Abstract
The effect of oral microbial composition on periodontal health and on systemic health has been, and is being established. The oral microbiome, in turn, can be altered by local and systemic diseases and conditions. Gastroesophageal reflux disease (GERD), has been associated with increased acidity in the oral cavity resulting in dental erosion, and controversially a reduced risk of periodontal disease. We hypothesized that presence of GERD was linked to a modified microbial profile in untreated GERD patients and that the use of proton pump inhibitor (PPI) drugs: potent disruptors of gut microbiome, in GERD patients might result in a salivary microbiome that is further distinct. Untreated GERD patients showed multiple differences in salivary microbiome as compared to healthy controls. Taxa found at lower levels related to the presence of GERD not treated by PPI included: Prevotella melaninogenica, Prevotella pallens, Leptotrichia, and Solobacterium moorei and thirteen others. In contrast, GERD patients chronically using PPI showed minimal differences in salivary taxa compared to healthy controls not using PPI.
Subject terms: Microbiology, Diseases, Medical research, Pathogenesis
Introduction
There are hundreds of species of bacteria that inhabit the mucosal and hard surfaces of the oral cavity, and upper respiratory and gastrointestinal tracts, that can be sampled by examining the saliva1,2. Oral bacteria of the mouth have been shown not only to play a role in oral disease such as caries, dental erosion, periodontal disease, and mucosal lesions, but also may contribute to systemic disease such as cardiovascular disease, diabetes mellitus, even pancreatic cancer1,3–5. A large number of bacteria genera and species contribute to a complex oral microbial ecosystem, thus it is difficult to differentiate a healthy microbiome or what used to be described as “normal oral flora” versus a pathogenic oral microbiome. A first step is identifying specific bacteria taxa that are associated with a disease or condition.
Gastroesophageal reflux disease (GERD) is a condition that is believed to occur due to dysfunction of the lower esophageal sphincter, which is responsible for preventing back flow of stomach contents into the esophagus6. This regurgitation of gastric juices into the esophagus, oropharyngeal and oral cavity lowers their pH7. In the esophagus, this can contribute to chronic inflammation, histological changes in the mucosal cells lining the distal esophagus, and increased risk for esophageal cancer8,9. Changes in distal esophageal microbiome have been characterized, including lower levels of Prevotella, Helicobacter, and Moraxella genera with GERD10–12. In the oral cavity, the presence of GERD is associated with acidic saliva (pH 4.9) versus the near neutral pH of 6.5 in healthy subjects13. Inflammation of the oral mucosa is often seen, as well as reduction in oral salivary flow14,15. This acidic environment favors loss of tooth structure through enamel erosion.
PPIs are drugs that work by inhibiting release of Hydrogen ions from mucosal cells of the GI tract, particularly those in the stomach7. While PPIs are indicated for usage against H. Pylori infection, erosive esophagitis, gastric ulcers, and for stress ulcer prevention in critically ill patients, they are available over the counter and are used long term for a number of other aliments9,16,17. Epidemiological studies reveal that usage of PPIs is associated with increased risk of Clostridium difficile infection in the gut, small intestinal bacterial overgrowth, pneumonia, and nutritional deficiencies18–20. These illnesses are thought to be linked to reduced levels of gastric acid release and increased pH in stomach and small intestine as well as other sites. PPI usage can have effects on upper GI sites such as esophagus and oral cavity even in healthy subjects. After 4 week of PPI administration, healthy subjects were seen to show changes in salivary microbiota, including reductions in Neisseria, Veillonella, possibly Haemophilus, and an increase in Streptococcus21.
The aim of this study was to determine the role of GERD in the makeup of the oral microbiome. This study examined the salivary microbiome in dental patients who reported with GERD. In addition, the salivary microbiome of patients with GERD chronically treated with PPI was also analyzed. In patients with GERD, usage of PPI has been shown to be associated with restoration of normal pH and possibly saliva flow7. In these patients one might expect the oral microbiome to be more similar to healthy subjects.
Results
Selected subjects
In the original study samples were collected from 208 subjects who came to the dental clinics for checkups. Of these 36 self-reported having GERD comprised of 20 patients with GERD who used PPI (GERD + PPI) group, and 16 who had GERD but did not use medication (GERD + No PPI) group. Negative controls did not have GERD and did not use PPI. This negative control group had a lower percentage of tobacco usage, fewer females and trended toward a higher level of periodontal disease than the group with GERD that did not use PPI (Supplemental Table 1).
