Abstract
Liver fibrosis is a consequence of chronic liver injury associated with chronic viral infection, alcohol abuse, and nonalcoholic fatty liver. The evidence from clinical and animal studies indicates that transforming growth factor-β (TGF-β) signaling is associated with the development of liver fibrosis. Krüppel-like factor 10 (KLF10) is a transcription factor that plays a significant role in TGF-β-mediated cell growth, apoptosis, and differentiation. In recent studies, it has been reported to be associated with glucose homeostasis and insulin resistance. In the present study, we investigated the role of KLF10 in the progression of liver disease upon a high-sucrose diet (HSD) in mice. Wild type (WT) and Klf10 knockout (KO) mice were fed either a control chow diet or HSD (50% sucrose) for eight weeks. Klf10 KO mice exhibited significant hepatic steatosis, inflammation, and liver injury upon HSD feeding, whereas the WT mice exhibited mild hepatic steatosis with no apparent liver injury. The livers of HSD-fed Klf10 KO mice demonstrated significantly increased endoplasmic reticulum stress, oxidative stress, and proinflammatory cytokines. Klf10 deletion led to the development of sucrose-induced hepatocyte cell death both in vivo and in vitro. Moreover, it significantly increased fibrogenic gene expression and collagen accumulation in the liver. Increased liver fibrosis was accompanied by increased phosphorylation and nuclear localization of Smad3. Here, we demonstrate that HSD-fed mice develop a severe liver injury in the absence of KLF10 due to the hyperactivation of the endoplasmic reticulum stress response and CCAAT/enhance-binding protein homologous protein (CHOP)-mediated apoptosis of hepatocytes. The current study suggests that KLF10 plays a protective role against the progression of hepatic steatosis into liver fibrosis in a lipogenic state.
Keywords: KLF10, NAFLD, NASH, sucrose diet, ER stress, hepatocyte cell death, fibrosis, Smad3
1. Introduction
Nonalcoholic fatty liver diseases (NAFLD), including nonalcoholic steatohepatitis (NASH), are frequent comorbidities that develop in patients with obesity, diabetes, hypertension, and dyslipidemia [1,2,3]. NAFLD represents a spectrum of disorders encompassing hepatic steatosis, which is a benign condition, to nonalcoholic steatohepatitis (NASH), which can cause fibrosis, cirrhosis, and hepatocellular carcinoma (HCC) [4,5,6]. During liver injury, transforming growth factor-beta (TGF-β), produced and secreted by inflammatory cells such as liver macrophages or Kupffer cells and platelets, activates hepatic stellate cells to promote fibrosis [7,8]. When chronic damage and inflammation persist, NASH progresses to liver fibrosis, with accumulation of scar tissue in the liver, thus limiting the ability of the liver to function and repair itself [9,10]. This could eventually lead to cirrhosis or hepatocellular carcinoma.
Fatty acids used in the synthesis of hepatic triglycerides (TG) come from various sources, including diet, fatty acids liberated via adipose tissue lipolysis, and de novo lipogenesis. An increase in fatty acids and reduced TG export as very-low-density lipoprotein (VLDL) or inefficient fatty acid oxidation can cause fat accumulation in the liver [11,12]. Dietary sugar is a primary stimulus for de novo hepatic lipogenesis, which may also contribute to fat accumulation in the liver [13]. The excess free fatty acids in the liver cells can induce reactive oxygen species (ROS) production, which causes lipotoxic stress in the mitochondria and endoplasmic reticulum (ER). This results in mitochondrial dysfunction, ER stress, and increased apoptotic activity [14,15].
Krüppel-like factor 10 (KLF10) is a zinc finger-containing transcription factor that is initially identified as the TGF-β-inducible early gene 1 (TIEG1) [16]. It plays an important role in TGF-β-mediated cell growth, differentiation, and apoptosis [17,18,19,20,21]. KLF10 is ubiquitously expressed, with high expression levels in muscle, pancreas, and liver. In the mouse liver, KLF10 is a circadian clock-controlled transcription factor that regulates genes implicated in glucose and lipid metabolism [22]. Moreover, the levels of KLF10 are increased in primary mouse hepatocytes upon glucose treatment by the carbohydrate response element-binding protein (ChREBP), which is another major transcription factor that regulates various genes involved in glucose and lipid metabolism [23,24,25,26]. The levels of KLF10 are increased in the mouse models of diabetes and obesity, and its gene variants exhibit a mild correlation with the development of type 2 diabetes [27,28]. KLF10 can also regulate hepatic glucose metabolism by directly regulating the expression of phosphoenolpyruvate carboxykinase, a rate-limiting enzyme of gluconeogenesis [22,27]. Previous studies suggested that KLF10 may regulate liver pathophysiology in a context-dependent manner [22,27]. However, whether KLF10 can regulate the progression of NAFLD in response to sugar intake has not yet been explored.
TGF-β is a potent and ubiquitous pro-fibrogenic cytokine that is increased in human patients with chronic liver diseases and in diverse animal models of liver fibrosis [7,8]. It transduces the signal through the Smad-dependent canonical and Smad-independent noncanonical signaling pathways, including c-Jun-N-terminal kinase (JNK), extracellular signal-regulated kinase (ERK), and p38 mitogen-activated protein kinase (MAPK). A recent publication has revealed the involvement of KLF10 in tissue fibrosis as one of the downstream effectors of TGF-β in tissue fibrosis. Klf10 deletion in mice led to the development of fibrosis in the skeletal muscle and heart [29,30]. These findings motivated us to study KLF10 as a target of ChREBP and TGF-β in the development of NASH during increased lipogenesis.
In this study, we investigated the role of KLF10 in the progression of NAFLD using Klf10 knockout (KO) mice. Klf10 KO mice developed hepatic fibrosis due to the induction of ER stress, inflammation, and Smad3 signaling in a lipogenic condition induced by a high-sucrose diet (HSD).
2. Results
2.1. Klf10 Deletion Induces High-Sucrose Diet-Mediated Liver Injury
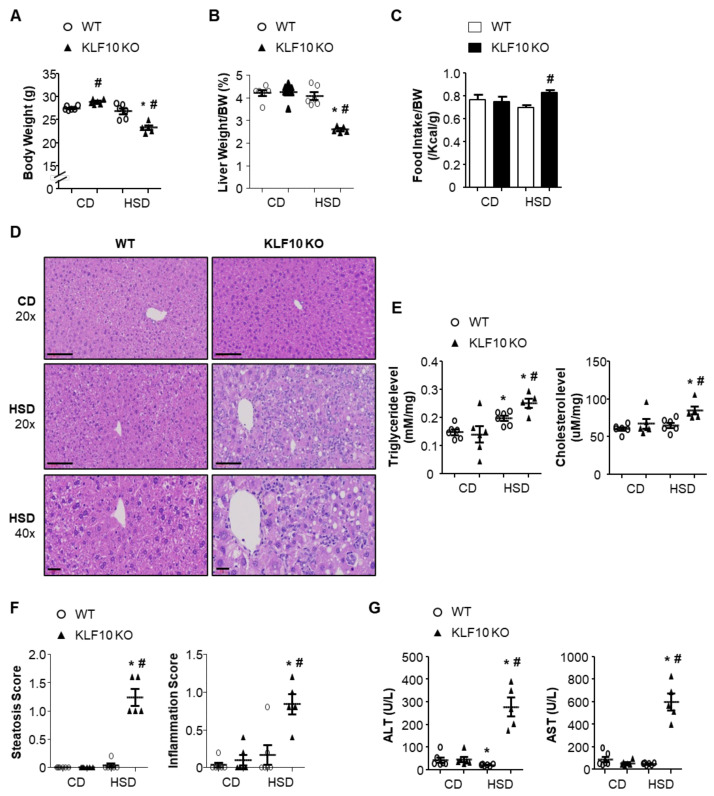
A long-term HSD induces lipogenesis and hepatic TG accumulation [13]. The role of KLF10 in the development of NAFLD was evaluated in 8-week-old male WT and Klf10 KO mice fed either a control chow diet (CD, 10% sucrose, 50% starch) or HSD (50% sucrose, 10% starch) for eight weeks. While the WT mice maintained a constant body weight on both diets, Klf10 KO mice lost weight significantly upon HSD compared with CD feeding (Figure 1A). The liver weights of Klf10 KO mice fed an HSD were significantly lower than those of Klf10 KO mice fed a CD, as well as those of the WT mice (Figure 1B). As presented in Figure 1C, the bodyweight loss in HSD-fed Klf10 KO mice was not a result of decreased food intake. Histologically, the hepatocytes of the livers of Klf10 KO mice fed an HSD contained more lipid droplets compared with WT mice, which is consistent with the increased liver TG and cholesterol contents (Figure 1D,E). The livers of Klf10 KO mice fed an HSD presented severe damage characterized by hepatocyte ballooning and inflammatory cell infiltration (Figure 1D,F). Consistent with the histological features, the markers of liver injury, such as alanine aminotransferase (ALT) and aspartate aminotransferase (AST), were significantly increased in the plasma of HSD-fed Klf10 KO mice (Figure 1G).
