Abstract
The acute sickness response (ASR) is a stereotyped set of symptoms including fatigue, pain, and disturbed mood, which are present in most acute infections. The immunological mechanisms of the ASR are conserved, with variations in severity determined partly by the pathogen, but also by polymorphisms in host genes. The ASR was characterised in three different serologically-confirmed acute infections in Caucasians (n = 484) across four symptom domains or endophenotypes (termed ‘Fatigue’, ‘Musculoskeletal pain’, ‘Mood disturbance’, and ‘Acute sickness’). Correlations were sought with functional single nucleotide polymorphisms in the NLRP3 inflammasone pathway and severity of the endophenotypes. Individuals with severe Fatigue, Musculoskeletal pain, or Mood endophenotypes were more likely to have prior episodes of significant fatigue (11.4 vs. 3.8%, p = 0.07), pain (14.3 vs. 1.2%, p = 0.001), or Mood disturbance (13 vs 1%, p=0.001), suggesting trait characteristics. The high functioning allele of the rs35829419 SNP in NLRP3 was more common in those with severe Fatigue (OR = 13.3, 95% CI: 1.7–104), particularly in a dominant inheritance pattern (OR = 13.4, 95% CI: 1.8–586.3). In a multivariable analysis assuming dominant inheritance, both rs35829419 and the rs4848306 SNP in Interleukin(IL)-1β, were independently associated with severe Fatigue (OR = 29.6, 95% CI: 2.6–330.9 and OR = 13, 95% CI: 2.7–61.8, respectively). The severity of fatigue in acute infection is influenced by genetic polymorphisms in NLRP3 and IL-1β.
Keywords: Acute sickness response, Fatigue, Severity, NLRP3, Inflammasome, Ross river virus, Epstein-Barr virus, Q fever
1. Introduction
Individual infectious diseases are associated with a wide variety of sickness manifestations ranging from brief and minimally symptomatic illness, through to severe, life-threatening conditions. This is well exemplified by infection with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which ranges in manifestations from asymptomatic infection through to severe acute respiratory distress syndrome (ARDS) (Su et al., 2020). This range in the severity of infectious disease manifestations is shaped by pathogen characteristics, and by the host immune response, which in turn is influenced by the genetic make-up of the host, as well as acquired co-morbidities, and environmental factors (Khera et al., 2018).
The innate immune responses of the host are largely conserved regardless of the infecting pathogen. Such responses are initiated via pathogen-associated molecular patterns (PAMPs) or tissue damage-associated molecular patterns (DAMPs), which are recognised by widely expressed pattern-recognition receptors (PRRs), such as the Toll-like receptor (TLR) family. Activation of PRRs induces type-1 Interferon (IFN) responses to rapidly control replication and prevent the dissemination of intracellular pathogens (Murphy et al., 2017). PRRs also trigger pyrogenic responses via the induction of pro-inflammatory cytokines required for the recruitment and activation of additional immune cells (Murphy et al., 2017). One such innate pathway is mediated by a nucleotide-binding oligomerization domain (NOD)-like receptor pyrin domain-containing 3 (NLRP3) inflammasome which activates caspase 1 (CASP1) through caspase recruitment domains (CARD) (Menu and Vince, 2011). CASP1 enhances the biological activity of several pro-inflammatory cytokines, particularly Interleukin-1β (IL-1β) and Interleukin-18 (IL-18), which promote both innate and adaptive responses (Menu and Vince, 2011). NLRP3 responses are triggered by the recognition of viral RNA, bacterial products or sub-particles, as well as by tissue injury factors such as release of extracellular ATP, or the efflux of K+ (Murphy et al., 2017). Interestingly, single nucleotide polymorphisms (SNPs) in different components, or regulators, of this pathway have been associated with heightened innate immune responses in different infective conditions, including dengue (Cansanção et al., 2016), hepatitis C (Estfanous et al., 2019), and tuberculosis (Abate et al., 2019), but also auto-immune conditions as rheumatoid arthritis (Jahid et al., 2018, Addobbati et al., 2018). In this study functional SNPs in the NLRP3 pathway were examined in relation to the severity and duration of the acute sickness response (ASR) to several different infectious diseases.
The clinical manifestations generated by these innate responses are also highly conserved. Fever, fatigue, musculoskeletal pain, disturbed mood, cognitive impairment, loss of appetite and increased sleep requirements are all typical manifestations during acute infections, and are collectively defined as the ASR (Hart, 1988, Dantzer and Kelley, 2007). Previous studies in the cohort reported here have identified associations between overall severity of the ASR, and functional single nucleotide polymorphsims (SNPs) in the pro- and anti-inflammatory cytokines, IFN-γ, tumour necrosis factor (TNF)-α, IL-6, and IL-10 (Piraino et al., 2012). In particular, the high-producing allele rs2430561 of pro-inflammatory cytokine IFN-ɣ, and the low-producing allele rs1800872 SNP of the anti-inflammatory cytokine IL-10 were associated with greater severity and prolonged duration of illness among n = 296 individuals individuals affected by Epstein-Barr virus (EBV; which causes infectious mononucleosis), Ross River virus (RRV; a mosquito borne infection causing polyarthritis), and Coxiella burnetii, the causative agent of Q fever (QF) (Vollmer-Conna et al., 2008). As C burnetii is a Gram negative, lipolpolysaccaride (LPS)-containing intracellular bacterium polymorphisms in the LPS receptors, TLR-2 and TLR-4 were also previously explored in n = 85 individuals affected by QF, although no association with illness severity or course was identified (Everett et al., 2007).
Although consistently present in the ASR, observations in this cohort revealed that individual manifestations often differed in intensity and duration among individuals infected with the same pathogen (Piraino et al., 2012). Accordingly, to better understand the genetic determinants of the ASR, these symptom manifestations have been grouped here into symptom domains, or endophenotypes, which encompass the set of self-reported symptoms of fatigue (e.g. feeling weak or tired, waking up tired), pain (e.g. myalgias, arthralgias), mood disturbance (e.g. irritability, or low mood), or acute sickness (e.g. fevers, anorexia) (Piraino et al., 2012). As originally described in psychiatry (Gottesman et al., 2003), the endophenotype concept stipulates that the characteristic should be reliably measurable, state independent (i.e., the characteristic is an underlying trait rather than simply being a feature of the current disease state), and heritable. In the psychiatric context, even if the endophenotype is genetically determined such as suicide risk (Chistiakov et al., 2012), it is clear that environmental factors (such as life stressors) modulate the expression. Accordingly, the present study aimed to explore the heritable nature of ASR endophenotypes and their association with nine SNPs in the NLRP3 inflammasome pathway among individuals acutely infected by EBV, RRV, or QF.