To facilitate comparison between the GERD subjects that did not use PPI and the negative control a subset of 92 negative control subjects was created so to minimize differences in clinical covariates. There was a total of 128 subjects in total in the final groups used for saliva microbiome analysis in this study (Table 1). Sixty patients were edentulous.
Table 1.
Demographic and clinical indices after matching.
| Feature | Control | GERD no PPI | GERD + PPI | Control + PPI | ||||
|---|---|---|---|---|---|---|---|---|
| Sex1 | Female | 54 | 14 | 13 | 4 | |||
| Male | 38 | 2 | p < 0.028 | 7 | p < 0.629 | 6 | p < 0.346 | |
| Age2 | Mean | 57.8 ± 1.66 | 54.4 ± 4.0 | p < 0.367 | 63.5 ± 2.2 | p < 0.120 | 67.4 ± 3.5 | p < 0.047 |
| Tobacco User1 | User | 31 | 7 | 7 | 2 | |||
| Nonuser | 61 | 9 | p < 0.571 | 13 | p < 0.419 | 8 | p < 1 | |
| Dentate1 | No | 21 | 4 | 6 | 8 | |||
| Yes | 71 | 12 | p < 1.0 | 14 | p < 1 | 2 | p < 0.0009 | |
| Periodontal Disease1 | No | 80 | 14 | 13 | 8 | |||
| Yes | 12 | 2 | p < 1.0 | 7 | p < 0.127 | 2 | p < 0.500 |
1Fisher Exact Test versus control.
2Student t Test versus control.
3Periodontal disease is Class III and IV by ADP/AAP.
Comparison of microbial communities among GERD patients with and without PPI usage
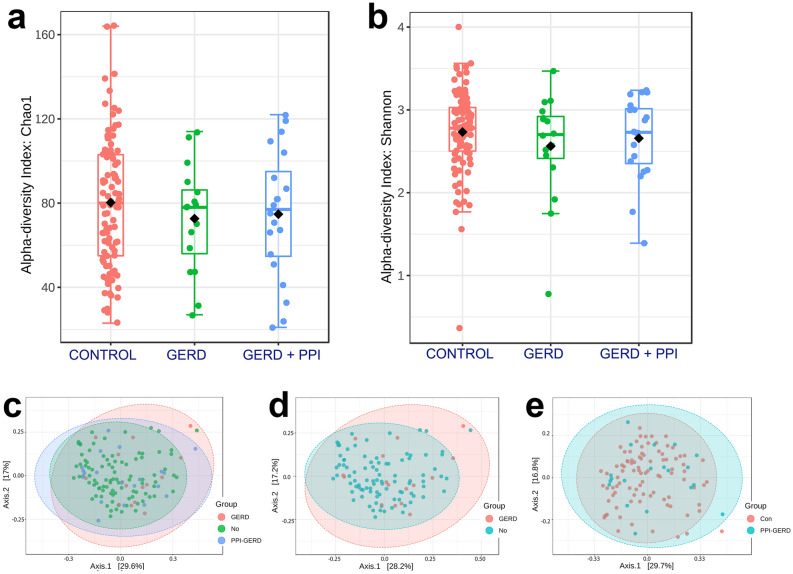
Differences in the microbial population in the large negative control group, the group with GERD that did not use PPI, and the group with GERD that did use PPI, were examined. In total 4,964,720 reads from 128 saliva samples were obtained. These were subjected to DADA2 for variant identification and followed by BLAST analysis so to be assigned to taxa of the Human Oral Microbiome Database (HOMD)27. Alpha diversity analysis revealed no significant differences among the groups (Fig. 1a,b). For beta diversity study, overall bacterial community structure was compared between the negative controls and the two other groups using Bray Curtis analysis (Fig. 1c–e). Principal coordinate analysis (PCoA) with Bray Curtis to calculate distances between the negative controls and GERD patients not using PPI showed the groups fell into clusters with some subtle differences. Due to the unbalanced nature of the datasets quantitation by PEMANOVA or ANOSIM of the differences was not possible29. A similar comparison of the GERD + PPI group to negative controls revealed the scatter plots were superimposable, and thus similar (Fig. 1e).