Figure 1.
Klf10 KO mice develop HSD-induced liver injury. Eight-week-old WT and Klf10 KO mice were fed either CD or HSD for eight weeks (n = 5–6 mice/group). (A) Body weights and (B) liver weights were determined after eight weeks of either diet. (C) Food intake (average over seven days) was measured in WT and Klf10 KO mice. (D) Representative histological images of H&E-stained liver sections (Scale bars: 60 μm, Magnification: 20×; enlarged images in the bottom panels of D, 40×). (E) Triglycerides and cholesterol in the liver were measured from liver samples. (F) The grade of the steatosis and inflammation score was calculated. (G) Plasma ALT and AST levels were measured as liver injury markers. All data are representative of at least three independent experiments and expressed as mean ± SEM. Charts were produced using GraphPad Prism 5.0. Statistical differences were determined by two-way ANOVA with Mann–Whitney U test using SPSS v17.0. * p < 0.05 vs. genotype-matched, CD-fed group and # p < 0.05 vs. diet-matched, genotype control. WT, wild type; KO, knockout; CD, control chow diet; HSD, high-sucrose diet.
2.2. Klf10 Deletion Impairs Triglyceride Packaging and Secretion upon HSD Feeding
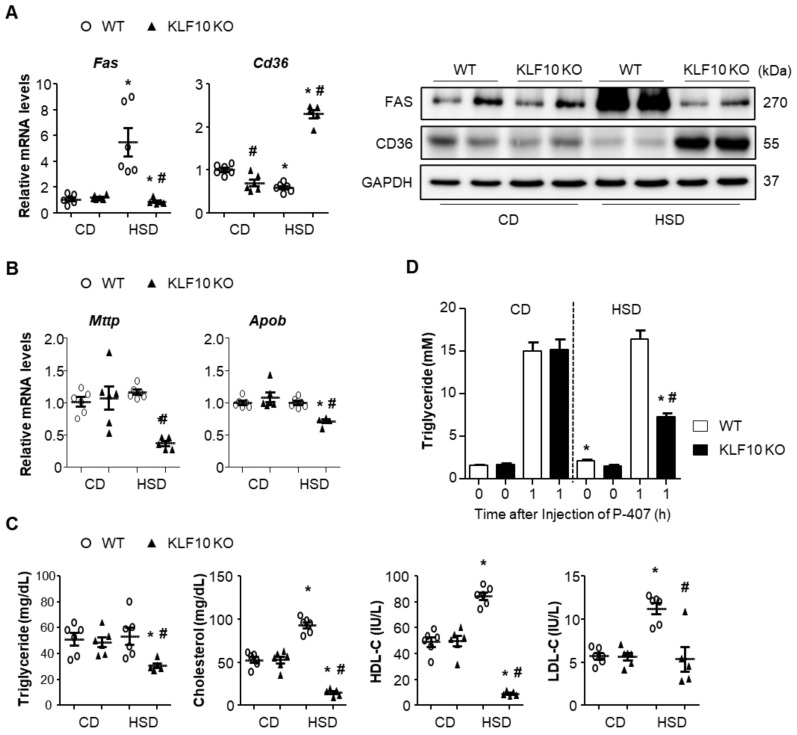
To elucidate the reasons behind fat accumulation in the liver upon HSD feeding of Klf10 KO mice, we evaluated the expression of genes related to lipid synthesis, uptake, and secretion in the liver. An HSD is known to induce hepatic lipogenesis via the expression of enzymes involved in the lipogenic pathway [11]. Fatty acid synthase (FAS), which encodes a rate-limiting enzyme of fatty acid synthesis, was induced by HSD in the WT mice but not in Klf10 KO mice (Figure 2A). Conversely, CD36, a scavenger receptor of fatty acid uptake, was significantly increased in HSD-fed Klf10 KO mice, whereas CD36 in HSD-fed WT mice was decreased compared with that of the CD-fed WT mice (Figure 2A). The genes involved in VLDL packaging and secretion, such as microsomal TG transfer protein (Mttp) and apolipoprotein B (Apob), were significantly reduced in the livers of HSD-fed Klf10 KO mice (Figure 2B). Consistent with these results, plasma TG, especially cholesterol, was significantly decreased in HSD-fed Klf10 KO mice compared with the WT mice (Figure 2C). To determine whether the decreased plasma TG and cholesterol resulted from the VLDL secretion from liver, we injected the nonionic surfactant poloxamer 407 (a lipoprotein lipase inhibitor) to inhibit TG hydrolysis and measured the entry of TG into plasma [31]. CD had no effect on the plasma TG levels in either WT or Klf10 KO mice. However, HSD significantly decreased the plasma TG levels in Klf10 KO mice compared with WT mice (Figure 2D). These results indicate that enhanced lipid uptake and decreased VLDL packaging and secretion may play roles in lipid accumulation in Klf10 KO mice. Fatty acid synthesis is unlikely to contribute to fat accumulation in HSD-fed Klf10 KO mice, as the expression of FAS was actually decreased in these mice.
Figure 2.
Klf10 deletion impairs triglyceride package and secretion upon high-sucrose feeding. Eight-week-old WT and Klf10 KO mice were fed either CD or HSD for eight weeks (n = 5–6 mice/group). Liver genes and proteins involved in (A) lipogenesis (fatty acid synthase, FAS), fatty acid uptake (fatty acid translocase, CD36), and (B) lipid secretion (microsomal triglyceride transfer protein, MTTP, and Apolipoprotein B, ApoB) were analyzed via qPCR and Western blotting. (C) Plasma triglyceride, cholesterol, HDL-cholesterol (HDL-C), and LDL-cholesterol (LDL-C) were measured. (D) In vivo VLDL secretion from livers of mice was determined. Eight-week-old WT and Klf10 KO mice were fed either CD or HSD for one week and then injected with poloxamer 407 (P-407) (n = 6 mice/group). Blood was collected at the indicated time points, and TG was measured. All data are representative of at least three independent experiments and expressed as mean ± SEM. Charts were produced using GraphPad Prism 5.0. Statistical differences were determined by two-way ANOVA with Mann–Whitney U test using SPSS v17.0. * p < 0.05 vs. genotype-matched, CD-fed group and # p < 0.05 vs. diet-matched, genotype control. WT, wild type; KO, knockout; CD, control chow diet; HSD, high-sucrose diet.
