Abstract
There are many varieties of Cannabis sativa that differ from each other by composition of cannabinoids, terpenes and other molecules. The medicinal properties of these cultivars are often very different, with some being more efficient than others. This report describes the development of a method and software for the analysis of the efficiency of various cannabis extracts to detect the anti-inflammatory properties of the various cannabis extracts. The method uses high-throughput gene expression profiling data but can potentially use other omics data as well. According to the signaling pathway topology, the gene expression profiles are convoluted into the signaling pathway activities using a signaling pathway impact analysis (SPIA) method. The method was tested by inducing inflammation in human 3D epithelial tissues, including intestine, oral and skin, and then exposing these tissues to various extracts and then performing transcriptome analysis. The analysis showed a different efficiency of the various extracts in restoring the transcriptome changes to the pre-inflammation state, thus allowing to calculate a different cannabis drug efficiency index (CDEI).
Keywords: cannabis drug efficiency index, signaling pathway impact analysis, anti-inflammatory properties
1. Introduction
In the twentieth century, enormous strides have been made in combatting various diseases. Treatment of chronic diseases still remains to be more challenging than acute ones. This is often due to substantial differences in individual responses to known drugs. Over the last few decades, with the advent of genomics and epigenomics, research has focused on the development of personalized medicine. A need has arisen to develop diagnostic tools for use in the characterization of personalized aspects of chronic diseases.
Intracellular signaling pathways (SPs) regulate numerous processes involved in normal and pathological conditions, including development, growth, aging and cancer. Many intracellular signaling pathways or maps are available at online websites. The information relating to signaling pathway activation (SPA) can be obtained from the massive proteomic or transcriptomic data [1]. Although the proteomics analysis may be somewhat closer to the biological function of SPA, the transcriptomics analysis is far more feasible in terms of performing experimental tests and analyzing the data.
The transcriptomic methods like next-generation sequencing (NGS) or microarray analysis of RNA can routinely determine the expression levels for all or virtually all human genes [2]. Transcriptome profiling may be performed from a minute amount of the tissue sample, which does not necessarily need to be fresh, as it can also be done for the clinical formalin-fixed, paraffin-embedded (FFPE) tissue blocks. For the molecular analysis of cancer, gene expression can be interpreted in terms of abnormal SPA features of various pro- and antimitotic signaling pathways [3]. Such an analysis may improve any decision-making process for treatment strategy selection by the clinician.
Pro- and antimitotic SPs that determine various stages of cell cycle progression remained in the spotlight of computational biologists for more than a decade [4,5]. Today, hundreds of SPs and related gene product interaction maps that show sophisticated relationships between the individual molecules are catalogued in various databases, such as UniProt [6], HPRD [7], QIAGEN SABiosciences, WikiPathways [8], Ariadne Pathway Studio [9], SPIKE [10], Reactome [11], KEGG [12], etc.
Many bioinformatics tools have been developed that analyze SPs. One group of bioinformatic approaches integrated the analysis of transcriptome-wide data with the models employing the mass action law and Michaelis–Menten kinetics [13]. However, these methods, which have developed during the last fifteen years, remained purely fundamental until recently, primarily because of the multiplicity of interaction domains in the signal transducer proteins that enormously increase the interactome complexity [14]. Secondly, a considerable number of unknown free parameters, such as kinetics constants and/or concentrations of protein molecules, significantly complicated the SPA analysis. Yizhak et al. (2013) suggested that the clinical efficiency of several drugs, e.g., geroprotectors, may be evaluated as the ability to induce the kinetic models of the pathways into the steady state [13]. However, protein–protein interactions were quantitatively characterized in detail only for a tiny fraction of SPs. This approach is also time-consuming, since to process each transcriptomic dataset, it requires extensive calculations for the kinetic models [13].
In addition, all the contemporary bioinformatical methods that were proposed for digesting large-scale gene expression data, followed by recognition and analysis of SPs, have an important disadvantage: they do not allow tracing the overall pathway activation signatures and quantitively estimate the extent of the SPA [13,15]. This may be due to a lack of the definition of the specific roles of the individual gene products in the overall signal transduction process, incorporated in the calculation matrix used to estimate the SPA.
Thus, there remains an unmet, urgent and increasing need to provide effective personalized non-toxic disease therapies, as well as models for selecting a personalized optimal therapy for an individual.
Here, we propose a method for quick, informative and large-scale screening of changes in SPA in cells and tissues. These changes may reflect various conditions, such as differences in physiological state, aging, disease, treatment with drugs, media composition, additives, etc. One of the potential applications of SPA studies may be to utilize mathematical algorithms to identify and rank the medicines based on their predicted efficacy. In this paper, we give examples of the analysis of transcriptomics data for three sets of experiments involving the analysis of the anti-inflammatory properties of cannabis extracts. Such a method can potentially be used for the detection of the efficiency of various other botanical extracts or single compounds, although the versatility of the method will have to be confirmed using other data sets.
2. Methods
2.1. Cannabis Drug Efficiency Index (CDEI)
2.1.1. Input and Output Data for the CDEI Metric
Several methods were proposed for the assessment of drug efficiency based on gene/protein expression [16,17,18,19] or mutation patterns [20,21,22]. Unfortunately, most such methods are either proprietary or employ machine learning on preceding cases [23,24,25,26]. So, for evaluating a cannabis drug’s individual action, we have suggested a novel approach, the cannabis drug efficiency index (CDEI).
CDEI is our original approach for assessing the efficiency of a cannabis drug’s application regarding individual persons. CDEI is calculated based on high-throughput gene expression profiling and a signaling pathway topology, by comparison of gene expression and signaling pathway profiles between case and normal reference states for prediction of drug action in individual cases.
As input data, the CDEI operates with the results of various “omics” data stemming from the cells of individual patients and the healthy individuals. These data may include transcriptomic (e.g., performed with either next-generation sequencing or microarray mRNA hybridization), non-coding-RNAomic, proteomic and epigenomic data, etc.
The data of the full mRNA/protein abundance are integrated by the CDEI into the assessment values for activation of different cellular pathways (signalome).
The topology for the signaling, metabolic and cytoskeleton pathways, etc., were obtained from the database of QIAGEN SABiosciences (URL: https://www.qiagen.com/ru/shop/genes-and-pathways/pathway-central/).
2.1.2. SPIA (Signaling Pathway Impact Analysis): A Method for Assessment of Signaling Pathway Activation
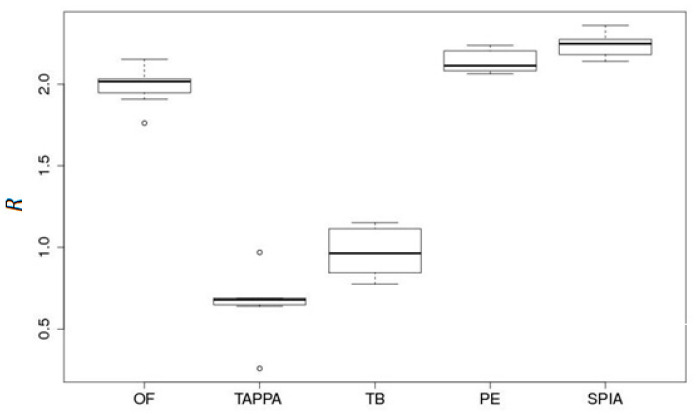
The high-throughput gene expression data were converted according to our CDEI approach to more aggregated values of the molecular pathway activities, using the signaling pathway impact analysis (SPIA) method [27]. Among the other methods for gene-expression-based assessment of signaling pathway activation, including TAPPA [28], topology-based score (TB) [29], Pathway-Express (PE) [1], and OncoFinder [1], the SPIA [27] approach showed the best statistical performance during the comparison between pathway-based and gene-based values (Figure 1; taken from [30]).
Figure 1.
Data aggregation effect R for five pathway activation scoring methods (OncoFinder (OF), TAPPA, TBScore (TB), Pathway-Express (PE) and signaling pathway impact analysis (SPIA)) on a renal carcinoma dataset [30].
Imagine a pathway graph, G(V, E) where is the set of graph nodes (vertices), and is the set of graph edges. The adjacency matrix is defined as , if or , and , if .
Consider then the values, called perturbation factors (PF), for the all genes g of the pathway K,
Here, is the signed log-fold-change of the gene g expression level in a given sample compared with the average value for the pool of normal samples. The latter term expresses the summation over all the genes γ that belong to the set Ug of the upstream genes for the gene g. The value of ndown (γ) denotes the number of downstream genes for gene γ. The weight factor indicates the interaction type between γ and g: if γ activates g, and when γ inhibits g. The search for upstream/downstream genes is performed according to the depth-first search method [31].
To obtain an estimator for pathway perturbation that is positive for an upregulated and negative for a downregulated pathway, use the second term in the formula for the perturbation factor (PF) from the previous paragraph, resulting in the accuracy value, . It can be shown that this accuracy vector may be expressed as follows: , where
I is the identity matrix, and
The overall score for the pathway perturbation was calculated as [27].
