Abstract
Mastocytosis is a rare and complex disease characterized by expansion of clonal mast cells (MC) in skin and/or various internal organ systems. Involvement of internal organs leads to the diagnosis of systemic mastocytosis (SM). The WHO classification divides SM into indolent SM, smoldering SM and advanced SM variants, including SM with an associated hematologic neoplasm, aggressive SM, and MC leukemia. Historically, genetic analysis of individuals with pure cutaneous mastocytosis (CM) and SM have focused primarily on cohort studies of inherited single nucleotide variants and acquired pathogenic variants. The most prevalent pathogenic variant (mutation) in patients with SM is KIT p.D816V, which is detectable in most adult patients. Other somatic mutations have also been identified—especially in advanced SM—in TET2, SRSF2, ASXL1, RUNX1, CBL and JAK2, and shown to impact clinical and cellular phenotypes. Although only small patient cohorts have been analyzed, disease associations have also been identified in several germline variants within genes encoding certain cytokines or their receptors (IL13, IL6, IL6R, IL31, IL4R) and toll-like receptors. More recently, an increased prevalence of hereditary alpha-tryptasemia (HαT) caused by increased TPSAB1 copy number encoding alpha-tryptase has been described in patients with SM. Whereas HαT is found in 3–6% of general Western populations, it is identified in up to 17% of patients with SM. In the current manuscript we review the prevalence, functional role and clinical impact of various germline and somatic genetic variants in patients with mastocytosis.
Keywords: mast cells, mast cell activation syndrome, gene polymorphisms, prognostication, hereditary alpha-tryptasemia, KIT variants
1. Introduction
Mastocytosis is a term used to define a group of rare myeloid neoplasms characterized by the expansion and accumulation of clonal (neoplastic) mast cells (MC) in the skin and/or in various internal organs. In common with normal MC, neoplastic MC are considered to derived from CD34+/CD38− stem cells [1,2]. However, in contrast to normal CD34+/CD38− stem cells, neoplastic stem cells in mastocytosis are transformed cells that undergo clonal evolution and sub-clone formation depending on the acquisition of somatic mutations in critical target genes [3,4,5].
In general, cutaneous forms of mastocytosis (CM) and variants of systemic mastocytosis (SM) have been described [6,7,8,9,10,11]. The World Health Organization (WHO) divides SM into indolent SM (ISM), smoldering SM (SSM), SM with an associated hematologic neoplasm (SM-AHN), aggressive SM (ASM), and MC leukemia (MCL)—Table 1 [9,10,11].
Table 1.
WHO classification of mastocytosis *.
| Cutaneous mastocytosis (CM) |
| maculopapular cutaneous mastocytosis (MPCM) = urticaria pigmentosa (UP) |
| diffuse cutaneous mastocytosis (DCM) |
| Mastocytoma of skin (cutaneous mastocytoma) |
| Systemic mastocytosis (SM) |
| Indolent systemic mastocytosis (ISM) |
| Smoldering systemic mastocytosis (SSM) |
| Systemic mastocytosis with associated hematologic neoplasm (SM-AHN) |
| Aggressive Systemic mastocytosis (ASM) |
| Mast cell leukemia (MCL) |
| Mast cell sarcoma (MCS) |
The diagnosis of SM is based on WHO criteria, including abnormal histopathological and cytological aspects of MC in the bone marrow (BM) or in other internal organs, an elevated basal serum tryptase (BST) level, an aberrant immunophenotype of MC (CD2+/CD25+), and/or identification of the somatic mutation (pathogenic variant) KIT p.D816V [9,10,11]. Most adult patients with SM presenting with ISM (>80% cases) and have a normal or near-normal life expectancy [6,7,8,9,10,11]. Only a few of these ISM patients (<5%) progress to advanced SM [9,10,11]. Patients with advanced SM, including SM-AHN, ASM and MCL have a poor prognosis with reduced overall and progression-free survival [9,10,11,12,13]. In these patients, MC infiltration leads to organ damage [9,10,11,12,13]. In many cases, the disease is resistant to most conventional and targeted drugs including tyrosine kinase inhibitors (TKI).
From a molecular point of view, SM can be divided into three groups: (i) SM with the KIT p.D816V variant detected only in MC and a few other cells, (ii) multi-lineage SM where KIT p.D816V is also detected in most or all other myeloid cells in the BM and blood, (iii) and multi-mutated SM where, in addition to KIT p.D816V, mutations in other myeloid malignancy-related genes, such as TET2 or ASXL1 are found [14,15,16,17,18].
The diagnosis of CM is based on typical cutaneous features, including typical macular or maculopapular skin lesions and a positive Darier’s sign, as well as an increase in MC in lesional skin. The WHO recognizes 3 forms of cutaneous mastocytosis (CM): maculopapular CM (MPCM), diffuse CM, and mastocytoma of skin [9,10,11,19]. The majority of CM patients are children. The prognosis in both children and adults with CM is excellent. In these patients, transition to SM may occur but is rare and progression of true CM into advanced SM is very unusual [19,20,21,22].
MC activation syndrome (MCAS) is a condition defined by severe systemic symptoms induced by MC-derived mediators. MCAS can be primary, such as in SM or clonal MCAS, secondary, or idiopathic. Most MCAS cases present with clinical signs of anaphylaxis which in the case of secondary MCAS may result from an underlying IgE-dependent allergy [23,24,25].
2. Studies of the Common Germline Genetic Variants in Patients with Mastocytosis
The presence of activating KIT mutations alone is not sufficient to explain the different clinical forms of mastocytosis, suggesting that other inherited or somatic genetic variants are important in the regulation of MC proliferation and/or activation.
Genetic studies in CM and SM have focused on genes encoding cytokines and their receptors (IL13, IL6, IL6R, IL31, IL4R) or on genes encoding Toll-like receptors (TLRs) [26,27,28,29,30].However, given the rarity of mastocytosis only small numbers of patients have been studied and more studies are required to draw definitive conclusions regarding the impact of these genetic variants on phenotypes and outcomes associated with mastocytosis.
2.1. IL4R and IL13 Variants
Interleukin 13 and 4 are pleiotropic cytokines that are 20–25% identical and have similar effector functions within the type 2 immune response [31,32]. Both IL-4 and IL-13 are capable of signaling through the shared type-II IL-4 receptor, which is a heterodimer consisting of IL-4 receptor α (IL-4Rα) encoded by IL4R, and IL-13 receptor α 1 (IL-13Rα1) [33,34]. In the context of MC, IL-13 has been shown to induce expression of SCF, IL-6, and MCP-1 (monocyte chemotactic protein-1) [35,36]. Results of linkage and GWAS studies indicated that polymorphisms of IL4, IL4R and IL13 genes could be associated with total serum IgE levels in patients with allergies such as asthma and atopic dermatitis [37]
One study examining the role of genetic variants in this pathway was conducted by Daley, Metcalfe and Akin. They identified an association between the gain-of-function variant p.Q576R (rs 1801275) in the IL4R gene and CM, wherein the presence of this missense variant was associated with a more favorable prognosis [26]—Table 2. The authors found that the presence of the IL4R p.Q576R missense was also associated with a lower MC burden as determined both by the extent of clinical disease and by circulating levels of surrogate disease markers (i.e., tryptase and soluble CD117) and suggested that increased IL-4 and/or IL-13 signaling may limit tissue MC numbers in patients with mastocytosis [26].
Table 2.
Summary of the published data on IL4R and IL13 genes polymorphisms in mastocytosis.
| Gene | Polymorhism | Effect on Gene/Protein Function | Effects on Risk of Mastocytosis and Prognosis | References |
|---|---|---|---|---|
| IL4R | p.Q576R (rs 1801275) |
gain-of-function | Association p.Q576R of IL4R gene with CM and more favorable prognosis | [26] |
| IL13 | −1112C > T (rs 18 00925) |
−1112T high transcription variant | Presence of −1112T variant increase the risk of SM and correlate with increased tryptase level | [27] |
In contrast to these findings, suggesting that increased IL-4 and/or IL-13 signaling may limit MC numbers and disease severity in patients with CM, Nedoszytko et al. [27] identified a distinct pattern associated with IL-13 in patients with mastocytosis—Table 2. In fact, when examining the promoter polymorphism −1112C > T (rs 1800925) in the IL13 gene in CM and SM patients [21], it appeared that the high transcription variant (−1112T) may be present more frequently in mastocytosis patients than in controls, and was significantly more common among patients with SM as compared to those with CM [27]. The −1112C > T polymorphism was also found to correlate with an elevated serum tryptase level and with adult-onset of the disease, both of which are almost invariably associated with SM. Examined patients with SM also exhibited elevated serum IL-13 levels [21]. Moreover, the human MC line HMC-1 was found to display a functional IL-13 receptor [27]. Together, these data suggest that the IL13 high transcription promoter variant and elevated IL-13 levels may contribute to MC expansion and may thus promote evolution of SM [27].
However, in all these studies only a limited number of patients with SM or CM have been examined. Therefore, it remains unknown whether these cytokine and cytokine receptor variants are indeed relevant clinically. Future studies with more patients are required to draw definitive conclusions on the role of altered signaling in the IL-4 and IL-13 signaling pathways in CM and SM variants.
2.2. IL31 Variants
Interleukin 31 is a member of the IL-6 cytokine superfamily and is secreted by a variety of cells including activated type 2 CD4+ T cells and MCs [38,39,40,41]. Interleukin 31 has been reported to play a role in the etiology of pruritus, chronic inflammation and regulation of innate and adaptive immunity in tissues exposed to environmental factors. Interleukin 31 has also been demonstrated to be involved in the pathogenesis of chronic skin inflammation and pruritus in patients with chronic spontaneous urticaria, atopic dermatitis, prurigo nodularis, primary cutaneous lymphomas and myeloproliferative disorders including mastocytosis [39,40,41,42,43,44].
Several variants of the IL31 gene have been reported in association with atopic dermatitis [45,46,47]. Recently, Hartmann et al. [44] have shown that patients with mastocytosis have increased serum IL-31 levels and that serum IL-31 levels correlate with the disease severity, tryptase levels, and the degree of BM MC infiltration in adult patients with mastocytosis. A study by Lange et al. confirmed increased serum levels of IL-31 in mastocytosis patients and reported that IL-31 levels in pruritic cases of mastocytosis were significantly higher than in non-pruritic cases [28]. Authors analyzed association of mastocytosis with two variants in promoter IL31 gene (−1066 G > A (rs 11608363), −2057 G > A (rs 6489188), and one in intronic region (IVS2 + 12A > G) and found that −2057AA genotype is associated with a high prevalence of mastocytosis (CM and SM) in adult patients. They also identified an association between non-coding IL31 variants and an increased risk for CM in adults and children (Table 3).