2.3. Klf10 Deletion Induces Oxidative Stress and ER Stress upon HSD Feeding
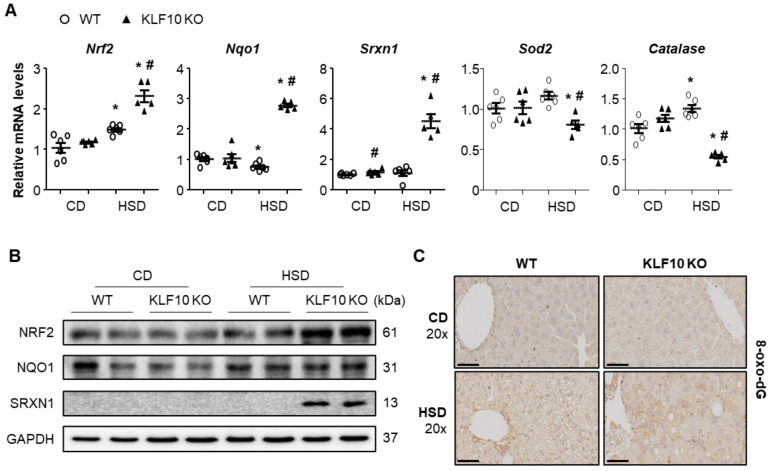
Impaired lipid secretion by MTTP inhibition was shown to increase free cholesterol in the mitochondria and ER, which resulted in oxidative stress and ER stress in the liver [32]. Therefore, we first measured the oxidative stress-related gene expression. The mRNA and protein levels of nuclear factor erythroid-derived 2-like 2 (Nrf2), NAD(P)H dehydrogenase quinone 1 (Nqo1), and sulfiredoxin 1 (Srxn1) were significantly increased in the livers of HSD-fed Klf10 KO mice compared with those of the WT mice (Figure 3A,B). The enzymes superoxide dismutase 2 (Sod2) and Catalase, which are involved in antioxidant defense, were significantly decreased in HSD-fed Klf10 KO mice (Figure 3A). 8-oxo-2′-deoxyguanosine (8-oxo-dG), a biomarker of oxidative DNA damage, was higher in Klf10 KO mice than in the WT mice upon HSD feeding (Figure 3C). These results suggest that Klf10 deletion induced oxidative DNA damage in HSD possibly via impaired detoxification of ROS.
Figure 3.
Klf10 deletion induces oxidative stress upon high-sucrose feeding. Eight-week-old WT and Klf10 KO mice were fed either CD or HSD for eight weeks (n = 5–6 mice/group). Changes in hepatic mRNA and protein expression involved in oxidative stress response were analyzed via (A) qPCR and (B) Western blotting. Antioxidant genes (Nrf2, Nqo1, and Srxn1) and ROS scavenger genes (Catalase and Sod2) were analyzed. Target gene expression was normalized to the expression of Rplp0. (C) Representative 8-oxo-2′-deoxyguanosine (8-oxo-dG)-stained livers (Scale bars: 60 μm, Magnification: 20×). All data are representative of at least three independent experiments and expressed as mean ± SEM. Charts were produced using GraphPad Prism 5.0. Statistical differences were determined by two-way ANOVA with Mann–Whitney U test using SPSS v17.0. * p < 0.05 vs. genotype-matched, CD-fed group and # p < 0.05 vs. diet-matched, genotype control. WT, wild type; KO, knockout; CD, control chow diet; HSD, high-sucrose diet.
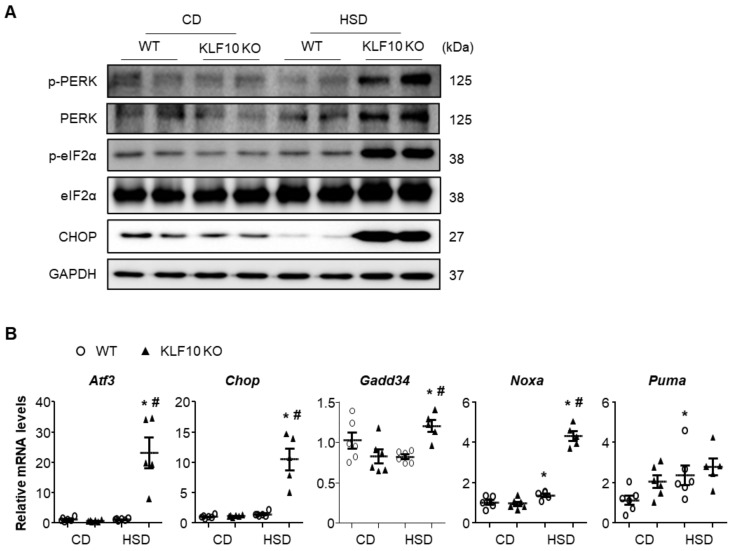
We also measured the levels of known ER stress markers. The livers of HSD-fed Klf10 KO mice exhibited increased phosphorylation of protein kinase R-like ER kinase (PERK) and eukaryotic translation initiation factor 2α (eIF2α) as well as C/EBP homologous protein (CHOP, a key component in ER stress-mediated apoptosis), which were not observed in WT mice (Figure 4A). As expected, the mRNA levels of eIF2α downstream target genes, such as Atf3, Chop, Gadd34, Noxa, and Puma, were significantly increased in the livers of HSD-fed Klf10 KO mice (Figure 4B). These results suggest that increased activation of PERK/CHOP signaling in the livers of Klf10 KO mice was responsible for the liver injury upon HSD feeding.
Figure 4.
Klf10 deletion activates the proapoptotic PERK-CHOP pathway upon high-sucrose feeding. Eight-week-old WT and Klf10 KO mice were fed either CD or HSD for eight weeks (n = 5–6 mice/group). (A) Western blot analysis of p-PERK, PERK, p-eIF2α, eIF2α, and CHOP in the livers of WT and Klf10 KO mice fed either CD or HSD. (B) Relative expression of genes involved in ER stress (Atf3, Chop, Gadd34, Noxa, and Puma) in the liver was analyzed via qPCR. Target gene expression was normalized to the expression of Rplp0. All data are representative of at least three independent experiments and expressed as mean ± SEM. Charts were produced using GraphPad Prism 5.0. Statistical differences were determined by two-way ANOVA with Mann–Whitney U test using SPSS v17.0. * p < 0.05 vs. genotype-matched, CD-fed group and # p < 0.05 vs. diet-matched, genotype control. WT, wild type; KO, knockout; CD, control chow diet; HSD, high-sucrose diet.
2.4. Klf10 Deletion Increases Stress-Mediated Hepatic Cell Death upon HSD Feeding
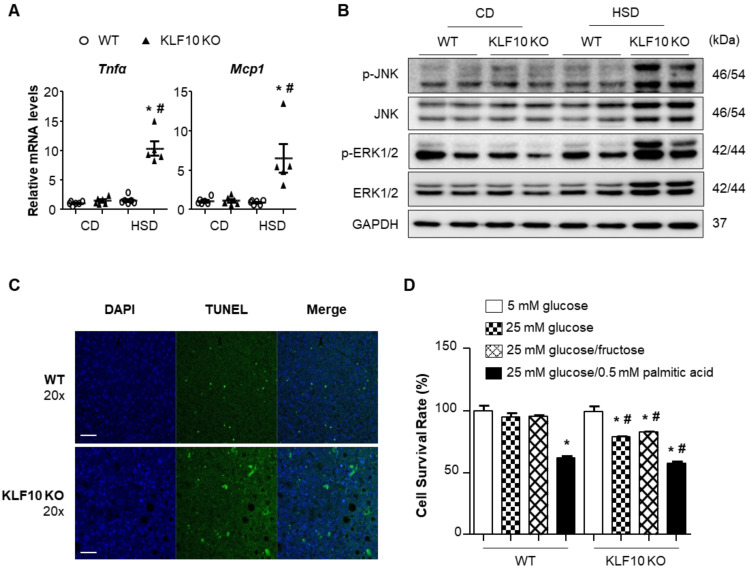
Recent findings have demonstrated that oxidative stress and ER stress are closely associated with inflammation and hepatocyte cell death [33,34]. Thus, we determined the levels of the genes involved in inflammation and cell death in our experimental system. The mRNA levels of proinflammatory cytokines, such as tumor necrosis factor α (Tnfα) and monocyte chemoattractant protein 1 (Mcp1), were significantly increased in the livers of HSD-fed Klf10 KO mice (Figure 5A). Moreover, stress-activated JNK and ERK1/2 MAP kinases were activated in the livers of HSD-fed Klf10 KO mice (Figure 5B). Then, we determined whether the HSD-induced ER stress and oxidative stress led to liver cell death in HSD-fed Klf10 KO mice. The livers of HSD-fed Klf10 KO mice contained more TUNEL-positive cells compared with those of WT mice (Figure 5C). The effects of Klf10 deletion on hepatic cell death were also determined in isolated primary hepatocytes via incubation in media containing glucose or fructose. The cell survival rates of the primary hepatocytes isolated from Klf10 KO mice were significantly reduced by high-glucose and high-fructose feeding (Figure 5D). These results suggest that Klf10 deletion induced stress-mediated cell death, which may explain the decreased liver weight observed in HSD-fed Klf10 KO mice.