2.1.3. Calculation of the Cannabis Drug Efficiency Index (CDEI)
The SPIA-based CDEI metric was then calculated according to the following algorithm:
Obtain the signaling pathway impact analysis (SPIA) for each drug for each biological pathway.
Calculate the values of the pathway weight (wp) factor as follows: For pathways with a positive mean SPIA score of the case samples, wp = ((number of case samples with positive SPIA score)/(total number of case samples)). For pathways with a negative mean SPIA score of the case samples, wp = ((number of case samples with negative SPIA score)/(total number of case samples)).
Adjust the mean SPIA score of each pathway by the weight factor, SPIAμ = mean(SPIA)·wp.
- Perform Student’s t-test if the values of SPIAμ for the pool of case samples are different from 0 (for the pool of control samples, the values of SPIAμ are clearly equal to 0). During the Student’s t-test, the following case classes are taken into account: the untreated case (U), i.e., the pathological state before drug application, should be far from the control (C).
-
-Treated case (T), i.e., the pathological state after drug application, should be close to the control;
-
-The following values are the results for such calculations: |tU| = absolute t-value for the Student’s t-test for U-vs.-C profiles;
-
-|tT| = absolute t-value for the Student’s test for T-vs.-C profiles.
-
-
Calculate the cannabis drug efficiency index (CDEI) for each drug for a specific disease, wherein CDEI = 2 ((|tU|/(|tT| + |tU|) − 0.5).
Rank the drugs according to highest CDEI for a group of individual patients.
Note that our value of CDEI has the following properties:
-
-
CDEI is a value between −1 and 1;
-
-
CDEI is 0 if |tT| and |tU| are the same, which means no drug efficiency;
-
-
CDEI is 1 if |tT| is 0, which means the perfect efficiency;
-
-
CDEI is a value greater than 0 if |tT| is smaller than |tU|, which means a positive efficiency;
-
-
CDEI is a value less than 0 if |tT| is larger than |tU|, which means a negative efficiency.
Note also that the mean score of the case samples in each pathway is first calculated/adjusted and then the set of the mean scores in each data set are t-tested for their difference from 0 (i.e., a one-sample t-test). So, a t-statistic is calculated for each dataset and the CDEI metric is one value for each case sample dataset.
The CDEI calculations based on the SPIA values were done using R script.
2.2. Experiments Used for Validation of the CDEI
The CDEI metric was tested on several datasets, as explained below in several examples.
2.2.1. Plant Growth
All cannabis plant hybrids were generated and grown in a licensed facility at the University of Lethbridge. Various cultivars, numbered #1, #4, #7, #8, #9, #12, #13, #45, #114, #129, #130, #157, #167, #169 and #274, were used for the analysis. All hybrids represent individual lineages with different levels of cannabinoids (data not shown). Cuttings from the mother plants of the same age (~6 months) were made and allowed to root. After rooting (~10 days), plantlets were acclimated for another week and then were transplanted to a larger pot. Four plants per variety were grown at 22 °C for 18 h in light and 6 h in the dark, for 4 weeks, and then transferred to the chambers for a 12 h light/12 h dark regime to promote flowering. Plants were grown to maturity and flowers were harvested and dried. Approximately 5 g of the dry, lower samples from each of four plants per variety were combined and then used for extraction.
2.2.2. Crude Extract Preparation Using Solvent
Three grams of plant tissue was ground to a powder using a fine coffee grinder and the powdered plant tissue were weighed using an analytical balance. Plant material was placed inside a 250 mL Erlenmeyer flask. A total of 100 mL of ethyl acetate was poured into the flask containing the plant material. The flasks were then wrapped with tin foil and shaken continuously (120 rpm) in an incubator at 21 °C overnight and in the dark.
After overnight solvent extraction, the extracts were filtered through cotton into a 100 mL round bottom flask. The extracts were concentrated to around 2–3 mL using a rotary vacuum evaporator. The extracts were then transferred to a tared 3-dram vial (cat# 60975L Kimble obtained from Fisher Scientific). The leftover solvent was evaporated to dryness in an oven overnight at 50 °C to eliminate the solvent completely. The mass of each crude extract was recorded. All extractions were repeated twice.
2.2.3. Analysis of Cannabinoid Content
The levels of cannabinoids were analyzed using an Agilent Technologies 1200 Series HPLC system. The extract stocks were prepared from the crude extracts whereby 3–6 mg of crude extract was dissolved in DMSO (dimethyl sulfoxide anhydrous, Life Technologies) to reach a 60 mg/mL final concentration and stored at −20 °C. The appropriate cell culture media (RPMI + 10% FBS or EMEM + 10% FBS) were used to dilute the 60 mg/mL stock to make a working medium containing 0.01 mg/mL. The extracts were sterilized using a 0.22 µm filter. The composition of each extract is shown in Table 1.
Table 1.
Level of single and total cannabinoids in the flowers and extracts of selected C. sativa cultivars.
| Flowers, % | THC | CBD | CBGA | CBN | TOTAL Cannabinoids | CBD:THC Ratio |
|---|---|---|---|---|---|---|
| #1 | 0.25 | 6.79 | 0.12 | 7.16 | 27.16 | |
| #7 | 0.21 | 7.2 | 0 | 7.41 | 34.29 | |
| #9 | 0.22 | 6.91 | 0.31 | 7.44 | 31.41 | |
| #45 | 0.03 | 1.61 | 1.64 | 53.67 | ||
| #115 | 0.3 | 9.54 | 9.84 | 31.80 | ||
| #129 | 0.28 | 6.75 | 0.66 | 7.69 | 24.11 | |
| #130 | 0.86 | 2.63 | 0.31 | 0.03 | 3.83 | 3.06 |
| #157 | 0.2 | 3.75 | 0.09 | 0.15 | 4.19 | 18.75 |
| #167 | 0.08 | 2.25 | 0.16 | 2.49 | 28.13 | |
| #169 | 0.2 | 1.88 | 0.14 | 2.22 | 9.40 | |
| #274 | 0.44 | 9.02 | 0.31 | 9.77 | 20.50 | |
| Extracts, % | THC | CBD | CBGA | CBN | TOTAL Cannabinoids | CBD:THC Ratio |
| #1 | 0.88 | 34.6 | 0.25 | 0.12 | 35.85 | 39.32 |
| #7 | 1.1 | 32.9 | 0.27 | 0.15 | 34.27 | 29.91 |
| #9 | 0.98 | 32.6 | 0.97 | 0.15 | 34.55 | 33.27 |
| #45 | 0.44 | 24.92 | 0.13 | 0.14 | 25.63 | 56.64 |
| #115 | 1.23 | 42.52 | 0.42 | 0.28 | 44.45 | 34.57 |
| #129 | 1.3 | 35.3 | 1.2 | 0.42 | 38.22 | 27.15 |
| #130 | 2.43 | 28.43 | 0.98 | 0.18 | 32.02 | 11.70 |
| #157 | 0.62 | 33.5 | 0.73 | 0.33 | 34.85 | 54.03 |
| #167 | 0.38 | 24.3 | 0.29 | 0.12 | 24.97 | 63.95 |
| #169 | 0.67 | 19.28 | 0.45 | 0.18 | 20.58 | 28.78 |
| #274 | 0.93 | 43.81 | 1.2 | 0.12 | 46.06 | 47.11 |
| Molarity/µM | THC | CBD | CBGA | CBN | TOTAL Cannabinoids | TOTAL Cannabinoids |
| #1 | 0.28 | 11.00 | 0.08 | 0.04 | ||
| #7 | 0.35 | 10.46 | 0.09 | 0.05 | ||
| #9 | 0.31 | 10.37 | 0.31 | 0.05 | ||
| #45 | 0.14 | 7.92 | 0.04 | 0.05 | ||
| #115 | 0.39 | 13.52 | 0.13 | 0.09 | ||
| #129 | 0.41 | 11.23 | 0.38 | 0.14 | ||
| #130 | 0.77 | 9.04 | 0.31 | 0.06 | ||
| #157 | 0.20 | 10.65 | 0.23 | 0.11 | ||
| #167 | 0.12 | 7.73 | 0.09 | 0.04 | ||
| #169 | 0.21 | 6.13 | 0.14 | 0.06 | ||
| #274 | 0.30 | 13.93 | 0.38 | 0.04 |
Concentration of cannabinoids is shown in percentage of total dry weight (%) or in moles.
