Abstract
Environmental stresses have driven plants to develop various mechanisms to acclimate in adverse conditions. Extensive studies have demonstrated that a significant reprogramming occurs in the plant transcriptome in response to biotic and abiotic stresses. The highly conserved and large multi-subunit transcriptional co-activator of eukaryotes, known as the Mediator, has been reported to play a substantial role in the regulation of important genes that help plants respond to environmental perturbances. CDK8 module is a relatively new component of the Mediator complex that has been shown to contribute to plants’ defense, development, and stress responses. Previous studies reported that CDK8 module predominantly acts as a transcriptional repressor in eukaryotic cells by reversibly associating with core Mediator. However, growing evidence has demonstrated that depending on the type of biotic and abiotic stress, the CDK8 module may perform a contrasting regulatory role. This review will summarize the current knowledge of CDK8 module as well as other previously documented Mediator subunits in plant cell signaling under stress conditions.
Keywords: CDK8 module, Mediator subunits, biotic stress, abiotic stress, cell signaling
1. Introduction
In order to perceive and respond effectively to environmental stresses, plants have developed sophisticated signaling transduction pathways that induce gene expression changes via a complex network of transcription factors (TFs). Phytohormones of abscisic acid (ABA), jasmonic acid (JA), ethylene (ET), and salicylic acid (SA) are known to have extensive crosstalk with other hormones of auxin, brassinosteroid (BR), cytokinins, and gibberellic acid in plant stress response [1,2]. More than 1500 TFs in Arabidopsis have been reported to orchestrate the transcriptional control of abiotic stress responses [3]. The regulation of transcription in plant involves RNA polymerase II (RNA Pol II), general TFs, transcriptional activators/repressors, as well as co-regulators such as Mediator. Mediator was initially studied in yeast and subsequently identified to be critical for RNA Pol II-regulated transcription in eukaryotes including mammalian cells and plants through biochemical purification and comparative genomics [4,5].
The Mediator is an evolutionarily conserved large protein complex with multiple components called subunits that transfers upstream regulatory information from activators and repressors to the basal transcriptional machinery in the downstream pathway. A number of Mediator subunits have been identified to play critical roles in plant defense, adaptation, growth and development [6]. More subunits (25–35) are constituted in the Mediator of plant and mammalian cells than in yeast [7]. Mediator subunits of plants have been revealed to involve in various stress-response pathways through genetic analyses. There is also a fourth regulatory kinase module consisting of two Mediator subunits known as MED12 and MED13 as well as a separable kinase unit made up of CDK8 (cyclin-dependent kinase 8) and a C-type cyclin (CycC). CDK8, as its full name implies is a cyclin-dependent kinase and it along with its associated cyclin, CycC, also referred to as the kinase or CDK8-cyclin C module. The whole kinase module is an integral part of the Mediator complex. Studies have indicated that the protein components of the CDK8 kinase module may act as a both positive and negative transcription regulator in cells since CDK8 module was reported to act as a repressor to Mediator when bound to the complex [8,9]. However, there were situations when the CDK8 module worked as an activator on certain genes [10,11]. In plants, the kinase module was initially viewed as a predominant transcriptional repressor in eukaryotic cells by reversibly associating with the core Mediator. Later, evidence that supports the positive regulatory roles of CDK8 module in plants’ transcription has also appeared. In fact, the subunits of MED12 and MED13 were reported recently to serve as conditional positive gene regulators in Arabidopsis [12,13,14]. Likewise, the CDK8 subunit is capable of recruiting different TFs to RNA Pol II to regulate multiple signaling pathways in yeast and eukaryotic cells including plant cells [15]. The general working model for the signaling role of CDK8 and other Mediator subunits in plants during biotic and abiotic stresses are illustrated in Figure 1.
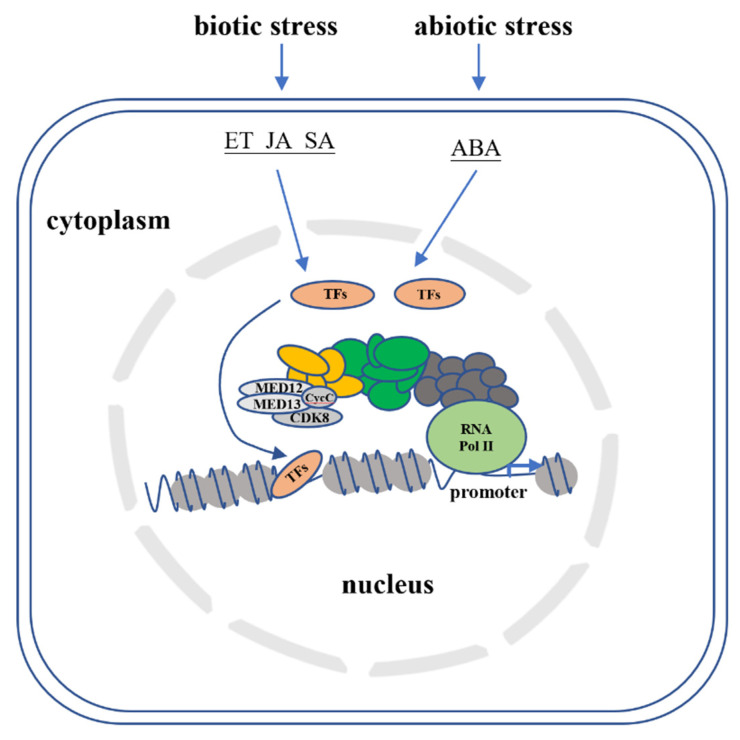
Figure 1.
The regulatory functions of Mediator complex in biotic and abiotic stress responses. Biotic and abiotic stresses trigger the plant hormones of ethylene (ET), jasmonic acid (JA), salicylic acid (SA) and abscisic acid (ABA) to activate signaling transduction pathway. A number of transcription factors (TFs) transmit these messages to the transcriptional machinery in the nucleus. Mediator complex functions as a bridge between TFs and RNA polymerase II to precisely regulate the transcription of stress responsive genes.