Figure 5.
High sucrose diet activates stress-mediated inflammation and cell death in Klf10 KO mouse livers. Eight-week-old WT and Klf10 KO mice were fed either CD or HSD for eight weeks (n = 5–6 mice/group). (A) Relative mRNA levels of proinflammatory cytokines Tnfα and Mcp1 in the liver were analyzed via qPCR. * p < 0.05 vs. genotype-matched, CD-fed group and # p < 0.05 vs. diet-matched, genotype control. (B) Western blot analysis of p-JNK, JNK, p-ERK1/2, and ERK1/2, in the livers of WT and Klf10 KO mice fed either CD or HSD. (C) Representative images of the TUNEL assay were used to confirm the induction of apoptosis in the liver sections (Scale bars: 50 μm, Magnification: 20×). (D) Primary hepatocytes were isolated from WT and Klf10 KO mice. The CCK-8 assay determined the cell viability of primary hepatocytes following treatment with 5 mM glucose, 25 mM glucose, 25 mM glucose/fructose, or 25 mM glucose with 0.5 mM palmitic acid for 24 h. * p < 0.05 vs. 5 mM glucose group. # p < 0.05 vs. treatment-matched. All data are representative of at least three independent experiments and expressed as mean ± SEM. Charts were produced using GraphPad Prism 5.0. Statistical differences were determined by two-way ANOVA with Mann–Whitney U test using SPSS v17.0. WT, wild type; KO, knockout; CD, control chow diet; HSD, high-sucrose diet, CCK-8, Cell Counting Kit-8.
2.5. Klf10 Deletion Induces Liver Fibrosis via the Activation of the TGF-β- Smad3 Pathway
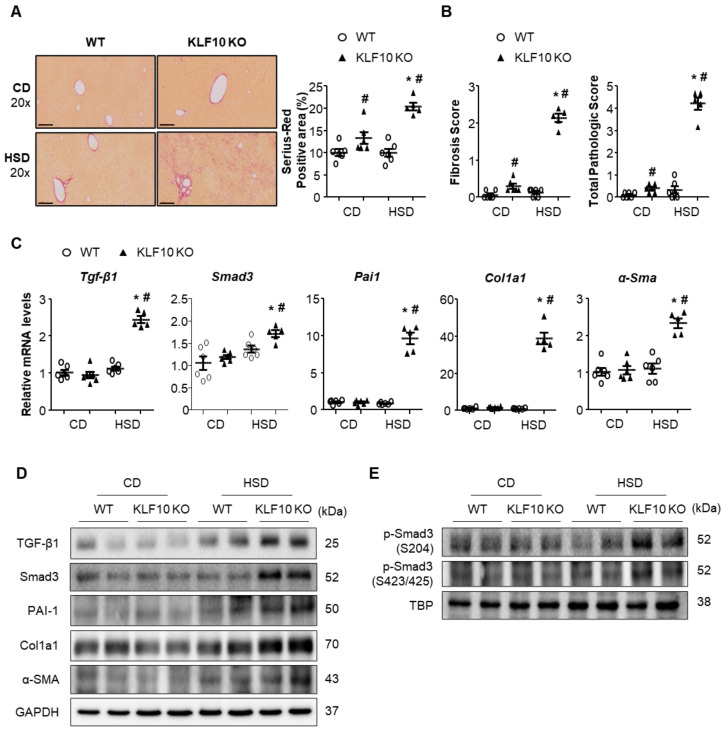
The increased liver inflammation and cell death in HSD-fed Klf10 KO mice motivated us to evaluate liver fibrosis. Hepatic fibrosis was determined by Sirius Red staining (Figure 6A). Interestingly, large quantities of collagen fibers accumulated in the livers of HSD-fed Klf10 KO mice, whereas fibrosis was hardly observed in HSD-fed WT mice. Histological analysis also revealed that HSD-fed Klf10 KO mice developed severe fibrosis (Figure 6B). We determined the mRNA and protein expression of genes involved in fibrosis, including TGF-β1, Smad3, plasminogen activator inhibitor type 1 (PAI-1), α-smooth muscle actin (α-SMA), and collagen type I alpha 1 chain (Col1a1). The expression of these genes was increased only in the livers of HSD-fed Klf10 KO mice (Figure 6C,D). Many previous studies have shown that TGF-β-mediated activation of Smad3 pathway plays a critical role during the process of liver fibrosis. During chronic liver injury, Smad3 is phosphorylated at the carboxyl terminal motif by TGF-β as well as phosphorylated at the linker region by MAPK pathway [35]. As shown in Figure 6C,D, the mRNA and protein levels of Smad3 were increased in HSD-fed Klf10 KO mice. Moreover, Smad3 phosphorylation at the linker region and carboxyl terminal motif was also significantly increased in the livers of HSD-fed Klf10 KO mice (Figure 6E). These results indicate that Klf10 deletion contributed to the activation of hepatic stellate cells, which worsened liver fibrosis in HSD-fed Klf10 KO mice.
Figure 6.
Klf10 deletion aggravates liver fibrosis through the activation of TGF-β-Smad3 pathway. Eight-week-old WT and Klf10 KO mice were fed either CD or HSD for eight weeks (n = 5–6 mice/group). (A) Collagen deposition was evaluated via Sirius Red staining of the liver sections (Scale bars: 60 μm, Magnification: 20×). Quantification indicates Sirius Red-positive area (percentage). (B) The grade of the fibrosis and NASH score was calculated. Changes in hepatic mRNA and protein expression involved in TGF-β (TGF-β1 and Smad3) and fibrogenesis (PAI-1, α-SMA, and Col1A1) pathway were analyzed via (C) qPCR and (D) Western blotting. (E) Nuclear extracts were used to detect the phosphorylation levels of Smad3 at S204 and S423/425. A TATA-binding protein (TBP) was used as a nuclear loading control. All data are representative of at least three independent experiments and expressed as mean ± SEM. Charts were produced using GraphPad Prism 5.0. Statistical differences were determined by two-way ANOVA with Mann–Whitney U test using SPSS v17.0. * p < 0.05 vs. genotype-matched, CD-fed group and # p < 0.05 vs. diet-matched, genotype control. WT, wild type; KO, knockout; CD, control chow diet; HSD, high-sucrose diet.
3. Discussion
In the present study, we demonstrated that the absence of Klf10 leads to significantly worse liver pathology upon HSD feeding. HSD-fed Klf10 KO mice developed severe liver injury due to the hyperactivation of the ER stress response and CHOP-mediated apoptosis of hepatocytes. Furthermore, Klf10 deletion activated hepatic stellate cells mediated by TGF-β/Smad3 signaling, which resulted in NASH progression to fibrosis.