2. Methods
2.1. Subjects
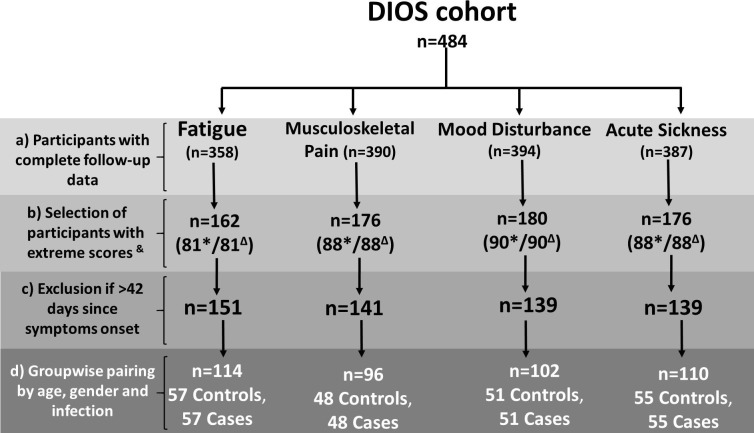
Longitudinally collected clinical datasets and DNA from participants of the Dubbo Infection Outcomes Study (DIOS) were used (Cvejic et al., 2019, Hickie et al., 2006). The DIOS cohort enroled n = 484 adults of Caucasian descent, who were acutely affected by a febrile illness which was provisionally diagnosed in primary care as being EBV, RRV or QF infection on the basis of screening by pathogen-specific IgM enzyme-linked immunosorbent assays (Fig. 1 ). These subjects were prospectively followed-up at 2–3 weeks, 4–6 weeks, and then three-monthly until 12 months post-infection. The EBV, RRV, or QF infections were confirmed by acute and convalesecent serological testing for pathogen-specific IgG antibodies. At baseline, a structured medical history interview was recorded to capture pre-morbid (i.e. pre-infection) physical and mental health status. At each follow-up visit, self-report questionnaires were administered to record the presence and severity of physical and psychological symptoms related to the ASR. For the purposes of the analysis reported here, demographic data was used in addition to that captured by the 34-item Somatic and Psychological Health Report (SPHERE) (Hickie et al., 2001) and the 19-items Physical Symptoms Checklist (PSC) (Hickie et al., 2006).
Fig. 1.
Study design and sequential steps for identification of phenotypic sub-groups to examine duration of illness and genetic associations. a) To establish the relation between baseline severity and duration of symptoms, only participants from the DIOS cohort without missing data from the longitudinal assessments were selected to describe the individual endophenotypes. b) Only individuals with extreme phenotypes (lower and upper quartiles of symptom scores) were included in the survival analysis. c) To ensure reliable reporting of acute-phase symptoms, the interval between symptom onset and enrolment was restricted to six weeks or less. d) Selection of participants for genetic associations testing utilised a case-control approach with severity groups matched by age, gender, and aetiological agent. & represents scores in the lower and upper quartile. *represents subjects with mild (lower quartile) symptoms. Δ represents subjects with severe (upper quartile) symptoms.
2.2. Ethics statement
This genetics study was approved by the institutional Human Research Ethics Committee at UNSW (HC190102). In DIOS, all subjects provided written informed consent for prospective clinical assessment and blood sampling for DNA storage and analysis under (HC 04257). These studies have adhered to the principles set out in the Declaration of Helsinki.
2.3. Endophenotype indices
Data from the 53 individual items of the SPHERE and PSC obtained at the baseline were used to derive indices of the endophenotypes of the ASR using a principal component analysis (PCA) with orthogonal rotation (Piraino et al., 2012). SPHERE (34 items) (Hickie et al., 2006) and PSC (19 items) (Hickie et al., 2001) are self-administered questionnaires recording the prevalence ‘’in the past few weeks” of a wide range of physical and psychological symptoms of acute infection (see Table S1 for details of the question items), recorded as “none of the time or some of the time” (scored as 0), “a good part of the time” (scored as 1), and “most of the time” (scored as 2). Six items duplicated between the questionnaires were removed. Only the first four principal components were retained because they satisfied the eigenvalue-one criteria, and each provided an independent contribution >3% to the overall variance (O'Rourke et al., 2013). These four PCs were labelled on the basis of the dominant theme captured by the items as (Fig. 1a): ‘Fatigue’, ‘Musculoskeletal pain’, ‘Mood disturbance’, and ‘Acute sickness’ (Hickie et al., 2006). The loading scores for each questionnaire item in these indices were calculated from the baseline dataset and then applied to the data from each of the follow-up assessments (see Supplementary material, Table 1).
2.4. Persistence of the endophenotypes
The SOMA subscale of the SPHERE questionnaire was used in the orginal study to designate the timepoint when the overall acute illness had resolved (with a score < 3 being indicative of illness resolution) (Piraino et al., 2012). Using data from the 12 month follow-up timepoint from subjects (n = 327) whose acute illness had resolved within 4 weeks of initial enrolment, a ‘threshold’ for background in the PC score for each endophenotype was designated as the 99th percentile of the recovered group. This threshold value was then used to identify the timepoint at which each subject first recorded scores below the threshold for each endophenotype. The midpoint in days between the last above-threshold and the first below-threshold timepoint was used to determine the duration of illness within each endophenotype from symptom onset to resolution (i.e. below background).
2.5. Evidence of endophenotypes as a trait characteristic
As trait characteristics are intrinsic and lifelong modulated by environmental factors (Dick and Kendler, 2012, Baud et al., 2017), the medical history interview was scrutinised for prior episodes of: severe acute infections (as a surrogate for a propensity to severe Acute sickness), severe and prolonged fatigue or pain (as a surrogate for a propensity to severe Fatigue or Musculoskletal pain), and for premorbid mood disorder (as a surrogate for a propensity to severe Mood disturbances).
