Abstract
High-resolution mass spectrometry equipped with electrospray ionization (ESI) and atmospheric pressure chemical ionization (APCI) sources was used to enhance the characterization of phytochemicals of ethanol extracts of Manilkara zapota L. leaves (ZLE). Sugar compounds, dicarboxylic acids, compounds of phenolic acids and flavonoids groups, and other phytochemicals were detected from the leaves. Antioxidant activity and inhibition potentiality of ZLE against α-glucosidase enzyme, and elastase enzyme activities were evaluated in in vitro analysis. ZLE significantly inhibited activities of α-glucosidase enzyme at a lower concentration (IC50 2.51 ± 0.15 µg/mL). Glucose uptake in C2C12 cells was significantly enhanced by 42.13 ± 0.15% following the treatment with ZLE at 30 µg/mL. It also exhibited potential antioxidant activities and elastase enzyme inhibition activity (IC50 27.51 ± 1.70 µg/mL). Atmospheric pressure chemical ionization mass spectrometry (APCI–MS) detected more m/z peaks than electrospray ionization mass spectrometry (ESI–MS), and both ionization techniques illustrated the biological activities of the detected compounds more thoroughly compared to single-mode analysis. Our findings suggest that APCI along with ESI is a potential ionization technique for metabolite profiling, and ZLE has the potential in managing diabetes by inhibiting α-glucosidase activity and enhancing glucose uptake.
Keywords: Manilkara zapota L., electrospray ionization (ESI), atmospheric pressure chemical ionization (APCI), antioxidant, α-glucosidase
1. Introduction
Nature is an abundant source of medicinal plants; various natural foodstuffs containing prospective bioactive components which support potential health care are traditionally used as herbal remedies for numerous life-threatening diseases [1]. Medicinal plants for their potential bioactive compounds are used for the ailment of numerous illness like inflammations, oxidative stresses, fatness and hypertensive conditions, diabetic mellitus, skin care, neurological complaints, and cancer, and to regulate expression of various genes [2]. Thus, the demand of discovering and developing new natural therapeutics is of growing interest to the researchers. Screening of plant extracts using different ionization techniques in mass spectrometry is one of the fastest and better ways for the discovery of new bioactive compounds [3].
Manilkara zapota L. is commonly referred to as sapodilla and also locally known as sofeda in Bangladesh, belonging to the Sapotaceae family. It is a long-living, evergreen plant commonly found in India, the south part of Mexico, and Central America [4]. Sapodilla is edible and has a sweet taste with rich flavor. There have been some studies on the fruits, seeds, barks, and leaves of M. zapota for phytochemical characterization and assessment of biological activities such as antioxidant, antimicrobial, antihyperglycemic, hypocholesterolemic, antityrosinase, and antidiabetic properties [4,5,6,7,8,9]. However, there have been few studies on thorough phytoconstituent identification based on advanced mass spectrometry and assessment of α-glucosidase enzyme inhibition activity of M. zapota leaves. Hence, characterization of bioactive secondary metabolites of M. zapota L. leaves and evaluation of their potential biological activities are of greater interest for further investigating the prospective use of sapodilla because of climacteric change, and as this becomes possible, the popularity of this fruit in many countries is rapidly increasing.
Mass spectrometry (MS)-based analytical methods are highly sensitive for the identification, quantification, and structural interpretation of metabolites. Sample ionization is one of the most important steps for mass spectrometry-based metabolomics. Electrospray ionization (ESI), atmospheric pressure chemical ionization (APCI), and electron impact ionization (EI) are typically used in the field of metabolomics [10]. ESI ionizes competently the strong and moderate polar metabolites, whereas APCI is typically considered for nonpolar and less-polar metabolites [11]. Mass spectrometers using different ionizations provide a mass-to-charge ratio (m/z peaks) of ions by generating mass spectrum, and tandem mass spectrometry (MS/MS) split precursor ions into smaller fragmented ions for the structural elucidation of unknown compounds [12,13].
In this study, direct injection negative-mode ESI–MS and APCI–MS were undertaken to explore a broad range of metabolites from M. zapota L. leaves. Furthermore, various in vitro tests were performed to assess its α-glucosidase inhibition activity, uptake of glucose in C2C12 myotubes, and its antioxidant and elastase inhibition activities. In this study, the experimental findings were also compared with those of other previous studies. Here, a systematic approach for metabolite profiling and bioactivity studies of ZLE was employed.
2. Results
2.1. Identification of Secondary Metabolites
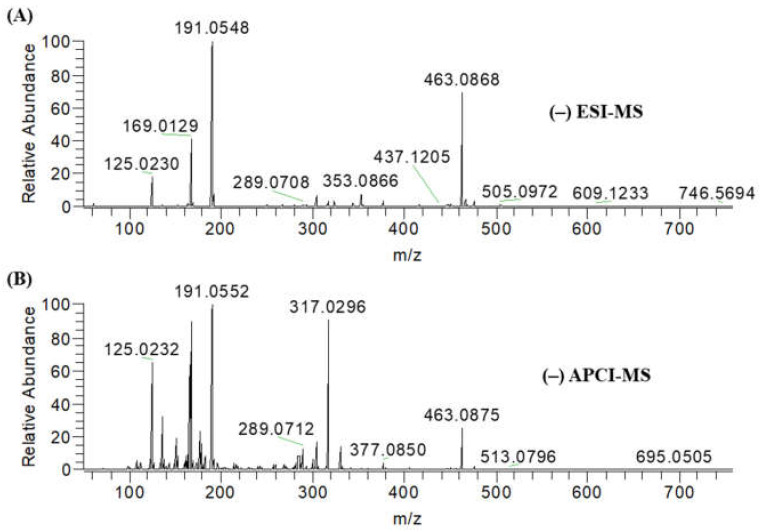
Obtained mass spectra of the extracts from negative-mode ESI–MS and APCI–MS are displayed in Figure 1. Using negative-mode ESI–MS and APCI–MS, 174 and 225 peaks were detected, respectively; 65 peaks were commonly found with both ionization techniques (Figure S1). Peak lists detected from mass spectra in APCI–MS and ESI–MS are provided in Supplementary Data (Table S1). Proposed compounds were first assumed by comparing their experimental masses in high-resolution mass spectrometry with theoretical calculated masses.
Figure 1.
(A) Negative-mode electrospray ionization mass spectrometry (ESI–MS), (B) negative-mode atmospheric pressure chemical ionization mass spectrometry (APCI–MS) mass spectra of ethanol extracts of M. zapota leaves (ZLE). Prepared sample was directly infused to ESI and APCI sources.
Identification of compounds was carried out using tandem mass spectrometry (MS/MS), which is an eminent technique to elucidate the structure of compounds and thereby very effective to identify unknown compounds comprising complex mixtures. MS/MS fragmentations of all targeted m/z peaks were carried out using the higher-energy collisional dissociation (HCD) activation mode at collision energy (CE) values of 10, 20, and 30 eV.
Gallic acid (m/z 169.0129), myricetin (m/z 317.0296), quinic acid (m/z 191.0548), chlorogenic acid (m/z 353.0866), and myricetin-3-O-rhamnoside (m/z 463.0868) peaks were observed abundant in negative-mode ESI–MS and APCI–MS spectra of ZLE (Figure 1). From ZLE analyzed in ESI–MS and APCI–MS, 39 compounds including 3 sugar compounds, 4 dicarboxylic acids, 8 phenolic acids, 9 flavonoids, and 15 other phytochemicals were identified (Table 1). Sugar compounds rhamnose, glucose, and sucrose were detected at m/z 163.0598, 179.0547, and 341.1077, respectively [14,15].
Table 1.
Secondary metabolites identified in leaves of Manilkara zapota using negative-mode electrospray ionization mass spectrometry (ESI–MS) and atmospheric pressure chemical ionization mass spectrometry (APCI–MS).