Arabidopsis CDK8, also named HUA ENHANCER3 (HEN3), was first reported for its regulation of floral organ identity. Thus, it was suggested to contribute to cell differentiation in a multicellular organism [16]. CDK8 was later found to interact with Arabidopsis LEUNIG, a transcription co-repressor [17]. Another name for Arabidopsis CDK8 is CDKE1 (cyclin-dependent kinase E1). Studies on the Arabidopsis regulator of alternative oxidase 1 (rao1) mutant which carries a mutation in CDKE1 further documented that CDKE1 regulates mitochondrial retrograde signaling under H2O2 and cold stress [18]. Moreover, CDK8 has been indicated to contribute in drought stress regulation recently. In terms of plant immune responses, Arabidopsis CDK8 was reported to perform a contrasting regulatory role [19]. Each subunit of the Mediator complex appears to specifically respond to various environmental conditions to different degrees. The significance of the reported subunits, particularly in the CDK8 module, are being explored next for their signaling involvement in biotic and abiotic stresses [20].
2. The Role of Mediator Subunits in Biotic Stress Regulation
When plants encounter biotic stress such as pathogen infection, they must either activate and/or suppress many genes. It has been documented that more than 620 genes were induced by common necrotrophic fungi such as Botrytis cinerea or Alternaria brassicicola [21,22]. Undeniably within minutes after infection, significant transcriptional reprogramming occurs [23,24]. In fact, an extensive number of genes such as lignin synthesis for cell wall fortification, defense proteins that attenuate infection, as well as antimicrobial secondary metabolite production, were induced in plants when a pathogen is sensed. TFs including WRKY33, MYC2, ethylene response factor1 (ERF1), ethylene-insensitive3 (EIN3) along with a few activators are important for defense gene induction as well as resistance to pathogens [25,26].
Since Bäckström et al. isolated the plant Mediator from Arabidopsis back in 2007 [27], Mediator has been established as an essential component of plant defense and development. Its subunit of MED21 was determined by Dhawan et al. to have a role in necrotrophic pathogen defense [28]. MED25, also called PHYTOCHROME AND FLOWERING TIME1, along with MED8 were further uncovered by Kidd et al. for their involvement in plant defense [29]. Moreover, MED25 was also identified to regulate JA and SA-induced gene expression. In 2011, Ou et al. identified eight other TFs that have a direct interaction with Arabidopsis MED25. Three of the identified eight genes directly recognize the GCC-box of the promoter of Plant Defensin 1.2 (PDF1.2) gene, a gene that involves in plant defense [30]. Other subunits including MED33a and MED33b, which correspond to the known Arabidopsis mutants REDUCED EPIDERMAL FLUORESCENCE4 (REF4) and REF4-RESEMBLING1 (RFR1), have been reported to regulate the phenylpropanoid pathway [31]. The phenylpropanoid pathway generates products of lignin and anthocyanin, which are essential for plant defense as lignin fortification of cell walls curb pathogen growth [32,33] and anthocyanins are plant pigments that possess antimicrobial activity along with free radical scavenging properties. In fact, during pathogen infection, anthocyanins synthesis incorporates compounds of jasmonates, ABA and sugars to support plant defense [34,35]. The accumulation of anthocyanins is affected by MED25 [29]. As can be seen from these studies, the involvement of Mediator subunits in biotic stress is a sophisticated network of highly interconnected systems. Mediator subunits interact with defense genes to regulate various signaling transduction pathways that involve phytohormones as well as other hormones that enable the strengthening of plants to fight off pathogen. At the time when these subunits were reported for their roles in biotic stress, the involvement of CDK8 module in environmental disturbances was still unspecified, until recently when more evidence began to emerge to show its importance [36].
3. Subunits of the CDK8 Module Play a Role in Regulating Biotic Stress
Along with the subunits of MED8, MED15, MED16, MED21, and MED25, recent evidence about the substantial role of CDK8 module subunits in the resistance against necrotrophic pathogens has appeared [28,37,38,39]. In response to pathogen infection, plants produce different hormones such as ET, JA and SA [40]. These hormones further activate the expression of defense-related genes. As mentioned previously, very few studies focused on the transcriptional regulation of the CDK8 kinase module in biotic stress. In 2014, Zhu et al. studied the roles of CDK8 subunit in immune responses of Arabidopsis [19]. Beside CDK8, they further noted that the two other kinase module mutants, med12 and med13, presented signs of disease responses and increased cuticle permeability that were similarly observed in cdk8 mutant. They proposed that a shared function and structural conservation exist among the kinase module subunits. Since their study was targeting the functional role of CDK8 subunit in biotic stress, they did not mine into the mechanistic roles of med12 and med13. Very recently, MED12 and MED13 have been reported for their roles in gene regulation. MED12 and MED13 were indicated to participate in the beginning steps of gene transcription and were identified as positive gene regulators under certain conditions. Liu et al. performed mutations of MED12 and MED13 and discovered that they suppress morc1-reactivated pSDC:GFP (SUPPRESSOR OF DRM1 DRM2 CMT3) [13] (Figure 2A). Microrchidia (MORC) is a GHKL (gyrase, Hsp90, histidine kinase, MutL)-type ATPase-containing protein that exists in both animal and plant species [41]. It was reported that the silenced DNA methylated genes are reactivated by the mutations of MORC proteins in Arabidopsis [42]. Apparently, MED12 and MED13 are necessary for the expression of genes depleted in active chromatin marks, a chromatin signature shared with morc1 reactivated loci [43].
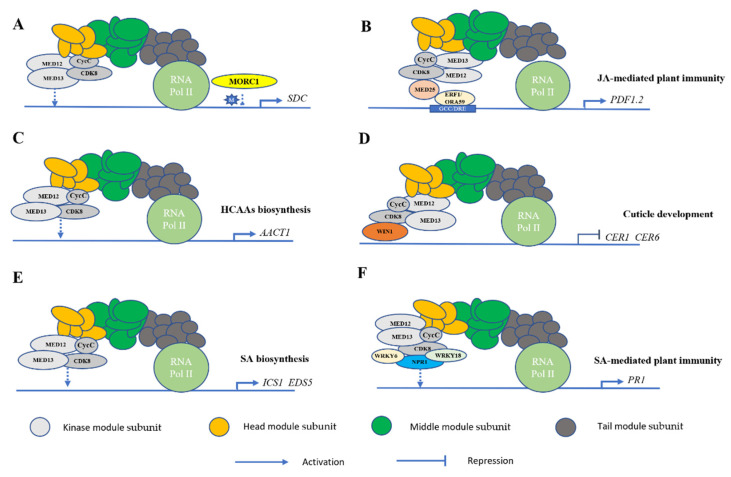
Figure 2.