2.6. Selection of comparison groups
Individuals with mild or severe manifestations were selected from the lower and upper quartiles of the severity scores across all endophenotypes (see Fig. 1b) to examine the relationship between acute phase severity and duration of illness. A reverse genetics approach, which starts from a trait without genetic information and then works backward to find a genetic variant, was then applied (Griffiths et al., 2005). To do this, of those subjects reporting mild or severe symptoms for each endophenotype, only who had a symptom onset <42 days prior to enrolment were retained in the analysis to ensure temporal proximity with the acute illness phase for reliable reporting of symptoms (Fig. 1c). Finally, for comparisons of allelic frequencies, cases with severe endophenotypes were matched groupwise by age (within 10 years), gender, and the pathogen causing the acute illness to controls (Fig. 1d).
2.7. Selection of functional SNPs
The SNPs within the NLRP3, CASP1, CARD8, IL-1β, and IL-18 genes were selected to test associations with the severity of ASR endophenotypes. Firstly, SNPs listed for each gene in non-Finnish European populations were download from the Genome Aggregation Database of the Broad Institute (Karczewski et al., 2020). In the case of NLRP3, 1193 SNPs were found, whereas 789, 1054, 363 and 22 SNPs were available for CASP1, CARD8, IL-1β, and IL-18. Of this list, SNPs with: i) a minor allele frequency (MAF) > 3% were then selected; and ii) published evidence of altered transcript or protein production in vitro; or iii) a reported association with an infectious or immunological disease (Database of Single Nucleotide Polymorphisms (dbSNP), https://www.ncbi.nlm.nih.gov/snp/) were selected. The resulting nine functional SNPs were: rs360719 (IL-18), rs1946519 (IL-18), rs5744292 (IL-18), rs4848306 (IL-1β), rs16944 (IL-1β), rs501192 (CASP1), rs35829419 (NLRP3), rs10754558 (NLRP3), and rs2043211 (CARD8).
2.8. Genotyping
Genomic DNA was extracted from stored peripheral blood mononuclear cells using the Wizard DNA kit (Promega, Madison, USA). DNA was quantified using NanoDropR ND-1000 (BioLab, Mulgrave, Australia), and the quality verified by agarose gel electrophoresis. Genotyping of selected SNPs was performed using the Sequenom MassARRAY platform (Gabriel et al., 2009) at the Australian Genome Research Facility.
2.9. Statistical analysis
Descriptive statistics, comparisons of groups (i.e. mild and severe cases among each endophenotype), generation of endophenotype indices by PCA, and survival analysis, were performed using Stata/IC version 14 (StataCorp, College Station, Texas, USA). Fisher exact and Chi-square were used for comparison of qualitative variables. Mann–Whitney–Wilcoxon was used for comparison of all quantitative variables due to their abnormal distribution. Survival distributions of the mild and severe illness groups were analysed by the logrank test comparing persistence of symptoms among each endophenotype according to baseline severity. These comparisons were adjusted by age, gender, and infecting pathogen by Cox proportional hazards. Right censoring for the complete dataset was defined at 90 days after the initiation of symptoms. Chi-square tests for independence and odds ratio (OR) calculations with their respective 95% confidence intervals (95% CI) were used to infer whether the alleles and genotypes were associated with severe endophenotypes in the case-control groups using PLINK (http://zzz.bwh.harvard.edu/plink/) (Purcell et al., 2007). A p-value of 0.05 was considered the threshold for statistical significance. p-values of allelic associations were subject to Bonferroni correction. All SNPs were in Hardy-Weinberg equilibrium in the study population.
3. Results
3.1. Demographic characteristics according to endophenotype
Participants in the cohort (n = 484) were predominantly female (n = 248/484; 51%), and their median age was 32 years (interquartile range 18–42, range 16–69). EBV was the most frequent infection (n = 144/484; 30%), followed by RRV (n = 98/484; 20%), and QF (n = 84/484; 17%). The pathogen provisionally implicated in the acute infectious disease was not serologically confirmed in 33% of participants (n = 158/484). Endophenotype scores were not available at some longitudinal timepoints in a minority of individuals, hence the Fatigue endophenotype subgroup was composed of n = 358 individuals, the Musculoskeletal pain endophenotype by n = 390, the Mood disturbance endophenotype by n = 394, and the Acute sickness endophenotype by n = 387 participants (Fig. 1). The baseline characteristics of endophenotype subgroups were all similar to the entire cohort (Table 1. ).
Table 1.
Demographics of the endophenotype subgroups according to severity.
| Total (n = 484) | Fatigue (n = 358) | Musculoskeletal pain (n = 390) | Mood disturbance (n = 394) | Acute sickness (n = 387) | |
|---|---|---|---|---|---|
| Age*(IQR) | 32 (19–44) | 33 (19–45) | 33 (19–45) | 33 (19–45) | 33 (19–45) |
| Female; n (%) | 248 (51) | 197 (55) | 218 (60) | 217 (55) | 210 (54) |
| Infection type; n (%) | |||||
| EBV | 144 (30) | 106 (30) | 117 (30) | 117 (30) | 116 (30) |
| RRV | 98 (20) | 76 (21) | 82 (21) | 84 (21) | 84 (22) |
| QF | 84 (17) | 61 (17) | 66 (17) | 67 (17) | 65 (17) |
| Undetermined | 158 (33) | 115 (32) | 125 (32) | 126 (32) | 122 (31) |
| Days since symptoms onset - median (IQR) | 30 (22–41) | 33 (24–42) | 33 (25–42) | 33 (25–42) | 32 (23–41) |
Median values are reported.