HSD induced hepatic steatosis via the induction of lipogenic genes in the liver, as demonstrated by WT mice in this study. However, it did not induce lipogenic gene expression in the livers of Klf10 KO mice, although the levels of TG and cholesterol in the livers were increased. Fat accumulation in the livers of Klf10 KO mice appeared to be a result of increased fatty acid uptake and decreased VLDL secretion, as demonstrated by the induction of the CD36 expression and decreased MTTP expression. Interestingly, hepatic steatosis and fibrosis observed in HSD-fed Klf10 KO mice were also reported in Mttp KO mice [36]. MTTP transfers TG to the newly synthesized ApoB, which is a critical step in VLDL assembly in hepatocytes. Pharmacological inhibition or genetic ablation of MTTP can result in the accumulation of cholesterol in the membranes of the mitochondria and ER, which causes ER stress and subsequent hepatocyte apoptosis and fibrosis [37,38], as observed in the livers of HSD-fed Klf10 KO mice. Hepatic inflammation and severe fibrosis in Klf10 KO mice were accompanied by a significant increase in proinflammatory cytokine expression and inflammatory cell infiltration, compared with Mttp KO mice. These discrepancies may be due to the difference in diet composition, i.e., a high-sucrose diet vs. a high-fat diet. The increased fatty acid import may represent another factor that induces inflammation. CD36 is a cell surface protein that enhances lipid accumulation in hepatocytes by facilitating fatty acid uptake, impairing fatty acid oxidation, which can activate the proinflammatory JNK/NF-κB signaling pathway. As the deletion of liver fatty acid binding protein (L-Fabp) in Mttp KO mice improved hepatic steatosis and fibrosis [36], increased fatty acid flux and intrahepatic cholesterol accumulation could be mediators of ER stress and fibrosis, although the mechanisms involved in this process remain to be elucidated.
Reduced Mttp expression is an important observation in the steatosis of HSD-fed Klf10 KO mice. Several transcription factors, such as small heterodimer partner (SHP), ChREBP, sterol regulatory element-binding protein (SREBP), and forkhead transcription factor1 (Foxo1), regulate Mttp gene transcription [39,40,41,42]. The proximal promoter region of Mttp also contains putative KLF10 binding sites, which strongly suggests that Mttp may be a direct target gene of KLF10, and the reduced Mttp expression in Klf10 KO mice could result from the reduction of Mttp transcription. However, we cannot rule out the possibility of a secondary effect in the damaged liver. Signal pathways responsible for controlling Mttp expression in response to an HSD should be investigated in the future.
We unexpectedly determined that KLF10 is required to protect hepatocytes from apoptosis and to preserve the liver function in response to sugar intake. It has been reported that transient hepatic ER stress alleviates misfolded protein stress without triggering hepatocyte cell death, whereas prolonged ER stress activates proapoptotic protein expression, inhibits prosurvival factors, and eventually triggers apoptosis via the PERK/CHOP pathway [43,44]. Our data provide evidence that hepatic ER stress dysregulation is a major cause of hepatocyte cell death in HSD-fed Klf10 KO mice. Sucrose feeding had no significant effect on the expression of ER stress markers in the livers of WT mice; however, it activated the proapoptotic PERK-CHOP pathway in the livers of Klf10 KO mice. Of note, recent data showed that Klf10 deficiency worsens hepatocyte death and fibrosis upon methionine and choline deficient fibrogenic diet that blocks VLDL secretion, causing oxidative stress, inflammation, and ER stress [45]. Fibroblast growth factor 21 (FGF21), which is a direct target of ChREBP, is required for normal hepatic metabolic response to fructose consumption, and the absence of FGF21 leads to liver inflammation and fibrosis in mice on a high-fructose or methionine- and choline-deficient diet [46,47]. Moreover, FGF21 is induced by ER stress in a PERK-eIF2α-ATF4-dependent manner, and its deletion accelerates ER stress-induced hepatic injury [48]. In HSD-fed Klf10 mice, hepatic expression of FGF21 is significantly increased (data not shown), suggesting that the hepatic injury present in Klf10 KO mice may not be because of the impaired FGF21 response elicited by ER stress on a HSD. These results suggest that impaired KLF10 signaling could be an underlying cause of steatosis progression to fibrosis in certain cases of human NAFLD.
The TGF-β signaling pathway plays a significant role in the activation of hepatic stellate cells in liver fibrosis. In HSD-fed Klf10 KO mice, we observed the activation of TGF-β/Smad signaling. In the context of hepatic fibrosis, Smad3 is known as a pro-fibrotic transcription factor [49]. Phosphorylation of the carboxyl terminal motif of Smad3 occurs via the TGF-β receptor, whereas phosphorylation of the linker region of Smad3 occurs via MAPK [35,50]. Phosphorylation of the linker region of Smad3 rather than carboxyl terminal motif seems to be dominant in the course of transdifferentiation of HSCs to myofibroblasts, which produce extracellular matrix molecules that contribute to liver fibrosis [51]. Indeed, increased Smad3 linker phosphorylation was observed in the livers of Klf10 KO mice. Understanding the molecular mechanism of how Smad3 is induced and the linker region is phosphorylated when KLF10 is absent is important for the identification of therapeutic targets to control the activation of hepatic stellate cells in fibrosis.
Further, there is evidence of an association between KLF10 and human fibrotic disease. A study by the Spelsberg group revealed a positive correlation between six missense mutations in KLF10 and hypertrophic cardiomyopathy [52]. Furthermore, these KLF10 variants are associated with increased pituitary tumor transforming gene 1 (PTTG1) promoter activity and protein expression. PTTG1 has also been reported to be functionally required for hepatic fibrosis progression in an animal model of chronic liver injury [53]. Collectively, these data suggest that KLF10 may be a potential biomarker of hepatic fibrosis progression in NAFLD or other types of chronic liver disease.
In conclusion, this study demonstrated that KLF10 is required for the protection against the progression of hepatic steatosis to NASH with fibrosis upon HSD. Our findings highlight the need for an in-depth understanding of the regulation and effects of the KLF10 pathway upon sucrose feeding and as well as the progression of other ER stress-related diseases.
4. Materials and Methods
4.1. Mice and Diets
The wild type (WT) C57BL/6J mice were purchased from Jackson Laboratory (Bar Harbor, ME, USA). Klf10 KO mice were kindly provided by Dr. Woon-Kyu Lee (Inha University, Incheon, Korea) [54]. The mice were maintained on a CD containing 60% carbohydrate (~10% sucrose, PicoLab Rodent Diet 20, Orient Bio, Gyeonggi Province, Korea) under a 12-h light/dark cycle. The 8-week-old male mice were fed either a CD or HSD (50% sucrose/60% carbohydrate, D10001 AIN-76A; Research Diets, Inc., New Brunswick, NJ, USA) for eight weeks. Their body weight and food intake were measured weekly. At the end of the experiment, mice fasted for 2 h to minimize any effects of immediate feeding on gene expression and plasma biochemistry, followed by euthanization. Blood was immediately collected via the hepatic portal vein, and plasma was obtained by centrifugation at 300 × g for 15 min at 4 °C. Plasma samples were stored at −80 °C for biochemical analysis. The livers were removed, weighed, and either formalin-fixed or snap-frozen in liquid nitrogen. All animal studies were performed in accordance with the protocols approved by the Institutional Animal Care and Use Committee of the Lee Gil Ya Cancer and Diabetes Institute, Gachon University (LCDI-2019-0003).
4.2. Plasma and Tissue Biochemical Analysis
The levels of plasma ALT, AST, TG, cholesterol, HDL-cholesterol, and LDL-cholesterol were determined via automated analysis (Model AU-480; Olympus, Tokyo, Japan). In addition, the liver TG levels were determined as previously described [55] using the TG Quantification Kit (BioVision, Inc., Milpitas, CA, USA), and the liver cholesterol levels were determined using a Multi-Calibrator Lipid (Fujifilm Wako Pure Chemical, Tokyo, Japan).
4.3. Histological Evaluation
The liver samples were fixed in neutral-buffered formalin and embedded in paraffin blocks according to standard procedures. Paraffin-embedded tissue sections were incubated in xylene for 15 min. After rehydration in graded ethanol, antigen retrieval was performed in IHC-TekTM Epitope Retrieval Solution (IHC World, Ellicott City, MA, USA) by heating in a retrieval steamer for 40 min. Then, the sections were allowed to cool for 20 min. Subsequently, the sections were incubated with 3% H2O2 at room temperature for 30 min, blocked for 5 min, and then incubated with anti-8-oxo-dG (8-oxo-2′-deoxyguanosine) antibodies (Trevigen, Gaithersburg, MD, USA) overnight. The next day, the sections were incubated with the goat anti-mouse IgG HRP-conjugated secondary antibody (Merck Millipore, Temecula, CA, USA) for 30 min, washed with Dulbecco’s phosphate-buffered saline containing Tween-20, and then stained with 3,3′-diaminobenzidine. Hematoxylin and eosin and Sirius Red staining were processed to confirm the pathological analysis of liver disease by the KPNT (Korea Pathology Technical Center, Cheongju, Korea). Histological scoring (Supplementary Materials Table S1) was performed as previously described [56].