Table 3.
Summary of the published data on IL31 gene polymorphisms in mastocytosis [28].
| Gene | Polymorhism | Effect on Gene/Protein Function | Effects on Risk of Mastocytosis and Prognosis |
|---|---|---|---|
| IL31 | −1066G > A (rs 11608363) |
Not known | Not increase mastocytosis risk |
| −2057G > A (rs 6489188) |
Not known | −2057AA genotype increase the risk of mastocytosis in adult patients with CM and SM | |
| IVS2 + 12A > G | Noncoding intronic variant | AA and AG genotypes increased the risk of mastocytosis in adults and children with CM |
2.3. IL6 and IL6R Variants
Interleukin 6 (IL-6) is a multifunctional inflammatory cytokine that plays a role in the regulation of cell proliferation, hematopoiesis, immune response, and inflammation. Interleukin 6 is a major MC-derived mediator and is elevated in patients with mastocytosis [48,49,50,51,52,53,54].
Interleukin 6 may also act as an autocrine growth factor for neoplastic MC [53,54]. Brockow et al. demonstrated that IL-6 plasma levels, but not sIL-6R levels, were elevated in patients with mastocytosis and correlated with BM pathology, organomegaly, and the extent of skin involvement. In plasma, there was a positive correlation of IL-6 levels with total tryptase levels, alkaline phosphatase, IgM, and white blood cell counts [48,49]. In addition, increased IL-6 serum levels correlated with severity of symptoms, the SCORMA (SCORing MAstocytosis) index, disease progression, and the presence of osteoporosis [49,51,53].
An examination of genetic variants in this pathway was undertaken by Rausz et al. [29] where the frequency of the −174G > C (rs 1800795) variant in the IL6 promoter and prevalence of the common p.D358A missense variant in IL6R were determined in mastocytosis patients. The authors showed that homozygous carriers of the missense variant of the IL6R gene (rs 9192284 AA genotype) had a 2.5-fold lower risk for mastocytosis than those with the AC or CC genotypes. No association with mastocytosis was found for the IL6 promoter variant—Table 4 [29].
Table 4.
Summary of the published data on IL6 and IL6R gene polymorphisms in mastocytosis.
| Gene | Polymorhism | Effect on Gene/Protein Function | Effects on Risk of Mastocytosis and Prognosis | References |
|---|---|---|---|---|
| IL6 | −174G > C (rs 1800795) |
−174G high transcription variant | No association with mastocytosis | [29] |
| IL6R | p.D358A (rs 9192284) |
Misense variant | homozygous AA carriers of the missense variant had lower risk for mastocytosis than those with the AC or CC genotypes | [29] |
2.4. TLR2 Gene Variants
Toll-like receptors (TLRs) are essential members of the pathogen recognition receptor family, expressed broadly by immune and non-immune cells including MC. All but two of the ten known TLR—the exceptions being TLR-8 and -9—have been reported to be expressed on human MC [55]. TLR-1/2, TLR-2/6, TLR-4, and TLR-5 receptors activate P13K-AKT, MAPK, and increase production of eicosanoid and pro-inflammatory cytokines, including TNF-α, IL-1β, and IL-6 [55,56,57]. Several variants in TLR genes that affect these signaling pathways have been described [52,53,54]. In certain infectious, inflammatory, and allergic diseases, the missense variants TLR2 p.R753Q and TLR4 p.D299E, and the TLR9 promoter variant −1237C > T have been associated with increased signaling [58,59,60,61].
In 2018, Nedoszytko et al. examined the frequency of the TLR2 p.R753Q (rs5743708) variant, an intronic TLR4 variant 896A > G (rs496790), and the −1237C > T (rs5743836) TLR9 promotor variant in CM and SM patients—Table 5 [30]. In this study, the TLR2 p.R753Q variant was significantly more common among patients with mastocytosis compared to healthy controls and in particular those with systemic disease compared to controls. The presence of the TLR2 p.R753Q variant was also associated with a 2-fold increased risk for mastocytosis, and 4-fold risk for systemic disease. In contrast, the TLR4 and TLR9 variants examined were not associated with disease [30].
Table 5.
Summary of the published data on TLR genes polymorphisms in mastocytosis.
| Gene | Polymorphism | Effect on Gene/Protein Function | Effects on Risk of Mastocytosis and Prognosis | References |
|---|---|---|---|---|
| TLR2 | p.R753Q (rs5743708) | 753Q variant encode non-functional receptor | Presence of 753Q variant of TLR-2 gene increased the risk of systemic mastocytosis | [30] |
| TLR4 | 896A > G (rs496790) | intronic variant | Not increase mastocytosis risk | [30] |
| TLR9 | −1237C > T (rs5743836) | CC lower transcription rate | No association with mastocytosis | [30] |
3. mRNA Expression Studies in Patients with Mastocytosis
While few studies published to date have examined differences in gene expression patterns in bone marrow (BM) and peripheral blood cells from mastocytosis patients, a study by d’Ambrosio et al. [62] first suggested that mastocytosis patients have altered RNA expression profiles in BM cells. The most over-expressed gene products included genes encoding secreted tryptases, carboxypeptidase A, and proteins involved in regulation of transcription, proliferation, and apoptosis. The expression level of 3 genes (TPSAB1 encoding α- and β-tryptases, ATF3 encoding Activating transcription factor type 3, and MAFF encoding the Muscle aponeurotic fibrosarcoma type F oncogene) were significantly associated with serum tryptase levels and these 3 genes have been suggested as candidate molecular markers for systemic disease.
In a second similar study, Teodosio et al. [63] analyzed the gene expression profile in highly purified BM MC from ISM and ASM mastocytosis patients with KIT p.D816V. In comparison to controls, 758 genes were deregulated in ISM/ASM patients. Patients with ISM had fewer deregulated genes (n = 479) than patients with ASM (n = 677); more than half (398/758) of these genes were shared by both groups of patients. The most deregulated genes were KIT, CD25, TPSAB1, and gene products involved in the innate and inflammatory immune response including IL-1 receptor type 1 (IL1R1), CCL23, and CD4, as well as interferon-induced gene products and gene products involved in cellular responses to viral antigens, together with complement inhibitory molecules and genes involved in lipid metabolism and protein processing.
Purified BM MC from patients with ISM have been found to have a number of genes upregulated that are classically associated with signaling pathways involved in MC degranulation, such as Linker for activation of T cells, member 2 (LAT2) and CD33 encoding Siglec-3, as well as significant enrichment in genes involved in cell adhesion including Cadherin 12 (CDH12) and CD81 encoding tetraspanin-28. By contrast, purified BM MCs from patients with ASM displayed upregulation of genes principally related to inflammation and apoptosis, including Caspase 1 (CASP1), Caspase 10 (CASP10), XIAP associated factor 1 (XAF1), Tumor necrosis factor receptor superfamily member 10B (TNFRSF10B), Interleukin 1 beta (IL-1B), and Oncostatin M (OSM) [63].
More recently, Niedoszytko et al. [64] have compared global gene expression in peripheral blood cells obtained from ISM patients to those from healthy controls. A total of 2330 mRNA transcripts were found to be differentially expressed in leukocytes from ISM patients compared to controls. Of these, 1951 gene products (84%) were upregulated in ISM patients and 379 (16%) were downregulated. The main pathways in which the differentially expressed mRNA were identified included ubiquitin-mediated proteolysis, as well as JAK-STAT, MAPK, and p53 signaling pathways. In another study significant differences were reported among ISM patient with insect venom anaphylaxis as compared to those without anaphylaxis in history [65].
Finally, in a study by Górska et al. [66] of gene expression in mastocytosis patients, increased expression of TRAF4 mRNA was associated with food hypersensitivity and decreased expression of B3GAT1 mRNA was associated with insect venom allergy.
In all these studies, the numbers of patients were limited. Therefore, the clinical impact of these gene products and their specific influence on clinical variables, presentations and courses remains uncertain. In addition, all these studies were retrospective in nature. Therefore, prospective studies including higher numbers of patients are required to draw definitive conclusions regarding the clinical and prognostic impact of these preliminary findings.
4. Missense MRGPRX2, ADGRE2, and PLCG2 Genes Variants and Mast Cell Activation
Two primary pathways have been identified in MC activation: Immunoglobulin E (IgE)-dependent and IgE-independent pathways. In the IgE-independent pathway, MCs are activated by stimulation of various receptors: IL6R, IL18R, ST2, TLR, KIT, histamine receptors, IgGR, C3aR, C5aR, and the G protein-coupled receptors (GPCRs) ADREG2 and MRGPRX2, though many of these pathways are insufficient to result in MC degranulation [56,67,68,69,70,71,72,73,74,75,76,77,78].
MRGPRX2 (Mas-related G protein-coupled receptor) is activated by different ligands including but not limited to: substance P, MC degranulating peptide, neuropeptide Y, VIP, somatostatin, compound 48/80, fluoroquinolone antibiotics, as well as the antimicrobial peptides LL37 and HBD, and insect venom peptides melittin and vespid mastoparan [69,70,71,72]. Recently, gain- and loss-of-function variants in MRGPRX2 were found to impact MC degranulation in vitro, in response to substance P and other peptides [73,74]. While it has been suggested that quinolones are potential triggers of anaphylaxis in mastocytosis [74,75], these variants have yet to be shown to impact clinical phenotypes in humans.
ADGRE2 (Adhesion G protein–coupled receptor E2) is a mechanoreceptor present on MC. The ADGRE2 p.C492Y missense has been shown to result in a severe autosomal dominant form of vibratory urticaria. In patients with this variant, vibration-induced activation of MC results in diffuse tryptase staining in skin upon histologic staining, and the rapid increase in histamine levels in venous blood from challenged areas [76].
Another study by Ombrello et al. [77] showed that variants of PLCG2 gene encoding Phospholipase Cγ2 (PLCγ2) are involved in the pathogenesis of cold-induced urticaria. Cold-induced urticaria is a unique inflammatory disorder that is characterized by MC degranulation and triggered by exposure to cold stimuli, a condition that can culminate in life-threatening anaphylaxis [77,78]. In three examined families with this disease the authors showed that deletions of auto-regulatory domains of PLCG2 resulted in protein products with constitutive phospholipase activity under cold temperature conditions, while they remained hippomorphic at physiologic conditions [78]. This unusual dichotomy resulted in impaired humoral immunity and enhanced MC and granulocyte-mediated cutaneous inflammation [79].
Variants in genes such as MRGPRX2, ADGRE2 and PLCG2 have not been extensively examined among individuals with mastocytosis. However, we consider these molecules and their pathologic variants as candidates and promising targets mediating MC activation in patients with CM or SM. Whether indeed these molecules play a major role in MC activation or MCAS and whether indeed these targets can be exploited to establish new therapies against MC activation in mastocytosis contexts remains to be examined in forthcoming studies.