3.2. Associations between baseline severity and duration of illness endophenotypes
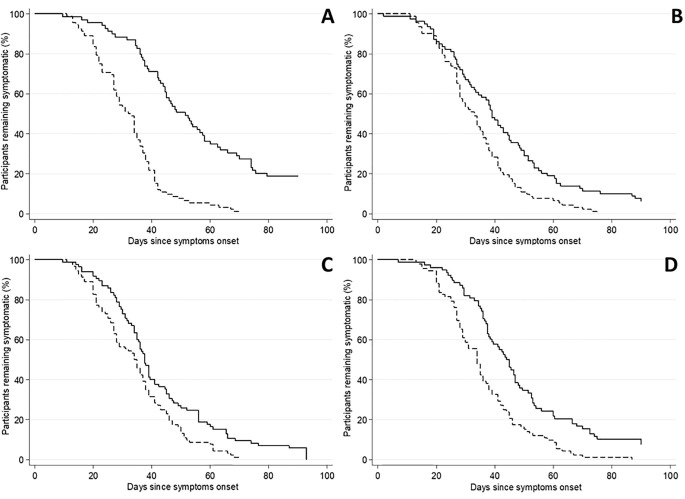
The kinetics of resolution of illness in relation to baseline illness severity (Fig. 1a) of the Fatigue (n = 358), Musculoskeletal pain (n = 390), Mood disturbance (n = 394), and Acute sickness (n = 387) endophenotypes are shown in Fig. 2 (the numbers vary with the availability of complete follow-up data). As expected, individuals with severe acute phase illness had a more prolonged illness course for each endophenotype compared to those with mild acute phase illness. The comparison of the survival distributions by illness severity (upper quartile versus lower quartile) demonstrated a difference in duration for each endophenotype, including Fatigue (median duration 51.5 vs. 32.5 days for severe and mild respectively, p < 0.001;), Musculo-skeletal pain (median duration 39 vs. 33 days, p = 0.003), Mood disorder (median duration 37.5 vs. 34.5 days, p = 0.003), and Acute sickness (median duration 44.5 vs. 34 days, p < 0.001).
Fig. 2.
Kinetics of resolution of individual endophenotypes over 90 days following onset of illness in relation to acute phase illness severity. (A) Fatigue (n = 358). (B) Musculoskeletal pain (n = 390). (C) Mood disturbance (n = 394). (D) Acute sickness (n = 387). The dashed lines represent individuals with mild acute phase endophenotype scores (lower quartile) and the solid lines represent individuals with severe acute phase endophenotype scores (upper quartile).
3.3. Associations between severity of endophenotypes and prior episodes
Subjects with more severe acute phase illness, reflected by high scores for the individual endophenotypes (Fig. 1b), were also more likely to have pre-morbid illness reports suggesting these same features, reflecting prior vulnerability or trait characteristics (Table 2 ). In particular, those with severe Musculoskeletal pain were more likely to have experienced prolonged pain lasting one month or more (p = 0.001), whereas those with severe Fatigue did not have a greater likelihood of experiencing prior prolonged fatigue lasting one month or more (p = 0.07). The data for Mood disturbance was available only for n = 197 subjects, but individuals with severe Mood disturbance endophenotype were more likely to have experienced prior mood disorder (p < 0.001). This was not observed in the Acute sickness endophenotype where past severe infectious conditions were similarly reported among mild and severe illness (p = 0.29).
Table 2.
Demographics of the endophenotype subgroups according to severity.
| Fatigue (F) |
Musculoskeletal pain (MP) |
Mood disturbance (MD) |
Acute sickness (AS) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MildΔ (n = 81) | SevereΔ (n = 81) | p value | MildΔ (n = 88) | SevereΔ (n = 88) | p value | MildΔ (n = 90) | SevereΔ (n = 90) | p value | MildΔ (n = 88) | SevereΔ (n = 88) | p value | |
| Age* (range) | 37 (19–47) | 34 (18–43) | 0.23 | 22 (18–39) | 37 (27–45) | 0.001 | 34 (20–49) | 30 (17–41) | 0.01 | 36 (24–47) | 30 (18–45) | 0.03 |
| Female€; n (%) | 32 (40) | 47 (58) | 0.18 | 49 (50) | 48 (49) | 0.88 | 46 (51) | 53 (59) | 0.29 | 49 (56) | 46 (52) | 0.65 |
| Infection type€; n (%) | 0.025 | 0.001 | 0.158 | 0.001 | ||||||||
| EBV | 26 (32) | 27 (33) | 46 (52) | 10 (11) | 31 (34) | 26 (29) | – | 18 (20) | 32 (36) | – | ||
| RRV | 26 (32) | 13 (16) | 8 (9) | 33 (38) | 22 (25) | 16 (18) | – | 42 (48) | 5 (6) | – | ||
| QF | 12 (15) | 9 (11) | 13 (15) | 17 (19) | 16 (18) | 13 (14) | – | 9 (10) | 23 (26) | – | ||
| Undeterminate | 17 (21) | 32 (40) | 21 (24) | 28 (32) | 21 (23) | 35 (39) | – | 19 (22) | 28 (32) | – | ||
| Prior severe episode&; n (%) | 3 (4) | 9 (11) | 0.07 | 1 (1) | 12 (14) | 0.001 | 1 (1) | 12 (13) | 0.001 | 6 (7) | 10 (11) | 0.29 |
*Median values and IQR are reported and compared by Mann-Whitney tests. €Frequencies are compared by Chi (Khera et al., 2018) test. &Defined in Fatigue and Musculoskeletal pain endophenotype subgroups as previous episodes of severe and prolonged fatigue or pain (>30 days); or previous severe acute infection for the Acute sickness endophenotype. For the Mood disturbance endophenotype, a history of premorbid mood disorder was available only for n = 71 individuals (n = 30 and n = 41 from the severe and mild illness groups respectively). Δ Described as mild or severe illness severity groups in Fig. 1b.
3.4. Associations between SNPs and the severe endophenotypes
As these differences between severe and mild endophenotypes were also potentially attributable to other features of the groups including age, gender and causative pathogen, the severe and mild comparison sub-groups for each endophenotype were identified from those with six weeks from illness onset at enrolment (Fig. 1c), and then matched by age, sex, and aetiological agent, for case-control testing of genetic associations (Fig. 1d). Demographic characteristics for these sub-groups were comparable across all endophenotypes, with no differences between cases and controls in key features as age, aetiological agent as well as the number of days between the initiation of symptoms and the baseline assessment (Table S2). By contrast, cases had a more prolonged period out of their usual role during the acute phase of the illness in the Fatigue (14 vs. 7 days in controls, p = 0.0009), Mood disturbance (14 vs. 8 days, p = 0.01), and Acute sickness endophenotypes (15 vs. 10 days, p = 0.0008). Those with severe symptoms in the Acute sickness endophenotype also had more days in bed due to illness (7 vs. 2 days, p = 0.0007).
The severity of the Fatigue endophenotype was associated with the minor allele of NLRP3 rs35829419 (Table 3 ). The A allele of rs35829419 was more frequent in all endophenotypes, but was significant only for the severe Fatigue endophenotype (OR = 13.3, 95% CI: 1.7–104, p = 0.01). The risk was comparable, assuming a dominant inheritance pattern of the A allele (OR = 13.4, 95% CI: 1.8–586.3, p = 0.002). A recessive inheritance pattern was not tested due to the lack of AA individuals in the matched control group.