4.4. In Vivo VLDL Secretion
Poloxamer 407 (P-407) was purchased from Sigma-Aldrich (St. Louis, MO, USA); 10% (w/v) P-407 solution was prepared using saline. Eight-week-old male WT and Klf10 KO mice were fed either a CD or HSD for 1-week. Then, mice were fasted for 5 h and intraperitoneally injected with P-407 (1 g/kg). Blood was collected from the tail vein before injection (0 h) and at 1 h after injection. Plasma was separated, and TG was determined.
4.5. RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction
Total RNA was isolated from the mouse liver tissue using RNAiso Plus (Takara, Shiga, Japan). Purified total RNA was reverse-transcribed using the PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara, Shiga, Japan). Gene-specific primers were designed using Primer Express Software (PerkinElmer Life Sciences, Waltham, MA, USA) and validated via analysis of template titration and dissociation curves. Quantitative real-time polymerase chain reaction (qPCR) was conducted on a CFX384 Touch™ Real-Time PCR Detection System (Bio-Rad Laboratories, Inc., Hercules, CA, USA) using SYBR® Premix Ex Taq™ II, ROX Plus (Takara, Shiga, Japan). Relative gene expression was determined using the 2–ΔΔCT method [57], with the gene encoding ribosomal protein, large, P0 (Rplp0), which serves as the internal control. The primer sequences used are presented in Supplementary Materials Table S2.
4.6. Preparation of Nuclear and Cytoplasmic Proteins from Cells and Tissues
Nuclear and cytoplasmic protein fractions were prepared using commercial reagents, NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, Waltham, MA, USA) and PRO-PREP™ Protein Extraction Solution (Intron biotechnology, Seongnam, Korea) according to the manufacturer’s instructions. Protein concentrations were determined using the Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific).
4.7. Western Blotting
Western blot analyses were conducted according to standard protocols. The following antibodies were used: anti-FAS, anti-Nrf2, anti-NQO1, anti-Srxn1, anti-PERK, anti-JNK, anti-TGFβ1, anti-Col1a1, anti-fibronectin, anti-α-SMA, and anti-PAI1 from Santa Cruz, (Santa Cruz, CA, USA); anti-eIF2α, anti-phospho-eIF2α (Ser51), anti-CHOP, anti-ERK, anti-phospho-ERK (Tyr204/Tyr187), anti-phospho-JNK (Thr183/Tyr185), anti-p38, anti-phospho-p38 (Thr180/Tyr182), anti-Smad3, and anti-phospho-Smad3 (Ser423/425) from Cell Signaling Technology (Beverly, MA, USA); anti-phospho-PERK (Thr980) from Thermo Fisher Scientific; anti-phospho-Smad3 (Thr179) and anti-phospho-Smad3 (Ser208) from Invitrogen (Carlsbad, CA, USA); anti-TATA-binding protein (TBP) from Abcam (Cambridge, MA, USA); and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from Merck Millipore. TBP and GAPDH were used as a loading control.
4.8. Preparation of Mouse Hepatocytes and Cell Viability Assay
Primary hepatocytes were isolated from 8–10-week-old WT and Klf10 KO mice by perfusion with collagenase, as previously described [58]. The primary hepatocyte viability was evaluated by colorimetric assay using the Cell Counting Kit-8 (Dojindo Molecular Technologies, Rockville, MD, USA). The isolated hepatocytes were placed on 96-well plates at a density of 0.25 × 105 cells/well and incubated in Hepatozyme (Life Technologies, Carlsbad, CA, USA) for 24 h, after which the media was changed to DMEM supplemented with 5 mM glucose, 25 mM glucose, 25 mM glucose/fructose, or 25 mM glucose with 0.5 mM palmitic acid. After 24 h, CCK-8 reagent (10 µL) was added to the wells and incubated for 3 h, followed by measurement of absorbance at 450 nm using a microplate reader.
4.9. TUNEL Assay
The TransDetect® In Situ Fluorescein TUNEL Apoptosis Detection Kit was purchased from Transgen Biotech (Beijing, China) and used according to the manufacturer’s instructions. The fluorescein-labeled DNA was detected via fluorescence microscopy.
4.10. Statistical Analysis
Data are expressed as mean ± standard error of the mean. Charts were generated using GraphPad Prism 5.0 (GraphPad Software, Inc., San Diego, CA, USA). Statistical analysis was conducted using the SPSS software (Version 17.0; SPSS Inc., Chicago, IL, USA). Data were analyzed using the Mann–Whitney U test, and p < 0.05 was considered statistically significant.
Acknowledgments
We thank Woon-Kyu Lee for providing Klf 10 KO mice and Shi-Young Park for technical advice.
Abbreviations
| CD | Control chow diet |
| CHOP | C/EBP homologous protein |
| eIF2α | Eukaryotic translation initiation factor 2α |
| ER | Endoplasmic reticulum |
| HSD | High-sucrose diet |
| JNK | c-Jun-N-terminal kinase |
| KLF10 | Krüppel-like factor 10 |
| KO | Knockout |
| MTTP | Microsomal triglyceride transfer protein |
| NAFLD | Nonalcoholic fatty liver diseases |
| NASH | Nonalcoholic steatohepatitis |
| PERK | Protein kinase R-like ER kinase |
| TGF-β | Transforming growth factor-beta |
| VLDL | Very-low-density lipoprotein |
| WT | Wild type |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/22/1/331/s1.
Author Contributions
Conceptualization, J.L. and J.-Y.C.; methodology, J.L. and A.-R.O.; validation, J.L., A.-R.O., and J.-Y.C.; formal analysis, A.-R.O. and H.-Y.L.; investigation, J.L., A.-R.O., H.-J.L., and J.-Y.C.; writing—original draft preparation, J.L., Y.-A.M., and J.-Y.C.; writing—Review and editing, Y.-A.M., H.-J.L., and J.-Y.C.; supervision, H.-J.L. and J.-Y.C.; project administration, J.-Y.C.; funding acquisition, H.-J.L. and J.-Y.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology, grant number NRF-2019R1A2C2006001 (to J-Y Cha) and NRF-2016R1D1A1B 03935001 (to H-J Lee); the Korea Mouse Phenotyping Project, grant number 2013M3A9D5072550 (to J-Y Cha); and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare, grant number HI14C1135 (to J-Y Cha).