2.2.4. Preparation of the Cannabis Extract for Experimental Analysis Using Human Cells and Tissues
The stocks were prepared weighing 3–6 mg of crude extract into a micro centrifuge tube. The crude extract was dissolved in DMSO (dimethyl sulfoxide anhydrous from Life technologies cat # D12345) to reach a 60 mg/mL final concentration and stored at −20 °C. For the assay, a different amount of stock material (from 1 to 20 µL) was added to 21 mL of medium to make a working extract. Different concentrations were tested for cell/tissue toxicity and one concentration was chosen for further work (data not shown). Specifically, to obtain “low” (0.007 mg/mL) and “high” (0.015 mg/mL) functional concentrations, 2.45 µL or 5.25 µL of stock extract (60 mg/mL) were added to 21 mL of medium, respectively. The final concentration of DMSO was 0.012% and 0.025% in the “low” and “high” extract concentrations, respectively. The extracts were sterilized using a 0.22 µm filter.
2.2.5. Example #1
Human EpiDermFT 3D skin tissues (MatTek Life Sciences, Ashland, MA, USA) were exposed to 7000 ergs UVC to induce inflammation and then, 24 h after exposure, treated with extracts of several cannabis cultivars (#4, #8, #12 and #13) via their addition to the tissue growth media and incubated for another 24 h. Untreated sample (U) had DMSO added to the media instead of extracts. The control (C) sample had not been exposed to UVC. All samples were collected 24 h after the extracts were added and were used for mRNA extraction. All samples were done in triplicate.
RNA samples were extracted using TRIzolTM (Sigma Aldrich, St. Louis, MO, USA) according to the manufacturer’s instructions. The cDNA fragment libraries were prepared using the TruSeq Stranded mRNA library preparation kit (Illumina, San Diego, CA, USA) with polyA selection, as described in the manual. The high-throughput gene expression profiles were obtained using the Illumina mRNA next-generation sequencing platform NextSeq500. All of the library preparations and sequencing runs were completed using the same protocol, by the same technician, and on the same sequencing instrument.
2.2.6. Example #2
Human MatTek’s 3D EpiOral tissues (MatTek Life Sciences, Ashland, MA, USA) were equilibrated for 24 h, then the culture medium was replaced and the tissues incubated for another 24 h. Tissues were then exposed for 24 h to TNFα (40 ng/mL) to promote inflammation or to DMSO only. Tissues were then treated with various cannabis extracts (#1–#9) that were added to the media for 24 h. The control sample was exposed to DMSO only. Samples were then harvested for mRNA extraction and sequenced as in Example #1. All samples were done in triplicate.
2.2.7. Example #3
Human MatTek’s 3D EpiIntestinal tissues (MatTek Life Sciences, MA) were equilibrated for 24 h, then the culture medium was replaced, and the tissues incubated for another 24 h. Tissues were then exposed for 24 h to TNFα (40 ng/mL) or to DMSO only. Tissues were then treated with various cannabis extracts (#1, #2, #3, #4, #5, #6, #9, #10 and #11) that were added to the media for 24 h. The control sample was exposed to DMSO only. Samples were then harvested for mRNA extraction. Sequencing and data analysis were performed as in Example #2. All samples were done in triplicate.
2.2.8. Bioinformatics Workflow
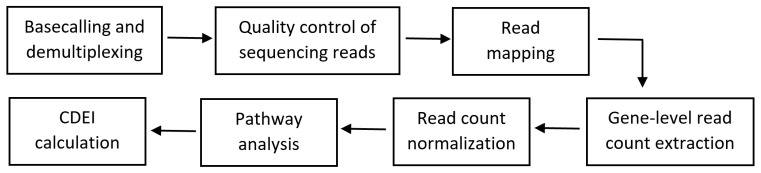
The bioinformatics workflow for the CDEI calculation is shown in Figure 2. Basecalling and demultiplexing were done using the CASAVA v.1.9 pipeline (Illumina, San Diego, CA, USA). The quality of the sequencing reads was assessed using FastQC v0.11.5 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The base qualities were over 30 on the Phred scale across all of the samples and no adapter contamination was noted. The reads were mapped to the human genome (GRCh37, Ensembl) downloaded from the Illumina iGenomes repository (https://support.illumina.com/sequencing/sequencing_software/igenome.html). The mapping was done using HISAT v2.0.5 [32] with the following command “hisat2-q—rna-strandness R—phred33-p 20—known-splicesite <known_splice_sites> -x <index> -U <fastq> -S <sam>”. The number of reads mapping to features (genes) was calculated using FeatureCounts v.1.6.1 [33], with the following command “featureCounts -T 20 -s 2 -a <genes.gtf> -o <counts> <sam>”.
Figure 2.
Bioinformatics workflow for the cannabis drug efficiency index (CDEI) calculation.
The gene-level read counts were loaded into the R v.3.6.1 statistical language environment. The raw count data were normalized using statistical methods implemented in the DESeq2 Bioconductor package v1.22.0 [34]. The DESeq normalized read counts were used to calculate the CDEI index using CDEI software. Table 2 shows the list of inflammation-related genes used for the CDEI calculation.
Table 2.
List of genes used for calculation of the level of gene expression in inflammation.
| UniProt | Gene Symbol | Gene Name |
|---|---|---|
| UniProtKB: Q12979 | ABR | Active breakpoint cluster region-related protein |
| UniProtKB: P13686 | ACP5 | Tartrate-resistant acid phosphatase type 5 |
| UniProtKB: Q04771 | ACVR1 | Activin receptor type-1 |
| UniProtKB: P00813 | ADA | Adenosine deaminase |
| UniProtKB: P30542 | ADORA1 | Adenosine receptor A1 |
| UniProtKB: P29274 | ADORA2A | Adenosine receptor A2a |
| UniProtKB: P29274 | ADORA2A | Adenosine receptor A2a |
| UniProtKB: P29275 | ADORA2B | Adenosine receptor A2b |
| UniProtKB: Q15109 | AGER | Advanced glycosylation end product-specific receptor |
| UniProtKB: Q15109 | AGER | Advanced glycosylation end product-specific receptor |
| UniProtKB: P50052 | AGTR2 | Type-2 angiotensin II receptor |
| UniProtKB: P50052 | AGTR2 | Type-2 angiotensin II receptor |
| UniProtKB: P23526 | AHCY | Adenosylhomocysteinase |
| UniProtKB: P31749 | AKT1 | RAC-alpha serine/threonine-protein kinase |
| UniProtKB: P09917 | ALOX5 | Arachidonate 5-lipoxygenase |
| UniProtKB: P02652 | APOA2 | Apolipoprotein A-II |
| UniProtKB: Q9NR48 | ASH1L | Histone-lysine N-methyltransferase ASH1L |
| UniProtKB: P00966 | ASS1 | Argininosuccinate synthase |
| UniProtKB: Q13315 | ATM | Serine-protein kinase ATM |
| UniProtKB: P30530 | AXL | Tyrosine-protein kinase receptor UFO |
| UniProtKB: P15291 | B4GALT1 | Beta-1,4-galactosyltransferase 1 |
| UniProtKB: Q9Y5Z0 | BACE2 | Beta-secretase 2 |
| UniProtKB: Q92560 | BAP1 | Ubiquitin carboxyl-terminal hydrolase BAP1 |
| UniProtKB: P11274 | BCR | Breakpoint cluster region protein |
| UniProtKB: P46663 | BDKRB1 | B1 bradykinin receptor |
| UniProtKB: P22004 | BMP6 | Bone morphogenetic protein 6 |
| UniProtKB: O00238 | BMPR1B | Bone morphogenetic protein receptor type-1B |
| UniProtKB: Q06187 | BTK | Tyrosine-protein kinase BTK |
| UniProtKB: Q06187 | BTK | Tyrosine-protein kinase BTK |
| UniProtKB: Q06187 | BTK | Tyrosine-protein kinase BTK |
| UniProtKB: Q5T7M4 | C1QTNF12 | Adipolin |
| UniProtKB: P01024 | C3 | Complement C3 |
| UniProtKB: P01024 | C3 | Complement C3 |