5. Hereditary Alpha-Tryptasemia in Clonal MC Disease
In 2014, it was first recognized by Lyons and colleagues that elevated basal serum tryptase (BST) levels could be inherited in humans in an autosomal dominant manner [80]. In that study, elevated BST was associated with multisystem complaints, including symptoms of MC mediator release such as abdominal pain, bloating, diarrhea, skin itching, flushing—and in some cases urticaria—as well as an increased prevalence of anaphylaxis in affected individuals. In a follow-up study in 2016 [81], the investigators went on to describe the basis for this finding, a common genetic trait they termed hereditary alpha-tryptasemia (HαT). HαT is caused by increased α-tryptase encoding TPSAB1 copy number [82] and is a common genetic cause for elevated BST in Western populations, affecting between 4–6% of all individuals [81,83,84,85]. Among these individuals with HαT a gene dosage effect has also been observed, where increasing TPSAB1 copy numbers are associated with higher BST levels and more prevalent clinical symptoms. Increasing ratios of α-tryptase to β-tryptase encoding genes also correspond to the portion of active tryptase in mast cells accounted for by α/β-tryptase heterotetramers; such heterotetramers (but not the homotetramers) are capable of activating protease-activated receptor-2 and ADGRE2, which in turn may contribute to aspects of anaphylactic severity [84,86].
In initial studies, 16% of affected individuals in families recruited with HαT reported systemic anaphylaxis to Hymenoptera envenomation (HVA). Moreover, in a small validation cohort of ostensibly healthy adult volunteers, the relative risk for HVA among individuals with HαT was found to be 9.1 [81]. In order to validate this association, two large cohorts of venom allergic patients from Italy and Slovenia were tryptase-genotyped, and in both cohorts, the prevalence of HαT among individuals with severe HVA (grade IV on the Mueller scale) was found to be at least twice that of the general population and that of those with less severe anaphylaxis (grades I–III) [84]. Importantly, the prevalence of HαT was not found to be increased among venom allergic patients en masse, indicating that HαT is not likely to affect sensitization or peripheral tolerance, rather, that among those who are allergic, reactions are more likely to be severe. Whether anaphylaxis resulting from other allergens is similarly modified by HαT in healthy individuals has not yet been determined. However, individuals with idiopathic anaphylaxis in the absence of clonal MC disease have been reported to be three times as likely to have HαT, suggesting that this may be a common phenomenon. Indeed, proposed mechanisms—involving specific cleavage and activation of EMR2 on MC cells and PAR2 on endothelial cells by mature α/β-tryptase heterotetramers—would appear to be generalizable [84,86].
The possibility that HαT might modify clonal MC disease was first suggested following the identification of a highly symptomatic family with HαT in whom the first TPSAB1 quintuplication was found [87]. In this family, one affected individual also had evidence of clonal MC disease, harboring the activating KIT p.D816V missense. The authors had previously noted that HαT was associated with increased MC numbers in the BM of affected individuals [82] and surmised that since a TPSAB1 quintuplication is a rare finding, also identifying similarly rare clonal MC disease in a single individual was unlikely to be happenstance. Thus, the investigators went on to examine the prevalence of HαT in clonal MC disease finding that 12.2% of individuals with SM followed at the U.S. National Institutes of Health (NIH) had concomitant HαT, a rate more than twice that of the general population [84]. In a follow-up study, performed in collaboration between the Gdansk and Vienna European Competence Network on Mastocytosis (ECNM) centers, this association was validated, and an even greater prevalence was seen [83]. A total of 17.2% of the 241 individuals with SM included in the study were found to have HαT. In both the U.S. and European studies, HαT appeared to modify the clinical course and phenotype associated with mastocytosis, making anaphylaxis more prevalent and more severe [83,84]. Moreover, in the latter study—much like what was reported in the initial description of HαT—investigators observed a gene dosage effect on clinical symptoms [83]. Using a validated mediator symptom severity grading system, Greiner and colleagues found that with increasing α-tryptase-encoding TPSAB1 copy, more pervasive symptoms and higher mediator-associated symptom grades were reported by their patients [83].
Data remain limited about HαT-associated augmentation of MC mediator production, release, and/or responsiveness in patients with allergies and MC disorders. However, IgE-mediated degranulation of primary MCs cultured from individuals with HαT has not been shown to be different from controls [81]. Given the finding of increased bone marrow MC in the absence of clonal disease among individuals with HαT, one could consider whether HαT may facilitate or promote the evolution of this clonal myeloid disorder. However, because HαT significantly modifies clinical phenotypes associated with clonal MC disease, its increased prevalence among patients with SM may also reflect a detection bias, wherein those with both HαT and mastocytosis are more likely to come to medical attention because of increased symptom severity. As additional clinical, epidemiologic, and basic research is conducted on this common genetic trait, more insights will be gained into the role that HαT plays in MC expansion and reactivity in SM and atopic disorders.
6. KIT Variants and Their Role in the Pathogenesis of Mastocytosis
KIT is activated by stem cell factor (SCF) and is a type III transmembrane receptor with intrinsic tyrosine kinase (TK) activity in its intracellular domain. KIT, which is expressed by germ cells, Cajal cells, melanoblasts, hematopoietic stem cells (HSC) and MC, is deeply involved in hematopoiesis and mastopoiesis. In particular, KIT activity plays a crucial role in the survival and proliferation of HSC and in the development of MC [88]. In the MC lineage, KIT is thus expressed throughout differentiation and appears to play a critical role in the differentiation, proliferation, survival and migration of MC and their progenitors [88].
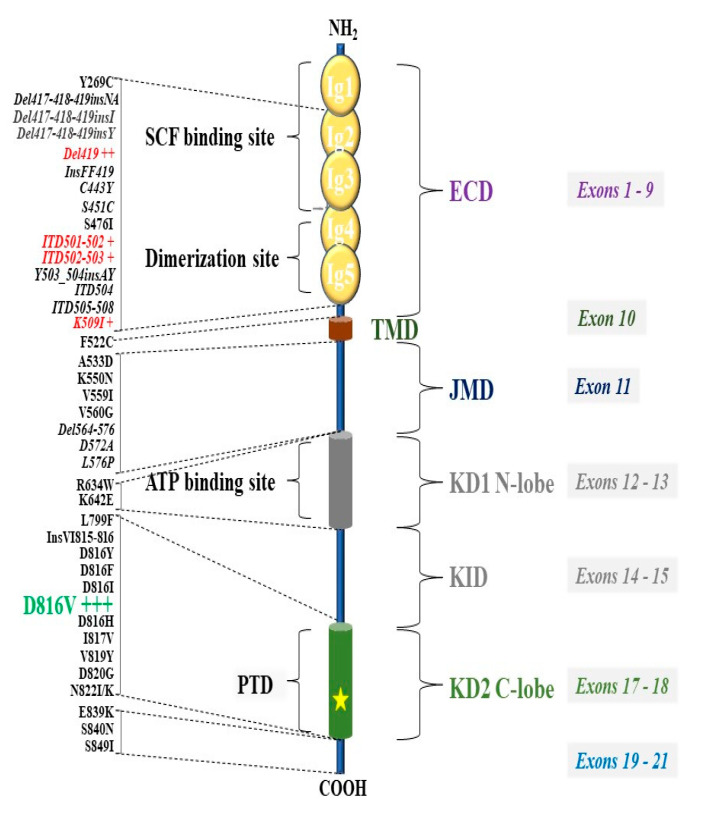
KIT mutations are thought to be involved in the pathogenesis of the majority—if not all—variants of mastocytosis. Indeed, in CM as well as in SM, disease pathogenesis is considered to be associated with the presence of acquired gain-of-function variants in the KIT gene. Most of these mutations lead to constitutive activation of the KIT receptor and thus autonomous (SCF-independent) development and accumulation of MC [89]. The KIT variants detected in mastocytosis can be classified into two categories. The first category includes regulatory variants, affecting the dimerization or conformational self-regulation of the receptor. These variants are often found in pediatric mastocytosis and are typically located in the extracellular domain (ECD) encoded by exons 2, 8 and 9 of the KIT gene [90]. The second category of variants occurs at the intracellular portion of the receptor, in the second kinase domain (KD2) encoded by exon 17 of the KIT gene [14].
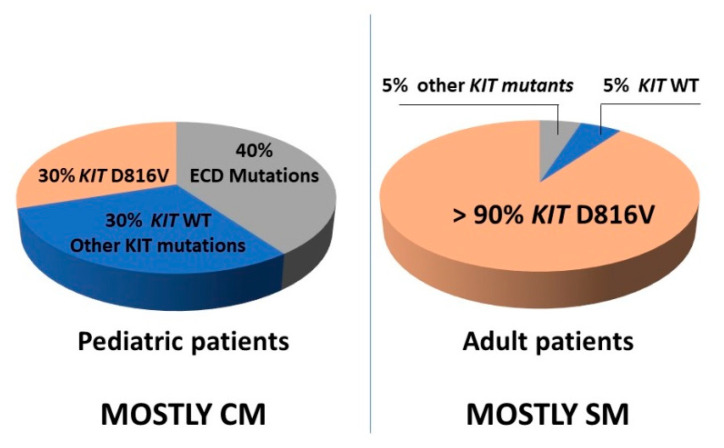
The KIT variant most frequently encountered in adults with SM (more than 80% of all SM cases and >90% of patients with ISM) is KIT p.D816V, which occurs in the KD2 of KIT [91]. Other KIT mutations are found less frequently in SM patients. Such alternative KIT mutations are particularly detected in patients with ASM, MCL, or MC sarcoma (MCS)—these variants are often located in the juxtamembrane domain (JMD), such as KIT p.V560G, or in the ECD of the receptor, such as KIT p.Del419 [92]. Finally, some patients with SM do not have identifiable variants in KIT (KIT wild type; KIT WT); this is particularly the case in the nearly 70% of patients who present with well-differentiated SM (WDSM), a subset of ISM characterized by the presence of compact multifocal infiltrates of round mature, CD2- and CD25-negative MC in BM [93].
The KIT p.D816V mutation is found in only ~30% of children with CM, while other KIT variants, located mainly in the ECD of the receptor, are found in ~40% of these cases [90]. An updated list of KIT variants described in mastocytosis is provided in Figure 1, and a comparison of the frequency of different KIT variants found in adults vs children is shown in Figure 2.
Figure 1.