Institutional Review Board Statement
All animal studies were performed in accordance with the protocols approved by the Institutional Animal Care and Use Committee of the Lee Gil Ya Cancer and Diabetes Institute, Gachon University.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Adams L.A., Lymp J.F., St Sauver J., Sanderson S.O., Lindor K.D., Feldstein A., Angulo P. The natural history of nonalcoholic fatty liver disease: A population-based cohort study. Gastroenterology. 2005;129:113–121. doi: 10.1053/j.gastro.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 2.Angulo P. Gi epidemiology: Nonalcoholic fatty liver disease. Aliment. Pharmacol. Ther. 2007;25:883–889. doi: 10.1111/j.1365-2036.2007.03246.x. [DOI] [PubMed] [Google Scholar]
- 3.Toledo F.G., Sniderman A.D., Kelley D.E. Influence of hepatic steatosis (fatty liver) on severity and composition of dyslipidemia in type 2 diabetes. Diabetes Care. 2006;29:1845–1850. doi: 10.2337/dc06-0455. [DOI] [PubMed] [Google Scholar]
- 4.Ludwig J., Viggiano T.R., McGill D.B., Oh B.J. Nonalcoholic steatohepatitis: Mayo clinic experiences with a hitherto unnamed disease. Mayo Clin. Proc. 1980;55:434–438. [PubMed] [Google Scholar]
- 5.Cotrim H.P., Parana R., Braga E., Lyra L. Nonalcoholic steatohepatitis and hepatocellular carcinoma: Natural history? Am. J. Gastroenterol. 2000;95:3018–3019. doi: 10.1111/j.1572-0241.2000.03241.x. [DOI] [PubMed] [Google Scholar]
- 6.Boutari C., Lefkos P., Athyros V.G., Karagiannis A., Tziomalos K. Nonalcoholic fatty liver disease vs. Nonalcoholic steatohepatitis: Pathological and clinical implications. Curr. Vasc. Pharmacol. 2018;16:214–218. doi: 10.2174/1570161115666170621075157. [DOI] [PubMed] [Google Scholar]
- 7.Dewidar B., Soukupova J., Fabregat I., Dooley S. Tgf-β in hepatic stellate cell activation and liver fibrogenesis: Updated. Curr. Pathobiol. Rep. 2015;3:291–305. doi: 10.1007/s40139-015-0089-8. [DOI] [Google Scholar]
- 8.Fabregat I., Moreno-Caceres J., Sanchez A., Dooley S., Dewidar B., Giannelli G., Ten Dijke P., Consortium I.-L. Tgf-beta signalling and liver disease. FEBS J. 2016;283:2219–2232. doi: 10.1111/febs.13665. [DOI] [PubMed] [Google Scholar]
- 9.Cha J.Y., Kim D.H., Chun K.H. The role of hepatic macrophages in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Lab. Anim. Res. 2018;34:133–139. doi: 10.5625/lar.2018.34.4.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bataller R., Brenner D.A. Liver fibrosis. J. Clin. Investig. 2005;115:209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Browning J.D., Horton J.D. Molecular mediators of hepatic steatosis and liver injury. J. Clin. Investig. 2004;114:147–152. doi: 10.1172/JCI200422422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen J.C., Horton J.D., Hobbs H.H. Human fatty liver disease: Old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donnelly K.L., Smith C.I., Schwarzenberg S.J., Jessurun J., Boldt M.D., Parks E.J. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J. Clin. Investig. 2005;115:1343–1351. doi: 10.1172/JCI23621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hauck A.K., Bernlohr D.A. Oxidative stress and lipotoxicity. J. Lipid Res. 2016;57:1976–1986. doi: 10.1194/jlr.R066597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rolo A.P., Teodoro J.S., Palmeira C.M. Role of oxidative stress in the pathogenesis of nonalcoholic steatohepatitis. Free Radic. Biol. Med. 2012;52:59–69. doi: 10.1016/j.freeradbiomed.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Fautsch M.P., Vrabel A., Rickard D., Subramaniam M., Spelsberg T.C., Wieben E.D. Characterization of the mouse tgfbeta-inducible early gene (tieg): Conservation of exon and transcriptional regulatory sequences with evidence of additional transcripts. Mamm. Genome. 1998;9:838–842. doi: 10.1007/s003359900878. [DOI] [PubMed] [Google Scholar]
- 17.Ribeiro A., Bronk S.F., Roberts P.J., Urrutia R., Gores G.J. The transforming growth factor beta(1)-inducible transcription factor tieg1, mediates apoptosis through oxidative stress. Hepatology. 1999;30:1490–1497. doi: 10.1002/hep.510300620. [DOI] [PubMed] [Google Scholar]
- 18.Subramaniam M., Gorny G., Johnsen S.A., Monroe D.G., Evans G.L., Fraser D.G., Rickard D.J., Rasmussen K., van Deursen J.M., Turner R.T., et al. Tieg1 null mouse-derived osteoblasts are defective in mineralization and in support of osteoclast differentiation in vitro. Mol. Cell Biol. 2005;25:1191–1199. doi: 10.1128/MCB.25.3.1191-1199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramaniam M., Harris S.A., Oursler M.J., Rasmussen K., Riggs B.L., Spelsberg T.C. Identification of a novel tgf-beta-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic. Acids Res. 1995;23:4907–4912. doi: 10.1093/nar/23.23.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyake M., Hayashi S., Iwasaki S., Chao G., Takahashi H., Watanabe K., Ohwada S., Aso H., Yamaguchi T. Possible role of tieg1 as a feedback regulator of myostatin and tgf-beta in myoblasts. Biochem. Biophys. Res. Commun. 2010;393:762–766. doi: 10.1016/j.bbrc.2010.02.077. [DOI] [PubMed] [Google Scholar]
- 21.Miyake M., Hayashi S., Iwasaki S., Uchida T., Watanabe K., Ohwada S., Aso H., Yamaguchi T. Tieg1 negatively controls the myoblast pool indispensable for fusion during myogenic differentiation of c2c12 cells. J. Cell Physiol. 2011;226:1128–1136. doi: 10.1002/jcp.22434. [DOI] [PubMed] [Google Scholar]
- 22.Guillaumond F., Grechez-Cassiau A., Subramaniam M., Brangolo S., Peteri-Brunback B., Staels B., Fievet C., Spelsberg T.C., Delaunay F., Teboul M. Kruppel-like factor klf10 is a link between the circadian clock and metabolism in liver. Mol. Cell Biol. 2010;30:3059–3070. doi: 10.1128/MCB.01141-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeong Y.S., Kim D., Lee Y.S., Kim H.J., Han J.Y., Im S.S., Chong H.K., Kwon J.K., Cho Y.H., Kim W.K., et al. Integrated expression profiling and genome-wide analysis of chrebp targets reveals the dual role for chrebp in glucose-regulated gene expression. PLoS ONE. 2011;6:e22544. doi: 10.1371/journal.pone.0022544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iizuka K., Takeda J., Horikawa Y. Kruppel-like factor-10 is directly regulated by carbohydrate response element-binding protein in rat primary hepatocytes. Biochem. Biophys. Res. Commun. 2011;412:638–643. doi: 10.1016/j.bbrc.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 25.Lee H.J., Cha J.Y. Recent insights into the role of chrebp in intestinal fructose absorption and metabolism. BMB Rep. 2018;51:429–436. doi: 10.5483/BMBRep.2018.51.9.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ortega-Prieto P., Postic C. Carbohydrate sensing through the transcription factor chrebp. Front. Genet. 2019;10:472. doi: 10.3389/fgene.2019.00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X., Chen Q., Sun L., Zhang H., Yao L., Cui X., Gao Y., Fang F., Chang Y. Klf10 transcription factor regulates hepatic glucose metabolism in mice. Diabetologia. 2017;60:2443–2452. doi: 10.1007/s00125-017-4412-2. [DOI] [PubMed] [Google Scholar]
- 28.Gutierrez-Aguilar R., Benmezroua Y., Balkau B., Marre M., Helbecque N., Charpentier G., Polychronakos C., Sladek R., Froguel P., Neve B. Minor contribution of smad7 and klf10 variants to genetic susceptibility of type 2 diabetes. Diabetes Metab. 2007;33:372–378. doi: 10.1016/j.diabet.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Rajamannan N.M., Subramaniam M., Abraham T.P., Vasile V.C., Ackerman M.J., Monroe D.G., Chew T.L., Spelsberg T.C. Tgfbeta inducible early gene-1 (tieg1) and cardiac hypertrophy: Discovery and characterization of a novel signaling pathway. J. Cell Biochem. 2007;100:315–325. doi: 10.1002/jcb.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DiMario J.X. Klf10 gene expression modulates fibrosis in dystrophic skeletal muscle. Am. J. Pathol. 2018;188:1263–1275. doi: 10.1016/j.ajpath.2018.01.014. [DOI] [PubMed] [Google Scholar]