Figure 1.
Box plot of (a) Chao1 index of control, GERD patients not using PPI, and GERD patients using PPI. Comparison of the control to GERD patients not using PPI, p < 0.302, and the control comparison to GERD using PPI, p < 0.513; and (b) Shannon index in controls versus GERD patients not using PPI, p < 0.330 and versus GERD using PPI, p < 0.571. Principal coordinate analysis (PCoA) of saliva microbiome profiles of (c) all 3 groups; (d) negative controls versus GERD patient not using PPI; and (e) negative controls versus GERD patients using PPI reveal some differences in microbiota community structure in the GERD group not using PPI.
Examination of individual taxa in the three groups
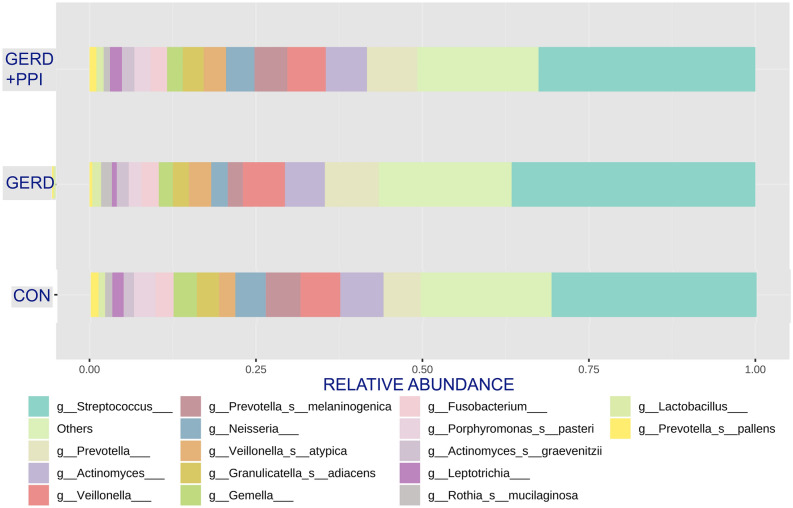
Analysis of the 16 s rDNA based HOMD database taxonomic assignments revealed in the barplot the 16 most common species and genera in GERD patients not using PPI and those found in the negative controls without GERD and not using PPI (Fig. 2). Abundant taxa that were significantly different in the two groups included Prevotella pallens, Prevotella melaninogenica and Leptotrichia (see below). Figure 2 also displays the distribution of major taxa for the GERD + PPI patients. The distribution of the major taxa looks remarkably similar to the controls without GERD and not using PPI.
Figure 2.
The bar plot reveals the 16 most common species and/or genus subgroups in saliva of the three groups studied (Control, without GERD and not using PPI, GERD, GERD patients not using PPI, and GERD + PPI, GERD patients using PPI) and allows comparison of relative taxa proportions.
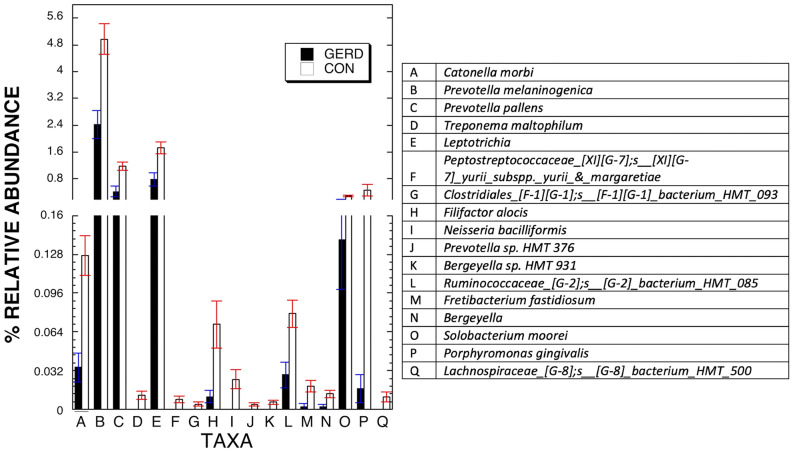
A key question is which of the apparent differences between the GERD with no PPI group, and the negative controls, were statistically significant. Two sample Welch’s t-test which is robust for imbalance of sample size between groups was employed to determine the specific difference between the two groups31. Of 176 taxa at the species and genus subset level in the saliva of the three patient groups, 17 were shown by Welch’s t-test, to be at different levels in the group of patients with GERD who did not use PPI versus the controls, FDR < 0.1 (Fig. 3). The same comparison of subjects with GERD using PPI versus the negative controls gave a different result. No taxa came close to showing a difference in levels between the two groups at FDR < 0.2.