The regulatory functions of CDK8 module in biotic stress response. (A) MED12 and MED13 reactivate the expression of a silenced DNA methylated SDC gene regulated by MORC1. (B) CDK8, MED12 and MED13 subunits interact with ERF1/ORA59 to regulate the expression of PDF1.2. (C) CDK8 is involved in the induced expression of AACT1 to regulate the biosynthesis of HCAAs for biotic stress response. (D) CDK8 represses the (ECERIFERUM) CER1 and CER6 gene expression to downregulate cuticle biosynthesis through interacting with WIN1, which binds to the GCC/DRE-box [45]. (E) CDK8 subunit integrates SA signaling through activating the expression of ICS1 and EDS5. (F) CDK8, WRKY6, WRKY18, and NPR1 form a complex to regulate the expression of pathogenesis-related (PR) genes. Light gray indicates the CDK8 kinase module, yellow indicates the head module, green indicates the middle module, and dark gray indicates the tail module.
As for the subunit of CDK8, Zhu et al. have elucidated the functions as well as the underlying molecular and biochemical mechanisms of the Mediator subunit CDK8 in plant defense. An extensive amount of work has been performed to unfold a contrasting defense function of CDK8 to two necrotrophic fungi of A. brassicicola and B. cinerea. Both fungi share similar mechanisms of pathogenesis, virulence, and modes of nutrition. In their investigation of Arabidopsis’ biotic response to A. brassicicola, they pointed out that CDK8 regulates the expression of defensin genes including PDF1.2 and several Ethylene Response Transcription Factors (ERFs) which are reported for disease resistance [19]. This finding was consistent with the observation that ERF1- and OCTADECANOID RESPONSIVE ARABIDOPSIS AP2/ERF59 (ORA59)-dependent activation of PDF1.2 expression requires CDK8 [44] (Figure 2B). Since an interaction was found between CDK8 and MED25, it was suggested that CDK8 regulates plant immunity through a JA-dependent pathway. Simultaneously, CDK8 contributes to Arabidopsis’ resistance to A. brassicicola through direct regulation of AGMATINE COUMAROYLTRANSFERASE (AACT1) transcription. AACT1 is involved in the biosynthesis of a class of secondary metabolites called hydroxycinnamic acid amides (HCAAs) (Figure 2C), which are known for their functions in fungal resistance [19]. Therefore, Arabidopsis would fail to induce critical defense responses without CDK8.
Despite their discovery of the positive regulatory role of CDK8 in plant immunity, they also uncovered another interesting finding that shows cdk8 mutant exhibits enhanced resistance to B. cinerea. Changes in the cuticle structure and permeability of the cdk8 mutant were noticed [19]. Therefore, they suggested that the increased cuticle permeability and altered cuticle structure are factors that influenced the cdk8 mutant’s biotic resistance as cuticles are related to plants’ improved resistance to B. cinerea [46,47]. To test this hypothesis, they investigated if CDK8 could interact with WAX INDUCER1 (WIN1) which is an ERF family protein known for cuticular wax biosynthesis regulation. In fact, they found that an interaction occurs between CDK8 and WIN1, which indicates that CDK8 is also involved in cuticle development (Figure 2D). Additionally, they observed that the expression of CDK8 that was defective in phosphorylation activity failed to rescue the susceptibility of the cdk8 mutant to A. brassicicola while the mutant’s resistance to B. cinerea was restored to the wild type (WT); indicating two differential functions of CDK8 in response to different strains of fungi. They also noted a strong interaction of CDK8 with two CycCs, thereby supporting an evolutionarily conserved structure of the kinase module in plants [19]. Their overall results from their study have revealed that the Mediator subunit CDK8 as well as the CDK8 module possess multiple regulatory roles in plant defense and development.
It has been revealed that CDK8 mutations diminished the plant’s resistance against the necrotrophic fungus B. cinerea. CDK8 was also demonstrated to interact with MED25 to play a role in the JA signaling [19]. In 2019, another piece of evidence about the subunit CDK8’s role in biotic stress has emerged. Huang et al. discovered CDK8 through a suppressor screen that used the triple mutant of camta1/2/3 (calmodulin-binding transcriptional activator). Similar to the Mediator complex, CAMTAs are evolutionarily conserved in multicellular eukaryotes in which their roles in transcriptional activity of plants are controversial [48]. They discovered that the mutation of cdk8 partially suppresses autoimmunity mediated by camta1/2/3 which includes enhanced resistance against Hpa Noco2 and SA accumulation levels. Hence, they suggested that CDK8 positively regulates SA accumulation and systemic acquired resistance (SAR) in Arabidopsis. SA is a phytohormone that participates in diverse immune responses including SAR, local defense, and effector-triggered immunity (ETI) whereas SAR is a form of plants’ systemic immune response that becomes activated in uninfected distal parts of plants when a local pathogen infection is detected [49]. SA accumulation is reported to be triggered in both infected and distal tissues after an infection occurs. The study further indicated that CDK8 subunit positively regulates these roles through increasing the expression of SA biosynthesis genes of ICS1 (Isochorismate Synthase 1) and EDS5 (Enhanced Disease Susceptibility 5) as the expression of these two genes was compromised in cdk8 mutants, preventing the plants to perform immune defense (Figure 2E) [50,51].
Chen et al. later showed that NONEXPRESSER OF PATHOGENESIS-RELATED GENES (NPR1) interacts with CDK8 as well as with WRKY DNA-BINDING PROTEINs such as WRKY18 to induce the expression of PATHOGENESIS-RELATED (PR) genes to promote defense responses in Arabidopsis [52]. Additionally, SA was indicated to substantially promote the interactions of these proteins to trigger immune response. NPR1 has been recognized as a master regulator of SA-mediated local and systemic plant immunity due to its control of the expression of over 2000 genes [53,54]. SAR was found to be included in the cdk8 and CDK8-associated Mediator mutants. The reduced expression of NPR1 and NPR1-dependent defense genes was observed as well in these mutants compared to WT. CDK8 positively regulates the expression of NPR1 through the interaction with both WRKY6 and WRKY18 and they are associated with the promoter of NPR1. Furthermore, CDK8 interacts with TGACG-Binding (TGA) TFs of TGA5 and TGA7; both are associated with the PR1 promoter to regulate PR1 gene expression. Another interesting finding about CDK8 from their study is that CDK8 recruits RNA Pol II to the promoters and coding regions of NPR1 and PR1 to promote their gene expression [52]. From their study, CDK8’s contribution to plant immunity such as SAR is further established. In fact, the study explained that CDK8 could essentially be the component that helps clarify the relationship between NPR1 and its activation of PR1 to initiate plant immune response (Figure 2F). Overall, they have demonstrated that under SA influence, CDK8 links TGA TFs and NPR1 (through interacting with WRKYs) with RNA Pol II to facilitate PR1 gene expression.