| Group | Compound Name | Elemental Formula | Observed m/z | Calculated m/z | Adducts | MS/MS Fragments | CE * (eV) |
Ionizations |
|---|---|---|---|---|---|---|---|---|
| Sugar | Rhamnose | C6H12O5 | 163.0598 | 163.0611 | [M–H]− | 145, 119 | 20 | ESI, APCI |
| Glucose | C6H12O6 | 179.0547 | 179.0561 | [M–H]− | 161, 143, 113, 101 | 10 | ESI, APCI | |
| 215.0316 | 215.0324 | [M+Cl]− | 179, 161, 143, 113, 101 | 10 | ESI | |||
| Sucrose | C12H22O11 | 341.1077 | 341.1083 | [M–H]− | 179, 161, 119, 101 | 10 | ESI, APCI | |
| Dicarboxylic acid | Succinic acid | C4H6O4 | 117.0179 | 117.0193 | [M–H]− | 99, 73 | 10 | APCI |
| Malic Acid | C4H6O5 | 133.0128 | 133.0133 | 115, 89, 71 | 10 | ESI, APCI | ||
| Adipic acid | C6H10O4 | 145.0492 | 145.0506 | 127, 101 | 10 | APCI | ||
| 3-Oxoadipic acid | C6H8O5 | 159.0285 | 159.0298 | 141, 115, 97 | 10 | APCI | ||
| Phenolic acids | Salicylic acid | C7H6O3 | 137.0230 | 137.0244 | [M–H]− | 93 | 20 | ESI, APCI |
| 3,4-Dihydroxybenzoic acid | C7H6O4 | 153.0179 | 153.0193 | 109 | 10 | ESI, APCI | ||
| Vanillic acid | C8H8O4 | 167.0336 | 167.0349 | 152, 123, 108 | 10 | ESI, APCI | ||
| Gallic acid | C7H6O5 | 169.0129 | 169.0142 | 125 | 10 | ESI, APCI | ||
| Caffeic acid | C9H8O4 | 179.0336 | 179.0349 | 161, 135 | 10 | ESI, APCI | ||
| Ferulic acid | C10H10O4 | 193.0591 | 193.0506 | 178, 149, 134 | 10 | ESI, APCI | ||
| Syringic acid | C9H10O5 | 197.0442 | 197.0455 | 182, 153, 125 | 10 | ESI, APCI | ||
| Chlorogenic acid | C16H18O9 | 353.0866 | 353.0878 | 191 | 10 | ESI, APCI | ||
| Flavonoids | Afzelechin | C15H14O5 | 273.0765 | 273.0768 | [M–H]− | 167 | 10 | APCI |
| Epicatechin | C15H14O6 | 289.0708 | 289.0719 | 245, 205, 179, 137 | 20 | ESI, APCI | ||
| Epigallocatechin | C15H14O7 | 305.0656 | 305.0666 | 287, 261, 219, 179 | 10 | ESI, APCI | ||
| Myricetin | C15H10O8 | 317.0296 | 317.0302 | 193, 165, 137 | 20 | APCI | ||
| Ampelopsin | C15H12O8 | 319.0447 | 319.0459 | 193, 178, 153, 125 | 10 | ESI | ||
| Laricitrin | C16H12O8 | 331.0454 | 331.0459 | 316, 178, 151 | 20 | ESI, APCI | ||
| Myricetin-3-O-rhamnoside | C21H20O12 | 463.0868 | 463.0882 | 316 | 30 | ESI, APCI | ||
| Laricitrin-3-O-rhamnoside | C22H22O12 | 477.1022 | 477.1038 | 331, 316, 287 | 20 | ESI, APCI | ||
| Prodelphinidin B | C30H26O14 | 609.1233 | 609.1249 | 441, 423, 305, 125 | 10 | ESI | ||
| Others | 2-Hydroxybenzaldehyde | C7H6O2 | 121.0280 | 121.0295 | [M–H]− | 30 | ESI, APCI | |
| Guaiacol | C7H8O2 | 123.0437 | 123.0451 | 108, 93 | 10 | APCI | ||
| Pyroglutamic acid | C5H7NO3 | 128.0386 | 128.0353 | 82 | 20 | ESI, APCI | ||
| Threonic acid | C4H8O5 | 135.0285 | 135.0298 | 117, 91, 75 | 10 | ESI, APCI | ||
| Vanillin | C8H8O3 | 151.0388 | 151.0407 | 136 | 10 | ESI, APCI | ||
| 3-Hydroxycoumarin | C9H6O3 | 161.0230 | 161.0244 | 133, 117 | 10 | ESI, APCI | ||
| Shikimic acid | C7H10O5 | 173.0442 | 173.0455 | 155, 129 | 10 | ESI, APCI | ||
| Esculetin | C9H6O4 | 177.0179 | 177.0193 | 159, 149, 133, 121 | 10 | ESI, APCI | ||
| Quinic acid | C7H12O6 | 191.0548 | 191.0561 | 173, 127, 93, 85 | 20 | ESI, APCI | ||
| Norathyriol | C13H8O6 | 259.0242 | 259.0247 | 231, 215, 187, 171 | 30 | APCI | ||
| Hydroquinone glucuronide | C12H14O8 | 285.0615 | 285.0615 | 152, 109, 108 | 10 | ESI | ||
| Leucodelphinidin | C15H14O8 | 321.0604 | 321.0616 | 303, 195, 125 | 10 | ESI | ||
| 3-Glucogallic acid | C13H16O10 | 331.0655 | 331.0670 | 287, 241, 169, 125 | 10 | ESI | ||
| 3-p-Coumaroylquinic acid | C16H18O8 | 337.0917 | 337.0928 | 191, 173,163 | 10 | ESI | ||
| 3-O-Galloylquinic acid | C14H16O10 | 343.0658 | 343.0670 | 191,173,169, 125 | 10 | ESI | ||
| Unknown | C6H6O3 | 125.0230 | 125.0244 | [M–H]− | 96 | 10 | ESI, APCI | |
| Unknown | C24H20O10 | 467.0967 | 467.0983 | 357, 303, 217 | 10 | ESI, APCI |
Compounds were identified through comparison of their observed mass values with the calculated values and also comparing with the online database and literature. * CE: Collision energy. We used 10–30 eV collision energy for tandem mass spectrometry (MS/MS). For most of the cases, fragmentations of the precursor peaks occurred well at collision energy of 10 eV. The fragmented peak’s abundance started to increase with the increasing value of CE. We considered the optimal CE value for MS/MS fragmentations of the target peaks.
Succinic acid was detected at m/z 117.0179 [M–H]− with fragmented peaks at m/z 99.0075 [M–H–H2O]− and 73.0282 [M–H–CH2O3]− (Figure S2) [16]. Malic acid was identified at m/z 133.0128 [M–H]− with fragmented peaks at m/z 115.0024 [M–H–H2O]−, 89.0231 [M–H–CO2]−, and 71.0125 [M–H–CH2O3]− (Figure S3) [17]. Adipic acid was proposed at m/z 145.0492 with fragmented peaks at m/z 127.0387 and 101.0594. 3-Oxoadipic acid was supposed at m/z 159.0285 with its yielded fragments at m/z 141.0180, 115.0387, and 97.0281 [15].
m/z 137.0230 was identified as salicylic acid as it produced fragments at m/z 93.0333. 3,4-Hihydroxybenzoic acid was suggested at m/z 153.0179 and it displayed fragment ion at m/z 109.0282 [18]. Vanillic acid, caffeic acid, ferulic acid, and syringic acid were detected at m/z 167.0336, 179.0336, 193.0591, and 197.0442, comparing the data found in the Human Metabolome Database (HMDB) and literatures [15,19,20]. m/z 169.0129 [M–H]− was characterized as gallic acid and yielded a fragmented ion at m/z 125.0232 corresponding to [M–H–CO2]− (Figure S4) [21].
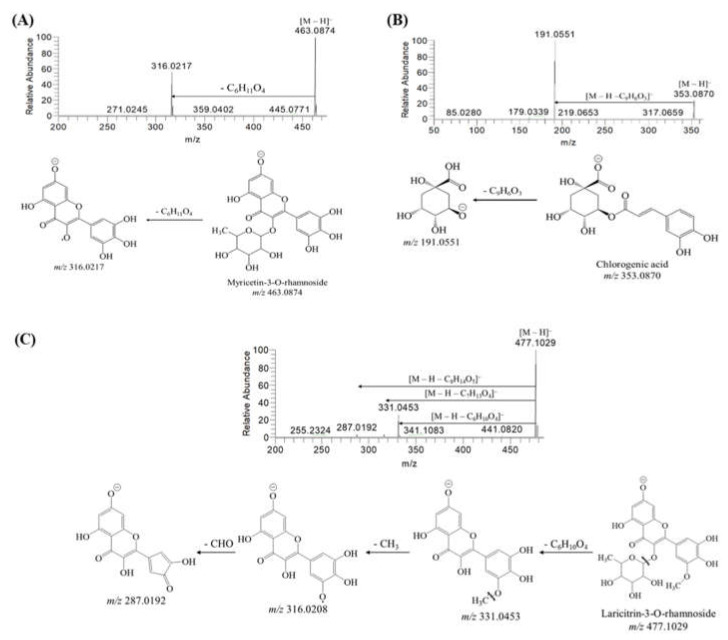
Afzelechin was supposed at m/z 273.0765 [15]. Epicatechin was detected at m/z 289.0708 and it generated fragmented peaks at m/z 245.0812 [M–H–CO2]−, m/z 205.0497 [M–H–C4H4O2]−, m/z 179.0339 [M–H–C6H6O2]−, and m/z 137.0232 [M–H–C8H8O3]− (Figure S5) [22]. Epigallocatechin had a peak at m/z 305.0656 and upon fragmentation exhibited peaks at m/z 287.0561, 261.0769, 219.0659, 179.0343, 165.0186, and 125.0235, respectively [23]. m/z 317.0296 [M–H]− was proposed to be myricetin following fragmented peaks at m/z 193.0135, 165.0185, and 137.0234 through the loss of C6H4O3, C7H4O4, and C8H4O5, respectively (Figure S6). m/z 319.0447 exhibited fragmented ions at m/z 193.0137, 178.9980, 153.0185, and 125.0234, accordingly, and identified as ampelopsin [24]. m/z 331.0454 was characterized as laricitrin due to the appearance of fragmented ions at m/z 316.0219, 178.9976, and 151.0028, accordingly [25]. In addition, myricetin-3-O-rhamnoside was supposed at m/z 463.0868 with the fragmented ion at m/z 316.0217 [M–H–C6H11O4]− (Figure 2A) [26]. m/z of 353.0866 [M–H]− produced a fragmented peak at m/z 191.0551 at collision energy 10 eV with losing C9H6O3, and the compound was suggested as chlorogenic acid (Figure 2B) [27]. m/z 477.1022 [M–H]− was suggested to be laricitrin-3-O-rhamnoside with fragmented ions of m/z 331.0453 [M–H–C6H10O4]−, m/z 316.0208 [M–H–C6H10O4–CH3]−, and m/z 287.0192 [M–H–C6H10O4–CH3–CHO]− (Figure 2C) [25]. Prodelphinidin B was detected at m/z 609.1233 based on literature data with similar MS/MS fragmented ions at m/z 441.0818, 423.0711, 305.0659, 191.0550, and 125.0231, accordingly [28].
Figure 2.
Negative-mode electrospray ionization tandem mass spectrometry (ESI–MS/MS) spectra and schematic fragmentation patterns of (A) myricetin-3-O-rhamnoside, (B) chlorogenic acid, and (C) laricitrin-3-O-rhamnoside; 10–30 eV collision energy (CE) was applied to produce fragmented peaks of the precursor ions.