Figure 3.
Differentially abundant species in saliva of GERD patients not using PPI versus negative controls without GERD and not using PPI. Shown are the 17 taxa that showed changes in relative abundance at FDR < 0.05 in separate analyses V3 16S rDNA. Bars represent relative abundance in the two groups.
While the GERD subjects not using PPI and negative controls were matched based on periodontal status, edentualism, and more or less for tobacco use, they still differed by gender. For that reason, multivariable analysis was used to re-examine the differences between this group and the controls after adjusting for differences in gender and tobacco usage in the two groups. Table 2 indicates that after adjustment for gender and tobacco 4 taxa were revealed to be significantly different between the two groups.
Table 2.
Summary of composition of the oral microbiome by two groups.
| Microbiome | Multivariate analysis results** | ||
|---|---|---|---|
| Outcome variables | |||
| Control | GERD | p value | |
| (N = 92) | (N = 16) | ||
| Catonella morbi | 0.129623 | 0.061025 | 0.086 |
| − 0.015568 | − 0.037048 | ||
| Prevotella melaninogenica | 4.889842 | 2.392873 | 0.031 |
| − 0.448333 | − 1.066951 | ||
| Prevotella pallens | 1.145807 | 0.308243 | 0.008 |
| − 0.121866 | − 0.290018 | ||
| Treponema maltophilum | 0.012477 | − 0.000206 | 0.126 |
| − 0.003232 | − 0.007691 | ||
| Leptotrichia | 1.625858 | 0.690662 | 0.023 |
| − 0.159576 | − 0.379762 | ||
| Peptostreptococcaceae_[XI][G-7];s__[XI][G-7]_yurii_subspp._yurii_&_margaretiae | 0.006525 | 6.97E−05 | 0.308 |
| − 0.002479 | − 0.005899 | ||
| Clostridiales_[F-1][G-1];s__[F-1][G-1]_bacterium_HMT_093 | 0.005791 | 0.001101 | 0.190 |
| − 0.001397 | − 0.003325 | ||
| Filifactor alocis | 0.089173 | 0.029482 | 0.193 |
| − 0.017922 | − 0.042651 | ||
| Neisseria bacilliformis | 0.026814 | 0.002862 | 0.230 |
| − 0.007802 | − 0.018568 | ||
| Prevotella sp. HMT 376 | 0.003765 | − 0.00108 | 0.135 |
| − 0.001265 | − 0.00301 | ||
| Bergeyella sp. HMT 931 | 0.006029 | 0.00092 | 0.306 |
| − 0.001954 | − 0.004649 | ||
| Ruminococcaceae_[G-2];s__[G-2]_bacterium_HMT_085 | 0.083247 | 0.031361 | 0.074 |
| − 0.011299 | − 0.026889 | ||
| Fretibacterium fastidiosum | 0.024701 | 0.008493 | 0.153 |
| − 0.004434 | − 0.010551 | ||
| Bergeyella | 0.0113 | − 0.000885 | 0.119 |
| − 0.003053 | − 0.007266 | ||
| Solobacterium moorei | 0.297201 | 0.135003 | 0.047 |
| − 0.0318 | − 0.075679 | ||
| Porphyromonas gingivalis | 0.572156 | 0.021349 | 0.158 |
| − 0.152375 | − 0.362623 | ||
| Lachnospiraceae_[G-8];s__[G-8]_bacterium_HMT_500 | 0.012903 | 0.001121 | 0.232 |
| − 0.003852 | − 0.009168 | ||
**The least square means and SE were estimated using general linear model (GLM) after adjusting for Gender and Tobacco User. Shown is range of values and p value.