4. Signaling Roles of Mediator Subunits in Abiotic Stress
ABA is a phytohormone that contributes significantly to various developmental processes in the plant’s life cycle. The hormone plays a vital role in the plant’s response to various abiotic stresses including drought, salt, and heat stresses. In addition to biotic stress, Mediator complex, has also been found to serve important roles in the ABA signaling transduction. MED25 was the first Mediator subunit that was documented to act in response to ABA. MED25 was found to negatively regulate the ABA signaling pathway as med25 mutants display an increased sensitivity to ABA during seed germination and early seedling growth. Consistent with its negative role in ABA signaling, med25 mutant was observed to have an increased expression of ABA-responsive genes in response to ABA treatment compared to WT plants. The transcription of ABA-INSENSITIVE5 (ABI5), a key TF regulating the ABA signaling during seed germination, is induced by ABA, and interestingly, the ABA-induced transcription of ABI5 was suppressed in med25 mutants compared to WT. ABI5 protein, however, accumulated at higher abundance in med25 mutants compared to WT, indicating that MED25 may negatively regulate ABI5 at post-transcriptional level. Chromatin immunoprecipitation (ChIP) experiments further indicated that MED25 was highly enriched at the promoters of ABI5 downstream genes but this enrichment was decreased upon ABA treatment. A direct interaction was noticed between MED25 and ABI5 and this interaction was attenuated by ABA, which was in accordance with the negative impacts of MED25 on the ABI5-regulated ABA responses [38]. MED25 may act as a critical regulator in hormones crosstalk between JA, ethylene and ABA signaling due to its interaction with MYC2 and several TFs in plants. In addition to MED25, the head module subunit MED18 has also been reported to have a role in ABA signaling. Opposite to med25, med18 mutants are more insensitive to ABA at seed germination and early growth stages, similar to abi4 and abi5 mutants. Interestingly, the ABA-induced expressions of ABI4 and ABI5 are much lower in med18 mutants than those in WT, suggesting that the transcription of ABI4 and ABI5 are positively regulated by MED18. ChIP-qPCR further revealed that MED18 is recruited to the ABI4 binding site on the ABI5 promoter under both mock and ABA treatments [55]. The physical interaction between MED18 and TF ABI4 further reinforces that MED18 regulates ABA response and expression of ABI5 by interacting with ABI4. MED12 of the CDK8 module was also found recently to contribute to abiotic stress by blocking transient gene upregulation after light treatment as it has interactions with genes that are responsive to environmental stimuli such as light and radiation [13].
5. Signaling Roles of CDK8 Subunit in Abiotic Stress
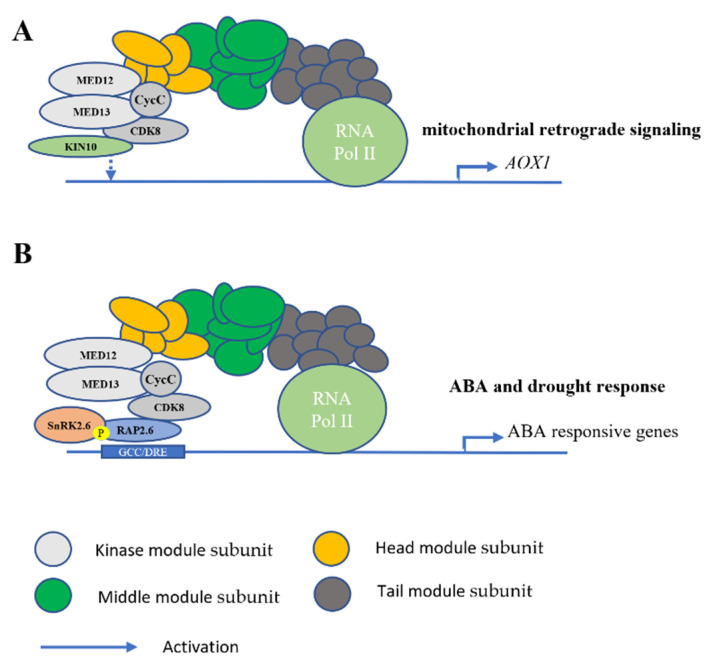
In 2013, Ng et al. studied cyclin-dependent kinase E1/Regulator of AOX1a 1 (CDKE1/RAO1) to understand how the induction of alternative oxidase (AOX) is integrated into the general regulatory context of the cell. To their surprise, CDKE1/RAO1 did not interact directly with any other cyclin components even though CDKE1 is a cyclin-dependent kinase. rao1 mutants in the study did not show an alternation in ABI4 gene expression under normal conditions but its gene expression was changed in a complex aox1a knock-out mutant. In fact, they noticed that only 119 transcripts genome-wide were significantly changed by 2.5-fold greater than the pAOX1a:LUC under normal conditions in both rao1–1 and rao1–2 [18]. Additionally, KIN10 was found to directly interact with CDK8 to coordinately regulate overlapping target genes in response to mitochondrial stress (Figure 3A). They suggested this change was due to the integrated network regulated by direct phosphorylation events associated with the Mediator complex and not any secondary effect caused by stress responsive TF dysregulation.
Figure 3.
The regulatory functions of CDK8 subunit in abiotic stress response. (A) CDK8 cooperates with KIN10 to control the expression of AOX1 to regulate mitochondrial retrograde signaling. (B) CDK8 along with phosphorylated proteins of RAP2.6 and SnRK2.6 bind to the GCC/DRE-box to initiate the transcription of ABA responsive genes in response to ABA and drought stress. Light gray indicates the CDK8 kinase module, yellow indicates the head module, green indicates the middle module and dark gray indicates the tail module.