- 31.Millar J.S., Cromley D.A., McCoy M.G., Rader D.J., Billheimer J.T. Determining hepatic triglyceride production in mice: Comparison of poloxamer 407 with triton wr-1339. J. Lipid Res. 2005;46:2023–2028. doi: 10.1194/jlr.D500019-JLR200. [DOI] [PubMed] [Google Scholar]
- 32.Josekutty J., Iqbal J., Iwawaki T., Kohno K., Hussain M.M. Microsomal triglyceride transfer protein inhibition induces endoplasmic reticulum stress and increases gene transcription via ire1alpha/cjun to enhance plasma alt/ast. J. Biol. Chem. 2013;288:14372–14383. doi: 10.1074/jbc.M113.459602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kandel-Kfir M., Almog T., Shaish A., Shlomai G., Anafi L., Avivi C., Barshack I., Grosskopf I., Harats D., Kamari Y. Interleukin-1alpha deficiency attenuates endoplasmic reticulum stress-induced liver damage and chop expression in mice. J. Hepatol. 2015;63:926–933. doi: 10.1016/j.jhep.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 34.Kaplowitz N., Than T.A., Shinohara M., Ji C. Endoplasmic reticulum stress and liver injury. Semin. Liver Dis. 2007;27:367–377. doi: 10.1055/s-2007-991513. [DOI] [PubMed] [Google Scholar]
- 35.Matsuzaki K. Smad phosphoisoform signals in acute and chronic liver injury: Similarities and differences between epithelial and mesenchymal cells. Cell Tissue Res. 2012;347:225–243. doi: 10.1007/s00441-011-1178-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newberry E.P., Xie Y., Kennedy S.M., Graham M.J., Crooke R.M., Jiang H., Chen A., Ory D.S., Davidson N.O. Prevention of hepatic fibrosis with liver microsomal triglyceride transfer protein deletion in liver fatty acid binding protein null mice. Hepatology. 2017;65:836–852. doi: 10.1002/hep.28941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wetterau J.R., Zilversmit D.B. Localization of intracellular triacylglycerol and cholesteryl ester transfer activity in rat tissues. Biochim. Biophys. Acta. 1986;875:610–617. doi: 10.1016/0005-2760(86)90084-6. [DOI] [PubMed] [Google Scholar]
- 38.Wetterau J.R., Zilversmit D.B. A triglyceride and cholesteryl ester transfer protein associated with liver microsomes. J. Biol. Chem. 1984;259:10863–10866. [PubMed] [Google Scholar]
- 39.Hirokane H., Nakahara M., Tachibana S., Shimizu M., Sato R. Bile acid reduces the secretion of very low density lipoprotein by repressing microsomal triglyceride transfer protein gene expression mediated by hepatocyte nuclear factor-4. J. Biol. Chem. 2004;279:45685–45692. doi: 10.1074/jbc.M404255200. [DOI] [PubMed] [Google Scholar]
- 40.Niwa H., Iizuka K., Kato T., Wu W., Tsuchida H., Takao K., Horikawa Y., Takeda J. Chrebp rather than shp regulates hepatic vldl secretion. Nutrients. 2018;10:321. doi: 10.3390/nu10030321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sato R., Miyamoto W., Inoue J., Terada T., Imanaka T., Maeda M. Sterol regulatory element-binding protein negatively regulates microsomal triglyceride transfer protein gene transcription. J. Biol. Chem. 1999;274:24714–24720. doi: 10.1074/jbc.274.35.24714. [DOI] [PubMed] [Google Scholar]
- 42.Kamagate A., Qu S., Perdomo G., Su D., Kim D.H., Slusher S., Meseck M., Dong H.H. Foxo1 mediates insulin-dependent regulation of hepatic vldl production in mice. J. Clin. Investig. 2008;118:2347–2364. doi: 10.1172/JCI32914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hetz C. The unfolded protein response: Controlling cell fate decisions under er stress and beyond. Nat. Rev. Mol. Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 44.Tabas I., Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 2011;13:184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leclere P.S., Rousseau D., Patouraux S., Guerin S., Bonnafous S., Grechez-Cassiau A., Ruberto A.A., Luci C., Subramaniam M., Tran A., et al. Mcd diet-induced steatohepatitis generates a diurnal rhythm of associated biomarkers and worsens liver injury in klf10 deficient mice. Sci. Rep. 2020;10:12139. doi: 10.1038/s41598-020-69085-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fisher F.M., Kim M., Doridot L., Cunniff J.C., Parker T.S., Levine D.M., Hellerstein M.K., Hudgins L.C., Maratos-Flier E., Herman M.A. A critical role for chrebp-mediated fgf21 secretion in hepatic fructose metabolism. Mol. Metab. 2017;6:14–21. doi: 10.1016/j.molmet.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watanabe M., Singhal G., Fisher F.M., Beck T.C., Morgan D.A., Socciarelli F., Mather M.L., Risi R., Bourke J., Rahmouni K., et al. Liver-derived fgf21 is essential for full adaptation to ketogenic diet but does not regulate glucose homeostasis. Endocrine. 2020;67:95–108. doi: 10.1007/s12020-019-02124-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim S.H., Kim K.H., Kim H.K., Kim M.J., Back S.H., Konishi M., Itoh N., Lee M.S. Fibroblast growth factor 21 participates in adaptation to endoplasmic reticulum stress and attenuates obesity-induced hepatic metabolic stress. Diabetologia. 2015;58:809–818. doi: 10.1007/s00125-014-3475-6. [DOI] [PubMed] [Google Scholar]
- 49.Zhang L., Liu C., Meng X.M., Huang C., Xu F., Li J. Smad2 protects against tgf-beta1/smad3-mediated collagen synthesis in human hepatic stellate cells during hepatic fibrosis. Mol. Cell Biochem. 2015;400:17–28. doi: 10.1007/s11010-014-2258-1. [DOI] [PubMed] [Google Scholar]
- 50.Foglia B., Cannito S., Bocca C., Parola M., Novo E. Erk pathway in activated, myofibroblast-like, hepatic stellate cells: A critical signaling crossroad sustaining liver fibrosis. Int. J. Mol. Sci. 2019;20:2700. doi: 10.3390/ijms20112700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshida K., Matsuzaki K. Differential regulation of tgf-beta/smad signaling in hepatic stellate cells between acute and chronic liver injuries. Front. Physiol. 2012;3:53. doi: 10.3389/fphys.2012.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bos J.M., Subramaniam M., Hawse J.R., Christiaans I., Rajamannan N.M., Maleszewski J.J., Edwards W.D., Wilde A.A., Spelsberg T.C., Ackerman M.J. Tgfbeta-inducible early gene-1 (tieg1) mutations in hypertrophic cardiomyopathy. J. Cell Biochem. 2012;113:1896–1903. doi: 10.1002/jcb.24058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buko V., Belonovskaya E., Naruta E., Lukivskaya O., Kanyuka O., Zhuk O., Kranc R., Stoika R., Sybirna N. Pituitary tumor transforming gene as a novel regulatory factor of liver fibrosis. Life Sci. 2015;132:34–40. doi: 10.1016/j.lfs.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 54.Song K.D., Kim D.J., Lee J.E., Yun C.H., Lee W.K. Klf10, transforming growth factor-beta-inducible early gene 1, acts as a tumor suppressor. Biochem. Biophys. Res. Commun. 2012;419:388–394. doi: 10.1016/j.bbrc.2012.02.032. [DOI] [PubMed] [Google Scholar]
- 55.Bae J.S., Park J.M., Lee J., Oh B.C., Jang S.H., Lee Y.B., Han Y.M., Ock C.Y., Cha J.Y., Hahm K.B. Amelioration of non-alcoholic fatty liver disease with npc1l1-targeted igy or n-3 polyunsaturated fatty acids in mice. Metabolism. 2017;66:32–44. doi: 10.1016/j.metabol.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 56.Kim E.H., Bae J.S., Hahm K.B., Cha J.Y. Endogenously synthesized n-3 polyunsaturated fatty acids in fat-1 mice ameliorate high-fat diet-induced non-alcoholic fatty liver disease. Biochem. Pharmacol. 2012;84:1359–1365. doi: 10.1016/j.bcp.2012.08.029. [DOI] [PubMed] [Google Scholar]
- 57.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 58.Matsuda M., Korn B.S., Hammer R.E., Moon Y.A., Komuro R., Horton J.D., Goldstein J.L., Brown M.S., Shimomura I. Srebp cleavage-activating protein (scap) is required for increased lipid synthesis in liver induced by cholesterol deprivation and insulin elevation. Genes Dev. 2001;15:1206–1216. doi: 10.1101/gad.891301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.