Compared to data from HMDB and the literature [15,16,18,20,29], m/z 121.0280, 123.0437, 128.0386, 135.0285, 151.0388, 161.0230, 173.0442, and 177.0179 were characterized as 2-Hydroxybenzaldehyde, guaiacol, pyroglutamic acid, threonic acid, vanillin, 3-Hydroxycumarin, shikimic acid, and esculetin. The peak at m/z 191.0548 [M–H]− was supposed as quinic acid by detecting fragmented peaks at m/z 173.0445, 127.0388, 109.0282, and 93.0333 with the loss of H2O, CH4O3, CH6O4, and CH6O5, accordingly (Figure S7) [30]. m/z 259.0242 [M–H]− was identified as norathyriol based on fragmentation peaks at m/z 231.0295, 215.0345, 187.0394, and 171.0444 with the loss of CO, CO2, C2O3, and C2O4 (Figure S8) [31]. m/z 285.0615, 321.0604, and 331.0655 were proposed to be hydroquinone glucuronide, leucodelphinidin, and 3-Glucogallic acid following the comparison with the HMDB database and literatures [15,32].
m/z 337.0917 showed fragmented ions at m/z 191.0555, 173.0449, and 163.0393 upon fragmentation at collision energy (CE) 10 eV and it was characterized as 3-p-Coumaroylquinic acid [20,33]. m/z 343.0658 was identified as 3-O-Galloylquinic acid as it produced fragmented ions at m/z 191.0555, 173.0449, 169.0136, and 125.0235, accordingly [15,34].
2.2. Antioxidant Effects of ZLE
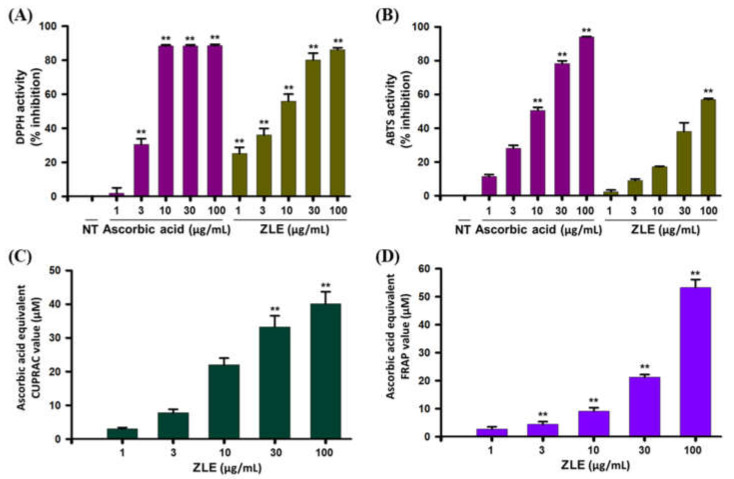
In this study, antioxidant activity of ZLE was evaluated following DPPH● and ABTS●+ scavenging assays, and the results of the assays are shown in Figure 3A,B. It was found that the percentage inhibition of DPPH● was 25.31%, 36.07%, 56.06%, 80.21%, and 86.28% at 1, 3, 10, 30, and 100 µg/mL, accordingly. There was a progressive radical scavenging trend with a higher concentration of ZLE. IC50 of ZLE was determined at 7.93 ± 1.43 µg/mL, where ascorbic acid had IC50: 5.34 ± 0.27 µg/mL, which indicated that the leaf extracts possessed a powerful DPPH● scavenging activity.
Figure 3.
Antioxidant potential of ethanol extracts of M. zapota L. leaves (ZLE). (A) 2,2-Diphenyl-1-picrylhydrazyl radical (DPPH•) scavenging, (B) 2,2′-Azino-bis(3-ethylbenzthiazoline-6-sulphonic acid radical cation (ABTS•+) scavenging, (C) Cupric-reducing antioxidant capacity (CUPRAC), and (D) Ferric-reducing antioxidant power (FRAP) assays. Ascorbic acid as a positive control was used in the assays. Results were as mean ± standard deviation of three experiments (** p < 0.05 versus control using one-way ANOVA).
Experimental results of the ABTS●+ scavenging assay are displayed in Figure 3B. At 100 µg/mL, the inhibition of ABTS●+ by the leaf extracts was 56.15% and IC50 was 72.85 µg/mL, which suggested that ZLE had a capacity to donate both electrons and hydrogen, resulting in having antioxidant potential. With the purpose of determining the reducing potentiality of ZLE, ferric-reducing antioxidant power (FRAP) and cupric-reducing antioxidant capacity (CUPRAC) assays were conducted. ZLE (100 µg/mL) demonstrated strong reducing power with 53.30 ± 2.85 µM ascorbic acid equivalent FRAP and 40.09 ± 3.61 µM ascorbic acid equivalent CUPRAC value, respectively. These results suggested that ZLE also had a strong electron donating capacity, resulting in having powerful antioxidant capacity (Figure 3C,D).
2.3. α-Glucosidase Inhibitory Activity of ZLE
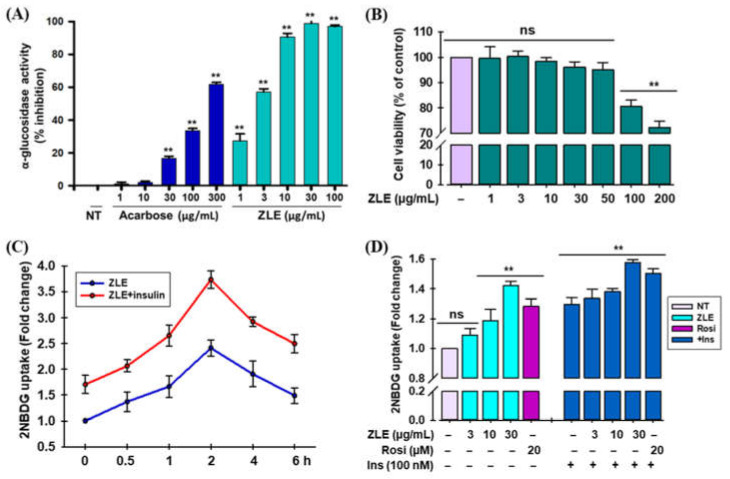
α-Glucosidase enzymes aid in hydrolysis of carbohydrates including starches during digestion into glucose in the intestine which, in turn, contribute to the increase of blood glucose levels. Inhibiting the α-glucosidase enzyme activity with type 2 diabetes patients may suppress postprandial hyperglycemia [35]. ZLE’s ability to inhibit α-glucosidase is presented in Figure 4A and the results showed that ZLE inhibited the activity of α-glucosidase in concentration dependently. ZLE significantly inhibited α-glucosidase activity even at 1 µg/mL and the calculated IC50 was found very low (2.51 ± 0.15 µg/mL), whereas acarbose had a higher value (IC50 216.26 ± 1.63 µg/mL).
Figure 4.
(A) α-Glucosidase inhibition activity of ethanol extracts of M. zapota leaves (ZLE). Acarbose as positive control was used in the assay. (B) Cell viability in C2C12 myotubes. C2C12 cells were seeded in a 96-well plate (density 1 × 105 cells/well) and carried out 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay as referred to in Materials and Methods. (C) Time-dependent uptake of glucose by C2C12 myotubes. Cells treated with ZLE (30 μg/mL) alone or ZLE (30 μg/mL) along with insulin (100 nM) were incubated for the time periods indicated. (D) Effects of ZLE on the uptake of glucose in C2C12 myotubes. Cells were incubated with ZLE alone and/or with insulin for 2 h. Rosiglitazone (20 μM) was employed as a positive control. (‘ns’ indicated non-significant; ‘–‘ indicated sample was not added and ‘+’ indicated sample was added). Results were as mean ± standard deviation of three analyses (** p < 0.05 versus control using one-way ANOVA).
2.4. Effects of ZLE on Cell Viability and Glucose Uptake in C2C12 Myotubes
The effects of ZLE on cell viability in C2C12 cells are presented in Figure 4B. ZLE exhibited moderate to high cytotoxicity at 100 and 200 μg/mL, respectively, whereas at up to 50 μg/mL, there were no cytotoxic effects observed. Glucose uptake potential of ZLE in C2C12 myotubes was measured following the treatment with ZLE at the designated time-points (Figure 4C). The effect of ZLE (at 30 μg/mL) on basal- or insulin-induced glucose uptake initiated at 0.5 h, which was utmost at 2 h then gradually reduced. Hence, we considered 2 h time period for ZLE administration in the sequential assays. We observed that basal- or insulin-administered glucose uptake augmented in a concentration proportional fashion (Figure 4D). Basal- and insulin-administered glucose uptake improved remarkably by 42.13 ± 0.27% and 57.74 ± 0.17%, respectively, at the highest concentration of ZLE (30 μg/mL).
Rosiglitazone, a commonly recognized thiazolidinedione group antidiabetic agent, was applied here in the form of positive control. It also substantially augmented basal- and insulin-applied glucose uptake levels by 28.49 ± 0.49% and 50.38 ± 0.32%, respectively.
2.5. Elastase Inhibition Activity of ZLE
Experimental results showed that elastase inhibition activity was increased with the increased concentration of ZLE (Figure S9). IC50 for elastase inhibition activity was measured at a concentration of 27.51 ± 1.70 µg/mL of ZLE, whereas less inhibition activity was observed for EGCG at a higher concentration.
3. Discussion
Previous studies found that ethanol extracts of plant materials exhibit good biological activities and contain more bioactive components [36,37,38]. In this study, we performed ethanol extraction of M. zapota leaves (ZLE) and used ZLE for the subsequent experiments.
Mass spectrometry analysis employing direct sample solution injection is a rapid and cost-effective technique compared to other separation procedures [39]. Determination of precise mass and its fragmentation peak using tandem mass spectrometry (MS/MS) aids in the elemental composition information for unknown compound identification. However, for accurate and isomeric identification, we needed to follow a sequential analysis using liquid chromatography mass spectrometry (LC MS), infrared (IR) spectroscopy, and nuclear magnetic resonance (NMR) spectroscopy [40,41].