| UniProtKB: Q96GV9 | C5orf30 | UNC119-binding protein C5orf30 |
| UniProtKB: P13671 | C6 | Complement component C6 |
| UniProtKB: P01258 | CALCA | Calcitonin |
| UniProtKB: P01258 | CALCA | Calcitonin |
| UniProtKB: Q16602 | CALCRL | Calcitonin gene-related peptide type 1 receptor |
| UniProtKB: P51671 | CCL11 | Eotaxin |
| UniProtKB: O00175 | CCL24 | C-C motif chemokine 24 |
| UniProtKB: P51679 | CCR4 | C-C chemokine receptor type 4 |
| UniProtKB: P10747 | CD28 | T-cell-specific surface glycoprotein CD28 |
| UniProtKB: P25942 | CD40 | Tumor necrosis factor receptor superfamily member 5 |
| UniProtKB: P40200 | CD96 | T-cell surface protein tactile |
| UniProtKB: Q9BWU1 | CDK19 | Cyclin-dependent kinase 19 |
| UniProtKB: Q9UNI1 | CELA1 | Chymotrypsin-like elastase family member 1 |
| UniProtKB: O15516 | CLOCK | Circadian locomoter output cycles protein kaput |
| UniProtKB: P21554 | CNR1 | Cannabinoid receptor 1 |
| UniProtKB: P21554 | CNR1 | Cannabinoid receptor 1 |
| UniProtKB: P34972 | CNR2 | Cannabinoid receptor 2 |
| UniProtKB: P25025 | CXCR2 | C-X-C chemokine receptor type 2 |
| UniProtKB: P11511 | CYP19A1 | Aromatase |
| UniProtKB: Q9NR63 | CYP26B1 | Cytochrome P450 26B1 |
| UniProtKB: Q9Y271 | CYSLTR1 | Cysteinyl leukotriene receptor 1 |
| UniProtKB: Q9NRR4 | DROSHA | Ribonuclease 3 |
| UniProtKB: Q1HG43 | DUOXA1 | Dual oxidase maturation factor 1 |
| UniProtKB: Q1HG44 | DUOXA2 | Dual oxidase maturation factor 2 |
| UniProtKB: Q9Y6W6 | DUSP10 | Dual specificity protein phosphatase 10 |
| UniProtKB: Q16610 | ECM1 | Extracellular matrix protein 1 |
| UniProtKB: P24530 | EDNRB | Endothelin receptor type B |
| UniProtKB: P00533 | EGFR | Epidermal growth factor receptor |
| UniProtKB: P00533 | EGFR | Epidermal growth factor receptor |
| UniProtKB: Q9BQI3 | EIF2AK1 | Eukaryotic translation initiation factor 2-alpha kinase 1 |
| UniProtKB: P08246 | ELANE | Neutrophil elastase |
| UniProtKB: P29317 | EPHA2 | Ephrin type-A receptor 2 |
| UniProtKB: P01588 | EPO | Erythropoietin |
| UniProtKB: P03372 | ESR1 | Estrogen receptor |
| UniProtKB: P14921 | ETS1 | Protein C-ets-1 |
| UniProtKB: P15090 | FABP4 | Fatty acid-binding protein, adipocyte |
| UniProtKB: O15360 | FANCA | Fanconi anemia group A protein |
| UniProtKB: Q9BXW9 | FANCD2 | Fanconi anemia group D2 protein |
| UniProtKB: P30273 | FCER1G | High affinity immunoglobulin epsilon receptor subunit gamma |
| UniProtKB: P30273 | FCER1G | High affinity immunoglobulin epsilon receptor subunit gamma |
| UniProtKB: P30273 | FCER1G | High affinity immunoglobulin epsilon receptor subunit gamma |
| UniProtKB: P30273 | FCER1G | High affinity immunoglobulin epsilon receptor subunit gamma |
| UniProtKB: Q12946 | FOXF1 | Forkhead box protein F1 |
| UniProtKB: Q9BZS1 | FOXP3 | Forkhead box protein P3 |
| UniProtKB: P22466 | GAL | Galanin peptides |
| UniProtKB: P23771 | GATA3 | Trans-acting T-cell-specific transcription factor GATA-3 |
| UniProtKB: Q13304 | GPR17 | Uracil nucleotide/cysteinyl leukotriene receptor |
| UniProtKB: P07203 | GPX1 | Glutathione peroxidase 1 |
| UniProtKB: P36969 | GPX4 | Phospholipid hydroperoxide glutathione peroxidase |
| UniProtKB: P81172 | HAMP | Hepcidin |
| UniProtKB: Q30201 | HFE | Hereditary hemochromatosis protein |
| UniProtKB: P14210 | HGF | Hepatocyte growth factor |
| UniProtKB: Q96A08 | HIST1H2BA | Histone H2B type 1-A |
| UniProtKB: P10809 | HSPD1 | 60 kDa heat shock protein, mitochondrial |
| UniProtKB: P05362 | ICAM1 | Intercellular adhesion molecule 1 |
| UniProtKB: P14902 | IDO1 | Indoleamine 2,3-dioxygenase 1 |
| UniProtKB: P14902 | IDO1 | Indoleamine 2,3-dioxygenase 1 |
| UniProtKB: P22692 | IGFBP4 | Insulin-like growth factor-binding protein 4 |
| UniProtKB: P22301 | IL10 | Interleukin-10 |
| UniProtKB: P29460 | IL12B | Interleukin-12 subunit beta |
| UniProtKB: P35225 | IL13 | Interleukin-13 |
| UniProtKB: P40933 | IL15 | Interleukin-15 |
| UniProtKB: Q9UHF5 | IL17B | Interleukin-17B |
| UniProtKB: Q96PD4 | IL17F | Interleukin-17F |
| UniProtKB: Q96F46 | IL17RA | Interleukin-17 receptor A |
| UniProtKB: Q9NRM6 | IL17RB | Interleukin-17 receptor B |
| UniProtKB: Q8NAC3 | IL17RC | Interleukin-17 receptor C |
| UniProtKB: P01583 | IL1A | Interleukin-1 alpha |
| UniProtKB: P14778 | IL1R1 | Interleukin-1 receptor type 1 |
| UniProtKB: P27930 | IL1R2 | Interleukin-1 receptor type 2 |
| UniProtKB: Q01638 | IL1RL1 | Interleukin-1 receptor-like 1 |
| UniProtKB: Q9HB29 | IL1RL2 | Interleukin-1 receptor-like 2 |
| UniProtKB: P60568 | IL2 | Interleukin-2 |
| UniProtKB: Q6UXL0 | IL20RB | Interleukin-20 receptor subunit beta |
| UniProtKB: Q6UXL0 | IL20RB | Interleukin-20 receptor subunit beta |
| UniProtKB: Q969J5 | IL22RA2 | Interleukin-22 receptor subunit alpha-2 |
| UniProtKB: Q9H293 | IL25 | Interleukin-25 |
| UniProtKB: P01589 | IL2RA | Interleukin-2 receptor subunit alpha |
| UniProtKB: P01589 | IL2RA | Interleukin-2 receptor subunit alpha |
| UniProtKB: Q8NI17 | IL31RA | Interleukin-31 receptor subunit alpha |
| UniProtKB: O95760 | IL33 | Interleukin-33 |
| UniProtKB: P05113 | IL5 | Interleukin-5 |
| UniProtKB: Q01344 | IL5RA | Interleukin-5 receptor subunit alpha |
| UniProtKB: P17301 | ITGA2 | Integrin alpha-2 |
| UniProtKB: P05107 | ITGB2 | Integrin beta-2 |
| UniProtKB: P18564 | ITGB6 | Integrin beta-6 |
| UniProtKB: O60674 | JAK2 | Tyrosine-protein kinase JAK2 |
| UniProtKB:Q9BX67 | JAM3 | Junctional adhesion molecule C |
| UniProtKB: P05412 | JUN | Transcription factor AP-1 |
| UniProtKB: O15054 | KDM6B | Lysine-specific demethylase 6B |
| UniProtKB: P04264 | KRT1 | Keratin, type II cytoskeletal 1 |
| UniProtKB: P18428 | LBP | Lipopolysaccharide-binding protein |
| UniProtKB: P01130 | LDLR | Low-density lipoprotein receptor |
| UniProtKB: P38571 | LIPA | Lysosomal acid lipase/cholesteryl ester hydrolase |
| UniProtKB: Q5S007 | LRRK2 | Leucine-rich repeat serine/threonine-protein kinase 2 |
| UniProtKB: P01374 | LTA | Lymphotoxin-alpha |
| UniProtKB: P07948 | LYN | Tyrosine-protein kinase Lyn |
| UniProtKB: P07948 | LYN | Tyrosine-protein kinase Lyn |
| UniProtKB: P46734 | MAP2K3 | Dual specificity mitogen-activated protein kinase kinase 3 |
| UniProtKB: P04201 | MAS1 | Proto-oncogene Mas |
| UniProtKB: Q8NEM0 | MCPH1 | Microcephalin |
| UniProtKB: P43490 | NAMPT | Nicotinamide phosphoribosyltransferase |
| UniProtKB: Q16236 | NFE2L2 | Nuclear factor erythroid 2-related factor 2 |
| UniProtKB: P19838 | NFKB1 | Nuclear factor NF-kappa-B p105 subunit |
| UniProtKB: Q9BYH8 | NFKBIZ | NF-kappa-B inhibitor zeta |
| UniProtKB: P59044 | NLRP6 | NACHT, LRR and PYD domains-containing protein 6 |
| UniProtKB: Q86UT6 | NLRX1 | NLR family member X1 |
| UniProtKB: P46531 | NOTCH1 | Neurogenic locus notch homolog protein 1 |
| UniProtKB: O15130 | NPFF | Pro-FMRFamide-related neuropeptide FF |
| UniProtKB: P01160 | NPPA | Natriuretic peptides A |
| UniProtKB: Q15761 | NPY5R | Neuropeptide Y receptor type 5 |
| UniProtKB: Q15761 | NPY5R | Neuropeptide Y receptor type 5 |