KIT variants reported in mastocytosis. Variants frequently observed in pediatric patients with CM are shown in red, whereas the KIT p.D816V mutation (pathogenic variant) recurrently found in adult SM patients is shown in green. Abbreviation: ECD: Extracellular domain; JMD: Juxtamembrane domain; KID: Kinase insert domain; PTD: phosphotransferase domain; KD1, tyrosine kinase domains; TMD: transmembrane domain.
Figure 2.
Frequency of KIT variants in pediatric and adult patients with mastocytosis. Abbreviation: CM: cutaneous mastocytosis; ECD: Extracellular domain; KIT WT: KIT wild-type; SM: systemic mastocytosis.
In adult SM patients, KIT variants may be found in hematopoietic cells other than MCs, suggesting involvement of other cell lineages and their progenitors in these cases [94]. This condition is more frequently seen in patients with smoldering SM (SSM) and advanced SM, and is associated with a higher risk of disease progression [95]. In patients with SM-AHN, the KIT p.D816V variant is expressed not only in MC but can also be detected in other cell lineages affected by the AHN [96].
Even today, the mechanism(s) of the constitutive activation of the KIT mutant receptors is (are) not yet fully understood, particularly for KIT p.D816V. However, several hypotheses have been proposed, such as: (a) homologous or heterologous spontaneous dimerization of the receptor in the absence of SCF; (b) triggering of the signaling cascade by a single monomer or; (c) dysfunction of the JMD and of its self-inhibitory action [97].
Interestingly, there are several discrepancies between wild-type (WT) KIT and mutant KIT p.D816V receptor signaling, which suggest the existence of molecular differences specific to oncogenic signaling. For instance, following the activation of WT KIT, a number of negative regulatory mechanisms and pathways are canonically activated. These regulatory feed-back mechanisms involve diverse pathways, such as certain phosphatases, endocytosis-related mechanisms, and degradation of receptor molecules [98]. In mastocytosis, many of these regulatory systems have been shown to no longer control KIT signaling [98]. Mutated KIT receptor—which is constitutively active—is usually a cytoplasmic molecule and in contrast to WT KIT, is not expressed on the cell surface. The KIT p.D816V mutant receptor has the ability to escape inhibitory effects of phosphatases. Indeed, the phosphatase SHP-1 (Src Homology region domain-containing Phosphatase-1), involved in the negative regulation of KIT, seems to be degraded rapidly in a mouse cell model exhibiting the Kit p.D814Y variant (equivalent to the human KIT p.D816V mutation) [99].
With regard to the oncogenic pathway evoked by KIT p.D816V, two pathways seem to play a central role in the pathogenesis of clonal disease in patients with KIT p.D816V SM, i.e., the STAT5 and the PI3K/AKT pathways [100,101]. STAT5 is constitutively activated in neoplastic cells from patients with KIT p.D816V-positive SM, as reflected by the presence of phospho-STAT5 (pSTAT5) at the nuclear and cytoplasmic level. Of note, pSTAT5 is found bound to p85 (PI3K regulatory subunit) by Gab2 in the cytoplasm of neoplastic MC [101]. In vitro, in two human neoplastic MC lines harboring the KIT p.D816V variant, namely HMC-1.2 and ROSAKIT D816V, not only STAT5 but also AKT have been found to be constitutively activated [102,103].
In addition, other kinases appear involved in the oncogenic signaling resulting from the KIT p.D816V mutation. Indeed, the FES tyrosine kinase (“FEline Sarcoma”) seems to be a major effector of proliferation through its role as a regulator of STAT and mTOR signaling, whereas it is not involved in the proliferation mediated by WT KIT [104,105]. Its activity is increased in malignant MC and it phosphorylates ribosomal protein kinase S6 (P70S6K), a serine/threonine kinase which impacts several genes regulating cell growth, survival, and metabolism [106].
Apart from KIT p.D816V, neoplastic MC may express a number of other molecules believed to be involved in their abnormal expansion and accumulation. Indeed, neoplastic MC may also overexpress LYN and BTK, Oncostatin M (OSM), NF-κB, and several anti-apoptotic molecules, such as Microphtalmia-associated Transcription Factor (MITF) and the BCL2 Family Member MCL-1 [107,108,109]. Finally, it has been reported that in neoplastic MC the expression of pro-apoptotic molecules such as BIM are decreased [110]. All these events may contribute to the pathological accumulation of MC in mastocytosis.
7. Role of Additional Genetic Variants (Mutations) as Drivers of Advanced SM
KIT p.D816V is considered to be a critical driver of SM. However, the presence of this mutation alone does not explain the diverse clinical levels of aggressiveness of clinical variants of SM [95].
Recent studies have shown that in >60% of all patients with advanced SM (ASM, SM-AHN or MCL), neoplastic cells harbor somatic variants in relevant genes other than KIT [111,112,113,114,115,116,117]. These additional mutations affect genes encoding transcription factors (e.g., RUNX1), signaling molecules (JAK, KRAS, NRAS, CBL), epigenetic regulators (TET2, ASXL1, DNMT3A, EZH2) or splicing factors (SRSF2, SF3B1, U2AF1). In the study by Schwaab et al. [3], the most frequently affected genes were TET2, SRSF2, ASXL1, CBL, and RUNX1. In advanced SM, 21/27 patients (78%) carried ≥3 variants, and 11/27 patients (41%) exhibited ≥5 variants. Overall survival was significantly shorter in patients with additional aberrations as compared to those identified as having KIT p.D816V alone.
Several groups have investigated the prognostic relevance of additional variants in myeloid malignancy-related genes [111,112,113,114,115,116,117]. Jawhar et al. [111,112] have reported that variants in 3 critical genes, namely SRSF2, ASXL1, and RUNX1 (S/A/R gene panel) can influence overall survival and are thus the most important predictive indicators of a poor outcome in patients with SM. Finally, the presence and number of mutated genes within the S/A/R panel are associated with the presence of advanced SM, especially SM-AHN [112,116].
Munoz-Gonzales et al. have since demonstrated that in addition to the S/A/R gene panel, somatic EZH2 gene variants might also provide prognostic information and have proposed an S/A/R/E panel [116]. In the Mayo prognostic score, ASXL1, RUNX1, and NRAS gene variants were found to be independent predictors of an inferior survival in SM [117]. Moreover, additional variants in the genes ASXL1, RUNX1, and DNMT3A have also been described as prognostic factors in indolent SM [16].
In conclusion, the clinical and morphological diversity of SM is associated with the presence and number of somatic variants in certain target genes, and the respective gene products are considered to play a role in the pathogenesis, aggressiveness and poor prognosis of patients with advanced SM. For this reason, the current consensus is that molecular signatures should be determined in all patients with SM.
8. Conclusions
Molecular studies in patients with mastocytosis have demonstrated a multifactorial genetic basis with disease being dictated by somatic mutations in critical driver genes—including recurrent gain-of-function mutations in KIT—and expressivity being significantly modified by heritable genetic modifiers, including hereditary alpha-tryptasemia. In more than 80% of all patients with SM, neoplastic cells harbor a somatic mutation in KIT at codon 816, mostly KIT p.D816V. In advanced SM, additional variants in other critical genes encoding transcription factors, signaling molecules, epigenetic regulators and splicing factors, may contribute to disease evolution and the aggressiveness of SM. Based on such additional somatic mutations, clinical variables and the WHO classification, new prognostic scores have been developed, which may help improve prognostication and management of individual patients with SM.
Abbreviations
| ADGRE2 | Adhesion G protein–coupled receptor E2 |
| ASM | Aggressive Systemic mastocytosis |
| ASXL1 | ASXL Transcriptional Regulator 1 |
| BST | basal serum tryptase |
| CBL | Cbl Proto-Oncogene |
| CM | Cutaneous mastocytosis |
| DNMT3A | DNA Methyltransferase 3 Alpha |
| ECD | Extracellular domain of KIT |
| EZH2 | Enhancer Of Zeste 2 Polycomb Repressive Complex 2 Subunit |
| HSC | hematopoietic stem cells |
| HαT | hereditary alpha-tryptasemia |
| HVA | Hymenoptera venom allergy |
| ISM | Indolent systemic mastocytosis |
| JMD: | Juxtamembrane domain of KIT |
| KID | Kinase insert domain of KIT |
| KIT | KIT Proto-Oncogene, Receptor Tyrosine Kinase |
| MCL | Mast cell leukemia |
| MC | Mast cells |
| PTD | phosphotransferase domain of KIT |
| SM | systemic mastocytosis |
| SM-AHN | Systemic mastocytosis with associated hematologic neoplasm |
| MCS | Mast cell sarcoma |
| TPSAB1 | Genes encoding α- and β-tryptases |
| MCAS | MC activation syndrome |
| MRGPRX2 | Mas-related G protein-coupled receptor |
| NRAS | NRAS Proto-Oncogene, GTPase |
| PLCG2 | Phospholipase Cγ2 |
| RUNX1 | RUNX Family Transcription Factor 1 |
| SCF | Stem cell factor |
| SF3B1 | Splicing Factor 3b Subunit 1 |
| SRSF2 | Serine And Arginine Rich Splicing Factor 2 |
| TET2 | Tet Methylcytosine Dioxygenase 2 |
| TLR | Toll-like receptor |
| U2AF1 | U2 Small Nuclear RNA Auxiliary Factor 1 |
| WDSM | well-differentiated systemic mastocytosis |
Author Contributions
All co-authors contributed by drafting parts of the manuscript. All co-authors approved the final version of the document. All authors have read and agreed to the published version of the manuscript.