Today, the ESI technique is widely used for the analysis of singly-charged small molecules to multiply charged large molecules in complex substances [42,43]. APCI is also applicable for the analysis of less polar compounds with a molecular weight of <1500 Da [44]. In this study, direct injection APCI was used along with the ESI technique equipped with high-resolution MS for the phytochemical screening of ZLE. We used negative-mode electrospray ionization (ESI) in mass spectrometry for the metabolite profiling of M. zapota leaves because it yielded enhanced sensitivity and more observable peaks in the mass spectra [45]. Negative-mode ESI-MS presented extensive insights of polyketide classes (flavones, flavanols, flavanones, isoflavones, flavonols, phloroglucinols, anthraquinones, bisanthraquinones, and stilbenes) compounds particularly through the loss of H2O, CO, CH2CO, and CO in tandem mass spectrometry (MS/MS) [46].
In this study, more m/z peaks were observed in the APCI technique (225 peaks) than ESI technique (174 peaks), and 65 m/z peaks were found common. Using both ESI and APCI, 334 m/z peaks of ZLE were detected, which specified the advantage of using both techniques for enhanced metabolite profiling. As a result, a higher number of compounds could be identified in this study compared to previous ones. One study reported the identification of six compounds, including apigenin-7-O-α-ʟ-rhamnoside, oleanolic acid, caffeic acid, lupeol acetate, myricetin-3-O-α-ʟ-rhamnoside, some hydrocarbons, and fatty acids [5], and another study reported the presence of myricetin-3-O-α-ʟ-rhamnoside and myricetin [47] in the M. zapota leaves. However, in this study, 39 compounds including sugar, dicarboxylic acids, phenolic acids, flavonoids, and other phytochemicals were identified in ZLE (Table 1).
DPPH● and ABTS●+ scavenging assays are widely practiced procedures for the determination of antioxidant prospective of natural supplements (e.g., foods, plants). DPPH● and ABTS●+ scavenging methods measure the transfer of hydrogen or electrons from potential antioxidants to free radicals. In this study, ZLE exhibited more significant DPPH● scavenging activity than the ABTS●+ scavenging activity (Figure 3A,B). FRAP and CUPRAC assays were performed to measure the reducing potentiality of antioxidants in which the reduction is performed with the assistance of compounds which break free-radical chain through donating hydrogen atom [48]. Experimental findings suggest that ZLE exhibited a noticeable antioxidant potential (ascorbic acid equivalent CUPRAC and FRAP values), increased with increasing concentration of the samples (Figure 3C,D). Taken together, the results demonstrated that ZLE has a strong potential antioxidant activity. The DPPH● and ABTS●+ scavenging abilities of Manilkara zapota fruit with IC50 37.63 ± 1.18 μg/mL and 73.14 ± 2.84 μg/mL, respectively, have been reported [49]. However, in our present study, we found the DPPH● and ABTS●+ scavenging activities of ZLE with IC50 7.93 ± 1.43 and 72.85 μg/mL, accordingly, which indicated that M. zapota leaves might be a promising bioactive agent.
Phenolic compounds exhibit antioxidant activities through donating hydrogen to the reactive oxygen or nitrogen species from the hydroxyl groups, which broke the radical generating cycle [50]. Identified phenolic compounds of M. zapota leaves such as 3,4-dihydroxybenzoic acid (DPPH●: IC50 1.88 µg/mL, ABTS●+: IC50 0.89 µg/mL) [51], gallic acid (DPPH●: IC50 3.53 ± 0.24 µg/mL, ABTS●+: IC50 8.85 ± 0.74 µg/mL), caffeic acid (DPPH●: IC50 6.34 ± 0.37 µg/mL, ABTS●+: IC50 18.04 ± 0.68 µg/mL), syringic acid (DPPH●: IC50 5.44 ± 0.53 µg/mL, ABTS●+: IC50 13.97 ± 0.59 µg/mL), ferulic acid (DPPH●: IC50 11.75 ± 0.45 µg/mL, ABTS●+: IC50 9.47 ± 0.69 µg/mL) [52], ampelopsin (DPPH●: IC50 4.94 µM), and myricetin-3-O-rhamnoside (DPPH●: IC50 5.14 µM) [53] have been reported to possess antioxidant activity.
More research is focusing on the functional food field especially on foods, and plants to prevent type-2 diabetes because of their α-glucosidase inhibition activity [54]. Various phytochemicals from herbs, barks, fruits, leaves, and different parts of medicinal plants have been recorded to exhibit inhibition against α-glucosidase activity and used as a remedy for type-2 diabetes [55]. Dichloromethane extracts of Croton bonplandianum [56]; ethanol extracts of Enhalus acoroides, Thalassia hemprichii, and Cymodocea rotundata [57]; ethanol extracts of Castanea mollissima [58]; acetone extracts of Undaria pinnatifida [59]; ethanol extracts of Vitis aestivalis [60]; and water extracts and ethanol extracts from Fucus vesiculosus [61] exhibited α-glucosidase inhibition abilities with IC50 values of 14.93, 168.15, 425.86, 429.28, 2.3, 80, 384, 0.32, and 0.49 µg/mL, respectively, and have traditionally been used for treatment of type 2 diabetes (T2D). Distilled water extraction of Achras sapota (M. zapota) fruit has been reported to inhibit α-glucosidase enzyme with IC50 56 µg/mL [62], whereas our results revealed that ZLE had significant potential for inhibiting α-glucosidase activity with IC50 2.51 ± 0.15 µg/mL, which suggested that ZLE might be as a prospective source in developing anti-diabetic agents.
It has been reported that caffeic acid and chlorogenic acid (compounds found in ZLE) have remarkable α-glucosidase inhibition ability with IC50 values of 4.98 and 9.24 µg/mL, respectively [63]. Other phenolic compounds like ferulic acid (IC50 10.80 ± 0.90 mg/mL) [64] and epicatechin (IC50 5.86 µg/mL) [65] also possess α-glucosidase inhibition activity. In this study, we identified norathyriol and myricetin from ZLE using negative-mode APCI–MS which possesses a potential α-glucosidase inhibition activity with IC50 values of 0.81 and 2.09 µg/mL, accordingly [66,67]. Therefore, it was assumed that ZLE showed a significant α-glucosidase inhibition potential due to the combined effect of these compounds.
As we found that ZLE possesses significant α-glucosidase inhibition activity, we carried out the uptake of glucose in C2C12 myotubes for determining the prospects of ZLE as an anti-diabetic agent. Skeletal muscle is an important site for the insulin-mediated glucose uptake, transport, and repository. Furthermore, an increase in glucose uptake in this cell is considered to play a key function in the control of glucose homeostasis and regulation of T2D [68]. ZLE significantly improved the uptake of glucose in C2C12 cells in concentration dependently. We observed synergistic effect of ZLE on glucose uptake in C2C12 cells on exposure to insulin, which suggested the role of ZLE to imitate the insulin-like actions (Figure 4D).
Collagen and elastin being the prominent proteins in the body modulate important biological and mechanical functions. Collagen is the principal part of the extracellular matrix (ECM), and it provides structural support and strength. On the other hand, elastin is the most prevalent constituent of ECM, which contributes to elasticity in living tissues [69,70,71]. Elastin is degraded by the activation of elastase upon UV irradiation [72]. We found in this study that ZLE strongly inhibited elastase enzyme activity (Supplementary Figure S9). In our previous study, it was found that different combinations of epicatechin, syringic acid, and vanillic acid had a substantial interactive effect on inhibitory ability of elastase [73]. Sirinya et al. found the elastase inhibition activity of Manilkara zapota fruit with IC50 35.73 ± 0.61 μg/mL [49], whereas in our study ZLE exhibited elastase inhibitory action with IC50 27.51 ± 1.70 µg/mL. Therefore, M. zapota leaves possessed more significant elastase inhibition activity than the fruits.
4. Materials and Methods
4.1. Chemicals
Neocuproine (≥98%), p-nitrophenyl-α-D-glucopyranoside (pNPG) (≥99%), 2,2-diphenyl-1-picrylhydrazyl (DPPH) (≥ 98%), 2,4,6-tris(2-pyridyl)-s-triazine (≥99%), 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid (ABTS) (≥98%), α-glucosidase (23 units/mg protein), N-succinyl-(Ala)-3-p-nitroanilide (≥98%), dimethylsulfoxide (DMSO), ascorbic acid (≥99%), elastase (7.6 units/ mg protein), and HPLC-grade ethanol were obtained from Sigma–Aldrich (St. Louis, MO, USA). We used all of these chemicals without further purification to conduct this research work.
4.2. Plant Materials and Extraction
Leaves of M. zapota L. were collected from the Khulna area of Bangladesh in June 2018. Taxonomic identification (accession number DACB-23801) was carried out at the Bangladesh National Herbarium. Shade-dried fresh leaves were ground into fine homogenized powder using a mortar and pestle. Of the fine power, 100 g was processed in three cycles with a reflux system of the extraction process of 6 h using ethanol. A Whatman filter paper (Schleicher and Schuell, Keene, NH, USA) was used to filter the extracts. Evaporation of the solvent was done using a rotary evaporator (SB-1000, Tokyo Rikakikai Co. Ltd., Tokyo, Japan). At last, we obtained powdered extracts of the sample after using a freeze-dryer (MCFD 8518, Ilshin Biobase Co. Ltd., Goyang, Korea).
4.3. Sample Preparation
Stock solution of ZLE was prepared by dissolving the extract in 100% ethanol (10 mg/mL), and subsequently, the stock solution was diluted using ethanol and water for MS analysis. Homogenization of the prepared samples solution was made following 1 min vortex and 5 min sonication in a sonication bath (Powersonic 410, Hwashin Technology Co., Gyeonggi, South Korea). For in vitro analysis, the extracts at a concentration of 30 mg/mL were dissolved in DMSO and deionized water (50:50, v/v), and subsequent diluted solutions were made using deionized water.