Discussion
This observational study is the first that we know of to characterize the salivary microbiome in GERD patients using PPI and in those with GERD who do not use PPI. Within the original population in this study, subjects who had GERD and did not use PPI were more frequently tobacco users and trended toward having no, or milder, periodontal disease (Supp. Table 1). Along with GERD patient groups a matched group negative control without GERD who did not use PPI was further analyzed for salivary microbiome. Even after matching for age, periodontal status, and edentualism, which may affect salivary bacteria1,32, there were 17 species and subgenera that were at different levels in the GERD patients not using PPI compared to negative controls. Four of the 17 bacteria taxa shown to be decreased in the GERD patients who did not use PPI compared to controls, Porphyromonas gingivalis, Filifactor alocis, Fretibacterium fastidiosum, and Lachnospiraceae_[G-8];s_[G-8]_bacterium_HMT_500, have been found to be elevated in the saliva and/or periodontia of patients with chronic or aggressive periodontitis (Fig. 3)33–35. This is consistent with the recent findings that GERD patients have lower risk for periodontal disease36. Multivariable analysis adjusting for a trend toward differences in tobacco usage, and an imbalance in gender, in the GERD group versus the negative controls, revealed none of these 4 taxa showed statistically significant differences in level. This relationship between GERD and the 4 periodontal disease-related taxa may well not be direct. Further exploration of the role of these periodontal pathogens in GERD effects on the oral cavity awaits a larger study.
Adverse effects of long-term use of PPI on other organ systems has been established and cautioned against19,20,37, presumably due to pH increases. By lowering acid release in the stomach, the pH there and in the intestine can be increased, which can change microbiome at those sites whether the patient is healthy or not11,12,38–40. Less attention has been paid to how PPI usage affects the oral or esophageal microbiota in GERD patients. Earlier it has been shown that healthy subjects without GERD who used PPIs for 4 weeks experienced changes in oral bacterial taxa21. In contrast, the subset of GERD patients chronically treated with PPI showed salivary taxa profiles quite similar to that of the negative controls (Fig. 1, 2). Finally, the GERD patients in this study who did not use PPI showed 17 different taxa at reduced levels versus negative controls. Four taxa, Prevotella melaninogenica, Prevotella pallens, Solobacterium moorei, and Leptotrichia remained at lower abundance in this group versus the negative control group after multivariable analysis suggesting the lower levels of these taxa in these subjects (Table 2) is directly related to the presence of GERD without PPI treatment. Together these studies revealed that these factors, GERD or PPI separately, were linked to differences in oral microbiome. When present together, the effects seem to be minimized. It has already been shown that usage of PPI can restore normal oral pH, and even salivary flow, in GERD patients13,14. One can speculate that in the oral cavity, a body site not normally subjected to gastric acid, the usage of a PPI to inhibit acid production in the gut would help maintain a healthy microbiota. Though not studied here it is expected to have a similar effect on the esophagus with long-term treatment of GERD with PPIs linked to a normal or healthy microbiome. While the current study included 36 subjects with GERD, the groups were small after stratification by PPI usage. It was unclear what caused the differences in salivary microbiome among the groups and whether the presence of GERD has a complex role in oral microbiome makeup. GERD patients who did not use PPI could do this for different reasons7. One cannot be certain that having GERD and not using PPI causes the differences in microbiome. To get a better understanding about GERD effects on oral microbiome and, in turn, PPI effects on GERD, it will be necessary to study patients before and after administration of these drugs.
Conclusion
The differences in bacteria taxa levels in untreated GERD patients may be a result of a pH change due to GERD and may have effects on oral conditions. The long-term effects on oral health due to the differences can only be guessed. Remarkably, in GERD subjects who use PPI, the salivary microbiome was quite similar to that in negative controls without the disease not using PPI. This would suggest at least some benefit in the oral cavity with usage of those drugs in people with GERD. Clinical studies looking at the effects of GERD on the oral cavity should take into consideration salivary oral microbial differences between treated and untreated GERD patients.
Material and methods
Study design
A cross-sectional study was conducted at the University of Illinois at Chicago College of Dentistry, General Practice and Denture clinics that compared the oral microbiome using stimulated saliva22 between subjects with GERD and those without, with an effort to match subjects based on age. All subjects were questioned on physician diagnosed medical conditions on each visit as part of routine clinical procedures. Subjects who indicated that they had GERD at multiple visits including time of sample collection were considered to have GERD. Patients who included GERD in the medical history at the time of collection but no other time, and vice versa, were reinterviewed to confirm GERD diagnosis. Twelve potential subjects who used PPI but did not report as having GERD were also reinterviewed, two reported as having GERD, the remaining ten did not and were not included in the study. All subjects provided written informed consent to participate in accordance with guidelines of the Office for the Protection of Research Subjects of the University of Illinois at Chicago, with formal approval of the study protocol, 2016-0696, by the Institutional Review Board 1 of the University of Illinois at Chicago.