Table 3.
Results of associations between ASR phenotypes and NLRP3 inflammasome pathway SNPs in the DIOS cohort.
| SNP | A1/A2 | MAF | Fatigue |
Musculoskeletal Pain |
Mood disturbances |
Acute sickness |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F_Ca* | OR | 95% CI | F_Ca* | OR | 95% CI | F_Ca* | OR | 95% CI | F_Ca* | OR | 95% CI | |||
| rs35829419 | A/C | 0.051 | 0.105 | 13.3 | 1.7–104£ | 0.083 | 1.36 | 0.5–4.1 | 0.068 | 1.8 | 0.5–6.4 | 0.063 | 3.67 | 0.7–18.1 |
| rs10754558 | G/C | 0.415 | 0.368 | 1.16 | 0.7–1.9 | 0.447 | 1.7 | 0.9–3.1 | 0.323 | 0.68 | 0.4–1.2 | 0.418 | 1.16 | 0.7–2 |
| rs4848306 | A/G | 0.450 | 0.368 | 0.52 | 0.3–0.8 | 0.385 | 0.71 | 0.4–1.3 | 0.372 | 0.66 | 0.4–1.1 | 0.381 | 0.59 | 0.34–1 |
| rs16944 | A/G | 0.343 | 0.421 | 1.71 | 0.9–2.9 | 0.364 | 0.95 | 0.5–1.7 | 0.362 | 1.36 | 0.8–2.5 | 0.381 | 1.17 | 0.7–2 |
| rs360719 | G/A | 0.274 | 0.368 | 1.31 | 0.8–2.3 | 0.291 | 0.95 | 0.5–1.7 | 0.303 | 1.41 | 0.8–2.6 | 0.336 | 1.12 | 0.6–1.9 |
| rs5744292 | C/T | 0.241 | 0.236 | 0.83 | 0.5–1.5 | 0.291 | 1.38 | 0.7–2.6 | 0.205 | 1 | 0.5–1.9 | 0.272 | 0.91 | 0.6–1.9 |
| rs1946519 | A/C | 0.406 | 0.438 | 1.15 | 0.7–1.9 | 0.354 | 0.62 | 0.3–1.1 | 0.382 | 0.88 | 0.5–1.5 | 0.445 | 1.07 | 0.5–1.6 |
| rs501192 | T/C | 0.170 | 0.193 | 1.57 | 0.8–3.2 | 0.166 | 1.17 | 0.5–2.6 | 0.187 | 0.88 | 0.4–1.7 | 0.2 | 0.94 | 0.5–1.8 |
| rs2043211 | T/A | 0.331 | 0.271 | 0.616 | 0.4–1.1 | 0.375 | 0.95 | 0.5–1.7 | 0.372 | 1.18 | 0.7–2.1 | 0.336 | 0.73 | 0.4–1.3 |
A1 : minor alleles, A2 : major alleles; MAF: minor allele frequency. *F_Ca : allele frequency among cases. £ p = 0.015 is adjusted to the number of SNPs by a Bonferroni correction.
In a multivariable analysis controlling for age, sex, and the infection type, both the A allele of NLRP3 rs35829419 and the G allele of IL-1β rs4848306 were simultaneously associated with the severe Fatigue endophenotype (OR = 29.6, 95% CI 2.6–330.9, p = 0.006 and OR = 13, 95% CI 2.7–61.8, p = 0.002 respectively) assuming a dominant inheritance mode. This effect was not found for the Acute sickness endophenotype.
Given the strong association between severity and duration of the endophenotypes, the potential role of these genetic variants in determining illness duration was also investigated. No genetic association with sex, age, or infection type was identified for duration of illness for any of these endophenotypes. Similarly, no associations with two SNPs associated with acute illness severity - NLRP3 rs35829419 and IL-1β rs4848306, were found with Fatigue duration (Cox proportional hazards model; p > 0.2).
4. Discussion
The severity of infectious diseases has been traditionally linked with pathogen virulence factors, as well as co-morbid conditions in the host, including immunosuppressed states and non-communicable conditions such as diabetes, obesity, or cardiovascular disease (Osterholm et al., 2015). Despite the major advances in genetics including high throughput sequencing technologies, the contribution of genetic variation to infectious diseases outcomes has been explored for relatively few pathogens. Using the notion that endophenotypes in the ASR are similarly triggered by different infections, we have shown that these symptom domains appear to represent underlying vulnerabilities repetitively affecting susceptible individuals. Further in a case-control design with a reverse genetics approach, two functional SNPs in the NLRP3 inflammasome pathway were found to be associated with the severity of fatigue, independent of the pathogen causing the infection or demographic characteristics. Thus, our study supports the concept of endophenotypes applied to the ASR, the notion of genetically-determined contributions to these symptom domains, and implicates the NLRP3 inflammasome in the genesis of the ASR.
The causative pathogens in this study (EBV, RRV and QF) generally cause self-limiting acute infections which are seldom associated with hospitalisation or mortality (Johannsen et al., 2015, Mylonas et al., 2002, Harley et al., 2001, Marrie et al., 2015). Hence, the study sought to better understand genetic determinants of the varied severity of the typical acute illness manifesations. The generation of endophenotype scores using PCA provided a quantitative characterisation of otherwise qualitative symptoms such as fatigue. These endophenotypes were constructed from individual symptom items (Table S1), but also captured their frequency and individual contribution within each broad domain, generating a solid phenotype conceptualisation. These scores additionally highlighted differences in independent measures of the functional impact of the different symptom domains including the number of days out of the usual role, and days in bed in the acute phase of the illness (Cvejic et al., 2019, Bennett et al., 1998, Cvejic et al., 2014). It should be noted that the original concept of endophenotypes from psychiatry research stipulated that the endophenotype should be ‘state’ independent (that is consistently present whether the particular illness is present or in remission) (Gottesman et al., 2003). In the context of acute infection, this may be taken to imply that individuals who experience severe fatigue associated with one of the infections in this study may similarly experience the same pattern with another acute infection or inflammatory insult. We corroborated this assumption with evidence that individuals experiencing severe pain or mood disturbance endophenotypes reported a higher frequency of similar episodes in the past than those with mild manifestations. It is also possible that the endophenotype may cross boundaries (i.e. be trans-diagnostic), so that the propensity to mood disorder associated with acute infection may predict the advent of mood disorder consequent upon more traditional psychosocial stressors. Further investigations are required to test these notions as well as to explore their usefulness in clinical settings where the identification of individuals with at-risk endophenotypes could preceed the development of severe manifestations.