4.4. Mass Spectrometry Analysis
Negative-mode ESI–MS and APCI–MS experiments were executed with Q-Exactive™ Orbitrap Mass Spectrometer (Thermo Fisher Scientific Inc., San Jose, CA, USA). A 500-µL syringe (Hamilton Company Inc., Reno, NV, USA) and a syringe pump (model 11, Harvard, Holliston, MA, USA) were used to infuse the sample solution into the ESI source at 20 µL/min. Typical negative-mode ESI–MS conditions were at mass resolution 140,000 (full width at half maximum, FWHM), sheath gas flow rate 6, auxiliary gas flow rate 2, sweep gas flow rate 0 (arbitrary units), spray voltage 4.00 kV, capillary temperature 400 °C, S-lens RF level 50, and automatic gain control 5 E6. Highly pure (99.99 %) nitrogen obtained from the evaporation of liquid nitrogen was applied as the sheath, auxiliary, and sweep gas. Negative-mode external calibration was performed using Pierce Velos solution (Thermo Fisher Scientific, Rockford, IL, USA) into the ESI interface.
Operating conditions for negative-mode APCI–MS were as follows: sample infusion rate 200 µL/min through a 5-mL syringe (Hamilton Company Inc., Reno, NV, USA), mass resolution (FWHM) 140,000, sheath gas flow rate 15, auxiliary gas flow rate 2, sweep gas flow rate 0 (arbitrary units), discharge current 2 µA, capillary temperature 300 °C, vaporizer temperature 400 °C, S-lens RF level 70, and automatic gain control 5 E6.
Mass spectra were acquisitioned from m/z 50 to m/z 750 for both ionizations and 10–30 eV collision energy was applied in the tandem mass spectrometry (ESI-MS/MS and APCI-MS/MS).
Experimental conditions for ESI and APCI mass spectrometry were optimized considering the most abundant peak’s intensity to obtain a good quality spectrum. Operating parameters for ESI–MS and APCI–MS were optimized and briefed in the Supplementary Data (Figures S10–12, Tables S2 and S3).
4.5. Data Processing
Obtained mass spectrometry data were handled using Xcalibur 3.1 with Foundation 3.1 (Thermo Fisher Scientific, USA). A signal-to-noise ratio (S/N) value of 100 was considered to detect m/z peaks, and exported peak lists were further processed using Microsoft Excel. Elemental formulas for the detected m/z peaks were assigned within typical conditions of CcHhNnOo, counting c unlimited, h unlimited, and 0 ≤ n ≤ 10, and 0 ≤ o ≤ 50 using MIDAS Formula Calculator. ChemDraw Professional 15.0 (PerkinElmer, Waltham, MA, USA) was used to draw the structures of the identified compounds. A Venn diagram for the overlapped peaks was generated by FunRich 3.1.3 [74].
Compound identification was executed by comparing theoretically calculated masses of [M–H]− and [M+Cl]− adducts with the observed m/z peak values. For the structural elucidation of the detected phytochemicals, ESI–MS/MS- and APCI–MS/MS-produced fragments of the target peaks were compared with the Human Metabolome Database (online database) [15] and literatures.
4.6. Antioxidant Activity Assays
The DPPH● scavenging capability of ZLE was determined following a previous methodology [75]. Into 2-µL sample solutions of different concentrations (1, 3, 10, 30, and 100 µg/mL), 198 µL of 0.2 mM solution of DPPH in 50% ethanol was added. The mixture was allowed to stand for 10 min at 25 °C, and the absorbance was recorded by a microplate reader (Multiskan GO 51119200; Thermo Fisher Scientific Oy, Ratastie, 01620 Vantaa, Finland) at 517 nm. Ascorbic acid was used as a reference antioxidant compound.
The ABTS●+ scavenging activity was determined adapting the method described by Re et al. [76] with slight modifications, and 2 µL of different concentrations of ZLE (1, 3, 10, 30, and 100 µg/mL) was mixed with 198 µL ABTS solution, and the absorbance was recorded at 734 nm. Ascorbic acid was used as a reference antioxidant compound, and both DPPH● and ABTS●+ scavenging activities were calculated using Equation (1).
| Radical-scavenging activity (% inhibition) = [(Acontrol − Asample) / Acontrol] × 100 | (1) |
where Acontrol is the control’s absorbance and Asample is the sample’s absorbance. Data were obtained for three samples.
The ferric-reducing antioxidant power (FRAP) assay and cupric-reducing antioxidant capacity (CUPRAC) assay were performed following the reported methods [77,78]. Then, 2 µL of the sample solution of ZLE at different concentrations (1, 3, 10, 30, and 100 µg/mL) was mixed with 198 µL of the FRAP reagent, and the absorbance was measured at 595 nm. Ascorbic acid was also used as a reference compound. In the case of the CUPRAC assay, the samples at different concentrations (1, 3, 10, 30, and 100 µg/mL) were added with the mixture of 10 mM CuCl2, 7.5 mM neocuproine, and 1 M ammonium acetate buffer (pH 7.0). The mixture was incubated for 1 h at 25 °C temperature, and the absorbance was determined at 450 nm. Ascorbic acid was used as a reference compound, and the reducing capability was marked as ascorbic acid equivalent (in micromolar).
4.7. Inhibitory Assay of α-Glucosidase
The capability of ZLE to restrain α-glucosidase activity was determined in accordance with the reported method [79]. With 0.2 U/mL of α-glucosidase in 0.1 M sodium phosphate buffer (pH 7.0), the sample solutions of 2 µL of different concentrations (1, 3, 10, 30, and 100 µg/mL) were mixed, and incubated for 10 min at 37 °C. Then, pNPG (substrate) was mixed into the prepared solution to terminate enzyme–substrate activity at 37 °C for 1 h. The enzyme–substrate reaction was carried out at 37 °C for 30 min. Absorbance of the mixture was figured out utilizing a 96-well plate microplate reader (Multiskan GO 51119200; Thermo Fisher Scientific Oy, Ratastie, 01620 Vantaa, Finland) at 405 nm. Acarbose as a positive control was employed and inhibitory percentage was calculated following the Equation (2).
| Inhibition of enzyme activity (%) = [(Acontrol − Asample)/Acontrol] × 100 | (2) |
where Acontrol is the control’s absorbance and Asample is the sample’s absorbance. Three sets of the sample’s data were obtained.
4.8. Cell Culture and Cell Viability Assay
C2C12 myotubes (American Type Culture Collection, Manassas, VA, USA) were grown in Dulbecco’s modified eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) and streptomycin-penicillin (100 µg/mL and 100 U/mL, accordingly; Hyclone, Mordialloc, VIC, Australia) at 37 °C under a 5% CO2 atmosphere. The viability of C2C12 cells were detected following the tetrazolium dye colorimetric test (MTT) assay. At first C2C12 cells were grown at a density of 1 × 105 cells/well in 96-well plates for 24 h. They were treated with different concentrations of ZLE after reaching 90% confluence. After 24 h of incubation, an MTT reagent was added to each well and the plate was incubated for 1 h at 37 °C. The media was removed and then the wells were washed two times with phosphate-buffered saline (PBS) (pH 7.4). The intracellular formazan was solubilized in 100% DMSO. The absorbance was recorded by microplate reader (Perkin Elmer, Wallac Victor 3, MA, USA) at 570 nm and the viability percentage was determined.
4.9. Muscle Differentiation and Glucose Uptake Assay
C2C12 cells were grown at a density of 1 × 105 cells/well in 96-well plates with DMEM supplemented with 10% fetal bovine serum (FBS) and 1% streptomycin–penicillin at 37 °C under 5% CO2 atmosphere. The cells were differentiated in DMEM containing 2% horse serum for 5 days after reaching confluence. The cells were allowed to starve in low glucose serum-free DMEM for 24 h. Thereafter, 2-deoxy-2-[(7-nitro-2,1,3-benzoxadiazol-4-yl) amino]-D-glucose (2-NBDG) assay was carried out to determine glucose uptake [80]. Briefly, cells were treated with different concentrations of ZLE and insulin (100 nM) for 30 min, followed by 20 µM of 2-NBDG for 6 h. The cells were rinsed three times using cold PBS after incubation and uptake of 2-NBDG uptake recorded with a microplate reader (Perkin Elmer, Wallac Victor 3, MA, USA) at an excitation wavelength of 490 nm and emission wavelengths of 535 nm, accordingly.
4.10. Elastase Inhibition Assay
ZLE’s potentiality to hinder elastase activity was employed based on a previous study [73]. The reaction was performed in a mixture of 0.04 U elastase and 0.1 M Tris–HCl buffer (pH 8.0) with 0.78 mM N-succinyl-(Ala)-3-p-nitroanilide (substrate). With 100 µL of the substrate solution, different concentrations of the sample solution (1, 3, 10, 30, and 100 µg/mL) were mixed and then added to 100-µL enzyme solution. Then, mixer absorbance was figured out using a microplate reader (Multiskan GO 51119200; Thermo Fisher Scientific Oy, Ratastie, 01620 Vantaa, Finland) at 405 nm. Epigallocatechin gallate (EGCG) as a positive control was used, and the elastase inhibition percentage was calculated following above mentioned Equation (2).
4.11. Statistical Analysis
All results were as mean ± standard deviation. Linear regression analyses were carried out to calculate half maximal inhibitory concentration values (IC50). Statistical analyses were accomplished through one-way analysis of variance (ANOVA), following Dennett’s test by SigmaPlot 12.5 (Systat Software Inc., San Jose, CA, USA). Differences were counted significant if p < 0.05.