| UniProtKB: P21589 | NT5E | 5’-nucleotidase |
| UniProtKB: O60356 | NUPR1 | Nuclear protein 1 |
| UniProtKB: O15527 | OGG1 | N-glycosylase/DNA lyase |
| UniProtKB: P35372 | OPRM1 | Mu-type opioid receptor |
| UniProtKB: P51575 | P2RX1 | P2X purinoceptor 1 |
| UniProtKB: Q99572 | P2RX7 | P2X purinoceptor 7 |
| UniProtKB: Q96KB5 | PBK | Lymphokine-activated killer T-cell-originated protein kinase |
| UniProtKB: O75594 | PGLYRP1 | Peptidoglycan recognition protein 1 |
| UniProtKB: Q96PD5 | PGLYRP2 | N-acetylmuramoyl-L-alanine amidase |
| UniProtKB: P48736 | PIK3CG | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform |
| UniProtKB: Q9Y263 | PLAA | Phospholipase A-2-activating protein |
| UniProtKB: P60201 | PLP1 | Myelin proteolipid protein |
| UniProtKB: P06746 | POLB | DNA polymerase beta |
| UniProtKB: P37231 | PPARG | Peroxisome proliferator-activated receptor gamma |
| UniProtKB: P42785 | PRCP | Lysosomal Pro-X carboxypeptidase |
| UniProtKB: P28070 | PSMB4 | Proteasome subunit beta type-4 |
| UniProtKB: P25105 | PTAFR | Platelet-activating factor receptor |
| UniProtKB: O14684 | PTGES | Prostaglandin E synthase |
| UniProtKB: O14684 | PTGES | Prostaglandin E synthase |
| UniProtKB: P35354 | PTGS2 | Prostaglandin G/H synthase 2 |
| UniProtKB: Q9ULZ3 | PYCARD | Apoptosis-associated speck-like protein containing a CARD |
| UniProtKB: O95267 | RASGRP1 | RAS guanyl-releasing protein 1 |
| UniProtKB: Q06330 | RBPJ | Recombining binding protein suppressor of hairless |
| UniProtKB: Q9Y3P4 | RHBDD3 | Rhomboid domain-containing protein 3 |
| UniProtKB: Q6R327 | RICTOR | Rapamycin-insensitive companion of mTOR |
| UniProtKB: P05109 | S100A8 | Protein S100-A8 |
| UniProtKB: P05109 | S100A8 | Protein S100-A8 |
| UniProtKB: Q99500 | S1PR3 | Sphingosine 1-phosphate receptor 3 |
| UniProtKB: Q15858 | SCN9A | Sodium channel protein type 9 subunit alpha |
| UniProtKB: P18827 | SDC1 | Syndecan-1 |
| UniProtKB: Q96EE3 | SEH1L | Nucleoporin SEH1 |
| UniProtKB: P16109 | SELP | P-selectin |
| UniProtKB: P01008 | SERPINC1 | Antithrombin-III |
| UniProtKB: P36955 | SERPINF1 | Pigment epithelium-derived factor |
| UniProtKB: Q86VZ5 | SGMS1 | Phosphatidylcholine:ceramide cholinephosphotransferase 1 |
| UniProtKB: P52569 | SLC7A2 | Cationic amino acid transporter 2 |
| UniProtKB: P52569 | SLC7A2 | Cationic amino acid transporter 2 |
| UniProtKB: Q15797 | SMAD1 | Mothers against decapentaplegic homolog 1 |
| UniProtKB: P84022 | SMAD3 | Mothers against decapentaplegic homolog 3 |
| UniProtKB: Q99835 | SMO | Smoothened homolog |
| UniProtKB: O14543 | SOCS3 | Suppressor of cytokine signaling 3 |
| UniProtKB: O75159 | SOCS5 | Suppressor of cytokine signaling 5 |
| UniProtKB: Q9NYA1 | SPHK1 | Sphingosine kinase 1 |
| UniProtKB: P10451 | SPP1 | Osteopontin |
| UniProtKB: P40763 | STAT3 | Signal transducer and activator of transcription 3 |
| UniProtKB: P51692 | STAT5B | Signal transducer and activator of transcription 5B |
| UniProtKB: Q9UEW8 | STK39 | STE20/SPS1-related proline-alanine-rich protein kinase |
| UniProtKB: Q9BXA5 | SUCNR1 | Succinate receptor 1 |
| UniProtKB: P20366 | TAC1 | Protachykinin-1 |
| UniProtKB: Q9NUY8 | TBC1D23 | TBC1 domain family member 23 |
| UniProtKB: Q9UP52 | TFR2 | Transferrin receptor protein 2 |
| UniProtKB: P21980 | TGM2 | Protein-glutamine gamma-glutamyltransferase 2 |
| UniProtKB: P01033 | TIMP1 | Metalloproteinase inhibitor 1 |
| UniProtKB: O60603 | TLR2 | Toll-like receptor 2 |
| UniProtKB: O15455 | TLR3 | Toll-like receptor 3 |
| UniProtKB: O00206 | TLR4 | Toll-like receptor 4 |
| UniProtKB: Q9Y2C9 | TLR6 | Toll-like receptor 6 |
| UniProtKB: Q9NYK1 | TLR7 | Toll-like receptor 7 |
| UniProtKB: Q9NR97 | TLR8 | Toll-like receptor 8 |
| UniProtKB: P01375 | TNF | Tumor necrosis factor |
| UniProtKB: P21580 | TNFAIP3 | Tumor necrosis factor alpha-induced protein 3 |
| UniProtKB: P20333 | TNFRSF1B | Tumor necrosis factor receptor superfamily member 1B |
| UniProtKB: P20333 | TNFRSF1B | Tumor necrosis factor receptor superfamily member 1B |
| UniProtKB: Q8NER1 | TRPV1 | Transient receptor potential cation channel subfamily V member 1 |
| UniProtKB: Q8NER1 | TRPV1 | Transient receptor potential cation channel subfamily V member 1 |
| UniProtKB: Q9HBA0 | TRPV4 | Transient receptor potential cation channel subfamily V member 4 |
| UniProtKB: O60636 | TSPAN2 | Tetraspanin-2 |
| UniProtKB: O75896 | TUSC2 | Tumor suppressor candidate 2 |
| UniProtKB: P55089 | UCN | Urocortin |
| UniProtKB: P22309 | UGT1A1 | UDP-glucuronosyltransferase 1-1 |
| UniProtKB: Q70J99 | UNC13D | Protein unc-13 homolog D |
| UniProtKB: P19320 | VCAM1 | Vascular cell adhesion protein 1 |
| UniProtKB: P19320 | VCAM1 | Vascular cell adhesion protein 1 |
| UniProtKB: Q9HC57 | WFDC1 | WAP four-disulfide core domain protein 1 |
| UniProtKB: Q15942 | ZYX | Zyxin |
3. Results
The CDEI software was tested on several datasets, explained below in several examples. We used the transcriptomic data from three different experiments. In the first experiment, we have used the data from human EpiDermFT 3D skin tissues exposed to UVC to induce inflammation and then treated with extracts of several cannabis cultivars. In the second experiment, human EpiOral tissues were treated with TNFα to induce inflammation and then treated with several different extracts. In the third experiment, human EpiIntestinal tissues were treated with TNFα and then treated with several different extracts.
The total number of reads calculated from the three experiments was in the range of 17,700,978 to 45,054,688 with a median of 24,613,834.5 reads per sample. The minimum mapping rate, calculated as a fraction of reads with at least one match to the genome, was at least 93.80% with a median of 97.65%. The fraction of mapped reads assigned to features (genes) was in the range of 72.93–76.00% with a median of 74.86%.
In the first experiment, we have induced inflammation in human EpiDermFT 3D skin tissues by exposing it to UVC. Analysis of the samples from the first experiment revealed that Extract #8 is the most efficient extract in restoring the transcriptome response after the UVC exposure (Table 3). Extract #4 was less efficient, whereas Extract #13 was not efficient. Extract #12 was actually harmful as it has increased the UVC-induced changes in the transcriptome.
Table 3.
CDEI testing results—EpiDermFT.
| Data Set | Sample | Number of Profiles | t-Value | p-Value | CDEI |
|---|---|---|---|---|---|
| DMSO | Control (C) | 3 | 0 | 1 | - |
| UV | Untreated (U) | 3 | 1.04 | 0.23 | 0.00 |
| Extract #4 | Treated (T) | 5 | 0.67 | 0.50 | 0.22 |
| Extract #12 | Treated (T) | 5 | 2.06 | 0.04 | −0.33 |
| Extract #8 | Treated (T) | 5 | −0.19 | 0.85 | 0.69 |
| Extract #13 | Treated (T) | 5 | −1.04 | 0.30 | 0.00 |
In the second experiment, inflammation in 3D EpiOral tissues was established using exposure to TNFα (40 ng/mL) (Table 4). The effect of the extracts on the reversal of the inflammation processes was evaluated using the CDEI.
Table 4.