Funding
This project was supported by the Polish Ministry of Science and Higher Education—grant no ST 02-0066/07/253 and by the Austrian Science Fund (FWF)—grants F4704-B20 and P32470-B. This research was also supported in part by the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases, NIH. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Conflicts of Interest
The authors declare that they have no conflict of interest in this study.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Födinger M., Fritsch G., Winkler K., Emminger W., Mitterbauer G., Gadner H., Valent P., Mannhalter C. Origin of human mast cells: Development from transplanted hematopoietic stem cells after allogeneic bone marrow transplantation. Blood. 1994;84:2954–2959. doi: 10.1182/blood.V84.9.2954.2954. [DOI] [PubMed] [Google Scholar]
- 2.Eisenwort G., Sadovnik I., Schwaab J., Jawhar M., Keller A., Stefanzl G., Berger D., Blatt K., Hoermann G., Bilban M., et al. Identification of a leukemia-initiating stem cell in human mast cell leukemia. Leukemia. 2019;33:2673–2684. doi: 10.1038/s41375-019-0460-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwaab J., Schnittger S., Sotlar K., Walz C., Fabarius A., Pfirrmann M., Kohlmann A., Grossmann V., Meggendorfer M., Horny H.-P., et al. Comprehensive mutational profiling in advanced systemic mastocytosis. Blood. 2013;122:2460–2466. doi: 10.1182/blood-2013-04-496448. [DOI] [PubMed] [Google Scholar]
- 4.Jawhar M., Schwaab J., Schnittger S., Sotlar K., Horny H.P., Metzgeroth G., Müller N., Schneider S., Naumann N., Walz C., et al. Molecular profiling of myeloid progenitor cells in multi-mutated advanced systemic mastocytosis identifies KIT D816V as a distinct and late event. Leukemia. 2015;29:1115–1122. doi: 10.1038/leu.2015.4. [DOI] [PubMed] [Google Scholar]
- 5.Yun H.D., Antony M.L., Linden M.A., Noble-Orcutt K.E., Eckfeldt C., Ustun C., Nelson A.C., Sachs Z. Evolution of clonal dynamics and differential response to targeted therapy in a case of systemic mastocytosis with associated myelodysplastic syndrome. Leuk. Res. 2020;95:106404. doi: 10.1016/j.leukres.2020.106404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valent P., Horny H.-P., Escribano L., Longley B., Li C.Y., Schwartz L.B., Marone G., Nuñez R., Akin C., Sotlar K., et al. Diagnostic criteria and classification of mastocytosis: A consensus proposal. Leuk. Res. 2001;25:603–625. doi: 10.1016/S0145-2126(01)00038-8. [DOI] [PubMed] [Google Scholar]
- 7.Valent P., Sperr W.R., Schwartz L.B., Horny H.-P. Diagnosis and classification of mast cell proliferative disorders: Delineation from immunologic diseases and non–mast cell hematopoietic neoplasms. J. Allergy Clin. Immunol. 2004;114:3–11. doi: 10.1016/j.jaci.2004.02.045. [DOI] [PubMed] [Google Scholar]
- 8.Valent P., Akin C., Escribano L., Födinger M., Hartmann K., Brockow K., Castells M., Sperr W.R., Kluin-Nelemans H.C., Hamdy N.A.T., et al. Standards and standardization in mastocytosis: Consensus Statements on Diagnostics, Treatment Recommendations and Response Criteria. Eur. J. Clin. Investig. 2007;37:435–453. doi: 10.1111/j.1365-2362.2007.01807.x. [DOI] [PubMed] [Google Scholar]
- 9.Horny H.-P., Sotlar K., Valent P. Mastocytosis: State of the Art. Pathobiology. 2007;74:121–132. doi: 10.1159/000101711. [DOI] [PubMed] [Google Scholar]
- 10.Horny H.P., Akin C., Arber D., Peterson L.A., Tefferi A., Metcalfe D.D., Bennett J.M., Bain B., Escribano L., Valent P. Mastocytosis. In: Swerdlow S.H., Campo E., Harris N.L., Jaffe E.S., Pileri S.A., Stein H., Thiele J., Arber D.A., Hasserjian R.P., Le Beau M.M., et al., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Volume 3. IARC Press; Lyon, France: 2017. pp. 62–69. [Google Scholar]
- 11.Valent P., Akin C., Metcalfe D.D. Mastocytosis: 2016 updated WHO classification and novel emerging treatment concepts. Blood. 2017;129:1420–1427. doi: 10.1182/blood-2016-09-731893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georgin-Lavialle S., Lhermitte L., Dubreuil P., Chandesris M.-O., Hermine O., Damaj G. Mast cell leukemia. Blood. 2013;121:1285–1295. doi: 10.1182/blood-2012-07-442400. [DOI] [PubMed] [Google Scholar]
- 13.Ustun C., Arock M., Kluin-Nelemans H.C., Reiter A., Sperr W.R., George T., Horny H.-P., Hartmann K., Sotlar K., Damaj G., et al. Advanced systemic mastocytosis: From molecular and genetic progress to clinical practice. Haematologica. 2016;101:1133–1143. doi: 10.3324/haematol.2016.146563. [DOI] [PubMed] [Google Scholar]
- 14.Arock M., Sotlar K., Akin C., Broesby-Olsen S., Hoermann G., Escribano L., Kristensen T.K., Kluin-Nelemans H.C., Hermine O., Dubreuil P., et al. KIT mutation analysis in mast cell neoplasms: Recommendations of the European Competence Network on Mastocytosis. Leukemia. 2015;29:1223–1232. doi: 10.1038/leu.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montero A.G., Jara-Acevedo M., Teodosio C., Sánchez-Muñoz L., Núñez R., Prados A., Aldanondo I., Domínguez M., Botana L.M., Sánchez-Jiménez F., et al. KIT mutation in mast cells and other bone marrow hematopoietic cell lineages in systemic mast cell disorders: A prospective study of the Spanish Network on Mastocytosis (REMA) in a series of 113 patients. Blood. 2006;108:2366–2372. doi: 10.1182/blood-2006-04-015545. [DOI] [PubMed] [Google Scholar]
- 16.Muñoz-González J.I., Álvarez-Twose I., Jara-Acevedo M., Henriques A., Viñas E., Prieto C., Sánchez-Muñoz L., Caldas C., Mayado A., Matito A., et al. Frequency and prognostic impact of KIT and other genetic variants in indolent systemic mastocytosis. Blood. 2019;134:456–468. doi: 10.1182/blood.2018886507. [DOI] [PubMed] [Google Scholar]
- 17.Teodosio C., García-Montero A.C., Jara-Acevedo M., Álvarez-Twose I., Sánchez-Muñoz L., Almeida J., Morgado J.M., Matito A., Escribano L., Orfao A. An immature immunophenotype of bone marrow mast cells predicts for multilineage D816V KIT mutation in systemic mastocytosis. Leukemia. 2011;26:951–958. doi: 10.1038/leu.2011.293. [DOI] [PubMed] [Google Scholar]
- 18.Kristensen T.K., Broesby-Olsen S., Vestergaard H., Bindslev-Jensen C., Moller M.B., on behalf of the Mastocytosis Centre Odense University Hospital (MastOUH) Circulating KITD816V mutation-positive non-mast cells in peripheral blood are characteristic of indolent systemic mastocytosis. Eur. J. Haematol. 2012;89:42–46. doi: 10.1111/j.1600-0609.2012.01789.x. [DOI] [PubMed] [Google Scholar]
- 19.Hartmann K., Escribano L., Grattan C., Brockow K., Carter M.C., Alvarez-Twose I., Matito A., Broesby-Olsen S., Siebenhaar F., Lange M., et al. Cutaneous manifestations in patients with mastocytosis: Consensus report of the European Competence Network on Mastocytosis; the American Academy of Allergy, Asthma & Immunology; and the European Academy of Allergology and Clinical Immunology. J. Allergy Clin. Immunol. 2016;137:35–45. doi: 10.1016/j.jaci.2015.08.034. [DOI] [PubMed] [Google Scholar]
- 20.Matito A., Azaña J.M., Torrelo A., Álvarez-Twose I. Cutaneous Mastocytosis in Adults and Children. Immunol. Allergy Clin. N. Am. 2018;38:351–363. doi: 10.1016/j.iac.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 21.Lange M., Nedoszytko B., Górska A., Zawrocki A., Sobjanek M., Kozlowski D. Mastocytosis in children and adults: Clinical disease heterogeneity. Arch. Med. Sci. 2012;8:533–541. doi: 10.5114/aoms.2012.29534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lange M., Niedoszytko M., Renke J., Gleń J., Nedoszytko B. Clinical aspects of paediatric mastocytosis: A review of 101 cases. J. Eur. Acad. Dermatol. Venereol. 2011;27:97–102. doi: 10.1111/j.1468-3083.2011.04365.x. [DOI] [PubMed] [Google Scholar]
- 23.Valent P., Akin C., Arock M., Brockow K., Butterfield J.H., Carter M.C., Castells M., Escribano L., Hartmann K., Lieberman P., et al. Definitions, Criteria and Global Classification of Mast Cell Disorders with Special Reference to Mast Cell Activation Syndromes: A Consensus Proposal. Int. Arch. Allergy Immunol. 2011;157:215–225. doi: 10.1159/000328760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valent P. Mast cell activation syndromes: Definition and classification. Allergy. 2013;68:417–424. doi: 10.1111/all.12126. [DOI] [PubMed] [Google Scholar]
- 25.Valent P., Akin C., Bonadonna P., Hartmann K., Brockow K., Niedoszytko M., Nedoszytko B., Siebenhaar F., Sperr W.R., Elberink J.N.O., et al. Proposed Diagnostic Algorithm for Patients with Suspected Mast Cell Activation Syndrome. J. Allergy Clin. Immunol. Pract. 2019;7:1125–1133.e1. doi: 10.1016/j.jaip.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daley T., Metcalfe D.D., Akin C. Association of the Q576R polymorphism in the interleukin-4 receptor α chain with indolent mastocytosis limited to the skin. Blood. 2001;98:880–882. doi: 10.1182/blood.V98.3.880. [DOI] [PubMed] [Google Scholar]
- 27.Nedoszytko B., Niedoszytko M., Lange M., Van Doormaal J., Gleń J., Zabłotna M., Renke J., Vales A., Buljubasic F., Jassem E., et al. Interleukin-13promoter gene polymorphism -1112C/T is associated with the systemic form of mastocytosis. Allergy. 2009;64:287–294. doi: 10.1111/j.1398-9995.2008.01827.x. [DOI] [PubMed] [Google Scholar]