In the analysis presented here, the participants in the lower and upper severity quartiles were selected for the matched case-control comparison. This approach was adopted firstly as the identification of risk or protective factors in case-control studies is more efficient when confounding factors are removed or controlled (Rose and Laan, 2009). This may be considered the main limitation of genome-wide association studies (GWAS) which often survey an enormous number of SNPs in subjects with broad and relatively heterogenous phenotypes, requiring very large sample sizes to manage confounding factors (Tam et al., 2019). Secondly, the phenotype-to-genotype, or reverse genetics approach, and the candidate pathway chosen here for investigation, rely upon an evidence base of biological plausibility that the pathway is implicated in the pathophysiology of the illness (Casanova et al., 2002). The NLRP3 and pro-inflammatory cytokine pathway chosen for investigation here and the process of selection of functional SNPs meets this expectation.
Fatigue is reported as a dominant element of the ASR (Piraino et al., 2012, Cvejic et al., 2019, Hickie et al., 2006), but is also prevalent in other conditions, including in association with cancer (Bower, 2014), and autoimmune diseases (Morris et al., 2015). In this context multiple biological mechanisms are proposed to underpin this symptom domain including pro- and anti-inflammatory cytokines, hypothalamic–pituitary–adrenal (HPA) axis changes, and central serotoninergic and other neurotransmitter systems (Landmark-Høyvik et al., 2010). From the immunology perspective, genetic variations in the pro-inflammatory cytokines, IL-1β, IL-6, TNF-α, IFN-γ and the anti-inflammmatory cytokine, IL-10, have been all been associated with the severity of fatigue (Bower, 2014, Landmark-Høyvik et al., 2010, Yamato and Kataoka, 2015). Several immunological therapies are also recognised to have fatigue and mood disturbances as a prominent adverse effects, with the most notable being IFN-α treatments which were previously used commonly for chronic hepatitis C infection (Scheiner et al., 2016). In the current SARS-CoV-2 pandemic, fatigue and myalgias are among the most frequent symptoms reported (Ghayda et al., 2020). Interestingly, a high frequency of post-infective fatigue has been reported after SARS-CoV-2 infection, independent of acute phase illness severity (Townsend et al., 2020).
The findings reported here that the NLRP3 rs35829419 is associated with severity of the fatigue endophenotype, and has a synergistic interaction with IL-1β rs4848306 extend the previous genetic association findings. The A allele of NLRP3 rs35829419 is a missense mutation associated with an overactive NLRP3 inflammasome generating increased CASP1 activity and excessive IL-1β and IL-18 levels (Verma et al., 2012). Although sometimes considered as a SNP of uncertain clinical significance, an increase in plasma IL-1β levels were found in association with the A allele of NLRP3 rs35829419 and the T allele of CARD8 rs2043211 (Sahdo et al., 2013). Further, NLRP3 rs35829419 has been associated with multiple auto-immune conditions (Zhang et al., 2015). The IL-1β rs4848306 is located in an NF-κβ binding site in the promote region, and the A allele has been associated with transcriptional factor binding and reduced IL-1β mRNA expression (Chen et al., 2006, Bank et al., 2014). In the data presented here an increased risk of severe fatigue was putatively associated with increased levels of IL-1β induced by an overactive NLRP3 inflammasome due to the A allele in the rs35829419, and this effect could be magnified by the presence of a G allele of the IL-1β rs4848306, which enhances IL-1β mRNA expression.
Even though these associations were significant, the study has limitations. Firstly, there were wide confidence intervals for each relevant association involving rs35829419. Wide confidence intervals are multi-factorial, but are generally associated with inadequate sample sizes and co-linear interactions among variables (Irala et al., 1997). We consider sample size is the main determinant in the data reported here. Secondly, although the endophenotypes may be influenced by psychosocial factors, these were not considered in the case-control matching – largely due to limitations in sample size. Accordingly, the findings require corroboration and replication in prospective studies with larger sample sizes, and careful characterisation of endophenotypes and outcomes of interest.
5. Conclusions
This study has identified the fatigue endophenotype as a likely genetic determinant of the severity of the ASR, particularly implicating the NLRP3 inflammasome and the pro-inflammatory cytokine, IL-1β. These findings warrant further longitudinal studies of the phenotype to understand whether it is a trans-diagnostic trait, and to explore other genetic contributions to the phenotype.