CDEI testing results—EpiOral.
| Data Set | Sample | t-Value | p-Value | CDEI |
|---|---|---|---|---|
| DMSO | Control (C) | - | - | - |
| TNFα | Untreated (U) | −2.78 | 0.006 | 0.00 |
| Extract #1 | Treated (T) | 0.86 | 0.39 | 0.53 |
| Extract #7 | Treated (T) | 0.19 | 0.85 | 0.87 |
| Extract #9 | Treated (T) | −0.03 | 0.98 | 0.98 |
| Extract #45 | Treated (T) | −2.02 | 0.04 | 0.16 |
| Extract #115 | Treated (T) | −0.15 | 0.88 | 0.90 |
| Extract #129 | Treated (T) | −0.63 | 0.53 | 0.63 |
| Extract #157 | Treated (T) | −0.29 | 0.77 | 0.81 |
| Extract #167 | Treated (T) | −2.27 | 0.02 | 0.10 |
| Extract #169 | Treated (T) | −0.17 | 0.86 | 0.88 |
Ranking of the CDEI scores revealed that Extract #3 was the most efficient; it has restored the TNF-induced transcriptome nearly completely—with a CDEI score of 0.98. Extracts #5, #9 and #2 were also quite efficient, with CDEI scores of 0.90, 0.88 and 0.87, respectively. Extracts #8 and #4 were not very efficient, with CDEI scores of 0.10 and 0.16, respectively.
Finally, in the 3rd experiment, EpiIntestinal tissues were exposed to TNFα and several extracts were used to reverse the transcriptome changes (Table 5). The CDEI score showed that Extract #5 was the most efficient, followed by Extract #6.
Table 5.
CDEI testing results—EpiIntestinal.
| Data Set | Sample | t-Value | p-Value | CDEI |
|---|---|---|---|---|
| DMSO | Control (C) | - | - | - |
| TNFα | Untreated (U) | 2.43 | 0.016 | 0.00 |
| Extract #1 | Treated (T) | −1.37 | 0.17 | 0.28 |
| Extract #7 | Treated (T) | 1.15 | 0.25 | 0.36 |
| Extract #9 | Treated (T) | 1.43 | 0.15 | 0.26 |
| Extract #45 | Treated (T) | 1.02 | 0.31 | 0.41 |
| Extract #115 | Treated (T) | 0.21 | 0.84 | 0.84 |
| Extract #129 | Treated (T) | −0.56 | 0.58 | 0.63 |
| Extract #130 | Treated (T) | 1.56 | 0.12 | 0.22 |
| Extract #169 | Treated (T) | 1.75 | 0.08 | 0.16 |
| Extract #274 | Treated (T) | 0.99 | 0.32 | 0.42 |
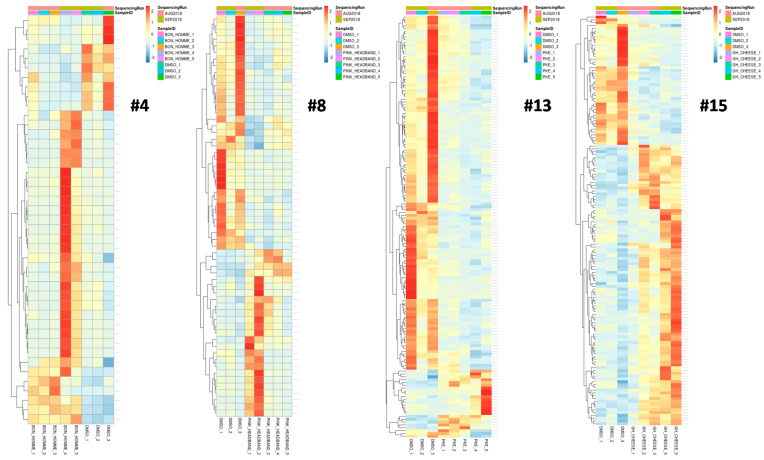
Examples of heatmaps, with the differentially expressed genes for Samples #4, #8, #13 and #15, are shown in Figure 3.
Figure 3.
Heatmaps of the differentially expressed genes for Extracts #4, #8, #13 and #15. Five independent samples for the extracts and three independent samples for DMSO (control) are shown. Only genes with a p-value < 0.05 are shown.
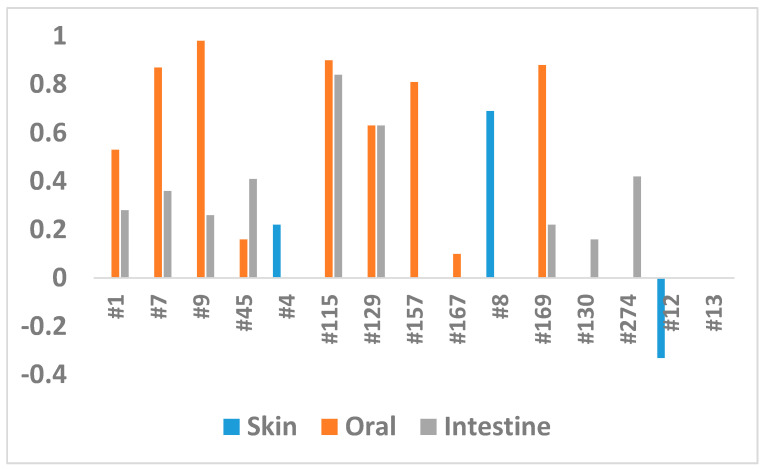
These experiments revealed that different extracts were efficient for different tissues. For example, a comparison of extracts for the reduction of inflammation in EpiOral and EpiIntestinal tissues showed that Extract #115 was equally and highly effective, while Extract #167 was not (Figure 4). At the same time, Extracts #7, #9 and #169 were effective for EpiOral tissues but not for EpiIntestinal tissues (Figure 4). Our attempts to correlate the level of cannabinoids and the efficiency of the extracts did not show any correlation. This is perhaps not surprising, as other molecules in the extracts likely have a strong modulatory effect.
Figure 4.
Comparison of the CDEI in three experiments. The Y axis shows the CDEI, while the X axis shows the extracts.
To analyze whether there is a correlation between the cannabinoid content and the activity of the extracts, we have analyzed the amount of THC, CBD, CBG, CBN, total cannabinoids and CBD to THC ratio (Table 1). The correlation analysis showed a moderate positive correlation between the CBD level and CDEI score (0.49) and a strong positive correlation between the CBN level and CDEI score (0.61). No correlation was found between THC and CDEI, CBG and CDEI or the CBD to THC ratio and CDEI.
4. Discussion
We developed a methodology to compute a cannabis drug efficiency index (CDEI) across the 241 pathways that contain the genes responding to the cannabis drug. The advantages of our methodology can be summarized as follows.
CDEI explicitly calculates the drug efficiency of cannabis on diseases, using the expression values of the genes. As far as we understand, this is a novel methodology and there is no other methodology or software similar to CDEI to do it.
CDEI evaluates the enrichment scores of the individual pathways in the control, untreated and treated cases. Then CDEI statistically integrates and compares the results from all samples. Finally, CDEI measures the overall drug efficiency of cannabis on diseases.
An important characteristic of the CDEI calculation is that CDEI is not affected by the properties of the input data chosen. CDEI purely measures the efficacy of the cannabis drug by statistically comparing the cannabis-treated and untreated cases with the control.
With the emergence of large-scale methods in genomics, the gene-level transcription changes became an obvious target in the search for biomarkers in complex diseases. Yet, in over a decade of research effort, only a handful of gene or protein biomarkers made its way into clinical practice. For example, as of 2017, 26 gene-based predictive and diagnostic biomarkers were used in cancer medicine [35]. Biomarker discovery based on gene expression signatures is challenging due to low reproducibility when presented with different datasets [36,37] and a high occurrence of stochastic associations [38]. As an alternative to a single gene or gene signature approach, methods that aggregate data across pathways or gene network modules are gaining traction, potentially being more effective tools for biomarker discovery [39,40,41]. Several studies demonstrated that the use of pathway-based biomarkers enables robust predictions of drug response [42,43] and accurate classification of disease types [44].
Considering the importance of pathway analysis, multiple methods were developed to address the task. These methods can be roughly classified as non-topology-based and topology-based [45]. Non-topological methods treat pathways as non-structured lists of genes (gene sets); they take a list of differentially expressed (DE) genes and attempt to determine the probability of observing a given number of DE genes within a pathway. More sophisticated non-topological methods rely on the analysis of gene rankings in the whole dataset, avoiding the selection of target gene lists according to arbitrary thresholds. Topology-based methods take into account the biological reality of pathways by incorporating the data on the type and direction of protein interactions. Not surprisingly, topology-based methods were shown to outperform their counterparts in benchmarking tests [45].
Recognizing the importance of pathway topology, CannSelect uses the SPIA algorithm to calculate pathway activation scores based on gene expression profiles. Previously, SPIA was shown to outperform other pathway analysis methods in terms of data aggregation [30] and in the ability to detect pathways, inducing a specific phenotype [45]. SPIA was also ranked third out of 10 methods used to generate pathway activation scores for machine learning classification in an iPanda benchmarking test [17].