They also conducted global transcriptional analyses and found that transcripts relating to both growth and stress are affected in the rao1 mutant background but in opposite ways. It appears that CDK8 plays a role in initiating cell wide stress responses, protein metabolism and protein synthesis, which are the essential building blocks for cell division and growth. Photosynthetic components, however, are switched off. Their study demonstrated that CDKE1/RAO1 has a role as it can integrate cellular responses to environmental signals for cell division or elongation. Furthermore, they have shown that CDKE1/RAO1 essentially serves as a sensitive relay between specific stress-induced TFs that are bound to the promoter and RNA Pol II, therefore it can directly regulate transcription under stress [18]. In general, the study has indicated that plants can utilize CDKE1/RAO1 as a mechanism to switch between growth and stress responses when responding to different environmental conditions.
Drought and cold stresses are some of the environmental challenges that prevent plant growth and development. In order to overcome stress, plants utilize various strategies including regulating signaling transduction pathways to respond to adverse environmental conditions. MED16, MED25, MED14, and MED2 are notable for their roles in cold stress regulation. At the time when these subunits were known for their involvement in abiotic stress responses, the function of CDK8 in abiotic stress was unidentified. It was not until 2020 that Zhu et al. reported that CDK8 positively modulates drought response in Arabidopsis. Since CDK8 is known for possessing kinase activity, they thought that it would be interesting to explore its potential in the ABA signaling pathway in which phosphorylation is involved. Through utilizing genetic, transcriptomic, and biochemical approaches, it was solidified that CDK8 associates with RAP2.6 and SnRK2.6 to positively regulate the transcription of ABA-responsive genes (Figure 3B). From their study, they have discovered that CDK8 mutation in Arabidopsis results in higher stomata density, impaired stomatal aperture as well as reduced tolerance to drought. Consistently, over-expression of CDK8 in Arabidopsis enhances drought tolerance. They have also observed improved cuticle permeability and thinner cutin in cdk8 mutants and, thus, they suggested that CDK8 may possibly regulate drought response through multiple mechanisms [56]. CDK8 was revealed to have a direct interaction with ERF/AP2 type TFs WIN1 (WAX INDUCER1) and RAP2.6 [19,56]. As mentioned previously, WIN1 is a key regulator of cuticle wax biosynthesis and RAP2.6 is an abiotic stress responsive gene. It was very likely that CDK8 upregulates cutin biosynthesis and wax accumulation through interacting with WIN1. Interestingly, WIN1 may also participate in abiotic stress response as its expression is significantly induced by various abiotic stresses. Their study suggested that WIN1 can bind to the GCC-box and DRE element sequences to activate several stress-responsive genes, suggesting a potential function of CDK8-WIN1 interaction in drought response. Their findings also uncovered another strategy which Arabidopsis utilizes CDK8 to cooperate with RAP2.6-SnRK2.6 complex to facilitate the immediate transcription of stress-responsive genes in drought.
6. Conclusions and Perspectives
When plants encounter biotic stress, they tackle the challenges using their built-in defense mechanisms and/or triggering defense-related signaling pathways. Activation of these responses often leads to the altered expression of various defense genes. Some of these defense genes are responsible for cuticle formation as well as for the biosynthesis and modification of cell wall [53,57]. Other defense genes are related to pathogenesis. Moreover, genes involved in various hormone signaling pathways have also been reported to change when plants respond to biotic stress [58,59]. As an essential player in the transcriptional regulation of gene expression, it was no surprise to find that the subunits of Mediator have connections with stress mitigation. The current reports of the core Mediator subunits along with the CDK8 module in plant cell signaling has revealed that Mediator has contrary roles in gene regulation when responding to stress.
The CDK8 module, consisting of CDK8, MED12, MED13 and C-type cyclins, is an important part of the Mediator complex. This kinase module is dissociable during transcription and thus initially considered to be a negative regulator of transcription [60]. Increasing evidence, however, indicated that the module can also stimulate transcription [61,62], thereby suggesting that CDK8 can both positively and negatively regulate transcription as indicated in Table 1. It is believed that the positive regulatory roles of CDK8 in transcription are either performed through the phosphorylation of TFs which would lead to protein degradation in some reported cases or through the promotion of RNA Pol II elongation [63]. CDK8 was revealed to regulate plant immunity through kinase dependent and independent functions in Arabidopsis. It is worth investigating if CDK8 may potentially interact with Arabidopsis signal responsive1 (AtSR1)/CAMTA3 to regulate plant growth during plant defense against pathogens as AtSR1 has been indicated to involve in the regulation of auxin- and BRs-related pathways as well as in the suppression of genes elicited by pathogen attack through binding to the “CGCG” containing CG-box in target gene promoters (51). Aside from serving a role in plant immunity, Arabidopsis CDK8 module also has been shown to play a role in abiotic stresses. Other subunits such as MED12 and MED13 in the CDK8 module are reportedly served as conditional positive regulators in biotic stress. Subunits in the Mediator complex including MED25 contributes to drought and salt tolerance in Arabidopsis by physically interacting with dehydration-responsive element-binding protein 2a (DREB2a), zinc-finger homeodomain protein 1 (ZFHD1), and a MYB TF through the ACID domain [64].
Table 1.
Genes regulated by the CDK8 module in Arabidopsis.
| Subunit | Genes | Regulation | Functions | Reference |
|---|---|---|---|---|
| CDK8 | ICS1, EDS5 | positive | SA biosynthesis | [12] |
| MED12, MED13 | SDC | positive | unknown | [13] |
| CDK8 | PDF1.2, ERF | positive | JA-mediated plant immunity | [19] |
| CDK8 | NPR1, PR1 | positive | SA-mediated plant immunity | [52] |
| CDK8 | AACT1 | positive | Biosynthesis of defense metabolites HCAAs | [19] |
| CDK8 | CER1, CER6 | negative | Cuticle development | [19] |
| CDK8 | AOX1 | positive | Mitochondrial retrograde signaling | [18] |
| CDK8 | COR15A, RD29B, DREB2A | positive | ABA and drought responses | [56] |
More evidence is emerging regarding the signaling roles of Mediator subunits in biotic and abiotic stresses. The involvement of these subunits in plant defense indicates the capability of the Mediator to accommodate new pathogen resistances in plants. Not only that, recent reports about these subunits in abiotic stresses also indicate the ability of Mediator to assist plants in acclimating in harsh environment [64,65]. The diverse roles depicted about the Mediator, especially its potential with various TFs interaction in regulating abiotic and biotic stress signaling, suggested a unique capacity of plants to recognize new factors to generate effective responses to adverse conditions. Since the subunit(s) of Mediator can interact with various TFs, it is comprehensible to see it serving as a hub for cross-linking many hormones and other signaling pathways to regulate stress responses. It is very possible that the subunits within the Mediator will cooperate with each other when the plants face new environmental challenges. Moreover, the function of Mediator subunits that respond to biotic and abiotic stresses may also be expanded with the possibility of them acquiring other new subunit(s) to recognize new regulatory proteins in plant survival against unfavorable circumstances.