Funding
This project was funded in part by National Health and Medical Research Council of Australia (NHMRC) Project Grants (157092 and 157062), Mason Foundation, and a Cooperative Research Agreement with the US Centers for Disease Control and Prevention (U50/CCU019851-01). AL is supported by a NHMRC Practitioner Fellowship (1137587). CR is supported by NHMRC Investigator Grant (1173666). BMV is supported by a Scientia PhD Scholarship from UNSW Sydney.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The support of the general practitioners, the public health unit, and the diagnostic pathology services in the Dubbo region, NSW, Australia are gratefully acknowledged, as well as the enduring cooperation of the subjects in this research.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbi.2021.01.005.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Abate E., Blomgran R., Verma D., et al. Polymorphisms in CARD8 and NLRP3 are associated with extrapulmonary TB and poor clinical outcome in active TB in Ethiopia. Sci. Rep. 2019;9:3126. doi: 10.1038/s41598-019-40121-8. 2019/03/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addobbati C., da Cruz H.L.A., Adelino J.E., et al. Polymorphisms and expression of inflammasome genes are associated with the development and severity of rheumatoid arthritis in Brazilian patients. Inflamm. Res. 2018;67:255–264. doi: 10.1007/s00011-017-1119-2. 2017/12/13. [DOI] [PubMed] [Google Scholar]
- Bank S., Andersen P.S., Burisch J., et al. Associations between functional polymorphisms in the NFκB signaling pathway and response to anti-TNF treatment in Danish patients with inflammatory bowel disease. Pharmacogenomics J. 2014;14:526–534. doi: 10.1038/tpj.2014.19. 2014/04/30. [DOI] [PubMed] [Google Scholar]
- Baud, A., Mulligan, M.K., Casale, F.P., et al. Genetic variation in the social environment contributes to health and disease. PLoS Genet. 13: e1006498. 2017/01/26. DOI: 10.1371/journal.pgen.1006498. [DOI] [PMC free article] [PubMed]
- Bennett B.K., Hickie I.B., Vollmer-Conna U.S., et al. The relationship between fatigue, psychological and immunological variables in acute infectious illness. Aust. New Zealand J. Psychiatry. 1998;32:180–186. doi: 10.3109/00048679809062727. 1998/05/20. [DOI] [PubMed] [Google Scholar]
- Bower J.E. Cancer-related fatigue–mechanisms, risk factors, and treatments. Nat. Rev. Clin. Oncol. 2014;11:597–609. doi: 10.1038/nrclinonc.2014.127. 2014/08/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cansanção I.F., do Carmo A.P., Leite R.D., et al. Association of genetic polymorphisms of IL1β -511 C>T, IL1RN VNTR 86 bp, IL6 -174 G>C, IL10 -819 C>T and TNFα -308 G>A, involved in symptomatic patients with dengue in Brazil. Inflamm. Res. 2016;65:925–932. doi: 10.1007/s00011-016-0975-5. 2016/07/21. [DOI] [PubMed] [Google Scholar]
- Casanova J.L., Schurr E., Abel L., et al. Forward genetics of infectious diseases: immunological impact. Trends Immunol. 2002;23:469–472. doi: 10.1016/s1471-4906(02)02289-5. 2002/09/26. [DOI] [PubMed] [Google Scholar]
- Chen H., Wilkins L.M., Aziz N., et al. Single nucleotide polymorphisms in the human interleukin-1B gene affect transcription according to haplotype context. Hum. Mol. Genet. 2006;15:519–529. doi: 10.1093/hmg/ddi469. [DOI] [PubMed] [Google Scholar]
- Chistiakov D.A., Kekelidze Z.I., Chekhonin V.P. Endophenotypes as a measure of suicidality. J. Appl. Genetics. 2012;53:389–413. doi: 10.1007/s13353-012-0113-1. [DOI] [PubMed] [Google Scholar]
- Cvejic E., Lemon J., Hickie I.B., et al. Neurocognitive disturbances associated with acute infectious mononucleosis, Ross River fever and Q fever: a preliminary investigation of inflammatory and genetic correlates. Brain Behav. Immun. 2014;36:207–214. doi: 10.1016/j.bbi.2013.11.002. 2013/11/12. [DOI] [PubMed] [Google Scholar]
- Cvejic E., Li H., Hickie I.B., et al. Contribution of individual psychological and psychosocial factors to symptom severity and time-to-recovery after naturally-occurring acute infective illness: The Dubbo Infection Outcomes Study (DIOS) Brain Behav. Immun. 2019;82:76–83. doi: 10.1016/j.bbi.2019.07.034. [DOI] [PubMed] [Google Scholar]
- Dantzer R., Kelley K.W. Twenty years of research on cytokine-induced sickness behavior. Brain Behav. Immun. 2007;21:153–160. doi: 10.1016/j.bbi.2006.09.006. 2006/11/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick, D.M., Kendler, K.S., 2012. The impact of gene-environment interaction on alcohol use disorders. Alcohol. Res. 34: 318–324. 2012/11/09. [DOI] [PMC free article] [PubMed]
- Estfanous S.Z.K., Ali S.A., Seif S.M., et al. Inflammasome genes' polymorphisms in egyptian chronic hepatitis C patients: influence on vulnerability to infection and response to treatment. Mediators Inflamm.. 2019;2019:3273645. doi: 10.1155/2019/3273645. 2019/02/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett B., Cameron B., Li H., et al. Polymorphisms in Toll-like receptors-2 and -4 are not associated with disease manifestations in acute Q fever. Genes Immun. 2007;8:699–702. doi: 10.1038/sj.gene.6364428. [DOI] [PubMed] [Google Scholar]
- Gabriel S, Ziaugra L and Tabbaa D. SNP genotyping using the sequenom massARRAY iPLEX platform. Curr. Protocols in Hum. Genetics 2009; 60: 2.12.11-12.12.18. DOI: 10.1002/0471142905.hg0212s60. [DOI] [PubMed]
- Ghayda, R.A., Lee, J., Lee, J.Y., et al., 2020. Correlations of clinical and laboratory characteristics of COVID-19: A systematic review and meta-analysis. Int. J. Environ. Res. Public Health 17 2020/07/17. DOI: 10.3390/ijerph17145026. [DOI] [PMC free article] [PubMed]
- Gottesman Irving I., Ph D., Hon F.R.C., Psych, Todd D., Gould M.D. The endophenotype concept in psychiatry: etymology and strategic intentions. Am. J. Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Griffiths J.F., Griffiths A.J.F., Wessler S.R., et al. In: An Introduction to Genetic Analysis. Griffiths J.F., Griffiths A.J.F., Wessler S.R., editors. Macmillan; New York: 2005. Applications of recombinant DNA technology. [Google Scholar]
- Harley D., Sleigh A., Ritchie S. Ross River virus transmission, infection, and disease: a cross-disciplinary review. Clin. Microbiol. Rev. 2001;14:909–932. doi: 10.1128/cmr.14.4.909-932.2001. 2001/10/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart B.L. Biological basis of the behavior of sick animals. Neurosci. Biobehav. Rev. 1988;12:123–137. doi: 10.1016/S0149-7634(88)80004-6. 1988/01/01. [DOI] [PubMed] [Google Scholar]