- 28.Lange M., Gleń J., Zabłotna M., Nedoszytko B., Sokołowska-Wojdyło M., Rębała K., Ługowska-Umer H., Niedoszytko M., Górska A., Sikorska M., et al. Interleukin-31 Polymorphisms and Serum IL-31 Level in Patients with Mastocytosis: Correlation with Clinical Presen-tation and Pruritus. Acta Derm. Venereol. 2017;97:47–53. doi: 10.2340/00015555-2474. [DOI] [PubMed] [Google Scholar]
- 29.Rausz E., Szilágyi Á., Nedoszytko B., Lange M., Niedoszytko M., Lautner-Csorba O., Falus A., Aladzsity I., Kókai M., Valent P., et al. Comparative analysis of IL6 and IL6 receptor gene polymorphisms in mastocytosis. Br. J. Haematol. 2013;160:216–219. doi: 10.1111/bjh.12086. [DOI] [PubMed] [Google Scholar]
- 30.Nedoszytko B., Lange M., Renke J., Niedoszytko M., Zabłotna M., Gleń J., Nowicki R. The Possible Role of Gene Variant Coding Nonfunctional Toll-Like Receptor 2 in the Pathogenesis of Mastocytosis. Int. Arch. Allergy Immunol. 2018;177:80–86. doi: 10.1159/000489343. [DOI] [PubMed] [Google Scholar]
- 31.Pulendran B., Artis D. New Paradigms in Type 2 Immunity. Science. 2012;337:431–435. doi: 10.1126/science.1221064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seyfizadeh N., Seyfizadeh N., Gharibi T., Babaloo Z. Interleukin-13 as an important cytokine: A review on its roles in some human diseases. Acta Microbiol. Immunol. Hung. 2015;62:341–378. doi: 10.1556/030.62.2015.4.2. [DOI] [PubMed] [Google Scholar]
- 33.Laporte S.L., Juo Z.S., Vaclavikova J., Colf L.A., Qi X., Heller N.M., Keegan A.D., Garcia K.C. Molecular and Structural Basis of Cytokine Receptor Pleiotropy in the Interleukin-4/13 System. Cell. 2008;132:259–272. doi: 10.1016/j.cell.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaur D., Hollins F., Woodman L., Yang W., Monk P., May R., Bradding P., Brightling C.E. Mast cells express IL-13R alpha 1: IL-13 promotes human lung mast cell proliferation and Fc epsilon RI expression. Allergy. 2006;61:1047–1053. doi: 10.1111/j.1398-9995.2006.01139.x. [DOI] [PubMed] [Google Scholar]
- 35.McLeod J.J., Baker B., Ryan J.J. Mast cell production and response to IL-4 and IL-13. Cytokine. 2015;75:57–61. doi: 10.1016/j.cyto.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toru H., Pawankar R., Ra C., Yata J., Nakahata T. Human mast cells produce IL-13 by high-affinity IgE receptor cross-linking: Enhanced IL-13 production by IL-4–primed human mast cells. J. Allergy Clin. Immunol. 1998;102:491–502. doi: 10.1016/S0091-6749(98)70140-X. [DOI] [PubMed] [Google Scholar]
- 37.Potaczek D.P., Kabesch M. Current concepts of IgE regulation and impact of genetic determinants. Clin. Exp. Allergy. 2012;42:852–871. doi: 10.1111/j.1365-2222.2011.03953.x. [DOI] [PubMed] [Google Scholar]
- 38.Cornelissen C.G., Lüscher-Firzlaff J., Baron J.M., Lüscher B. Signaling by IL-31 and functional consequences. Eur. J. Cell Biol. 2012;91:552–566. doi: 10.1016/j.ejcb.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 39.Rabenhorst A., Hartmann K. Interleukin-31: A Novel Diagnostic Marker of Allergic Diseases. Curr. Allergy Asthma Rep. 2014;14:1–7. doi: 10.1007/s11882-014-0423-y. [DOI] [PubMed] [Google Scholar]
- 40.Bilsborough J., Leung D.Y., Maurer M., Howell M., Boguniewicz M., Yao L., Storey H., LeCiel C., Harder B., Gross J.A. IL-31 is associated with cutaneous lymphocyte antigen–positive skin homing T cells in patients with atopic dermatitis. J. Allergy Clin. Immunol. 2006;117:418–425. doi: 10.1016/j.jaci.2005.10.046. [DOI] [PubMed] [Google Scholar]
- 41.Ishii T., Wang J., Zhang W., Mascarenhas J., Hoffman R., Dai Y., Wisch N., Xu M. Pivotal role of mast cells in pruritogenesis in patients with myeloproliferative disorders. Blood. 2009;113:5942–5950. doi: 10.1182/blood-2008-09-179416. [DOI] [PubMed] [Google Scholar]
- 42.Niyonsaba F., Ushio H., Hara M., Yokoi H., Tominaga M., Takamori K., Kajiwara N., Saito H., Nagaoka I., Ogawa H., et al. Antimicrobial Peptides Human β-Defensins and Cathelicidin LL-37 Induce the Secretion of a Pruritogenic Cytokine IL-31 by Human Mast Cells. J. Immunol. 2010;184:3526–3534. doi: 10.4049/jimmunol.0900712. [DOI] [PubMed] [Google Scholar]
- 43.Raap U., Wichmann K., Bruder M., Ständer S., Wedi B., Kapp A., Werfel T. Correlation of IL-31 serum levels with severity of atopic dermatitis. J. Allergy Clin. Immunol. 2008;122:421–423. doi: 10.1016/j.jaci.2008.05.047. [DOI] [PubMed] [Google Scholar]
- 44.Hartmann K., Wagner N., Rabenhorst A., Pflanz L., Leja S., Förster A., Gehring M., Kapp A., Raap U. Serum IL-31 levels are increased in a subset of patients with mastocytosis and correlate with disease severity in adult patients. J. Allergy Clin. Immunol. 2013;132:232–235. doi: 10.1016/j.jaci.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 45.Schulz F., Marenholz I., Fölster-Holst R., Chen C., Sternjak A., Baumgrass R., Esparza-Gordillo J., Gruber C., Nickel R., Schreiber S., et al. A common haplotype of the IL-31 gene influencing gene expression is associated with nonatopic eczema. J. Allergy Clin. Immunol. 2007;120:1097–1102. doi: 10.1016/j.jaci.2007.07.065. [DOI] [PubMed] [Google Scholar]
- 46.Sokołowska-Wojdyło M., Gleń J., Zabłotna M., Rębała K., Sikorska M., Florek A., Trzeciak M., Barańska-Rybak W., Malek M., Nedoszytko B. Association of distinct IL-31 polymorphisms with pruritus and severity of atopic dermatitis. J. Eur. Acad. Dermatol. Venereol. 2013;27:662–664. doi: 10.1111/j.1468-3083.2012.04649.x. [DOI] [PubMed] [Google Scholar]
- 47.Sokołowska-Wojdyło M., Gleń J., Zabłotna M., Rębała K., Trzeciak M., Sikorska M., Ruckemann-Dziurdzińska K., Msc B.N., Florek A., Nowicki R. The frequencies of haplotypes defined by three polymorphisms of theIL-31gene: −1066, −2057, and IVS2+12 in Polish patients with atopic dermatitis. Int. J. Dermatol. 2014;54:62–67. doi: 10.1111/ijd.12666. [DOI] [PubMed] [Google Scholar]
- 48.Brockow K., Akin C., Huber M., Scott L.M., Schwartz L.B., Metcalfe D.D. Levels of mast-cell growth factors in plasma and in suction skin blister fluid in adults with mastocytosis: Correlation with dermal mast-cell numbers and mast-cell tryptase. J. Allergy Clin. Immunol. 2002;109:82–88. doi: 10.1067/mai.2002.120524. [DOI] [PubMed] [Google Scholar]
- 49.Brockow K., Akin C., Huber M., Metcalfe D.D. IL-6 levels predict disease variant and extent of organ involvement in patients with mastocytosis. Clin. Immunol. 2005;115:216–223. doi: 10.1016/j.clim.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 50.Mayado A., Teodosio C., Garcia-Montero A.C., Matito A., Rodriguez-Caballero A., Morgado J.M., Muñiz C., Jara-Acevedo M., Álvarez-Twose I., Sanchez-Muñoz L., et al. Increased IL6 plasma levels in indolent systemic mastocytosis patients are associated with high risk of disease progression. Leukemia. 2016;30:124–130. doi: 10.1038/leu.2015.176. [DOI] [PubMed] [Google Scholar]
- 51.Theoharides T.C., Boucher W., Spear K. Serum interleukin-6 reflects disease severity and osteoporosis in mastocytosis patients. Int. Arch. Allergy Immunol. 2002;128:344–350. doi: 10.1159/000063858. [DOI] [PubMed] [Google Scholar]
- 52.Lange M., Renke J., Gleń J., Niedoszytko M., Nedoszytko B. Serum tryptase, interleukin 6 and SCORMA Index as disease severity parameters in childhood mastocytosis. Postępy Dermatol. Alergol. 2010;27:238. [Google Scholar]
- 53.Tobío A., Bandara G., Morris D.A., Kim D.K., O’Connell M.P., Komarow H.D., Carter M.C., Smrz D., Metcalfe D.D., Olivera A. Oncogenic D816V-KIT signaling in mast cells causes persistent IL-6 production. Haematologica. 2020;105:124–135. doi: 10.3324/haematol.2018.212126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valent P. KIT D816V and the cytokine storm in mastocytosis: Production and role of interleukin-6. Haematologica. 2020;105:5–6. doi: 10.3324/haematol.2019.234864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sandig H., Bulfone-Paus S. TLR signaling in mast cells: Common and unique features. Front. Immunol. 2012;3:185. doi: 10.3389/fimmu.2012.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olivera A., Beaven M.A., Metcalfe D.D. Mast cells signal their importance in health and disease. J. Allergy Clin. Immunol. 2018;142:381–393. doi: 10.1016/j.jaci.2018.01.034. [DOI] [PubMed] [Google Scholar]
- 57.Lin M., Yu B., Zhang W., Zhang W., Xiao Z., Mao Z., Lai Y., Lin D., Ma Q., Pan E., et al. Toll-like receptor 2-mediated MAPKs and NF-κB activation requires the GNAO1- dependent pathway in human mast cells. Integr. Biol. 2016;12:968–975. doi: 10.1039/c6ib00097e. [DOI] [PubMed] [Google Scholar]
- 58.Salpietro C., Rigoli L., Del Giudice M., Cuppari C., Di Bella C., Salpietro A., Maiello N., La Rosa M., Marseglia G.L., Leonardi S., et al. TLR2 and TLR4 gene polymorphisms and atopic dermatitis in Italian children: A multicenter study. Int. J. Immunopathol. Pharmacol. 2011;24:33–40. doi: 10.1177/03946320110240S408. [DOI] [PubMed] [Google Scholar]
- 59.Niebuhr M., Langnickel J., Draing C., Renz H., Kapp A., Werfel T. Dysregulation of toll-like receptor-2 (TLR-2)-induced effects in monocytes from patients with atopic dermatitis: Impact of the TLR-2 R753Q polymorphism. Allergy. 2008;63:728–734. doi: 10.1111/j.1398-9995.2008.01721.x. [DOI] [PubMed] [Google Scholar]
- 60.Mrabet-Dahbi S., Dalpke A.H., Niebuhr M., Frey M., Draing C., Brand S., Heeg K., Werfel T., Renz H. The Toll-like receptor 2 R753Q mutation modifies cytokine production and Toll-like receptor expression in atopic dermatitis. J. Allergy Clin. Immunol. 2008;121:1013–1019. doi: 10.1016/j.jaci.2007.11.029. [DOI] [PubMed] [Google Scholar]