Funding
National Natural Science Foundation of China (NSFC 31900238 and NSFC 32070307) to Y.Z. and 111 Project #D16014.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Verma V., Ravindran P., Kumar P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016;16:1–10. doi: 10.1186/s12870-016-0771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang J., Duan G., Li C., Liu L., Han G., Zhang Y., Wang C. The Crosstalks Between Jasmonic Acid and Other Plant Hormone Signaling Highlight the Involvement of Jasmonic Acid as a Core Component in Plant Response to Biotic and Abiotic Stresses. Front. Plant Sci. 2019;10:1349. doi: 10.3389/fpls.2019.01349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qu L.-J., Zhu Y.-X. Transcription factor families in Arabidopsis: Major progress and outstanding issues for future research. Curr. Opin. Plant Biol. 2006;9:544–549. doi: 10.1016/j.pbi.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Sato S., Tomomori-Sato C., Banks C.A.S., Sorokina I., Parmely T.J., Kong S.E., Jin J., Cai Y., Lane W.S., Brower C.S., et al. Identification of Mammalian Mediator Subunits with Similarities to Yeast Mediator Subunits Srb5, Srb6, Med11, and Rox3. J. Biol. Chem. 2003;278:15123–15127. doi: 10.1074/jbc.C300054200. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y., Ranish J.A., Aebersold R., Hahn S. Yeast nuclear extract contains two major forms of RNA polymerase II mediator complexes. J. Biol. Chem. 2001;276:7169–7175. doi: 10.1074/jbc.M009586200. [DOI] [PubMed] [Google Scholar]
- 6.Hasan A.S.M.M., Schoor J.K.V., Hecht V., Weller J.L. The CYCLIN-DEPENDENT KINASE Module of the Mediator Complex Promotes Flowering and Reproductive Development in Pea1[OPEN] Plant Physiol. 2020;182:1375–1386. doi: 10.1104/pp.19.01173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y., Li L., Qu L.-J. Plant Mediator complex and its critical functions in transcription regulation. J. Integr. Plant Biol. 2015;58:106–118. doi: 10.1111/jipb.12377. [DOI] [PubMed] [Google Scholar]
- 8.Akoulitchev S., Chuikov S., Reinberg D. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nat. Cell Biol. 2000;407:102–106. doi: 10.1038/35024111. [DOI] [PubMed] [Google Scholar]
- 9.Elmlund H., Baraznenok V., Lindahl M., Samuelsen C.O., Koeck P.J.B., Holmberg S., Hebert H., Gustafsson C.M. The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc. Natl. Acad. Sci. USA. 2006;103:15788–15793. doi: 10.1073/pnas.0607483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donner A.J., Ebmeier C.C., Taatjes D.J., Espinosa J.M. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat. Struct. Mol. Biol. 2010;17:194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donner A.J., Szostek S., Hoover J.M., Espinosa J.M. CDK8 Is a Stimulus-Specific Positive Coregulator of p53 Target Genes. Mol. Cell. 2007;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J., Sun Y., Orduna A.R., Jetter R., Li X. The Mediator kinase module serves as a positive regulator of salicylic acid accumulation and systemic acquired resistance. Plant J. 2019;98:842–852. doi: 10.1111/tpj.14278. [DOI] [PubMed] [Google Scholar]
- 13.Liu Q., Bischof S., Harris C.J., Zhong Z., Zhan L., Nguyen C., Rashoff A., Barshop W.D., Sun F., Feng S., et al. The characterization of Mediator 12 and 13 as conditional positive gene regulators in Arabidopsis. Nat. Commun. 2020;11:1–13. doi: 10.1038/s41467-020-16651-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raya-González J., López-Bucio J.S., Prado-Rodríguez J.C., Ruiz-Herrera L.F., Guevara-García Á.A., López-Bucio J. The MEDIATOR genes MED12 and MED13 control Arabidopsis root system configuration influencing sugar and auxin responses. Plant Mol. Biol. 2017;95:141–156. doi: 10.1007/s11103-017-0647-z. [DOI] [PubMed] [Google Scholar]
- 15.Galbraith M.D., Allen M.A., Bensard C.L., Wang X., Schwinn M.K., Qin B., Long H.W., Daniels D.L., Hahn W.C., Dowell R.D., et al. HIF1A Employs CDK8-Mediator to Stimulate RNAPII Elongation in Response to Hypoxia. Cell. 2013;153:1327–1339. doi: 10.1016/j.cell.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W.-M., Chen X. HUA ENHANCER3 reveals a role for a cyclin-dependent protein kinase in the specification of floral organ identity in Arabidopsis. Development. 2004;131:3147–3156. doi: 10.1242/dev.01187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez D., Bowen A.J., Carroll T.S., Conlan R. The Transcription Corepressor LEUNIG Interacts with the Histone Deacetylase HDA19 and Mediator Components MED14 (SWP) and CDK8 (HEN3) To Repress Transcription. Mol. Cell. Biol. 2007;27:5306–5315. doi: 10.1128/MCB.01912-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng S., Giraud E., Duncan O., Law S.R., Wang Y., Xu L., Narsai R., Carrie C., Walker H., Day D.A., et al. Cyclin-dependent Kinase E1 (CDKE1) Provides a Cellular Switch in Plants between Growth and Stress Responses. J. Biol. Chem. 2013;288:3449–3459. doi: 10.1074/jbc.M112.416727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Y., Schluttenhoffer C.M., Wang P., Fu F., Thimmapuram J., Zhu J., Lee S.Y., Yun D., Mengiste T. CY-CLIN-DEPENDENT KINASE8 Differentially Regulates Plant Immunity to Fungal Pathogens through Kinase-Dependent and -Independent Functions inArabidopsis. Plant Cell. 2014;26:4149–4170. doi: 10.1105/tpc.114.128611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agrawal R., Jiří F., Thakur J.K. The kinase module of the Mediator complex: An important signalling processor for the development and survival of plants. J. Exp. Bot. 2020 doi: 10.1093/jxb/eraa439. [DOI] [PubMed] [Google Scholar]
- 21.AbuQamar S., Chen X., Dhawan R., Bluhm B., Salmeron J., Lam S., Dietrich R.A., Mengiste T. Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. Plant J. 2006;48:28–44. doi: 10.1111/j.1365-313X.2006.02849.x. [DOI] [PubMed] [Google Scholar]
