi: 10.1053/j.gastro.2009.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dusheiko G, Wang B. Hepatitis B surface antigen loss: too little, too late and the challenge for the future. Gastroenterology. 2019;156:548–551. doi: 10.1053/j.gastro.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 70.Lu M, Ma Z, Zhang E, Gao S, Xiong Y. Toward a functional cure for hepatitis B: the rationale and challenges for therapeutic targeting of the B cell immune response. Front. Immunol. 2019;10:2308. doi: 10.3389/fimmu.2019.02308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gish RG, et al. Synthetic RNAi triggers and their use in chronic hepatitis B therapies with curative intent. Antivir. Res. 2015;121:97–108. doi: 10.1016/j.antiviral.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 72.Vaillant A. Nucleic acid polymers: broad spectrum antiviral activity, antiviral mechanisms and optimization for the treatment of hepatitis B and hepatitis D infection. Antivir. Res. 2016;133:32–40. doi: 10.1016/j.antiviral.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 73.Al-Mahtab M, Bazinet M, Vaillant A. Safety and efficacy of nucleic acid polymers in monotherapy and combined with immunotherapy in treatment-naive Bangladeshi patients with HBeAg+ chronic hepatitis B infection. PLoS ONE. 2016;11:e0156667. doi: 10.1371/journal.pone.0156667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bazinet M, et al. Safety and efficacy of 48 weeks REP 2139 or REP 2165, tenofovir disoproxil, and pegylated interferon alfa-2a in patients with chronic HBV infection naïve to nucleos (t) ide therapy. Gastroenterology. 2020;158:2180–2194. doi: 10.1053/j.gastro.2020.02.058. [DOI] [PubMed] [Google Scholar]
- 75.Dembek C, Protzer U, Roggendorf M. Overcoming immune tolerance in chronic hepatitis B by therapeutic vaccination. Curr. Opin. Virol. 2018;30:58–67. doi: 10.1016/j.coviro.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 76.Zhang T-Y, et al. A unique B cell epitope-based particulate vaccine shows effective suppression of hepatitis B surface antigen in mice. Gut. 2020;69:343–354. doi: 10.1136/gutjnl-2018-317725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Le Bert N, et al. Comparative characterization of B cells specific for HBV nucleocapsid and envelope proteins in patients with chronic hepatitis B. J. Hepatol. 2020;72(1):34–44. doi: 10.1016/j.jhep.2019.07.015. [DOI] [PubMed] [Google Scholar]
- 78.Lada O, Benhamou Y, Poynard T, Thibault V. Coexistence of hepatitis B surface antigen (HBsAg) and anti-HBs antibodies in chronic hepatitis B virus carriers: influence of “a” determinant variants. J. Virol. 2006;80:2968–2975. doi: 10.1128/JVI.80.6.2968-2975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shapira MY, Zeira E, Adler R, Shouval D. Rapid seroprotection against hepatitis B following the first dose of a pre-S1/pre-S2/S vaccine. J. Hepatol. 2001;34:123–127. doi: 10.1016/S0168-8278(00)00082-9. [DOI] [PubMed] [Google Scholar]
- 80.Le Hoa PT, et al. Randomized controlled study investigating viral suppression and serological response following pre-S1/pre-S2/S vaccine therapy combined with lamivudine treatment in HBeAg-positive patients with chronic hepatitis B. Antimicrob. Agents Chemother. 2009;53:5134–5140. doi: 10.1128/AAC.00276-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yuen R, et al. Short term rna interference therapy in chronic hepatitis B using JNJ-3989 brings majority of patients to HBsAg 100 IU/ml threshold. J. Hepatol. 2019;70:e51–e52. doi: 10.1016/S0618-8278(19)30092-1. [DOI] [Google Scholar]
- 82.Gane, E. J. et al. Dose response with the RNA interference (RNAi) therapy JNJ-3989 combined with nucleos (t) ide analogue (NA) treatment in expanded cohorts of patients (PTS) with chronic hepatitis B (CHB). In Poster Abstract, AASLD The Liver Meeting, Boston, USA, November 9–13 2019. Hepatoogy (2019).