Estimation of pathway-level activities opens a possibility of selecting drugs or other bioactive compounds that could alter the physiological state of the system in the desired direction by changing the activities of perturbed pathways.
This idea is at the core of several computational methods, including Oncofinder [1], GeroScope [46] and iPanda [17]; this is also true for the method presented in this study. Oncofinder is based on the unique computational algorithm that calculates pathway activation strength based on the activating or repressive actions of proteins in the pathway. Pathway activation strength reflects the activation or inhibition of the pathway in a pathological state compared to the healthy controls. It also incorporates the data on drug–target interactions to predict the action of the drugs on specific pathways and rank the drugs based on a predicted efficiency score. The output of Oncofinder is a list of drugs ranked by the predicted efficiency meant to assist in selecting a treatment strategy for a specific patient. GeroScope is an extension of Oncofinder to analyze aging-related pathways and screen chemical compounds that act as geroprotectors. iPanda is a topology-based method that assigns importance coefficient to genes in a pathway derived using a combination of statistical and topological methods [17]. iPanda can be adapted to estimate drug scores following similar principles as in Oncofinder.
In theory, the CDEI approach mechanistically resembles Oncofinder, Geroscope, or iPanda; however, it differs from them in terms of pathway score and drug score algorithms and, more importantly, it is designed with a different purpose in mind. Oncofinder and other similar methods require two conditions—disease and normal state—and serve to facilitate the selection of drugs that could act on pathways altered between these two conditions. The data can be obtained experimentally or downloaded from public repositories. CDEI, on the other hand, requires three conditions—an untreated control (healthy state), pathological state, and pathological state treated with a drug of interest. The output of CDEI is a numerical measure of the ability of a drug to reverse gene expression changes in a pathological state to mimic those in the healthy control. In contrast to Oncofinder, CDEI is useful as a computational step that follows laboratory testing of panels of drugs or other bioactive compounds to select prospective candidates for further investigation.
There are different methodologies to analyze the enrichment of individual pathways. GSEA (Gene Set Enrichment Analysis) finds the enrichment of pathways by focusing on gene sets [47]. SPIA measures the perturbation score on pathways [26]. NEA (Network Enrichment Analysis) outputs the network enrichment scores of the altered gene sets per pathway [48]. However, these methodologies do not further derive an integrated meaningful result (i.e., a drug efficiency index) from the analysis of the individual pathways.
As detailed in the accompanying manual of the software, CDEI provides a convenient graphical user interface software available to users. The CDEI software works on both Windows and Linux systems. It validates the user inputs of the gene expression values and computes the scores of the individual pathways as an intermediate output. Then the software generates the drug efficiency index of cannabis on diseases as the final output.
One of the recent studies using the SPIA method was the report by Franco et al. (2019) [48]. The authors presented a method to identify the biomarkers of drug response and survival in proliferative disease using enrichment over the gene networks. Unlike the CDEI algorithm, it does not calculate drug scores that reflect the overall shift in pathway activity towards a normal (non-diseased) state; also, in their study, the SPIA scores were calculated for individual pathways and correlated with clinical variables of interest (drug response and survival). In the CDEI, the SPIA scores are weighted and summarized following a specific application of a drug screen to the individual sample, which allows a calculation of a drug score and the ranking of drugs by efficiency. In this, respect the CDEI is a tool that allows a selection of a drug for an individual patient. This functionality is not present in Franco’s network approach [48].
Pathway analysis has the potential to improve performance by applying machine learning methods. For example, the representative genes that represent a pathway can be selected by feature selection methods and pathways can be further ranked or weighted based on the representative genes using classification methods [49].
To conclude, the CDEI algorithm represents a multistage process that includes the specific application of a drug screen, integration of pathway databases, weighted summarization of the pathway activation scores and identification of the most efficient drugs/extracts. The bioinformatics part of the CDEI process is wrapped into user-friendly graphical interface software. Currently, the CDEI algorithm was only used for the analysis of the efficacy of cannabis extracts in the reduction of inflammation. It remains to be shown whether it would also be able to rank the cannabis extracts (or extracts of any other medicinal herb or synthetic drug) by their efficacy for other molecular processes, diseases, or conditions. The CDEI algorithm may also be an efficient tool to stratify patients in the clinical studies or clinical trials by their response to a drug, thus providing selection criteria upon transition from Phase 2 to Phase 3 or Phase 4 clinical trials. It will also be very useful in pre-clinical experiments using various cell, tissue or animal models, like recently used analysis of the efficiency of cannabis extracts for the reduction of inflammation [50].
Author Contributions
I.K. came up with the idea of the development of new software for the analysis of cannabis efficiency; N.B., Y.I. and B.B. wrote the codes for the software; Y.I. and B.B. analyzed transcriptomic data; I.K., O.K. analyzed the data and prepared the manuscript; All authors were involved in writing and proofreading the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Pathway Rx. Inc.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Buzdin A., Zhavoronkov A., Korzinkin M.B., Venkova L., Zenin A.V., Smirnov P.Y., Borisov N.M. Oncofinder, a new method for the analysis of intracellular signaling pathway activation using transcriptomic data. Front. Genet. 2014;5 doi: 10.3389/fgene.2014.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang Z., Zhou X., Li R., Michal J.J., Zhang S., Dodson M.V., Zhang Z., Harland R.M. Whole transcriptome analysis with sequencing: Methods, challenges and potential solutions. Cell Mol. Life Sci. 2015;72:3425–3439. doi: 10.1007/s00018-015-1934-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kholodenko B.N. Spatially distributed cell signalling. FEBS Lett. 2009;583:4006–4012. doi: 10.1016/j.febslet.2009.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borisov N., Aksamitiene E., Kiyatkin A., Legewie S., Berkhout J., Maiwald T., Kaimachnikov N.P., Timmer J., Hoek J.B., Kholodenko B.N. Systems-level interactions between insulin–EGF networks amplify mitogenic signaling. Mol. Syst. Biol. 2009;5:256. doi: 10.1038/msb.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dimmer E.C., Huntley R.P., Alam-Faruque Y., Sawford T., O’Donovan C., Martin M.J., Bely B., Browne P., Chan W.M., Eberhardt R., et al. The UniProt-GO Annotation database in 2011. Nucleic Acids Res. 2011;40:D565–D570. doi: 10.1093/nar/gkr1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mishra G.R. Human protein reference database--2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kutmon M., Riutta A., Nunes N., Hanspers K., Willighagen E.L., Bohler A., Mélius J., Waagmeester A., Sinha S.R., Miller R.A., et al. WikiPathways: Capturing the full diversity of pathway knowledge. Nucleic Acids Res. 2016;44:D488–D494. doi: 10.1093/nar/gkv1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikitin A., Egorov S., Daraselia N., Mazo I. Pathway studio--the analysis and navigation of molecular networks. Bioinformatics. 2003;19:2155–2157. doi: 10.1093/bioinformatics/btg290. [DOI] [PubMed] [Google Scholar]
- 9.Elkon R., Vesterman R., Amit N., Ulitsky I., Zohar I., Weisz M., Mass G., Orlev N., Sternberg G., Blekhman R., et al. SPIKE—A database, visualization and analysis tool of cellular signaling pathways. BMC Bioinform. 2008;9:110. doi: 10.1186/1471-2105-9-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haw R., Hermjakob H., D’Eustachio P., Stein L. Reactome pathway analysis to enrich biological discovery in proteomics data sets. Proteomics. 2011;11:3598–3613. doi: 10.1002/pmic.201100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakaya A., Katayama T., Itoh M., Hiranuka K., Kawashima S., Moriya Y., Okuda S., Tanaka M., Tokimatsu T., Yamanishi Y., et al. KEGG OC: A large-scale automatic construction of taxonomy-based ortholog clusters. Nucleic Acids Res. 2012;41:D353–D357. doi: 10.1093/nar/gks1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yizhak K., Gabay O., Cohen H., Ruppin E. Model-based identification of drug targets that revert disrupted metabolism and its application to ageing. Nat. Commun. 2013;4:2632. doi: 10.1038/ncomms3632. [DOI] [PubMed] [Google Scholar]