- 22.Van Wees S.C.M., Chang H.-S., Zhu T., Glazebrook J. Characterization of the Early Response of Arabidopsis to Alternaria brassicicola Infection Using Expression Profiling[w] Plant Physiol. 2003;132:606–617. doi: 10.1104/pp.103.022186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hahlbrock K., Bednarek P., Ciolkowski I., Hamberger B., Heise A., Liedgens H., Logemann E., Nürnberger T., Schmelzer E., Somssich I., et al. Non-self recognition, transcriptional reprogramming, and secondary metabolite accumulation during plant/pathogen interactions. Proc. Natl. Acad. Sci. USA. 2003;100:14569–14576. doi: 10.1073/pnas.0831246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cormack R.S., Hahlbrock K., Somssich I. Isolation of putative plant transcriptional coactivators using a modified two-hybrid system incorporating a GFP reporter gene. Plant J. 1998;14:685–692. doi: 10.1046/j.1365-313x.1998.00169.x. [DOI] [PubMed] [Google Scholar]
- 25.Zheng Z., Qamar S.A., Chen Z., Mengiste T. Arabidopsis WRKY33 transcription factor is required for resistance to ne-crotrophic fungal pathogens. Plant J. 2006;48:592–605. doi: 10.1111/j.1365-313X.2006.02901.x. [DOI] [PubMed] [Google Scholar]
- 26.Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. JASMONATE-INSENSITIVE1 Encodes a MYC Transcription Factor Essential to Discriminate between Different Jasmonate-Regulated Defense Responses in Arabidopsis. Plant Cell. 2004;16:1938–1950. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bäckström S., Elfving N., Nilsson R., Wingsle G., Björklund S. Purification of a Plant Mediator from Arabidopsis thaliana Identifies PFT1 as the Med25 Subunit. Mol. Cell. 2007;26:717–729. doi: 10.1016/j.molcel.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Dhawan R., Luo H., Foerster A.M., Abuqamar S., Du H.N., Briggs S.D., Mittelsten S.O., Mengiste T. HISTONE MON-OUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell. 2009;21:1000–1019. doi: 10.1105/tpc.108.062364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kidd B.N., Edgar C.I., Kumar K.K., Aitken E.A., Schenk P.M., Manners J.M., Kazan K. The Mediator Complex Subunit PFT1 Is a Key Regulator of Jasmonate-Dependent Defense in Arabidopsis. Plant Cell. 2009;21:2237–2252. doi: 10.1105/tpc.109.066910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ou B., Yin K.Q., Liu S.N., Yang Y., Gu T., Wing H.J., Zhang L., Miao J., Kondou Y., Matsui M., et al. A high-throughput screening system for Arabidopsis transcription factors and its application to Med25-dependent transcrip-tional regulation. Mol. Plant. 2011;4:546–555. doi: 10.1093/mp/ssr002. [DOI] [PubMed] [Google Scholar]
- 31.Stout J., Romero-Severson E., Ruegger M.O., Chapple C. Semidominant Mutations inReduced Epidermal Fluorescence 4Reduce Phenylpropanoid Content in Arabidopsis. Genetics. 2008;178:2237–2251. doi: 10.1534/genetics.107.083881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schenke D., Bottcher C., Scheel D. Crosstalk between abiotic ultraviolet-B stress and biotic (flg22) stress signalling in Ara-bidopsis prevents flavonol accumulation in favor of pathogen defence compound production. Plant Cell Environ. 2011;34:1849–1864. doi: 10.1111/j.1365-3040.2011.02381.x. [DOI] [PubMed] [Google Scholar]
- 33.Bhuiyan N.H., Selvaraj G., Wei Y., King J. Role of lignification in plant defense. Plant Signal. Behav. 2009;4:158–159. doi: 10.4161/psb.4.2.7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loreti E., Povero G., Novi G., Solfanelli C., Alpi A., Perata P. Gibberellins, jasmonate and abscisic acid modulate the su-crose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytol. 2008;179:1004–1016. doi: 10.1111/j.1469-8137.2008.02511.x. [DOI] [PubMed] [Google Scholar]
- 35.Teng S., Keurentjes J., Bentsink L., Koornneef M., Smeekens S. Sucrose-Specific Induction of Anthocyanin Biosynthesis in Arabidopsis Requires the MYB75/PAP1 Gene. Plant Physiol. 2005;139:1840–1852. doi: 10.1104/pp.105.066688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mao X., Kim J.I., Wheeler M.T., Heintzelman A.K., Weake V.M., Chapple C. Mutation of Mediator subunit CDK8 coun-teracts the stunted growth and salicylic acid hyperaccumulation phenotypes of an Arabidopsis MED5 mutant. New Phytol. 2019;223:233–245. doi: 10.1111/nph.15741. [DOI] [PubMed] [Google Scholar]
- 37.Wathugala D.L., Hemsley P.A., Moffat C.S., Cremelie P., Knight M.R., Knight H. The Mediator subunit SFR6/MED16 controls defence gene expression mediated by salicylic acid and jasmonate responsive pathways. New Phytol. 2012;195:217–230. doi: 10.1111/j.1469-8137.2012.04138.x. [DOI] [PubMed] [Google Scholar]
- 38.Chen R., Jiang H., Li L., Zhai Q., Qi L., Zhou W., Liu X., Li H., Zheng W., Sun J., et al. The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcrip-tion factors. Plant Cell. 2012;24:2898–2916. doi: 10.1105/tpc.112.098277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X., Wang C., Zhang Y., Sun Y., Mou Z. The Arabidopsis mediator complex subunit16 positively regulates salicy-late-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell. 2012;24:4294–4309. doi: 10.1105/tpc.112.103317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glazebrook J. Contrasting Mechanisms of Defense Against Biotrophic and Necrotrophic Pathogens. Annu. Rev. Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 41.Pastor W.A., Stroud H., Nee K., Liu W., Pezic D., Manakov S., Lee S.A., Moissiard G., Zamudio N., Bourc’His D., et al. MORC1 represses transposable elements in the mouse male germline. Nat. Commun. 2014;5:5795. doi: 10.1038/ncomms6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moissiard G., Cokus S.J., Cary J., Feng S., Billi A.C., Stroud H., Husmann D., Zhan Y., Lajoie B.R., Mccord R.P., et al. MORC Family ATPases Required for Heterochromatin Condensation and Gene Silencing. Science. 2012;336:1448–1451. doi: 10.1126/science.1221472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imura Y., Kobayashi Y., Yamamoto S., Furutani M., Tasaka M., Abe M., Araki T. CRYPTIC PRECOCIOUS/MED12 is a Novel Flowering Regulator with Multiple Target Steps in Arabidopsis. Plant Cell Physiol. 2012;53:287–303. doi: 10.1093/pcp/pcs002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C., Ding Y., Yao J., Zhang Y., Sun Y., Colee J., Mou Z. Arabidopsis Elongator subunit 2 positively contributes to resistance to the necrotrophic fungal pathogens Botrytis cinerea and Alternaria brassicicola. Plant J. 2015;83:1019–1033. doi: 10.1111/tpj.12946. [DOI] [PubMed] [Google Scholar]