Study inclusion criteria were: Subjects 18 years and older; complete medical record; current medication list; and full periodontal exam. Study exclusion criteria were: presence of restored dental implants; removable partial dentures; scaling of teeth within the past 3 months; acute disease that requires urgent care; less than twenty (20) natural teeth for the dentate subjects; antibiotic use within the past month; use of antimicrobial mouthwash within the past 48 h; and food consumption within the past 1 h. Periodontal health was assessed using the ADA/AAP classification system based on clinical and radiographic findings. Subjects were classified into groups based on the severity of the disease. Classes I and II are considered healthy or have mild form of periodontal disease, Class III have moderate and Class IV have severe periodontal disease. The focus is active, not historical periodontal disease. Edentulous patients without periodontia are considered to lack active periodontal disease. For the purpose of microbiome analysis, subjects in the negative control group were selected to maximize matching with the GERD without PPI group in regard to periodontal disease status, tobacco usage, presence of teeth and age.
Sampling procedure
After rinsing with water, patients chewed on paraffin wax for 5 min and then stimulated saliva was collected. All samples were centrifuged at 6000 × g for 5 min prior to 2× washing of pellet with cold PBS and storage frozen prior to DNA extraction23.
Characterization of microbial community structure
Genomic DNA was extracted from saliva samples as described earlier23. Amplicon assay targeted the V1-V3 variable region of bacterial 16S ribosomal RNA rRNA genes as described23. The second PCR reactions with barcoding and the Illumina miSeq sequencing were performed at the University of Illinois at Chicago Sequencing Core.
Bioinformatics analysis
For taxa assignment and measurement, reverse sequences from the FASTQ files were analyzed using the software package QIIME224,25. Sequences were trimmed if the average quality was lower than 25. As a result, the read sequences were truncated at 258nt. DADA2-plugin in QIIME2 was used to sequence, denoise, and generate feature data and feature tables for the dataset26. Taxonomy assignments were done by classify-consensus-blast function with 98% match identity to the Human Oral Microbiome Database27. Of the 208 samples, there were 13,383 to 63,154 reads per sample. There were 8,349,860 reads total. Data from 3 additional subject samples were discarded due to low read numbers.
Alpha diversity analysis were performed using MicrobiomeAnalyst28. This analysis included calculation of Shannon’s diversity index of both species number and their distribution, and Chao1′s of richness. The significance of the differences between each group and the controls was derived using an unpaired Welch’s t-test. Beta diversity analysis was visulalized using Bray Curtis dissimilarity (non-phylogenetic) metric. PERMANOVA or ANOSIM test to determine differences between groups were not run due to the unbalanced datasets used29.
STAMP was used to determine differences in specific microbiota taxonomic abundance between the groups using Welch’s t-test and with determination of false discovery rate (FDR) using the Benjamini Hochberg test30. This was performed after eliminating taxa that appeared in 10% or fewer subjects at < 2 reads30.
General linear model (GLM) was used in the multivariable analyses to investigate the adjusted difference in Taxa between different groups after adjusting for significant patient’s characteristic and clinical factors. The best predictive GLM model was used to explore the significant predictors of Taxa using a backward selection with a retaining criteria of 0.2. All multivariable analyses were done in SPSS and SAS.
Supplementary Information
Acknowledgements
The Research Open Access Publishing (ROAP) fund of the University of Illinois at Chicago paid in part the open access fees for this article.
Author contributions
G.R.A., J.L.S., N.K., S.G.P. designed the research; J.L.S., N.K., G.R.A., S.G.P., N.C., A.O., and B.Y., conducted the research; G.R.A., S.G.P. and Z.C. analyzed the data; and G.R.A., N.K., A.O. wrote the manuscript. G.R.A., N.K. and J.L.S. had primary responsibility for the final content. All authors read and approved the final manuscript.
Data availability
The sequencing data from this study is deposited as PRJNA674379 in the National Center for Biotechnology Information Sequence Read Archive.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-80170-y.