- 13.Borisov N.M., Kholodenko B.N., Faeder J.R., Chistopolsky A.S. Domain-oriented reduction of rule-based network models. IET Syst. Biol. 2008;2:342–351. doi: 10.1049/iet-syb:20070081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwang S. Comparison and evaluation of pathway-level aggregation methods of gene expression data. BMC Genom. 2012;13:S26. doi: 10.1186/1471-2164-13-S7-S26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borisov N., Sorokin M., Garazha A., Buzdin A. Quantitation of Molecular Pathway Activation Using RNA Sequencing Data. Methods Mol. Biol. 2020;2063:189–206. doi: 10.1007/978-1-0716-0138-9_15. [DOI] [PubMed] [Google Scholar]
- 16.Buzdin A., Sorokin M., Garazha A., Sekacheva M., Kim E., Zhukov N., Wang Y., Li X., Kar S., Hartmann C., et al. Molecular pathway activation—New type of biomarkers for tumor morphology and personalized selection of target drugs. Semin. Cancer Biol. 2018;53:110–124. doi: 10.1016/j.semcancer.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Ozerov I.V., Lezhnina K.V., Izumchenko E., Artemov A.V., Medintsev S., Vanhaelen Q., Aliper A., Vijg J., Osipov A.N., Labat I., et al. In silico Pathway Activation Network Decomposition Analysis (iPANDA) as a method for biomarker development. Nat. Commun. 2016;7:13427. doi: 10.1038/ncomms13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tkachev V., Sorokin M., Garazha A., Borisov N., Buzdin A. Oncobox Method for Scoring Efficiencies of Anticancer Drugs Based on Gene Expression Data. Methods Mol. Biol. 2020;2063:235–255. doi: 10.1007/978-1-0716-0138-9_17. [DOI] [PubMed] [Google Scholar]
- 19.Zolotovskaia M., Sorokin M., Garazha A., Borisov N., Buzdin A. Molecular Pathway Analysis of Mutation Data for Biomarkers Discovery and Scoring of Target Cancer Drugs. Methods Mol. Biol. 2020;2063:207–234. doi: 10.1007/978-1-0716-0138-9_16. [DOI] [PubMed] [Google Scholar]
- 20.Zolotovskaia M.A., Sorokin M.I., Emelianova A.A., Borisov N.M., Kuzmin D.V., Borger P., Garazha A.V., Buzdin A.A. Pathway Based Analysis of Mutation Data Is Efficient for Scoring Target Cancer Drugs. Front. Pharmacol. 2019;10:1. doi: 10.3389/fphar.2019.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zolotovskaia M.A., Sorokin M.I., Roumiantsev S.A., Borisov N.M., Buzdin A.A. Pathway Instability Is an Effective New Mutation-Based Type of Cancer Biomarkers. Front. Oncol. 2019;8:658. doi: 10.3389/fonc.2018.00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borisov N., Buzdin A. New Paradigm of Machine Learning (ML) in Personalized Oncology: Data Trimming for Squeezing More Biomarkers From Clinical Datasets. Front. Oncol. 2019;9:658. doi: 10.3389/fonc.2019.00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borisov N., Tkachev V., Suntsova M., Kovalchuk O., Zhavoronkov A., Muchnik I., Buzdin A. A method of gene expression data transfer from cell lines to cancer patients for machine-learning prediction of drug efficiency. Cell Cycle. 2018;17:486–491. doi: 10.1080/15384101.2017.1417706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tkachev V., Sorokin M., Mescheryakov A., Simonov A., Garazha A., Buzdin A., Muchnik I., Borisov N. FLOating-Window Projective Separator (FloWPS): A Data Trimming Tool for Support Vector Machines (SVM) to Improve Robustness of the Classifier. Front. Genet. 2019;9:717. doi: 10.3389/fgene.2018.00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tkachev V., Sorokin M., Borisov C., Garazha A., Buzdin A., Borisov N. Flexible Data Trimming Improves Performance of Global Machine Learning Methods in Omics-Based Personalized Oncology. Int. J. Mol. Sci. 2020;21:713. doi: 10.3390/ijms21030713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarca A.L., Draghici S., Khatri P., Hassan S.S., Mittal P., Kim J.-S., Kim C.J., Kusanovic J.P., Romero R. A novel signaling pathway impact analysis. Bioinformatics. 2008;25:75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao S., Wang X. TAPPA: Topological analysis of pathway phenotype association. Bioinformatics. 2007;23:3100–3102. doi: 10.1093/bioinformatics/btm460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ibrahim M.A.-H., Jassim S., A Cawthorne M., Langlands K. A Topology-Based Score for Pathway Enrichment. J. Comput. Biol. 2012;19:563–573. doi: 10.1089/cmb.2011.0182. [DOI] [PubMed] [Google Scholar]
- 29.Draghici S., Khatri P., Tarca A.L., Amin K., Done A., Voichita C., Georgescu C., Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borisov N., Suntsova M., Sorokin M., Garazha A., Kovalchuk O., Aliper A., Ilnitskaya E., Lezhnina K., Korzinkin M., Tkachev V., et al. Data aggregation at the level of molecular pathways improves stability of experimental transcriptomic and proteomic data. Cell Cycle. 2017;16:1810–1823. doi: 10.1080/15384101.2017.1361068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Even S. Graph Algorithms. 2nd ed. Cambridge University Press; Cambridge, UK: 2011. [Google Scholar]
- 32.Kim D., Langmead B., Salzberg S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao Y., Smyth G.K., Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2013;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 34.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selleck M.J., Senthil M., Wall N.R. Making Meaningful Clinical Use of Biomarkers. Biomark. Insights. 2016;12 doi: 10.1177/1177271917715236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ein-Dor L., Zuk O., Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc. Natl. Acad. Sci. USA. 2006;103:5923–5928. doi: 10.1073/pnas.0601231103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ein-Dor L., Kela I., Getz G., Givol D., Domany E. Outcome signature genes in breast cancer: Is there a unique set? Bioinformatics. 2004;21:171–178. doi: 10.1093/bioinformatics/bth469. [DOI] [PubMed] [Google Scholar]
- 38.Pan Y., Neuss S., Leifert A., Fischler M., Wen F., Simon U., Schmid G., Brandau W., Jahnen-Dechent W. Size-Dependent Cytotoxicity of Gold Nanoparticles. Small. 2007;3:1941–1949. doi: 10.1002/smll.200700378. [DOI] [PubMed] [Google Scholar]
- 39.Ben-Hamo R., Berger A.J., Gavert N., Miller M., Pines G., Oren R., Pikarsky E., Benes C.H., Neuman T., Zwang Y., et al. Predicting and affecting response to cancer therapy based on pathway-level biomarkers. Nat. Commun. 2020;11:3296. doi: 10.1038/s41467-020-17090-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Z., Su Z., Wen Z., Shi L., Chen T. Microarray platform consistency is revealed by biologically functional analysis of gene expression profiles. BMC Bioinform. 2009;10:S12. doi: 10.1186/1471-2105-10-S11-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J., Zhang Y., Marian C., Ressom H.W. Identification of aberrant pathways and network activities from high-throughput data. Briefings Bioinform. 2012;13:406–419. doi: 10.1093/bib/bbs001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ben-Hamo R., Efroni S. Biomarker robustness reveals the PDGF network as driving disease outcome in ovarian cancer patients in multiple studies. BMC Syst. Biol. 2012;6:3. doi: 10.1186/1752-0509-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ben-Hamo R., Efroni S. Gene expression and network-based analysis reveals a novel role for hsa-miR-9 and drug control over the p38 network in glioblastoma multiforme progression. Genome Med. 2011;3:77. doi: 10.1186/gm293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su J., Yoon B.-J., Dougherty E.R. Accurate and Reliable Cancer Classification Based on Probabilistic Inference of Pathway Activity. PLoS ONE. 2009;4:e8161. doi: 10.1371/journal.pone.0008161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen T.-M., Shafi A., Nguyen T., Draghici S. Identifying significantly impacted pathways: A comprehensive review and assessment. Genome Biol. 2019;20:1–15. doi: 10.1186/s13059-019-1790-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alexander A., Aleksey V.B., Andrew G., Leslie J., Artem A., Maria S., Alena I., Larisa V., Nicolas B., Anton B., et al. In search for geroprotectors: In silico screening and in vitro validation of signalome-level mimetics of young healthy state. Aging. 2016;8:2127–2141. doi: 10.18632/aging.101047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aravind S., Pablo T., Vamsi K.M., Sayan M., Benjamin L.E., Michael A.G., Amanda P., Scott L.P., Todd R.G., Eric S.L., et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franco M., Jeggari A., Peuget S., Bottger F., Selivanova G., Alexeyenko A. Prediction of response to anti-cancer drugs becomes robust via network integration of molecular data. Sci. Rep. 2019;9:2379. doi: 10.1038/s41598-019-39019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang W., Emrich S., Zeng E. A two-stage machine learning approach for pathway analysis; Proceedings of the IEEE International Conference on Bioinformatics and Biomedicine 2010; Hong Kong, China. 18–21 December 2010; pp. 274–279. [Google Scholar]
- 50.Wang B., Kovalchuk A., Li D., Rodriguez-Juarez R., Ilnytskyy Y., Kovalchuk I., Kovalchuk O. In search of preventive strategies: Novel high-CBD Cannabis sativa extracts modulate ACE2 expression in COVID-19 gateway tissues. Aging. 2020;12:22425–22444. doi: 10.18632/aging.202225. [DOI] [PMC free article] [PubMed] [Google Scholar]