- Hickie I.B., Davenport T.A., Hadzi-Pavlovic D., et al. Development of a simple screening tool for common mental disorders in general practice. The Med. J. Australia. 2001;175:S10–S17. doi: 10.5694/j.1326-5377.2001.tb143784.x. 2001/09/15. [DOI] [PubMed] [Google Scholar]
- Hickie I., Davenport T., Wakefield D., et al. Post-infective and chronic fatigue syndromes precipitated by viral and non-viral pathogens: prospective cohort study. BMJ (Clinical research ed) 2006;333:575. doi: 10.1136/bmj.38933.585764.AE. 2006/09/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irala Jd., Fernandez-Crehuet Navajas R., Serrano del Castillo A. Abnormally wide confidence intervals in logistic regression: interpretation of statistical program results. Revista Panamericana de Salud Pública. 1997;2:268–271. [PubMed] [Google Scholar]
- Jahid M., Rehan Ul H., Chawla D., et al. Association of polymorphic variants in IL1B gene with secretion of IL-1β protein and inflammatory markers in north Indian rheumatoid arthritis patients. Gene. 2018;641:63–67. doi: 10.1016/j.gene.2017.10.051. 2017/10/22. [DOI] [PubMed] [Google Scholar]
- Johannsen E.C., Kaye K.M. In: Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases. Bennett J.E., Dolin R., Blaser M.J., editors. Elsevier Health Sciences; Philadelphia: 2015. Epstein-barr virus (infectious mononucleosis, epstein-barr virus-associated malignant diseases, and other diseases) pp. 1754–1771. [Google Scholar]
- Karczewski, K.J., Francioli, L.C., Tiao, G., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. bioRxiv 2020: 531210. DOI: 10.1101/531210. [DOI] [PMC free article] [PubMed]
- Khera A.V., Chaffin M., Aragam K.G., et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. 2018/08/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmark-Høyvik, H., Reinertsen, K.V., Loge, J.H., et al. The genetics and epigenetics of fatigue. Pm r 2010; 2: 456–465. 2010/07/27. DOI: 10.1016/j.pmrj.2010.04.003. [DOI] [PubMed]
- Marrie T.J., Raoult D. In: Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases. Bennett J.E., Dolin R., Blaser M.J., editors. Elsevier Health Sciences; Philadelphia: 2015. Coxiella burnetii (Q Fever) pp. 2208–2216. [Google Scholar]
- Menu P., Vince J.E. The NLRP3 inflammasome in health and disease: the good, the bad and the ugly. Clin. Exp. Immunol. 2011;166:1–15. doi: 10.1111/j.1365-2249.2011.04440.x. 2011/07/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, G., Berk, M., Walder, K., et al., 2015. Central pathways causing fatigue in neuro-inflammatory and autoimmune illnesses. BMC Med. 13: 28. 2015/04/10. DOI: 10.1186/s12916-014-0259-2. [DOI] [PMC free article] [PubMed]
- Murphy K., Weaver C. In: Janeway's Immunobiology. Murphy K., Weaver C., editors. Garland Science; New York: 2017. The induced responses of innate immunity; pp. 77–134. [Google Scholar]
- Mylonas A.D., Brown A.M., Carthew T.L., et al. Natural history of Ross River virus-induced epidemic polyarthritis. Med. J. Australia. 2002;177:356–360. doi: 10.5694/j.1326-5377.2002.tb04837.x. 2002/10/03. [DOI] [PubMed] [Google Scholar]
- O'Rourke N., Hatcher L. In: A step-by-step approach to using SAS for factor analysis and structural equation modeling. O'Rourke N., Hatcher L., editors. SAS Institute Inc; North Carolina: 2013. Principal component analysis; pp. 1–42. [Google Scholar]
- Osterholm M.T., Hedberg C.W. In: Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases. Bennett J.E., Dolin R., Blaser M.J., editors. Elsevier Health Sciences; Philadelphia: 2015. Epidemiologic principles; pp. 146–157. [Google Scholar]
- Piraino B., Vollmer-Conna U., Lloyd A.R. Genetic associations of fatigue and other symptom domains of the acute sickness response to infection. Brain Behav. Immun. 2012;26:552–558. doi: 10.1016/j.bbi.2011.12.009. 2012/01/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. 2007/08/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, S., Laan, M.J., 2009. Why match? Investigating matched case-control study designs with causal effect estimation. Int. J. Biostat. 5: Article 1. 2009/01/01. DOI: 10.2202/1557-4679.1127. [DOI] [PMC free article] [PubMed]
- Sahdo B., Fransén K., Asfaw Idosa B., et al. Cytokine profile in a cohort of healthy blood donors carrying polymorphisms in genes encoding the NLRP3 inflammasome. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0075457. e75457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiner, B., Schwabl, P., Steiner, S., et al., 2016. Interferon-free regimens improve health-related quality of life and fatigue in HIV/HCV-coinfected patients with advanced liver disease: A retrospective study. Medicine 95: e4061. 2016/07/12. DOI: 10.1097/md.0000000000004061. [DOI] [PMC free article] [PubMed]
- Su L., Ma X., Yu H., et al. The different clinical characteristics of corona virus disease cases between children and their families in China - the character of children with COVID-19. Emerging Microbes Infect. 2020;9:707–713. doi: 10.1080/22221751.2020.1744483. 2020/03/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam V., Patel N., Turcotte M., et al. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019;20:467–484. doi: 10.1038/s41576-019-0127-1. [DOI] [PubMed] [Google Scholar]
- Townsend, L., Dyer, A.H., Jones, K., et al., 2020. Persistent fatigue following SARS-CoV-2 infection is common and independent of severity of initial infection. medRxiv 2020.2007.2029.20164293. DOI: 10.1101/2020.07.29.20164293. [DOI] [PMC free article] [PubMed]
- Verma, D., Särndahl, E., Andersson, H., et al., 2012. The Q705K polymorphism in NLRP3 is a gain-of-function alteration leading to excessive interleukin-1β and IL-18 production. PloS One 7: e34977. 2012/04/25. DOI: 10.1371/journal.pone.0034977. [DOI] [PMC free article] [PubMed]
- Vollmer-Conna U., Piraino B.F., Cameron B., et al. Cytokine polymorphisms have a synergistic effect on severity of the acute sickness response to infection. Clin. Infect. Dis.: An Official Publ. Infect. Dis. Soc. Am. 2008;47:1418–1425. doi: 10.1086/592967. 2008/10/22. [DOI] [PubMed] [Google Scholar]
- Yamato, M., Kataoka, Y., 2015. Fatigue sensation following peripheral viral infection is triggered by neuroinflammation: who will answer these questions? Neural Regeneration Res. 10: 203–204. Perspective. DOI: 10.4103/1673-5374.152369. [DOI] [PMC free article] [PubMed]
- Zhang, Q., Fan, H.W., Zhang, J.Z., et al., 2015. NLRP3 rs35829419 polymorphism is associated with increased susceptibility to multiple diseases in humans. Genet. Mol. Res. 14: 13968-13980. 2015/11/05. DOI: 10.4238/2015.October.29.17. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.