- 61.Zabłotna M., Sobjanek M., Purzycka-Bohdan D., Szczerkowska-Dobosz A., Nedoszytko B., Nowicki R.J. The significance of Toll-like receptor (TLR) 2 and 9 gene polymorphisms in psoriasis. Adv. Dermatol. Allergol. 2017;34:85–86. doi: 10.5114/ada.2017.65628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.D’Ambrosio C., Akin C., Wu Y., Magnusson M.K., Metcalfe D.D. Gene expression analysis in mastocytosis reveals a highly consistent profile with candidate molecular markers. J. Allergy Clin. Immunol. 2003;112:1162–1170. doi: 10.1016/j.jaci.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 63.Teodosio C., García-Montero A.C., Jara-Acevedo M., Sánchez-Muñoz L., Pedreira C.E., Álvarez-Twose I., Matarraz S., Morgado J.M., Bárcena P., Matito A., et al. Gene expression profile of highly purified bone marrow mast cells in systemic mastocytosis. J. Allergy Clin. Immunol. 2013;131:1213–1224.e4. doi: 10.1016/j.jaci.2012.12.674. [DOI] [PubMed] [Google Scholar]
- 64.Niedoszytko M., Elberink J.N.G.O., Bruinenberg M., Nedoszytko B., De Monchy J.G.R., Meerman G.J.T., Weersma R.K., Mulder A.B., Jassem E., Van Doormaal J.J. Gene expression profile, pathways, and transcriptional system regulation in indolent systemic mastocytosis. Allergy. 2010;66:229–237. doi: 10.1111/j.1398-9995.2010.02477.x. [DOI] [PubMed] [Google Scholar]
- 65.Niedoszytko M., Bruinenberg M., Van Doormaal J.J., De Monchy J.G.R., Nedoszytko B., Koppelman G.H., Nawijn M.C., Wijmenga C., Jassem E., Elberink J.N.G.O. Gene expression analysis predicts insect venom anaphylaxis in indolent systemic mastocytosis. Allergy. 2010;66:648–657. doi: 10.1111/j.1398-9995.2010.02521.x. [DOI] [PubMed] [Google Scholar]
- 66.Górska A., Gruchala-Niedoszytko M., Niedoszytko M., Maciejewska A., Chełmińska M., Skrzypski M., Wasąg B., Kaczkan M., Lange M., Nedoszytko B., et al. The Role of TRAF4 and B3GAT1 Gene Expression in the Food Hypersensitivity and Insect Venom Allergy in Mastocytosis. Arch. Immunol. Ther. Exp. 2016;64:497–503. doi: 10.1007/s00005-016-0397-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bulfone-Paus S., Nilsson G., Draber P., Blank U., Levi-Schaffer F. Positive and Negative Signals in Mast Cell Activation. Trends Immunol. 2017;38:657–667. doi: 10.1016/j.it.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 68.McNeil B.D., Pundir P.P., Meeker S., Han L., Undem B.J., Kulka M., Dong X. Identification of a mast-cell-specific receptor crucial for pseudo-allergic drug reactions. Nat. Cell Biol. 2015;519:237–241. doi: 10.1038/nature14022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Subramanian H., Gupta K., Ali H. Roles of Mas-related G protein-coupled receptor X2 on mast cell-mediated host defense, pseudoallergic drug reactions, and chronic inflammatory diseases. J. Allergy Clin. Immunol. 2016;138:700–710. doi: 10.1016/j.jaci.2016.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tatemoto K., Nozaki Y., Tsuda R., Konno S., Tomura K., Furuno M., Ogasawara H., Edamura K., Takagi H., Iwamura H., et al. Immunoglobulin E-independent activation of mast cell is mediated by Mrg receptors. Biochem. Biophys. Res. Commun. 2006;349:1322–1328. doi: 10.1016/j.bbrc.2006.08.177. [DOI] [PubMed] [Google Scholar]
- 71.Alkanfari I., Gupta K., Jahan T., Ali H. Naturally Occurring Missense MRGPRX2 Variants Display Loss of Function Phenotype for Mast Cell Degranulation in Response to Substance P, Hemokinin-1, Human β-Defensin-3, and Icatibant. J. Immunol. 2018;201:343–349. doi: 10.4049/jimmunol.1701793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Na Ayudhya C.C., Roy S., Alkanfari I., Ganguly A., Ali H. Identification of Gain and Loss of Function Missense Variants in MRGPRX2’s Transmembrane and Intracellular Domains for Mast Cell Activation by Substance P. Int. J. Mol. Sci. 2019;20:5247. doi: 10.3390/ijms20215247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tatemoto K., Nozaki Y., Tsuda R., Kaneko S., Tomura K., Furuno M., Ogasawara H., Edamura K., Takagi H., Iwamura H., et al. Endogenous protein and enzyme fragments induce immunoglobulin E-independent activation of mast cells via a G protein-coupled receptor, MRGPRX2. Scand. J. Immunol. 2018;87:e12655. doi: 10.1111/sji.12655. [DOI] [PubMed] [Google Scholar]
- 74.Giavina-Bianchi P., Gonçalves D.G., Zanandréa A., De Castro R.B., Garro L.S., Kalil J., Castells M. Anaphylaxis to quinolones in mastocytosis: Hypothesis on the mechanism. J. Allergy Clin. Immunol. Pract. 2019;7:2089–2090. doi: 10.1016/j.jaip.2019.01.059. [DOI] [PubMed] [Google Scholar]
- 75.Weiler C.R. Mastocytosis, Quinolones, MRGPRX2, and Anaphylaxis. J. Allergy Clin. Immunol. Pract. 2019;7:2091–2092. doi: 10.1016/j.jaip.2019.02.015. [DOI] [PubMed] [Google Scholar]
- 76.Boyden S.E., Desai A., Cruse G., Young M.L., Bolan H.C., Scott L.M., Eisch A.R., Long R.D., Lee C.-C.R., Satorius C.L., et al. Vibratory Urticaria Associated with a Missense Variant in ADGRE2. N. Engl. J. Med. 2016;374:656–663. doi: 10.1056/NEJMoa1500611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ombrello M.J., Remmers E.F., Sun G., Freeman A.F., Datta S., Torabi-Parizi P., Subramanian N., Bunney T.D., Baxendale R.W., Martins M.S., et al. Cold Urticaria, Immunodeficiency, and Autoimmunity Related toPLCG2Deletions. N. Engl. J. Med. 2012;366:330–338. doi: 10.1056/NEJMoa1102140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gandhi C., Healy C., Wanderer A.A., Hoffman H.M. Familial atypical cold urticaria: Description of a new hereditary disease. J. Allergy Clin. Immunol. 2009;124:1245–1250. doi: 10.1016/j.jaci.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aderibigbe O.M., Priel D.L., Lee C.R., Ombrello M.J., Prajapati V.H., Liang M.G., Lyons J.J., Kuhns D.B., Cowen E.W., Milner J.D. Distinct Cutaneous Manifestations and Cold-Induced Leukocyte Activation Associated With PLCG2 Mutations. JAMA Dermatol. 2015;151:627–634. doi: 10.1001/jamadermatol.2014.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lyons J.J., Sun G., Stone K.D., Nelson C.M., Wisch L.B., O’Brien M., Jones N., Lindsley A., Komarow H.D., Bai Y., et al. Mendelian inheritance of elevated serum tryptase associated with atopy and connective tissue abnormalities. J. Allergy Clin. Immunol. 2014;133:1471–1474. doi: 10.1016/j.jaci.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lyons J.J., Yu X., Hughes J.D., Le Q.T., Jamil A., Bai Y., Ho N., Zhao M., Liu Y., O’Connell M.P., et al. Elevated basal serum tryptase identifies a multisystem disorder associated with increased TPSAB1 copy number. Nat. Genet. 2016;48:1564–1569. doi: 10.1038/ng.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lyons J.J. Hereditary Alpha Tryptasemia: Genotyping and Associated Clinical Features. Immunol. Allergy Clin. N. Am. 2018;38:483–495. doi: 10.1016/j.iac.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Greiner G., Sprinzl B., Górska A., Ratzinger F., Gurbisz M., Witzeneder N., Schmetterer K.G., Gisslinger B., Uyanik G., Hadzijusufovic E., et al. Hereditary alpha tryptasemia is a valid genetic biomarker for severe mediator-related symptoms in mastocytosis. Blood. 2020 doi: 10.1182/blood.2020006157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lyons J.J., Chovanec J., O’Connell M.P., Liu Y., Šelb J., Zanotti R., Bai Y., Kim J., DiMaggio T., Schwartz L.B., et al. Heritable risk for severe anaphylaxis associated with increased α-tryptase-encoding germline copy number at TPSAB1. J. Allergy Clin. Immunol. 2020 doi: 10.1016/j.jaci.2020.06.035. [DOI] [PubMed] [Google Scholar]
- 85.Robey R.C., Wilcock A., Bonin H., Beaman G., Myers B., Grattan C., Briggs T.A., Arkwright P.D. Hereditary Alpha-Tryptasemia: UK Prevalence and Variability in Disease Expression. J. Allergy Clin. Immunol. Pract. 2020;8:3549–3556. doi: 10.1016/j.jaip.2020.05.057. [DOI] [PubMed] [Google Scholar]
- 86.Le Q.T., Lyons J.J., Naranjo A.N., Olivera A., Lazarus R.A., Metcalfe D.D., Milner J.D., Schwartz L.B. Impact of naturally forming human alpha/beta-tryptase heterotetramers in the pathogenesis of hereditary alpha-tryptasemia. J. Exp. Med. 2019;216:2348–2361. doi: 10.1084/jem.20190701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sabato V., Chovanec J., Faber M.A., Milner J.D., Ebo D., Lyons J.J. First Identification of an Inherited TPSAB1 Quintuplication in a Patient with Clonal Mast Cell Disease. J. Clin. Immunol. 2018;38:457–459. doi: 10.1007/s10875-018-0506-y. [DOI] [PubMed] [Google Scholar]
- 88.Lennartsson J., Rönnstrand L. Stem Cell Factor Receptor/c-Kit: From Basic Science to Clinical Implications. Physiol. Rev. 2012;92:1619–1649. doi: 10.1152/physrev.00046.2011. [DOI] [PubMed] [Google Scholar]
- 89.Cruse G., Metcalfe D.D., Olivera A. Functional deregulation of KIT: Link to mast cell proliferative diseases and other neoplasms. Immunol. Allergy Clin. N. Am. 2014;34:219–237. doi: 10.1016/j.iac.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bodemer C., Hermine O., Palmérini F., Yang Y., Grandpeix-Guyodo C., Leventhal P.S., Hadj-Rabia S., Nasca L., Georgin-Lavialle S., Cohen-Akenine A., et al. Pediatric Mastocytosis Is a Clonal Disease Associated with D816V and Other Activating c-KIT Mutations. J. Investig. Dermatol. 2010;130:804–815. doi: 10.1038/jid.2009.281. [DOI] [PubMed] [Google Scholar]