5. Conclusions
In this study, we found that combining ESI and APCI techniques with mass spectrometry maximized the coverage of m/z peak detection rather than using a single ionization technique. The combination of ESI and APCI ionization techniques in mass spectrometry provided us with more detailed characteristic features of metabolites and biological activities of natural compounds. We rapidly determined the secondary metabolites of M. zapota leaves using direct injection mode mass spectrometry. We identified 39 different phytochemicals of M. zapota leaves using negative-mode ESI–MS and APCI–MS. It was found that the leaf extracts of M. zapota had a significant α-glucosidase enzyme inhibition activity and also dose-dependently augmented glucose uptake alone or in combination with insulin in C2C12 myotubes. M. zapota leaves extracts also showed strong antioxidant and elastase inhibition potential. Compounds identified using negative-mode APCI–MS together with negative-mode ESI–MS strongly demonstrated the α-glucosidase activity of ZLE. It can be concluded that combining ESI and APCI with high-resolution mass spectrometry is an enhanced approach for metabolite profiling and biological activity studies. The findings of this work suggest that M. zapota leaves can be a potential resource for antidiabetic agents, antioxidants, and elastase enzyme inhibitors. Furthermore, we need to carry out liquid chromatography mass spectrometry (LC-MS) to determine the concentration of bioactive compounds. In vivo biological assays are also needed to be performed for the better characterization of antidiabetic ability of M. zapota leaves. Finally, we are interested to continue research on selective bioactive compounds for enhanced antidiabetic activity.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/22/1/132/s1. Figure S1: Venn diagram showing the overlapping peaks detected in (–) mode electrospray ionization (ESI) and atmospheric pressure chemical ionization (APCI) mass spectrometry (MS). Figure S2: (–) mode electrospray ionization tandem mass spectrometry (ESI MS/MS) fragmentation pattern of succinic acid. Figure S3: (–) mode electrospray ionization tandem mass spectrometry (ESI MS/MS) fragmentation pattern of malic acid. Figure S4: (–) mode electrospray ionization tandem mass spectrometry (ESI MS/MS) fragments of gallic acid. Figure S5: (–) mode electrospray ionization tandem mass spectrometry (ESI MS/MS) fragments of epicatechin. Figure S6: (–) mode atmospheric pressure chemical ionization tandem mass spectrometry (APCI MS/MS) fragments of myricetin. Figure S7: (–) mode electrospray ionization tandem mass spectrometry (ESI MS/MS) fragments of quinic acid. Figure S8: (–) mode atmospheric pressure chemical ionization tandem mass spectrometry (APCI MS/MS) fragments of norathyriol. Figure S9: Elastase-inhibition activities of Manilkara zapota leaves ethanol extracts (ZLE). Figure S10: Optimization of negative-mode electrospray ionization mass spectrometry (ESI–MS) operating parameters. Figure S11: Optimization of (–) mode atmospheric pressure chemical ionization mass spectrometry (APCI-MS) operating parameters. Figure S12: (–) mode atmospheric pressure chemical ionization (APCI) mass spectra at different vaporizer temperatures. Table S1. Detected peak lists in APCI and ESI MS. Table S2. Optimized operational conditions for negative-mode electrospray ionization mass spectrometry (ESI–MS). Table S3. Optimized operational conditions for (–) mode atmospheric pressure chemical ionization mass spectrometry (APCI–MS).
Author Contributions
Conceptualization, S.-H.L. and S.K.; methodology, S.I. and M.B.A.; validation, S.I. and M.B.A.; formal analysis, S.I., M.B.A., H.-J.A., and J.-H.P.; investigation, S.I., M.B.A., H.-J.A., and J.-H.P.; resources, S.-H.L. and S.K.; data curation, S.I. and M.B.A.; writing—original draft preparation, S.I.; writing—review and editing, S.I., M.B.A., S.K., and S.-H.L.; visualization, M.B.A.; supervision, S.K. and S.-H.L.; project administration, S.K.; funding acquisition, S.K. and S.-H.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was carried out with the assistance of National Research Foundation of Korea (NRF) grant, financed by Ministry of Science, ICT and Future Planning (MSIP), Government of South Korea (2017R1A2B3003455), Korea Basic Science Institute (KBSI), and a National Research Facilities and Equipment Center (NFEC) grant financed by Ministry of Education, Government of South Korea (2019R1A6C1010001 and 2019R1A6C1030012).
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cragg G.M., Newman D.J. Natural product drug discovery in the next millennium. Pharm. Biol. 2001;39:8–17. doi: 10.1076/phbi.39.s1.8.0009. [DOI] [PubMed] [Google Scholar]
- 2.Ortega A.M.M., Campos M.R.S. Bioactive compounds as therapeutic alternatives. In: Campos M.R.S., editor. Bioactive Compounds: Health Benefits and Potential Applications. Woodhead Publishing; Sawston, UK: 2019. pp. 247–264. Chapter 13. [Google Scholar]
- 3.Hostettmann K., Wolfender J.L., Terreaux C. Modern screening techniques for plant extracts. Pharm. Biol. 2001;39:18–32. doi: 10.1076/phbi.39.s1.18.0008. [DOI] [PubMed] [Google Scholar]
- 4.Kaneria M., Chanda S. Evaluation of antioxidant and antimicrobial properties of Manilkara zapota L. (chiku) leaves by sequential soxhlet extraction method. Asian Pac. J. Trop. Biomed. 2012;2:S1526–S1533. doi: 10.1016/S2221-1691(12)60448-1. [DOI] [Google Scholar]
- 5.Fayek N.M., Monem A.R., Mossa M.Y., Meselhy M.R., Shazly A.H. Chemical and biological study of Manilkara zapota (L.) Van Royen leaves (Sapotaceae) cultivated in Egypt. Pharmacogn. Res. 2012;4:85–91. doi: 10.4103/0974-8490.94723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chunhakant S., Chaicharoenpong C. Antityrosinase, Antioxidant, and Cytotoxic Activities of Phytochemical Constituents from Manilkara zapota L. Bark. Molecules. 2019;24:2798. doi: 10.3390/molecules24152798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chanda S., Nagani K. Antioxidant capacity of Manilkara zapota L. leaves extracts evaluated by four in vitro methods. Nat. Sci. 2010;8:260–266. [Google Scholar]
- 8.Ma J., Luo X.D., Protiva P., Yang H., Ma C.Y., Basile M.J., Weinstein I.B., Kennelly E.J. Bioactive novel polyphenols from the fruit of Manilkara zapota (Sapodilla) J. Nat. Prod. 2003;66:983–986. doi: 10.1021/np020576x. [DOI] [PubMed] [Google Scholar]
- 9.Barbalho S.M., Bueno P.C.D., Delazari D.S., Guiguer E.L., Coqueiro D.P., Araujo A.C., de Souza M.D.S., Farinazzi-Machado F.M.V., Mendes C.G., Groppo M. Antidiabetic and Antilipidemic Effects of Manilkara zapota. J. Med. Food. 2015;18:385–391. doi: 10.1089/jmf.2013.0170. [DOI] [PubMed] [Google Scholar]
- 10.Gowda G.A., Djukovic D. Overview of mass spectrometry-based metabolomics: Opportunities and challenges. Methods Mol. Biol. 2014;1198:3–12. doi: 10.1007/978-1-4939-1258-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Commisso M., Anesi A., Dal Santo S., Guzzo F. Performance comparison of electrospray ionization and atmospheric pressure chemical ionization in untargeted and targeted liquid chromatography/mass spectrometry based metabolomics analysis of grapeberry metabolites. Rapid Commun. Mass Spectrom. 2017;31:292–300. doi: 10.1002/rcm.7789. [DOI] [PubMed] [Google Scholar]
- 12.Sleeman R., Carter J.F. Mass spectrome try|Overview. In: Worsfold P., Townshend A., Poole C., editors. Encyclopedia of Analytical Science. 2nd ed. Elsevier; Oxford, UK: 2005. pp. 337–344. [Google Scholar]
- 13.Lynn K.S., Cheng M.L., Chen Y.R., Hsu C., Chen A., Lih T.M., Chang H.Y., Huang C.J., Shiao M.S., Pan W.H., et al. Metabolite Identification for Mass Spectrometry-Based Metabolomics Using Multiple Types of Correlated Ion Information. Anal. Chem. 2015;87:2143–2151. doi: 10.1021/ac503325c. [DOI] [PubMed] [Google Scholar]