References
- 1.Belstrom D. The salivary microbiota in health and disease. J. Oral Microbiol. 2020;12:1723975. doi: 10.1080/20002297.2020.1723975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen T, et al. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010 doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graves DT, Correa JD, Silva TA. The oral microbiota is modified by systemic diseases. J. Dent. Res. 2019;98:148–156. doi: 10.1177/0022034518805739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Kolltveit KM, Tronstad L, Olsen I. Systemic diseases caused by oral infection. Clin. Microbiol. Rev. 2000;13:547–558. doi: 10.1128/cmr.13.4.547-558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakano K, et al. The collagen-binding protein of Streptococcus mutans is involved in haemorrhagic stroke. Nat. Commun. 2011;2:485. doi: 10.1038/ncomms1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vakil N, van Zanten SV, Kahrilas P, Dent J, Jones R. The Montreal definition and classification of gastroesophageal reflux disease: a global evidence-based consensus. Am. J. Gastroenterol. 2006;101:1900–1920. doi: 10.1111/j.1572-0241.2006.00630.x. [DOI] [PubMed] [Google Scholar]
- 7.Azzam RS. Are the persistent symptoms to proton pump inhibitor therapy due to refractory gastroesopahgeal reflux disease or to other disorders? Arq Gastroenterol. 2018;55(Suppl 1):85–91. doi: 10.1590/s0004-2803.201800000-48. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Brady P. Gastroesophageal reflux disease: pathophysiology, diagnosis, and treatment. Gastroenterol. Nurs. 2019;42:20–28. doi: 10.1097/sga.0000000000000359. [DOI] [PubMed] [Google Scholar]
- 9.Katz PO, Gerson LB, Vela MF. Guidelines for the diagnosis and management of gastroesophageal reflux disease. Am. J. Gastroenterol. 2013;108:308–328. doi: 10.1038/ajg.2012.444. [DOI] [PubMed] [Google Scholar]
- 10.Blackett KL, et al. Oesophageal bacterial biofilm changes in gastro-oesophageal reflux disease, Barrett's and oesophageal carcinoma: association or causality? Aliment Pharmacol. Ther. 2013;37:1084–1092. doi: 10.1111/apt.12317. [DOI] [PubMed] [Google Scholar]
- 11.Liu N, et al. Characterization of bacterial biota in the distal esophagus of Japanese patients with reflux esophagitis and Barrett's esophagus. BMC Infect. Dis. 2013;13:130. doi: 10.1186/1471-2334-13-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu Y, Gao F, Chen X, Zheng S, Zhang J. Changes in the distal esophageal microbiota in Chinese patients with reflux esophagitis. J. Dig. Dis. 2019;20:18–24. doi: 10.1111/1751-2980.12692. [DOI] [PubMed] [Google Scholar]
- 13.Caruso AA, et al. Relationship between gastroesophageal reflux disease and Ph nose and salivary: proposal of a simple method outpatient in patients adults. Open Med. (Wars) 2016;11:381–386. doi: 10.1515/med-2016-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckley CA, Costa HO. Comparative study of salivary pH and volume in adults with chronic laryngopharyngitis by gastroesophageal reflux disease before and after treatment. Braz. J. Otorhinolaryngol. 2006;72:55–60. doi: 10.1016/s1808-8694(15)30035-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe M, et al. Oral soft tissue disorders are associated with gastroesophageal reflux disease: retrospective study. BMC Gastroenterol. 2017;17:92. doi: 10.1186/s12876-017-0650-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaynes M, Kumar AB. The risks of long-term use of proton pump inhibitors: a critical review. Ther. Adv. Drug Saf. 2019;10:2042098618809927. doi: 10.1177/2042098618809927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mejia A, Kraft WK. Acid peptic diseases: pharmacological approach to treatment. Expert Rev. Clin. Pharmacol. 2009;2:295–314. doi: 10.1586/ecp.09.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moayyedi P, et al. Safety of proton pump inhibitors based on a large, multi-year, randomized trial of patients receiving rivaroxaban or aspirin. Gastroenterology. 2019;157:682–691.e682. doi: 10.1053/j.gastro.2019.05.056. [DOI] [PubMed] [Google Scholar]