- 91.Orfao A., Montero A.G., Sánchez-Muñoz L., Escribano L. Recent advances in the understanding of mastocytosis: The role of KIT mutations. Br. J. Haematol. 2007;138:12–30. doi: 10.1111/j.1365-2141.2007.06619.x. [DOI] [PubMed] [Google Scholar]
- 92.Bibi S., Langenfeld F., Jeanningros S., Brenet F., Soucie E., Hermine O., Damaj G., Dubreuil P., Arock M. Molecular defects in mastocytosis: KIT and beyond KIT. Immunol. Allergy Clin. N. Am. 2014;34:239–262. doi: 10.1016/j.iac.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 93.Alvarez-Twose I., Jara-Acevedo M., Morgado J.M., Garcia-Montero A., Sanchez-Munoz L., Teodosio C., Matito A., Mayado A., Caldas C., Mollejo M., et al. Clinical, immunophenotypic, and molecular characteristics of well-differentiated systemic mastocytosis. J. Allergy Clin. Immunol. 2016;137:168–178.e1. doi: 10.1016/j.jaci.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 94.Grootens J., Ungerstedt J.S., Ekoff M., Ronnberg E., Klimkowska M., Amini R.M., Arock M., Soderlund S., Mattsson M., Nilsson G., et al. Single-cell analysis reveals the KIT D816V mutation in haematopoietic stem and progenitor cells in systemic mastocytosis. EBioMedicine. 2019;43:150–158. doi: 10.1016/j.ebiom.2019.03.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pardanani A. Systemic mastocytosis in adults: 2017 update on diagnosis, risk stratification and management. Am. J. Hematol. 2016;91:1146–1159. doi: 10.1002/ajh.24553. [DOI] [PubMed] [Google Scholar]
- 96.Lasho T., Finke C., Zblewski D., Hanson C.A., Ketterling R.P., Butterfield J.H., Tefferi A., Pardanani A. Concurrent activating KIT mutations in systemic mastocytosis. Br. J. Haematol. 2016;173:153–156. doi: 10.1111/bjh.13560. [DOI] [PubMed] [Google Scholar]
- 97.de Beauchene I.C., Allain A., Panel N., Laine E., Trouve A., Dubreuil P., Tchertanov L. Hotspot mutations in KIT receptor differentially modulate its allosterically coupled conformational dynamics: Impact on activation and drug sensitivity. PLoS Comput. Biol. 2014;10:e1003749. doi: 10.1371/journal.pcbi.1003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Laine E., de Beauchene I.C., Perahia D., Auclair C., Tchertanov L. Mutation D816V alters the internal structure and dynamics of c-KIT receptor cytoplasmic region: Implications for dimerization and activation mechanisms. PLoS Comput. Biol. 2011;7:e1002068. doi: 10.1371/journal.pcbi.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Piao X., Paulson R., van der Geer P., Pawson T., Bernstein A. Oncogenic mutation in the Kit receptor tyrosine kinase alters substrate specificity and induces degradation of the protein tyrosine phosphatase SHP-1. Proc. Natl. Acad. Sci. USA. 1996;93:14665–14669. doi: 10.1073/pnas.93.25.14665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bibi S., Arslanhan M.D., Langenfeld F., Jeanningros S., Cerny-Reiterer S., Hadzijusufovic E., Tchertanov L., Moriggl R., Valent P., Arock M. Co-operating STAT5 and AKT signaling pathways in chronic myeloid leukemia and mastocytosis: Possible new targets of therapy. Haematologica. 2014;99:417–429. doi: 10.3324/haematol.2013.098442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Harir N., Boudot C., Friedbichler K., Sonneck K., Kondo R., Martin-Lannerée S., Kenner L., Kerenyi M., Yahiaoui S., Gouilleux-Gruart V., et al. Oncogenic Kit controls neoplastic mast cell growth through a Stat5/PI3-kinase signaling cascade. Blood. 2008;112:2463–2473. doi: 10.1182/blood-2007-09-115477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Baumgartner C., Cerny-Reiterer S., Sonneck K., Mayerhofer M., Gleixner K.V., Fritz R., Kerenyi M., Boudot C., Gouilleux F., Kornfeld J.W., et al. Expression of activated STAT5 in neoplastic mast cells in systemic mastocytosis: Subcellular distribution and role of the transforming oncoprotein KIT D816V. Am. J. Pathol. 2009;175:2416–2429. doi: 10.2353/ajpath.2009.080953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Peter B., Bibi S., Eisenwort G., Wingelhofer B., Berger D., Stefanzl G., Blatt K., Herrmann H., Hadzijusufovic E., Hoermann G., et al. Drug-induced inhibition of phosphorylation of STAT5 overrides drug resistance in neoplastic mast cells. Leukemia. 2018;32:1016–1022. doi: 10.1038/leu.2017.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Voisset E., Lopez S., Dubreuil P., De Sepulveda P. The tyrosine kinase FES is an essential effector of KITD816V proliferation signal. Blood. 2007;110:2593–2599. doi: 10.1182/blood-2007-02-076471. [DOI] [PubMed] [Google Scholar]
- 105.Smrz D., Kim M.S., Zhang S., Mock B.A., Smrzova S., DuBois W., Simakova O., Maric I., Wilson T.M., Metcalfe D.D., et al. mTORC1 and mTORC2 differentially regulate homeostasis of neoplastic and non-neoplastic human mast cells. Blood. 2011;118:6803–6813. doi: 10.1182/blood-2011-06-359984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gabillot-Carre M., Lepelletier Y., Humbert M., de Sepuvelda P., Hamouda N.B., Zappulla J.P., Liblau R., Ribadeau-Dumas A., Machavoine F., Letard S., et al. Rapamycin inhibits growth and survival of D816V-mutated c-kit mast cells. Blood. 2006;108:1065–1072. doi: 10.1182/blood-2005-06-2433. [DOI] [PubMed] [Google Scholar]
- 107.Gleixner K.V., Mayerhofer M., Cerny-Reiterer S., Hormann G., Rix U., Bennett K.L., Hadzijusufovic E., Meyer R.A., Pickl W.F., Gotlib J., et al. KIT-D816V-independent oncogenic signaling in neoplastic cells in systemic mastocytosis: Role of Lyn and Btk activation and disruption by dasatinib and bosutinib. Blood. 2011;118:1885–1898. doi: 10.1182/blood-2010-06-289959. [DOI] [PubMed] [Google Scholar]
- 108.Hoermann G., Cerny-Reiterer S., Perne A., Klauser M., Hoetzenecker K., Klein K., Mullauer L., Groger M., Nijman S.M., Klepetko W., et al. Identification of oncostatin M as a STAT5-dependent mediator of bone marrow remodeling in KIT D816V-positive systemic mastocytosis. Am. J. Pathol. 2011;178:2344–2356. doi: 10.1016/j.ajpath.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee Y.N., Brandal S., Noel P., Wentzel E., Mendell J.T., McDevitt M.A., Kapur R., Carter M., Metcalfe D.D., Takemoto C.M. KIT signaling regulates MITF expression through miRNAs in normal and malignant mast cell proliferation. Blood. 2011;117:3629–3640. doi: 10.1182/blood-2010-07-293548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Aichberger K.J., Mayerhofer M., Gleixner K.V., Krauth M.T., Gruze A., Pickl W.F., Wacheck V., Selzer E., Mullauer L., Agis H., et al. Identification of MCL1 as a novel target in neoplastic mast cells in systemic mastocytosis: Inhibition of mast cell survival by MCL1 antisense oligonucleotides and synergism with PKC412. Blood. 2007;109:3031–3041. doi: 10.1182/blood-2006-07-032714. [DOI] [PubMed] [Google Scholar]
- 111.Jawhar M., Schwaab J., Schnittger S., Meggendorfer M., Pfirrmann M., Sotlar K., Horny H.P., Metzgeroth G., Kluger S., Naumann N. Additional mutations in SRSF2, ASXL1 and/or RUNX1 identify ahigh-risk group of patients with KIT D816V+ advanced systemic mastocytosis. Leukemia. 2016;30:136. doi: 10.1038/leu.2015.284. [DOI] [PubMed] [Google Scholar]
- 112.Jawhar M., Schwaab J., Hausmann D., Clemens J., Naumann N., Henzler T., Horny H.P., Sotlar K., Schoenberg S.O., Cross N.C.P. Splenomegaly, elevated alkaline phosphatase and mutations in the SRSF2/ASXL1/RUNX1 gene panel are strong adverse prognostic markers in patients with systemic mastocytosis. Leukemia. 2016;30:2342–2350. doi: 10.1038/leu.2016.190. [DOI] [PubMed] [Google Scholar]
- 113.Damaj M.J.G., Chandesris O., Hanssens K., Soucie E., Canioni D., Kolb B., Durieu I., Gyan E., Livideanu C., Chèze S., et al. ASXL1 but not TET2 mutations adversely impact overall survival of patients suffering systemic mastocytosis with associated clonal hematologic non-mast-cell diseases. PLoS ONE. 2014;9:e85362. doi: 10.1371/journal.pone.0085362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pardanani A., Lasho T.L., Finke C., Zblewski D., Abdelrahman R., Wassie E., Gangat N., Hanson C.A., Ketterling R.P., Tefferi A. ASXL1 and CBL mutations are independently predictive of inferior survival in advanced systemic mastocytosis. Br. J. Haematol. 2015;175:534–536. doi: 10.1111/bjh.13865. [DOI] [PubMed] [Google Scholar]
- 115.Jawhar M., Schwaab J., Álvarez-Twose I., Shoumariyeh K., Naumann N., Lübke J., Perkins C., Muñoz-González J.I., Meggendorfer M., Kennedy V., et al. MARS: Mutation-Adjusted Risk Score for Advanced Systemic Mastocytosis. J. Clin. Oncol. 2019;37:2846–2856. doi: 10.1200/JCO.19.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Muñoz-González J.I., Jara-Acevedo M., Alvarez-Twose I., Merker J.D., Teodosio C., Hou Y., Henriques A., Roskin K.M., Sanchez-Muñoz L., Tsai A.G., et al. Impact of somatic and germline mutations on the outcome of systemic mastocytosis. Blood Adv. 2018;2:2814–2828. doi: 10.1182/bloodadvances.2018020628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pardanani A., Shah S., Mannelli F., Elala Y.C., Guglielmelli P., Lasho T.L., Patnaik M.M., Gangat N., Ketterling R.P., Reichard K.K., et al. Mayo alliance prognostic system for mastocytosis: Clinical and hybrid clinical-molecular models. Blood Adv. 2018;2:2964–2972. doi: 10.1182/bloodadvances.2018026245. [DOI] [PMC free article] [PubMed] [Google Scholar]