- 14.Jin J., Lao J., Zhou R., He W., Qin Y., Zhong C., Xie J., Liu H., Wan D., Zhang S., et al. Simultaneous Identification and Dynamic Analysis of Saccharides during Steam Processing of Rhizomes of Polygonatum cyrtonema by HPLC⁻QTOF⁻MS/MS. Molecules. 2018;23:2855. doi: 10.3390/molecules23112855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wishart D.S., Feunang Y.D., Marcu A., Guo A.C., Liang K., Vazquez-Fresno R., Sajed T., Johnson D., Li C., Karu N., et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018;46:D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou D., Wang J., Zhang B., Xie S., Wang Q., Xu K., Lin R. Analysis of Chemical Constituents in Wuzi-Yanzong-Wan by UPLC-ESI-LTQ-Orbitrap-MS. Molecules. 2015;20:21373–21404. doi: 10.3390/molecules201219765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang T.M., Fu Y., Yu W.J., Chen C., Di X., Zhang H., Zhai Y.J., Chu Z.Y., Kang T.G., Chen H.B. Identification of Polar Constituents in the Decoction of Juglans mandshurica and in the Medicated Egg Prepared with the Decoction by HPLC-Q-TOF MS2. Molecules. 2017;22:1452. doi: 10.3390/molecules22091452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J., Jia Z., Zhang Z., Wang Y., Liu X., Wang L., Lin R. Analysis of Chemical Constituents of Melastoma dodecandrum Lour. by UPLC-ESI-Q-Exactive Focus-MS/MS. Molecules. 2017;22:476. doi: 10.3390/molecules22030476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Y.H., Kim B., Kim S., Kim M.S., Kim H., Hwang S.R., Kim K., Lee J.H. Characterization of metabolite profiles from the leaves of green perilla (Perilla frutescens) by ultra high performance liquid chromatography coupled with electrospray ionization quadrupole time-of-flight mass spectrometry and screening for their antioxidant properties. J. Food Drug Anal. 2017;25:776–788. doi: 10.1016/j.jfda.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ismail H., Gillespie A.L., Calderwood D., Iqbal H., Gallagher C., Chevallier O.P., Elliott C.T., Pan X., Mirza B., Green B.D. The Health Promoting Bioactivities of Lactuca sativa can be Enhanced by Genetic Modulation of Plant Secondary Metabolites. Metabolites. 2019;9:97. doi: 10.3390/metabo9050097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu M.H., Zhang Q., Zhang Y.H., Lu X.Y., Fu W.M., He J.Y. Chemical Analysis of Dietary Constituents in Rosa roxburghii and Rosa sterilis Fruits. Molecules. 2016;21:1204. doi: 10.3390/molecules21091204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding M., Jiang Y., Yu X., Zhang D., Li J., Wang H., Shen J., Gao X.M., Chang Y.X. Screening of Combinatorial Quality Markers for Natural Products by Metabolomics Coupled with Chemometrics: A Case Study on Pollen Typhae. Front. Pharmacol. 2018;9:691. doi: 10.3389/fphar.2018.00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh D., Siew Y.Y., Chong T.I., Yew H.C., Ho S.S., Lim C.S.E., Tan W.X., Neo S.Y., Koh H.L. Identification of Phytoconstituents in Leea indica (Burm. F.) Merr. Leaves by High Performance Liquid Chromatography Micro Time-of-Flight Mass Spectrometry. Molecules. 2019;24:714. doi: 10.3390/molecules24040714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abu-Reidah I.M., Ali-Shtayeh M.S., Jamous R.M., Arráez-Román D., Segura-Carretero A. HPLC–DAD–ESI-MS/MS screening of bioactive components from Rhus coriaria L. (Sumac) fruits. Food Chem. 2015;166:179–191. doi: 10.1016/j.foodchem.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 25.Ancillotti C., Ciofi L., Rossini D., Chiuminatto U., Stahl-Zeng J., Orlandini S., Furlanetto S., Del Bubba M. Liquid chromatographic/electrospray ionization quadrupole/time of flight tandem mass spectrometric study of polyphenolic composition of different Vaccinium berry species and their comparative evaluation. Anal. Bioanal. Chem. 2017;409:1347–1368. doi: 10.1007/s00216-016-0067-y. [DOI] [PubMed] [Google Scholar]
- 26.Taamalli A., Iswaldi I., Arraez-Roman D., Segura-Carretero A., Fernandez-Gutierrez A., Zarrouk M. UPLC-QTOF/MS for a rapid characterisation of phenolic compounds from leaves of Myrtus communis L. Phytochem. Anal. 2014;25:89–96. doi: 10.1002/pca.2475. [DOI] [PubMed] [Google Scholar]
- 27.Tahri W., Chatti A., Romero-González R., López-Gutiérrez N., Frenich A.G., Landoulsi A. Phenolic profiling of the aerial part of Chrysanthemum trifurcatum using ultra high performance liquid chromatography coupled to Orbitrap high resolution mass spectrometry. Anal. Methods. 2016;8:3517–3527. doi: 10.1039/C6AY00365F. [DOI] [Google Scholar]
- 28.Candela L., Formato M., Crescente G., Piccolella S., Pacifico S. Coumaroyl Flavonol Glycosides and More in Marketed Green Teas: An Intrinsic Value beyond Much-Lauded Catechins. Molecules. 2020;25:1765. doi: 10.3390/molecules25081765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanz M., de Simón B.F., Cadahía E., Esteruelas E., Muñoz A.M., Hernández T., Estrella I., Pinto E. LC-DAD/ESI-MS/MS study of phenolic compounds in ash (Fraxinus excelsior L. and F. americana L.) heartwood. Effect of toasting intensity at cooperage. J. Mass Spectrom. 2012;47:905–918. doi: 10.1002/jms.3040. [DOI] [PubMed] [Google Scholar]
- 30.Fathoni A., Saepudin E., Cahyana A.H., Rahayu D.U.C., Haib J. Identification of nonvolatile compounds in clove (Syzygium aromaticum) from Manado. AIP Conf. Proc. 2017;1862:030079. [Google Scholar]
- 31.Guo X., Cheng M., Hu P., Shi Z., Chen S., Liu H., Shi H., Xu Z., Tian X., Huang C. Absorption, Metabolism, and Pharmacokinetics Profiles of Norathyriol, an Aglycone of Mangiferin, in Rats by HPLC-MS/MS. J. Agric. Food Chem. 2018;66:12227–12235. doi: 10.1021/acs.jafc.8b03763. [DOI] [PubMed] [Google Scholar]
- 32.Abu-Reidah I.M., Arráez-Román D., Segura-Carretero A., Fernández-Gutiérrez A. Profiling of phenolic and other polar constituents from hydro-methanolic extract of watermelon (Citrullus lanatus) by means of accurate-mass spectrometry (HPLC–ESI–QTOF–MS) Food Res. Int. 2013;51:354–362. doi: 10.1016/j.foodres.2012.12.033. [DOI] [Google Scholar]
- 33.Orlando G., Zengin G., Ferrante C., Ronci M., Recinella L., Senkardes I., Gevrenova R., Zheleva-Dimitrova D., Chiavaroli A., Leone S., et al. Comprehensive Chemical Profiling and Multidirectional Biological Investigation of Two Wild Anthemis Species (Anthemis tinctoria var. Pallida and A. cretica subsp. tenuiloba): Focus on Neuroprotective Effects. Molecules. 2019;24:2582. doi: 10.3390/molecules24142582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Álvarez-Fernández M.A., Hornedo-Ortega R., Cerezo A.B., Troncoso A.M., García-Parrilla M.C. Determination of Nonanthocyanin Phenolic Compounds Using High-Resolution Mass Spectrometry (UHPLC-Orbitrap-MS/MS) and Impact of Storage Conditions in a Beverage Made from Strawberry by Fermentation. J. Agric. Food Chem. 2016;64:1367–1376. doi: 10.1021/acs.jafc.5b05617. [DOI] [PubMed] [Google Scholar]
- 35.Kumar S., Narwal S., Kumar V., Prakash O. α-glucosidase inhibitors from plants: A natural approach to treat diabetes. Pharmacogn. Rev. 2011;5:19–29. doi: 10.4103/0973-7847.79096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christhudas I.N., Kumar P.P., Sunil C., Vajravijayan S., Sundaram R.L., Siril S.J., Agastian P. In vitro studies on α-glucosidase inhibition, antioxidant and free radical scavenging activities of Hedyotis biflora L. Food Chem. 2013;138:1689–1695. doi: 10.1016/j.foodchem.2012.11.051. [DOI] [PubMed] [Google Scholar]
- 37.Sun C., Wu Z., Wang Z., Zhang H. Effect of Ethanol/Water Solvents on Phenolic Profiles and Antioxidant Properties of Beijing Propolis Extracts. Evid. Based Complement. Altern. Med. 2015;2015:595393. doi: 10.1155/2015/595393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borges A., José H., Homem V., Simões M. Comparison of Techniques and Solvents on the Antimicrobial and Antioxidant Potential of Extracts from Acacia dealbata and Olea europaea. Antibiotics. 2020;9:48. doi: 10.3390/antibiotics9020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia-Flores M., Juarez-Colunga S., Montero-Vargas J.M., Lopez-Arciniega J.A.I., Chagolla A., Tiessena A., Winkler R. Evaluating the physiological state of maize (Zea mays L.) plants by direct-injection electrospray mass spectrometry (DIESI-MS) Mol. Biosyst. 2012;8:1658–1660. doi: 10.1039/c2mb25056j. [DOI] [PubMed] [Google Scholar]
- 40.van Leerdam J.A., Vervoort J., Stroomberg G., de Voogt P. Identification of Unknown Microcontaminants in Dutch River Water by Liquid Chromatography-High Resolution Mass Spectrometry and Nuclear Magnetic Resonance Spectroscopy. Environ. Sci. Technol. 2014;48:12791–12799. doi: 10.1021/es502765e. [DOI] [PubMed] [Google Scholar]
- 41.Liotta L.J., James-Pederson M. Identification of an Unknown Compound by Combined Use of IR, 1H NMR, 13C NMR, and Mass Spectrometry: A Real-Life Experience in Structure Determination. J. Chem. Educ. 2008;85:832. doi: 10.1021/ed085p832. [DOI] [Google Scholar]
- 42.Da Silva L.C., Pereira I., De Carvalho T.C., Allochio Filho J.F., Romão W., Vaz B.G. Paper spray ionization and portable mass spectrometers: A review. Anal. Methods. 2019;11:999–1013. doi: 10.1039/C8AY02270D. [DOI] [Google Scholar]
- 43.Siuzdak G. An Introduction to Mass Spectrometry Ionization: An Excerpt from The Expanding Role of Mass Spectrometry in Biotechnology, 2nd ed.; MCC Press: San Diego, 2005. J. Assoc. Lab. Autom. 2004;9:50–63. doi: 10.1016/j.jala.2004.01.004. [DOI] [Google Scholar]
- 44.Das C. Fundamentals of Contemporary Mass Spectrometry. John Wiley & Sons; Hoboken, NJ, USA: 2007. [Google Scholar]