- 45.Yang C.-K., Huang B.-H., Ho S.-W., Huang M.-Y., Wang J.-C., Gao J., Liao P.-C. Molecular genetic and biochemical evidence for adaptive evolution of leaf abaxial epicuticular wax crystals in the genus Lithocarpus (Fagaceae) BMC Plant Biol. 2018;18:196. doi: 10.1186/s12870-018-1420-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bessire M., Chassot C., Jacquat A.-C., Humphry M., Borel S., Petétot J.M.-C., Métraux J.-P., Nawrath C. A permeable cuticle in Arabidopsis leads to a strong resistance to Botrytis cinerea. EMBO J. 2007;26:2158–2168. doi: 10.1038/sj.emboj.7601658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurdyukov S., Faust A., Nawrath C., Bar S., Voisin D., Efremova N., Franke R., Schreiber L., Saedler H., Metraux J.P., et al. The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Ara-bidopsis. Plant Cell. 2006;18:321–339. doi: 10.1105/tpc.105.036079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finkler A., Ashery-Padan R., Fromm H. CAMTAs: Calmodulin-binding transcription activators from plants to human. FEBS Lett. 2007;581:3893–3898. doi: 10.1016/j.febslet.2007.07.051. [DOI] [PubMed] [Google Scholar]
- 49.Vlot A.C., Dempsey D.A., Klessig D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- 50.Yuan P., Yang T., Poovaiah B.W. Calcium Signaling-Mediated Plant Response to Cold Stress. Int. J. Mol. Sci. 2018;19:3896. doi: 10.3390/ijms19123896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan P., Du L., Poovaiah B.W. Ca(2+)/Calmodulin-Dependent AtSR1/CAMTA3 Plays Critical Roles in Balancing Plant Growth and Immunity. Int. J. Mol. Sci. 2018;19:1764. doi: 10.3390/ijms19061764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen J., Mohan R., Zhang Y., Li M., Chen H., Palmer I.A., Chang M., Qi G., Spoel S.H., Mengiste T., et al. NPR1 Promotes Its Own and Target Gene Expression in Plant Defense by Recruiting CDK8. Plant Physiol. 2019;181:289–304. doi: 10.1104/pp.19.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fu Z.Q., Dong X. Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 2013;64:839–863. doi: 10.1146/annurev-arplant-042811-105606. [DOI] [PubMed] [Google Scholar]
- 54.Wang D., Amornsiripanitch N., Dong X. A Genomic Approach to Identify Regulatory Nodes in the Transcriptional Network of Systemic Acquired Resistance in Plants. PLOS Pathog. 2006;2:e123. doi: 10.1371/journal.ppat.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lai Z., Schluttenhofer C.M., Bhide K., Shreve J., Thimmapuram J., Lee S.Y., Yun D.-J., Mengiste T. MED18 interaction with distinct transcription factors regulates multiple plant functions. Nat. Commun. 2014;5:3064. doi: 10.1038/ncomms4064. [DOI] [PubMed] [Google Scholar]
- 56.Zhu Y., Huang P., Guo P., Chong L., Yu G., Sun X., Hu T., Li Y., Hsu C., Tang K., et al. CDK8 is associated with RAP2.6 and SnRK2.6 and positively modulates abscisic acid signaling and drought response in Arabidopsis. New Phytol. 2020 doi: 10.1111/nph.16787. [DOI] [PubMed] [Google Scholar]
- 57.Boller T., Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pat-tern-recognition receptors. Annu. Rev. Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 58.Chen F., Dixon R. Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 2007;25:759–761. doi: 10.1038/nbt1316. [DOI] [PubMed] [Google Scholar]
- 59.Jones J.D., Dangl J.L. The plant immune system. Nat. Cell Biol. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 60.Gillmor C.S., Silva-Ortega C.O., Willmann M.R., Buendía-Monreal M., Poethig R. The Arabidopsis Mediator CDK8 module genes CCT (MED12) and GCT (MED13) are global regulators of developmental phase transitions. Development. 2014;141:4580–4589. doi: 10.1242/dev.111229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nemet J., Jeličić B., Rubelj I., Sopta M. The two faces of Cdk8, a positive/negative regulator of transcription. Biochimie. 2014;97:22–27. doi: 10.1016/j.biochi.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 62.Conaway R.C., Conaway J.W. Function and regulation of the Mediator complex. Curr. Opin. Genet. Dev. 2011;21:225–230. doi: 10.1016/j.gde.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Allen B.L., Taatjes D.J. The Mediator complex: A central integrator of transcription. Nat. Rev. Mol. Cell Biol. 2015;16:155–166. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elfving N., Davoine C., Benlloch R., Blomberg J., Brännström K., Müller D., Nilsson A., Ulfstedt M., Ronne H., Wingsle G., et al. The Arabidopsis thaliana Med25 mediator subunit integrates environmental cues to control plant development. Proc. Natl. Acad. Sci. USA. 2011;108:8245–8250. doi: 10.1073/pnas.1002981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Crawford T., Karamat F., Lehotai N., Rentoft M., Blomberg J., Strand Å., Björklund S. Specific functions for Mediator complex subunits from different modules in the transcriptional response of Arabidopsis thaliana to abiotic stress. Sci. Rep. 2020;10:1–18. doi: 10.1038/s41598-020-61758-w. [DOI] [PMC free article] [PubMed] [Google Scholar]