- 19.Sheen E, Triadafilopoulos G. Adverse effects of long-term proton pump inhibitor therapy. Dig. Dis. Sci. 2011;56:931–950. doi: 10.1007/s10620-010-1560-3. [DOI] [PubMed] [Google Scholar]
- 20.Yang YX, Lewis JD, Epstein S, Metz DC. Long-term proton pump inhibitor therapy and risk of hip fracture. JAMA. 2006;296:2947–2953. doi: 10.1001/jama.296.24.2947. [DOI] [PubMed] [Google Scholar]
- 21.Mishiro T, et al. Oral microbiome alterations of healthy volunteers with proton pump inhibitor. J. Gastroenterol. Hepatol. 2018;33:1059–1066. doi: 10.1111/jgh.14040. [DOI] [PubMed] [Google Scholar]
- 22.Navazesh M. Methods for collecting saliva. Ann. N. Y. Acad. Sci. 1993;694:72–77. doi: 10.1111/j.1749-6632.1993.tb18343.x. [DOI] [PubMed] [Google Scholar]
- 23.Gazdeck RK, et al. Diversity of the oral microbiome between dentate and edentulous individuals. Oral Dis. 2019;25:911–918. doi: 10.1111/odi.13039. [DOI] [PubMed] [Google Scholar]
- 24.Bolyen E, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall M, Beiko RG. 16S rRNA Gene Analysis with QIIME2. Methods Mol. Biol. 2018;1849:113–129. doi: 10.1007/978-1-4939-8728-3_8. [DOI] [PubMed] [Google Scholar]
- 26.Callahan BJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dewhirst FE, et al. The human oral microbiome. J. Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chong J, Liu P, Zhou G, Xia J. Using microbiome analyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020;15:799–821. doi: 10.1038/s41596-019-0264-1. [DOI] [PubMed] [Google Scholar]
- 29.Anderson MJ, Walsh DCI. PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: what null hypothesis are you testing? Ecol. Monogr. 2013;83:8. doi: 10.1890/12-2010.1. [DOI] [Google Scholar]
- 30.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruxton GD. The unequal variance t-test is an underused alternative to Student's t-test and the Mann-Whitney U test. Behav. Ecol. 2006;17:3. doi: 10.1093/beheco/ark016. [DOI] [Google Scholar]
- 32.Wu J, et al. Cigarette smoking and the oral microbiome in a large study of American adults. ISME J. 2016;10:2435–2446. doi: 10.1038/ismej.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui X, Liu J, Xiao W, Chu Y, Ouyang X. Subgingival microbiome in Chinese patients with generalized aggressive periodontitis compared to healthy controls. Arch. Oral. Biol. 2019;101:92–99. doi: 10.1016/j.archoralbio.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 34.Darby IB, Hodge PJ, Riggio MP, Kinane DF. Microbial comparison of smoker and non-smoker adult and early-onset periodontitis patients by polymerase chain reaction. J. Clin. Periodontol. 2000;27:417–424. doi: 10.1034/j.1600-051x.2000.027006417.x. [DOI] [PubMed] [Google Scholar]
- 35.Kumar PS, et al. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J. Clin. Microbiol. 2006;44:3665–3673. doi: 10.1128/jcm.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chatzopoulos GS, Cisneros A, Sanchez M, Wolff LF. Systemic medical conditions and periodontal status in older individuals. Spec. Care Dent. 2018;38:373–381. doi: 10.1111/scd.12319. [DOI] [PubMed] [Google Scholar]
- 37.Hojo M, et al. Gut microbiota composition before and after use of proton pump inhibitors. Dig. Dis. Sci. 2018;63:2940–2949. doi: 10.1007/s10620-018-5122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imhann F, et al. Proton pump inhibitors affect the gut microbiome. Gut. 2016;65:740–748. doi: 10.1136/gutjnl-2015-310376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson MA, et al. Proton pump inhibitors alter the composition of the gut microbiota. Gut. 2016;65:749–756. doi: 10.1136/gutjnl-2015-310861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seto CT, Jeraldo P, Orenstein R, Chia N, DiBaise JK. Prolonged use of a proton pump inhibitor reduces microbial diversity: implications for Clostridium difficile susceptibility. Microbiome. 2014;2:42. doi: 10.1186/2049-2618-2-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data from this study is deposited as PRJNA674379 in the National Center for Biotechnology Information Sequence Read Archive.