- 45.Farag M.A., Otify A., Porzel A., Michel C.G., Elsayed A., Wessjohann L.A. Comparative metabolite profiling and fingerprinting of genus Passiflora leaves using a multiplex approach of UPLC-MS and NMR analyzed by chemometric tools. Anal. Bioanal. Chem. 2016;408:3125–3143. doi: 10.1007/s00216-016-9376-4. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt J. Negative ion electrospray high-resolution tandem mass spectrometry of polyphenols. J. Mass Spectrom. 2016;51:33–43. doi: 10.1002/jms.3712. [DOI] [PubMed] [Google Scholar]
- 47.Subramanian S.S., Nair A.G.R. Myricetin and myricetin-3-O-l-rhamnoside from the leaves of Madhuca indica and Achras sapota. Phytochemistry. 1972;11:3090–3091. doi: 10.1016/0031-9422(72)80119-5. [DOI] [Google Scholar]
- 48.Othman A., Ismail A., Ghani N.A., Adenan I. Antioxidant capacity and phenolic content of cocoa beans. Food Chem. 2007;100:1523–1530. doi: 10.1016/j.foodchem.2005.12.021. [DOI] [Google Scholar]
- 49.Pientaweeratch S., Panapisal V., Tansirikongkol A. Antioxidant, anti-collagenase and anti-elastase activities of Phyllanthus emblica, Manilkara zapota and silymarin: An in vitro comparative study for anti-aging applications. Pharm. Biol. 2016;54:1865–1872. doi: 10.3109/13880209.2015.1133658. [DOI] [PubMed] [Google Scholar]
- 50.Pereira D.M., Valentao P., Pereira J.A., Andrade P.B. Phenolics: From Chemistry to Biology. Molecules. 2009;14:2202–2211. doi: 10.3390/molecules14062202. [DOI] [Google Scholar]
- 51.Li X., Wang X., Chen D., Shuzhi C. Antioxidant Activity and Mechanism of Protocatechuic Acid in vitro. Funct. Foods Health Dis. 2011;7:232–244. doi: 10.31989/ffhd.v1i7.127. [DOI] [Google Scholar]
- 52.Singh D.P., Verma S., Prabha R. Investigations on Antioxidant Potential of Phenolic Acids and Flavonoids: The Common Phytochemical Ingredients in Plants. J. Plant Biochem. Physiol. 2018;6:1000219. doi: 10.4172/2329-9029.1000219. [DOI] [Google Scholar]
- 53.Hsu F.-L., Huang W.-J., Wu T.-H., Lee M.-H., Chen L.-C., Lu H.-J., Hou W.-C., Lin M.-H. Evaluation of antioxidant and free radical scavenging capacities of polyphenolics from pods of Caesalpinia pulcherrima. Int. J. Mol. Sci. 2012;13:6073–6088. doi: 10.3390/ijms13056073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alongi M., Celayeta J.M.F., Vriz R., Kinsella G.K., Rulikowska A., Anese M. In vitro digestion nullified the differences triggered by roasting in phenolic composition and α-glucosidase inhibitory capacity of coffee. Food Chem. 2020:128289. doi: 10.1016/j.foodchem.2020.128289. [DOI] [PubMed] [Google Scholar]
- 55.Assefa S.T., Yang E.Y., Chae S.Y., Song M., Lee J., Cho M.C., Jang S. Alpha Glucosidase Inhibitory Activities of Plants with Focus on Common Vegetables. Plants. 2019;9:2. doi: 10.3390/plants9010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maitla S. Evaluation of α-Glucosidase Inhibitory Activity of Dichloromethane and Methanol Extracts of Croton bonplandianum Baill. Trop. J. Pharm. Res. 2014;13:1833–1836. [Google Scholar]
- 57.Agung W., Effionora A., Tati N. In vitro Assay of Alpha-Glucosidase Inhibitor Activities of Three Seagrasses from Banten Bay, Indonesia. Pharmacogn. J. 2018;10:907–910. [Google Scholar]
- 58.Zhang Y., Yang Z., Liu G., Wu Y., Ouyang J. Inhibitory effect of chestnut (Castanea mollissima Blume) inner skin extract on the activity of α-amylase, α-glucosidase, dipeptidyl peptidase IV and in vitro digestibility of starches. Food Chem. 2020;324:126847. doi: 10.1016/j.foodchem.2020.126847. [DOI] [PubMed] [Google Scholar]
- 59.Zaharudin N., Staerk D., Dragsted L.O. Inhibition of α-glucosidase activity by selected edible seaweeds and fucoxanthin. Food Chem. 2019;270:481–486. doi: 10.1016/j.foodchem.2018.07.142. [DOI] [PubMed] [Google Scholar]
- 60.Zhang L., Hogan S., Li J., Sun S., Canning C., Zheng S.J., Zhou K. Grape skin extract inhibits mammalian intestinal α-glucosidase activity and suppresses postprandial glycemic response in streptozocin-treated mice. Food Chem. 2011;126:466–471. doi: 10.1016/j.foodchem.2010.11.016. [DOI] [Google Scholar]
- 61.Lordan S., Smyth T.J., Soler-Vila A., Stanton C., Ross R.P. The α-amylase and α-glucosidase inhibitory effects of Irish seaweed extracts. Food Chem. 2013;141:2170–2176. doi: 10.1016/j.foodchem.2013.04.123. [DOI] [PubMed] [Google Scholar]
- 62.Das S., De B. In Vitro Inhibition of Key Enzymes Related to Diabetes by the Aqueous Extracts of Some Fruits of West Bengal, India. Curr. Nutr. Food Sci. 2012;8:19–24. [Google Scholar]
- 63.Oboh G., Agunloye O.M., Adefegha S.A., Akinyemi A.J., Ademiluyi A.O. Caffeic and chlorogenic acids inhibit key enzymes linked to type 2 diabetes (in vitro): A comparative study. J. Basic Clin. Physiol. Pharm. 2015;26:165–170. doi: 10.1515/jbcpp-2013-0141. [DOI] [PubMed] [Google Scholar]
- 64.Deng N., Zheng B., Li T., Liu R.H. Assessment of the Phenolic Profiles, Hypoglycemic Activity, and Molecular Mechanism of Different Highland Barley (Hordeum vulgare L.) Varieties. Int. J. Mol. Sci. 2020;21:1175. doi: 10.3390/ijms21041175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Subramoniam A. Plants with Anti-Diabetes Mellitus Properties. 1st ed. CRC Press; Boca Raton, FL, USA: 2016. [Google Scholar]
- 66.Islam S., Alam M.B., Ahmed A., Lee S., Lee S.-H., Kim S. Identification of secondary metabolites in Averrhoa carambola L. bark by high-resolution mass spectrometry and evaluation for α-glucosidase, tyrosinase, elastase, and antioxidant potential. Food Chem. 2020;332:127377. doi: 10.1016/j.foodchem.2020.127377. [DOI] [PubMed] [Google Scholar]
- 67.He C., Liu X., Jiang Z., Geng S., Ma H., Liu B. Interaction Mechanism of Flavonoids and α-Glucosidase: Experimental and Molecular Modelling Studies. Foods. 2019;8:355. doi: 10.3390/foods8090355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koehn F.E., Carter G.T. The evolving role of natural products in drug discovery. Nat. Rev. Drug Discov. 2005;4:206–220. doi: 10.1038/nrd1657. [DOI] [PubMed] [Google Scholar]
- 69.Miranda-Nieves D., Chaikof E.L. Collagen and Elastin Biomaterials for the Fabrication of Engineered Living Tissues. ACS Biomater. Sci. Eng. 2017;3:694–711. doi: 10.1021/acsbiomaterials.6b00250. [DOI] [PubMed] [Google Scholar]
- 70.Linsenmayer T.F. Collagen. Cell Biology of Extracellular Matrix. Springer US; New York, NY, USA: 1991. pp. 7–44. [Google Scholar]
- 71.Mithieux S.M., Weiss A.S. Elastin. Adv. Protein Chem. 2005;70:437–461. doi: 10.1016/S0065-3233(05)70013-9. [DOI] [PubMed] [Google Scholar]
- 72.Seite S., Zucchi H., Septier D., Igondjo-Tchen S., Senni K., Godeau G. Elastin changes during chronological and photo-ageing: The important role of lysozyme. J. Eur. Acad. Dermatol. Venereol. 2006;20:980–987. doi: 10.1111/j.1468-3083.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 73.Kwon K.-R., Alam M.B., Park J.-H., Kim T.-H., Lee S.-H. Attenuation of UVB-Induced Photo-Aging by Polyphenolic-Rich Spatholobus Suberectus Stem Extract Via Modulation of MAPK/AP-1/MMPs Signaling in Human Keratinocytes. Nutrients. 2019;11:1341. doi: 10.3390/nu11061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pathan M., Keerthikumar S., Ang C.S., Gangoda L., Quek C.Y., Williamson N.A., Mouradov D., Sieber O.M., Simpson R.J., Salim A., et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- 75.Alam M.B., Ju M.K., Lee S.H. DNA Protecting Activities of Nymphaea nouchali (Burm. f) Flower Extract Attenuate t-BHP-Induced Oxidative Stress Cell Death through Nrf2-Mediated Induction of Heme Oxygenase-1 Expression by Activating MAP-Kinases. Int. J. Mol. Sci. 2017;18:2069. doi: 10.3390/ijms18102069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Re R., Pellegrini N., Proteggente A., Pannala A., Yang M., Rice-Evans C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999;26:1231–1237. doi: 10.1016/S0891-5849(98)00315-3. [DOI] [PubMed] [Google Scholar]
- 77.Benzie I.F., Strain J.J. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal. Biochem. 1996;239:70–76. doi: 10.1006/abio.1996.0292. [DOI] [PubMed] [Google Scholar]
- 78.Apak R., Guclu K., Ozyurek M., Karademir S.E. Novel total antioxidant capacity index for dietary polyphenols and vitamins C and E, using their cupric ion reducing capability in the presence of neocuproine: CUPRAC method. J. Agric. Food Chem. 2004;52:7970–7981. doi: 10.1021/jf048741x. [DOI] [PubMed] [Google Scholar]
- 79.Alam M.B., Bajpai V.K., Ra J.S., Lim J.Y., An H., Shukla S., Quan K.T., Khan I., Huh Y.S., Han Y.K., et al. Anthraquinone-type inhibitor of alpha-glucosidase enhances glucose uptake by activating an insulin-like signaling pathway in C2C12 myotubes. Food Chem. Toxicol. 2019;129:337–343. doi: 10.1016/j.fct.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 80.Xiong M., Huang Y., Liu Y., Huang M., Song G., Ming Q., Ma X., Yang J., Deng S., Wen Y., et al. Antidiabetic Activity of Ergosterol from Pleurotus Ostreatus in KK-A(y) Mice with Spontaneous Type 2 Diabetes Mellitus. Mol. Nutr. Food Res. 2018;62 doi: 10.1002/mnfr.201700444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.